Abstract
Since the early 2000’s, much of the neuroimaging work at Washington University (WU) has been facilitated by the Central Neuroimaging Data Archive (CNDA), an XNAT-based imaging informatics system. The CNDA is uniquely related to XNAT, as it served as the original codebase for the XNAT open source platform. The CNDA hosts data acquired in over 1000 research studies, encompassing 36,000 subjects and more than 60,000 imaging sessions. Most imaging modalities used in modern human research are represented in the CNDA, including magnetic resonance (MR), positron emission tomography (PET), computed tomography (CT), nuclear medicine (NM), computed radiography (CR), digital radiography (DX), and ultrasound (US). However, the majority of the imaging data in the CNDA are MR and PET of the human brain. Currently, about 20% of the total imaging data in the CNDA is available by request to external researchers. CNDA’s available data includes large sets of imaging sessions and in some cases clinical, psychometric, tissue, or genetic data acquired in the study of Alzheimer’s disease, brain metabolism, cancer, HIV, sickle cell anemia, and Tourette syndrome.
Keywords: Central Neuroimaging Data Archive, CNDA, Washington University, Imaging, Informatics, Neuroinformatics, Data, Sharing, Database, Archive, Multi-site, Multi-center, De-identify, Anonymization, Pipeline, JAAT, XNAT, Alzheimer’s Disease, Brain Metabolism, Controls, Cancer, HIV, Sickle-Cell Anemia, Tourette Syndrome
Introduction
Washington University has a long and rich history of neuroimaging research, including seminal contributions in the development of positron emission tomography (PET), fMRI, resting state fMRI, and human connectomics (Raichle, 1998; Snyder and Raichle, 2012; Van Essen et al., 2013). Since the early 2000’s, much of this work has been facilitated by the Central Neuroimaging Data Archive (CNDA), an XNAT-based imaging informatics system. The CNDA is uniquely related to XNAT, as it served as the original codebase for the XNAT open source platform.
History
The CNDA was initiated in 2002 to manage the Washington University (WU) Charles F. and Joanne Knight Alzheimer’s Disease Research Center (Knight ADRC) imaging data. It was designed to be both an active study management tool as well as a long term imaging archive. The system was implemented as an XML database that provided a streamlined mechanism for developers to implement new clinical and behavioral data types. Its rudimentary imaging workflow depended largely on FTP-based data transfer and a semi-structured directory and file naming convention. A web application provided basic data navigation and reporting. By 2004, the CNDA had evolved into a Java web application with a unique hybrid XML/relational database. With its increasing sophistication, additional WU research studies began relying on the CNDA, and its core software was extracted into the generalized open source Extensible Neuroimaging Archive Toolkit (XNAT). XNAT is now widely used within neuroimaging (see reports in this issue by Alpert et al and Yvernault et al for example systems) and the broader imaging community (Doran et al., 2012; Gao et al., 2013; Winslow et al., 2011). For several years, the CNDA and XNAT continued to grow in lock step, with the XNAT development team also functioning as the development, operations, and user support team for the CNDA. Support for DICOM was incorporated by 2006 and project-based access control was developed soon after. Since 2012, the CNDA has served as the official repository for all research imaging at Washington University with automated archiving of scans obtained on all the University’s standard dedicated research scanners (see Hodge et al, this issue, for management of data collected by the Human Connectome Project). In addition to hosting WU-generated data, the CNDA also supports a number of multi-center studies, including the Dominantly Inherited Alzheimer Network (DIAN) (Morris et al., 2012) and the INTRuST Post-traumatic Stress Disorder/Traumatic Brain Injury Clinical Consortium (http://intrust.sdsc.edu). The CNDA continues to serve as a primary test bed for ongoing XNAT development. Most recently, scripting tools have been incorporated into the CNDA infrastructure to enable de-identification and import of large batches of retrospective patient data from the WU Radiology Department’s diagnostic division.
Current Services
The CNDA provides investigators with a permanent archive for imaging data. All data is securely stored on a modern ZFS file system with a full offsite backup. New research imaging sessions acquired at Washington University can either be sent directly to the CNDA from the scanner or uploaded through the website using a Java-based upload wizard. To ensure secure transmission of all imaging sessions, outside institutions are restricted to uploads through the CNDA website unless specialized DICOM relay hardware has been installed at their scan facility.
All data in the CNDA is organized into projects. A project can be marked as belonging to one or multiple investigators. Generally one project is created for each new study, except in the case of multi-center studies, for which one project is often created for each institution contributing data.
Researchers can configure their projects to automatically apply session anonymization to scrub PHI or recode their DICOM data before it is permanently stored. As the data is archived, CNDA extracts and stores to the database a limited set of information about the session, including time and date of scan, patient name, patient id, and the scanner make and model, which can be used later for searching or generating reports.
Automated imaging data analysis is implemented in CNDA using XNAT pipelines (Table 1). Investigators can add a pipeline to a project by submitting a short form on the pipeline configuration tab to provide default processing variable values for their images. Currently, the processing pipelines available to researchers in the CNDA are all neuroimaging-based. Data analysis pipelines are available for structural and functional MR as well as for PET. Also available is a set of utilitarian pipelines for tasks like atlas registration, DICOM conversion, structural MR defacing and quality control.
Table 1.
CNDA Pipelines
| Pipeline | Description | Input |
|---|---|---|
| FreeSurfer v5.1 | Runs FreeSurfer v5.1 recon-all with - qcache |
MR with T1 |
| FreeSurfer v5.3 | Runs Freesurfer v5.3 recon-all with configurable parameters |
MR with T1 Optional: Flair or other T2 |
| Generic Bold Preprocessing |
Prepares (BOLD) images for seed-based data analysis |
MR with BOLD, T1 and one of the following: T1W, TSE, or PDT2 Optional: resting state scans |
| Benice | Computes the Resting State Networks from BOLD studies for pre-surgical use |
MR with BOLD and T1 |
| HOF | Prepares sessions from clinical tumor protocol studies for ROI analysis |
MR with T1, T2 and DWI |
| PET Unified Pipeline | Generates binding potentials for regions of interest (ROI) |
PET Recommended: FreeSurfer or hand-drawn ROIs |
| Register MR To Atlas | Registers MR scans to specified atlas | MR with T1 |
| Dicom To Nifti | Uses dcm2nii to convert some or all DICOM series to NIfTI |
Any neuroimaging DICOM of modalities MR, PET, CT or SPECT |
| Face Masking | De-identifies a subject's face by blurring recognizable facial characteristics |
MR with T1 or T2 |
| Protocol Validation | Checks the experiment for pre-defined project acquisition parameters |
Any DICOM session |
| FBIRN Phantom | Creates Automated Phantom QA data using 64bit version 1.9.5 of BIRN tools |
MR with BOLD acquired on fBIRN phantom |
The CNDA offers a variety of services to support multi-center projects. Access to data can be restricted by site for study coordinators and site investigators, while the overall investigator and administrative core are provided with a pooled view of all the sites’ data. To enable image data transfer directly from an external site’s scanner, the CNDA can provide a relay device, a Mini PC running RSNA Clinical Trial Processor (CTP) (http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor) which receives images directly from a scanner via the unsecure DICOM protocol and forwards them to the CNDA over secure HTTPS protocol, while also de-identifying and recoding the image headers. CNDA’s Protocol Validation pipeline and Manual QC form can help to ensure that study imaging quality is maintained.
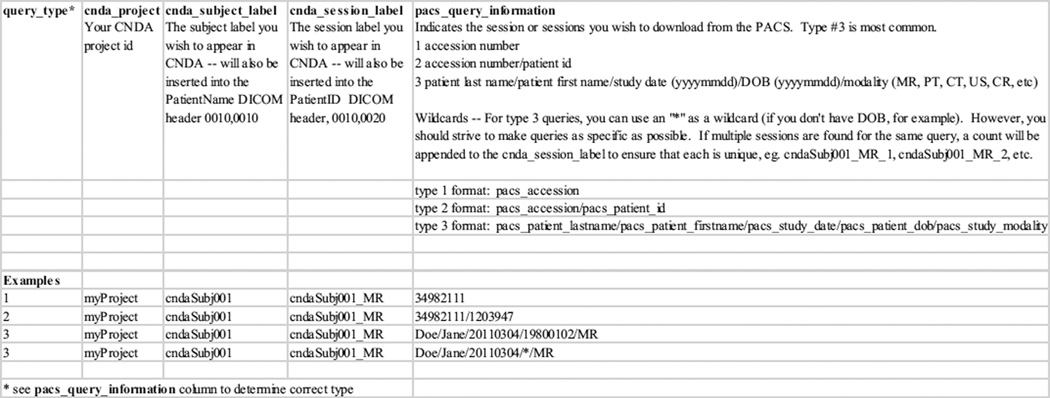
For researchers conducting retrospective studies, the CNDA provides an automated way to retrieve clinical scans from the Washington University Medical Center’s clinical image archive (often referred to as a PACS). The Joint Anonymization and Archive Tool (JAAT) (http://nrg.wustl.edu/software/jaat) simplifies the transfer of DICOM for the researcher, shifting the download and de-identification responsibilities to CNDA staff (Figure 1). Once IRB approval for the study has been established, the investigator only needs to provide a spreadsheet specifying the patient exams to be pulled from the clinical PACS, the CNDA subject and session labels to be used in recoding and the researcher’s project. To date, the CNDA has retrieved close to 5000 clinical scans using JAAT. A project currently underway will allow qualified CNDA users to deploy the JAAT at remote institutions and securely transfer anonymized patient exams to the CNDA.
Figure 1.
JAAT Clinical Pull Template
The CNDA provides numerous routes for the upload of non-imaging data. The DIAN and ADRC projects regularly retrieve and store data into the CNDA using automated custom tools built upon the XNAT RESTful Web Services (REST) interface. DIAN coordinators use a custom uploader to unpack computerized battery assessments into the CNDA data archive. Other studies use the XNAT CSV upload tool to conveniently bulk upload entire spreadsheets of data via the CNDA website. Finally, many projects choose to just enter data directly into web forms created for individual assessments. The choice in upload method usually depends on the regularity of data acquisition, the expected length of the project, and funding available for custom tools development.
Users can download large sets of image sessions either via the multi-session downloader available on the CNDA website or through the REST interface. For large data transfers, the JAAT can be used in conjunction with the XNAT Gateway (http://nrg.wustl.edu/software/xnat-gateway-2) to pull sessions from the CNDA and send them to another imaging repository (including another XNAT), assuming it has a DICOM receiver. To export non-imaging data from the CNDA website, investigators can use the spreadsheet downloader available from any listing.
Hosted Data Sets
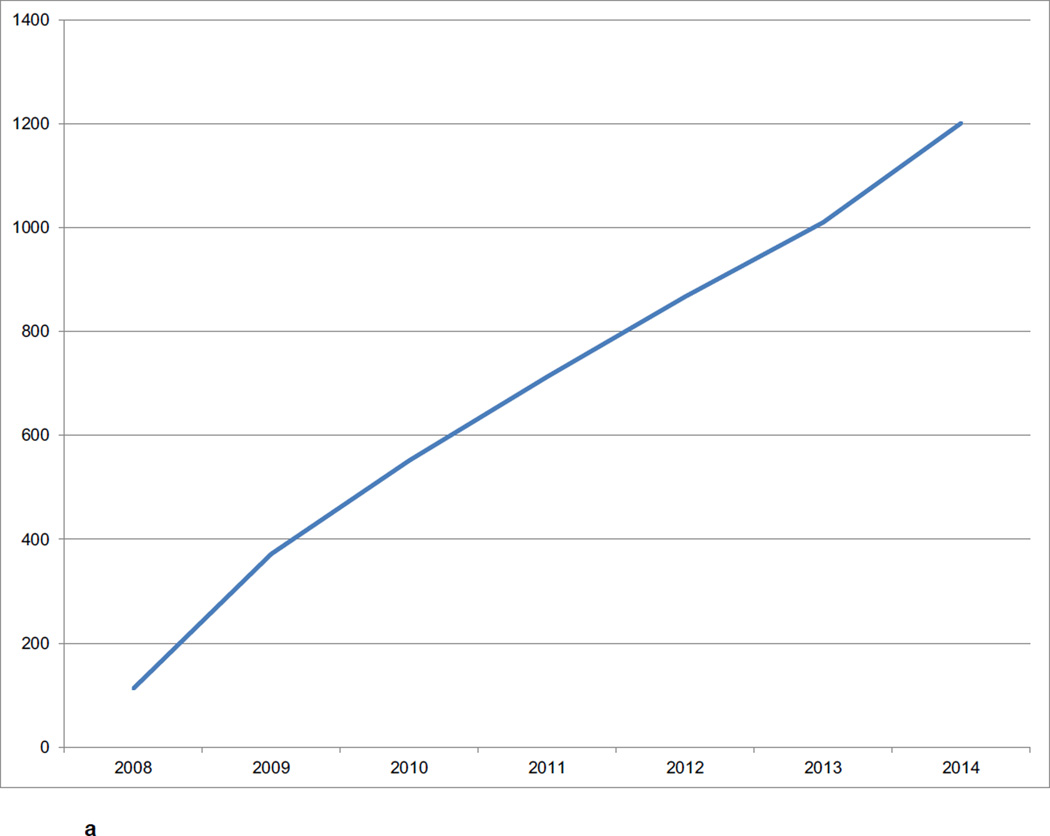
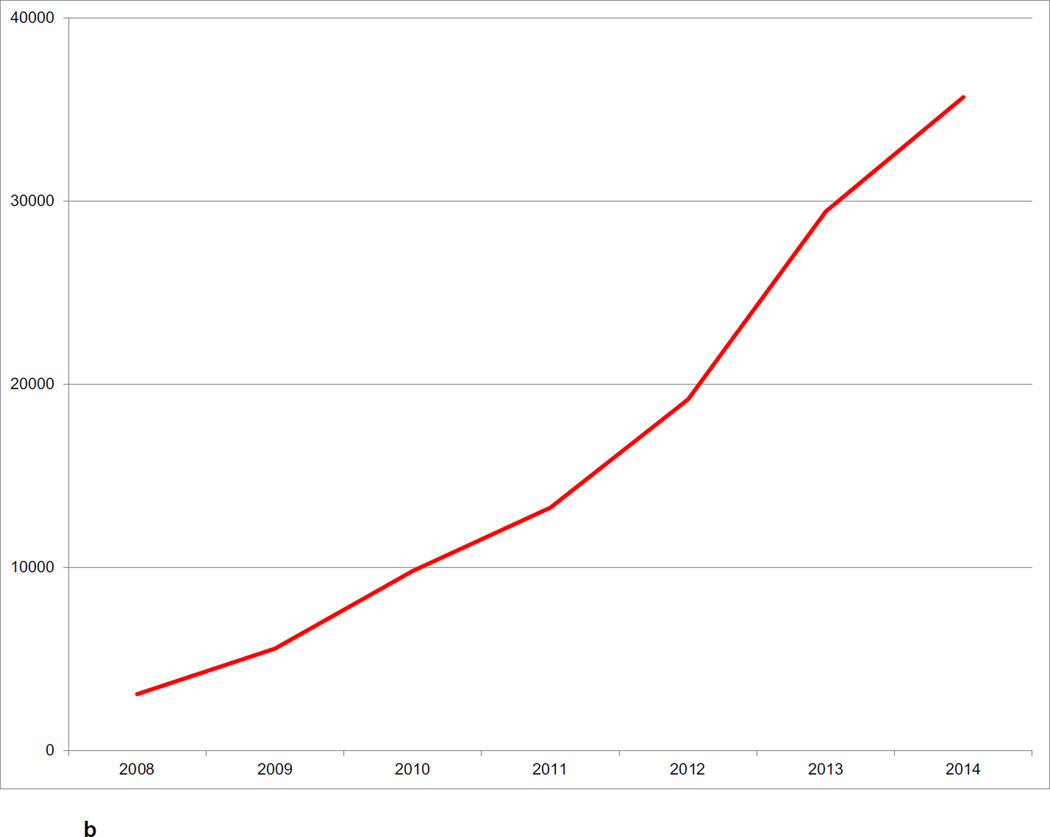
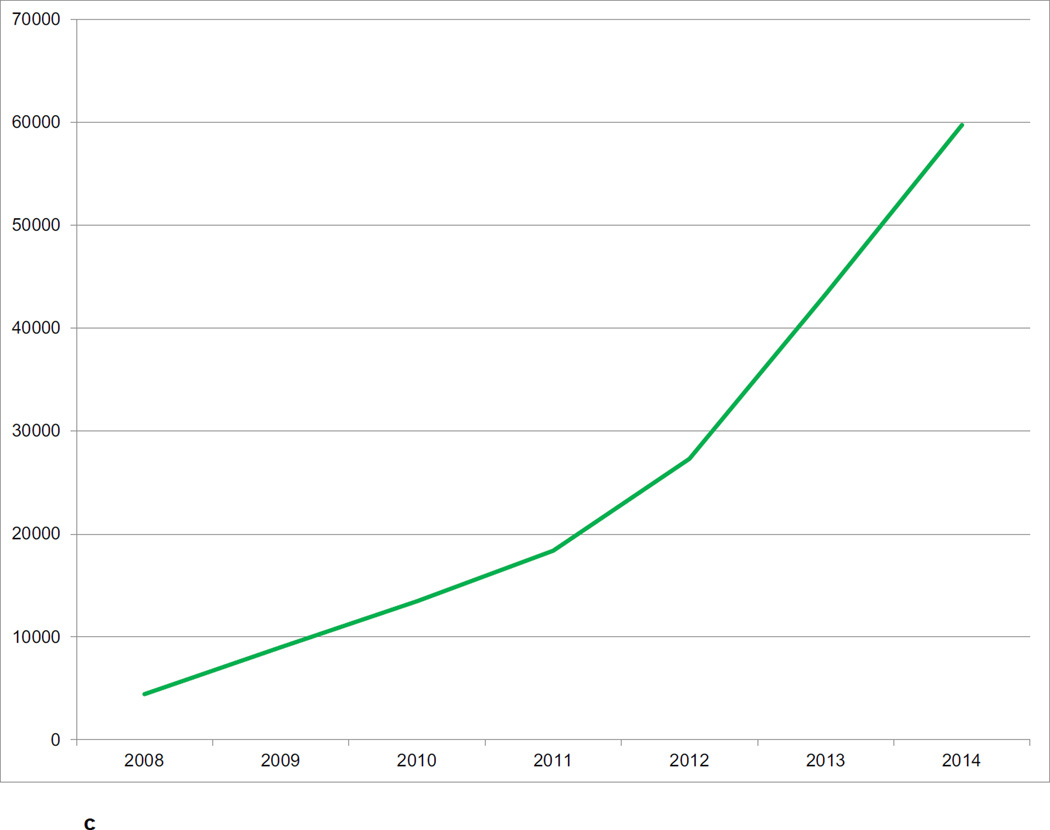
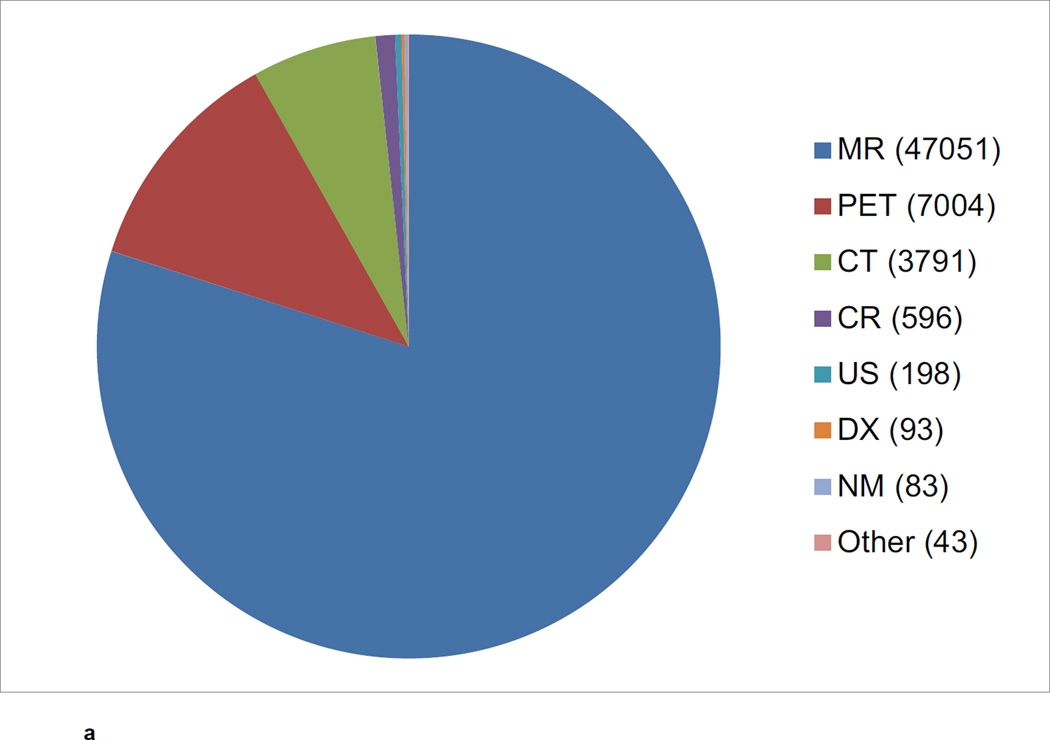
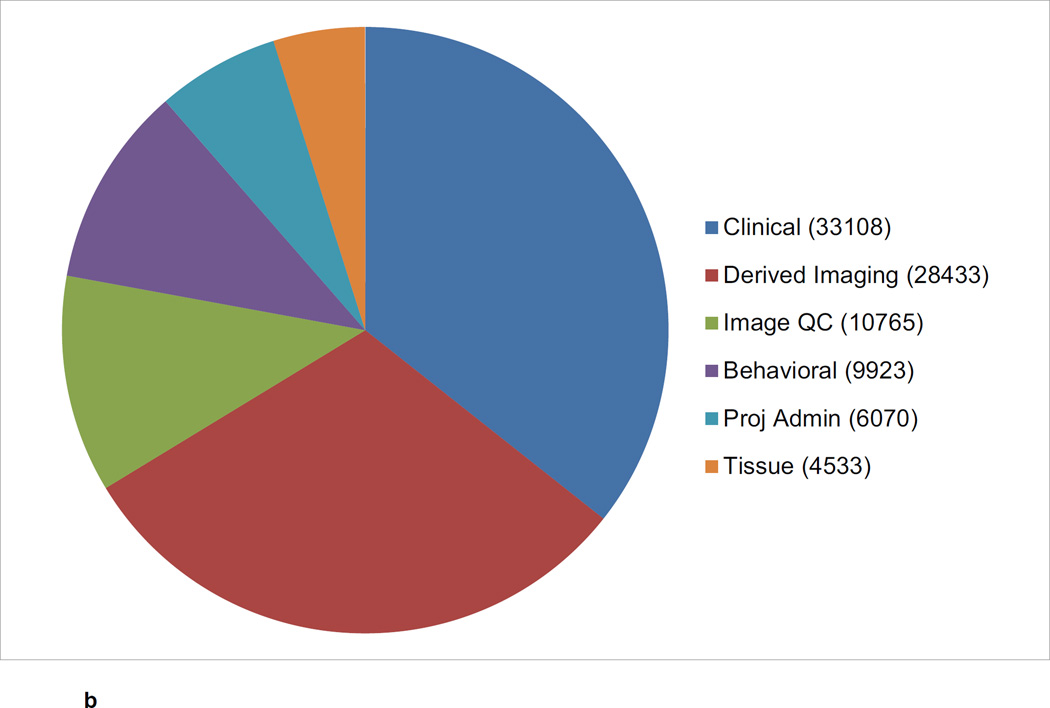
The CNDA hosts data acquired in over 1000 research studies, encompassing 36,000 subjects and more than 60,000 imaging sessions (Figure 2a, Figure 2b, Figure 2c). Additional data is received on a daily basis. Most imaging modalities used in modern human research are represented in the CNDA, including 1.5T and 3T magnetic resonance (MR), positron emission tomography (PET) (with a variety of tracers including glucose, amyloid, and Tau-binding compounds), computed tomography (CT), nuclear medicine (NM), computed radiography (CR), digital radiography (DX), and ultrasound (US). However, the majority of the imaging data in the CNDA are MR and PET of the human brain (Figure 3a). The CNDA also stores almost 100,000 instances of non-imaging data collected for neuroimaging studies, including clinical, psychometric, tissue, and derived imaging data. (Figure 3b). Although these non-imaging forms are usually based on standard instruments, they are often customized for the unique needs of the study of a particular disorder. For convenience, several studies have aggregate non-imaging data types, in which clinical, behavior, biomarkers, and genetics data can be stored together.
Figure 2.
a CNDA Projects by Year
b CNDA Subjects by Year
c CNDA Image Sessions by Year
Figure 3.
a CNDA Image Sessions by Modality
b CNDA Non-Imaging Data Types
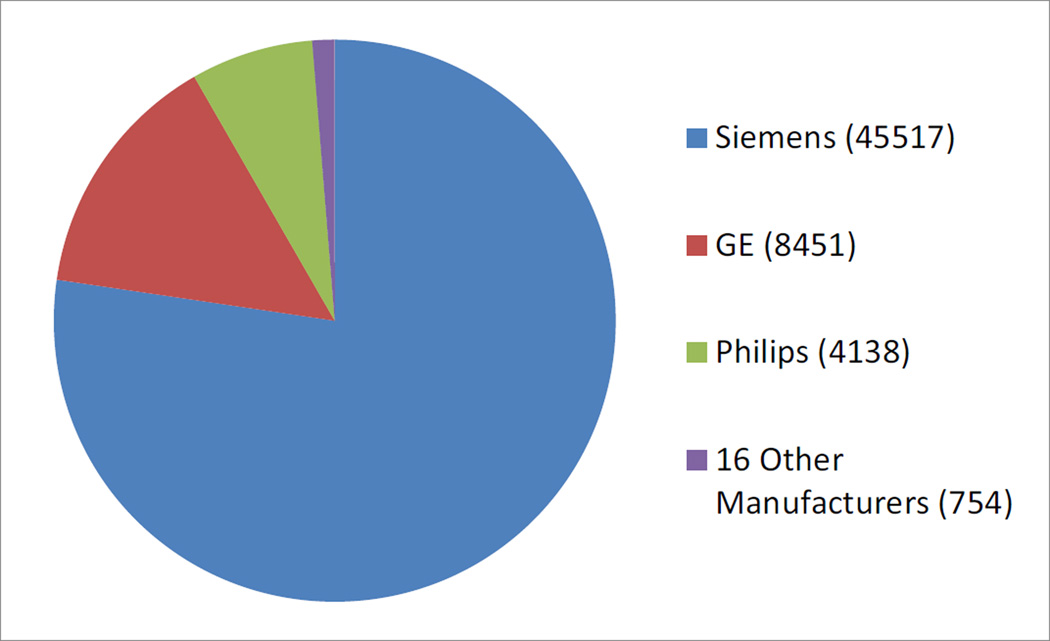
The CNDA primarily archives studies from the research scanners at WUSM. All studies acquired at five of the school’s six research scanners are sent to directly to the CNDA for archive. The sixth scanner is used almost exclusively to acquire data for the Human Connectome Project (HCP), and so the data is sent to the HCP XNAT instance (see Hodge et al, this issue). Nonetheless because of multi-center projects and WUSM investigator collaborations, the CNDA houses imaging data from close to 100 different academic and government research institutions. Although the majority of data comes from Siemens scanners, the CNDA also contains a diverse set of imaging from other scanner manufacturers and models (Figure 4). Most of the image sessions were acquired for research, limiting the amount of protected health information (PHI) contained in the metadata, however a growing number of retrospective clinical studies, for which (with institutional review board (IRB) approval) scans are pulled from a hospital PACS, de-identified, and coded for research in the CNDA.
Figure 4.
CNDA Image Sessions by Scanner Manufacturer
There are large result sets for processing done with FreeSurfer v5.0 and v5.1. FreeSurfer v5.3 was just added as a pipeline. CNDA researchers also frequently use a 4dfp-based BOLD pre-processing pipeline (Shulman et al., 2010; Fox et al., 2009) to generate BOLD QC and prepared files, which can be downloaded to do offline seed-based analysis. For PET data, the Knight ADRC imaging core has created a processing pipeline to generate binding potentials for either FreeSurfer regions (if available) or manually drawn regions (Su et al., 2013). These data are available via request to the respective studies.
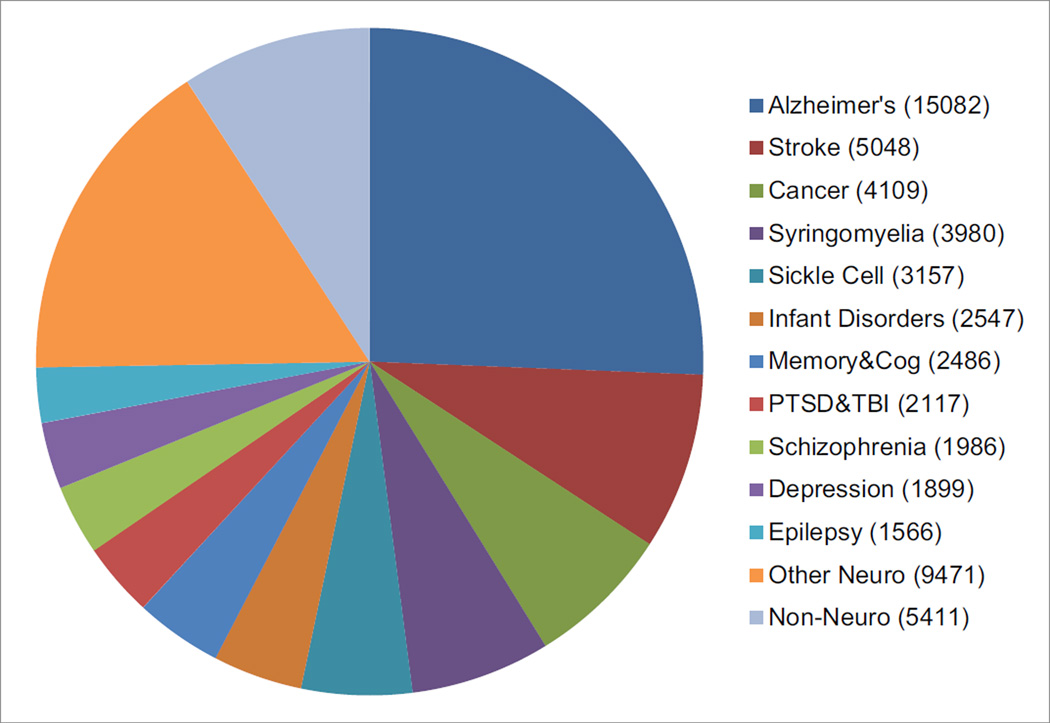
The CNDA contains images acquired for most neuroimaging topics of study. Approximately one quarter of all CNDA images belong to Alzheimer’s disease (AD) studies, the largest of which is the ongoing DIAN. DIAN is a worldwide 15-site project to investigate genetic mutations known to cause AD in individuals under the age of 60. All DIAN clinical, behavioral, biomarkers and imaging data is stored in the CNDA. Projects studying stroke account for almost another 10% of the CNDA’s imaging data. The CNDA stores CT and MR images for the Stroke Genetics Network (SiGN) (Meschia et al., 2013) project, a genetics study to determine ischemic stroke subtypes by using imaging to categorize the strokes. Cancer is the subject of study for about 7% of the CNDA’s investigators. Hosted by the CNDA, the Comprehensive Neuro-Oncology Data Repository (CONDR) (Fouke et al., 2014) is a project to develop advanced imaging biomarkers, and an informatics platform to facilitate neuro research related to these markers. The CNDA stores MR and PET imaging for the CONDR project as well as clinical data and tumor data in forms specially designed for the project. The study of syringomyelia also makes up about 7% of all CNDA imaging studies. All of this data belongs to one study, the Park-Reeves Syringomyelia Research Consortium (Park Reeves) (https://park-reeves.wustl.edu) a multi-institutional North American research effort founded to improve the medical and surgical care of children with syringomyelia related to Chiari I malformation. Park Reeves stores in the CNDA, MR, CT, CR and DX data from 31 institutions across the US. (Figure 5).
Figure 5.
CNDA Image Sessions by Topic of Study
Quality Control
For quality control (QC), the CNDA uses XNAT’s standard features in conjunction with a number of customized forms and pipelines. At the most basic level, XNAT provides a quality field for each scan in an imaging session. These fields are often used in CNDA by scanner technicians and study coordinators to indicate when movement or other issues may have rendered a scan unusable for reading or processing. For several CNDA projects, a QC expert assesses by hand each individual imaging session and records the results in the XNAT Manual QC form, which is also used to inform processing decisions. Several studies in the CNDA use XNAT’s automated DICOM protocol validation tool. This QC method allows studies to vary specific parameters to be used in the acquisition of imaging studies. Projects can then set the pipeline to launch automatically upon the upload of a new imaging session. The actual acquisition protocol parameters recorded in the DICOM metadata is compared to the expected protocol, and a verification report is generated and distributed to designated individuals.
QC support is also provided for two widely used processing pipelines, FreeSurfer (Fischl, 2012) and 4dfp BOLD (Shulman et al., 2010). Because changes in the brain caused by a few diseases can actually interfere with FreeSurfer’s segmentation algorithm, the results are almost always manually QC’ed. Since for BOLD processing movement is the cause of issues, the 4dfp BOLD pipeline actually performs its own automated QC report on movement and excess variation in field. The QC results are stored in XNAT’s QC form.
In addition to all the imaging QC measures mentioned above, other types of data such as clinical or psychometric assessments often undergo QC outside the CNDA.
Data Access
The CNDA requires all users to create password-protected system accounts. Data within the CNDA are managed within XNAT projects, which limit access to users who have been explicitly granted permission to access the data within them. Anyone may create a CNDA user id by navigating to cnda.wustl.edu and clicking the “Register” link, however, ids are only enabled for those users who are expected to be granted access to project data. Access is controlled by the project owner, typically the study’s principal investigator or a designated proxy. To further restrict access, project owners can use the XNAT sharing feature to expose subsets of data into a secondary project.
Once granted access to a project, users can download the data using a variety of mechanisms. Imaging sessions can be downloaded as a zip or straight to a directory. CNDA provides a single session zip downloader, a multi-session bulk downloader through the website, and an interface to download data via REST. The multi-session bulk downloader provides support to stop and resume downloads and is able to restart interrupted downloads from previous sessions. All other data in CNDA, clinical, psychometrics, tissue lab results, image processing results, etc. are most typically downloaded via the website into a CSV file. Via the XNAT REST interface, non-imaging data can be downloaded not only in CSV format, but also HTML, XML or JavaScript Object Notation (JSON). Users can use the XNAT Advanced Search capability to create their own custom data sets for download.
Investigators wishing to cite data in the CNDA can use URLs provided by the standard XNAT programming interface. The CNDA is currently exploring methods to streamline data distribution for our projects so that study collaborators do not need special knowledge of the system to download a predefined data set. Our hope is that XNAT will ultimately provide this support, including unique DOIs for data sets. Were this feature implemented, the CNDA would ultimately like to provide a de-identification pipeline for published studies to easily export their data to the public imaging repository, XNAT Central (Herrick et al, this issue).
Data Requests
All data requests and Data Usage Agreements for data housed in the CNDA are administered external to the CNDA by the individual studies. Observed data requests in the CNDA have ranged between 50 and 1000 imaging sessions. However, because individual studies have great autonomy over access to their data (including the ability to prepare access to a limited data set using XNAT’s data sharing feature), in many cases, the CNDA staff may not be aware that a data request has been executed. The CNDA does not currently track data download metrics.
Many factors dictate whether data may be available to outside researchers, including the consent signed by the participants, the terms of IRB approval, and the resources available to thoroughly de-identify data. Also many researchers choose not to make their data available until after publication. Currently, about 20% of the total imaging data in the CNDA is available to external researchers.
Almost any research imaging data acquired at WU was acquired on a Siemens scanner. Imaging data is generally made available in DICOM or NIfTI format. Most of the available studies offer to share not only imaging data, but often clinical, psychometric, tissue and genetics data if available. In some cases only a project’s imaging data will be stored within the CNDA while the other data resides in the WU REDCap instance. In all cases, the individual studies determine the mechanism for distributing the non-imaging portions of the data.
In addition to the DIAN project mentioned above (scanning at 15 sites on Siemens, Philips, and GE scanners), the Knight ADRC also makes available data from three other long term, longitudinal Alzheimer’s and dementia-related studies, including The Healthy Aging and Senile Dementia Study (HASD), the core ADRC study, and the Adult Children Study (ACS) (Coats and Morris, 2005), all of which were conducted at WU on Siemens scanners of various magnet strengths and models. A number of additional CNDA data sets are also available. Healthy control data is available from the Regional Aerobic Glycolysis in the Human Brain study (Vaishnavi et al., 2010). The CONDR study, mentioned above, makes available a large data set of pre-surgical, intra-surgical and follow-up scans acquired at two sites on Siemens and GE scanners. A large HIV-related data set is offered by a number of studies including the Chronic Co-Morbid Conditions in HIV+ U.S. Adults on Highly-Active Anti-Retroviral Therapy (HAART) (Ances et al., 2012) and Neuroimaging and Neurobehavioral Basis of Risky Decision-Making in Adolescents (Baker et al., 2014). The Silent Cerebral Infarct Transfusion Trial (SITT) (Vendt et al., 2009) offers a data set for the 29 site sickle cell study which ran from 2004 through 2013, acquiring MR data on Siemens, Philips and GE scanners. Finally, the Tourette Syndrome Association Neuroimaging Consortium (TSANIC), a four site study, will be making its Siemens and Philips MR data available sometime in the near future.
Although the feature deserves serious future consideration, no automated system currently exists within the CNDA to alert users to data set withdrawals, revisions or additions. The ongoing Alzheimer’s studies all prepare “data freezes” once or twice a year to provide researchers with a recent, quality-controlled snapshot of the cumulative data set. Researchers may work with the projects’ respective administrative cores to arrange access to a specific data freeze.
Intended Use
The CNDA accepts data from all WU investigators and by contract to non-WU studies. The CNDA is intended primarily to support data collection, management, collaborative research, and controlled data sharing. Non-Washington University studies are considered by CNDA on a case-by-case basis. Considerations in accepting external studies into the CNDA include the study’s compatibility with the existing CNDA infrastructure and availability of CNDA resources. There are fees associated with non-Washington University studies. A contract is established between the CNDA and the outside study based on the specific services to be provided and length of service. Full open access data sharing is better served by alternative systems such as XNAT Central, NITRC IR (Kennedy, et al., this issue), and Open fMRI (Poldrack et al., 2013).
Long term plans
The CNDA will continue to serve as the primary WUSM imaging archive. A major focus of ongoing work is to address overall performance to better handle the rapid expansion of the system over the last several years. Another ongoing IT challenge is the question of how to store and maintain large imaging data sets, particularly as they are no longer in active use. Ideally, tiered data storage would provide lower cost storage for infrequently used data. We also intend to introduce improved de-identification and data sharing capabilities, so that anonymized data sets can be efficiently contributed to open access data sharing platforms such as those listed above. Finally, we intend to integrate XNAT and the WU REDCap (Harris et al., 2009) system to provide streamlined access to data for the growing number of CNDA projects with imaging and clinical data sets that span the two systems.
Table 2.
CNDA Data Sets Available by Request. Updated table available at http://cnda-help.wustl.edu/data-sharing. “X” denotes that data of that type is available. CT data indicated is often acquired in the form of attenuation data on a PET-CT scanner.)
| Topic of Study |
Study | Description | Subj | MR | PET | CT | Clin | Psych | Tissue | Gene | To Request Data |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alzheimer’s Disease |
Healthy Aging and Senile Dementia (HASD), P01AG03991 Knight ADRC |
Since 1984, HASD has longitudinally studied older adult (65y+) with annual clinical and cognitive assessments and biomarker measures at longer intervals to distinguish normal aging from dementia due to AD, with emphasis on pre-clinical AD. |
417 | 739 | 373 | X | X | X | X | X | http://knightadrc.wustl.edu/ResourceDB/default2.htm |
| Alzheimer’s Disease |
Alzheimer's Disease Research Center (ADRC), P50AG05681 Knight ADRC |
Since 1985, ADRC has longitudinally studied older adult (65y+) with annual clinical and cognitive assessments and biomarker measures at longer intervals to distinguish normal aging from dementia due to AD. Support for all aging/dementia research but with emphasis on pre-clinical AD. |
478 | 974 | 498 | X | X | X | X | X | http://knightadrc.wustl.edu/ResourceDB/default2.htm |
| Alzheimer’s Disease |
Antecedent Biomarkers of AD: the Adult Children Study (ACS), P01AG026276) Knight ADRC |
Since 2005, ACS has longitudinally studied participants (ages 45 to 74y at entry with increased AD risk due to a having parent diagnosed with AD) in order to help detect the earliest signs or markers of dementing illnesses, even pre-clinical disease, such as AD. |
342 | 927 | 859 | X | X | X | X | X | http://knightadrc.wustl.edu/ResourceDB/default2.htm |
| Alzheimer’s Disease |
Dominantly Inherited Alzheimer's Network (DIAN), UF1AG032438 |
Since 2008, DIAN has enrolled adult children (18y+) at 50% risk of inheriting a causative mutation for AD. Mutation carriers and non-carriers are comprehensively assessed longitudinally with clinical, cognitive, imaging and biofluid assessments. |
351 | 696 | 1308 | X | X | X | X | X | http://www.dian-info.org/resourcedb/Data/default.asp |
| Brain Metabolism |
Regional Aerobic Glycolysis in the Human Brain NS08633 |
Study scanned 33 neurologically normal young adults at rest. |
33 | 33 | 33 | https://cnda-help.wustl.edu/data-sharing-brain-met | |||||
| Cancer |
Comprehensive Neuro-Oncology Data Repository (CONDR), R01NS066905 |
CONDR focuses on subjects with brain tumors, comparing clinical information, biopsy pathology results, and MRI imaging for each subject. |
157 | 1396 | 15 | X | X | X | http://nrg.wustl.edu/nrg-projects/condr | ||
| HIV |
Multiple Studies, including R01NR012907, R01NR012657 |
Data acquired in HIV-related studies over the last 7 years on affected and non-affected individuals over a wide age distribution. |
617 | 745 | 28 | X | X | https://cnda-help.wustl.edu/data-sharing-HIV | |||
| Sickle Cell Anemia |
Silent Cerebral Infarct Transfusion Trial (SITT) |
Now concluded, SITT studied subjects ages 5–14y with sickle cell anemia. The primary aim of the trial was to determine the effectiveness of blood transfusion therapy for the prevention of silent strokes in children. |
1074 | 1575 | X | https://cnda-help.wustl.edu/data-sharing-sickle-cell | |||||
| Tourette Syndrome |
Tourette Syndrome Association Neuroimaging Consortium (TSANIC), WU-12–40-MOD-2 NCTE, WU via Tourette Syndrome Association |
TSANIC studies subjects 7 to 17y with a diagnosis at any time of a chronic primary tic disorder or TS and at least one tic present in the past 3 months. Control subjects meet the same criterion except with no diagnosis of a lifetime tic disorder, OCD or ADHD. |
418 | 643 | X | https://cnda-help.wustl.edu/data-sharing-tourette |
Highlights.
Central Neuroimaging Data Archive houses Washington University's research imaging.
Initiated in 2002, CNDA served as the original codebase for the XNAT platform.
CNDA holds over 1000 research studies, 36,000 subjects and 60,000 image sessions.
CNDA stores MR, PET, and CT scans, clinical, behavioral and tissue-derived data.
Alzheimer’s, cancer, HIV, sickle cell anemia, and Tourette's data can be requested.
Acknowledgments
Funded in part by the Neuroimaging Informatics and Analysis Center (Principal Investigator: Daniel S. Marcus; P30 NS048056), the Dominantly Inherited Alzheimer Network (Principal Investigator: John C. Morris; U01 AG032438), ACS, Healthy Aging and Senile Dementia (Principal Investigator: Tammie Benzinger; 5P01AG00399130), Amyloid Imaging in the Adult Children Study (Principal Investigator: John C. Morris; 5P01AG026276-02), Washington University's Intellectual and Developmental Disabilities Research Center (Principal Investigator: Terrie E. Inder; 1P30HD062171), and The XNAT Imaging Informatics Platform (Principal Investigator: Daniel S. Marcus; 5R01EB009352).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ances BM, Ortega M, Vaida F, Heaps J, Paul R. Independent Effects of HIV, Aging, and HAART on Brain Volumetric Measures. J Acquir Immune Defic Syndr. 2012;59:469–477. doi: 10.1097/QAI.0b013e318249db17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LM, Paul RH, Heaps JM, Westerhaus E, Chang JY, Williams S, Brier MR, Plax K, Ances BM. Impact of human immunodeficiency virus on neurocognition and risky behaviors in young adults. J Neurovirol. 2014;20:466–473. doi: 10.1007/s13365-014-0264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats M, Morris JC. Antecedent Biomarkers of Alzheimer’s Disease: The Adult Children Study. J Geriatr Psychiatry Neurol. 2005;18:242–244. doi: 10.1177/0891988705281881. [DOI] [PubMed] [Google Scholar]
- Doran SJ, d’ Arcy J, Collins DJ, Andriantsimiavona R, Orton M, Koh D-M, Leach MO. Informatics in radiology: development of a research PACS for analysis of functional imaging data in clinical research and clinical trials. Radiographics. 2012;32:2135–2150. doi: 10.1148/rg.327115138. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouke SJ, Benzinger TL, Milchenko M, Lamontagne P, Shimony JS, Chicoine MR, Rich KM, Kim AH, Leuthardt EC, Keogh B, Marcus DS. The Comprehensive Neuro-Oncology Data Repository (CONDR): A Research Infrastructure to Develop and Validate Imaging Biomarkers. Neurosurgery. 2014;74:88–98. doi: 10.1227/NEU.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Burns SS, Lauzon CB, Fong AE, James TA, Lubar JF, Thatcher RW, Twillie DA, Wirt MD, Zola MA, Logan BW, Anderson AW, Landman BA. Integration of XNAT/PACS, DICOM, and Research Software for Automated Multi-modal Image Analysis. Proc SPIE. 2013;8674 doi: 10.1117/12.2007621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschia JF, Arnett DK, Ay H, Brown RD, Benavente OR, Cole JW, de Bakker PIW, Dichgans M, Doheny KF, Fornage M, Grewal RP, Gwinn K, Jern C, Conde JJ, Johnson JA, Jood K, Laurie CC, Lee J-M, Lindgren A, Markus HS, McArdle PF, McClure LA, Mitchell BD, Schmidt R, Rexrode KM, Rich SS, Rosand J, Rothwell PM, Rundek T, Sacco RL, Sharma P, Shuldiner AR, Slowik A, Wassertheil-Smoller S, Sudlow C, Thijs VNS, Woo D, Worrall BB, Wu O, Kittner SJ, NINDS SiGN Study Stroke Genetics Network (SiGN) study: design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–2702. doi: 10.1161/STROKEAHA.113.001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Aisen PS, Bateman RJ, Benzinger TLS, Cairns NJ, Fagan AM, Ghetti B, Goate AM, Holtzman DM, Klunk WE, McDade E, Marcus DS, Martins RN, Masters CL, Mayeux R, Oliver A, Quaid K, Ringman JM, Rossor MN, Salloway S, Schofield PR, Selsor NJ, Sperling RA, Weiner MW, Xiong C, Moulder KL, Buckles VD. Developing an international network for Alzheimer research: The Dominantly Inherited Alzheimer Network. Clin Investig (Lond) 2012;2:975–984. doi: 10.4155/cli.12.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Barch DM, Mitchell JP, Wager TD, Wagner AD, Devlin JT, Cumba C, Koyejo O, Milham MP. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinform. 2013;7:12. doi: 10.3389/fninf.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. Behind the scenes of functional brain imaging: a historical and physiological perspective. Proc. Natl. Acad. Sci. U.S.A. 1998;95:765–772. doi: 10.1073/pnas.95.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, Pope DLW, Astafiev SV, McAvoy MP, Snyder AZ, Corbetta M. Right hemisphere dominance during spatial selective attention and target detection occurs outside the dorsal frontoparietal network. J Neurosci. 2010;30:3640–3651. doi: 10.1523/JNEUROSCI.4085-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AZ, Raichle ME. A brief history of the resting state: the Washington University perspective. Neuroimage. 2012;62:902–910. doi: 10.1016/j.neuroimage.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, D’Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, Blazey TM, Christensen JJ, Vora S, Morris JC, Mintun MA, Benzinger TLS. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS ONE. 2013;8:e73377. doi: 10.1371/journal.pone.0073377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. Regional aerobic glycolysis in the human brain. PNAS. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: An overview. NeuroImage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendt BA, McKinstry RC, Ball WS, Kraut MA, Prior FW, Barton B, Casella JF, DeBaun MR. Silent Cerebral Infarct Transfusion (SIT) Trial Imaging Core: Application of Novel Imaging Information Technology for Rapid and Central Review of MRI of the Brain. J Digit Imaging. 2009;22:326–343. doi: 10.1007/s10278-008-9114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow R, Saltz JH, Foster I, Carr J, Ge Y, Miller M, Younes L, Geman D, Granite S, Kurc T, Madduri R, Rananather T, Larkin J, Ardkani S, Brown T, Kolasny A, Reynolds K, Shipway M, Toerper M. The CardioVascular Research Grid (CVRG) Project. Proceedings of the AMIA Summit on Translational Bioinformatics. 2011:77–81. [Google Scholar]