Abstract
Sarcomas are mesenchymal tumors characterized by blocked differentiation process. In Ewing sarcoma (EWS) both CD99 and EWS-FLI1 concur to oncogenesis and inhibition of differentiation. Here we demonstrate that uncoupling CD99 from EWS-FLI1 by silencing the former, NF-kB signaling is inhibited and the neural differentiation program is re-established. NF-kB inhibition passes through miR-34a-mediated repression of Notch pathway. CD99 counteracts EWS-FLI1 in controlling NF-kB signaling through the miR-34a, which is increased and secreted into exosomes released by CD99-silenced EWS cells. Delivery of exosomes from CD99-silenced cells was sufficient to induce neural differentiation in recipient EWS cells through miR-34a inhibition of Notch-NF-kB signaling. Notably, even the partial delivery of CD99 siRNA may have a broad effect on the entire tumor cell population thanks to the spread operated by their miR-34a-enriched exosomes, a feature opening to a new therapeutic option.
Keywords: CD99, Ewing sarcoma, neural differentiation, NF-kB signaling, miR-34a, exosome
Introduction
Ewing sarcoma (EWS) is the second most common type of primary bone tumor and mainly affects children and adolescents. It is an aggressive, poorly differentiated tumor, with elevated tendency to give lung and/or bone metastases. Despite the use of intensive, multimodality therapy 1, the prognosis of patients with metastatic EWS remains grim (survival less than 40% even with intensive chemotherapy) 2 and few treatments can be offered to those who relapse after first-line therapies. Even for children who are cured, the long-term morbidity of cytotoxic treatment is substantial 3, indicating the need of new therapeutic strategies for this disease.
From a genetic point of view, EWS is characterized by highly recurrent translocations involving ETS transcription factors, with EWS–FLI1 and EWS–ERG being the most common 4, 5. EWS-FLI1 functions as an aberrant transcription factor that regulates crucial processes like cell growth, apoptosis and differentiation through induction or repression of specific target genes and it is the oncogenetic driver of EWS 6. Forced expression of EWS-FLI1 in human mesenchymal stem cells (hMSCs), the closest EWS related normal cell type, was demonstrated to be sufficient to transform cells and induce a gene expression profile similar to that observed in EWS cells 7, while deprivation of EWS-FLI1 in EWS cells resulted in a gene expression signature that overlapped with mesenchymal progenitor cells 8 and decreased in vivo and in vitro tumor growth 9, 10. The genomic landscape of EWS, which has been recently explored by three different groups through multiple next-generation sequencing methods 11–13 clearly showed that the somatic mutation rate in EWS is low. Apart from the already known copy-number gains in whole chromosomes 8, 12 and the q arm of chr 1 and the loss in the long arm of chr 16 and the CDKN2A locus on chr 9p 14, 15, no recurrent somatic mutations have been reported in EWS, with the notable exceptions of TP53 (5%–20%) and the cohesin complex family member STAG2 (15-21%) 11–13. Although these alterations are reported to influence patient’s prognosis and serve as potential biomarkers for patient’s risk stratification 11, 16, their therapeutic value is limited. The sequencing of EWS genomes further sustains the pivotal role of EWS-FLI1 in the pathogenesis and progression of EWS and points to EWS-FLI1 as the best target. Nevertheless, EWS-FLI1 provides necessary but insufficient condition for tumor transformation requiring a permissive cellular background. Although the list of critical adjuvant transforming factors is still incomplete, disruption of the p53 and RB pathways, the presence of intact IGF signaling and of CD99 have been confirmed 17–19. CD99 is a cell surface molecule of 32 KDa 20 involved in crucial biological processes like migration, cell death and differentiation 21–23. CD99 is constantly present at high levels in EWS cells and its detection is routinely used for differential diagnosis. The EWS-FLI1 oncogenic activity 6 is facilitated by CD99 (ref. 19) and consistently, EWS-FLI1 maintains high levels of CD99 expression 19, 24–26 either directly through binding of CD99 promoter 19 or indirectly through miRNA regulation 27. CD99 knockdown in EWS cells induces terminal neural differentiation and reduces tumor growth and bone metastasis upon transplantation into immunodeficient mice 19, supporting a central role for CD99 in the pathogenesis of EWS. In this article, we analyze the relationship between CD99 and EWS-FLI1 in an effort to identify the mechanisms that reversing the tumorigenicity of EWS cells would take them back to the road of normal differentiation. Differently from other solid tumors, sarcomas could be reprogrammed to resume normal differentiation 28, an intriguing approach that offers new options for the treatment of these tumors. Both CD99 and EWS-FLI1 appear to impact on EWS cell differentiation with opposite effects: while EWS-FLI1 may induce neural differentiation, CD99 prevents it 19. The net effect is a cell that exhibits some neural features while maintaining cell growth capacity. Additional activation or repression of other molecular pathways would be necessary to alter this equilibrium in favor of the achievement of a complete neuronal differentiation process. With this study we aimed at the identification of the missing molecules, providing mechanistic proof of principle for potential novel therapeutic intervention in EWS.
Results
CD99 affects EWS cell differentiation through NF-kB transcriptional activity
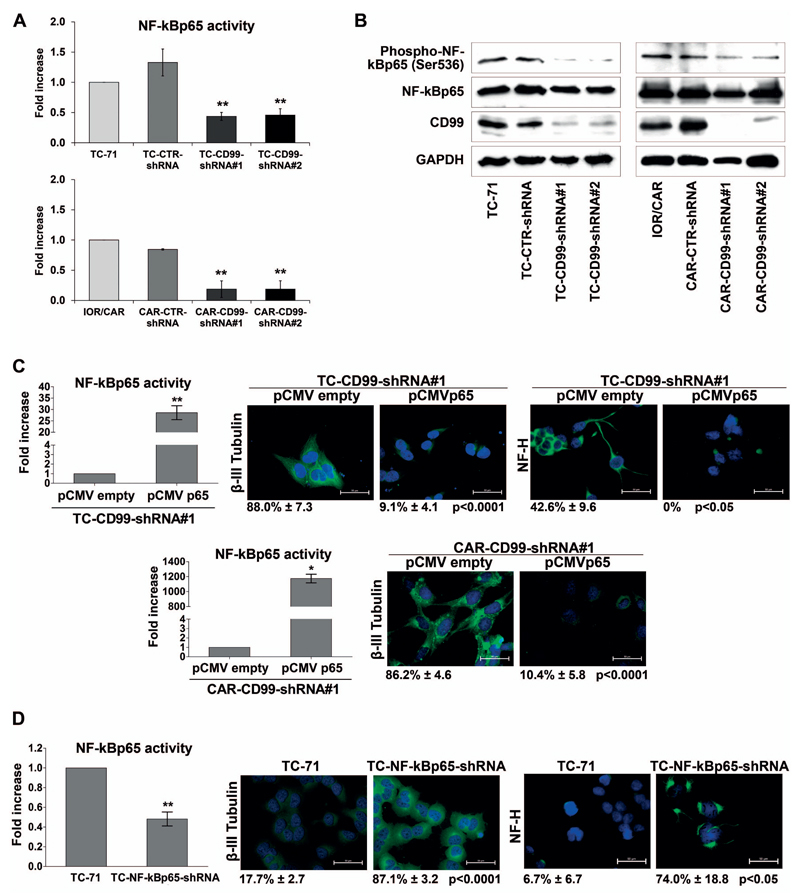
Deprivation of CD99 drives EWS cells toward neural differentiation 19 (Supplementary Figure 1). To understand the mechanism behind we studied the transcriptional activity of five transcription factors (SRE, E2F, CREB, NF-kB and AP1) involved in cell growth and differentiation. Using luciferase reporter assay, upon CD99 silencing we observed a significant down-regulation of Nuclear Factor kappa B (NF-kB) activity (Figure 1A), but not of SRE, E2F, CREB and AP1 (Supplementary Figure 2). In general, the activation of NF-kB occurs when NF-kB members are transported from the cytoplasm to the nucleus upon uncoupling NF-kB factors from inhibitory IkB proteins 29. Optimal induction of NF-kB target genes also requires phosphorylation of NF-kB proteins, such as p65, whose phosphorylation at Ser536 enhances its transactivation potential 30. Accordingly, knock-down of CD99 reduced the levels of Ser536 phosphorylation in EWS cells (Figure 1B). To establish a functional relationship between NF-kB activity and EWS cell differentiation, we used a gain- and loss-of-function approach. Overexpression of NF-kB in TC-CD99-shRNA#1 and CAR-CD99-shRNA#1 cells through transient transfection of the pCMVp65 plasmid induced a loss of the neural differentiation, shifting the expression of β-III Tubulin and NF-H back to baseline (Figure 1C). Conversely, stable silencing of NF-kBp65 in the parental, CD99-high expressing TC-71 cell line (Supplementary Figure 3A) induced neural differentiation (Figure 1D). All together, these evidences indicate that the pro-differentiative effect due to CD99-silencing occurs via inhibition of NF-kB activity in EWS cells.
Figure 1. CD99–induced inhibition of NF-kB transcriptional activity favors EWS cell neural differentiation.
(A) Transcriptional activity of NF-kBp65 was assessed by luciferase assay. Fold increase of the transcriptional activity of the reporter is expressed as ratio between the luciferase activity of clones and luciferase activity of parental cell line. The graphs summarize the results of at least three independent experiments performed in triplicate (Mean ± SEM, **p Value <0.01, Student’s t test). (B) Knockdown of CD99 in TC-71 and IOR/CAR induces a decrease in phospho-NF-kBp65 (Ser536) levels compared with empty vector-transfected cells (CTR-shRNA) or parental cell lines. The blots are representative of two independent experiments. GAPDH is shown as loading control. (C) Over expression of NF-kB (pCMVp65) in TC-CD99-shRNA and CAR-CD99-shRNA cells induces a strong reduction of the neural differentiation marker β-III Tubulin and NF-H immunofluorescence. Bars graphs represent the fold increase of NF-kBp65 activity of three independent experiments performed in triplicate (Mean ± SEM, *p Value <0.05, **p Value <0.01 Student’s t test). (D) Silencing of NF-kB (NF-kBp65-shRNA) (Supplementary Figure 3A) in TC-71 induces an upregulation of β-III Tubulin and Neurofilament-H (NF-H). Digital images were taken under identical conditions, at the same time and using the same image analysis software as described in Material and Methods. Scale bars: 50μm. Bars graph represents the fold increase of NF-kBp65 activity of three independent experiments performed in triplicate (Mean ± SEM, **p Value <0.01 Student’s t test). Percentage of β-III Tubulin and NF-H positive cells are reported.
CD99 antagonizes EWS-FLI1 in controlling NF-kB-mediated EWS differentiation
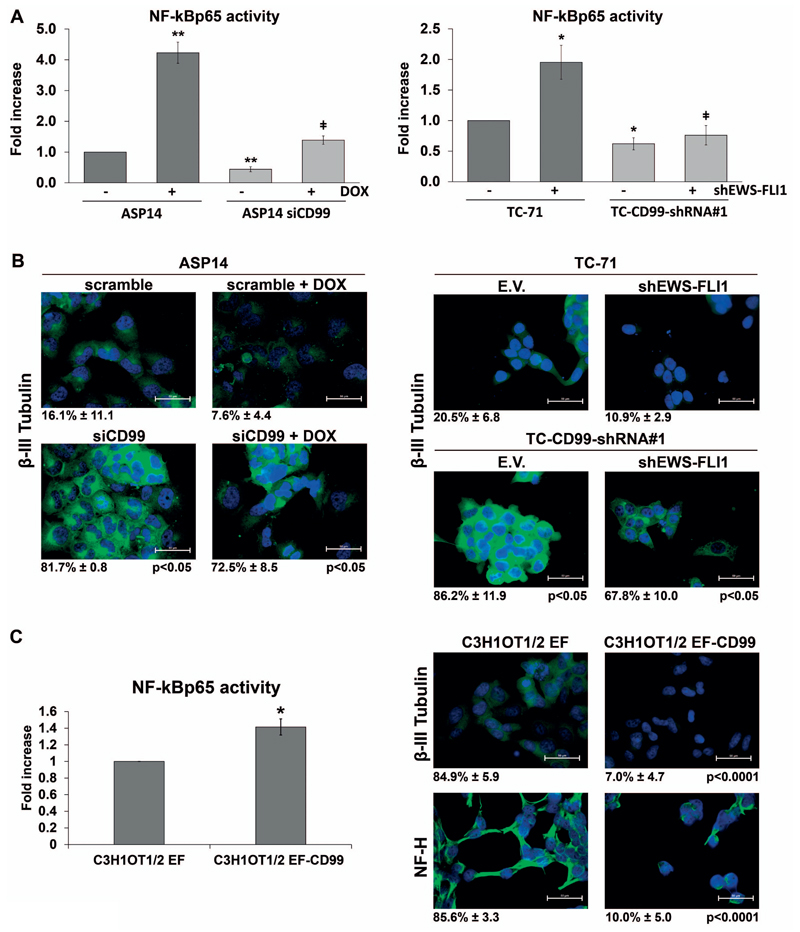
Previous studies have suggested that EWS-FLI1 induces a neural phenotype in a variety of cellular contexts 25, 31, 32. EWS-FLI1 also upregulates CD99 expression 17, 19, 25, 27 which in turn antagonizes EWS-FLI1-induced neural differentiation 19. We examined the functional interplay between CD99 and EWS-FLI1 with respect to the neural differentiation of EWS cells and the modulation of NF-kB transcriptional activity. We took advantage of the ASP14 cell line, harnessed for the doxycyclin-inducible shRNA targeting of EWS-FLI1. In these cells, induced silencing of EWS-FLI1 resulted in a strong activation of NF-kB (Figure 2A), without significantly changing the expression of the neural marker β-III Tubulin (Figure 2B). When CD99 was simultaneously silenced (Supplementary Figure 3B), the above increase of NF-kB activity was completely lost (Figure 2A). The parallel analysis of neural differentiation showed that ASP14 cells deprived of CD99 but not of EWS-FLI1 displayed a significant increase of β-III Tubulin immunostaining an effect that was still maintained when EWS-FLI1 was concomitantly silenced (Figure 2B). The same result was obtained with the TC-71 cell line (Figure 2A, B). Transient silencing of EWS-FLI1 in TC-71 cells (Supplementary Figure 3B) induced up-regulation of NF-kB transcriptional activity that did not occur in case CD99 was also silenced with NF-kB remaining at baseline level (Figure 2A). In silenced CD99 cells with low NF-kB transcriptional activity, the expression of β-III Tubulin was increased independently from EWS-FLI1 expression (Figure 2B), suggesting that CD99 efficiently contrasts EWS-FLI1 in term of neuronal differentiation. To confirm this finding, we used the C3H1OT1/2 murine mesenchymal multipotent cell line that loses the ability to differentiate into the osteoblastic and adipocytic, while gains neural phenotype and tumorigenic properties upon EWS-FLI1 transfection 33 (Supplementary Figure 3C). In this cell line, CD99 cDNA transfection repressed the feature of neural differentiation induced by EWS-FLI1 (ref. 19). Here we show that the forced expression of CD99 enhances NF-kB activity, in parallel with reduction of β-III Tubulin and NF-H expression (Figure 2C). On these evidences, we propose that CD99 expression in EWS cells sustains NF-kB transcriptional activity, prevents terminal neural differentiation and counteracts the effects of EWS-FLI1.
Figure 2. CD99 has a predominant role on NF-kB-mediated neural differentiation with respect to EWS-FLI1.
(A) NF-kB driven luciferase activity was tested in response to CD99 and EWS-FLI1. Both genes were transiently silenced in ASP14 cells with siCD99 (40nM) and DOX. EWS-FLI1 was transiently silenced in TC-71 and TC-CD99-shRNA#1 cells with shEWS-FLI1 (50ng) (Supplementary Figure 3B). The graphs reported the mean of three independent experiments performed in triplicate. (Mean ± SEM, *p Value <0.05, **p Value <0.01 Student’s t test) (ǂ p Value <0.05 ASP14 +DOX vs ASP14siCD99 +DOX; TC-71+shEWS-FLI1 vs TC-CD99-shRNA +shEWS-FLI1). (B) Immunofluorescence analysis of β-III Tubulin after simultaneous silencing of EWS-FLI1 and CD99. Only upon CD99 knockdown, the induction of neural differentiation was observed, independently from EWS-FLI1 status. Scale bars: 50μm. Percentage of β-III Tubulin positive cells are reported. (C) Murine mesenchymal C3H1OT1/2 cell line transfected with EWS-FLI1 (C3H1OT1/2 EF) and with CD99 (C3H1OT1/2 EF-CD99) (Supplementary Figure 3C). Forced expression of CD99 in C3H1OT1/2 EF increased NF-kB transactivation (bars graph represents Mean ± SEM of two independent experiment performed in triplicate, *p Value <0.05 Student’s t test) and repressed neural differentiation, as shown by β-III Tubulin and NF-H immunostaining. Digital images were taken under identical conditions, at the same time and using the same image analysis software as described in Material and Methods. Scale bars: 50 μm. Percentage of β-III Tubulin and NF-H positive cells are reported.
CD99 regulation of NF-kB transcriptional activity is through miR-34a-induced control of Notch pathway
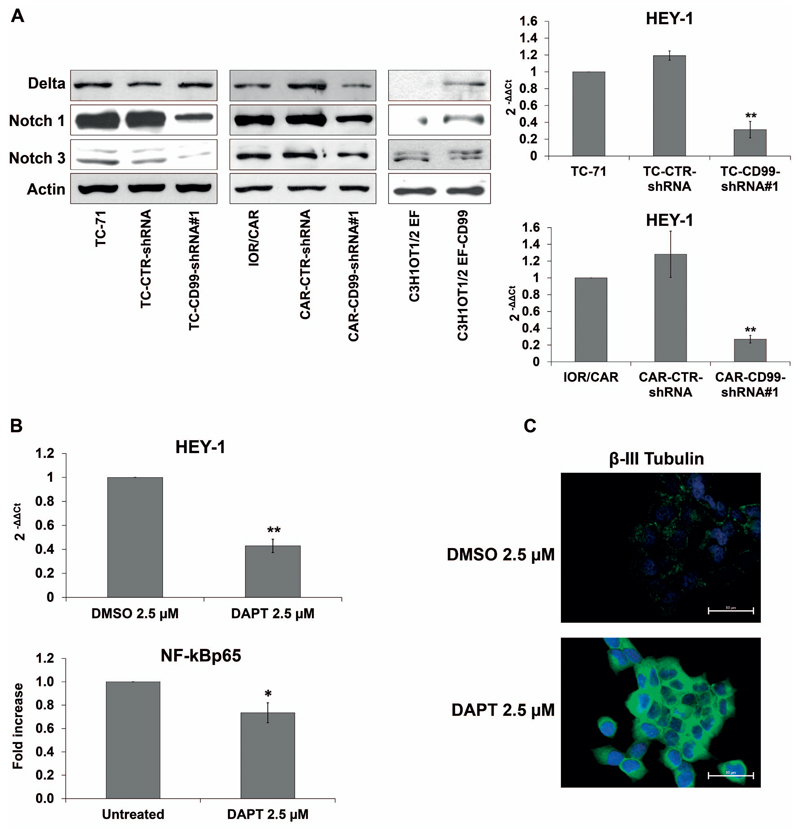
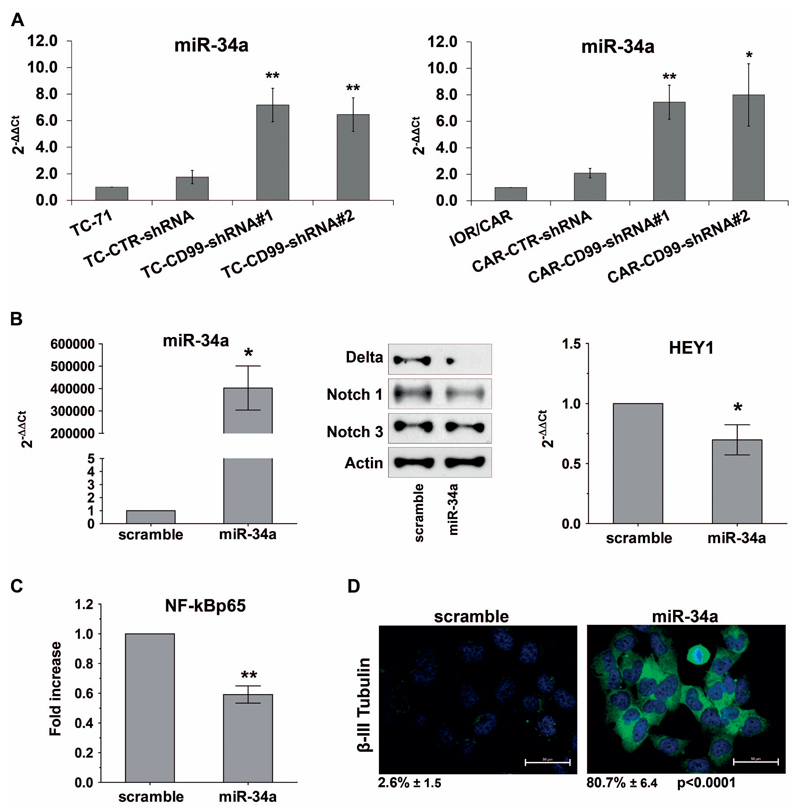
CD99 does not regulate NF-kB either at mRNA or protein levels (Figure 1B; Supplementary Figure 4A), but rather its signalling machinery. Here we showed that treatment of the EWS cell lines with TNFα, a prototype inducer of NF-kB signaling 34, translocated NF-kBp65 in the nucleus and induced NF-kB-dependent transcription regardless CD99 was either present or absent (Supplementary Figure 4B, C). This suggests that CD99 regulation is outside the NF-kB signalling machinery responding to TNFα and may be independent of IkBα regulation 35. Thus, we evaluated the involvement of other, still incompletely understood mechanisms that may act as alternative transcriptional regulators of NF-kB 36. In particular, we investigated the role of Notch, a developmental pathway involved in the regulation of cell fate. NF-kB and Notch reciprocal modulation has been described in specific cellular contexts, such as during T-cell, neuron and osteoblast differentiation and in pancreatic cancer cells 37, 38. In our experimental model, CD99-shRNA EWS cells showed lower levels of Notch receptor and ligand proteins than parental cells and reduced expression of the Notch-target gene HEY-1. Conversely, the forced expression of CD99 in C3H1OT1/2 EF cells increased the protein levels of the Notch ligand Delta and Notch 1 receptor (Figure 3A). Inhibition of Notch pathway in parental EWS cells by treatment with the γ-secretase inhibitor DAPT (Figure 3B), decreased the levels of NF-kB p65 luciferase activity (Figure 3B) and concomitantly increased the expression of β-III Tubulin (Figure 3C). This indicates that Notch inhibition is associated with the NF-kB-mediated neural differentiation of EWS cells. We then examined whether the miR-34a, which regulates the Notch pathway and has a role in EWS malignancy 39, 40, could be the molecule bridging CD99 with Notch and NF-kB. According to our hypothesis, miR-34a was significantly up-regulated in CD99-silenced EWS cells (Figure 4A). Exposure of EWS cells to miR-34a mimic decreased the expression of Notch 1 and Delta as well as of the Notch-target gene HEY-1 (Figure 4B), decreased NF-kB transcriptional activity (Figure 4C) and increased neural differentiation (Figure 4D), indicating that the sole miR-34a can reproduce all the CD99 effects on EWS cell differentiation. As confirmation, stable ectopic expression of miR-34a was induced in TC-71 cells (TCpMIF miR-34a) and associated with reduced expression of Delta and Notch 1 protein levels (Supplementary Figure 5).
Figure 3. CD99 regulation of NF-kB activity is mediated by Notch pathway.
(A) Delta, Notch 1, and Notch 3 expression was assessed by western blotting in TC-CD99-shRNA, CAR-CD99-shRNA and C3H1OT1/2 EF-CD99 cellular models. Actin was used as loading control. Blots are representative of three independent experiments. mRNA expression level of HEY-1 expressed as 2−ΔΔCt, in CD99-shRNA cells respect to parental cells TC-71 and IOR/CAR. Parental cell lines were used as calibrator (2−ΔΔCt=1). Reduction of Notch pathway was observed in CD99-silenced cells. (Graphs represent Mean ± SEM of two independent experiment, **p Value <0.01 Student’s t test). (B) Inhibition of Notch pathway by γ-secretase inhibitor (DAPT), as detected by reduction of HEY-1 mRNA expression, resulted in inhibition of NF-kB activity. Histograms showing IOR/CAR cells treated or not with DAPT 2.5 μM for 48 hours. Data indicate Mean ± SEM of two independent experiments. Differences between control and treated cells were compared using Student’s t test *p Value <0.05, **p Value <0.01. (C) Down regulation of Notch pathway in IOR/CAR cells induce neural differentiation as shown by β-III Tubulin. Representative photomicrographs are shown. Scale bars: 50 μm. Percentage of β-III Tubulin positive cells are reported.
Figure 4. Down-regulation of Notch pathway in CD99-shRNA cells is mediated by miR-34a.
(A) Expression levels of miR-34a in TC-CD99-shRNA and CAR-CD99-shRNA models in basal condition. Graphs indicate Mean ± SEM of three independent experiments (*p Value <0.05, **p Value <0.01 Student’s t test). (B) Expression levels of miR-34a in IOR/CAR cells transfected with miR-34a mimics. Graph represents Mean ± SEM of two independent experiments (*p Value <0.05 Student’s t test). Ectopic expression of miR-34a induces down regulation of Notch pathway. Delta, Notch 1 and Notch 3 expressions were detected by western blot. Actin was used as loading control. mRNA expression of HEY-1 was detected by qRT-PCR and is expressed as 2−ΔΔCt, in miR-34a-transfected cells respect to IOR/CAR cells transfected with scramble (Mean ± SEM of two independent experiments *p Value <0.05 Student’s t test). (C) Overexpression of miR-34a resulted in inhibition of NF-kB activity. Data represents Mean ± SEM of three independent experiments performed in triplicate. (**p Value <0.01 Student’s t test). (D) Representative immunofluorescence pictures showing β-III Tubulin expression in IOR/CAR cells transfected with miR-34a mimic. Cells were treated as indicated in Material and Methods. Scale bars: 50 μm. Percentage of β-III Tubulin positive cells are reported.
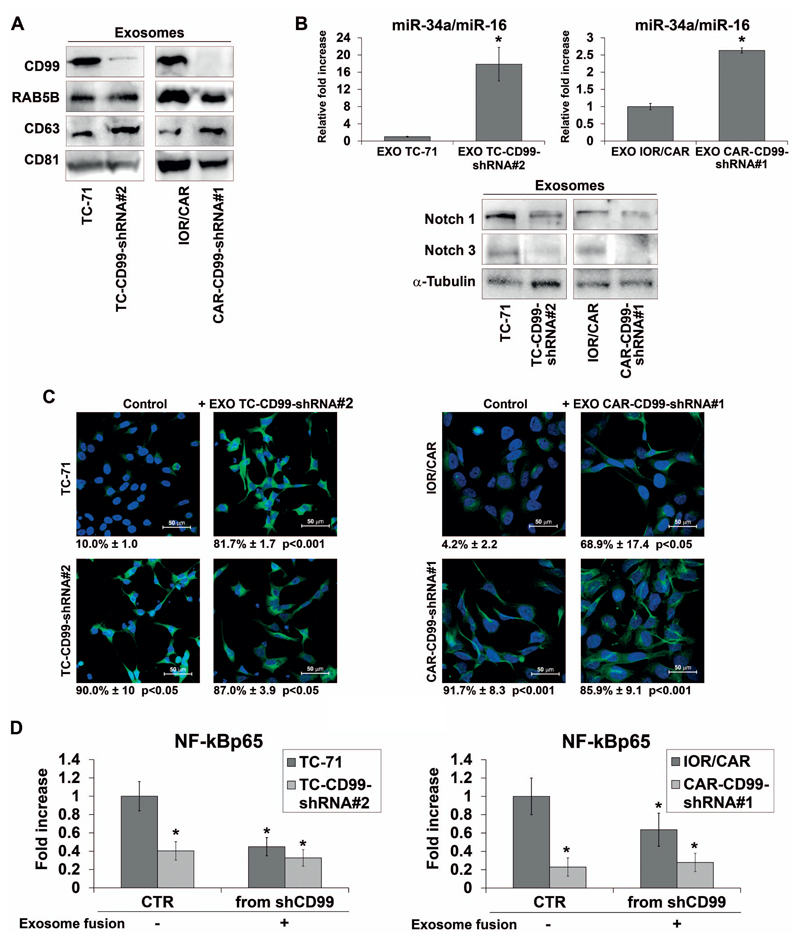
CD99 is released via exosomes inducing differentiation of EWS recipient cells by recapitulating miR-34a-Notch dependent regulation of NF-kB activity
CD99 was found to be secreted by EWS cells (data not shown), shuttled by exosomes (Figure 5A). Exosomes were purified from either parental or CD99-shRNA EWS cell lines and characterized by western blot for the enrichment of proteins commonly utilized as microvesicle markers, such as RAB5B, CD63 and CD81 (Figure 5A). In addition, the expression analysis of the CD99-shRNA-derived exosomal cargo showed, besides the obvious lack of CD99 (Figure 5A), a higher level of miR-34a coincident with Notch 1 and Notch 3 repression (Figure 5B). We then evaluated whether the uptake of exosomes from CD99-silenced cells by parental EWS cells might be sufficient to mimic CD99 silencing in virtue of miR-34a content. Evidence of effective exosome internalization was indirectly obtained by revealing CD99 expression variation in CD99-shRNA-cells after exposure to exosomes derived from parental and silenced cells (Supplementary Figure 3D). The neural differentiation of EWS parental cells induced by the internalization of exosomes lacking CD99 was detected as increase of β-III Tubulin (Figure 5C). Concordantly, this effect mimics that of miR-34a transfer, such as repression of Notch 1 and 3 and in turn down-regulation of NF-kB transcriptional activity (Figure 5D and data not shown). These findings support the notion that exosomes can convey the same pro-differentiative signals in EWS recipient cells as induced by stable silencing of CD99.
Figure 5. Exosomes released by CD99-silenced EWS cells are able to recapitulate mechanisms and phenotype obtained through stable CD99 silencing.
(A) Representative western blot showing CD99, RAB5B, CD63 and CD81 in exosomes of TC-CD99-shRNA and CAR-CD99-shRNA experimental models. (B) Exosomal miR-34a expression is shown for TC-71, TC-CD99-shRNA#2 cells and IOR/CAR, CAR-CD99-shRNA#1 cells. Results are referred to miR-16 as a constantly expressed internal standard (Mean ± SEM of two independent experiments *p Value <0.05 Student’s t test). Exosomal protein expression was detected by western blot. Blots of Notch 1 and Notch 3 are representative of three independent experiments. α-Tubulin was used as a loading control. (C) Immunofluorescence staining of β-III Tubulin in TC-71, IOR/CAR and CD99-shRNA clones incubated with CD99-shRNA derived exosomes. Scale bars: 50 μm (D) NF-kB transcriptional activity in TC-71, IOR/CAR and CD99-shRNA clones after fusion with CD99-shRNA derived exosomes (data indicate Mean ± SEM of two experiment performed in triplicate. *p Value <0.05 Student’s t test). Percentage of β-III Tubulin positive cells are reported.
Discussion
NF-kB plays a pivotal role in inflammation, immunity, development and cancer. Indeed, NF-kB transcription factors control the expression of genes active in cell proliferation, survival, differentiation, invasion, angiogenesis, and metastasis and it is constitutively active in most tumor cells 36, 41. Of interest here is the role of the NF-kB pathway in the differentiation of normal and malignant cells of the central nervous system, myeloid lineage and osteoclasts 42–46. EWS cells have low NF-kB activity 47. Nevertheless, its inhibition sensitized EWS cells to TNFα-induced killing 48, 49 and reduced their ability to form tumors when transplanted into immunodeficient mice 50. Inhibition of NF-kB was also shown to enhance the apoptotic activity of histone deacetylase inhibitors 51 and of nutlin-3 (ref. 52), thus supporting the concept of targeting NF-kB as antitumor strategy. In this paper, we demonstrate that although CD99 co-operates with EWS-FLI1 to sustain EWS malignancy 19, it contrasts EWS-FLI1 for maintaining active NF-kB. Indeed, we show that EWS-FLI1 has a negative influence on NF-kB basal activity, in line with a previous report showing EWS-FLI1 impairing TNFα-induced NF-kB-driven transcription, partly through inhibition of NF-kB binding to DNA 53. We also show that silencing CD99 in human EWS cell lines reduces NF-kB signaling regardless EWS-FLI1 is either active or silenced and oriented the cells toward a terminal neural differentiation. This is particularly important because other studies have suggested that EWS-FLI1 itself induces neural differentiation in a variety of cellular context 54 and that CD99 participates in the transformed phenotype by preventing neural differentiation of EWS cells 19. Here we unveil the role of NF-kB in blocking the terminal differentiation. This effect may result from a direct activity of NF-kB in controlling proliferation and differentiation of neural stem cells 55, 56, or from indirect mechanisms due to a physical interaction between NF-kB and EWS-FLI1 (ref. 53). NF-kB activation promotes osteogenesis in human mesenchymal stem cells 57 or in adipose tissue-derived stromal cells 58. In contrast, EWS-FLI1 expression in murine MSCs blocked their differentiation into osteocytic or adipocytic lineages in favor of neural differentiation 33. It might be possible that the interaction between these two transcription factors contribute to the maintenance of an undifferentiated state of the EWS cells through reciprocal impairment. Silencing of CD99 by abolishing NF-kB signaling led to unrestricted EWS-FLI1 activity, which may drive EWS cells toward neural differentiation. Our previous study has also shown how EWS-FLI1 and CD99 display opposite effects with respect to MAPK pathway 19. In particular, silencing of CD99 prolonged nuclear ERK1/2 phosphorylation and this seems to be crucial for shifting the biological functions of ERK1/2 toward neural differentiation. This suggests a model according to which EWS-FLI1 and CD99 have opposite effects with respect to several crucial transcriptional factors. Suppression of CD99 may thus serve to fine-tune levels of transcriptional gene regulation, shifting the equilibrium in favor of cell differentiation rather than proliferation. Further studies are required to test the functional cross-talk between EWS-FLI1 and ERK1/2 in the absence of NF-kB. Among the transcriptional regulators of NF-kB there is the Notch pathway, another major cell fate regulatory network. Multiple cross-talk mechanisms between Notch and NF-kB in diverse experimental models have been described 59. Conventional Notch signaling has been reported to be off in EWS 60, 61. However, the expression of Notch receptors was found to be high either in EWS cell lines or in clinical samples 61. Thus, Notch may still have a role in EWS cells through non-canonical signaling. In this paper we indeed suggest that Notch may have an activatory role on NF-kB signaling, as previously described in hippocampal neurons 62. Of note, Baliko et al 63 reported that inhibition of Notch induced neural differentiation in two EWS cell lines, without affecting tumor growth in vivo. In our hands, inhibition of Notch pathway by the gamma-secretase inhibitor DAPT restrains NF-kB transcriptional activity and increases neural differentiation, thus providing an explanation to the contradictory findings on the presence and the role of Notch signaling in EWS. Moreover, EWS cells deprived of CD99 showed diminished expression of Notch 1 and 3 as well as of their downstream target HEY-1. We provide evidence that this effect could be miR-34a-mediated. Notch 1 and other components of the pathway are in fact validated targets of miR-34a 64. We show that the level of miR-34a is higher in CD99 silenced cells and that miR-34a mimic can both reduce Notch 1 and NF-kB and it increases neural differentiation of EWS cells. The same outcome was obtained by exposing EWS cells to exosomes derived from cells silenced for CD99. Exosomes are small vesicles (40–130 nm), naturally produced by many cell types, that entrap key elements necessary for intercellular communication. They are emerging as crucial messengers that can regulate physiological and pathological processes through the delivery of their content to recipient cells 65. The composition of exosomes is related to their site of biogenesis. Due to their endosomal origin, all exosomes showed an enrichment of certain protein families such as Rab GTPases, heat shock proteins, tetraspanins, MVB biogenesis proteins. CD99 can directly interact and form complex with tetraspanin CD81 (ref. 66), a well-characterized exosomal marker. Exosomes released by CD99-silenced EWS cells contain levels of miR-34a higher than parental counterpart and successfully down-regulate Notch 1 and Notch 3 expression as well as NF-kB transcriptional activity in recipient EWS cells, which in turn differentiate toward a neural phenotype. The composition of exosomes from cells deprived of CD99 is therefore sufficient to induce in recipient parental cells the same phenotype obtained by stable CD99 silencing. Confirming the sequential path miR-34a/Notch/NF-kB that follows CD99 silencing, our results suggest that exosomes released from CD99-silenced EWS cells have the capacity to interfere with the fate of human EWS cells. A schematic representation of the molecular mechanisms associating CD99 deprivation with neural differentiation of EWS cells is shown (Figure 6). Our findings indicate that EWS patients may benefit from combinations of agents targeting both Notch and NF-kB pathways. Furthermore, exosome-mediated diffusion of the effects initiated by CD99 abrogation suggests that even partial delivery of CD99 siRNA may have a broad effect on the entire tumor cell population, a feature that can be exploited for therapeutic application.
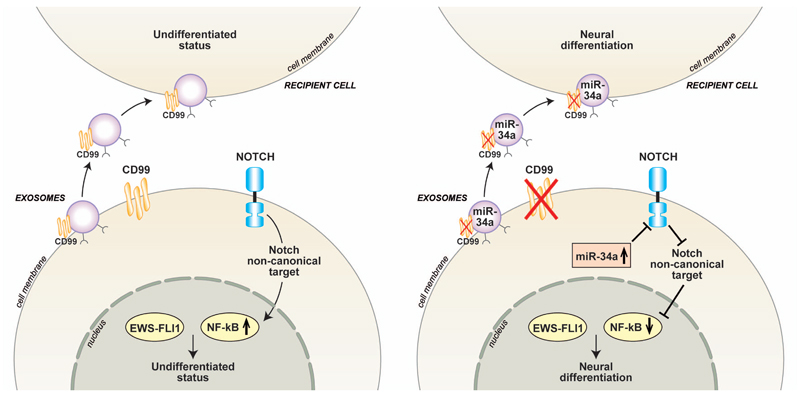
Figure 6. Schematic representation of the mechanistic relationship between CD99 silencing and EWS neural differentiation.
This figure depicts the capability of sh-CD99 exosomes, containing high miR-34a and low Notch levels, to induce neural differentiation by transferring their cargo in recipient EWS cells.
Material and Methods
Cell lines
Patient-derived EWS cell lines were grown as described previously 19. ASP14 cell line, a stable cell line generated from A673 EWS cell line transfected with doxycyclin-inducible shRNA targeting EWS-FLI1 (ref. 67) was cultured in complete medium supplemented with 20 μg/ml Blasticidin (Invitrogen, Grand Island, NY, USA) and 50 μg/ml Zeocin (Sigma Aldrich, St Louis, MO, USA). Stable silencing of CD99 was obtained in TC-71 (ref. 19) and IOR/CAR EWS cell lines (Supplementary Figure 1). TC-CTR-shRNA cells and TC-CD99-shRNA clones were established and cultured as previously described 19. CAR-CTR-shRNA and CAR-CD99-shRNA clones were maintained in complete medium supplemented with 1000 μg/ml G418 disulfate salt (Sigma Aldrich). C3H1OT1/2 murine mesenchymal cells transfected with EWS-FLI1 (C3H1OT1/2 EF) and with CD99 (C3H1OT1/2 EF-CD99) were cultured as described previously 19. TC-71 cells stably expressing miR-34a (TCpMIF miR-34a) were obtained by infection with the feline immunodeficiency lentivirus-based construct pMIF-cGFPZeo-miR-34a (System Biosciences, Mountain View, CA, USA), as previously described 68. Empty lentivirus (pMIF-cGFPZeo EV) was used as a control; cells were selected with 100 μg/ml Zeocin (Sigma Aldrich) before performing functional studies (Supplementary Figure 5). All cell lines were tested for mycoplasma contamination every three months by MycoAlert mycoplasma detection kit (Lonza, Allendale, NJ, USA) and were recently authenticated by STR PCR analysis using genRESVR MPX-2 and genRESVR MPX-3 kits (Serac, Bad Homburg, Germany). The following loci were verified: D16S539, D18S51, D19S433, D21S11, D2S1338, D3S1358, D5S818, D8S1179, FGA, SE33, TH01, TPOX VWA.
Plasmid and gene transfection
For stable silencing of CD99, a shRNA plasmid (pSilencer 2.1-U6 Neo vector; Ambion, Grand Island, NY, USA) expressing CD99 siRNA-1 was transfected in IOR/CAR cell line as previously reported 19. Transient suppression of CD99 was performed by using siRNA-CD99 (Ambion) sense strand 5′-GGCUGGCCAUUAUUAAGUCtt-3′ and antisense strand 5′-GACUUAAUAAUGGCCAGCCtt-3′; ON-TARGETplus™ Non-Targeting siRNAs was used as a control (GE Healthcare, Piscataway, NJ, USA). Transient silencing of EWS-FLI1 was performed with siRNA sequences for EWS-FLI1–specific EF30 (ref. 60). Stable silencing of NF-kBp65 was obtained with plasmid shNF-kBp65(a), kindly provided by Dr. Johannes Schmid, University of Vienna. Transient overexpression of NF-kBp65 was induced by pCMVp65 construct (Obtained from Prof. Alain Israel, Departement de Biologie Cellulaire et Infection, Institute Pasteur, Paris, France). Plasmids and siRNAs were transfected into cells by Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. Cells were transfected with miR-34a mimics and non-specific control miRNAs (miRNC) (Applied Biosystem, Grand Island, NY, USA; assays # AM17101, #AM17110) 24 hours after cell seeding as previously described 39.
Flow cytometry
Cell lines were analyzed on a FACSCalibur flow cytometer (BD Transduction Labs, Lexington, KY, USA). Following antibody was used: anti-CD99 O13 (Signet Laboratories, Dedham, MA, USA).
Immunoblot analysis
Western blotting experiments were performed as previously described 19. The following antibodies were used: anti-Actin (Millipore, Billerica, MA, USA; cat. no. MAB1501); anti-CD99 (sc-53148), anti-Delta (sc-9102), anti-Delta-3 (sc-67270), anti-FLI1 (sc-356), anti-GAPDH (sc-25778), anti-Lamin-B (sc-6216), anti-NF-kBp65 (sc-372), anti-Notch 1 (sc-6014_R) and anti-Notch 3 (sc-5593) (Santa Cruz Biotechnology, Dallas, TX, USA); anti-Phopsho-NF-kBp65 (Ser536) (Cell Signaling Technology, Beverly, MA, USA; cat. no. 3031); anti-α-Tubulin (Sigma Aldrich; cat. no. T5168); anti-rabbit or anti-mouse antibodies conjugated to horseradish peroxidase (GE Healthcare; cat. no. NA934V, NA931V) were used as secondary antibodies. Proteins were visualized by incubating with ECL (EuroClone, Milan, Italy).
Immunofluorescence analysis
Immunofluorescence was performed on adherent cells grown on coverslips for 72 hours and fixed in 4% para-formaldehyde or in methanol/acetone 3:7, and permeabilized with 0.15% Triton X-100 in phosphate buffered saline (PBS), and incubated with the following antibodies: anti-β-III Tubulin (Sigma Aldrich; cat. no. T5076), anti-Neurofilament-H (Cell Signaling Technology; cat. no. 2836), anti-NF-kBp65 (Santa Cruz Biotechnology; cat. no. sc-372). Nuclei were counterstained by bisbenzimide Hoechst 33258 (Sigma Aldrich). Cell fluorescence was then evaluated by microscope Nikon Eclipse 90i (Nikon Instruments, Firenze, Italy).
Luciferase assay
30 000 cells/well were plated in triplicate and grown for 24 hours before transfection in a 24 well plate coated with 3μg/cm2 Fibronectin (Sigma Aldrich). Cells were transfected with 50 ng of the appropriate following responsive reporter: IgkB-conA-luciferase, conA-luciferase [all obtained from Prof. Alain Israel, Departement de Biologie Cellulaire et Infection, Institute Pasteur, Paris, France]; pRL-TK (Promega, Madison, WI, USA); SRE, E2F, CREB and AP1 cignal reporters (Qiagen, Hilden, Germany) by Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. Firefly luciferase activity was normalized to that of Renilla luciferase included as an internal control. Luciferase activity was measured by the Dual-Glo Luciferase Assay System according to manufacturer’s protocol (Promega) with a GloMax®-Multi+ Microplate Multimode Reader (Promega). When indicated, cells were treated with 1 ng/ml TNFα (Sigma Aldrich) 4 hours before measuring luciferase signal. When luciferase assays were performed in presence of exosomes, 20 000 cells/well were plated and grown as reported before. Then, the cells were transfected with 200 ng of the appropriate responsive reporter (IgkB-conA-luciferase, conA-luciferase, pRL-TK) by using Lipofectamine 2000 according to manufacturer’s protocol. After 24 hours, the same amounts of control (CTR) or CD99-null purified exosomes were incubated with recipient cells for 30 min at 37°C before performing luciferase assay.
Neural differentiation
EWS cells were seeded at low density (30 000/dish 60 mm) in standard medium. 72 hours later, immunofluorescence analysis was carried out to stain the markers of neurons (β-III Tubulin and Neurofilament-H).
DAPT treatment
EWS cells were treated with dimethyl sulfoxide (DMSO) (Sigma Aldrich) carrier alone or the γ-secretase inhibitor DAPT (Selleck Chemicals, Houston, TX, USA) at 2.5 μM for 48 hours.
RNA extraction, PCR and Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted by TRIzol (Invitrogen) following the manufacturer’s instructions. Reverse transcription, PCR amplification, and 1% agarose gel electrophoresis were done according to standard protocols. Primer pairs sequences used for PCR amplifications were as follows: (C-Rel Sense: 5′-TTGAAGACTGCGACCTCAATG-3′ , Antisense: 5′-GGGGCACGGTTATCATAAATTGG-3′ ; RelB Sense: 5′-TGATGTCCTTGGAGAGTTGAG-3′ , Antisense: 5′-GCAGGTCCAACATAGTGAAGA-3′ ; RelA Sense: 5′-AGCCTCCAGCCCCAGTGAAG-3′ , Antisense: 5′-CTCGGTAAAGCTGAGTTTGCG-3′ ; NF-kB1 Sense: 5′-TCCACTGTCTGCCTCTCTCGTC-3′ , Antisense: 5′-GCCTTCAATAGGTCCTTCCTGC-3′ ; NF-kB2 Sense: 5′-TCCACCTTTAAGTTGCCCTG-3′ , Antisense: 5′-TCTGCTCTCGTCATGTCACC-3′ ; ϐ-actin Sense: 5′-GCCGGGAAATCGTGCGTG-3′, Antisense: 5′-GGGTACATGGTGGTGCCG-3′). Quantitative Real-Time PCR was performed on ABI Prism 7900 (Applied Biosystems) using TaqMan HEY-1 assay (Applied Biosystems; cat. no. Hs00232618_m1 ) as previously reported 69. Primer Express software (Applied Biosystems) was used to design appropriate primer pairs for reference gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) 69. qRT-PCR analysis of miR-34a was performed using a TaqMan miRNAassay kit (Applied Biosystems) as previously described 39. Two replicates were considered. Relative quantification analysis was performed on ΔΔCt method 70.
Exosome isolation, characterization and functional assays
Exosomes (EXO) were isolated from 24 hours cell culture media by ultracentrifugation (UC) or Exoquick-TC (EQ) (System Biosciences) methods according to standards procedures or manufacturer’s instruction, with minor modifications. For exosome purification, serum was depleted of bovine exosomes by ultracentrifugation at 100 000 × g for 6 hours, followed by passing it through 0.2 μm filter prior to use. EXO protein concentration was determined using a protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). RNA was isolated from exosomes using the “Total RNA Purification micro Kit” (NorgenBioteK Corp, Thorold, ON Canada) and analyzed by qRT-PCR for miR-34a and miR-16 (Applied Biosystems; assays #000426 and #000391, respectively). Expression of miR-16 was used for normalizing secreted miRNAs as previously described 71, 72. For western blot analysis exosome samples were lysed in buffer (0.5% Triton; 300 mM NaCl; 50 mM Tris-NaCl) supplemented with protease inhibitor cocktail. Antibodies listed below were used in accordance to the manufacturer’s instructions: RAB5B (Santa Cruz Biotechnology; cat. no. sc-598); CD63 (cat. no. EXOAB_CD63A-1) and CD81 (cat. no. EXOAB_CD81A-1) from System Biosciences. The expression levels were evaluated by the AlphaView or Image Quant Software. For in vitro experimental model of fusion, the same amounts of exosomes, either purified from parental or CD99-shRNA EWS cell lines, were incubated with recipient cells for 24 hours at 37°C before performing expression studies and functional assays. Exosome-induced differentiation was evaluated by immunofluorescence after 3 days of culture in low serum condition.
Statistics
Differences among means were analyzed using 2-tailed Student’s t test. Differences were considered significant if P-values were < 0.05 (*) and 0.01 (**).
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Acknowledgements
We are in debt to Cristina Ghinelli for editing the manuscript. We wish to thank Dr. Gianfranco Mattia for the assessment of immunofluorescence analysis on the exosome fusion. We are also grateful to Prof. Alain Israel and Dr. Johannes Schmid for sharing NF-kB reporter and plasmids. This work was supported by grants from the Italian Association for Cancer Research (AIRC project: IG2013_14049 to KS; IG2012_13247 to AC), Ministry of Education, Research and Universities (FIRB project: RBAP11884 M_005 to KS), Austrian Science Fund (project: P24708-B21 and PROVABES project: I-1225-B19 to HK). Selena Ventura is in receipt of the fellowship “Guglielmina Lucatello e Gino Mazzega” granted by Fondazione Italiana per la Ricerca sul Cancro (FIRC project code: 13811).
Footnotes
Conflict of interest: All the authors declare no competing interests.
References
- 1.Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children’s Oncology Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magnan H, Goodbody CM, Riedel E, Pratilas CA, Wexler LH, Chou AJ. Ifosfamide dose-intensification for patients with metastatic Ewing sarcoma. Pediatr Blood Cancer. 2015;62:594–597. doi: 10.1002/pbc.25373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsberg JP, Goodman P, Leisenring W, Ness KK, Meyers PA, Wolden SL, et al. Long-term survivors of childhood Ewing sarcoma: report from the childhood cancer survivor study. Journal of the National Cancer Institute. 2010;102:1272–1283. doi: 10.1093/jnci/djq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nature genetics. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 6.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annual review of pathology. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer research. 2008;68:2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 8.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing’s sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11:1049–1054. [PubMed] [Google Scholar]
- 10.Maksimenko A, Malvy C. Oncogene-targeted antisense oligonucleotides for the treatment of Ewing sarcoma. Expert opinion on therapeutic targets. 2005;9:825–830. doi: 10.1517/14728222.9.4.825. [DOI] [PubMed] [Google Scholar]
- 11.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer discovery. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer discovery. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 13.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS genetics. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armengol G, Tarkkanen M, Virolainen M, Forus A, Valle J, Bohling T, et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridization. British journal of cancer. 1997;75:1403–1409. doi: 10.1038/bjc.1997.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Guerrero JA, Pellin A, Noguera R, Carda C, Llombart-Bosch A. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Laboratory investigation; a journal of technical methods and pathology. 2001;81:803–814. doi: 10.1038/labinvest.3780290. [DOI] [PubMed] [Google Scholar]
- 16.Huang HY, Illei PB, Zhao Z, Mazumdar M, Huvos AG, Healey JH, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 17.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 18.Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ. The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. The Journal of biological chemistry. 1997;272:30822–30827. doi: 10.1074/jbc.272.49.30822. [DOI] [PubMed] [Google Scholar]
- 19.Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. The Journal of clinical investigation. 2010;120:668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelin C, Aubrit F, Phalipon A, Raynal B, Cole S, Kaczorek M, et al. The E2 antigen, a 32 kd glycoprotein involved in T-cell adhesion processes, is the MIC2 gene product. The EMBO journal. 1989;8:3253–3259. doi: 10.1002/j.1460-2075.1989.tb08485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA. CD99 plays a major role in the migration of monocytes through endothelial junctions. Nat Immunol. 2002;3:143–150. doi: 10.1038/ni749. [DOI] [PubMed] [Google Scholar]
- 22.Bernard G, Breittmayer JP, de Matteis M, Trampont P, Hofman P, Senik A, et al. Apoptosis of immature thymocytes mediated by E2/CD99. J Immunol. 1997;158:2543–2550. [PubMed] [Google Scholar]
- 23.Alberti I, Bernard G, Rouquette-Jazdanian AK, Pelassy C, Pourtein M, Aussel C, et al. CD99 isoforms expression dictates T cell functional outcomes. FASEB J. 2002;16:1946–1948. doi: 10.1096/fj.02-0049fje. [DOI] [PubMed] [Google Scholar]
- 24.Miyagawa Y, Okita H, Nakaijima H, Horiuchi Y, Sato B, Taguchi T, et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing’s family tumor-like phenotypes to human mesenchymal progenitor cells. Mol Cell Biol. 2008;28:2125–2137. doi: 10.1128/MCB.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing’s family of tumors. Cancer Res. 2005;65:4633–4644. doi: 10.1158/0008-5472.CAN-04-2857. [DOI] [PubMed] [Google Scholar]
- 26.Amaral AT, Manara MC, Berghuis D, Ordonez JL, Biscuola M, Lopez-Garcia MA, et al. Characterization of human mesenchymal stem cells from ewing sarcoma patients. Pathogenetic implications. PloS one. 2014;9:e85814. doi: 10.1371/journal.pone.0085814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2013;32:3915–3921. doi: 10.1038/onc.2012.403. [DOI] [PubMed] [Google Scholar]
- 28.Charytonowicz E, Terry M, Coakley K, Telis L, Remotti F, Cordon-Cardo C, et al. PPARgamma agonists enhance ET-743-induced adipogenic differentiation in a transgenic mouse model of myxoid round cell liposarcoma. The Journal of clinical investigation. 2012;122:886–898. doi: 10.1172/JCI60015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Rorie CJ, Thomas VD, Chen P, Pierce HH, O’Bryan JP, Weissman BE. The Ews/Fli-1 fusion gene switches the differentiation program of neuroblastomas to Ewing sarcoma/peripheral primitive neuroectodermal tumors. Cancer research. 2004;64:1266–1277. doi: 10.1158/0008-5472.can-03-3274. [DOI] [PubMed] [Google Scholar]
- 32.Teitell MA, Thompson AD, Sorensen PH, Shimada H, Triche TJ, Denny CT. EWS/ETS fusion genes induce epithelial and neuroectodermal differentiation in NIH 3T3 fibroblasts. Lab Invest. 1999;79:1535–1543. [PubMed] [Google Scholar]
- 33.Gonzalez I, Vicent S, de Alava E, Lecanda F. EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. J Mol Med (Berl) 2007;85:1015–1029. doi: 10.1007/s00109-007-0202-5. [DOI] [PubMed] [Google Scholar]
- 34.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin Immunol. 2014;26:253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki CY, Barberi TJ, Ghosh P, Longo DL. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J Biol Chem. 2005;280:34538–34547. doi: 10.1074/jbc.M504943200. [DOI] [PubMed] [Google Scholar]
- 36.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Inhibition of nuclear factor kappab activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer. 2006;118:1930–1936. doi: 10.1002/ijc.21589. [DOI] [PubMed] [Google Scholar]
- 39.Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- 40.Marino MT, Grilli A, Baricordi C, Manara MC, Ventura S, Pinca RS, et al. Prognostic significance of miR-34a in Ewing sarcoma is associated with cyclin D1 and ki-67 expression. Ann Oncol. 2014;25:2080–2086. doi: 10.1093/annonc/mdu249. [DOI] [PubMed] [Google Scholar]
- 41.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem. 2010;336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogueira L, Ruiz-Ontanon P, Vazquez-Barquero A, Lafarga M, Berciano MT, Aldaz B, et al. Blockade of the NFkappaB pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene. 2011;30:3537–3548. doi: 10.1038/onc.2011.74. [DOI] [PubMed] [Google Scholar]
- 43.Sabolek M, Herborg A, Schwarz J, Storch A. Dexamethasone blocks astroglial differentiation from neural precursor cells. Neuroreport. 2006;17:1719–1723. doi: 10.1097/01.wnr.0000236862.08834.50. [DOI] [PubMed] [Google Scholar]
- 44.Ozeki A, Suzuki K, Suzuki M, Ozawa H, Yamashita S. Acceleration of astrocytic differentiation in neural stem cells surviving X-irradiation. Neuroreport. 2012;23:290–293. doi: 10.1097/WNR.0b013e3283509a79. [DOI] [PubMed] [Google Scholar]
- 45.De Molfetta GA, Luciola Zanette D, Alexandre Panepucci R, Dos Santos AR, da Silva WA, Jr, Antonio Zago M. Role of NFKB2 on the early myeloid differentiation of CD34+ hematopoietic stem/progenitor cells. Differentiation. 2010;80:195–203. doi: 10.1016/j.diff.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Vaira S, Johnson T, Hirbe AC, Alhawagri M, Anwisye I, Sammut B, et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc Natl Acad Sci U S A. 2008;105:3897–3902. doi: 10.1073/pnas.0708576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed AA, Sherman AK, Pawel BR. Expression of therapeutic targets in Ewing sarcoma family tumors. Hum Pathol. 2012;43:1077–1083. doi: 10.1016/j.humpath.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Javelaud D, Wietzerbin J, Delattre O, Besancon F. Induction of p21Waf1/Cip1 by TNFalpha requires NF-kappaB activity and antagonizes apoptosis in Ewing tumor cells. Oncogene. 2000;19:61–68. doi: 10.1038/sj.onc.1203246. [DOI] [PubMed] [Google Scholar]
- 49.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 50.Javelaud D, Poupon MF, Wietzerbin J, Besancon F. Inhibition of constitutive NF-kappa B activity suppresses tumorigenicity of Ewing sarcoma EW7 cells. Int J Cancer. 2002;98:193–198. doi: 10.1002/ijc.10192. [DOI] [PubMed] [Google Scholar]
- 51.Sonnemann J, Dreyer L, Hartwig M, Palani CD, Hong le TT, Klier U, et al. Histone deacetylase inhibitors induce cell death and enhance the apoptosis-inducing activity of TRAIL in Ewing’s sarcoma cells. J Cancer Res Clin Oncol. 2007;133:847–858. doi: 10.1007/s00432-007-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnemann J, Palani CD, Wittig S, Becker S, Eichhorn F, Voigt A, et al. Anticancer effects of the p53 activator nutlin-3 in Ewing’s sarcoma cells. Eur J Cancer. 2011;47:1432–1441. doi: 10.1016/j.ejca.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Lagirand-Cantaloube J, Laud K, Lilienbaum A, Tirode F, Delattre O, Auclair C, et al. EWS-FLI1 inhibits TNFalpha-induced NFkappaB-dependent transcription in Ewing sarcoma cells. Biochem Biophys Res Commun. 2010;399:705–710. doi: 10.1016/j.bbrc.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 54.von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One. 2011;6:e19305. doi: 10.1371/journal.pone.0019305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widera D, Mikenberg I, Kaltschmidt B, Kaltschmidt C. Potential role of NF-kappaB in adult neural stem cells: the underrated steersman? Int J Dev Neurosci. 2006;24:91–102. doi: 10.1016/j.ijdevneu.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 56.Yang C, Atkinson SP, Vilella F, Lloret M, Armstrong L, Mann DA, et al. Opposing putative roles for canonical and noncanonical NFkappaB signaling on the survival, proliferation, and differentiation potential of human embryonic stem cells. Stem Cells. 2010;28:1970–1980. doi: 10.1002/stem.528. [DOI] [PubMed] [Google Scholar]
- 57.Hess K, Ushmorov A, Fiedler J, Brenner RE, Wirth T. TNFalpha promotes osteogenic differentiation of human mesenchymal stem cells by triggering the NF-kappaB signaling pathway. Bone. 2009;45:367–376. doi: 10.1016/j.bone.2009.04.252. [DOI] [PubMed] [Google Scholar]
- 58.Cho HH, Shin KK, Kim YJ, Song JS, Kim JM, Bae YC, et al. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J Cell Physiol. 2010;223:168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- 59.Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- 60.Ban J, Bennani-Baiti IM, Kauer M, Schaefer KL, Poremba C, Jug G, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 2008;68:7100–7109. doi: 10.1158/0008-5472.CAN-07-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennani-Baiti IM, Aryee DN, Ban J, Machado I, Kauer M, Muhlbacher K, et al. Notch signalling is off and is uncoupled from HES1 expression in Ewing's sarcoma. J Pathol. 2011;225:353–363. doi: 10.1002/path.2966. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Chan SL, Miele L, Yao PJ, Mackes J, Ingram DK, et al. Involvement of Notch signaling in hippocampal synaptic plasticity. Proc Natl Acad Sci U S A. 2004;101:9458–9462. doi: 10.1073/pnas.0308126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baliko F, Bright T, Poon R, Cohen B, Egan SE, Alman BA. Inhibition of notch signaling induces neural differentiation in Ewing sarcoma. Am J Pathol. 2007;170:1686–1694. doi: 10.2353/ajpath.2007.060971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pang RT, Leung CO, Lee CL, Lam KK, Ye TM, Chiu PC, et al. MicroRNA-34a is a tumor suppressor in choriocarcinoma via regulation of Delta-like1. BMC Cancer. 2013;13:25. doi: 10.1186/1471-2407-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pata S, Otahal P, Brdicka T, Laopajon W, Mahasongkram K, Kasinrerk W. Association of CD99 short and long forms with MHC class I, MHC class II and tetraspanin CD81 and recruitment into immunological synapses. BMC Res Notes. 2011;4:293. doi: 10.1186/1756-0500-4-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I, et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res. 2007;13:2429–2440. doi: 10.1158/1078-0432.CCR-06-1762. [DOI] [PubMed] [Google Scholar]
- 68.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garofalo C, Mancarella C, Grilli A, Manara MC, Astolfi A, Marino MT, et al. Identification of common and distinctive mechanisms of resistance to different anti-IGF-IR agents in Ewing’s sarcoma. Mol Endocrinol. 2012;26:1603–1616. doi: 10.1210/me.2012-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Kok MG, Halliani A, Moerland PD, Meijers JC, Creemers EE, Pinto-Sietsma SJ. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J. 2015;29:3853–3862. doi: 10.1096/fj.15-271312. [DOI] [PubMed] [Google Scholar]
- 72.McDermott AM, Kerin MJ, Miller N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One. 2013;8:e83718. doi: 10.1371/journal.pone.0083718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.