The present studies reveal for the first time that thioredoxin-interacting protein (TXNIP) regulates myocardial fatty acid oxidation, identify a novel TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB signaling pathway, provide new insight into the physiological role of TXNIP and miR-33a in the heart, and discover the upstream signaling mechanisms that control miR-33a expression.
Keywords: thioredoxin-interacting protein, microRNA, nuclear factor Y, β-oxidation, heart
Abstract
Myocardial fatty acid β-oxidation is critical for the maintenance of energy homeostasis and contractile function in the heart, but its regulation is still not fully understood. While thioredoxin-interacting protein (TXNIP) has recently been implicated in cardiac metabolism and mitochondrial function, its effects on β-oxidation have remained unexplored. Using a new cardiomyocyte-specific TXNIP knockout mouse and working heart perfusion studies, as well as loss- and gain-of-function experiments in rat H9C2 and human AC16 cardiomyocytes, we discovered that TXNIP deficiency promotes myocardial β-oxidation via signaling through a specific microRNA, miR-33a. TXNIP deficiency leads to increased binding of nuclear factor Y (NFYA) to the sterol regulatory element binding protein 2 (SREBP2) promoter, resulting in transcriptional inhibition of SREBP2 and its intronic miR-33a. This allows for increased translation of the miR-33a target genes and β-oxidation-promoting enzymes, carnitine octanoyl transferase (CROT), carnitine palmitoyl transferase 1 (CPT1), hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase-β (HADHB), and AMPKα and is associated with an increase in phospho-AMPKα and phosphorylation/inactivation of acetyl-CoA-carboxylase. Thus, we have identified a novel TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB pathway that is conserved in mouse, rat, and human cardiomyocytes and regulates myocardial β-oxidation.
NEW & NOTEWORTHY
The present studies reveal for the first time that thioredoxin-interacting protein (TXNIP) regulates myocardial fatty acid oxidation, identify a novel TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB signaling pathway, provide new insight into the physiological role of TXNIP and miR-33a in the heart, and discover the upstream signaling mechanisms that control miR-33a expression.
thioredoxin-interacting protein (TXNIP) has emerged as an important player in myocardial pathology, including ischemia/reperfusion injury (43), and cardiac hypertrophy (44, 45), but the physiological role of TXNIP in the heart is still not well understood. TXNIP is known to promote oxidative stress by binding to, and inhibiting, thioredoxin (22, 27), and TXNIP overexpression suppresses thioredoxin activity and induces apoptosis (33, 40, 45). On the other hand, we recently found that downregulation of TXNIP inhibits diabetes-induced cardiomyocyte apoptosis (5, 6). TXNIP also appears to influence cardiac metabolism, including both mitochondrial function and glucose uptake/utilization (43). However, despite the fact that fatty acid β-oxidation is essential for the heart's ability to generate energy and for the maintenance of myocardial energy homeostasis (26, 37), the effects of cardiomyocyte TXNIP on β-oxidation have remained largely unexplored. Using a new cardiomyocyte-specific TXNIP knockout (cTKO) mouse and working mouse heart perfusions, the present studies were, therefore, aimed at determining the effects of TXNIP deficiency on myocardial β-oxidation and elucidating the molecular mechanisms involved.
MATERIALS AND METHODS
Animal studies.
All mouse studies were approved by the University of Alabama at Birmingham Animal Care and Use Committee and conform to the National Academy of Sciences' Guide for the Care and Use of Laboratory Animals. The cTKO was generated using the Cre-loxP system. The floxed TXNIP mouse generated by flanking exons 2–8 of the TXNIP gene (total of 8 exons) by loxP sites has been described previously (7, 20) and backcrossed for 10 generations into the C57BL/6J background using mice obtained from JAX. Crossing these mice with the αMyHC-Cre mice also on the C57BL/6J background expressing the Cre recombinase under the cardiac-specific α-myosin heavy-chain (αMyHC) promoter yielded the cTKO mice. Genotyping for Txniplox/lox and αMyHC-cre+/− was performed by PCR using the primers listed in Supplemental Table S1.
Working mouse heart perfusions.
Isolated working mouse heart perfusions were performed on 20-wk-old male control lox/lox and cTKO mouse hearts. [For consistency and to avoid variability due to sex-related differences in cardiac physiology and metabolism (10, 16, 41), male mice were used throughout the studies.] Heart rates were 334 ± 33 in lox/lox and 371 ± 5 in cTKO. Metabolism and function were assessed at preload of 15 mmHg and afterload of 50 mmHg for 30 min. Concentrations of glucose, oleate, and insulin in the perfusate were 8 mM, 0.4 mM, and 10 μU/ml, respectively, and oleate was conjugated to 3% BSA. Indices of metabolism, including oleate and glucose oxidation and contractile function, such as cardiac power and rate pressure product, were measured, as described previously (12, 38). Reliance on exogenous (glucose and oleate) and endogenous substrate utilization was calculated as a percentage of oxygen consumption, similar to previously published studies (46, 47).
Echocardiography.
Echocardiography was performed for assessment of contractile function in vivo, as described previously (13, 38). In brief, mice were anesthetized with 0.5–2% isoflurane in a 100% oxygen mix, and m-mode images were acquired using a Vevo 2100 imaging system. Images were taken at heart rates >400 beats/min to obtain measurements that are physiologically relevant.
Tissue culture.
H9C2 rat cardiomyocytes (American Type Culture Collection, Manassas, VA) were maintained in DMEM modified to contain 4 mM l-glutamine, 4,500 mg/l glucose, 1 mM sodium pyruvate, 1,500 mg/l sodium bicarbonate, 1.8 mM CaCl2, and 0.8 mM MgCl2, pH 7.3, and supplemented with 10% FBS and 1% penicillin-streptomycin. The AC16 adult human ventricular cardiomyocyte cell line was maintained in DMEM-F12 (GIBCO cat. no. 11330-032) supplemented with 10% FBS and 1% penicillin-streptomycin. Both cell lines are widely used as in vitro models of ventricular cardiomyocytes and share multiple metabolic features including β-oxidation (21, 28).
Quantitative real-time RT-PCR.
Total RNA was extracted using RNeasy kit (Qiagen), according to the manufacturer's instructions. One microgram of RNA was reverse-transcribed to cDNA using the first-strand cDNA synthesis kit (Roche, Indianapolis, IN). Quantitative real-time PCR was performed as described previously (7) using SYBR Green (Applied Biosystems, Foster City, CA), a LightCycler 480 System (Roche) and the primers listed in Supplemental Table S1. miR-33 expression was quantified using a TaqMan microRNA Assay (Applied Biosystems). All gene and microRNA expression results were corrected for 18S and U6 run as internal standards, respectively, or transcript numbers were assessed and compared with cyclophilin, as described previously (14, 35, 48).
RNA interference and microRNA overexpression.
Cells were grown in six-well plates and transfected with specific siRNA oligos for TXNIP [siTXNIP; Dharmacon (Lafayette, CO), siGENOME SMAPRTpool gene id:117514 cat. no. M-089121-01-0005], nuclear factor Y (NFYA; Dharmacon ON-TARGETplus SMARTpool cat. no. L-090481-02-0005), or scrambled oligo (Dharmacon, D-001810-01-20) using DharmaFECT transfection reagent 1 (Dharmacon/Thermo Scientific, Chicago, IL) (4 μl per well in 2 ml medium). The final concentration of oligos used was 25 nM. Cells were harvested after 48 h for RNA and protein extraction. For overexpression of miR-33a, miR-33a mimic (final concentration 25 nM; Life Technologies, assay ID MC12410) was transfected using DharmaFECT transfection reagent 1. Twenty-four hours after transfection, the medium was changed to fresh medium containing oleate (0.2 mM) conjugated to 1% BSA, and cells were harvested 48 h after transfection for protein extraction.
Immunoblotting.
Protein extracts were prepared using a lysis buffer containing HEPES (50 mM), Nonidet P-40 (10%), sodium fluoride (100 mM), sodium pyrophosphate (10 mM), EDTA (4 mM), PMSF (1 mM), leupeptin (2 μM), activated sodium orthovanadate (2 mM), and okadaic acid (100 nM). Proteins were separated by 4–20% SDS-PAGE, blotted onto PVDF membranes, and detected using the following primary antibodies: TXNIP (1:400) (JY2, MBL International, Woburn, MA), AMPKα (1:1,000), p-AMPKα (1:1,000), p-ACC (1:600) (Cell Signaling Technology, Beverly, MA), CROT (1:400) (Novus Biologicals, Littleton, CO), CPT1a (1:500) and CPT1b (1:1,000) (ProteinTech, Chicago, IL), HADHB (1:200) (Aviva Systems Biology, San Diego, CA), actin (1:2,000) (Abcam, Cambridge, MA), and the secondary antibodies: anti-mouse IgG (1:5,000) (Santa Cruz Biotechnology, Dallas, TX) and anti-rabbit IgG (Santa Cruz Biotechnology). Bands were visualized by ECL detection reagent (GE Healthcare Lifesciences Pittsburgh, PA) and were quantified by ImageQuant version TL (GE Healthcare Lifesciences Pittsburgh, PA).
Chromatin immunoprecipitation.
ChIP assays were performed as described previously (5). In brief, 500 μg of cross-linked H9C2 protein extracts (by BCA protein assay) were incubated overnight at 4°C with 10 μg of NFYA antibody (sc-10779X) (Santa Cruz Biotechnology), or purified polyclonal IgG as a negative control (sc-2027) (Santa Cruz Biotechnology).
Statistics.
Student's t-tests were used to calculate the significance of a difference between two groups. For data sets of more than two groups, we performed one-way-ANOVA calculations.
RESULTS
Myocardial fatty acid oxidation is increased in cardiomyocyte-specific TXNIP knockout mice.
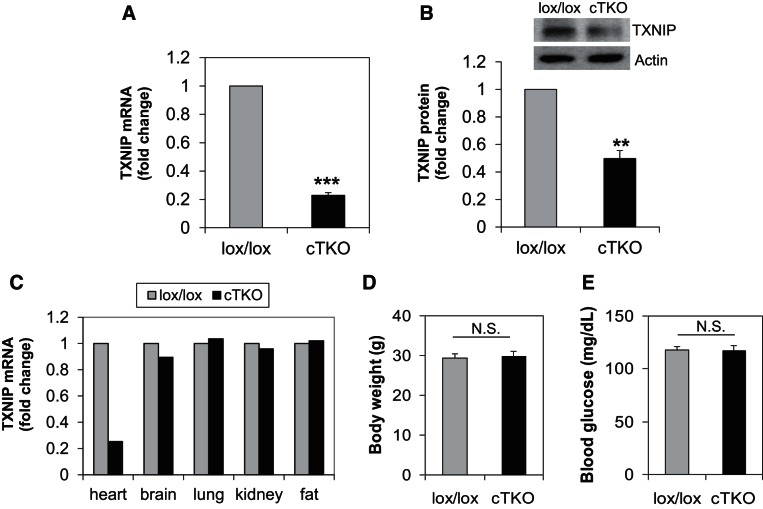
To define the specific physiological role of TXNIP in cardiomyocytes and cardiac metabolism, we generated a cTKO mouse by crossing floxed TXNIP mice (7) with αMyHC-Cre mice (2). Effective reduction of TXNIP expression and tissue specificity of the knockout were confirmed in the cTKO mice compared with control lox/lox littermates (Fig. 1, A–C). cTKO mice seemed to be overall healthy and were not distinguishable from control littermates. Also, in contrast to whole body TXNIP-deficient HcB-19 mice, which have increased adiposity and decreased blood glucose levels (7), the weight and blood glucose of cTKO mice was not significantly different from their control littermates, consistent with the cardiomyocyte-specific nature of this model (Fig. 1, D–E).
Fig. 1.
Characterization of cardiomyocyte-specific TXNIP knockout (cTKO) mice. TXNIP mRNA level (A) and TXNIP protein level (B) in primary ventricular samples of cTKO and lox/lox control mice, comparison of TXNIP expression levels in different tissues of cTKO and control mice (C), body weight (D), and blood glucose (E) in cTKO and control mice. Twenty-week-old male mice (n = 3 per group) were used; values are expressed as means ± SE. **P < 0.005; ***P < 0.001; N.S., not significant (Student's t-test).
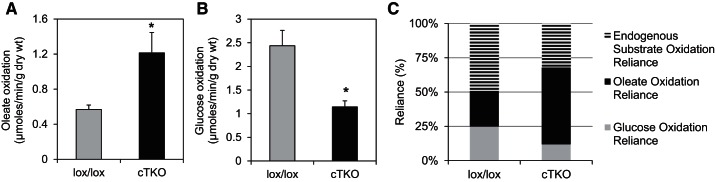
Next, we used isolated working mouse heart perfusions to assess cardiac metabolism and discovered that cTKO hearts exhibited dramatically higher rates of fatty acid oxidation compared with control lox/lox littermates (Fig. 2A), indicating that TXNIP deficiency promotes myocardial β-oxidation. The observed increase in fatty acid oxidation in cTKO hearts was also associated with a significant decrease in glucose oxidation (Fig. 2B), consistent with previous reports of decreased pyruvate dehydrogenase activity in hearts from TXNIP-null mice (43).
Fig. 2.
Myocardial fatty acid oxidation and substrate utilization in cTKO and control mice. Oleate oxidation (A), glucose oxidation (B), and substrate reliance (percentage of oxygen consumption) (C) in cardiomyocyte-specific TXNIP knockout (cTKO) vs. control (lox/lox) hearts as assessed by isolated working mouse heart perfusions. Heart rate in lox/lox was 334 ± 33 and in cTKO 371 ± 5. Bars are expressed as means ± SE; n = 4 mice per group. *P < 0.05 (Student's t-test).
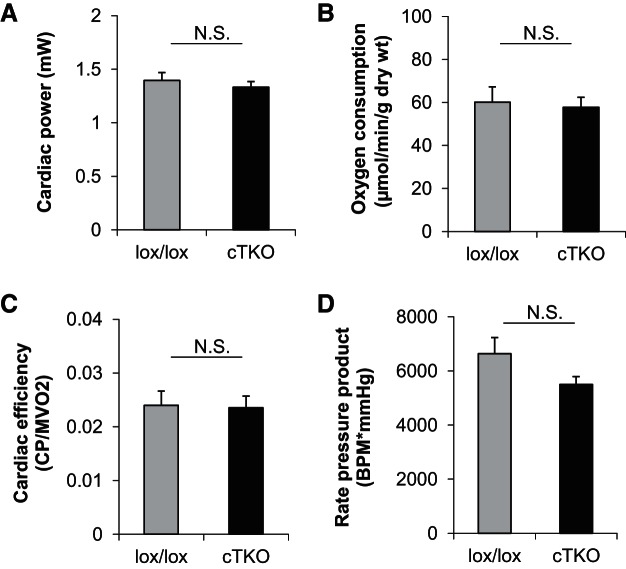
Importantly, the observed changes in substrate utilization appeared to be independent of global alterations in contractile and/or mitochondrial function, as evidenced by the lack of significant differences in cardiac power, rate pressure product, myocardial oxygen consumption, and cardiac efficiency between cTKO and control hearts (Fig. 3, A–D). Moreover, echocardiography did not reveal any notable differences in cardiac function or hypertrophy in vivo between cTKO and lox/lox control mice (Table 1). Combined, these observations reveal that lack of cardiomyocyte TXNIP results in an altered metabolic status, wherein reliance on fatty acid oxidation increases concomitant with decreased reliance on glucose oxidation (Fig. 2C), with no notable adverse effects on function.
Fig. 3.
Contractile function in cTKO and control mice. Cardiac power (A), oxygen consumption (B), cardiac efficiency (C), rate pressure product in cTKO vs. lox/lox control hearts (D), as measured by isolated working mouse heart perfusions. Bars represent means ± SE; n = 4 mice per group. N.S., not significant (Student's t-test).
Table 1.
Echocardiography data of control (lox/lox) and cardiomyocyte-specific TXNIP knockout mice
| lox/lox (n = 9) | cTKO (n = 6) | ||
|---|---|---|---|
| Left ventricular enddiastolic diameter (LVEDD), mm | 3.99 ± 0.08 | 3.92 ± 0.11 | NS |
| Left ventricular endsystolic diameter (LVESD), mm | 2.75 ± 0.10 | 2.74 ± 0.16 | NS |
| Left ventricular posterior wall thickness (LVPW), mm | 1.06 ± 0.03 | 1.16 ± 0.07 | NS |
| Left ventricular volume, diastolic (LV Vol; d), μl | 71 ± 3 | 67 ± 5 | NS |
| Left ventricular volume, systolic (LV Vol; s), μl | 29 ± 2 | 29 ± 4 | NS |
| Left ventricular ejection fraction (LV EF), % | 60 ± 3 | 58 ± 4 | NS |
| Left ventricular fractional shortening (LV FS), % | 32 ± 2 | 30 ± 2 | NS |
| Left ventricular cardiac output (LV CO), ml/min | 21 ± 1 | 20 ± 1 | NS |
Numbers represent means ± SE. All mice analyzed were age-matched, 16–20 wk old males. All measurements were taken at heart rates >400 beats/min. cTKO, cardiomyocyte-specific TXNIP knockout mice; NS, not significant.
TXNIP deficiency decreases cardiomyocyte mir-33a expression.
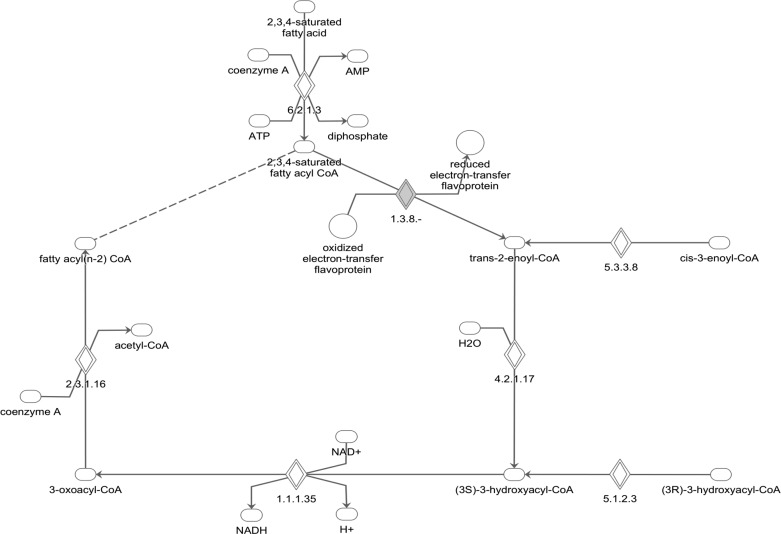
To investigate the potential mechanisms responsible for the striking increase in β-oxidation in cTKO hearts, we performed an unbiased gene expression microarray (Supplemental Table S2). However, on the basis of an Ingenuity Pathway Analysis, the differentially expressed genes between cTKO and control littermate hearts did not seem to explain the observed increase in myocardial β-oxidation and the only gene found in the microarray that was related to the canonical β-oxidation pathway, isovaleryl-CoA dehydrogenase, was downregulated (Fig. 4). Furthermore, quantitative real-time RT-PCR analysis of additional candidate genes known to promote cardiac β-oxidation [i.e., cytosolic acyl-CoA thioesterase (cte1) (also known as acyl-CoA thioesterase 1; ACOT1), medium-chain acyl-CoA dehydrogenase (mcad), and pyruvate dehydrogenase kinase isoenzyme 4 (pdk4) (14, 35)] revealed that mRNA expression of these genes was not increased in cTKO hearts (Table 2). Analysis of the microarray data for downstream targets of TXNIP signaling again showed that ACOT1 expression was downregulated rather than increased, consistent with the RT-PCR results, but did not reveal any additional clues in regard to gene expression and β-oxidation (Fig. 5). Together, these observations suggested that TXNIP may influence fatty acid uptake and/or oxidation at a posttranscriptional level. Interestingly, we recently discovered that TXNIP acts as a powerful regulator of microRNA expression in pancreatic β-cells (42), raising the possibility that it may also do so in cardiomyocytes. MicroRNAs are small, noncoding RNAs of roughly 22 nucleotides that bind to complementary 3′UTR sequences and affect mRNA translation or stability of target genes (24, 36).
Fig. 4.
Ingenuity pathways analysis canonical pathway of fatty acid β-oxidation. The array data set was overlaid on this canonical pathway. Gray diamond denotes only differentially regulated (downregulated) gene in cTKO hearts. 1.3.8.- denotes isovaleryl-CoA dehydrogenase (IVD).
Table 2.
Expression of genes known to promote cardiac β-oxidation as assessed by quantitative RT-PCR in control (lox/lox) and cTKO mice
| lox/lox* (n = 4) | cTKO* (n = 4) | |
|---|---|---|
| Cytosolic acyl-CoA thioesterase (CTE1) | 17830 ± 2720 | 8560 ± 1282 |
| Medium-chain acycl-CoA dehydrogenase (MCAD) | 527686 ± 10719 | 473164 ± 9966 |
| Pyruvate dehydrogenase kinase isoenzyme 4 (PDK4) | 15288 ± 2084 | 9634 ± 1495 |
| Cyclophilin control | 28339 ± 3072 | 29469 ± 1398 |
Numbers represent transcript copies ± SE; Cyclophilin was run as the control housekeeping gene.
Fig. 5.
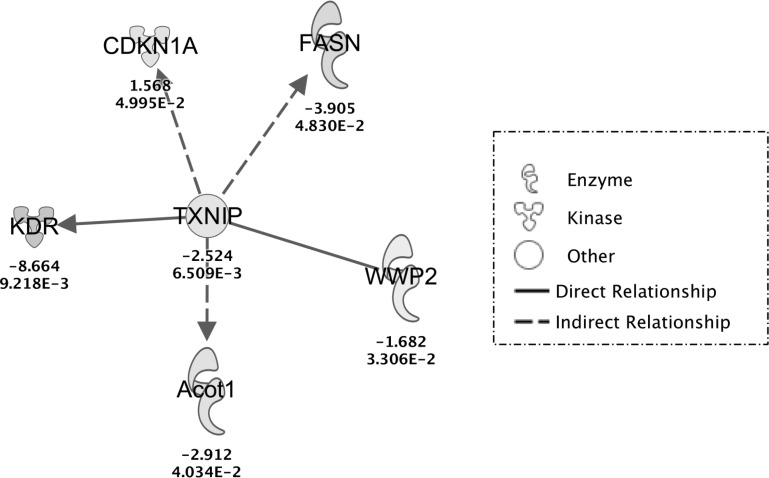
Ingenuity Pathways Analysis of the effects of TXNIP deletion. Ingenuity Pathways Analysis was used to generate this pathway by growing all downstream interactions of TXNIP that were identified in the array data set. Upregulated and downregulated genes in cTKO hearts are represented by positive and negative numbers, respectively. CDKN1A, cyclin-dependent kinase inhibitor 1; FASN, fatty acid synthase, WWP2, E3 ubiquitin-protein ligase; Acot1, acyl-CoA thioesterase; KDR, kinase insert domain receptor, a type III receptor tyrosine kinase.
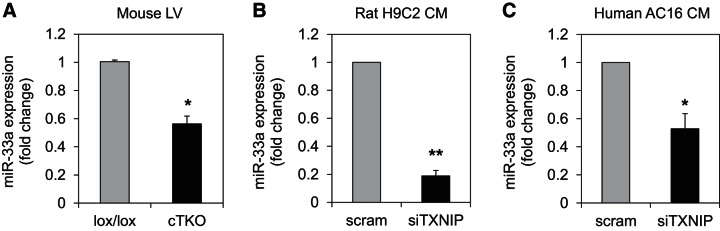
Therefore, we next investigated whether any particular microRNA(s) had been shown to be involved in β-oxidation and, indeed, found very strong evidence that in liver cell lines miR-33a controls β-oxidation by targeting and downregulating three key enzymes: 1) carnitine octanoyl transferase (CROT), which is responsible for the transport of medium- and long-chain Acyl CoAs out of the peroxisome and into the cytoplasm and mitochondria; 2) carnitine palmitoyltransferase 1a (CPT1a), which is associated with the outer mitochondrial membrane and mediates the transport of long-chain fatty acids across the membrane; and 3) hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase β-subunit (HADHB) (also known as acetyl-CoA acyl transferase) the β-subunit of the mitochondrial trifunctional protein that catalyzes the last three steps of long-chain fatty acid beta-oxidation (9, 17, 31). Furthermore, miR-33a has also been shown to target AMP-activated protein kinase (AMPKα) (15, 29, 31), a major regulator of cellular energy homeostasis (11) that promotes β-oxidation by phosphorylating and inactivating acetyl-CoA carboxylase (ACC2), leading to decreased malonyl-CoA and, thereby, relieving the inhibition on CPT1 (39). Therefore, we hypothesized that TXNIP might control β-oxidation by modulating miR-33a expression. Consistent with this hypothesis, we discovered that miR-33a expression was significantly lower in left ventricular samples of cTKO mice compared with lox/lox controls (Fig. 6A). Moreover, specific knockdown of TXNIP in rat H9C2 cardiomyocytes, as well as in human ventricular AC16 cardiomyocytes, also led to a dramatic decrease in miR-33a expression (Fig. 6, B and C), confirming that the effects were truly mediated by the decrease in TXNIP expression and occurred in other species, including humans. (While there is also a miR-33b in larger mammals and humans, no miR-33b expression was detectable in our human cardiomyocytes, and we, therefore, continued to focus on miR-33a.).
Fig. 6.
Effects of cardiomyocyte TXNIP deficiency on miR-33a. Expression of miR-33a as assessed by quantitative RT-PCR (qRT-PCR) in left ventricular samples of cTKO and lox/lox control mice (A), rat H9C2 cardiomyocytes (B), and human ventricular AC16 cardiomyocytes (C). Values are expressed as means ± SE of three independent experiments. *P < 0.05; **P < 0.005 (Student's t-test).
Decreased myocardial mir-33a expression in cTKO mice results in upregulation of mir-33a targets that promote fatty acid oxidation.
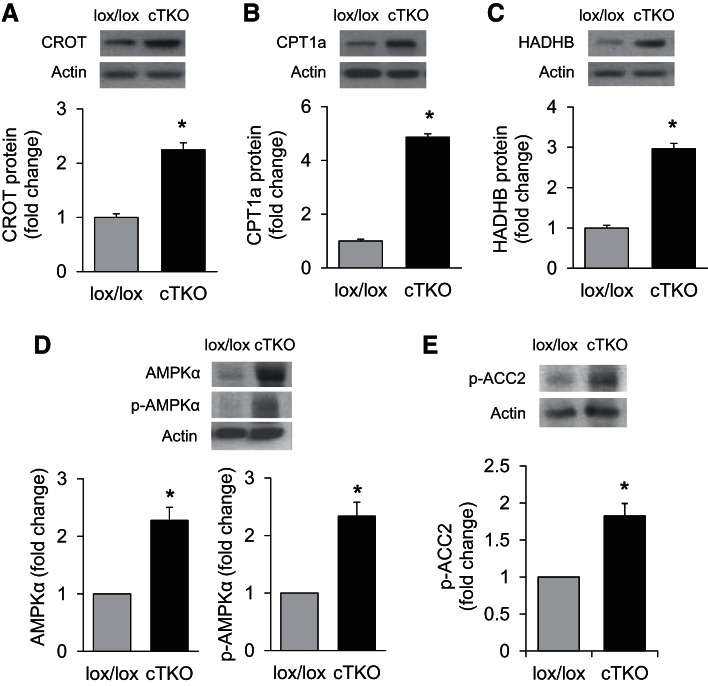
Consistent with the decrease in miR-33a, deletion of cardiomyocyte TXNIP expression led to elevated protein levels of the miR-33a targets CROT, CPT1a, and HADHB in ventricular samples of cTKO mice compared with lox/lox control littermates (Fig. 7, A–C). On the other hand, CROT, CPT1a, and HADHB mRNA expression was not significantly altered in cTKO hearts or H9C2 cells with TXNIP knockdown (data not shown), consistent with the lack of hits in our gene expression microarray study and the notion that miR-33a is regulating target gene expression primarily at the translational level (17). Moreover, the lack of cardiomyocyte TXNIP was also associated with significantly higher protein levels of another miR-33a target, AMPKα, and this was associated with increased levels of activated/phosphorylated AMPKα (p-AMPKα; Fig. 7D). In addition, the increase in p-AMPKα also enhanced phosphorylation/inactivation of ACC2 (Fig. 7E). Of note, the 3′UTRs of AMPKα, CROT, CPT1a, and HADHB contain a highly conserved miR-33a binding site (Fig. 8), and 3′UTR regulated luciferase reporter assays previously confirmed direct targeting of AMPKα, CROT, CPT1a, and HADHB by miR-33a (9, 17, 18). Taken together, this further supported our hypothesis and suggested that the lack of cardiomyocyte TXNIP was leading to decreased miR-33a expression, which, in turn, released CROT, CPT1a, HADHB, and AMPKα from their translational inhibition by this microRNA and, thereby, could explain the increase in β-oxidation observed in cTKO hearts.
Fig. 7.
Protein levels of miR-33a target genes in cTKO and control mice. miR-33a target protein levels as assessed by immunoblotting and corrected for actin in left ventricular samples of cTKO and control mice: CROT (A), CPT1a (B), HADHB (C), AMPKα and p-AMPKα (D), as well as p-ACC2 (E), the downstream signaling target phosphorylated and inactivated by p-AMPKα. Values are expressed as means ± SE; n = 3–5 mice per group. *P < 0.05 (Student's t-test).
Fig. 8.
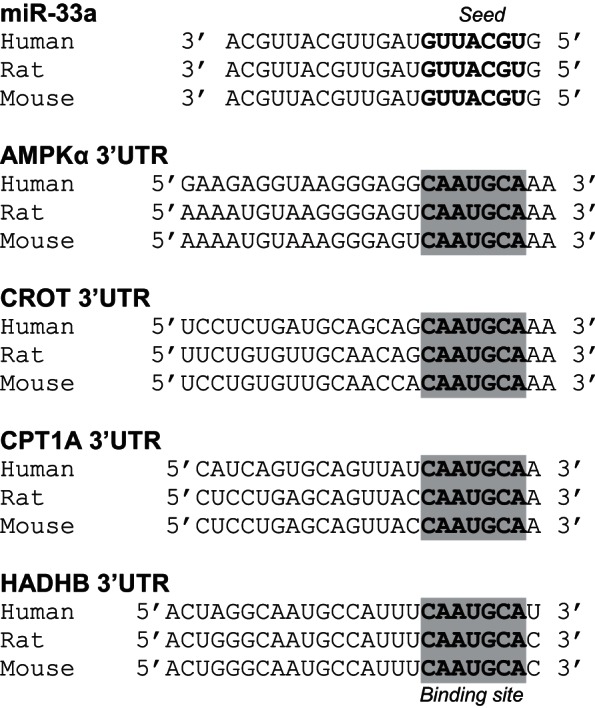
Alignment of conserved miR-33a seed sequence and targeted AMPKα, CROT, CPT1a, and HADHB 3′UTR binding sites. The mature sequence of miR-33a of human, rat and mouse, including the seed sequence (bold) was aligned with the 3′UTR regions of human, rat and mouse AMPKα, CROT, CPT1a, and HADHB (NCBI) containing the complementary miR-33a binding site (highlighted).
TXNIP knockdown upregulates fatty acid oxidation enzymes, and mir-33a overexpression blunts this effect.
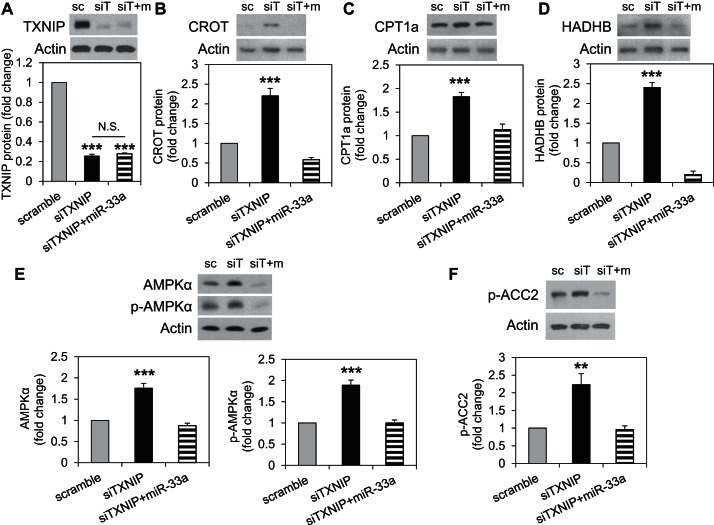
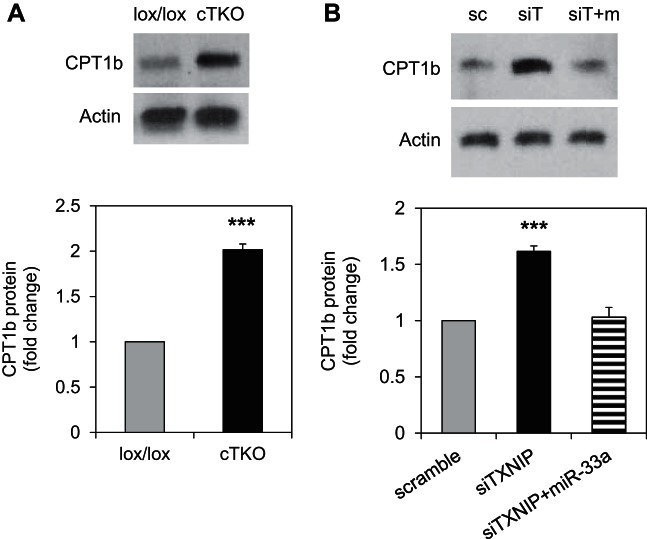
To further confirm the role of miR-33a downregulation in the induction of fatty acid oxidation enzymes in response to TXNIP deficiency, we tested whether miR-33a overexpression could attenuate the effects of TXNIP knockdown. Transfection of H9C2 cells with siTXNIP resulted in effective knockdown of TXNIP protein levels, and this was not affected by miR-33a overexpression (Fig. 9A). Interestingly, while knockdown of TXNIP in H9C2 cardiomyocytes resulted in a highly significant increase in CROT, CPT1a, and HADHB protein levels, similar to that observed in cTKO ventricular samples, simultaneous overexpression of miR-33a completely blunted these effects (Fig. 9, B–D). Similarly, TXNIP knockdown also led to an increase in AMPKα, p-AMPKα, and p-ACC2, and miR-33a overexpression prevented these effects, underlining the importance of this microRNA (Fig. 9, E and F). While CPT1a plays an important role in β-oxidation in the newborn heart, the major CPT1 isoform playing a rate-limiting role in mitochondrial fatty acid oxidation in the adult heart is CPT1b (3, 19). Therefore, we also assessed the effects of TXNIP deficiency on CPT1b and interestingly found that CPT1b levels were significantly increased in hearts of cTKO mice (Fig. 10A). Furthermore, TXNIP knockdown in H9C2 cardiomyocytes resulted in a very similar increase in CPT1b levels, and miR-33a overexpression again completely blunted this effect (Fig. 10B). Together, these results suggest that miR-33a downregulation in response to TXNIP knockdown was, indeed, conferring the observed changes promoting fatty acid oxidation. Moreover, they reveal the existence of a novel TXNIP-miR-33a-CROT/CPT1/HADHB/AMPKα pathway in cardiomyocytes.
Fig. 9.
Role of miR-33a in TXNIP deficiency-mediated induction of β-oxidation enzymes. A: confirmation of effective and persistent TXNIP knockdown in H9C2 cardiomyocytes with and without miR-33a overexpression. Protein expression levels of the fatty acid oxidation enzymes CROT (B), CPT1a (C), and HADHB (D), as well as of AMPKα and p-AMPKα (E) and p-ACC2 (F), as assessed by immunoblotting and corrected for actin in H9C2 cardiomyocytes with TXNIP knockdown (siTXNIP) or TXNIP knockdown and miR-33a overexpression (siTXNIP+miR-33a) and compared with cells transfected with scrambled control (scramble). Representative immunoblots and means ± SE of three independent experiments are shown. **P < 0.005; ***P < 0.001; N.S., not significant (one-way ANOVA).
Fig. 10.
Regulation of cardiac CPT1b protein levels by TXNIP and miR-33a. A: CPT1b protein levels in left ventricular samples of cTKO and control mice, as assessed by immunoblotting. B: CPT1b protein levels in response to TXNIP knockdown (siTXNIP) or TXNIP knockdown and miR-33a overexpression (siTXNIP+miR-33a) and compared with cells transfected with scrambled control (scramble). Representative immunoblots and means ± SE of three or four independent experiments are shown. ***P < 0.001 (A: Student's t-test; B: one-way ANOVA).
TXNIP controls mir-33a expression by promoting transcription of its host gene SREBP2.
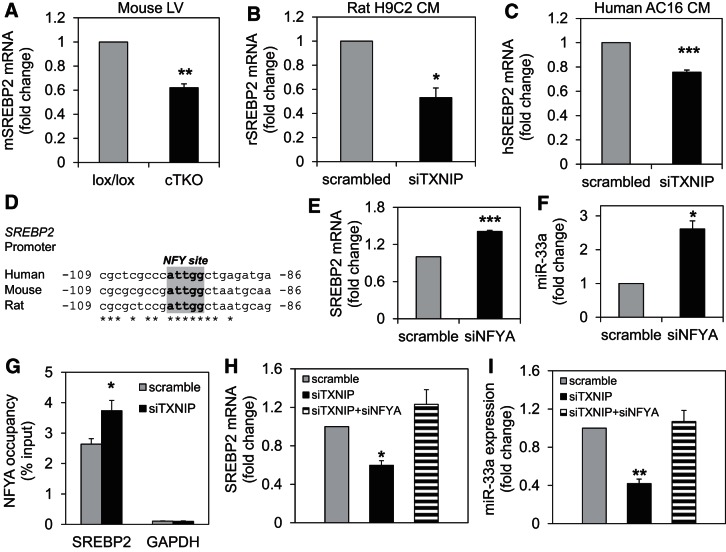
miR-33a is encoded in intron 16 of the human and rat (and intron 17 of the mouse) SREBP2 gene, a basic-helix-loop-helix leucine zipper transcription factor involved in sterol biosynthesis (31). The transcription of miR-33a is, therefore, under the control of a shared SREBP2/miR-33a promoter. To help determine the mechanism by which TXNIP induced miR-33a expression, we next investigated whether TXNIP deficiency also downregulated SREBP2 mRNA expression. We found that SREBP2 expression was indeed significantly lower in ventricular samples of cTKO mice compared with control lox/lox animals (Fig. 11A) and was also significantly decreased by TXNIP knockdown in rat H9C2 cardiomyocytes (Fig. 11B), as well as in human ventricular AC16 cardiomyocytes (Fig. 11C). These results suggested that TXNIP regulation of miR-33a occurs at the transcriptional level and in parallel to the regulation of its host gene SREBP2, indicating that the shared SREBP2/miR-33a promoter might be conferring the effects.
Fig. 11.
Role of NFYA in TXNIP deficiency-mediated effects on SREBP2 and miR-33a. A–C: expression of SREBP2, as assessed by qRT-PCR in left ventricular samples of cTKO and control lox/lox mice (A), rat H9C2 cardiomyocytes (B), and human ventricular AC16 cardiomyocytes (C). D: conserved NYF binding site in the SREBP2 promoter of human, mouse, and rat; gray highlight: inverted CCAAT box. Effects of NFYA knockdown on the expression of SREBP2 (E) and miR-33a (F) in H9C2 cardiomyocytes. G: NFYA occupancy of the SREBP2 promoter (or GAPDH negative control) in response to TXNIP knockdown compared with scrambled control and measured by ChIP in H9C2 cardiomyocytes. SREBP2 (H) and miR-33a expressions (I) in H9C2 cardiomyocytes in response to TXNIP knockdown (siTXNIP) or combined TXNIP and NFYA knockdown (siTXNIP+siNFYA) compared with transfection with scrambled control (scramble) and assessed by qRT-PCR. Values are expressed as means ± SE of three independent experiments. *P < 0.05; **P < 0.005; ***P < 0.001 (A–F: Student's t-test; G–I: one-way ANOVA).
NFYA mediates the TXNIP effects on SREBP2 and mir-33a.
While posttranslational modification and nuclear localization of SREBP2 have been widely studied, the understanding of its transcriptional control is limited to older gel shift experiments, which have suggested binding of the transcription factor nuclear factor-Y (NFY) to the SREBP2 promoter region (32). Consistent with these earlier observations, our MatInspector analysis revealed a highly conserved inverted CCAAT box NFY-binding site in the SREBP2 promoter (Fig. 11D). NFY is a ubiquitously expressed, bifunctional, trimeric transcription factor, and sequence specificity of its interaction with DNA is conferred by the α-subunit, NFYA, which is, therefore, considered the regulatory subunit (4, 49). NFYA knockdown in our cardiomyocytes resulted in significantly higher mRNA expression of SREBP2 and miR-33a (Fig. 11, E and F) supporting the notion that NFYA was acting as a transcriptional inhibitor at the SREBP2 promoter. This is also in alignment with our recent findings, in which NFY was found to function as a transcriptional repressor in cardiomyocytes (5). If NFYA were, indeed, mediating the effects of TXNIP deficiency on miR-33a, we would expect TXNIP knockdown to increase NFYA binding to the SREBP2 promoter, which would then lead to inhibition of SREBP2 and miR-33a transcription. In fact, this is exactly what we observed in our chromatin immunoprecipitation (ChIP) studies (Fig. 11G). Not only did we find strong NFYA binding at the SREBP2 promoter, providing proof of this interaction, but NFYA occupancy was also significantly increased in response to TXNIP knockdown, whereas GAPDH and IgG negative controls showed no enrichment.
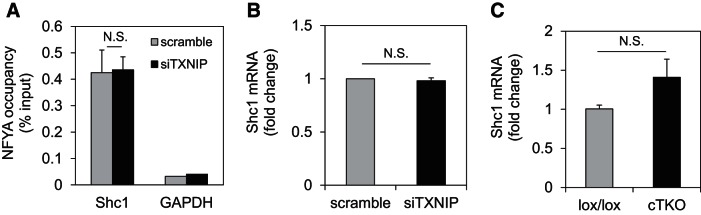
To further assess whether the effects of TXNIP knockdown on NFYA binding to the SREBP2 promoter were specific, we also investigated the effects on another NFYA target gene, Shc transforming protein (Shc1). Shc1 was initially discovered as a NFY target in a liver cell ChIP-genome tiling array study (30), and we previously reported that NFY binds to the Shc1 promoter in cardiomyocytes (5). Consistent with this notion, our ChIP studies showed NFYA occupancy of the Shc1 promoter, while GAPDH and IgG negative controls showed no enrichment (Fig. 12A). However, in contrast to SREBP2, TXNIP knockdown did not alter NFYA binding to the Shc1 promoter and did not affect Shc1 expression in H9C2 cardiomyocytes (Fig. 12B). Also, unlike SREBP2 expression, which was downregulated in mouse left ventricular cTKO samples (Fig. 11A), Shc1 expression was not at all decreased in cTKO hearts (Fig. 12C). These results suggest that TXNIP deficiency does not lead to a general increase in NFYA binding to other transcriptional targets and that the observed effects might be specific to SREBP2.
Fig. 12.
Effects of TXNIP deficiency on NFYA-mediated transcription of Shc1. A: ChIP showing NFYA occupancy of the Shc1 promoter (or GAPDH negative control) in response to TXNIP knockdown compared with scrambled. B: Shc1 mRNA expression in H9C2 cardiomyocytes in response to TXNIP knockdown (siTXNIP). C: Shc1 mRNA expression in hearts of lox/lox and cTKO mice. Values are expressed as means ± SE of three independent experiments. N.S., not significant (Student's t-test).
We next investigated whether NFYA knockdown could prevent the effects of TXNIP deficiency on SREBP2 and miR-33a expression. While TXNIP knockdown again led to a significant decrease in SREBP2 and miR-33a expression, simultaneous knockdown of NFYA completely blunted these effects and normalized SREBP2 and miR-33a expression levels (Fig. 11, H and I). Similarly, in human ventricular AC16 cardiomyocytes, TXNIP knockdown also significantly lowered SREBP2 and miR-33a expression, while NFYA knockdown abolished these effects completely (P < 0.05) (data not shown). These findings strongly suggest that TXNIP deficiency downregulates miR-33a by promoting binding of inhibitory NFYA to the shared SREBP2/miR-33a promoter and underline the critical role that this transcription factor plays in these effects and in linking TXNIP to miR-33a. They, thereby, also provide for the first time insight into the upstream signaling mechanisms that control miR-33a expression.
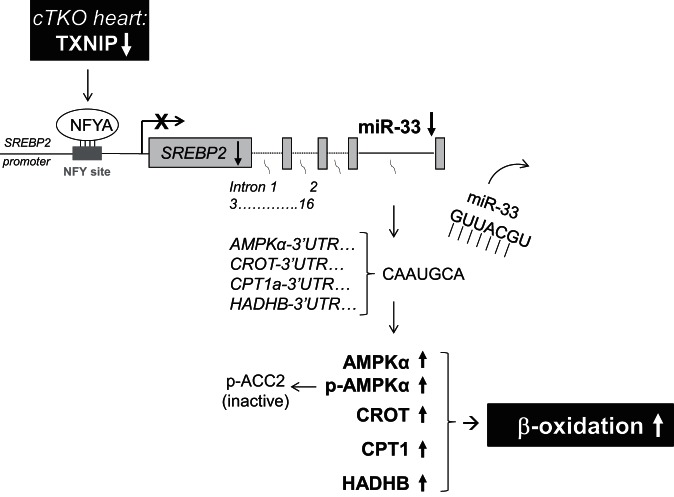
Combined, these findings reveal a novel pathway, in which cardiomyocyte TXNIP deficiency leads to increased binding of NFYA to the SREBP2 promoter, resulting in transcriptional inhibition of SREBP2 and intronic miR-33a. The decrease in miR-33a levels, in turn, releases the 3′UTRs of AMPKα, CROT, CPT1, and HADHB from the inhibition by this microRNA, allowing for increased translation of these critical β-oxidation enzymes and activation of myocardial fatty acid oxidation as observed in the working heart perfusions of the cTKO mice. The increase in AMPKα is also associated with an increase in phosphorylated/activated AMPKα, which promotes phosphorylation and inactivation of ACC2, further supporting an increase in β-oxidation (Fig. 13).
Fig. 13.
Schematic summary of novel TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB signaling pathway conferring TXNIP regulation of myocardial fatty acid oxidation.
DISCUSSION
In summary, we have discovered that TXNIP regulates myocardial fatty acid oxidation via miR-33a signaling and have identified of a novel, regulatory TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB pathway.
While microRNAs are increasingly recognized as important regulators of gene expression in many organ systems, including the heart (1, 23, 25), this is the first demonstration that miR-33a is involved in myocardial biology and is part of a previously unappreciated signaling cascade that is highly conserved in mouse, rat, as well as human cardiomyocytes and that links TXNIP to myocardial β-oxidation. On the other hand, our results showing that alterations in miR-33a expression control myocardial levels of the fatty acid oxidation enzymes CROT, CPT1, and HADHB are consistent with previous findings in liver cells, where miR-33a has been shown to target and modulate cholesterol homeostasis and β-oxidation (9, 17, 31). miR-33a has also been shown to directly target AMPKα (9, 15, 17, 18, 29, 31), and in alignment with this notion, we found AMPKα to be activated in TXNIP-deficient cTKO hearts and to phosphorylate and inactivate its downstream target ACC2. By inactivating ACC2 and leading to decreased malonyl-CoA, AMPKα also promotes β-oxidation (39). Furthermore, AMPKα has been shown to promote glucose uptake and glycolysis (39) and AMPKα activation, as found in our present studies in response to TXNIP deficiency, may also help explain the increased glucose uptake recently reported in hearts of TXNIP-null mice (43).
The study of downstream targets of microRNAs has evolved rapidly, but the analysis of their upstream regulation has proven to be very difficult and has been lagging behind. In this regard, our results also provide a major advance, as they demonstrate for the first time that miR-33a expression is controlled by TXNIP and that this effect is mediated by NFYA. As demonstrated by the NFYA ChIP studies, NFYA binding to the SREBP2/miR-33a promoter was increased in response to TXNIP knockdown, resulting in decreased SREBP2 and miR-33a expression, consistent with NFYA acting again as a transcriptional repressor (5). This effect seems further to be specific for SREBP2/miR-33a, as TXNIP knockout did not affect NFYA-mediated transcription of other bona fide target genes, such as Shc1. The fact that NFYA knockdown completely reversed the effects of TXNIP deficiency further supports the role that this transcription factor plays in conferring the TXNIP effects on miR-33a.
Interestingly, oxidative stress has recently been shown to induce SREBP2 (8), the host gene of miR-33a, raising the possibility that miR-33a would also be upregulated by oxidative stress. This notion is consistent with our current findings as TXNIP is known to induce oxidative stress (22, 27), and in our case, TXNIP deficiency resulted in decreased SREBP2 and miR-33a expression.
While previous studies using HcB-19 mice harboring a TXNIP nonsense mutation failed to reveal a significant effect on cardiac β-oxidation (34), our studies using cTKO mice with cardiomyocyte-specific TXNIP deletion now clearly demonstrated an improvement in myocardial fatty acid oxidation. This difference could be explained by confounding effects, such as the hyperinsulinemia, found in HcB-19 mice with whole body TXNIP deficiency (7), yet absent in our cTKO mice with cardiomyocyte-specific deletion of TXNIP. On the other hand, the decrease in glucose oxidation observed in our cTKO hearts is consistent with findings in HcB-19 mice (34), as well as with recent reports of decreased pyruvate dehydrogenase activity in hearts from TXNIP-null mice (43).
Taken together, our results have identified a novel TXNIP-NFYA-SREBP2/miR-33a-AMPKα/CROT/CPT1/HADHB signaling pathway that regulates myocardial fatty acid oxidation, a process that is essential for the maintenance of myocardial energy homeostasis. They, thereby, shed new light on cardiac physiology and on the role of TXNIP and microRNA signaling in the heart.
GRANTS
This work was supported by grants to A. Shalev from the National Institutes of Health (NIH) (Grants R01DK-078752 and UC4DK-104204) and JDRF (3-SRA-2014-302-M-R), and M. E. Young received an NIH Grant (R01 HL-074259), as well as by a Multi-Investigator Planning Grant from the University of Alabama at Birmingham Department of Medicine to A. Shalev, M. E. Young, and J. C. Chatham.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.C., M.E.Y., and L.J.D. performed experiments; J.C., M.E.Y., D.K.C., and L.J.D. analyzed data; J.C., M.E.Y., J.C.C., and L.J.D. interpreted results of experiments; J.C., M.E.Y., and D.K.C. prepared figures; J.C., M.E.Y., J.C.C., D.K.C., L.J.D., and A.S. approved final version of manuscript; A.S. conception and design of research; A.S. drafted manuscript; A.S. edited and revised manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The alphaMyHC-Cre mice were generously provided by Dr. Michael Schneider and the AC16 cells by Dr. Mercy Davidson. We would like to thank Truman B. Grayson, John W. Thomas, Betty M. Pat, and Wayne E. Bradley for their excellent technical assistance.
REFERENCES
- 1.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res 110: 638–650, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest 100: 169–179, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown NF, Weis BC, Husti JE, Foster DW, McGarry JD. Mitochondrial carnitine palmitoyltransferase I isoform switching in the developing rat heart. J Biol Chem 270: 8952–8957, 1995. [DOI] [PubMed] [Google Scholar]
- 4.Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol 28: 2047–2058, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha-Molstad H, Xu G, Chen J, Jing G, Young ME, Chatham JC, Shalev A. Calcium channel blockers act through nuclear factor Y to control transcription of key cardiac genes. Mol Pharmacol 82: 541–549, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Cha-Molstad H, Szabo A, Shalev A. Diabetes induces and calcium channel blockers prevent cardiac expression of pro-apoptotic thioredoxin-interacting protein. Am J Physiol Endocrinol Metab 296: E1133–E1139, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, Attie AD, Lusis AJ, Davis RA, Shalev A. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta cell mass and protects against diabetes. FASEB J 22: 3581–3594, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Wen L, Martin M, Hsu CY, Fang L, Lin FM, Lin TY, Geary MJ, Geary GG, Zhao Y, Johnson DA, Chen JW, Lin SJ, Chien S, Huang HD, Miller YI, Huang PH, Shyy JY. Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA-92a. Circulation 131: 805–814, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, Fisher EA, Penalva LO, Moore KJ, Suarez Y, Lai EC, Fernandez-Hernando C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA 108: 9232–9237, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djouadi F, Weinheimer CJ, Saffitz JE, Pitchford C, Bastin J, Gonzalez FJ, Kelly DP. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor α-deficient mice. J Clin Invest 102: 1083–1091, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolinsky VW, Dyck JR. Role of AMP-activated protein kinase in healthy and diseased hearts. Am J Physiol Heart Circ Physiol 291: H2557–H2569, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, Zou C, Zou L, Johnson RL Jr, Dyck JR, Bray MS, Gamble KL, Chatham JC, Young ME. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 286: 44,606–44,619, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durgan DJ, Pulinilkunnil T, Villegas-Montoya C, Garvey ME, Frangogiannis NG, Michael LH, Chow CW, Dyck JR, Young ME. Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Circ Res 106: 546–550, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durgan DJ, Smith JK, Hotze MA, Egbejimi O, Cuthbert KD, Zaha VG, Dyck JR, Abel ED, Young ME. Distinct transcriptional regulation of long-chain acyl-CoA synthetase isoforms and cytosolic thioesterase 1 in the rodent heart by fatty acids and insulin. Am J Physiol Heart Circ Physiol 290: H2480–H2497, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Hernando C, Moore KJ. MicroRNA modulation of cholesterol homeostasis. Arterioscler Thromb Vasc Biol 31: 2378–2382, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foryst-Ludwig A, Kreissl MC, Sprang C, Thalke B, Bohm C, Benz V, Gurgen D, Dragun D, Schubert C, Mai K, Stawowy P, Spranger J, Regitz-Zagrosek V, Unger T, Kintscher U. Sex differences in physiological cardiac hypertrophy are associated with exercise-mediated changes in energy substrate availability. Am J Physiol Heart Circ Physiol 301: H115–H122, 2011. [DOI] [PubMed] [Google Scholar]
- 17.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 285: 33,652–33,661, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramirez CM, Mattison JA, de Cabo R, Suarez Y, Fernandez-Hernando C. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol 33: 2339–2352, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Kim T, Long Q, Liu J, Wang P, Zhou Y, Ding Y, Prasain J, Wood PA, Yang Q. Carnitine palmitoyltransferase-1b deficiency aggravates pressure overload-induced cardiac hypertrophy caused by lipotoxicity. Circulation 126: 1705–1716, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, Davis RA. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA 105: 3921–3926, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R, Dludla P, Joubert E, February F, Mazibuko S, Ghoor S, Muller C, Louw J. Aspalathin, a dihydrochalcone C-glucoside, protects H9c2 cardiomyocytes against high glucose-induced shifts in substrate preference and apoptosis. Mol Nutrit Food Res 60: 922–934, 2016. [DOI] [PubMed] [Google Scholar]
- 22.Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG, Choi I. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol 164: 6287–6295, 2000. [DOI] [PubMed] [Google Scholar]
- 23.Karunakaran D, Rayner KJ. MicroRNAs in cardiovascular health: from order to disorder. Endocrinology 154: 4000–4009, 2013. [DOI] [PubMed] [Google Scholar]
- 24.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol 6: 419–429, 2009. [DOI] [PubMed] [Google Scholar]
- 26.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev 90: 207–258, 2010. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama A, Masutani H, Nakamura H, Nishinaka Y, Yodoi J. Redox regulation by thioredoxin and thioredoxin-binding proteins. IUBMB Life 52: 29–33, 2001. [DOI] [PubMed] [Google Scholar]
- 28.Palomer X, Alvarez-Guardia D, Rodriguez-Calvo R, Coll T, Laguna JC, Davidson MM, Chan TO, Feldman AM, Vazquez-Carrera M. TNF-α reduces PGC-1α expression through NF-κB and p38 MAPK leading to increased glucose oxidation in a human cardiac cell model. Cardiovasc Res 81: 703–712, 2009. [DOI] [PubMed] [Google Scholar]
- 29.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 478: 404–407, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet 4: e1000133, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13: 239–250, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem 271: 26,461–26,464, 1996. [DOI] [PubMed] [Google Scholar]
- 33.Schulze PC, Liu H, Choe E, Yoshioka J, Shalev A, Bloch KD, Lee RT. Nitric oxide-dependent suppression of thioredoxin-interacting protein expression enhances thioredoxin activity. Arterioscler Thromb Vasc Biol 26: 2666–2672, 2006. [DOI] [PubMed] [Google Scholar]
- 34.Sheth SS, Castellani LW, Chari S, Wagg C, Thipphavong CK, Bodnar JS, Tontonoz P, Attie AD, Lopaschuk GD, Lusis AJ. Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. J Lipid Res 46: 123–134, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Stavinoha MA, Rayspellicy JW, Hart-Sailors ML, Mersmann HJ, Bray MS, Young ME. Diurnal variations in the responsiveness of cardiac and skeletal muscle to fatty acids. Am J Physiol Endocrinol Metab 287: E878–E887, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res 32: e188, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taegtmeyer H, Stanley WC. Too much or not enough of a good thing? Cardiac glucolipotoxicity versus lipoprotection. J Mol Cell Cardiol 50: 2–5, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, Villegas-Montoya C, Jahoor A, Gonzalez R, Garvey ME, Boland B, Blasier Z, McElfresh TA, Nannegari V, Chow CW, Heird WC, Chandler MP, Dyck JR, Bray MS, Young ME. Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem 285: 2918–2929, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, Beauloye C, Foretz M, Andreelli F, Ventura-Clapier R, Bertrand L. AMPK: Lessons from transgenic and knockout animals. Front Biosci 14: 19–44, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, De Keulenaer GW, Lee RT. Vitamin D3-up-regulated protein-1 is a stress-responsive gene that regulates cardiomyocyte viability through interaction with thioredoxin. J Biol Chem 277: 26,496–26,500, 2002. [DOI] [PubMed] [Google Scholar]
- 41.Wittnich C, Tan L, Wallen J, Belanger M. Sex differences in myocardial metabolism and cardiac function: an emerging concept. Pflügers Arch 465: 719–729, 2013. [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 19: 1141–1146, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshioka J, Chutkow WA, Lee S, Kim JB, Yan J, Tian R, Lindsey ML, Feener EP, Seidman CE, Seidman JG, Lee RT. Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the myocardium from ischemia-reperfusion injury. J Clin Invest 122: 267–279, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshioka J, Imahashi K, Gabel SA, Chutkow WA, Burds AA, Gannon J, Schulze PC, MacGillivray C, London RE, Murphy E, Lee RT. Targeted deletion of thioredoxin-interacting protein regulates cardiac dysfunction in response to pressure overload. Circ Res 101: 1328–1338, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Yoshioka J, Schulze PC, Cupesi M, Sylvan JD, MacGillivray C, Gannon J, Huang H, Lee RT. Thioredoxin-interacting protein controls cardiac hypertrophy through regulation of thioredoxin activity. Circulation 109: 2581–2586, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Young ME, Guthrie PH, Razeghi P, Leighton B, Abbasi S, Patil S, Youker KA, Taegtmeyer H. Impaired long-chain fatty acid oxidation and contractile dysfunction in the obese Zucker rat heart. Diabetes 51: 2587–2595, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Young ME, Laws FA, Goodwin GW, Taegtmeyer H. Reactivation of peroxisome proliferator-activated receptor-α is associated with contractile dysfunction in hypertrophied rat heart. J Biol Chem 276: 44,390–44,395, 2001. [DOI] [PubMed] [Google Scholar]
- 48.Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res 89: 1199–1208, 2001. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Chen B, Li Y, Chen J, Lou G, Chen M, Zhou D. Transcriptional regulation of the human PNRC promoter by NFY in HepG2 cells. J Biochem 143: 675–683, 2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.