Summary
Using mouse models of breast cancer, we show that carcinogen exposures introduce driver mutations that are directed to preferred oncogenes with surprising specificity. In follow-up studies, we show that these oncogene preferences reflect strikingly distinct, carcinogen-specific, strand-biased mutation patterns.
Abstract
Carcinogen exposures inscribe mutation patterns on cancer genomes and sometimes bias the acquisition of driver mutations toward preferred oncogenes, potentially dictating sensitivity to targeted agents. Whether and how carcinogen-specific mutation patterns direct activation of preferred oncogenes remains poorly understood. Here, mouse models of breast cancer were exploited to uncover a mechanistic link between strand-biased mutagenesis and oncogene preference. When chemical carcinogens were employed during Wnt1-initiated mammary tumorigenesis, exposure to either 7,12-dimethylbenz(a)anthracene (DMBA) or N-ethyl-N-nitrosourea (ENU) dramatically accelerated tumor onset. Mammary tumors that followed DMBA exposure nearly always activated the Ras pathway via somatic HrasCAA61CTA mutations. Surprisingly, mammary tumors that followed ENU exposure typically lacked Hras mutations, and instead activated the Ras pathway downstream via BrafGTG636GAG mutations. HrasCAA61CTA mutations involve an A-to-T change on the sense strand, whereas BrafGTG636GAG mutations involve an inverse T-to-A change, suggesting that strand-biased mutagenesis may determine oncogene preference. To examine this possibility further, we turned to an alternative Wnt-driven tumor model in which carcinogen exposures augment a latent mammary tumor predisposition in Apcmin mice. DMBA and ENU each accelerated mammary tumor onset in Apcmin mice by introducing somatic, “second-hit” Apc mutations. Consistent with our strand bias model, DMBA and ENU generated strikingly distinct Apc mutation patterns, including stringently strand-inverse mutation signatures at A:T sites. Crucially, these contrasting signatures precisely match those proposed to confer bias toward HrasCAA61CTA versus BrafGTG636GAG mutations in the original tumor sets. Our findings highlight a novel mechanism whereby exposure history acts through strand-biased mutagenesis to specify activation of preferred oncogenes.
Introduction
Cancers frequently acquire mutations that increase signaling through the mitogen-activated protein kinase (MAPK) pathway (1). Aberrant activation of MAPK signaling can be triggered by mutations affecting a variety of pathway components, but the mechanisms favoring mutation of specific oncogenes in specific tumor types remain poorly understood. For example, nearly one-third of lung cancers acquire KRAS mutations, whereas fewer than 12% instead acquire mutations in EGFR, an upstream signaling component (2). By contrast, half of all melanomas acquire mutations in BRAF, a downstream signaling component, whereas a minority instead acquire mutations in NRAS (3). MAPK pathway mutations rarely co-occur within these cancers, implying that they act in either a functionally redundant or synthetic lethal manner (4). Determining which MAPK pathway component has been activated by mutation can be clinically important, since the response to drugs designed to antagonize mutated EGFR or BRAF strongly depends upon cancers carrying the corresponding driver mutations.
Notably, carcinogen exposures skew MAPK pathway-activating mutations toward specific oncogenes, thereby influencing which targeted agents are likely to be effective against carcinogen-induced cancers. For example, KRAS mutations are over-represented (and EGFR mutations are under-represented) in lung cancers linked to cigarette smoke (5,6), whereas BRAF mutations are over-represented in melanomas linked to sunlight exposure (7). Consequently, lung cancers from patients without a smoking history more frequently carry EGFR mutations heralding sensitivity to drugs targeting EGFR, whereas melanomas that arise on sunlight-exposed skin more frequently carry BRAF mutations heralding sensitivity to drugs targeting BRAF. Whether and how carcinogen-specific mutation signatures direct pathogenic mutations to preferred driver genes remains unclear. The most prevalent KRAS mutations in lung cancer arise through base substitutions replacing guanines on the sense strand, consistent with known mutational consequences of carcinogens found in cigarette smoke (1,6). However, the recurring BRAFV600E mutation in melanoma arises through a T:A-to-A:T transversion that is absent from the canonical mutation spectrum for ultraviolet light (3,8), suggesting that mutation signatures are not the sole mechanism linking specific carcinogen exposures to specific oncogene mutations. Presumably, oncogene preferences reflect a complex interplay among numerous factors, including host genetics, cell lineage programming, environmental exposure history and DNA repair pathways.
Decades before cancer genome re-sequencing became technically feasible, targeted re-sequencing of well-known cancer genes revealed mutation patterns indicative of antecedent DNA damage and repair. For example, classic molecular epidemiology studies identified carcinogen-specific mutation signatures within the TP53 tumor suppressor gene, thereby directly implicating cigarette smoke and ultraviolet light exposures in the pathogenesis of lung (9) and skin cancer (10), respectively. These stereotyped, carcinogen-specific driver mutations recurred across a range of preferred codons, providing strong confirmation of causation (11). Mutation spectra assembled by re-sequencing of gain-of-function oncogenes can yield even narrower codon specificities, owing to profound evolutionary constraints that restrict the range of mutations capable of yielding activated alleles. Notably, carcinogen-induced Hras mutations identified in rodent models typically are confined to a single codon. In chemically induced rat mammary cancers, tumor initiation via N-methyl-N-nitrosourea exposure preferentially introduces HrasGGA12GAA mutations (12), whereas using 7,12-dimethylbenz(a)anthracene (DMBA) in place of N-methyl-N-nitrosourea in the protocol preferentially introduces HrasCAA61CTA mutations instead (13). Remarkably, classic mouse models of multistep skin carcinogenesis precisely recapitulate these carcinogen-specific Hras mutation preferences, and the preferred base substitution for each exposure matches well with known modes of DNA damage and repair (14,15). Here, we show that the influence of carcinogen-specific mutation patterns can extend beyond the targeting of particular codons within a given oncogene to include the targeting of particular oncogenes within a signaling pathway. In this way, we identify strand-biased mutagenesis as a novel mechanism capable of imparting dramatic oncogene preferences in vivo.
Materials and methods
Mice and chemical carcinogenesis
Mice were housed at the Pennsylvania State University College of Medicine pathogen-free rodent facility with free access to water and chow. All experimental protocols were approved by the Pennsylvania State University College of Medicine’s Institutional Animal Care and Use Committee. The MMTV-rtTA and TetO-Wnt1 transgenic lines (iWnt) were maintained in an FVB/N background. Apcmin mice were obtained from Jackson Laboratories (C57BL/6J-Apcmin/J stock no. 002020). For Dox treatment, standard mouse chow was replaced with medicated chow containing 2g/kg drug (Bio Serv). For carcinogen exposures, mice were given DMBA (1mg) via oral gavage or N-ethyl-N-nitrosourea (ENU) (150mg/kg) via intraperitoneal (i.p.) injection. DMBA (Sigma-Aldrich D3254) was dissolved in sesame oil (Sigma-Aldrich S3547) at 5mg/ml. ENU solution was 1g ENU (Sigma-Aldrich N3385) dissolved in 10ml 95% ethanol and 90ml phospho-citrate buffer (Sigma-Aldrich P4809). Mice were euthanized and necropsies were performed when the diameter of the largest single mammary tumor reached 2cm (all iWnt and some Apcmin mice), or when mice became moribund due to their underlying intestinal adenoma predisposition (remaining Apcmin mice). Some mammary tumors, though non-palpable prior to necropsy, were readily apparent at necropsy. These macroscopic tumors were subjected to histolopathologic confirmation, were counted toward overall tumor multiplicity and were included in DNA sequencing analyses where they yielded an incidence of mutations comparable with that of palpable tumors. Mice were genotyped using PCR-based assays performed on genomic DNA derived from tail snips as described.
DNA preparation
Genomic DNA was isolated from tumor fragments and tail snips using Promega Maxwell 16 Tissue DNA Purification Kit (Promega AS1030). For tumors, primers specific for Hras (NCBI mRNA: NM_008284), Braf (NCBI mRNA: NM_13294.5) and Apc (NCBI mRNA: NM_007462.3) were used for PCR amplification. Hras: 5′-GGTCAGGCATCTATTAGCCGTC, 5′-GCCGAGACTCAACAGTGCGAG. Braf: 5′-GGGCCAAATCAAATTAGAACGTCC, 5′-GCCTGGCTTACAATGTTATTCCTG. Apc: 5′-CCTCCTCCACAGACAGTGC, 5′-AGCTGACTTGGTTTCCTTGC. PCR products were purified using a Qiaquick PCR purification kit (28104). Purified PCR fragments were Sanger sequenced using an ABI 3130XL Capillary sequencer. Sequences were analyzed using AB DNA Sequencing Analysis Software v5.2 and AB Sequence Scanner v1.0.
Results
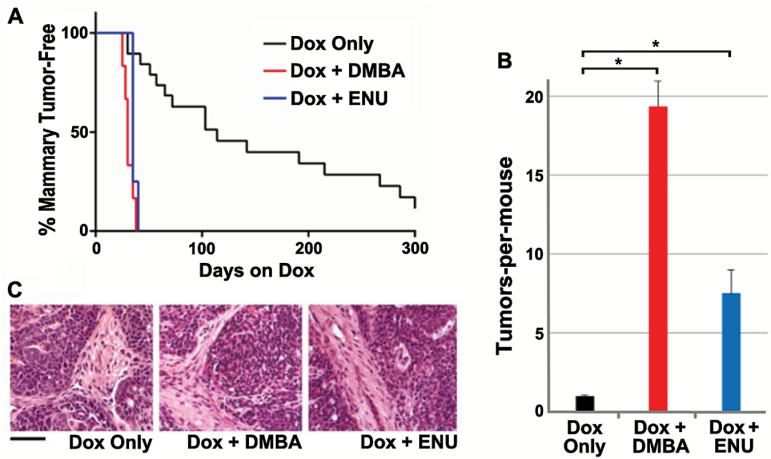
To investigate mechanisms of mammary tumor initiation by chemical carcinogens, iWnt mice (16) (engineered for inducible-Wnt1 expression) were started on chronic inducer treatment with doxycycline (Dox) at 5 weeks of age, then left unexposed or subjected to a one-time carcinogen exposure (either DMBA or ENU) 1 week later. Both carcinogen exposures reduced the time-to-onset of palpable mammary tumors nearly 6-fold (mean T50 of 25 days dox for either DMBA or ENU versus 140 days for carcinogen-naive mice; Figure 1A). In addition, carcinogen treatments markedly increased tumor multiplicity from a mean of 1.2 tumors-per-mouse in carcinogen-naive mice to a mean of 19.5 tumors and 7.5 tumors following DMBA and ENU exposure, respectively (Figure 1B). Tumors arising in the context of carcinogen exposure were indistinguishable from carcinogen-naive tumors in histopathology and invariably showed mixed-lineage differentiation (Figure 1C and data not shown).
Figure 1.
Mammary tumorigenesis enhanced by carcinogen exposures in iWnt mice. (A) Rapid onset of mammary tumors in carcinogen-exposed iWnt mice. One week after starting Dox treatment, cohorts of iWnt mice were left unexposed (Dox only, n = 18) or subjected to one-time exposure to either DMBA (n = 6) or ENU (n = 4), then monitored for mammary tumors. Tumor onset was more rapid in carcinogen-exposed versus unexposed cohorts (P < 0.0001, log rank test). (B) Increased tumor multiplicity following carcinogen exposure. Nearly all iWnt mice developed solitary mammary tumors in the absence of carcinogen exposure, whereas carcinogen-exposed mice developed numerous mammary tumors synchronously. Error bars depict standard error of the mean. *Denotes P < 0.0001, t-test. (C) Uniform iWnt mammary tumor histopathology in the presence and absence of carcinogen exposure. Tumors were mixed-lineage adenocarcinomas with prominent tumor cell nests and intervening stroma. Scale bar, 50 µm.
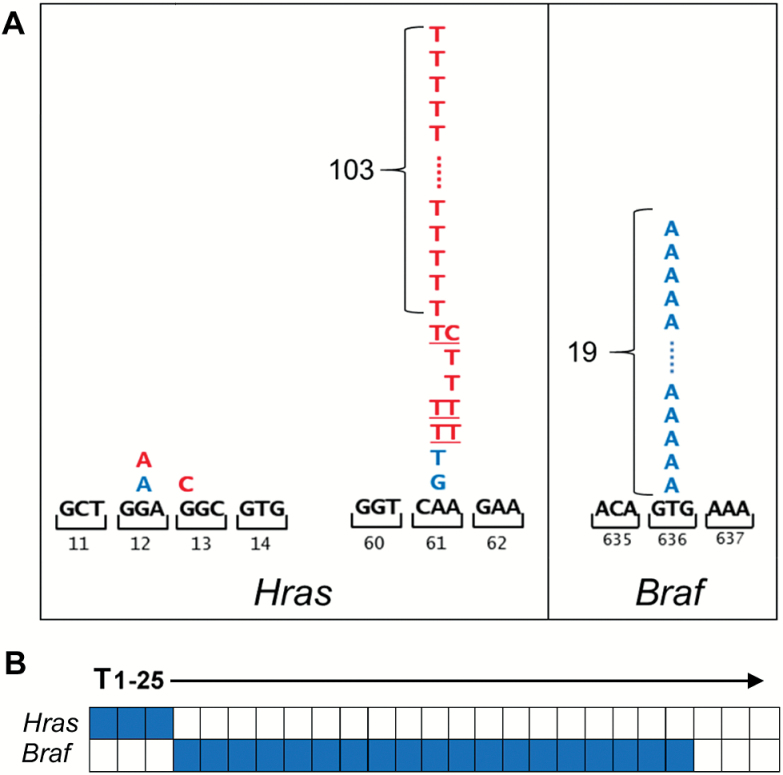
We reasoned that carcinogen-induced driver mutations likely explain the robust co-operation between carcinogen exposures and Wnt pathway activation during mammary tumorigenesis. In carcinogen-naive mice, Wnt-driven mammary tumors frequently acquire spontaneous co-operating Hras mutations (17,18). Accordingly, we sequenced Hras alleles derived from carcinogen-exposed iWnt tumors looking for carcinogen-specific mutation patterns. Mammary tumors arising following DMBA exposure invariably acquired activating Hras mutations (110 of 110, 100%). Nearly all of these mutations were the same HrasCAA61CTA base substitution (103 of 110, 94%; Figure 2A), precisely recapitulating the signature DMBA-induced mutation acquired as an obligate initiating step during multistage skin carcinogenesis modeling in mice (15). Since ENU, like DMBA, preferentially induces mutations at A:T base pairs, we expected that most iWnt tumors arising in the context of ENU exposure likewise would harbor HrasCAA61CTA mutations. Instead, we found that ENU tumors acquired Hras mutations only rarely (3 of 25, 12%), and only one of three Hras mutations identified in ENU tumors matched the signature HrasCAA61CTA change (Figure 2A).
Figure 2.
Carcinogen exposures direct MAPK pathway mutations to different oncogenes. (A) Somatic mutations in MAPK pathway oncogenes by carcinogen exposure. Tumor sets generated in the context of each carcinogen exposure were surveyed for mutations in candidate MAPK pathway genes at known sites of recurrent cancer-associated mutations (Hras codons 12, 13 and 61; Braf codon 636, corresponding to human BRAF codon 600). Color-coded letters indicate base substitutions identified in individual tumors (red for DMBA-induced mutations; blue for ENU-induced mutations). All 110 DMBA-induced tumors acquired activating Hras mutations, and 103 of these 110 mutations generated the HrasCAA61CTA allele. Only 3 of 25 ENU-induced tumors acquired an Hras mutation, whereas 19 of these tumors instead acquired a BrafGTG636GAG mutation. Three ENU-induced tumors acquired neither an Hras nor a Braf mutation. Underlined letters indicate mutations at both bases detected in a single tumor. (B) Mutual exclusivity of Hras and Braf mutations. Co-occurrence plot for somatic mutations acquired in the context of ENU exposure is depicted. No Braf mutations were detected in a survey of 20 DMBA-induced tumors, all of which carried Hras mutations.
To account for the dearth of ENU-induced Hras mutations, we considered whether ENU recurrently mutated an alternative oncogene within one of the effector pathways known to signal downstream of Hras. Accordingly, we examined Wnt tumors from ENU-exposed mice for an activating BrafV636E mutation analogous to the BRAFV600E mutation found in many human cancers (19). Indeed, most ENU tumors acquired a BrafGTG636GAG mutation (19 of 25, 76%; Figure 2A). Notably, Braf mutations never co-occurred with Hras mutations in the ENU tumor set, implying that the two mutations show either functional redundancy or synthetic lethality (Figure 2B). Concordantly, no BrafGTG636GAG mutations were detected in a survey of 20 tumors that arose in the setting of DMBA exposure, all of which had acquired Hras mutations. Taken together, each carcinogen activated a preferred oncogene through an opposite transversion mutation (HrasCAA61CTA via an A-to-T change on the sense strand for DMBA, versus BrafGTG636GAG via a T-to-A change for ENU). Assuming that DMBA and ENU preferentially form adducts with adenine and thymine respectively, each signature mutation replaces a damage-prone base residing on the sense strand. From these data, we inferred that strand-biased mutagenesis offers a plausible mechanism linking each carcinogen to its preferred oncogene.
For both Hras and Braf, cancer-associated mutations recur in just a few codons, reflecting powerful constraints on the DNA sequence changes capable of yielding activated alleles in a single step. Therefore, carcinogen-specific mutation signatures derived from these genes provide a skewed readout of carcinogen-induced DNA sequence changes, markedly altered by selection pressure. To assess strand bias in a less constrained genetic context, we turned to an alternative Wnt-driven mammary tumorigenesis model based on carcinogen treatment of Apcmin mutant mice. Best known for their highly penetrant predisposition toward intestinal tumors, Apcmin mice also show a less penetrant mammary tumor predisposition that can be enhanced by carcinogen exposure (20,21). Although intestinal and mammary tumors both acquire second “hits” that relieve Apc-mediated repression of Wnt signaling, mammary tumors uniquely select for stereotyped, hypomorphic Apc mutation. In mice engineered for mammary-specific knockout of one Apc allele, mammary tumors acquired second-hit nonsense mutations that clustered within a hotspot region of Apc spanning codons 1512 to 1579, hereafter designated Apcmmcr for mammary tumor mutation cluster region (22). Apcmmcr mutations encode truncated proteins with partially preserved capacity to repress the Wnt pathway due to retention of 3 out of 7 repeats of a conserved β-catenin binding domain (Supplementary Figure 1, available at Carcinogenesis Online), thereby conferring a modest increase in Wnt signaling conducive to mammary tumorigenesis (23). Consistent with this model, germline inheritance of a single Apcmmcr allele, such as Apc1572stop or Apc1576stop, imparts a robust mammary tumor predisposition (24,25).
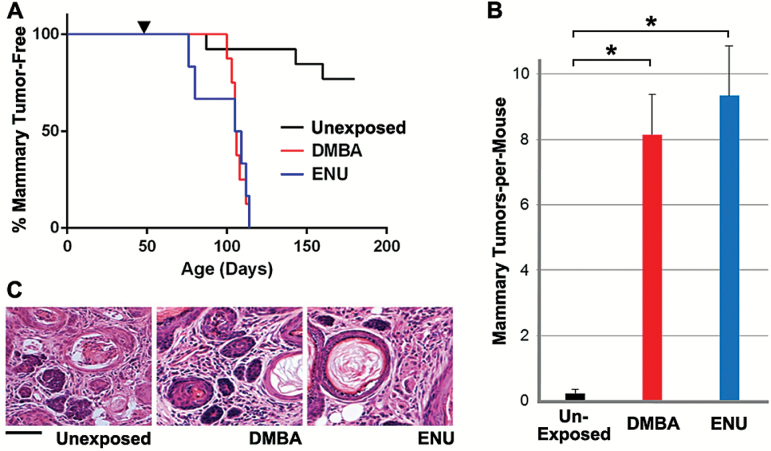
We reasoned that mammary tumors arising in Apcmin mice following carcinogen exposures likewise might acquire Apcmmcr mutations, enabling facile determination of carcinogen-specific mutation spectra. Crucial to our goal of detecting and quantifying strand bias, such mutations ought to arise through a variety of base changes affecting a wide range of codons, yet they ought to generate functionally equivalent, hypomorphic Apc alleles that confer a comparable selective advantage. To implement this strategy, cohorts of female Apcmin mice were left unexposed or were subjected to a one-time carcinogen exposure (either DMBA or ENU), then monitored for mammary tumorigenesis. In line with previous reports (21), carcinogen exposures hastened the onset of mammary tumors and dramatically increased mammary tumor incidence (20% incidence for unexposed mice versus 100% incidence for each exposure cohort; Figure 3A). In addition, carcinogen exposures markedly increased mammary tumor multiplicity from a mean of 0.2 tumors-per-mouse in unexposed mice to a mean of 8.2 tumors and 9.3 tumors following DMBA and ENU exposure, respectively (Figure 3B), without discernibly altering tumor histopathology (Figure 3C). By contrast, identical carcinogen exposures failed to yield mammary tumors in control mice inheriting two wild-type Apc alleles (data not shown), further implicating second-hit Apc mutations as a rate-limiting step during mammary tumorigenesis in Apcmin mice.
Figure 3.
Mammary tumorigenesis enhanced by carcinogen exposures in Apcmin mice. (A) Increased mammary tumor incidence in Apcmin mice. Carcinogen-naive Apcmin mice typically remained mammary tumor free and only rarely developed solitary mammary tumors after a long latency (3 of 13 mice; 23%). By contrast, all DMBA-exposed (n = 8) and ENU-exposed (n = 6) Apcmin mice developed mammary tumors within 10 weeks of carcinogen exposure. Arrowhead indicates the time of mutagen exposure. (B) Increased tumor multiplicity following carcinogen exposure. Each carcinogen exposure resulted in approximately a 40-fold increase in mammary tumor multiplicity. Error bars represent standard error of the mean. *Denotes P < 0.0001, t-test. (C) Uniform Apcmin mammary tumor histopathology in the presence and absence of carcinogen exposure. All tumors showed hallmark features of adenocarcinoma interspersed with areas of squamous differentiation and keratin pearls. Scale bar, 50 µm.
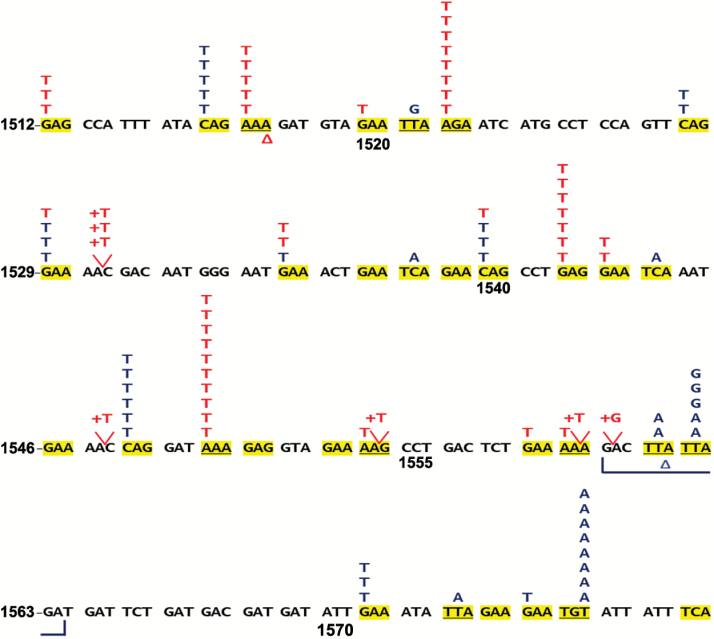
Carcinogen-induced tumors from Apcmin mice frequently acquired somatic Apcmmcr mutations as expected, and these in turn yielded carcinogen-specific mutation spectra. Overall, somatic Apcmmcr mutations were found in 83% of the mammary tumors arising in carcinogen-exposed Apcmin mice (51 of 59 DMBA tumors, 86%; 44 of 56 ENU tumors, 79%), with the great majority arising from single base substitutions (86 of 95 total mutations, 91%; Figure 4A). DMBA and ENU generated strikingly distinct, largely non-overlapping spectra of Apcmmcr point mutations, in good agreement with previous studies. Of the 68 codons spanning the Apcmmcr (Apc1512 through Apc1579), 32 harbor nonsense-prone base pairs (NPBPs), defined as those capable of yielding a stop codon in a single step via base substitution. In aggregate, DMBA and ENU tumors acquired mutations in 25 of the 32 available NPBPs, yet only three NPBPs were targeted by both carcinogens (Figure 4). Furthermore, eight “hotspot” NPBPs acquired five or more independent mutations across the tumor set, and in all eight cases these mutations were restricted to a specific carcinogen exposure (four DMBA-restricted hotspot NPBPs, each with 5–10 mutations and four ENU-restricted hotspot NPBPs, each with 5–8 mutations; Figure 4). These hotspots are not an artifact of repeat sampling of multifocal or disseminated tumor clones, since hotspot mutations consistently were distributed across multiple animals. Moreover, individual tumors that arose on the same Apcmin mouse consistently acquired a range of distinct Apcmmcr mutations, confirming their status as independent clones (Supplementary Table 1, available at Carcinogenesis Online).
Figure 4.
Carcinogen exposures generate distinct spectra of Apcmmcr mutations. Distribution of DMBA- and ENU-induced mutations across codons Apc1512 through Apc1579. The sense strand of the indicated segment of the Apc gene is depicted, with each color-coded letter indicating a base substitution identified in a single tumor (red for DMBA-induced mutations, n = 51; blue for ENU-induced mutations, n = 44). Δ denotes a deletion and + denotes an insertion. Note +G at codon 1560 could be either upstream or downstream of the endogenous G. Yellow highlight indicates an at-risk codon, which are codons that harbor a NPBP (n = 32; see main text for details). A subset of DMBA-induced tumors acquired single base pair insertion leading to a frame shift (7 of 51; 14%). Most of these insertions (6 out of 7) added a thymine (+T) immediately 3′ to a 5′-GAAAA-3′ run on the sense strand, indicating a mechanism favoring insertion downstream of a polyadenine tract.
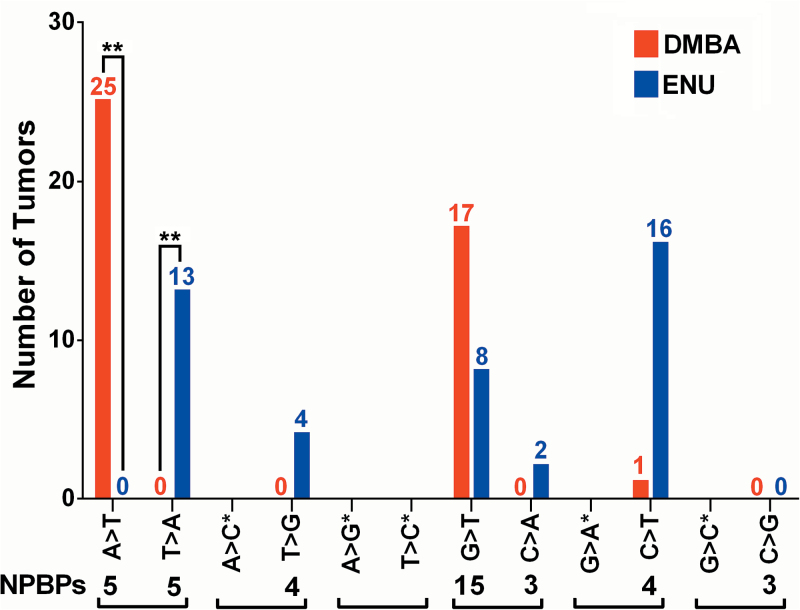
To assess strand bias in each carcinogen-induced mutation spectrum, we tallied Apcmmcr base substitutions replacing sense strand adenines versus thymines, which correspond to the recurring HrasCAA61CTA versus BrafGTG636GAG mutations from our iWnt model. The Apcmmcr is particularly well suited for this comparison, since the 10 nonsense-prone A:T base pairs residing therein are evenly split between sense strand adenines versus thymines (five instances each). As such, no pre-existing numerical bias favors detection of A-to-T versus T-to-A substitutions. Nonetheless, DMBA and ENU generated Apcmmcr mutation spectra with profound and opposing strand biases, mirroring the strand biases proposed to underlie each carcinogen’s oncogene preference in iWnt tumors. As summarized in Figure 5, in the setting of DMBA exposure, A-to-T Apcmmcr mutations were acquired by 25 independent tumors, whereas no tumors acquired the opposite T-to-A change. Inversely, in the setting of ENU exposure, T-to-A Apcmmcr mutations were acquired by 13 independent tumors, whereas no tumors acquired an A-to-T change. Four additional ENU tumors (but no DMBA tumors) acquired a T-to-G Apcmmcr mutation, consistent with ENU preferentially forming mutagenic adducts on sense strand thymines. Apcmmcr mutations were distributed unevenly among the NPBPs (e.g. DMBA hotspots at codons 1522, 1542 and 1550; ENU hotspots at codons 1562 and 1576), indicating that additional factors besides strand bias, such as sequence context, may play a role in directing carcinogen-induced mutations to particular codons. It is fair to note that none of the NPBPs within the Apcmmcr reside in a sequence context that precisely matches either Hras codon 61 (CAA) or Braf codon 636 (GTG). That said, of the carcinogen-induced Apcmmcr mutations acquired at A:T base pairs, all 42 were consistent with invariant and opposite, carcinogen-specific strand biases. Moreover, all 10 nonsense-prone A:T pairs in the Apcmmcr were mutated at least once, arguing that strand-biased mutagenesis was pervasive across a variety of sequence contexts (Supplementary Table 2, available at Carcinogenesis Online). Mechanistically, transcription most likely contributes to strand-biased mutation signatures through strand-specific DNA repair, as when mutagenic adducts are preferentially removed from the antisense strand during transcription-coupled repair (Figure 6). Alternatively, transcription may promote strand-specific DNA damage by leaving the non-transcribed sense strand relatively “exposed” to chemical mutagens.
Figure 5.
Apcmmcr mutation spectra generated by DMBA and ENU exposure show profound and inverse strand biases. Summary of carcinogen-induced base changes categorized by substitution type. Base change mutation spectra. Base changes on the sense strand are shown. **P < 0.0001 (analysis of variance). NPBP numerals denote the number of instances where the indicated base change would generate a stop codon within the Apcmmcr. Brackets pair together mutations that would be equivalent in the absence of a strand bias. *Indicates base changes that are incapable of generating a stop codon, either due to constraints inherent in the genetic code or due to the specific sequence context of the Apcmmcr.
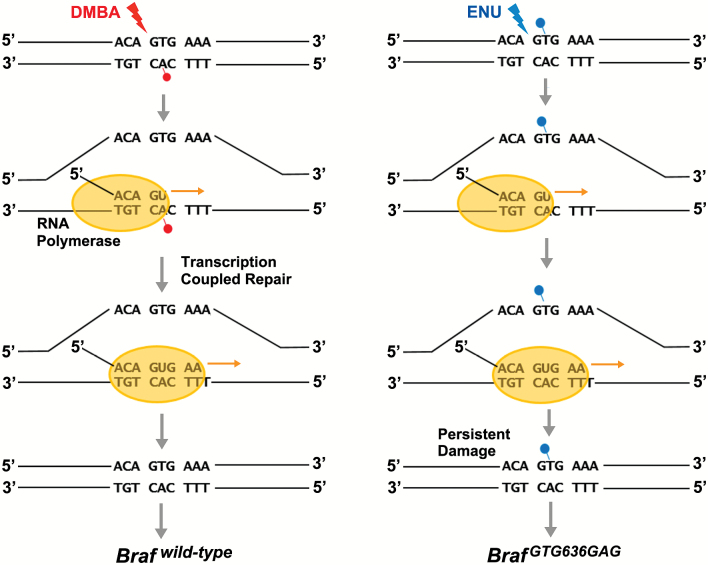
Figure 6.
A proposed mechanism leading to carcinogen-specific oncogene preferences. The schematic depicts carcinogen-induced damage to a relevant segment of the Braf gene centered on codon 636. Transcription-coupled repair removes potentially mutagenic DMBA adducts from the antisense strand (left), whereas mutagenic ENU adducts on the sense strand persist, culminating in ENU-induced BrafGTG636GAG mutations. An inverse scenario is proposed to play out at Hras codon 61 (not shown), such that ENU adducts on the antisense strand are repaired, whereas mutagenic DMBA adducts on the sense strand persist, culminating in DMBA-induced HrasCAA61CTA mutations.
Discussion
Carcinogens typically elicit tumor formation by introducing DNA damage, culminating in driver mutations that enable aberrant overgrowth of mutant cells. Whether and how carcinogen exposures direct driver mutations to specific oncogenes is poorly understood. Closing this knowledge gap might help to explain why certain oncogene mutations are favored in specific tumor types and in the setting of specific exposures to exogenous and endogenous carcinogens. Using mouse models of breast cancer, we show that chemical carcinogens can direct driver mutations to preferred oncogenes with surprising specificity. In follow-up studies, we find that these oncogene preferences reflect strikingly distinct, carcinogen-specific mutation patterns. Specifically, we show that DMBA versus ENU exposures generate stringently strand-inverse mutation signatures at A:T base pairs, providing a compelling mechanistic explanation for the preferred acquisition of DMBA-induced HrasCAA61CTA mutations versus ENU-induced BrafGTG636GAG mutations. It remains to be determined whether strand-biased mutagenesis contributes to oncogene preferences in human cancers, which arise in the context of more complex carcinogen exposures and selection pressures.
The robust carcinogen-specific oncogene preferences identified in our study almost certainly require evolutionary constraints operating at multiple levels. Small molecule mutagens, such as DMBA and ENU, would seem to lack sufficient structural complexity to damage DNA in a highly gene-specific manner, meaning that oncogene preferences paradoxically arise from carcinogen-specific mutation patterns inscribed genome wide. This paradox is resolved if one assumes that only a few of these myriad mutations confer a selective advantage sufficient to create a driver oncogene. By this reasoning, carcinogen-specific mutation patterns need only to discriminate among a handful of potently fitness-enhancing codon changes to impart an oncogene preference. One level of evolutionary constraint mentioned earlier involves the highly restricted set of mutations capable of activating a proto-oncogene. At another level, oncogene preference ought to be strongly influenced by the potential for co-operative interactions across oncogenic signaling pathways. In this regard, findings from our iWnt model underscore the strong selective advantage conferred when Ras-Raf pathway activation is superimposed on prior Wnt pathway activation. Notably, we identified mutually exclusive Hras and Braf mutations in iWnt mammary tumors, strongly implicating MAPK signaling as the key downstream Ras effector pathway enabling co-operation between mutant Hras and Wnt1. At the same time, our findings leave other constraints shaping carcinogen-specific oncogene preference unexplained. For example, DMBA-exposed iWnt tumors uniformly acquired HrasCAA61CTA mutations, but never KrasCAA61CTA or NrasCAA61CTA mutations (data not shown), despite the fact that Q61L alleles of these closely related family members possess documented oncogenic activity in other tissue compartments.
To reasonably attribute the strong carcinogen-specific oncogene preferences observed in our study to strand-biased mutations, the magnitude of the underlying strand bias itself must, likewise, be strong. For both DMBA and ENU exposure, we detected strand-biased mutagenesis at A:T base pairs indicating at least 10-fold strand specificity. By contrast, much lower levels of strand bias (on the order of 2-fold) are detected when genome-wide DNA sequencing is used to assemble collections of the somatic mutations acquired during human carcinogenesis (26) or the ENU-induced germline mutations acquired during forward genetic screens in mice (27,28). Here, reduced strand bias presumably reflects reduced transcription-coupled repair. Concordantly, more potent strand biases typically emerge when mutation analyses are confined to genes under strong selection pressure. As mentioned earlier, surveys of TP53 mutations in smoking-associated lung cancer reveal that 90% of mutations acquired at G:C base pairs involve substitution for damage-prone guanines on the sense strand (11,29). More recently, aristocholic acid was identified as a potent carcinogen that resembles DMBA in its predilection for damaging adenines and introducing substitutions at A:T base pairs. Whereas genome sequencing of cancers attributed to aristocholic acid exposure identified only a modest strand bias in the mutations affecting A:T sites exome wide, those substitutions altering bona fide “drivers” of carcinogenesis were strongly biased toward replacement of sense strand adenines (e.g. 13 of 14 TP53 mutations; 4 of 4 NRAS mutations) (30,31).
Similarly, previous studies using model systems indicate robust strand bias for those mutations that provide a selective growth advantage. With respect to the mutagens employed in our work, when selection for Hprt-mutant rodent cells was used to define mutation spectra following DMBA or ENU exposure, the resulting mutation signatures at A:T base pairs showed nearly invariant and opposite strand biases, which closely match those reported here (32–34). Intriguingly, comparison of DMBA-induced mutations generated at an endogenous, selectable gene (Hprt) versus an unselected reporter transgene (LacI) revealed nearly identical mutation signatures at A:T base pairs, except that strand bias was absent from mutations acquired within the unexpressed LacI gene (34). Our study extends these findings by demonstrating that these strand-inverse mutation signatures are inscribed not only on reporter genes from normal tissues but also on driver genes crucial to the growth of carcinogen-exposed mammary tumors.
Supplementary material
Supplementary Tables 1 and 2 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported by a grant from the National Cancer Institute (R01 CA152222) and funding received from the benefactors of the Jake Gittlen Laboratories for Cancer Research. Animal housing was provided through a facility constructed with support from a Research Facilities Improvement Grant (C06 RR-15428-01) from the National Center for Research Resources.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- DMBA
7,12-dimethylbenz(a)anthracene
- ENU
N-ethyl-N-nitrosourea
- MAPK
mitogen-activated protein kinase
- NPBP
nonsense-prone base pair
References
- 1. Kandoth C., et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature, 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network. (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hodis E., et al. (2012) A landscape of driver mutations in melanoma. Cell, 150, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unni A.M., et al. (2015) Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife, 4, e06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slebos R.J., et al. (1991) Relationship between K-ras oncogene activation and smoking in adenocarcinoma of the human lung. J. Natl Cancer Inst., 83, 1024–1027. [DOI] [PubMed] [Google Scholar]
- 6. Imielinski M., et al. (2012) Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell, 150, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Curtin J.A., et al. (2005) Distinct sets of genetic alterations in melanoma. New Engl. J. Med., 353, 2135–2147. [DOI] [PubMed] [Google Scholar]
- 8. Tsao H., et al. (2012) Melanoma: from mutations to medicine. Genes Dev., 26, 1131–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki H., et al. (1992) p53 mutations in non-small cell lung cancer in Japan: association between mutations and smoking. Cancer Res., 52, 734–736. [PubMed] [Google Scholar]
- 10. Brash D.E., et al. (1991) A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl Acad. Sci. USA, 88, 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greenblatt M.S., et al. (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res., 54, 4855–4878. [PubMed] [Google Scholar]
- 12. Zarbl H., et al. (1985) Direct mutagenesis of Ha-ras-1 oncogenes by N-nitroso-N-methylurea during initiation of mammary carcinogenesis in rats. Nature, 315, 382–385. [DOI] [PubMed] [Google Scholar]
- 13. Kito K., et al. (1996) Incidence of p53 and Ha-ras gene mutations in chemically induced rat mammary carcinomas. Mol. Carcinog., 17, 78–83. [DOI] [PubMed] [Google Scholar]
- 14. Brown K., et al. (1990) Carcinogen-induced mutations in the mouse c-Ha-ras gene provide evidence of multiple pathways for tumor progression. Proc. Natl Acad. Sci. USA, 87, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balmain A., et al. (2014) Milestones in skin carcinogenesis: the biology of multistage carcinogenesis. J. Invest. Dermatol., 134, E2–E7. [DOI] [PubMed] [Google Scholar]
- 16. Gunther E.J., et al. (2003) Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev., 17, 488–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Podsypanina K., et al. (2004) Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med., 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jang J.W., et al. (2006) Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol. Cell Biol., 26, 8109–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies H., et al. (2002) Mutations of the BRAF gene in human cancer. Nature, 417, 949–954. [DOI] [PubMed] [Google Scholar]
- 20. Moser A.R., et al. (1993) ApcMin, a mutation in the murine Apc gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc. Natl Acad. Sci. USA, 90, 8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moser A.R., et al. (1995) ApcMin: a mouse model for intestinal and mammary tumorigenesis. Eur. J. Cancer, 31A, 1061–1064. [DOI] [PubMed] [Google Scholar]
- 22. Kuraguchi M., et al. (2009) Genetic mechanisms in Apc-mediated mammary tumorigenesis. PLoS Genet., 5, e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bakker E.R., et al. (2013) beta-Catenin signaling dosage dictates tissue-specific tumor predisposition in Apc-driven cancer. Oncogene, 32, 4579–4585. [DOI] [PubMed] [Google Scholar]
- 24. Gaspar C., et al. (2009) A targeted constitutive mutation in the APC tumor suppressor gene underlies mammary but not intestinal tumorigenesis. PLoS Genet., 5, e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toki H., et al. (2013) Novel mouse model for Gardner syndrome generated by a large-scale N-ethyl-N-nitrosourea mutagenesis program. Cancer Sci., 104, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexandrov L.B., et al. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahasi K.R., et al. (2007) Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Mol. Biol., 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnold C.N., et al. (2012) ENU-induced phenovariance in mice: inferences from 587 mutations. BMC Res. Notes, 5, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfeifer G.P., et al. (2002) Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene, 21, 7435–7451. [DOI] [PubMed] [Google Scholar]
- 30. Hoang M.L., et al. (2013) Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med., 5, 197ra02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poon S.L., et al. (2013) Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med., 5, 197ra01. [DOI] [PubMed] [Google Scholar]
- 32. Skopek T.R., et al. (1992) Mutational spectrum at the Hprt locus in splenic T cells of B6C3F1 mice exposed to N-ethyl-N-nitrosourea. Proc. Natl Acad. Sci. USA, 89, 7866–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jansen J.G., et al. (1994) Molecular analysis of hprt gene mutations in skin fibroblasts of rats exposed in vivo to N-methyl-N-nitrosourea or N-ethyl-N-nitrosourea. Cancer Res., 54, 2478–2485. [PubMed] [Google Scholar]
- 34. Mittelstaedt R.A., et al. (1998) Comparison of the types of mutations induced by 7,12-dimethylbenz[a]anthracene in the lacI and hprt genes of Big Blue rats. Environ. Mol. Mutagen., 31, 149–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.