Summary
These occupational cholangiocarcinoma cases shared a high mutation burden, strand bias and unique trinucleotide mutational signatures, suggesting that the patients might have been exposed to a common strong mutagen. The underlying mechanisms of mutagenesis should be further investigated.
Abstract
Cholangiocarcinoma is a relatively rare cancer, but its incidence is increasing worldwide. Although several risk factors have been suggested, the etiology and pathogenesis of the majority of cholangiocarcinomas remain unclear. Recently, a high incidence of early-onset cholangiocarcinoma was reported among the workers of a printing company in Osaka, Japan. These workers underwent high exposure to organic solvents, mainly haloalkanes such as 1,2-dichloropropane (1,2-DCP) and/or dichloromethane. We performed whole-exome analysis on four cases of cholangiocarcinoma among the printing workers. An average of 44.8 somatic mutations was detected per Mb in the genome of the printing workers’ cholangiocarcinoma tissues, approximately 30-fold higher than that found in control common cholangiocarcinoma tissues. Furthermore, C:G-to-T:A transitions with substantial strand bias as well as unique trinucleotide mutational changes of GpCpY to GpTpY and NpCpY to NpTpY or NpApY were predominant in all of the printing workers’ cholangiocarcinoma genomes. These results were consistent with the epidemiological observation that they had been exposed to high concentrations of chemical compounds. Whole-genome analysis of Salmonella typhimurium strain TA100 exposed to 1,2-DCP revealed a partial recapitulation of the mutational signature in the printing workers’ cholangiocarcinoma. Although our results provide mutational signatures unique to occupational cholangiocarcinoma, the underlying mechanisms of the disease should be further investigated by using appropriate model systems and by comparison with genomic data from other cancers.
Introduction
Cholangiocarcinoma has been recognized as a relatively rare cancer, but its incidence is increasing worldwide. The incidence rates are higher in the Japanese population (5.2 cases per 100000 individuals) and Asian populations (including Japanese; 3.3 cases per 100000 individuals) than in Western populations (2.1 cases per 100000 individuals among non-Hispanic whites and blacks) (1–6). Cholangiocarcinoma is an aging-associated cancer, with its reported average age of diagnosis being between the sixth and seventh decades of life (7). Although several risk factors, such as primary sclerosing cholangitis, bile duct cystic disorders, liver fluke infection, hepatolithiasis, cirrhosis, hepatitis B or C virus infection, diabetes, obesity and exposure to the chemical carcinogen ‘Thorotrast’ (banned in the 1960s), have been suggested, the majority of cholangiocarcinomas arise sporadically (4,5). Thus, the etiology and pathogenesis of cholangiocarcinoma remain to be elucidated, and the emergence of other risk factors is a likely supposition.
An outbreak of cholangiocarcinoma among workers in an offset color proof-printing company in Osaka, Japan, was recently reported, and this disease was newly classified as an occupational disease by the Ministry of Health, Labour and Welfare of Japan in 2013 (http://www.mhlw.go.jp/english/policy/employ-labour/labour-standards/Occupational.html). Between 1996 and 2012, 17 of the 111 former or current workers of the company were diagnosed with cholangiocarcinoma (8). In addition, 13 workers in 11 other printing companies in Japan were acknowledged as exhibiting occupational cholangiocarcinoma by April 2014 (9,10). A detailed investigation of the 17 patients at the first reported company in Osaka revealed that the printing workers’ cholangiocarcinoma exhibited an unusually early onset, ranging from 25 to 45 years. All of the patients had been exposed to high concentrations of chemical compounds, including 1,2-dichloropropane (1,2-DCP) and/or dichloromethane (DCM) (8). The International Agency for Research on Cancer (IARC) re-evaluated the carcinogenic risks of these compounds, recategorizing 1,2-DCP as a Group 1 (carcinogenic to humans) carcinogen instead of Group 3 (not classifiable as to its carcinogenicity to humans) and DCM as Group 2A (probably carcinogenic to humans) instead of Group 2B (possibly carcinogenic to humans) (11,12). However, previous in vitro and in vivo studies suggested that both 1,2-DCP and DCM have limited mutagenic and tumorigenic capabilities, and bile duct tumorigenesis was not induced by 1,2-DCP or DCM in rodent models (13–16). Thus, the relevance of 1,2-DCP and/or DCM exposure to cholangiocarcinoma carcinogenesis remains to be further elucidated. We performed a genome-wide mutation analysis of four cases of occupational cholangiocarcinoma using genomic DNA samples derived from surgically resected specimens. We found a significant mutational landscape, including a high mutation burden, strand bias and unique trinucleotide mutation signatures that were commonly observed in all of the investigated cases, suggesting that the workers had been exposed to a common strong mutagen. We further investigated the mutational signatures in the genomic DNA of Salmonella typhimurium strain TA100 and human epithelial cells exposed to 1,2-DCP.
Materials and methods
Preparation of clinical samples
Tumor and matched normal formalin-fixed paraffin-embedded (FFPE) tissue samples were obtained from Osaka National Hospital, Osaka City University Hospital and National Cancer Center Hospital East. This study was approved by the Institutional Review Board of each institution. Four occupational cholangiocarcinoma samples were selected for analysis from 17 patients who were workers at a printing company in Osaka and diagnosed with cholangiocarcinoma between 1996 and 2012. Among the 17 occupational cholangiocarcinoma patients worked at the same printing company in Osaka Japan between 1996 and 2012, 12 were surgically treated. The resected tumor specimens of 8 of the 12 patients were available. According to the quality and quantity of the genomic DNA samples, four cases were selected for further analysis. Control common cholangiocarcinoma samples were randomly selected from patients who were surgically treated between 2012 and 2014 at National Cancer Center Hospital East and whose tissues were abundant enough for sequencing analysis. Twenty sections of 10-µm-thick FFPE tissue samples were subjected to laser-capture microdissection or to manual microdissection. The Absolutely RNA FFPE kit (modified protocol for DNA extraction, Agilent Technologies, Santa Clara, CA) was used to prepare the DNA. DNA quality was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), the Quant-iT PicoGreen dsDNA Reagent and Kit (Life Technologies, Carlsbad, CA) and the Infinium HD FFPE DNA Sample QC Kit (Illumina, San Diego, CA).
Whole-exome sequencing
Using 0.25–1.00 µg of double-stranded DNA, we prepared whole-exome sequencing libraries. The exomes were captured using the SureSelect Human All Exon V4+UTRs or the V5+UTRs Kit (Agilent Technologies) according to the manufacturer’s instructions. The exome capture libraries were then sequenced using a HiSeq 2000 system (Illumina) to generate 100bp paired-end data.
Identification of somatic mutations
Sequence reads were aligned to the human reference genome UCSC hg19 using the Burrows–Wheeler Aligner program (BWA, http://bio-bwa.sourceforge.net/). Single-nucleotide variants (SNVs) and insertions and deletions (INDELs) were called and annotated using the Genome Analysis Toolkit software package (GATK, http://www.broadinstitute.org/gatk/). Sequencing artifacts were filtered out using custom filters (GATK confidence score ≥50, number of variant reads in each direction ≥1, variant allele frequency ≥10%) and by visual inspection. Germline variants were filtered out using data from dbSNP build 131, the 1000 Genomes Project (Phase 1 exome data, released 21 May 2011), 1 Japanese genome, 299 in-house Japanese exomes and matched normal-tissue exomes of the cholangiocarcinoma cases.
Confirmation of somatic mutations
Somatic mutations in the ARID1A, BRAF, CDKN2A and MLL3 genes, which were detected in the case 1 patient, were confirmed using Sanger sequencing. PCR primers were designed using the Primer3Plus software (www.bioinformatics.nl/primer3plus/) and are listed in Supplementary Table 1, available at Carcinogenesis Online. PCR was performed using HotStarTaq DNA Polymerase (Qiagen, Valencia, CA), and the samples were incubated at 95°C for 15min, 95°C for 30s, 55°C or 61°C for 30s, and 72°C for 1min for 40 cycles and then at 72°C for a final 10min extension. PCR amplicons were sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and a 3500 Genetic Analyzer (Life Technologies).
Mutagenicity assay
The 1,2-DCP (>98.0% pure) and DCM (>99.5% pure) were purchased from Wako Pure Chemical Industries, Osaka, Japan and Nacalai Tesque, Kyoto, Japan, respectively. The standard plate-incorporation method in the absence of a metabolic activation system was performed to test the mutagenicity of 1,2-DCP and DCM in S.typhimurium TA100 with a slight modification for testing volatile chemicals (17). In brief, appropriately sized filter papers absorbed multiple doses of 1,2-DCP or DCM and were put into plastic bags. Plates in which bacteria had been plated in top agar were placed in the plastic bags without covers and sealed tightly. By this method, the bacteria were exposed to the evaporating haloalkanes. The chemical vapor concentration in the plastic bags was calculated using calibration curves determined by gas chromatography mass spectrometry analysis. After 2h of vapor exposure at 37°C, the plates were removed from the bags and incubated for another 48h. hisG+ revertant colonies were counted to evaluate mutagenicity. The mutagenic activity of the samples was calculated from the linear portions of the dose–response curves, which were obtained using three doses in duplicate plates on at least two independent experiments.
Analysis of global mutational profiles of 1,2-DCP and DCM in a Salmonella strain
The hisG+ colonies were randomly isolated, and genomic DNA was extracted using a Puregene Cell and Tissue kit (Qiagen). The mutational profiles induced by haloalkanes were subsequently examined using whole-genome sequencing with reference to a previous report (18). Using 3 µg of DNA and the SureSelect XT Library Prep Kit (Agilent Technologies), we prepared whole-genome sequencing libraries according to the manufacturer’s instructions. The prepared libraries were sequenced using a HiSeq 1500 system (Illumina) to generate 100bp paired-end data. The sequence reads were aligned to the reference sequence NC_003197 (Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 chromosome, complete genome, 4857432bp) using the Burrows–Wheeler Aligner program (BWA, http://bio-bwa.sourceforge.net/), and the SNVs were called and annotated using the Genome Analysis Toolkit software package (GATK, http://www.broadinstitute.org/gatk/). We selected ‘PASS’ variants annotated by the GATK program as high-confidence acquired variants.
Analysis of mutational profile of 1,2-DCP in mammalian cells
NCC-CC1 cells, which were originally established from human biliary tract carcinoma tissue (19), were kindly provided by Dr. Hidenori Ojima at the National Cancer Center Research Institute in 2013. The cells were maintained in RPMI-1640 medium (Wako Pure Chemicals, Japan) containing 10% fetal bovine serum (Thermo Fisher Scientific). These cells were authenticated by DNA microarray and quantitative RT-PCR and last checked in 2010. Human embryonic kidney 293 (HEK293) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and have never been passaged longer than 6 months after receipt or resuscitation. This cell line was not authenticated as it came from national repositories. Cloned HEK293 cells stably expressing an open reading frame of the pQCXIP vector (Clontech Laboratories, A Takara Bio Company, Mountain View, CA) were cultured in Dulbecco’s modified Eagle medium (Nissui Pharmaceutical, Tokyo, Japan) containing 10% fetal bovine serum (Biowest, Nuaillé, France). We used an in vitro vapor exposure system with slight modifications (20). In brief, appropriately sized filter papers were put into two rhombus-shaped holes in the center of a 6-well culture plate, and a toxic dose of 1,2-DCP (250 μL/plate for single exposure, once with 180 μL/plate and four times with 120 μL/plate for multiple exposure for NCC-CC1; and 50 μL/plate for single exposure for HEK293) was absorbed onto the filter papers. The culture plates were put into plastic bags without covers and sealed tightly. Using this method, the haloalkanes evaporated and dissolved into the culture medium (20). After 2 and 4h of vapor exposure for NCC-CC1 and HEK293, respectively, the cells were reseeded and cultured for 4 to 6 weeks to isolate individual clones. For multiple exposures, the cells were exposed to 1,2-DCP subsequently after recovery from cytotoxicity, and this procedure was repeated up to five times. Genomic DNA was extracted from 2 to 6 clones per exposure group using a DNeasy blood and tissue kit (Qiagen), and the mutational profile induced by the haloalkanes was subsequently examined using whole-exome sequencing.
Statistical analyses
The statistical significance of observed differences between the number of somatic SNVs in the printing worker cholangiocarcinoma cases and the control cholangiocarcinoma cases was evaluated using Student’s t-test. For strand bias analysis, the significance of the differences in the number of SNVs between the un-transcribed (sense) and transcribed (antisense) strand in the cases and controls was evaluated using the chi-square test. The test evaluated whether the proportion of SNVs in each strand differed from 0.5, which is the value expected by chance. For trinucleotide mutational signature analysis, the significance of differences in the number of mutations between the trinucleotide sequence of interest and any sequence except the one of interest was evaluated by the chi-square test. The test evaluated whether the proportion of mutations in each context differed from the frequency of the context in the targeted genome, which is the expected value by chance.
Results
Patient characteristics and quality of whole-exome sequencing
The characteristics of the printing worker patients upon admission are described in Table 1. The printing worker cholangiocarcinoma patients (cases 1–4) were 31- to 40-year-old males. The case 1 patient used both 1,2-DCP and DCM, and the case 2–4 patients used 1,2-DCP for 1–11 years. The characteristics of the control common cholangiocarcinoma patients are described in Supplementary Table 2, available at Carcinogenesis Online. The patients with common late-onset cholangiocarcinoma (controls 1–4) were 55- to 79-year-old males and females, and all patients had intrahepatic cholangiocarcinoma. The patients with common early-onset bile duct carcinomas (controls 5–7) were 26- to 39-year-old males and females, and the subtypes were gallbladder cancer, duodenum papilla cancer and extrahepatic cholangiocarcinoma.
Table 1.
Patient characteristics of printing workers’ cholangiocarcinoma cases
| Printing worker | ||||
|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 4 | |
| ID | CHCOSK001 | CHCOSK003 | CHCOSK004 | CHCOSK005 |
| Anatomical subtype | Intrahepatic | Intrahepatic | Intrahepatic | Intrahepatic |
| Age (years) | 40 | 39 | 31 | 34 |
| Sex | Male | Male | Male | Male |
| Duration of exposure | ||||
| 1,2-DCP | 11 years 11 months | 7 years 4 months | 6 years 6 months | 6 years 1 month |
| DCM | 1 year 5 months | None | None | None |
| Smoking habit | 20 cigarettes/day | 20 cigarettes/day | None | None |
| Alcohol consumption | 1.5–1.8L sake/week | 4.4L beer/week | None | Occasionally |
Tumor cells were condensed to approximately 50% by laser-capture microdissection or manual microdissection. Genomic DNA was retrieved and used for whole-exome sequencing. The average base coverage of the targeted regions in the tumor and normal samples of the printing workers’ cholangiocarcinomas was 139.7-fold (range: 89.8–197.2) and 105.7-fold (range: 66.8–131.2), respectively. The proportion of the targeted regions with 20 reads or higher was 92.1% (range: 76.3–98.5%) and 86.5% (range: 69.0–96.9%) for the tumor and normal samples, respectively. Among the common late-onset cholangiocarcinomas, the average base coverage in the tumor and normal samples was 241.9-fold (range: 210.7–272.8) and 289.4-fold (range: 267.6–309.0), respectively, and 98.2% (range: 97.2–98.6%) and 98.6% (range: 98.4–98.7%) of the target regions in the tumor and normal samples, respectively, had 20 reads or higher. Among the common early-onset bile duct carcinomas, the average base coverage in the tumor and normal samples was 142.2-fold (range: 131.4–155.5) and 68.3-fold (range: 64.0–75.6), respectively, and 96.5% (range: 96.2–96.8%) and 84.5% (range: 81.9–85.8%) of the target regions in the tumor and normal samples, respectively, had 20 reads or higher (Supplementary Table 3, available at Carcinogenesis Online).
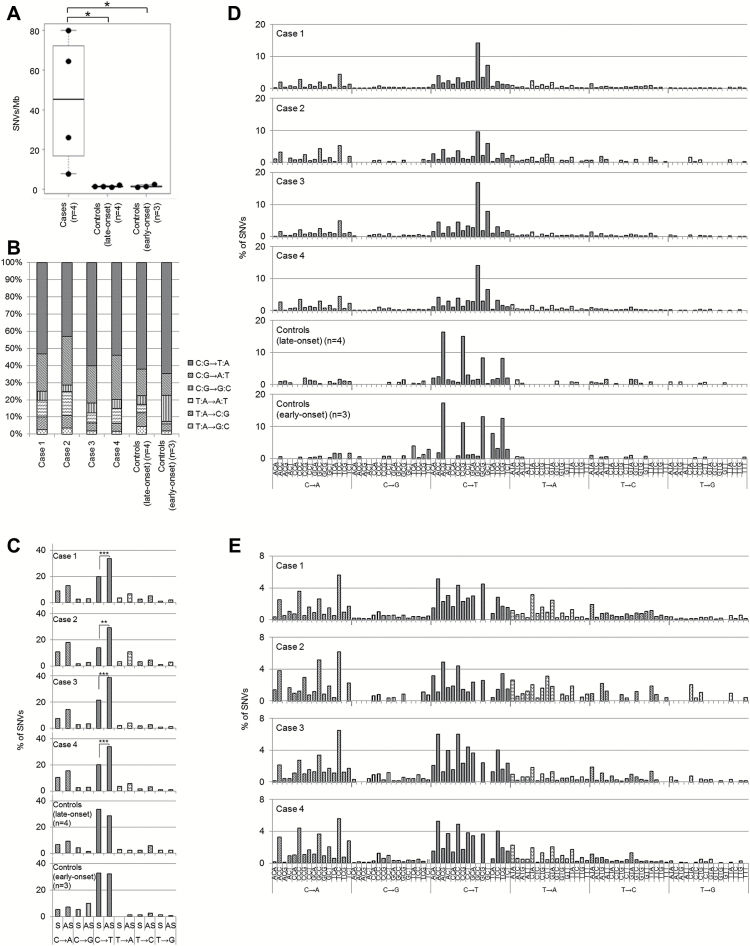
The printing workers’ cholangiocarcinomas are hypermutated tumors
We identified 1451±1089 (44.6±33.5/Mb) somatic SNVs and 6.8±5.0 (0.2±0.2/Mb) apparent somatic INDELs in the four printing workers’ cholangiocarcinomas (cases 1–4) (Table 2). The number of somatic SNVs in these cases was significantly higher than that in the exomes of the four cases of common late-onset intrahepatic cholangiocarcinoma (controls 1–4) or in the three cases of common early-onset bile duct carcinoma (controls 5–7), which showed an average of 44.8±11.9 (1.4±0.4/Mb) and 50.0±23.4 (1.5±0.7/Mb) SNVs, respectively (P = 0.04 and 0.04, respectively; Student’s t-test) (Table 2 and Figure 1A). All somatic variants detected in the patients are listed in Supplementary File 1, available at Carcinogenesis Online. The number of SNVs in the printing worker cholangiocarcinoma cases was also larger than that in the exomes of liver fluke infection-related and infection-unrelated cholangiocarcinoma cases (21,22) and that in 260 common biliary tract cancers (23). Notably, the number of INDELs was significantly smaller than the number of SNVs in the printing worker cholangiocarcinoma cases. This smaller number of INDELs is a unique characteristic compared with other hypermutated solid tumors such as microsatellite-instable colorectal cancers (24).
Table 2.
Number of somatic mutations identified in printing workers’ cholangiocarcinomas (cases) and common cholangiocarcinomas (controls)
| Number of SNVs | Number of INDELs | Mutation rate (/Mb) | ||
|---|---|---|---|---|
| Non-synonymous | Synonymous | |||
| Printing worker | ||||
| Case 1 | 1505 | 558 | 4 | 64.6 |
| Case 2 | 178 | 76 | 3 | 7.8 |
| Case 3 | 601 | 252 | 6 | 26.1 |
| Case 4 | 1862 | 770 | 14 | 80.5 |
| Common (late-onset) | ||||
| Control 1 | 28 | 12 | 5 | 1.4 |
| Control 2 | 35 | 12 | 3 | 1.5 |
| Control 3 | 20 | 12 | 8 | 1.2 |
| Control 4 | 45 | 15 | 4 | 1.9 |
| Common (early-onset) | ||||
| Control 5 | 26 | 9 | 4 | 1.2 |
| Control 6 | 31 | 7 | 4 | 1.3 |
| Control 7 | 52 | 25 | 3 | 2.4 |
Figure 1.
The printing workers’ cholangiocarcinomas share a high mutation burden, strand bias and a unique trinucleotide mutational signature. (A) Number of exome SNVs in the printing workers’ and control cholangiocarcinoma cases (*P < 0.05; Student’s t-test). (B) Single-nucleotide DNA substitution profiles in the printing workers’ and control cholangiocarcinoma cases. (C) Substantial strand bias in mutation counts in the sense (S) and antisense (AS) strands of the printing workers’ cholangiocarcinoma cases. Significant strand bias for C:G to T:A mutations was observed in the printing workers’ cholangiocarcinoma cases (**P < 0.01, ***P < 0.001; chi-square test). (D) Trinucleotide mutational patterns in the printing workers’ and control cholangiocarcinoma samples. The mutational signatures were normalized using the trinucleotide frequency in the genome. (E) Secondary characteristic trinucleotide mutational pattern in the printing workers’ cholangiocarcinoma samples that emerged after elimination of the prominent mutational pattern GpCpY to GpTpY. The mutational signatures were normalized using the trinucleotide frequency in the genome.
The printing workers’ cholangiocarcinoma genomes harbor mutations in genes frequently mutated in common bile tract carcinomas
We selected 20 genes that are frequently mutated in bile tract carcinoma (registered in Catalogue of Somatic Mutations in Cancer (COSMIC) v74). Each of the printing workers’ cholangiocarcinoma had amino acid-altering mutations in two to six genes (Table 3). Among the mutations detected in case 1, we confirmed mutations in ARID1A, BRAF, CDKN2A and MLL3 by Sanger sequencing (Supplementary Table 1 and Supplementary Figure 1, available at Carcinogenesis Online).
Table 3.
Identified mutations in commonly mutated genes in BTC
| Gene symbol | Frequency (%) | CCDS ID | Printing worker | Common (late-onset) | Common (early-onset) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COSMIC v74 | 260 Japanese BTCs | Case 1 | Case 2 | Case 3 | Case 4 | Control 1 | Control 2 | Control 3 | Control 4 | Control 5 | Control 6 | Control 7 | ||
| TP53 | 35 | 26 | CCDS11118 | N131Y | R213* | S241Y | R213* | |||||||
| KRAS | 23 | 18 | CCDS8702.1 | G12V | G12D | G12V | A146V | G12D | ||||||
| CDKN2A | 15 | 5 | CCDS56565.1 | P75T, E120* | W15*, E120* | |||||||||
| MLL3 | 15 | 3 | CCDS5931.1 | E141*, A2323Ta, L4157F | N2787fs | |||||||||
| ARID1A | 11 | 11 | CCDS285.1 | Q586* | Q528H, G1293fsa | E119*, S662G | K2189* | L2088del | ||||||
| IDH1 | 9 | 3 | — | |||||||||||
| BAP1 | 8 | 8 | CCDS2853.1 | D535fs | ||||||||||
| SMAD4 | 8 | 9 | CCDS11950.1 | G508Da | N107fs | D351V | G386R | G270fs | I527fs | |||||
| AXIN1 | 7 | 2 | CCDS10405.1 | S782Na | G676V | K165*, E717* | Q386* | |||||||
| CTNNB1 | 7 | 2 | — | |||||||||||
| GNAS | 7 | 6 | — | |||||||||||
| PBRM1 | 7 | 5 | — | |||||||||||
| PIK3CA | 7 | 7 | CCDS43171.1 | H665La | E545K | |||||||||
| ERBB3 | 6 | 4 | — | |||||||||||
| ATM | 5 | 4 | CCDS31669.1 | W1933*a | ||||||||||
| BRAF | 5 | 2 | CCDS5863.1 | D594G | ||||||||||
| FBXW7 | 5 | 3 | CCDS34078.1 | Q124L | R347H | |||||||||
| ZNF521 | 5 | 2 | — | |||||||||||
| TERT | 4 | 3 | — | |||||||||||
| IDH2 | 3 | 1 | — | |||||||||||
BTC, biliary tract carcinoma.
aRescued by visual inspection.
Predominant DNA substitution patterns in the printing workers’ cholangiocarcinomas: C:G-to-T:A transition
We next analyzed the single-nucleotide substitution patterns in the printing worker cholangiocarcinoma cases and found that C:G-to-T:A transitions were predominant, among the somatic SNVs, followed by C:G to A:T transversion. This mutational spectrum is similar to that in the control cholangiocarcinoma cases (Figure 1B).
C:G-to-T:A transitions in the printing workers’ cholangiocarcinomas indicate strand bias
We then examined the mutational signatures for transcriptional strand bias. In all of the printing workers’ cholangiocarcinomas, a substantial difference in the prevalence of mutations between the sense and antisense strands was observed for the C:G-to-T:A transition, whereas no significant difference was detected in the control cholangiocarcinoma cases (Figure 1C).
The printing workers’ cholangiocarcinomas share characteristic trinucleotide mutational signatures
We further analyzed the mutational signature of the base substitutions by incorporating information regarding the 5′ and 3′ neighboring sites of each mutated base (25). The most significant trinucleotide mutational pattern in the printing workers’ cholangiocarcinomas was GpCpY to GpTpY (P < 0.01, chi-square test) (Figure 1D), a novel signature that was not reported in the International Cancer Genome Consortium (ICGC) project, which classified more than 20 distinct mutational signatures from a mutational catalogue of 7042 primary cancers (25). This signature was identical among the four printing worker cases. Following the GpCpY to GpTpY signature, NpCpY to NpTpY and NpApY changes were the next most characteristic mutational signatures (Figure 1E). By contrast, the control late-onset cholangiocarcinoma cases harbored 5-methylcytosine deamination signature mutations (NpCpG to NpTpG changes; classified as Signature 1A and 1B in the ICGC project), and the control early-onset bile duct carcinoma cases harbored both 5-methylcytosine deamination signature mutations (NpCpG to NpTpG changes; classified as Signature 1A and 1B in the ICGC project) and APOBEC signature mutations (TpCpW to TpTpW or TpGpW changes; classified as Signatures 2 and 13 in the ICGC project) (Figure 1D).
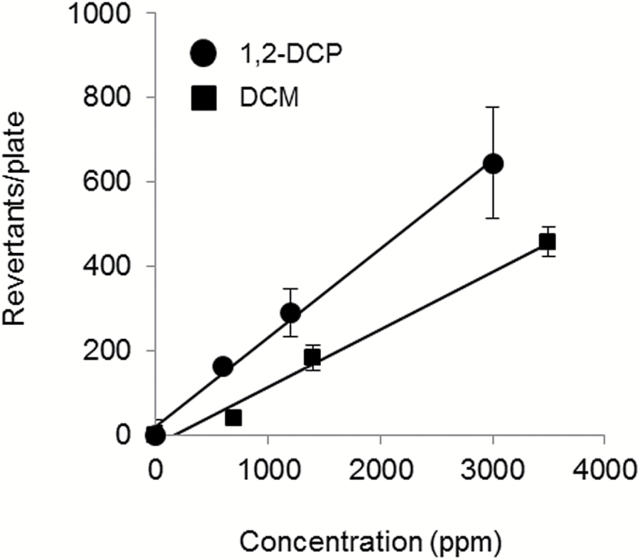
1,2-DCP and DCM show mutagenicity in S.typhimurium strain TA100
We examined the mutagenic activity of 1,2-DCP and DCM on S.typhimurium strain TA100 by Ames assay. Both haloalkanes showed mutagenicity toward TA100 in a dose-dependent manner. The mutagenic potency of 1,2-DCP was slightly higher than that of DCM (Figure 2).
Figure 2.
Both 1,2-DCP and DCM show mutagenic activity in S.typhimurium strain TA100. The mutagenic activity levels of 1,2-DCP (closed circle) and DCM (closed square) were estimated from the number of revertant colonies.
1,2-DCP and DCM preferentially induced SNVs at C:G residues
To determine the global mutational profiles of 1,2-DCP and DCM, we further performed whole-genome analysis of S.typhimurium TA100 with or without haloalkane exposure. Revertant mutations occurring within the hisG gene were excluded from the analysis because these mutations are affected by selection bias. The number of mutagenic events in the other bacterial DNA increased depending on the dose of 1,2-DCP. The average mutation rate in clones exposed to 1,2-DCP at 3000 ppm (n = 76) and 6000 ppm (n = 19) was 0.2 and 0.6/Mb, respectively. These mutation rates are 1.4- and 5.1-fold higher than that for the non-exposed control (P = 0.05 and P < 0.01, respectively, Student’s t-test). C:G-to-T:A transitions were the most predominant single-nucleotide substitutions (57.6 and 77.1% of total SNVs in the genomic DNA exposed to 3000 and 6000 ppm of 1,2-DCP, respectively). In contrast, the average mutation rate of clones exposed to 3500 ppm of DCM (n = 47) was 0.5/Mb, which is significantly higher than that for the non-exposed control (P < 0.01, Student’s t-test), and C:G to A:T transversions were predominant (46.6% of total SNVs), followed by C:G-to-T:A transitions (44.1% of total SNVs) (Figure 3A and B).
Figure 3.
The mutational signature of 1,2-DCP in S.typhimurium TA100 partially recapitulates the printing workers’ cholangiocarcinoma signature. (A) Number of SNVs in 1,2-DCP- and DCM-exposed TA100, except for the hisG gene target site. (B) Single-nucleotide DNA substitution profiles in TA100 exposed to 1,2-DCP or DCM except for the hisG gene target site. (C) Trinucleotide mutational pattern of TA100 exposed to 1,2-DCP or DCM except for the hisG gene target site. Clones exposed to 1,2-DCP harbored NpCpC to NpTpC changes (indicated by the asterisks). This signature partially recapitulates the printing workers’ cholangiocarcinoma signature as shown below the 1,2-DCP-exposed TA100 panel. The printing workers’ signature is the mean of the frequency of secondary characteristic mutational changes in four patients shown in Figure 1E. The mutational signatures were normalized using the trinucleotide frequency in the genome. The signature of 1,2-DCP was obtained from the sum of the 3000 and 6000 ppm exposure data.
The mutational signature of 1,2-DCP in S.typhimurium TA100 partially recapitulates the printing workers’ cholangiocarcinoma signature
We further analyzed the trinucleotide mutational pattern. Clones exposed to 1,2-DCP harbored NpCpC to NpTpC changes (Figure 3C, indicated by the asterisks), and these changes were more remarkable at 6000 ppm than at 3000 ppm (Supplementary Figure 2, available at Carcinogenesis Online). This signature is similar to the second most dominant signature of the printing workers’ cholangiocarcinoma. With DCM exposure, the clones harbored broad changes in C:G to A:T and C:G to T:A mutations, and no specific trinucleotide mutational patterns were observed (Figure 3C).
1,2-DCP has limited mutagenic capability in mammalian cells
To confirm the mutational signature of 1,2-DCP in mammalian cells, we examined the base substitution profiles using cholangiocarcinoma cell line NCC-CC1 and HEK293 cells. The mutation rate of the two independent NCC-CC1 clones after a single exposure to 1,2-DCP at a cytotoxic dose was 0.1 and 0.5/Mb, and they were not significantly higher than the mutation rate of the non-exposed control. The mutation rate of clones repeatedly exposed to 1,2-DCP was 0.5 to 1.3/Mb, and no significant difference was observed here either (Supplementary Table 4 and Supplementary Figure 3A, available at Carcinogenesis Online). Among the SNVs, C:G to T:A transitions and C:G to A:T transversions were predominant (60.0 and 59.4% of total SNVs in genomic DNA with single and repeated exposure to 1,2-DCP, respectively) (Supplementary Figure 3B, available at Carcinogenesis Online). Moreover, no specific trinucleotide signatures were observed (Supplementary Figure 3C, available at Carcinogenesis Online). The mutation rate in 1,2-DCP-exposed HEK293 cells was 0.3±0.1/Mb, and it was not significantly increased (Supplementary Table 5 and Supplementary Figure 4A, available at Carcinogenesis Online). Similarly, in NCC-CC1 cells, C:G to T:A transitions and C:G to A:T transversions were observed to predominate (59.6% of total SNVs) (Supplementary Figure 4B, available at Carcinogenesis Online), and the trinucleotide signatures observed in the printing workers’ cholangiocarcinomas were not recapitulated (Supplementary Figure 4C, available at Carcinogenesis Online).
Discussion
The representative characteristic mutational profile, including a high somatic mutation burden, substantial strand bias in C:G to T:A mutations and unique trinucleotide mutational changes (GpCpY to GpTpY and NpCpY to NpTpY or NpApY), shared in all of the investigated printing workers’ cholangiocarcinomas suggests that the patients might have been exposed to a common strong mutagen and that these conditions might increase the chance for mutations in cholangiocarcinoma driver genes. Mutations with transcriptional strand bias are known to occur in cancer genomes as a result of exposure to abundant mutagens and the formation of bulky DNA adducts, such as in smoking-related lung cancer and ultraviolet-associated melanoma (25,26). We thus suspected that some agents make abundant adducts on G residues and that the transcription-coupled DNA repair machinery preferentially repairs transcribed G residues, that is, sense C residues (27), inducing strand-biased mutations. These results are consistent with the epidemiological observation that these workers had been exposed to high concentrations of chemical compounds. Based on reconstructed experimental data, Kumagai et al. estimated the concentration of volatile solvent in a proof-printing room to be 100–670 ppm for 1,2-DCP and 80–540 ppm for DCM (28). Both 1,2-DCP and DCM have been used in industrial processes and household products (such as paint stripper) worldwide. However, as mentioned previously, 1,2-DCP and DCM reportedly have very limited tumorigenic capabilities in animal models (13–16). Suzuki et al. recently reported that co-exposure to 1,2-DCP and DCM induced an increase in mutations in a mouse gpt mutation model. However, they detected mutations predominantly at A:T pairs rather than C:G pairs, in contrast to the mutations observed in the printing workers’ cholangiocarcinomas in this study (29).
To verify the mutagenic features of 1,2-DCP and DCM more directly, we applied a model system using S.typhimurium strain TA100. Our data suggest that both 1,2-DCP and DCM exhibited mutagenic activity on the S.typhimurium TA100 genome, especially at C:G residues.
However, three of the four printing workers’ cholangiocarcinoma cases examined in this study were exposed only to 1,2-DCP and not to DCM. Prior epidemiological studies suggested that 1,2-DCP is a more suspicious causative agent than DCM, because all 17 of the patients in the first cohort in Osaka were exposed to 1,2-DCP, whereas 11 were exposed to DCM (8). Based on these findings, IARC designated 1,2-DCP as a higher rank carcinogen than DCM. Whole-genome analysis of 1,2-DCP- or DCM-exposed S.typhimurium TA100 might support these epidemiological assumption. The trinucleotide mutational signature of S.typhimurium TA100 exposed to 1,2-DCP revealed preferential mutational changes of NpCpC to NpTpC, which overlaps with the second most predominant trinucleotide mutational signature observed in the printing workers’ cholangiocarcinomas, namely, NpCpY to NpTpY or NpApY changes. These results suggest a contribution of 1,2-DCP to mutagenesis and carcinogenesis in the printing workers’ cholangiocarcinomas, at least in some capacity.
In mammalian species, inhaled DCM is supposed to be metabolized via a GSTT1-dependent pathway (27,30–33). S-(chloromethyl)glutathione from DCM is involved in mutagenesis by forming guanine adducts, inducing C:G to A:T and C:G to T:A substitutions, in bacteria and mammalian cells (17,34,35). In contrast to DCM, the mode of mutagenicity of 1,2-DCP or its potential metabolites remains unclear. NpCpY (the complement to RpGpN) sites are reported to be targets of electrophilic agents such as alkylating agents and platinum-derived drugs (25,36–38). The N7 and O6 positions of guanine are the most reactive nucleophilic sites (39–41), and electrophilic agents react with these sites to form alkyl-DNA adducts or intra- and inter-strand cross-linked DNA adducts, inducing cytotoxicity and mutagenicity. Whether 1,2-DCP or its potential metabolites possess electrophilic features and form alkyl-DNA adducts should be further investigated.
To strengthen the above findings, we examined the mutagenic profile of 1,2-DCP in human epithelial cell-derived cell lines. However, neither single nor repeated exposure of cultured cells to 1,2-DCP induced significant mutagenesis, and the mutation profiles in the in vitro system did not recapitulate the specific trinucleotide signatures observed in the clinical samples and S.typhimurium TA100 strain model.
Another remaining riddle regarding the specific mutational landscape of the printing workers’ cholangiocarcinomas is the most prominent trinucleotide mutational signature, GpCpY to GpTpY. Because this signature was not recapitulated in the S.typhimurium TA100 strain model, host factors might have contributed. Although the GpCpY to GpTpY signature has not been identified in any human clinical genome sequencing data, a similar signature was recently reported by Olivier et al. They reported that acquired mutations caused by overexpression of activation-induced cytidine deaminase (AID) in immortalized human TP53 knock-in mouse fibroblasts were predominantly C:G to T:A substitutions with a GpCp(A/C/T) to GpTp(A/C/T) trinucleotide signature (42). AID was originally identified as an inducer of somatic hypermutation in the variable region of immunoglobulin genes in activated B cells (43,44). In addition, the aberrant expression of AID induces lymphoid and nonlymphoid malignancies with accumulated mutations in known cancer-related genes in animal models (45). AID is reportedly induced by chronic tissue injury in parenchymal cells such as hepatocytes and gastric epithelial cells (46,47). Komori et al. reported significantly high AID expression in the epithelial cells of cholangiocarcinoma, with massive inflammation in the surrounding liver tissue (48). Although the tissue toxicity of 1,2-DCP and DCM has been reported mainly in animal models, and whether they affect biliary epithelia remains unclear (49), massive fibrosis was observed to surround cancerous and non-cancerous bile ducts in the printing workers’ cholangiocarcinoma samples (8). Our preliminary immunohistochemistry data suggested that AID was expressed in the transformed epithelial cells of the printing workers’ cholangiocarcinomas (data not shown). Whether the GpCpY to GpTpY changes in the printing workers’ cholangiocarcinomas were elicited by inflammation-induced AID expression must be further examined using in vitro and in vivo model systems.
The above-mentioned discrepancies in mutation signature between the clinical samples and the in vitro models suggest that the carcinogenic processes of the printing workers’ cholangiocarcinomas might be more complex than we had supposed. To elucidate the mechanisms, we should consider the tumor microenvironment including the metabolism of the inhaled chemical compounds at the biliary epithelia and interactions of tumor cells with surrounding non-tumor cells and possibly bacterial flora. To address these questions, model systems reproducing the actual conditions in biliary epithelial cells and the surrounding microenvironment of the printing worker cholangiocarcinoma patients are necessary.
Although the lack of model systems hampered further investigation, the particular mutational profiles, especially the specific trinucleotide mutation signatures, will be a clue to help elucidate the hidden mechanism of this disease. The mutational profiles of various cancer genomes are systematically collected and the mutational signatures are classified by ICGC. Although the signature presented in this study has not been reported, the accrual of further data may give us a chance to find other cases of biliary and other organ cancers with similar signatures. Referring to their epidemiological and clinicopathological background will provide information that the model systems hardly address.
Supplementary material
Supplementary Tables 1–5, Supplementary Figures 1–4 and Supplementary File 1 can be found at http://carcin.oxfordjournals.org/
Funding
This study was supported by the National Cancer Center Research and Development Fund (23-A-8, 25-A-5 and 25-A-6) and performed as a research program of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), the Japan Agency for Medical Research and Development.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- 1,2-DCP
1,2-dichloropropane
- AID
activation-induced cytidine deaminase
- COSMIC
Catalogue of Somatic Mutations in Cancer
- DCM
dichloromethane
- GSTT1
glutathione S-transferase theta 1
- HEK293
human embryonic kidney 293
- IARC
The International Agency for Research on Cancer
- ICGC
The International Cancer Genome Consortium
- INDELs
insertions and deletions
- SNVs
single-nucleotide variants
References
- 1. Matsuda A., et al. (2014) Cancer incidence and incidence rates in Japan in 2008: a study of 25 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J. Clin. Oncol., 44, 388–396. [DOI] [PubMed] [Google Scholar]
- 2. Everhart J.E., et al. (2009) Burden of digestive diseases in the United States Part III: liver, biliary tract, and pancreas. Gastroenterology, 136, 1134–1144. [DOI] [PubMed] [Google Scholar]
- 3. West J., et al. (2006) Trends in the incidence of primary liver and biliary tract cancers in England and Wales 1971-2001. Br. J. Cancer, 94, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Razumilava N., et al. (2014) Cholangiocarcinoma. Lancet, 383, 2168–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan S.A., et al. (2005) Cholangiocarcinoma. Lancet, 366, 1303–1314. [DOI] [PubMed] [Google Scholar]
- 6. Khan S.A., et al. (2003) DNA adducts, detected by 32P postlabelling, in human cholangiocarcinoma. Gut, 52, 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miyakawa S., et al. (2009) Biliary tract cancer treatment: 5,584 results from the Biliary Tract Cancer Statistics Registry from 1998 to 2004 in Japan. J. Hepatobiliary Pancreat. Surg., 16, 1–7. [DOI] [PubMed] [Google Scholar]
- 8. Kubo S., et al. (2014) Case series of 17 patients with cholangiocarcinoma among young adult workers of a printing company in Japan. J. Hepatobiliary Pancreat. Sci., 21, 479–488. [DOI] [PubMed] [Google Scholar]
- 9. Yamada K., et al. (2014) Chemical exposure levels in printing workers with cholangiocarcinoma. J. Occup. Health, 56, 332–338. [DOI] [PubMed] [Google Scholar]
- 10. Yamada K., et al. (2015) Chemical exposure levels in printing workers with cholangiocarcinoma (second report). J. Occup. Health, 57, 245–252. [DOI] [PubMed] [Google Scholar]
- 11. IARC. (2014) Perfluoro-octanoic acid, tetrafluoroethylene, dichloromethane, 1,2-dichloropropane, 1,3-propane sultone. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 110 IARC Working Group, Lyon. [Google Scholar]
- 12. Benbrahim-Tallaa L., et al. (2014) Carcinogenicity of perfluorooctanoic acid, tetrafluoroethylene, dichloromethane, 1,2-dichloropropane, and 1,3-propane sultone. Lancet Oncol., 15, 924–925. [DOI] [PubMed] [Google Scholar]
- 13. National Toxicology Program. (1986) NTP toxicology and carcinogenesis studies of 1,2-dichloropropane (propylene dichloride) (CAS No. 78-87-5) in F344/N rats and B6C3F1 mice (Gavage Studies). Natl Toxicol. Program Tech. Rep. Ser., 263, 1–182. [PubMed] [Google Scholar]
- 14. Umeda Y., et al. (2010) Inhalation carcinogenicity and toxicity of 1,2-dichloropropane in rats. Inhal. Toxicol., 22, 1116–1126. [DOI] [PubMed] [Google Scholar]
- 15. National Toxicology Program. (1986) NTP toxicology and carcinogenesis studies of dichloromethane (methylene chloride) (CAS No. 75-09-2) in F344/N rats and B6C3F1 mice (Inhalation Studies). Natl Toxicol. Program Tech. Rep. Ser., 306, 1–208. [PubMed] [Google Scholar]
- 16. Burek J.D., et al. (1984) Methylene chloride: a two-year inhalation toxicity and oncogenicity study in rats and hamsters. Fundam. Appl. Toxicol., 4, 30–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeMarini D.M., et al. (1997) Glutathione S-transferase-mediated induction of GC–>AT transitions by halomethanes in Salmonella. Environ. Mol. Mutagen., 30, 440–447. [PubMed] [Google Scholar]
- 18. Matsuda T., et al. (2013) A pilot study for the mutation assay using a high-throughput DNA sequencer. Genes Environ, 35, 53–56. [Google Scholar]
- 19. Ojima H., et al. (2010) Establishment of six new human biliary tract carcinoma cell lines and identification of MAGEH1 as a candidate biomarker for predicting the efficacy of gemcitabine treatment. Cancer Sci., 101, 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J.L., et al. (2001) An in vitro model for evaluation of vaporous toxicity of trichloroethylene and tetrachloroethylene to CHO-K1 cells. Chem. Biol. Interact., 137, 139–154. [DOI] [PubMed] [Google Scholar]
- 21. Ong C.K., et al. (2012) Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet., 44, 690–693. [DOI] [PubMed] [Google Scholar]
- 22. Chan-On W., et al. (2013) Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet., 45, 1474–1478. [DOI] [PubMed] [Google Scholar]
- 23. Nakamura H., et al. (2015) Genomic spectra of biliary tract cancer. Nat. Genet., 47, 1003–1010. [DOI] [PubMed] [Google Scholar]
- 24. Timmermann B., et al. (2010) Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One, 5, e15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alexandrov L.B., et al. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pleasance E.D., et al. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature, 463, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marsch G.A., et al. (2001) Characterization of nucleoside and DNA adducts formed by S-(1-acetoxymethyl)glutathione and implications for dihalomethane-glutathione conjugates. Chem. Res. Toxicol., 14, 600–608. [DOI] [PubMed] [Google Scholar]
- 28. Kumagai S., et al. (2013) Cholangiocarcinoma among offset colour proof-printing workers exposed to 1,2-dichloropropane and/or dichloromethane. Occup. Environ. Med., 70, 508–510. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki T., et al. (2014) Assessment of the genotoxicity of 1,2-dichloropropane and dichloromethane after individual and co-exposure by inhalation in mice. J. Occup. Health, 56, 205–214. [DOI] [PubMed] [Google Scholar]
- 30. Kim D.H., et al. (1990) Characterization of S-[2-(N1-adenyl)ethyl]glutathione as an adduct formed in RNA and DNA from 1,2-dibromoethane. Chem. Res. Toxicol., 3, 587–594. [DOI] [PubMed] [Google Scholar]
- 31. Inskeep P.B., et al. (1986) Covalent binding of 1,2-dihaloalkanes to DNA and stability of the major DNA adduct, S-[2-(N7-guanyl)ethyl]glutathione. Cancer Res., 46, 2839–2844. [PubMed] [Google Scholar]
- 32. Marsch G.A., et al. (2004) Formation and mass spectrometric analysis of DNA and nucleoside adducts by S-(1-acetoxymethyl)glutathione and by glutathione S-transferase-mediated activation of dihalomethanes. Chem. Res. Toxicol., 17, 45–54. [DOI] [PubMed] [Google Scholar]
- 33. Sherratt P.J., et al. (2002) Direct comparison of the nature of mouse and human GST T1-1 and the implications on dichloromethane carcinogenicity. Toxicol. Appl. Pharmacol., 179, 89–97. [DOI] [PubMed] [Google Scholar]
- 34. Graves R.J., et al. (1996) DNA sequence analysis of methylene chloride-induced HPRT mutations in Chinese hamster ovary cells: comparison with the mutation spectrum obtained for 1,2-dibromoethane and formaldehyde. Mutagenesis, 11, 229–233. [DOI] [PubMed] [Google Scholar]
- 35. Graves R.J., et al. (1996) Mouse liver glutathione S-transferase mediated metabolism of methylene chloride to a mutagen in the CHO/HPRT assay. Mutat. Res., 367, 143–150. [DOI] [PubMed] [Google Scholar]
- 36. Fichtinger-Schepman A.M., et al. (1985) Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification, and quantitation. Biochemistry, 24, 707–713. [DOI] [PubMed] [Google Scholar]
- 37. Saris C.P., et al. (1996) In vitro formation of DNA adducts by cisplatin, lobaplatin and oxaliplatin in calf thymus DNA in solution and in cultured human cells. Carcinogenesis, 17, 2763–2769. [DOI] [PubMed] [Google Scholar]
- 38. Woynarowski J.M., et al. (1998) Sequence- and region-specificity of oxaliplatin adducts in naked and cellular DNA. Mol. Pharmacol., 54, 770–777. [DOI] [PubMed] [Google Scholar]
- 39. Lawley P.D., et al. (1963) Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J., 89, 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beranek D.T. (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res., 231, 11–30. [DOI] [PubMed] [Google Scholar]
- 41. Lawley P.D., et al. (1996) DNA adducts from chemotherapeutic agents. Mutat. Res., 355, 13–40. [DOI] [PubMed] [Google Scholar]
- 42. Olivier M., et al. (2014) Modelling mutational landscapes of human cancers in vitro. Sci. Rep., 4, 4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Honjo T., et al. (2002) Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol., 20, 165–196. [DOI] [PubMed] [Google Scholar]
- 44. Honjo T., et al. (2004) AID: how does it aid antibody diversity? Immunity, 20, 659–668. [DOI] [PubMed] [Google Scholar]
- 45. Okazaki I.M., et al. (2003) Constitutive expression of AID leads to tumorigenesis. J. Exp. Med., 197, 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kou T., et al. (2007) Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int. J. Cancer, 120, 469–476. [DOI] [PubMed] [Google Scholar]
- 47. Matsumoto Y., et al. (2007) Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med., 13, 470–476. [DOI] [PubMed] [Google Scholar]
- 48. Komori J., et al. (2008) Activation-induced cytidine deaminase links bile duct inflammation to human cholangiocarcinoma. Hepatology, 47, 888–896. [DOI] [PubMed] [Google Scholar]
- 49. Trevisan A., et al. (1993) In-vitro mechanisms of 1,2-dichloropropane nephrotoxicity using the renal cortical slice model. Hum. Exp. Toxicol., 12, 117–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.