Developing metabolic syndrome leads to impaired cardiac autophagy and larger infarcts after myocardial ischemia and reperfusion. Quantitative proteomic analysis of hearts from diet-induced obese mice subjected to fasting stress reveal global changes in the proteome related to metabolism, redox, vesicular, and structural homeostasis that may increase vulnerability to ischemia-reperfusion injury.
Keywords: metabolic syndrome, peroxisome proliferator-activated receptor-α/γ, mammalian target of rapamycin, proteomics
Abstract
Autophagy is regulated by nutrient and energy status and plays an adaptive role during nutrient deprivation and ischemic stress. Metabolic syndrome (MetS) is a hypernutritive state characterized by obesity, dyslipidemia, elevated fasting blood glucose levels, and insulin resistance. It has also been associated with impaired autophagic flux and larger-sized infarcts. We hypothesized that diet-induced obesity (DIO) affects nutrient sensing, explaining the observed cardiac impaired autophagy. We subjected male friend virus B NIH (FVBN) mice to a high-fat diet, which resulted in increased weight gain, fat deposition, hyperglycemia, insulin resistance, and larger infarcts after myocardial ischemia-reperfusion. Autophagic flux was impaired after 4 wk on a high-fat diet. To interrogate nutrient-sensing pathways, DIO mice were subjected to overnight fasting, and hearts were processed for biochemical and proteomic analysis. Obese mice failed to upregulate LC3-II or to clear p62/SQSTM1 after fasting, although mRNA for LC3B and p62/SQSTM1 were appropriately upregulated in both groups, demonstrating an intact transcriptional response to fasting. Energy- and nutrient-sensing signal transduction pathways [AMPK and mammalian target of rapamycin (mTOR)] also responded appropriately to fasting, although mTOR was more profoundly suppressed in obese mice. Proteomic quantitative analysis of the hearts under fed and fasted conditions revealed broad changes in protein networks involved in oxidative phosphorylation, autophagy, oxidative stress, protein homeostasis, and contractile machinery. In many instances, the fasting response was quite discordant between lean and DIO mice. Network analysis implicated the peroxisome proliferator-activated receptor and mTOR regulatory nodes. Hearts of obese mice exhibited impaired autophagy, altered proteome, and discordant response to nutrient deprivation.
NEW & NOTEWORTHY
Developing metabolic syndrome leads to impaired cardiac autophagy and larger infarcts after myocardial ischemia and reperfusion. Quantitative proteomic analysis of hearts from diet-induced obese mice subjected to fasting stress reveal global changes in the proteome related to metabolism, redox, vesicular, and structural homeostasis that may increase vulnerability to ischemia-reperfusion injury.
metabolic syndrome (MetS) is characterized by obesity, hyperglycemia, insulin resistance, dyslipidemia, and, in many cases, hypertension. It is associated with accelerated atherosclerosis and worse outcome after myocardial infarction (7, 10, 13, 21). Efforts to increase the heart's tolerance to ischemia-reperfusion (I/R) injury through ischemic or pharmacological preconditioning or postconditioning that have been effective in animal studies were unsuccessful in clinical trials (3). One proposed explanation is that the presence of comorbid conditions such as MetS may interfere with cardioprotective interventions (44). The presence of MetS abrogates the protective effects conferred by ischemic preconditioning (IPC) and postconditioning against I/R injury (4, 11) and is associated with increased incidence and severity of acute myocardial infarction and heart failure (30, 32). We had previously shown that autophagy is essential for cardioprotection by IPC (14, 15, 31), and others had reported impaired autophagy in the setting of diet-induced obesity (DIO) (17). We previously reported that autophagy is upregulated as part of the homeostatic intracellular repair response in human hearts during cardiac surgery (16). There was an inverse correlation between the magnitude of the autophagy response during cardiac surgery and the presence of risk factors including features of MetS (16).
Autophagy is responsible for degradation of protein aggregates and removal of excess or damaged organelles (25). In the heart, autophagy is induced by cellular stresses such as starvation or ischemia (14, 31). Nutrient depletion (such as during ischemia) and nutrient excess (as in DIO) act on key energy- and nutrient-sensing pathways, namely, the AMP-activated kinase (AMPK) and mammalian target of rapamycin (mTOR) signaling pathways, that regulate autophagy. An understanding of how MetS impacts these pathways in the setting of nutrient deprivation is essential to develop new approaches for increasing the heart's tolerance to ischemia and reperfusion. Accordingly, we proposed to study the regulation of cardiac autophagy in the mouse model of DIO, using fasting as a cardiometabolic stress.
METHODS
Animals and experimental protocols.
All animal studies were approved by the San Diego State University Institutional Animal Care and Use Committee. Groups of aged-matched male FVBN mice (Jackson Labs) were maintained on a 12:12-h dark-light schedule, housed 3–4/cage, and fed ad libitum with normal chow (Teklad 2014, 13% kcal/100 kcal fat) or a lard-based high-fat diet (HFD, Research Diets D12492, fat 60% kcal/100 kcal fat) for 4–20 wk. For overnight fasting experiments, mice were placed in a fresh cage without food 3 h before lights out at 6:00 PM. At the end of the study, hearts from fed and fasted animals were freeze-clamped in liquid nitrogen and stored at −80°C for mRNA and protein analysis.
Myocardial I/R model and infarct size determination.
I/R was performed as previously described (15) with a few modifications. Briefly, lean or DIO FVBN mice were anesthetized with an intraperitoneal injection of pentobarbital sodium (70 mg/kg). Isoflurane anesthesia (0.5%) was used throughout the procedure. A volume-controlled ventilator (model 687; Harvard Apparatus) was used and maintained at a rate of 120–140 beats/min and a pressure of 2–5 cmH2O. The chest was then opened, and a snare composed of an 8-0 silk suture was used to occlude the left anterior descending artery. At 60 min occlusion, the snare was released but left in position, and the heart was reperfused for 120 min. Area-at-risk and infarct size were defined using standard dye delineation and triphenyltetrazolium chloride staining. Mortality was scored for animals that did not survive the 2-h reperfusion; hearts were included for infarct size analysis if the mice survived at least 80 min of reperfusion.
Measurements of leptin, insulin, and glucose.
Serum samples from lean and DIO mice were obtained from the tail vein blood and analyzed for leptin and insulin levels by ELISA (Mercodia). Blood glucose was measured with a hand-held glucometer (OneTouch Ultra; Lifescan). Measurements were performed in duplicate.
Measurement of insulin sensitivity: oral glucose tolerance test.
After overnight fasting, animals were administered d-glucose (2 mg/g, 25% solution) by oral gavage. Blood glucose was sequentially measured via tail vein samples taken at baseline and 30, 60, 90, and 120 min following administration. The homeostatic model assessment (HOMA-IR) was calculated using the standard international formula [fasting glucose (mmol/l) × fasting insulin (mU/l)/22.5].
Assessment of autophagic flux.
Chloroquine (CQ, 10 mg/kg in 50–200 μl saline; Sigma) or vehicle was administered via intraperitoneal injection to evaluate basal autophagic flux in the lean and DIO animals. A pilot study to assess the appropriate interval between CQ administration and tissue Western blot analysis revealed a significant increase in LC3-II (compared with vehicle control animals) as early as 2 h after CQ administration, indicating a robust basal autophagic flux (data not shown). Thereafter, 10 mg/kg CQ or saline injections were administered at 8:00 AM, before 10:00 AM harvest. Hearts were freeze-clamped, and tissue was obtained for measurement of LC3 by Western blot (35, 37) in age-matched mice maintained on HFD for 8 wk (after weaning) and compared with their controls (lean mice).
Assessment of LC3B, p62/SQSTM1, 18S rRNA gene expression.
RNA was isolated from 30 mg heart tissue using a Nucleospin RNA kit according to the manufacturer's protocol (Macherey Nagel). cDNA was prepared from 1 μg RNA using the iScript cDNA synthesis kit according to the manufacturer's protocol (Bio-Rad). Quantitative RT-PCR of LC3B and p62/SQSTM1 was performed in triplicate using the iTaq Universal SYBR Green Kit (Bio-Rad) on a Bio-Rad CFX96 Real-Time System with 18S rRNA serving as an endogenous control. Primer pairs were as follows: 1) LC3B forward: GCGACTGGAGAGCTGTTTCT; reverse: GTGAGCCAAGTCTGGAGCAT; 2) p62/SQSTM1 forward: AGCCCTCTAGGCATTGAGGTTGA; reverse: GCCTGTGCTGGAACTTTCTGGG; and 3) 18S rRNA forward: AGTCCCTGCCCTTTGTACACA; reverse: CGATCCGAGGGCCTCACTA.
PCR conditions were as follows: 95°C for 10 min (95°C for 10 s, 61°C for 30 s) for 40 cycles. Relative mRNA expressions were calculated using the ΔΔCT method using CFX manager software version 3.1 (Bio-Rad).
Assessment of autophagy and nutrient-sensing pathways.
Proteins were extracted from the frozen tissue samples by homogenization in ice-cold RIPA buffer with freshly added Protease Inhibitor Cocktail Complete and PhosSTOP (Roche) followed by centrifugation (1,000 g for 5 min, 4°C), recovery of supernatants, and determination of protein concentrations (Bio-Rad DC Protein assay). Equal amounts of protein (20 μg) were solubilized in sample buffer with 10% β-mercaptoethanol, resolved on 10–20% Tris-glycine SDS-PAGE (Life Technologies), and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk for 1 h (room temperature) and then incubated overnight at 4°C with 1:1,000 diluted primary antibodies against Rho GDIα (sc-373724) (Santa Cruz Biotechnology), LC3 (4108), phospho-AMPK (Thr172) (2653), AMPK (2532), pS6 (Ser235/236) (2211), S6 (2217), pUlk1 (Ser757) (6888), and Ulk1 (Cell Signaling) and p62/SQSTM1 (ab46516) (Abcam). Membranes were washed (3 × 10 min) in 150 mM NaCl buffered with 50 mM Tris (pH 7.6) with 0.1% Tween 20 (TBST) at room temperature and incubated with the appropriate peroxidase-conjugated secondary antibodies (1:5,000; Kirkegaard & Perry Laboratories) for 1 h. Following washing in TBST, blots were developed with SuperSignal West Dura Extended Duration Substrate (Thermo-Pierce), and immunoreactive bands were visualized within the linear range on the Bio-Rad ChemiDoc XRS system. Captured images were quantified by densitometry using NIH ImageJ.
Statistical analysis.
All quantified results are expressed as means ± SD. Two-tailed Student's t-test and one-way ANOVA followed by post hoc Tukey's test were used where appropriate, with a P value <0.05 being considered statistically significant.
Proteomic sample preparation.
Frozen hearts (n = 4/group) obtained from lean nonfasting, lean fasting (overnight), DIO nonfasting, and DIO fasting (overnight) animals were ground while frozen in a liquid N2 basin-cooled mortar and then prepared for IN-Sequence analysis to enrich for the 1) myofilament and 2) soluble proteins, including mitochondrial proteins (19). Briefly, 200 μg of protein were denatured and reduced in a solution (20 mM DTT, 50 mM ammonium bicarbonate, pH 8, and 8 M urea) at 60°C for 1 h, alkylated with iodoacetamide (50 mM) for 20 min at room temperature (in the dark) and then 75 μg of protein digested in 1 μg of Trypsin/Lys-C mix (Promega) using the Filter-Aided Sample Preparation kit (Expedion). Samples were desalted and cleaned using HLB plates (Oasis HLB 30 μm, 5 mg sorbent; Waters).
Liquid chromatography MS/MS analysis.
Liquid chromatography MS/MS was carried out on a Dionex Ultimate 3000 NanoLC connected to an Orbitrap Elite (Thermo Fisher) equipped with an EasySpray ion source. Mobile phase A was comprised of 0.1% aqueous formic acid, and mobile phase B was 0.1% formic acid in acetonitrile. Peptides were loaded on the analytical column (PepMap RSLC C18 2 μm, 100 Å, 50 μm ID × 15 cm) at a flow rate of 300 nl/min using a linear AB gradient composed of 2–25% mobile phase A for 120 min, 25–90% mobile phase B for 5 min, and then isocratic hold at 90% for 5 min with re-equilibrating at 2% mobile phase A for 10 min (20). Temperature was set to 40°C for both columns. Nanosource capillary temperature was set to 275°C, and spray voltage was set to 2 kV. MS1 scans were acquired in the Orbitrap Elite at a resolution of 60,000 full width at half-maximum with an AGC target of 1 × 106 ions over a maximum of 500 ms. MS2 spectra were acquired for the top 15 ions from each MS1 scan in normal scan mode in the ion trap with a target setting of 1 × 104 ions, an accumulation time of 100 ms, and an isolation width of 2 Da. Normalized collision energy was set to 35%, and one microscan was acquired for each spectra.
MS analysis and bioinformatics.
The raw MS files were converted to mzXZML using MS Convert and searched against the Swiss-Prot-reviewed mouse FASTA database (33,330 proteins and decoys) using the COMET search algorithm (9). Target-decoy modeling of peptide spectral matches was performed with peptide prophet (22), and peptides with a probability score of >95% from the entire experimental dataset were imported into Skyline software (29) to establish a library for quantification of precursor-extracted ion intensities (XICs). Hemoglobin and carbonic anhydrases were excluded due to their representation of erythrocyte background contamination (18). Precursor XICs from each experimental file were extracted against the Skyline library, and peptide XICs with isotope dot product scores 0.8 were filtered for final statistical analysis of proteomic differences (43). Normalization of raw peptide intensities and protein level abundance inference were calculated using the linear mixed-effects model built into the open sources MSSTATs (version 3.2.2) software suite (6). With the exception of initial network building with autophagy-associated proteins, only the results from MSSTATs output were used in further analysis if a nine-degrees-of-freedom cutoff and the two-peptide-identifier requirement were met. Protein isoforms were confirmed based on observed peptide with an amino acid sequence that was unique to the isoform. Pairwise comparisons between experimental groups were calculated in MSSTATs, and protein abundance differences with a nominal P value <0.05 were considered significantly different. Functional grouping and pathway analysis of the differentially abundant proteins (P < 0.05 or 0.10 as indicated) that were detected in all experimental groups were performed in Ingenuity Pathway Analysis (IPA). Selected networks were exported from IPA and visualized with customized network building in Cytoscape. MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD003345.
RESULTS
High-fat diet feeding leads to features of the metabolic syndrome and increased susceptibility to I/R injury.
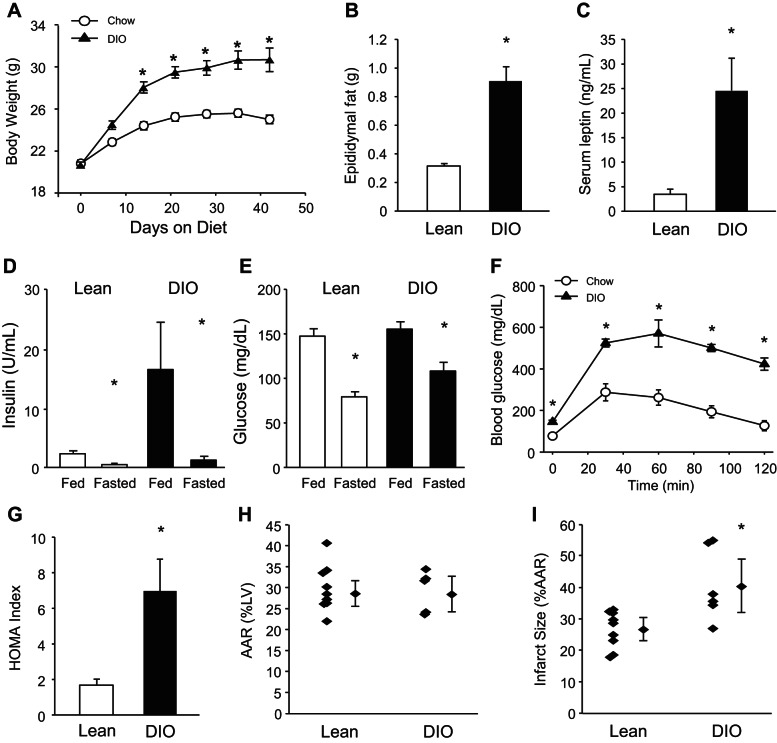
The C57BL/6 mouse strain placed on a high-fat diet (HFD) is an accepted rodent model of MetS. The mice develop obesity, elevated fasting blood glucose, and insulin resistance, in contrast to age-matched chow-fed animals (5, 23, 36, 39). Fewer studies of DIO have employed the FVBN strain albeit this strain was recently reported to resemble the DBA/2, 129X1, and C57BL/6 strains in that body weights and whole body fat mass increased and glucose tolerance was impaired in response to HFD (33). This strain is also the genetic background for the mice that we previously used to evaluate the role of autophagy in ischemic preconditioning (15). To validate this strain for our planned investigation of MetS and its impact on cardiac stress response, we maintained male FVBN mice on standard chow or HFD ad libitum for up to 20 wk. FVBN mice on HFD developed significantly increased body weight over time (Fig. 1A). Body fat was increased (Fig. 1B) and was accompanied by an increase in the adipose-derived hormone, leptin, which is known to circulate at levels directly proportional to body fat (Fig. 1C). Moreover, HFD-fed animals exhibited abnormal glucose homeostasis reflected by elevated insulin (Fig. 1D) and glucose (Fig. 1E) levels, abnormal oral glucose tolerance test (Fig. 1F), and an elevated HOMA index compared with chow-fed mice (Fig. 1G). In line with others (17), development of MetS in our model leads to increased susceptibility to I/R injury (Fig. 1, H and I).
Fig. 1.
High-fat diet feeding leads to features of the metabolic syndrome and increased ischemia-reperfusion (I/R) injury. Lean and diet-induced obesity (DIO) mice were assessed for time-dependent changes in body weight (*P < 0.001, A), epididymal fat pad weight (*P < 0.01, B), fasting serum leptin level (*P < 0.01, C), insulin levels (*P < 0.05, D), serum glucose levels (*P < 0.05, E), oral glucose tolerance test (*P < 0.05, F), and homeostatic model assessment (HOMA) index calculated from fasting insulin and serum glucose levels (*P < 0.01, G) (n = 4–6 mice/group in A–G). Lean and DIO mice were subjected to I/R to determine the percentage cardiac area at risk (AAR) after I/R (H) and infarct size as percentage of AAR (I) (n = 6–8/group, *P < 0.05).
HFD-induced metabolic syndrome blunts cardiac autophagic flux.
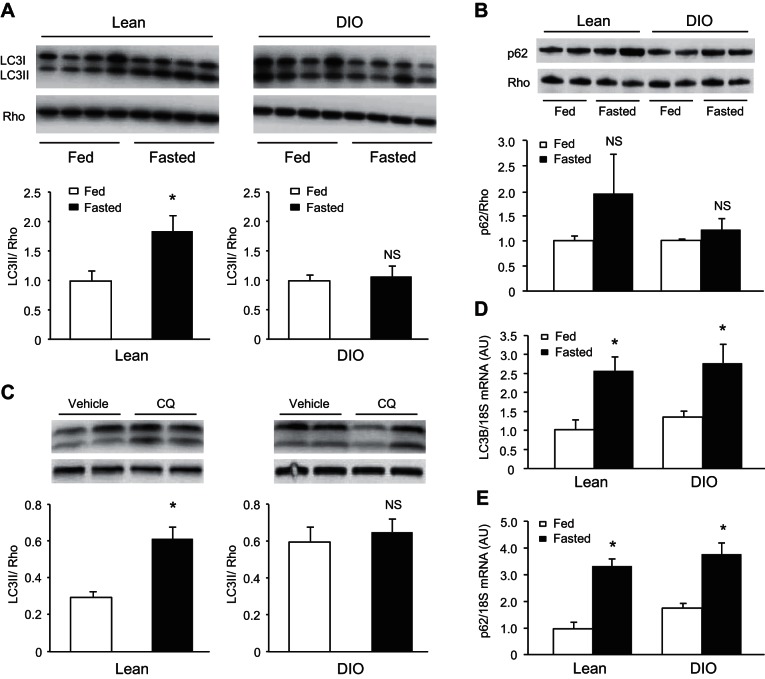
We investigated autophagy induction by comparing levels of LC3-II with Rho at baseline or 24 h after fasting. In lean mice there was an increased LC3-I conversion to LC3-II upon fasting; DIO mice failed to do so (Fig. 2A). Fasting increased the abundance of autophagy adapter protein p62/SQSTM1 that did not reach statistical significance; no such increase was seen in fasted DIO mice (Fig. 2B). To examine autophagic flux, we administered CQ 2 h before death to prevent autophagosome-lysosome fusion. DIO mice (after 8 wk of HFD feeding) did not show increased LC3-II after lysosomal blockade, consistent with diminished autophagic flux (Fig. 2C).
Fig. 2.
Impaired cardiac autophagy in diet-induced obese mice. Western blot analysis of LC3 (A) and p62/SQSTM1 (B) in lean and DIO mice fasted overnight (n = 4/group, *P < 0.05). C: autophagic flux of fasting animals (on control or HFD for 8 wk postweaning) was determined by LC3-II accumulation after administration of vehicle or chloroquine (CQ, 10 mg/kg ip) 2 h before tissue harvest (n = 4/group). mRNA levels of LC3B (D) and p62/SQSTM1 (E) were measured in lean and DIO mice fasted overnight (n = 6/group, *P < 0.05). NS, not significant.
Autophagy can be regulated at both the transcriptional and posttranslational level (12, 38). To assess transcriptional regulation of autophagy, we examined changes in mRNA for LC3B and p62/SQSTM1 in response to fasting. Both groups of animals showed similar upregulation of mRNA for LC3B and p62/SQSTM1 in response to fasting, suggesting that upstream signaling responses to induce autophagy at the transcriptional level remained intact (Fig. 2, D and E). These results combined support the likelihood that development of MetS blunts cardiac autophagy at the posttranscriptional level.
Autophagy-related energy- and nutrient-sensing pathways remain intact in hearts of diet-induced obese mice.
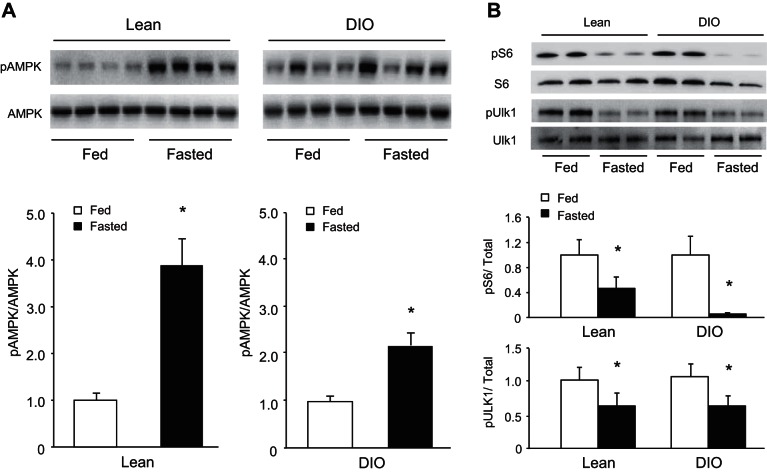
We next evaluated the autophagy-related energy- and nutrient-sensing pathways regulated by AMPK and mTOR, respectively. AMPK and mTOR control posttranslational induction of autophagy via phosphorylation of the critical autophagy regulator/initiator. Phosphorylation of Ulk1 by AMPK promotes autophagy (46), whereas mTOR action leads to repression (31, 46). Fasting elicited the expected increase in phosphorylation of AMPK in both lean and DIO mice (Fig. 3A). As an indicator of mTORC1 activity, the phosphorylation state of ribosomal S6 and Ulk1 was regulated (Fig. 3B). Fasting led to robust dephosphorylation of S6 in both groups of animals; however, the effect was exaggerated in DIO mice. mTOR has been demonstrated to directly target the Ser757 site of Ulk1, which leads to potent inhibition of autophagy (24). Fasting resulted in the appropriate dephosphorylation of Ulk1 at this site in both groups. These findings show that the major energy- and nutrient-sensing pathways that control the induction of autophagy at the posttranslational level remain largely intact in the heart despite the development of the metabolic syndrome.
Fig. 3.
Cardiac fasting energy- and nutrient-sensing pathways remain intact in obese animals. Western blot analysis of phospho (p) AMP-activated kinase (AMPK) Thr172 and total AMPK (n = 4/group, *P < 0.05, A) and ribosomal pS6 Ser235/236, total ribosomal S6, pUlk1 Ser757, and total Ulk1 (n = 4–6/group, *P < 0.05, B) in lean and DIO overnight-fasted mice.
Proteomes affecting energy metabolism, protein quality control, and redox status are discordant in diet-induced obese mice.
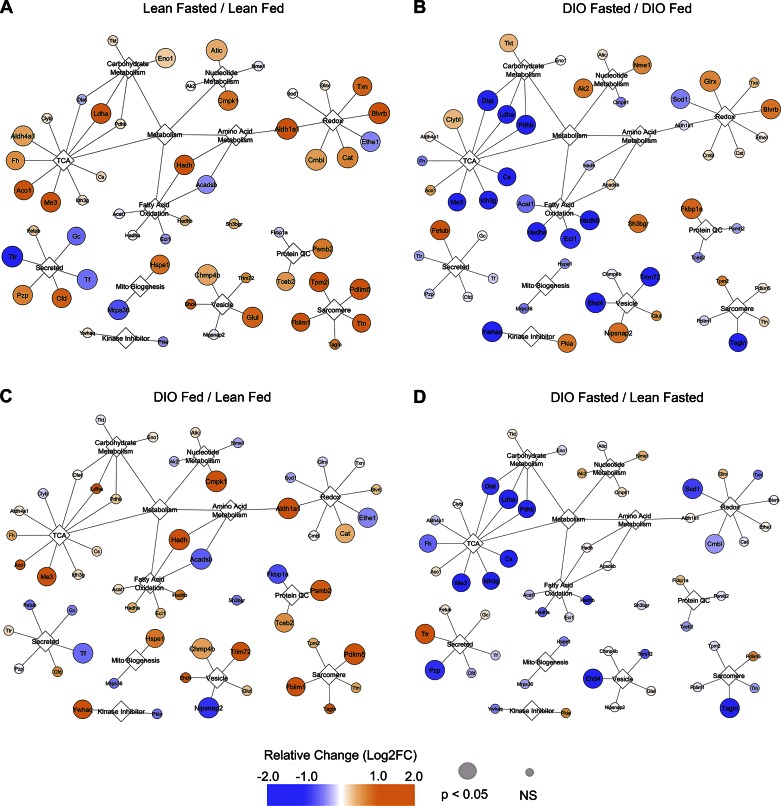
To gain insight as to why the diet-induced obese mice failed to induce autophagy despite intact energy and nutrient signaling, we turned to data-dependent acquisition MS-based proteomics for clues to rationalize this phenomenon. Overall, we identified 1,102 nonredundant proteins from 5,262 peptides (counting modifications as unique), among which 513 proteins were consistently detected across all experimental groups. The proteins with significant changes between groups (P < 0.05) and their exact P values can be found in Supplemental Table 1 (Supplemental material for this article is available online at the Journal website.). A full list of proteins quantified (Supplemental Table 2) and peptide/protein assignments (Supplemental Table 3) can also be found as online data supplements. The interconnections and major networks formed by the protein differences between lean and DIO animals and their response to the fasting challenge are shown in Fig. 4.
Fig. 4.
Proteomic analysis of cardiac fasting response. Cytoscape (Institute of Systems Biology) was used to display gene names of proteins reaching significant statistical differences between groups comparing lean fasted/lean fed (A), DIO fasted/DIO fed (B), DIO fed/lean fed (C), and DIO fasted/lean fasted (D). For complete proteomic dataset of proteins found here, see Supplemental Table 2.
We first examined the proteome change of hearts obtained from lean fasted vs. lean fed mice (Fig. 4A). We noted significant upregulation of enzymes involved in metabolism, as well as several enzymes responding to oxidative stress (Txn, Blvrb, Cat, Cmbl, and Aldh1a1), protein quality control (Tceb2, Psmb2), and vesicle traffic (Chmp4b, Glul). There were also increases in several sarcomeric proteins (Fblim1, Tpm2, Pdlim4, Ttn) in the fasted heart that were not seen in the DIO animals. On the other hand, the fasting response in DIO mice revealed a dramatically different profile (Fig. 4B). Tricarboxylic acid (TCA) and fatty acid oxidation (FAO) enzymes were decreased, as were vesicle transport (Ehd4, Trim72) and a myofilament-related protein (Tagln). In the baseline fed state, DIO mice (compared with lean) upregulated the expression of enzymes in the TCA cycle and those responsible for FAO, an appropriate response to a high-fat diet (Fig. 4C). We also noted an increase in sarcomeric proteins (Fblim1, Pdlim5), vesicle transport (Chmp4b, Trim72), protein quality control (Psmb2, Tceb2), and oxidative stress (Aldh1a1, Cat).
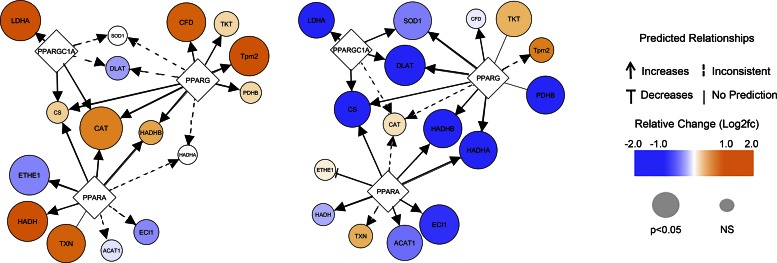
Because the DIO animals started out with higher baseline levels of TCA and FAO enzymes, it is important to compare DIO vs. lean animals in the fasting state (Fig. 4D). This comparison revealed a profound suppression of TCA cycle and FAO enzymes in the fasted DIO hearts compared with the lean. Thus, discordant regulation of metabolism emerged as the key distinguishing feature between lean and obese mice at baseline and in response to fasting. Analysis of the upstream regulators capable of explaining these protein abundance patterns implicated peroxisome proliferator-activated receptor-γ coactivator-1α (PCG-1α), peroxisome proliferator-activated receptor (PPAR)-α, and PPARγ, largely based on the changes of the oxidative phosphorylation and redox-related proteins; additional proteins from the redox, vesicle transport, and myofilament categories also supported these predictions (Fig. 5 and Supplemental Table 4).
Fig. 5.
Peroxisome proliferator-activated receptor (PPAR)-related fasting response in heart. Cytoscape was used to display predicted upstream PPAR-related changes in the heart in response to fasting. Predicted changes were determined through Ingenuity Pathway Analysis (IPA) (Qiagen) using the information from Supplemental Table 4.
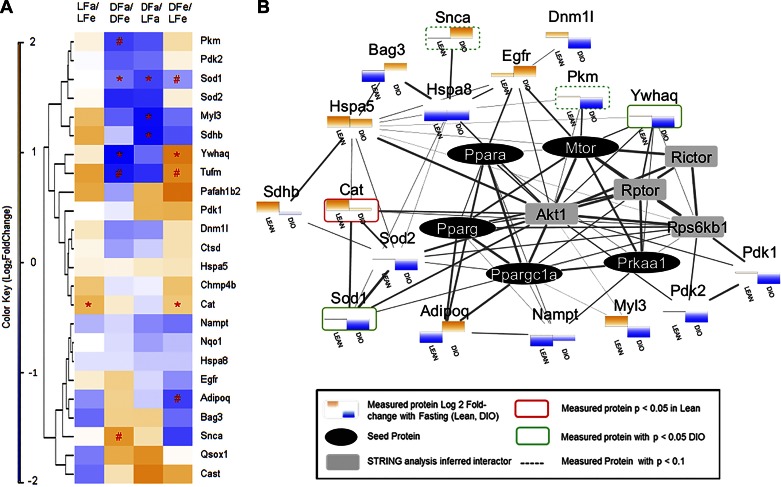
To gain a deeper understanding of why autophagy was differentially regulated between lean and DIO mice, we used IPA to identify the autophagy-related proteins present in the full dataset of 1,102 proteins (Fig. 6). Of these autophagy-related proteins, 24 proteins were considered as “confidently” identified with 2 or more peptide identifications, and an additional 18 proteins were considered more “putative” with a single high-quality proteotypic peptide detected for their identification (Supplemental Table Table 5). A heatmap representing the fold-change values observed for the 24 high-confident autophagy proteins across each of the major experimental comparisons revealed notable discordance in the trend of protein abundance changes between lean and DIO mice in response to fasting, which is emphasized by the grouping of proteins by hierarchical clustering (Fig. 6A). Further functional network analysis indicated that the discordancy of these abundance changes could be explained by the upstream activity of PPARα, PPARγ, PGC-1α, AMPK (Prkaa1), and/or mTOR, as indicated by a high degree of connectivity between measured autophagy protein nodes, these five seeded regulatory nodes, and their first-order interaction partners (e.g., Akt, Rictor, Raptor) (Fig. 6B). This autophagy-specific analysis of the proteomic data support the contention that the DIO mice have an inappropriate autophagy response to fasting and support the findings of our biochemical experiments.
Fig. 6.
Autophagy-focused proteomic analysis. The dataset comparing lean and DIO mice under fed and fasted conditions (1,102 proteins, Supplemental Table 2) was interrogated for autophagy-related proteins using IPA. Autophagy-related proteins identified by 2 or more peptides were used for subsequent clustering and network analysis. A: heatmap with hierarchical clustering of all 24 autophagy-related proteins and their observed Log2 fold-change [expressed as a color gradient from blue (negative) to orange (positive)] from the MSSTATs comparison. Hierarchical clustering was applied to the rows of the dataset to reveal groupings of autophagy proteins with similar patterns of abundance changes across biological comparisons. LFa, lean fasted; LFe, lean fed; OFa, obese fasted; OFe, obese fed. *P < 0.05 and #P < 0.1. B: string network analysis of functional protein-protein interactions among the observed autophagy-related proteins into which PPARa, PPARg, PGC1a, AMPK, and mammalian target of rapamycin (mTOR) were seeded based on findings from Western blot and IPA. Measured proteins were identified by mass spectrometry (colored shading). Differences in the fasting response between lean and DIO mice are plotted as bars with orange (above the axis), indicating an increase in protein abundance in the fasted state, whereas blue (below the axis) denotes a decrease. No change is indicated by a thin gray line. Proteins showing a statistically significant change (P < 0.05) are outlined by a red (Lean) or green (DIO) border, with dashed borders indicative of a trend toward significance (P < 0.1). Seeded regulators are shown in black ovals, whereas secondary regulators are indicated in gray squares. Edge width is scaled to the combined score assigned by the STRING network analysis software indicating the strength of the evidence supporting an interaction between any two nodes (range 0.404–0.999).
DISCUSSION
FVBN mice placed on a high-fat diet developed features of MetS, including obesity, hyperglycemia, and insulin resistance as well as increased I/R injury. These animals also exhibited impaired autophagy. Cardiac autophagy impairment in DIO mice occurred at the level of autophagic flux, thus implicating impaired autophagosome-lysosome fusion or inadequate lysosomal function (acidification or hydrolytic enzymes). Our results in mice are consistent with the findings of Jaishy et al. (17) who reported that fatty acid overload in mice and in rat H9C2 cardiomyocytes suppresses autophagosome clearance by attenuating lysosome activity. We suspect that this results in feedback inhibition of autophagosome initiation. Impaired autophagosome formation would therefore limit the ability to sequester damaged mitochondria, and impaired autophagic flux would interfere with energy substrate production from lysosomal activity, exacerbating the energetic crisis of the cardiomyocytes. The observed blunted autophagic flux likely contributes to the greater vulnerability of the MetS heart to ischemic stress.
In our study, both lean and DIO mice were able to upregulate key autophagy targets, LC3B and p62/SQSTM1, demonstrating that the transcriptional response to fasting remained intact. We noted differences in the magnitude of the signal transduction response to fasting between lean and DIO mice. For instance, the increase in AMPK phosphorylation was greater in lean mice than in DIO mice. Conversely, the fasting-induced dephosphorylation of the downstream mTOR target, ribosomal S6, was more profound in the DIO mice. This may indicate that, in the lean animals, lysosomal function was intact and enabled partial reactivation of mTOR (40), whereas, in DIO mice, mTOR remained supersuppressed due to lysosomal dysfunction. This raises the possibility that survival signaling through Akt could be compromised, which would carry consequences for I/R injury. Ulk1 Ser757 dephosphorylation during fasting was similar in lean and DIO mice, despite differences in the level of activity of the key upstream regulators AMPK and mTOR. It appears that, in lean mice, mTOR was less suppressed while AMPK was strongly activated, whereas, in DIO mice, mTOR was strongly suppressed while AMPK was only moderately activated. The net outcome was similar levels of Ulk1 phosphorylation, which should have given rise to similar levels of autophagy activation. There is evidence for feedback inhibition of autophagy when lysosome function is impaired. Thus, the lysosomal impairment (reflected by impaired autophagic flux) seen in the DIO mice might be responsible for the failure to upregulate autophagosome formation despite appropriate signaling. mTOR regulates lysosome biogenesis, and amino acids derived from lysosomal proteolysis reactivate mTOR, thereby downregulating autophagy and restoring protein synthesis (34, 40, 41, 45, 47, 48). Aside from limiting lysosomal regeneration, the sustained suppression of mTOR would limit the ability of cells to initiate repair processes after nutrient deprivation or ischemic injury and reflects a form of metabolic inflexibility that would increase vulnerability to ischemic stress in the setting of MetS. Supporting this concept are the findings by Ma et al. that impaired autophagosome clearance exacerbates I/R injury (27, 28).
Using bioinformatics algorithms to interrogate the 1,102 proteins identified in the heart samples, by proteomic analysis we identified 24 proteins that were autophagy related (Fig. 6). A majority of these proteins changed in opposite directions when comparing the fasting response in lean animals with that in obese animals. This suggests a widespread discordant regulation of autophagy in animals with DIO. Autophagy is primarily regulated by posttranslational modifications rather than changes in quantity, so it is not surprising that many of these autophagy-related proteins exhibited only modest changes (<1.5-fold) in abundance. Without further sample fractionation, MS has a limited dynamic range and limited observation of cellular low-abundance proteins compared with antibody-mediated detection, and therefore we were only able to quantify a limited number of the most widely recognized autophagy proteins by MS but emphasize the broader cellular responses. Among the autophagy proteins that were identified by MS, several are noteworthy. Dnm1l (Drp1) is a known regulator of mitochondrial autophagy and was decreased in the DIO fasting response, consistent with inappropriate suppression of autophagy. α-Synuclein (Snca) is a suppressor of autophagy and was increased in the hearts of DIO fasting mice, indicating dysregulation of autophagy (8). We also noted discordant regulation of myosin light chain 3 (Myl3). Myl3 gene mutations are found in familial hypertrophic cardiomyopathy (1) and was recently reported to upregulate autophagy (26). Interestingly, Bag3, which plays a role in clearing degradation-prone substrates via ubiquitin-independent chaperone-mediated autophagy (2), is increased in hearts of fasting DIO mice, suggesting that chaperone-mediated autophagy, rather than conventional macroautophagy, is upregulated. These findings are consistent with our finding of impaired autophagic flux by Western blot of LC3 +/- chloroquine (Fig. 2C).
There were many quantitative changes in proteins associated with FAO and components of the TCA cycle between lean and DIO mice subjected to fasting. During fasting, it is well known that fatty acids are liberated from peripheral adipose tissues and delivered to the heart, activating PPAR and thereby upregulating enzymes involved in the TCA cycle and FAO, which are needed to metabolize the fatty acids. This was reflected by appropriate changes in the cardiac proteome of the lean fasted animals. IPA, a bioinformatics tool used to interrogate our proteomics dataset, predicted that PGC-1α, PPARα, and PPARγ upstream signaling was responsible for many of the changes in the proteome (Fig. 5 and Supplemental Table 4). Surprisingly, we observed opposite changes to the PPAR-regulated proteome of the fasting DIO mice, although triglycerides would have been mobilized in both lean and obese mice as illustrated in the network diagram (Fig. 6B). This raises the possibility that, in DIO mice, an interacting signal transduction pathway, possibly mTOR, impinges on the PGC-1α/PPARγ/PPARα regulatory network.
Proteomic analysis of lean and DIO mice also revealed differences in the abundance of key redox-related proteins. At baseline, DIO mice had a higher abundance of catalase, suggesting ongoing H2O2-related stress due to a high rate of FAO. During fasting, lean mice upregulated catalase robustly to levels that matched DIO at the basal state. On the other hand, DIO mice did not increase catalase levels further during fasting, which suggests either an inability to handle a higher H2O2 load during stress or a potential attempt to compensate for a reduced H2O2 production due to decreased superoxide dismutase 1. Reactive oxygen species, specifically superoxide and H2O2, are well known for their involvement in the regulation of autophagy (42), although the precise mechanisms whereby these radicals modulate autophagic flux are not entirely clear. The proteomic data suggest that differences in the enzymes involved in ROS scavenging and conversion may be important components in the altered autophagy observed in the hearts of starved DIO mice.
Some limitations in this study need to be noted. The response to fasting is profoundly dysregulated in DIO mice, which also sustained larger infarcts. Although fasting results in nutrient depletion and the latter is a hallmark of ischemia, it does not mimic hypoxia or the added element of reperfusion. This study does not establish a causal relationship between MetS, impaired autophagy, proteome remodeling, and larger infarcts in the mouse. Restoring autophagy in the setting of MetS and showing that ischemic injury is reduced will provide stronger evidence. Work remains to identify a specific dysfunctional site(s) responsible for impaired autophagic flux, although the lysosome is implicated by our work and others (17). We chose not to conduct Western blotting or proteomic analysis of hearts subjected to ischemia and reperfusion because tissue necrosis would deteriorate protein quality, resulting in unpredictable artifacts.
Taken together, the Western blot and proteomics findings reveal a stark difference in the fasting response of lean and obese mice. Fasting obese mice exhibited impaired autophagic flux, reduced fatty acid oxidation, and dysregulated redox regulation. Mice with DIO have an abnormal response to nutrient deprivation that could explain the increased injury associated with ischemia and reperfusion.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant P01-HL-112730 (to R. A. Gottlieb) and American Heart Association SDG Grant 15SDG23230013 (to A. M. Andres).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.M.A., B.R.I., J.V.E., R.A.G., and R.M. conception and design of research; A.M.A., J.K., N.R., C.H., and V.V. performed experiments; A.M.A., J.K., S.P., K.T., N.R., B.R.I., C.H., V.V., J.V.E., R.A.G., and R.M. analyzed data; A.M.A., J.K., S.P., K.T., N.R., B.R.I., C.H., J.V.E., R.A.G., and R.M. interpreted results of experiments; A.M.A., J.K., S.P., K.T., N.R., C.H., R.A.G., and R.M. prepared figures; A.M.A., J.K., S.P., K.T., B.R.I., J.V.E., R.A.G., and R.M. drafted manuscript; A.M.A., J.K., S.P., K.T., J.V.E., R.A.G., and R.M. edited and revised manuscript; A.M.A., J.K., S.P., K.T., N.R., B.R.I., C.H., V.V., J.V.E., R.A.G., and R.M. approved final version of manuscript.
Supplementary Material
REFERENCES
- 1.Andersen PS, Hedley PL, Page SP, Syrris P, Moolman-Smook JC, McKenna WJ, Elliott PM, Christiansen M. A novel myosin essential light chain mutation causes hypertrophic cardiomyopathy with late onset and low expressivity. Biochem Res Int 2012: 685108, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behl C. BAG3 and friends: co-chaperones in selective autophagy during aging and disease. Autophagy 7: 795–798, 2011. [DOI] [PubMed] [Google Scholar]
- 3.Bolli R, Becker L, Gross G, Mentzer R Jr, Balshaw D, Lathrop DA. Myocardial protection at a crossroads: the need for translation into clinical therapy. Circ Res 95: 125–134, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Bouhidel O, Pons S, Souktani R, Zini R, Berdeaux A, Ghaleh B. Myocardial ischemic postconditioning against ischemia-reperfusion is impaired in ob/ob mice. Am J Physiol Heart Circ Physiol 295: H1580–H1586, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buettner R, Scholmerich J, Bollheimer LC. High-fat diets: modeling the metabolic disorders of human obesity in rodents. Obesity 15: 798–808, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, Vitek O. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30: 2524–2526, 2014. [DOI] [PubMed] [Google Scholar]
- 7.Clavijo LC, Pinto TL, Kuchulakanti PK, Torguson R, Chu WW, Satler LF, Kent KM, Suddath WO, Pichard AD, Waksman R. Metabolic syndrome in patients with acute myocardial infarction is associated with increased infarct size and in-hospital complications. Cardiovasc Revasc Med 7: 7–11, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Colasanti T, Vomero M, Alessandri C, Barbati C, Maselli A, Camperio C, Conti F, Tinari A, Carlo-Stella C, Tuosto L, Benincasa D, Valesini G, Malorni W, Pierdominici M, Ortona E. Role of alpha-synuclein in autophagy modulation of primary human T lymphocytes. Cell Death Dis 5: e1265, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics 13: 22–24, 2013. [DOI] [PubMed] [Google Scholar]
- 10.Galassi A, Reynolds K, He J. Metabolic syndrome and risk of cardiovascular disease: a meta-analysis. Am J Med 119: 812–819, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Giricz Z, Mentzer RM Jr, Gottlieb RA. Autophagy, myocardial protection, and the metabolic syndrome. J Cardiovasc Pharmacol 60: 125–132, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlieb RA, Pourpirali S. Lost in translation: miRNAs and mRNAs in ischemic preconditioning and ischemia/reperfusion injury. J Mol Cell Cardiol In press. [DOI] [PMC free article] [PubMed]
- 13.Grundy SM. Obesity, metabolic syndrome, and coronary atherosclerosis. Circulation 105: 2696–2698, 2002. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One 6: e20975, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM Jr, Gottlieb RA. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res 3: 365–373, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahania SM, Sengstock D, Vaitkevicius P, Andres A, Ito BR, Gottlieb RA, Mentzer RM Jr. Activation of the homeostatic intracellular repair response during cardiac surgery. J Am Coll Surg 216: 719–726, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaishy B, Zhang Q, Chung HS, Riehle C, Soto J, Jenkins S, Abel P, Cowart LA, Van Eyk JE, Abel ED. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J Lipid Res 56: 546–561, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakhniashvili DG, Bulla LA Jr, Goodman SR. The human erythrocyte proteome: analysis by ion trap mass spectrometry. Mol Cell Proteomics 3: 501–509, 2004. [DOI] [PubMed] [Google Scholar]
- 19.Kane LA, Neverova I, Van Eyk JE. Subfractionation of heart tissue: the “in sequence” myofilament protein extraction of myocardial tissue. Methods Mol Biol 357: 87–90, 2007. [DOI] [PubMed] [Google Scholar]
- 20.Kaushik G, Spenlehauer A, Sessions AO, Trujillo AS, Fuhrmann A, Fu Z, Venkatraman V, Pohl D, Tuler J, Wang M, Lakatta EG, Ocorr K, Bodmer R, Bernstein SI, Van Eyk JE, Cammarato A, Engler AJ. Vinculin network-mediated cytoskeletal remodeling regulates contractile function in the aging heart. Sci Trans Med 7: 292–299, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazlauskiene L, Butnoriene J, Norkus A. Metabolic syndrome related to cardiovascular events in a 10-year prospective study. Diabetol Metab Syndr 7: 102, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy AJ, Ellacott KL, King VL, Hasty AH. Mouse models of the metabolic syndrome. Disease Mod Mech 3: 156–166, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci USA 107: 14164–14169, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy 8: 1394–1396, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation 125: 3170–3181, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik S, Wong ND, Franklin SS, Kamath TV, L'Italien GJ, Pio JR, Williams GR. Impact of the metabolic syndrome on mortality from coronary heart disease, cardiovascular disease, and all causes in United States adults. Circulation 110: 1245–1250 2004. [DOI] [PubMed] [Google Scholar]
- 31.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 100: 914–922, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Miura T, Miki T. Limitation of myocardial infarct size in the clinical setting: current status and challenges in translating animal experiments into clinical therapy. Basic Res Cardiol 103: 501–513, 2008. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery MK, Hallahan NL, Brown SH, Liu M, Mitchell TW, Cooney GJ, Turner N. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia 56: 1129–1139, 2013. [DOI] [PubMed] [Google Scholar]
- 34.Nakadera E, Yamashina S, Izumi K, Inami Y, Sato T, Fukushima H, Kon K, Ikejima K, Ueno T, Watanabe S. Inhibition of mTOR improves the impairment of acidification in autophagic vesicles caused by hepatic steatosis. Biochem Biophys Res Commun 469: 1104–1110, 2016. [DOI] [PubMed] [Google Scholar]
- 35.Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 7: 188–204, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panchal SK, Brown L. Rodent models for metabolic syndrome research. J Biomed Biotechnol 2011: 351982, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol 453: 325–342, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pourpirali S, Valacca C, Merlo P, Rizza S, D'Amico S, Cecconi F. Prolonged Pseudohypoxia Targets Ambra1 mRNA to P-Bodies for Translational Repression. PLoS ONE 10: e0129750, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reuter TY. Diet-induced models for obesity and type 2 diabetes. Drug Disc Today 4: 3–8, 2007. [Google Scholar]
- 40.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saric A, Hipolito VE, Kay JG, Canton J, Antonescu CN, Botelho RJ. mTOR controls lysosome tubulation and antigen presentation in macrophages and dendritic cells. Mol Biol Cell 27: 321–333, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 36: 30–38, 2011. [DOI] [PubMed] [Google Scholar]
- 43.Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics 11: 202–214, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz Longacre L, Kloner RA, Arai AE, Baines CP, Bolli R, Braunwald E, Downey J, Gibbons RJ, Gottlieb RA, Heusch G, Jennings RB, Lefer DJ, Mentzer RM, Murphy E, Ovize M, Ping P, Przyklenk K, Sack MN, Vander Heide RS, Vinten-Johansen J, Yellon DM. New horizons in cardioprotection: recommendations from the 2010 National Heart, Lung, and Blood Institute Workshop. Circulation 124: 1172–1179, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stransky LA, Forgac M. Amino acid availability modulates vacuolar H+-ATPase assembly. J Biol Chem 290: 27360–27369, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weikel KA, Cacicedo JM, Ruderman NB, Ido Y. Glucose and palmitate uncouple AMPK from autophagy in human aortic endothelial cells. Am J Physiol Cell Physiol 308: C249–C263, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334: 678–683, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.