Abstract
A map of the transcriptional organization of genes of an organism is a basic tool that is necessary to understand and facilitate a more accurate genetic manipulation of the organism. Operon maps are largely generated by computational prediction programs that rely on gene conservation and genome architecture and may not be physiologically relevant. With the widespread use of RNA sequencing (RNAseq), the prediction of operons based on actual transcriptome sequencing rather than computational genomics alone is much needed. Here, we report a validated operon map of Mycobacterium tuberculosis, developed using RNAseq data from both the exponential and stationary phases of growth. At least 58.4% of M. tuberculosis genes are organized into 749 operons. Our prediction algorithm, REMap (RNA Expression Mapping of operons), considers the many cases of transcription coverage of intergenic regions, and avoids dependencies on functional annotation and arbitrary assumptions about gene structure. As a result, we demonstrate that REMap is able to more accurately predict operons, especially those that contain long intergenic regions or functionally unrelated genes, than previous operon prediction programs. The REMap algorithm is publicly available as a user-friendly tool that can be readily modified to predict operons in other bacteria.
Keywords: operon, Mycobacterium tuberculosis, RNAseq
1. Introduction
The success of Mycobacterium tuberculosis (M. tuberculosis) as the etiological agent of tuberculosis disease is largely due to its ability to adapt to the wide range of stresses it faces upon entering its host [1]. These include hypoxia in granulomatous lesions [2], low nutrient conditions [3] and nitric oxide exposure upon macrophage activation [4]. In addition to inhabiting both intracellular environments within host macrophages and extracellular environments within lesions, M. tuberculosis bacilli are able to switch from periods of active growth to dormancy, giving rise to latent tuberculosis disease [5]. The ability of M. tuberculosis to thrive in diverse microenvironments and modulate growth accordingly is due to complex transcriptional regulation that is often unique to mycobacterial species [6].
Although transcriptional studies are widely used in M. tuberculosis research, a map of operon structure based on the M. tuberculosis transcriptome remains unavailable. To date, the M. tuberculosis operon databases in use have been computationally generated based on statistical modelling. For example, predictions of M. tuberculosis operons have been made by the Database for prOkaryotic OpeRons (DOOR) [7]. However, the algorithm used in DOOR is optimized for Escherichia coli (E. coli) and Bacillus subtilis data. Transcription in mycobacteria has been shown to differ from these models as M. tuberculosis does not consistently harbour consensus −35 element in 5’ UTRs and also shows a heavy reliance on alternative sigma factors, suggesting the use of alternative promoters for ORFs [8,9]. In addition, a recent study demonstrated that a quarter of genes in M. tuberculosis are expressed as leaderless transcripts which lack a 5’UTR and a classical ribosomal binding site [10]. These findings suggest that the use of computational methods built on classical transcription mechanisms may not be accurate for determining operon structure in M. tuberculosis.
Four operon maps have been predicted specifically for M. tuberculosis. Each map used a different prediction approach, including an annotation-based method using gene pathways and length of intergenic region (IGR) (BioCyc Pathway/Genome Database, MtbRvCyc) [11], a comparative method using cross-species conservation of gene proximity (The Institute of Genomic Research, TIGR) [12], a method that combined IGR length and M. tuberculosis microarray coexpression data [13] and a method that predicted operons based on IGR length, conserved gene clusters and transcriptional termination predictions (MycoperonDB) [14]. A recent review on the transcriptome of M. tuberculosis revealed significant discrepancies and little overlap between these four databases, suggesting that these methods of predicting operons are not optimal for M. tuberculosis [15]. Furthermore, the MtbRvCyc and TIGR databases, both based on methods in E. coli, showed the least overlap, indicating the importance of species-specific transcriptome analysis in the determination of transcriptional units of M. tuberculosis.
Experimental methods, such as RNase protection assays, have been used to delineate operon boundaries [16]. Despite the advantages of specificity and physiological relevance of these methods compared to computational predictions, they are not feasible for the prediction of transcriptional units on a genome-wide scale. RNA-sequencing technology (RNAseq) provides researchers with the ability to analyze the transcriptome at the resolution of a single nucleotide. This has revolutionized the mapping of ORFs and transcriptional units as well as the study of gene expression across all fields. A recent publication updated annotations in the TubercuList database, resulting in the correction of multiple ORF annotations in M. tuberculosis based on alternative start codon usage in RNAseq data [17]. RNAseq studies in Helicobacter pylori and M. tuberculosis have also revealed a wide prevalence of alternative transcriptional start sites within operons, suggesting the uncoupling of polycistrons under different environmental conditions [10,18]. Another currently available transcriptome-based operon prediction algorithm is included in the Rockhopper RNAseq analysis package, which bases its classification on intergenic distances and expression level correlation across experiments [19]. These studies highlight the importance of transcriptional unit mapping based on transcriptome analysis and suggest that an operon map cannot be generalized for expression of an organism under all conditions, but is highly specific for the conditions during which the transcriptome is analyzed.
However, given the widespread use of RNAseq by researchers who are not trained bioinformaticians, and the limits of current operon prediction programs for M.tuberculosis, we developed a user-friendly program, REMap (RNA Expression Mapping of operons), to predict operons based on RNAseq data. A similar method has also been used to study the transcriptome of Mycobacterium marinum [20]. In this study, we report an operon map of M. tuberculosis during both exponential and stationary phases of growth and validate the REMap predictions through comparisons to previously published operons as well as confirmation of newly predicted operons. This study reveals extensive co-transcription in the M. tuberculosis genome, with operons up to 14 genes in length and 58.4% of genes being transcribed in 749 operons during the exponential phase of growth. This map is also presented in the form of a heat map that provides researchers with a whole genome view of both operonic structure and expression within operons in M. tuberculosis. REMap and the heat map output enable researchers to analyze RNAseq data for transcriptional units and compare expression under the different conditions being tested.
2. Methods
2.1 Strains and Growth Conditions
A M. tuberculosis clinical isolate CDC1551 [21] carrying a plasmid, pMH94 [22], integrated at the attB site, was used in this study. By carrying an empty vector that is often used for introducing DNA into M. tuberculosis, this strain would be an ideal parent strain for gene deletion and complemented strains. M. tuberculosis was grown in Middlebrook 7H9 broth (Difco) supplemented with 0.5% glycerol, 10% oleic acid-albumin-dextrose-catalase (OADC), 0.05% Tween 80 under constant shaking at 37°C and total RNA was isolated from exponential (optical density of culture A600nm = 0.8) and stationary phases of growth (two days following peak optical density).
2.2 RNA Isolation and RNA Sequencing
Total RNA was extracted from duplicate 150 ml cultures of M. tuberculosis at both exponential and stationary phases. M. tuberculosis cultures were centrifuged and the bacterial pellet was resuspended in Trizol (Invitrogen). This mixture was transferred to 1.8ml O-ring tubes containing 0.5 ml of 0.1 mm zirconia beads (BioSpec Products). Cells were incubated at 25°C for 10 minutes, lyzed by six cycles of bead-beating for 30 seconds and cooling on ice for 1 minute, using a mini-beadbeater at 4,800 RPM. Lysed cells were centrifuged for 5 minutes at 13,000 RPM, the supernatant was transferred to a fresh microfuge tube and RNA was then extracted as described [23], followed by column purification with the RNeasy Mini Kit (Qiagen). 16S and 23S rRNA were removed from the sample with MICROBExpress Bacterial mRNA Enrichment Kit (Invitrogen). The quality of RNA was assessed using a Nanodrop (ND-1000, Labtech) and Agilent 2100 Bioanalyzer (Agilent Technologies) after each step of processing. cDNA library preparations, fragment library protocols, and all parameters followed standard SOLiD Applied Biosystems protocols. Sequence reads were aligned to M. tuberculosis CDC1551 reference sequence (EMBL Accession number AE000516) using the Bioscope v1.3 Whole Transcriptome plugin. Sequencing data was visualised using the Integrated Genome Viewer V2.2. RNA-sequencing and analysis was carried out at the Next Generation Sequencing Center, Johns Hopkins University.
2.3 Reverse-Transcription and Polymerase Chain Reaction
For reverse-transcription PCR (RT-PCR), 5 µg total RNA was treated with 2U of TURBO™ DNA-free DNase (Applied Biosystems) according to manufacturer’s instructions for 30 minutes, followed by addition of 2U of DNase for another 30 minutes. Reverse transcription was carried out using SuperScriptIII Reverse Transcriptase (Invitrogen) and a gene specific reverse primer complementary to the gene at the 3’ end of the IGR. Approximately 350ng of DNase-treated RNA, dNTPs and the reverse primer were incubated at 65°C for 5 minutes, then on ice for 1 minute. This was followed by the addition of FS buffer, DTT and either reverse transcriptase or water (negative control) to each tube according to manufacturer’s instructions. The final concentration of dNTPs was 1mM and the final concentration of the reverse primer was 1uM. Reverse transcription was conducted in an isothermal cycler at 55°C for 30 minutes, followed by 70°C for 15 minutes to generate cDNA. cDNA was amplified using Taq DNA Polymerase, Recombinant (Invitrogen). PCR samples were run on a 1.5% agarose gel for analysis. PCR amplification from gDNA and a cDNA sample without reverse transcriptase were used as a positive and negative control respectively. Gene specific primers were designed to amplify intergenic region between two flanking genes of an operon. Primers were designed to be at least 100bp upstream of the 3’ end of the 5’ gene and 100bp downstream from the 5’ end of the 3’ gene to avoid false positives that could arise from untranslated regions (UTR). Sequences of the primers used in this study are listed in Supplementary Table S1.
2.4 Algorithm Overview
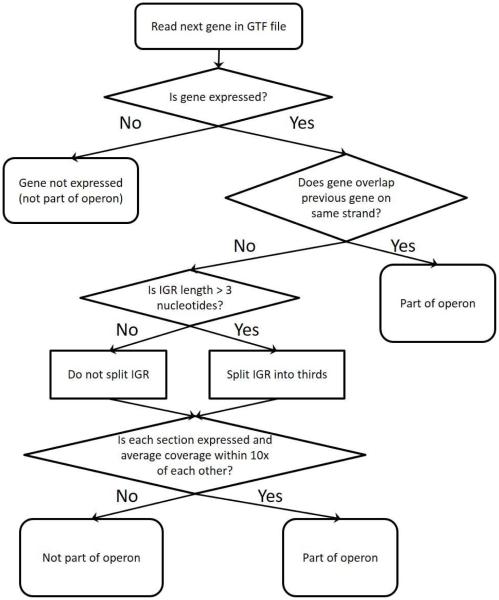
Figure 1 shows the decision tree used by REMap to group genes into operons. REMap takes as input a bam file containing the aligned RNAseq reads, some basic data on the reference genome, a user-input cutoff for expression level, a GTF file listing the genes in the reference genome (sorted by increasing position in the genome), and a location to write the results to (Supplementary Figure S1). The cutoff supplied by the user is used to define which genes are expressed. From these files, REMap determines if genes are in operons by calculating the average coverage for each gene and intergenic region (IGR). The IGR is defined as the region between the end of the previous gene and the beginning of the next gene. If a is defined as the start of the gene, and b as the end of the gene, coverage is calculated as
and
Figure 1. REMap algorithm.
A decision tree depicting how REMap determines if a gene is part of an operon.
where R is the set of reads whose primary alignment overlaps the region between a and b and are on the same strand as the gene. lr is the length of the portion of read r that aligns between a and b. In other words, the coverage is the sum of the section of each read whose primary alignment maps between a and b and nowhere else.
Two genes are considered part of an operon if (1) both genes are on the same strand, (2) both genes are expressed (average coverage is above the cutoff), and (3) the IGR is expressed. Regions of the genome are considered expressed if they are above the user supplied cutoff level. To determine if the IGR is expressed, if the IGR length is greater than 3 base pairs (bp), we divided the IGR into thirds and required that each third be expressed (average coverage is above the cutoff) and within 10x coverage of each other third. This helps prevent false positives resulting from high expression in the 5’ and 3’ untranslated regions (UTRs). If the IGR length is less than 3 bp, the IGR was not divided, and we just determined if the IGR is above the cutoff. If two genes on the same strand are overlapping, we automatically considered them part of the same operon. REMap also wraps around the genome, to account for the circular nature of most bacterial genomes, by determining whether any genes annotated at the beginning of the genome are part of any operons at the end of the annotated genome.
2.5 Histograms and Heatmaps
The histograms and heatmaps were generated in R using the ggplot2 package[24].
3. RESULTS
3.1 Algorithm
We have developed an algorithm, REMap, to analyze strand specific RNAseq data and predict co-transcribed genes. The algorithm coded in the REMap software is illustrated in Figure 1. REMap uses RNAseq data and a gene annotation file for an organism to identify and map operons. REMap generates an output file containing a list of operons, identities of genes that constitute each operon, and their levels of expression. In this study, we used RNAseq data obtained from sequencing ribodepleted RNA of M. tuberculosis CDC1551 isolated from cultures at exponential and stationary phases and its genome map [25] as input data. We tested multiple expression cutoffs to determine the optimal read density used to define whether a gene is expressed. This was based, first, on the accurate prediction of a positive control operon, the PDIM operon of M. tuberculosis [26], at exponential and stationary phases at a cutoff as low as 5 and second, the consistency of predicting the same proportion of independently transcribed genes in the genome when the cutoff was altered (Supplementary Table S2). Based on this analysis, we determined that 10 or more sequencing reads per nucleotide would ensure a conservative prediction of operons with limited false positives.
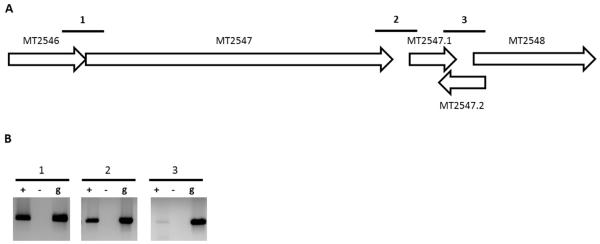
The IGR for two adjacent genes that are transcribed from the same strand was defined as the nucleotide sequence between the stop codon of the upstream gene and the putative translation start site of the downstream gene. While previous studies have used a short IGR length as a parameter to predict operons [27], REMap analysis does not require such assumptions and thereby eliminates bias resulting from arbitrary IGR length cutoffs. A further complex situation occurs when two adjacent genes on one strand are separated by a short IGR which encodes a gene on the complementary strand. This situation is further illustrated in Figure 2. MT2547.2 is encoded in the IGR between genes MT2547.1 and MT2548, but on the complementary strand. Existing algorithms would consider MT2547.1 and MT2548 to be transcribed separately because of the presence of MT2547.2. However, based on the RNAseq data we obtained, there is clear expression of the IGR on the same strand between MT2547.1 and MT2548, indicating that they are part of the same transcript. Consequently, REMap predicts both these genes to be part of the same operon. A detailed description of the parameters and method of analysis of REMap is described in the methods section.
Figure 2. Validation of the MT2546-MT2548 operon.
(A) Genetic locus of the predicted operon that spans MT2546-MT2548. Numbered bold lines indicate IGRs that are PCR amplified. The lengths of IGRs are IGR1:-1nt, IGR2:76n and IGR3: 41nt. (B) PCR amplified cDNA from IGRs run on a 1.0% agarose gel (Lane +). cDNA samples without reverse transcriptase (Lane −) and gDNA (Lane g) were used as negative and positive controls respectively.
3.2 REMap is validated by accurately predicting previously published and new operons.
To validate REMap, we first compared 15 published and extensively studied operons of M. tuberculosis to their DOOR and REMap predictions (Table 1). The majority of these published operons have been experimentally verified through co-transcription experiments, while a few (MT1014-MT1017, ABC fluoroquinolone efflux pump, narGHJI and inhA operons) have been determined through genomic conservation studies. As the expression of a significant proportion of the genome is reduced during stationary phase [13,28], we selected the REMap output obtained from analyzing RNA isolated from M. tuberculosis cultures at the exponential phase of growth. Predictions made by REMap and DOOR for 8 of the 15 operons were identical and matched the published data that initially described these operons. REMap predictions for two operons (MT3898-MT3900 and MT0509-MT0510) were identical to published data, but differed from DOOR predictions. We chose to study the five remaining operons that REMap predicted differently from published data in further detail.
Table 1.
Comparison of previously published operons to DOOR and REMap predictions.
| Published Operon |
Operon gene annotation/function |
DOOR Prediction | REMap Prediction | Ref |
|---|---|---|---|---|
| MT1455- MT1454 |
P27/P55 | MT1455-MT1454 | MT1455-MT1454 | [38] |
| MT2427- MT2428 |
smtB/furB | MT2427-MT2428 | MT2427-MT2428 | [39] |
| MT2506- MT2505 |
PE/PPE | MT2506-MT2505 | MT2506-MT2505 | [40] |
| MT3220- MT3218 |
Rv3134c/devR/devS | MT3220-MT3218 | MT3220-MT3218 | [37] |
| MT3168- MT3174 |
mymA operon | MT3168-MT3174 | MT3168-MT3174 | [41] |
| MT0960- MT0963 |
Phosphate transport operon | MT0960-MT0963 | MT0960-MT0963 | [42] |
| MT3988- MT3989 |
ESAT-6/CFP10 | MT3988-MT3989 | MT3988-MT3989 | [43] |
| MT1014- MT1017 |
Virulence operon | MT1014-MT1017 | MT1014-MT1017 | [44] |
| MT0509- MT0510 |
SenX3/RegX3 two component system |
MT0509,MT0510 | MT0509-MT0510 | [45] |
| MT3898- MT3900 |
dprE1/dprE2/aftA/embC | MT3897-MT3902 | MT3898-MT3900 | [46] |
| MT1524- MT1525 |
iip locus (macrophage invasion and intracellular persistence) |
MT1524-MT1525 | MT1523-MT1528 | [47] |
| MT2760- MT2762 |
ABC fluoroquinolone efflux pump |
MT2760-MT2762 |
MT2762 (MT2760 and
MT2721 expression below cutoff) |
[48] |
| MT0175- MT0187 |
mce1 (host cell invasion) operon |
MT0176,MT0178-
MT0183 |
MT0172-MT0187 | [31] |
| MT1198- MT1201 |
narGHJI (nitrate reductase) operon |
MT1198-MT1202 | MT1198-MT1202 | [49] |
| MT1530- MT1531 |
inhA operon | MT1530-MT1532 | MT1530-MT1532 | [30] |
Operons in bold indicate operons that differ from the published operon. Commas separate independently transcribed units.
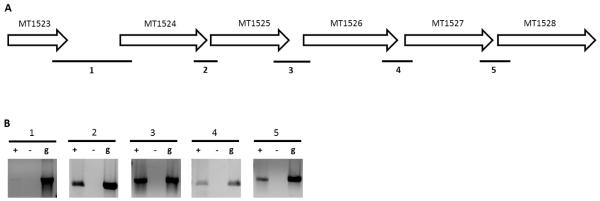
For two of these five operons, namely the iip locus and the ABC efflux pump operon, the prediction by DOOR was identical to the published operon, but REMap predictions differed. The ABC efflux pump operon is published as MT2760-MT2762, however REMap predicted MT2762 to be independently transcribed as expression of both MT2760 and MT2761 fell below the expression cut off and consequently, these genes were not considered by the program. REMap predicted the operon at the iip locus to be longer (MT1523-MT1528) than the operon currently published (MT1524-MT1525). To establish whether REMap prediction of this operon was more accurate, we carried out RT-PCR to determine if all the IGRs between MT1523 to MT1528 were co-transcribed. Our results (Figure 3) demonstrate that all the IGRs from MT1523 to MT1528 are expressed, indicating that the REMap prediction was more accurate than both the DOOR prediction and the currently published operon. RT-PCR of the IGR between MT1523 and MT1524 produces a faint band, suggesting that it is either an unstable transcript or is expressed at low levels under the exponential phase of growth.
Figure 3. Validation of the MT1523-MT1528 operon.
(A) Genetic locus of the MT1523-MT1528 predicted operon. Numbered bold lines indicate IGRs that are PCR amplified. The lengths of IGRs are IGR1:806nt, IGR2: 11nt, IGR3: 139nt. IGR4:49nt and IGR5: 11nt. (B) PCR amplified cDNA from IGRs run on a 1.0% agarose gel (Lane “+”). cDNA samples without reverse transcriptase (Lane ”−“) and gDNA (Lane “g”) were used as negative and positive controls respectively.
For another two of the five differentially predicted operons (narGHJI and inhA operons), predictions from REMap and DOOR were identical. The narGHJI operon is published as a well-studied nitrate reductase operon consisting of genes MT1198-MT1201 [29]. REMap and DOOR predictions extend this operon to include the downstream gene, MT1202, which encodes a tyrosine phosphorylated protein A and is functionally unrelated to the upstream nitrate reduction-associated genes. This may have been missed by the previous publications, which focused on the functional aspect of the nitrate reductase operon and consequently, may have omitted studying a downstream gene that is functionally unrelated. A similar explanation can be provided for the inhA operon, published as MT1530-MT1531, which encodes two genes involved in lipid metabolism but is predicted to also contain a third functionally unrelated, downstream gene (MT1532 encoding hemZ) by both DOOR and REMap. Both these operons have been determined through genome conservation, genetic manipulation and functional studies of the loci[29,30], but lack co-transcription data of all genes within the operons. Consequently, when both DOOR and REMap predict genes to be in operons based on transcriptional data, it can be expected that operons are predicted to be longer and to include functionally unrelated genes.
For the final differentially predicted operon (mce1), the prediction made by DOOR did not match that of REMap. The mce1 operon encodes genes involved in host cell invasion [31]. Consequently, this operon contains genes that function under conditions that differ from the conditions under which our RNAseq data was obtained. As a result, it is possible that the REMap prediction does not match published data, as transcripts from these operons were not being studied under physiologically relevant conditions.
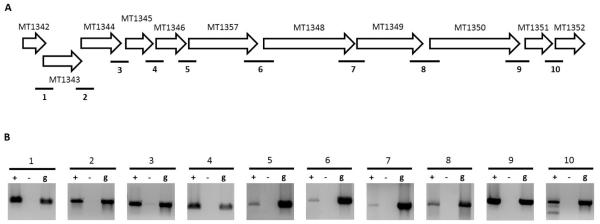
In addition to comparing REMap predictions to published operons, we also compared predictions to 10 operons predicted by a study by Roback et al. (Table 2) [13]. The latter study used M. tuberculosis microarray expression data in their bioinformatics prediction of operons. REMap predicted all operons to be longer than their Roback et al. predictions aside from MT3617-MT3618. The RNAseq read density for MT3618 was too low (<10 reads per nucleotide) for it to be considered as expressed by the REMap algorithm and consequently, the gene was not predicted to be part of an operon. Operon predictions based on algorithms such as that of Roback et al., may be shorter due to the inherent bias of using gene structure, such as IGR length, in the algorithm, causing them to ignore some gene pairs that are analysed by REMap. To determine if the REMap predictions were more accurate than the Roback et al. predictions, we selected the MT1344-MT1345 operon for further analysis. While the Roback et al. algorithm predicted a two gene operon for this locus, REMap predicted an 11 gene operon from MT1342-MT1352. Eight of these genes comprise the ATP synthase gene cluster [32] while the other three encode hypothetical proteins. The DOOR prediction for this locus split the 11 genes into three different transcripts, predicting them to be MT1342-MT1344, MT1345 and MT1346-MT1352. As a result, all three predictions for this locus differed. We carried out RT-PCR across the all the IGRs from MT1342-MT1352 and were able to identify expression from all IGRs (Figure 4). As a result, the REMap prediction appeared to be the most accurate.
Table 2.
Comparison of operons predicted by the Roback et al. study corresponding DOOR and REMap predictions.
| Roback et al. | DOOR | REMap |
|---|---|---|
| MT3004-MT3007 | MT2999-MT3009 | MT2998-MT3009 |
| MT1512-MT1513 | MT1507-MT1513 | MT1507-MT1513 |
| MT0300-MT0301 | MT0295-MT0303 | MT0293-MT0305 |
| MT0053-MT0052 | MT0052-MT0053 | MT0048-MT0053 |
| MT1344-MT1345 | MT1342-MT1344,MT1345,MT1346- MT1352 |
MT1342-MT1352 |
| MT1376-MT1376.1 | MT1373-MT1382 | MT1373-MT1382 |
| MT1874-MT1875 | MT1874,MT1875-MT1876 | MT1868- MT1874,MT1875-MT1877 |
| MT2816-MT2815 | MT2816,MT2814-MT2815,MT2817 | MT2814-MT2817 |
| MT3240-MT3241 | MT3233-MT3246 | MT3233-MT3246 |
| MT3617-MT3618 | MT3617,MT3618 | MT3617, MT3618 expression below cutoff |
Operons in bold indicate operons that are predicted identically between 2 programs. Commas separate independently transcribed units.
Figure 4. Validation of the MT1342-MT1352 operon.
(A) Genetic locus of the predicted operon that spans MT1342-MT1352. Numbered bold lines indicate IGRs that are PCR amplified. The lengths of IGRs are IGR1:-4nt, IGR2:-8nt, IGR3: 48nt. IGR4:29nt, IGR5: 6nt, IGR6: 44nt, IGR7:6nt, IGR8:39nt, IGR9:13nt and IGR10:7nt. Negative values indicate an overlap between genes flanking the IGR.(B) PCR amplified cDNA from IGRs run on a 1.0% agarose gel (Lane +). cDNA samples without reverse transcriptase (Lane −) and gDNA (Lane g) were used as negative and positive controls respectively.
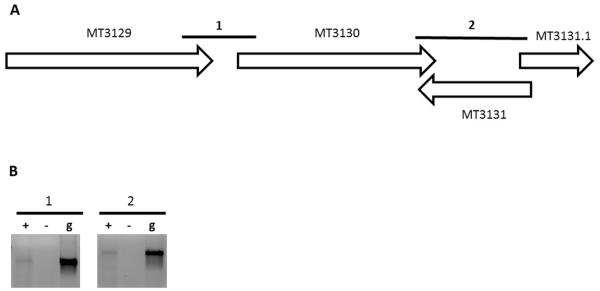
Finally, we validated the ability of REMap to accurately predict new operons for which there are no published reports. The first operon we selected was MT3129-MT3131.1. This was selected as, firstly, all three genes are functionally unrelated according to annotations in the TubercuList database [33] (MT3129 encodes an ATP synthase, MT3130 encodes an NADP-dependent alcohol dehydrogenase and MT3131.1 encodes a hypothetical protein), and secondly, the IGR between MT3130 and MT3131.1 contains a gene, MT3131, in the reverse orientation (Figure 5A). Consequently, this operon is one that would have defied operon parameters from most algorithms and is an example of an operon that would require transcriptional information to be predicted. Using RT-PCR, we found that all of the IGRs within this predicted operon are expressed, indicating that MT3129-MT3131.1 is transcribed as an operon (Figure 5B). Thus, REMap was able to accurately predict the operon at this locus. We selected a second predicted operon, MT2546-MT2548, for further validation. This operon is a four gene operon consisting of genes encoding two hypothetical proteins (MT2547.1 and MT2548) and two genes that are functionally unrelated (MT2546 encodes a globin and MT2647 encodes a probable alpha-glucosidase). The IGR between MT2547.1 and MT2548 also contains a gene in the reverse orientation (MT2547.2) (Figure 2A). We were able to amplify all IGRs through RT-PCR (Figure 2B), demonstrating that the REMap-predicted operon was expressed as a single transcript.
Figure 5. Validation of the MT3129-MT3131.1 operon.
(A) Genetic locus of the predicted operon that spans MT3129-MT3131.1. Numbered bold lines indicate IGRs that are PCR amplified. The lengths of IGRs are IGR1:69nt and IGR2: 336nt. (B) PCR amplified cDNA from IGRs run on a 1.0% agarose gel (Lane +). cDNA samples without reverse transcriptase (Lane −) and gDNA (Lane g) were used as negative and positive controls respectively.
3.3 The M. tuberculosis genome consists of at least 749 operons in exponential growth
REMap identified 2,448 genes (58.4% of all annotated genes) to be organized and expressed as 749 operons during the exponential phase of growth (Table 3; Supplementary Figure S2). A total of 762 genes were independently transcribed, and transcripts for the remaining 979 genes could not be detected during this phase of growth at an expression cutoff level of 10. During the stationary phase of growth, 860 genes are transcribed as independent units, 1,377 (32.9%) genes are co-transcribed as 494 operons, and 1,952 genes are not expressed using a cutoff of 10 (Supplementary Figure S2). Therefore, we conclude that at least 749 operons exist in M. tuberculosis. All operons predicted during the exponential phase of growth are listed in Supplementary Table S3, along with genomic coordinates, length, gene composition and average coverage. A similar table is provided for operons predicted during the stationary phase of growth (Supplementary Table S4). Operon predictions by gene are listed in Supplementary Tables S5 and S6 for exponential and stationary phases respectively.
Table 3.
Summary of operons identified by REMap for M. tuberculosis at stationary and exponential phase of growth using an expression cutoff level of 10.
| Exponential | Stationary | |||
|---|---|---|---|---|
| Number | % | Number | % | |
| Genes Not Expressed | 979 | 23.4 | 1952 | 46.6 |
| Independent Transcript | 762 | 18.2 | 860 | 20.5 |
| Genes Co-transcribed | 2448 | 58.4 | 1377 | 32.9 |
| Operons | 749 | 20.5 | 494 | |
Co-transcribed genes are defined as any genes that are in an operon (has met the criteria for being in an operon with at least one other gene).
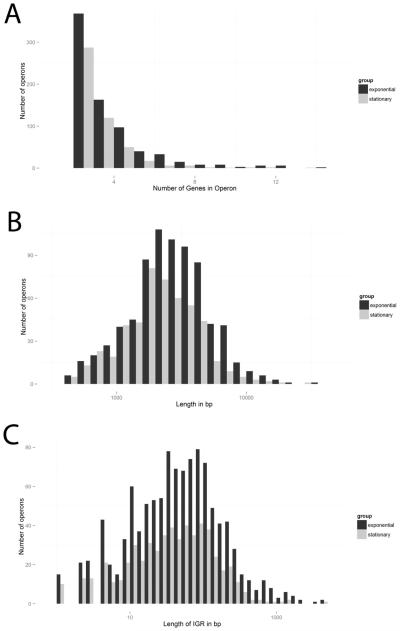
Gene densities in M. tuberculosis operons follow a skewed distribution, with the smallest operons, consisting of two genes, being the most numerous (Figure 6A). The two largest operons predicted in exponential phase consisted of 14 genes. The first contains the mce1 operon discussed earlier and the second is MT3233-MT3246. The latter operon was predicted to be a two gene operon by Roback et al. (MT3240-MT3241, Table 2) but was identically predicted to contain 14 genes by the DOOR algorithm. All 14 genes comprise the nuo gene cluster that encodes the subunits for NADH dehydrogenase I [34]. This gene cluster is an important component of the bacterial respiratory chain and has been demonstrated to be transcribed as an operon in Salmonella typhimurium [35].
Figure 6. Statistical analysis of operons of M. tuberculosis at exponential and stationary phases of growth identified by REMap.
(A) Histogram of the number of genes in an operon. (B) Histogram of the length of the operons in base pairs (bp). (C) Histogram of the length of the intergenic region (IGR) in bp for each pair of genes in all operons. For all histograms, an operon must have at least 2 genes, thus these histograms exclude individually transcribed genes and un-expressed genes. Black bars are exponential phase and gray bars are stationary phase.
We explored the distribution of the lengths of different operons and also observed a skewed distribution (Figure 6B). The largest proportion of operons fell in the 3-7kb range. The longest operon predicted is the PDIM operon (MT2998-MT3009) which we used to optimize the program. This consists of 12 genes and is approximately 34kb in length. The second longest operon predicted is MT2441-MT2451, which consists of 11 genes and is 23.8kb in length. The latter operon contains the mycobactin biogenesis gene cluster (mbt) as well as four downstream genes [36].
Finally, we explored the distribution of IGR length in predicted operons. The earlier study of operons in M. tuberculosis found the median of IGR length distribution of known operons to be around zero nucleotides in length [13]. The IGR length distribution we obtained (Figure 6C) shows that the largest proportion of operons contain IGRs that vary from 10-200 nucleotides in length. A few co-transcribed IGRs extend over 1kb in length. An example is the 2.47kb long IGR between MT0696 and MT0699, which are both encoded in the forward direction (Supplementary Figure S3). This IGR also contains a gene, MT0697 in the reverse direction. Consequently, the IGR could potentially encode an MT0697-complementary regulatory element, such as a small, non-coding RNA, that could account for the transcription in that direction. Alternatively, it could encode an ORF that is yet to be discovered or missing in the current annotation. In conclusion, there is greater co-transcription of IGRs than initially observed and assumed, with many regions requiring further exploration to resolve confusing annotations.
3.4 Expression can be highly variable within operons
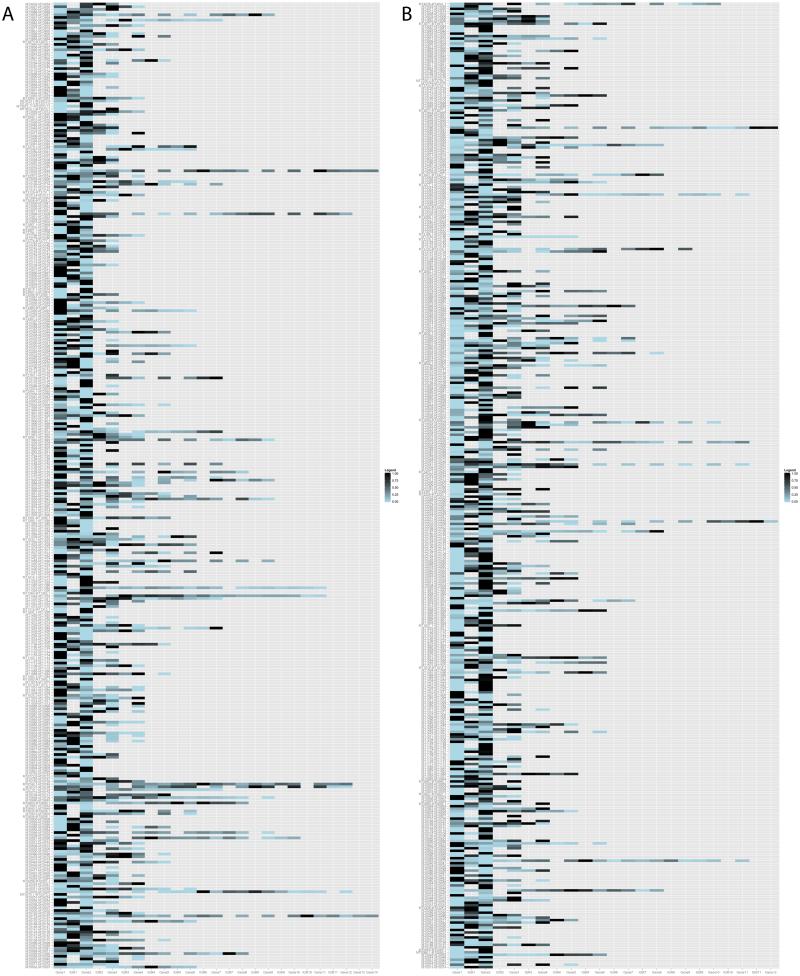
We also analyzed levels of expression of genes within each operon, normalized their levels to the most highly expressed gene within the operon and generated a heat map of all operons in M. tuberculosis expressed in exponential and stationary phases of growth (Figure 7 and Supplementary Figure S4, respectively). As an operon is transcribed as a single polycistronic message, we could expect the 5’ end to be more highly expressed than the 3’ end due to RNA polymerase processivity. In contrast, we did not observe a consensus trend for variation in expression, as the most highly expressed gene within the operon could be found at the 5’, 3’ or middle of an operon. This variation suggests that although these genes are transcriptionally coupled, post-transcriptional mechanisms, such as degradation of part of an mRNA, or internal promoters and suboperons, exist to regulate abundance of each transcript.
Figure 7. Variation in expression levels of ORFs within an operon in exponential phase of growth.
Heatmap of gene expression for all operons on the (A) forward or (B) reverse strand. The intergenic region (IGR) is blank if the two flanking genes overlap each other.
4.0 DISCUSSION
REMap is a robust algorithm that uses strand-specific RNA sequence reads to map operons. The length of an IGR does not affect analysis by REMap, and therefore it is able to identify co-transcribed genes that are separated by IGRs longer than the normally accepted length for an IGR within an operon [27]. It also does not rely on genomic conservation or gene function, and therefore avoids bias resulting from poor gene annotation. Another important feature of REMap, that sets it apart from other RNAseq based algorithms such as Rockhopper, is its ability to map and cluster transcripts on each DNA strand at a locus, and consequently identify operons even when an ORF exists in the complementary strand. In other words, REMap considers each strand of the chromosome as a discrete entity, aligns experimentally obtained RNA sequences directly onto the coding strand, and assesses if a polycistronic transcript exists, allowing it to combine many operons that are normally split in two due to the presence of a gene on the opposite strand. Additionally, REMap can generate operon maps for individual RNAseq data sets, and therefore is able to identify if operonic structure varies under different conditions such as growth, stress and physiology. For example, in this study we were able to assess differences in operon arrangements in exponential and stationary phases of growth and conclude that complex transcriptional regulation of operons occur at these growth phases. While we have focused on M. tuberculosis, REMap could also be applied to RNAseq data from other bacterial organisms.
Despite these strengths, an error in annotation of ORFs affects REMap output, just as it affects many other prediction algorithms. The program predicted 19 IGRs longer than 1kb in length to be co-transcribed with their flanking genes. On inspection, many of these IGRs did not possess a uniform expression and instead had peaks of higher abundance. These IGRs could potentially encode an ORF that is missing in the gene annotation files used. This is the case with the IGR between MT1547-MT1553, which is the longest transcribed IGR in a predicted operon. This IGR is 4.9kb in length but the annotation file is missing a gene, MT1552. Consequently, MT1547 and MT1553 are predicted to be co-transcribed due to the high expression of MT1552. These long, co-transcribed IGRs could also contain new ORFs that have yet to be annotated or non-coding RNAs that have yet to be discovered. An example would be the 3.19kb IGR between MT0562 and MT0566, which contains a spike in expression in the centre of the IGR (Supplementary Figure S5).
Inherent to using RNAseq data is that REMap will identify operons specific to the conditions in which the RNA was extracted. Since we used ribodepleted RNA isolated from exponential and stationary phases of growth, REMap identified operons that are expressed under these conditions. This also explains the differences in some REMap predictions of operons that have been published with different gene compositions. If an operon exists but is not expressed in the exponential or stationary phases of growth, REMap did not identify the operon in this study, and therefore it is not reported here. However, use of RNA isolated under multiple physiological and growth conditions should afford a comprehensive identification of operons in M. tuberculosis by REMap, and may provide interesting insights into gene expression under varying conditions.
In summary, this study represents the first attempt to map operons in M. tuberculosis based on empirical RNAseq data. We carried out extensive validation of the program and showed that REMap is able to accurately predict previously published operons, refine and correct previously published operons, and accurately identify new operons in the M. tuberculosis genome. In addition, the operon predictions generated by REMap highlight more extensive co-transcription that initially assumed. REMap predicts longer ORFs and longer co-transcribed IGRs. Furthermore, many ORFs that are not functionally related were found to be co-transcribed. These findings challenge the current assumptions of operon organization and highlight the importance of the transcriptome in mapping operons. In addition, many known operons, such as the Rv3134c/devR/devS[37] and P27/P55 operons[38], are transcribed from multiple promoters. The heatmap we generated shows great variation in expression within operons, which indicates a potentially important role for internal promoters and sub-operons in the regulation of gene expression in M. tuberculosis. REMap is thus a direct and simple yet novel approach to map operons that will aid in refining the genome annotation of M. tuberculosis and potentially other organisms as well.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health award DP2OD008459 to GL.
Abbreviations
- REMap
RNA Expression Mapping of operons
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SP, KW and GL contemplated the study. SP undertook microbiology and genetics studies. KW coded the algorithm. SP, KW, FX, RLS and GL analyzed the data. WRB provided material support. SP, KW and GL wrote the manuscript.
Supplementary data related to this article can be accessed at
References
- 1.Zumla A, George A, Sharma V, Herbert N, Baroness Masham of I WHO's 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–1767. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 2.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dannenberg AM., Jr. Immunopathogenesis of pulmonary tuberculosis. Hosp Pract (Off Ed) 1993;28:51–58. doi: 10.1080/21548331.1993.11442738. [DOI] [PubMed] [Google Scholar]
- 4.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manabe YC, Kesavan AK, Lopez-Molina J, Hatem CL, Brooks M, et al. The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis (Edinb) 2008;88:187–196. doi: 10.1016/j.tube.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton-Foot M, Gey van Pittius NC. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 2013;93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37:D459–463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashyam MD, Kaushal D, Dasgupta SK, Tyagi AK. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole ST. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology. 2002;148:2919–2928. doi: 10.1099/00221287-148-10-2919. [DOI] [PubMed] [Google Scholar]
- 10.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, et al. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero PR, Karp PD. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics. 2004;20:709–717. doi: 10.1093/bioinformatics/btg471. [DOI] [PubMed] [Google Scholar]
- 12.Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roback P, Beard J, Baumann D, Gille C, Henry K, et al. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res. 2007;35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranjan S, Gundu RK, Ranjan A. MycoperonDB: a database of computationally identified operons and transcriptional units in Mycobacteria. BMC Bioinformatics. 2006;7(Suppl 5):S9. doi: 10.1186/1471-2105-7-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haller R, Kennedy M, Arnold N, Rutherford R. The transcriptome of Mycobacterium tuberculosis. Appl Microbiol Biotechnol. 2010;86:1–9. doi: 10.1007/s00253-009-2399-4. [DOI] [PubMed] [Google Scholar]
- 16.Lynch D, O'Brien J, Welch T, Clarke P, Cuiv PO, et al. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lew JM, Mao C, Shukla M, Warren A, Will R, et al. Database resources for the tuberculosis community. Tuberculosis (Edinb) 2013;93:12–17. doi: 10.1016/j.tube.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 19.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Dong X, Zhu Y, Wang C, Sun G, et al. Revealing of Mycobacterium marinum transcriptome by RNA-seq. PLoS One. 2013;8:e75828. doi: 10.1371/journal.pone.0075828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, et al. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- 22.Lee MH, Pascopella L, Jacobs WR, Jr., Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatfull GF, Jacobs WR., Jr. Some Common Methods in Mycobacterial Genetics. In: Hatfull GF, Jacobs WR Jr., editors. Molecular Genetics of Mycobacteria. ASM; Washington DC: 2000. pp. 313–320. [Google Scholar]
- 24.Wickham H. ggplot2: elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- 25.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camacho LR, Constant P, Raynaud C, Laneelle MA, Triccas JA, et al. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 27.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci U S A. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Goh KS, Rastogi N, Berchel M, Huard RC, Sola C. Molecular evolutionary history of tubercle bacilli assessed by study of the polymorphic nucleotide within the nitrate reductase (narGHJI) operon promoter. J Clin Microbiol. 2005;43:4010–4014. doi: 10.1128/JCM.43.8.4010-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 31.Casali N, White AM, Riley LW. Regulation of the Mycobacterium tuberculosis mce1 operon. J Bacteriol. 2006;188:441–449. doi: 10.1128/JB.188.2.441-449.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sala C, Haouz A, Saul FA, Miras I, Rosenkrands I, et al. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol Microbiol. 2009;71:1102–1116. doi: 10.1111/j.1365-2958.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- 33.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 35.Archer CD, Elliott T. Transcriptional control of the nuo operon which encodes the energy-conserving NADH dehydrogenase of Salmonella typhimurium. J Bacteriol. 1995;177:2335–2342. doi: 10.1128/jb.177.9.2335-2342.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 37.Bagchi G, Chauhan S, Sharma D, Tyagi JS. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology. 2005;151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 38.Bigi F, Alito A, Romano MI, Zumarraga M, Caimi K, et al. The gene encoding P27 lipoprotein and a putative antibiotic-resistance gene form an operon in Mycobacterium tuberculosis and Mycobacterium bovis. Microbiology. 2000;146:1011–1018. doi: 10.1099/00221287-146-4-1011. Pt 4. [DOI] [PubMed] [Google Scholar]
- 39.Milano A, Branzoni M, Canneva F, Profumo A, Riccardi G. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res Microbiol. 2004;155:192–200. doi: 10.1016/j.resmic.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Tundup S, Akhter Y, Thiagarajan D, Hasnain SE. Clusters of PE and PPE genes of Mycobacterium tuberculosis are organized in operons: evidence that PE Rv2431c is co- transcribed with PPE Rv2430c and their gene products interact with each other. FEBS Lett. 2006;580:1285–1293. doi: 10.1016/j.febslet.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 41.Singh A, Jain S, Gupta S, Das T, Tyagi AK. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol Lett. 2003;227:53–63. doi: 10.1016/S0378-1097(03)00648-7. [DOI] [PubMed] [Google Scholar]
- 42.Torres A, Juarez MD, Cervantes R, Espitia C. Molecular analysis of Mycobacterium tuberculosis phosphate specific transport system in Mycobacterium smegmatis. Characterization of recombinant 38 kDa (PstS-1) Microb Pathog. 2001;30:289–297. doi: 10.1006/mpat.2001.0434. [DOI] [PubMed] [Google Scholar]
- 43.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. Pt 11. [DOI] [PubMed] [Google Scholar]
- 44.Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, et al. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol. 2006;23:1129–1135. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- 45.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 46.Goude R, Amin AG, Chatterjee D, Parish T. The critical role of embC in Mycobacterium tuberculosis. J Bacteriol. 2008;190:4335–4341. doi: 10.1128/JB.01825-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao LY, Pak M, Kish R, Kajihara K, Brown EJ. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun. 2006;74:1757–1767. doi: 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasca MR, Guglierame P, Arcesi F, Bellinzoni M, De Rossi E, et al. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2004;48:3175–3178. doi: 10.1128/AAC.48.8.3175-3178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stermann M, Bohrssen A, Diephaus C, Maass S, Bange FC. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J Clin Microbiol. 2003;41:3252–3259. doi: 10.1128/JCM.41.7.3252-3259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.