Abstract
An individual’s risk of developing a common disease typically depends on an interaction of genetic and environmental factors. Epigenetic research is uncovering novel ways through which environmental factors such as diet, air pollution, and chemical exposure can affect our genes. DNA methylation and histone modifications are the most commonly studied epigenetic mechanisms. The role of long non-coding RNAs (lncRNAs) in epigenetic processes has been more recently highlighted. LncRNAs are defined as transcribed RNA molecules greater than 200 nucleotides in length with little or no protein-coding capability. While few functional lncRNAs have been well characterized to date, they have been demonstrated to control gene regulation at every level, including transcriptional gene silencing via regulation of the chromatin structure, and DNA methylation. This review aims to provide a general overview of lncRNA function with a focus on their role as key regulators of health and disease, and as biomarkers of environmental exposure.
Keywords: lncRNA, disease, chemicals, toxicology, epigenetics, smoking
Introduction
Several common diseases, such as cardiovascular disease, diabetes, and cancer, as well as neurodegenerative disorders, have genetic components, but are rarely caused by single genes or chromosomal abnormalities. Instead, a combination of genetic and environmental factors usually interact to influence an individual’s risk of disease [1–5]. Epigenetics represents a promising research approach to understand this interaction. Epigenetic alterations modify the activation of certain genes, without directly affecting the DNA sequence. These modifications change in response to environmental stimuli, and can cause our cells to alter their gene expression. Epigenetic research is uncovering novel paths through which environmental factors such as diet, air pollution, and chemical exposure can affect our genes [6–8]. DNA methylation and histone modifications are the most commonly studied epigenetic mechanisms. The role of non-coding RNA (ncRNAs), and especially long non-coding RNAs (lncRNAs), in epigenetic processes has been more recently highlighted.
LncRNAs are defined as transcribed RNA molecules greater than 200 nucleotides in length with little or no protein-coding capability [9]. As opposed to microRNAs (miRNAs) which are involved in transcriptional and post-transcriptional gene silencing via specific base pairing with their targets, lncRNAs regulate gene expression by diverse mechanisms that are not yet fully understood [9–12]. Although only a few functional lncRNAs have been well characterized they have been demonstrated to control gene regulation at every level including transcriptional gene silencing via DNA methylation and regulation of the chromatin structure [10, 13–16]. This review aims to provide a general overview of epigenetic control regulated by lncRNAs with a focus on their role as key regulators of health and disease, and as novel biomarkers of environmental exposure.
LncRNAs
The central dogma of molecular biology is centered on protein-coding genes, with DNA as carriers of the genetic information and RNA mediating information transfer from DNA to proteins that play important structural or functional roles essential for all aspects of life. This limited view of RNA function has been revised over the past decade with the discovery that animal genomes are subjected to pervasive transcription resulting in a variety of ncRNAs that play key roles in cellular functions beyond those previously attributed to RNA [17–20]. In fact, protein-coding genes represent only a small portion of the human genome (<2%), whereas the major part is transcribed into ncRNAs [18, 21]. NcRNA research has primarily focused on miRNAs, a small subclass of ncRNAs that regulate gene expression. More recently the attention has shifted to the larger, in both number and size, lncRNAs. More than 80% of the mammalian genome transcription is estimated to result in lncRNA generation, but molecular mechanisms and functional importance have been described only for a few [20, 22, 23]. LncRNAs, in contrast to many miRNAs, do not show strong evolutionary conservation and were originally classified as non-functional, transcriptional noise [24]. However, accumulating evidence has shown that lncRNAs are important players in diverse biological processes including: gene regulation, genome packing, chromatin organization, dosage compensation, and genomic imprinting [25–29]. They perform crucial roles in the control of gene expression during development and differentiation processes, and the number of lncRNA transcripts increases with the complexity of organisms—indicating the importance of lncRNA-based regulatory mechanisms in the evolution of multicellular organisms [30, 31].
Classification
LncRNAs are a heterogeneous class of RNAs, varying in nucleotide length (200 nucleotides to over 100 kilobases), cellular location (nucleus, cytoplasm, or both), and functional role in various biological processes. LncRNAs are often expressed at low levels and are generally more cell type-specific than the expression of protein-coding genes [30, 32, 33]. Similar to mRNA, many lncRNAs are transcribed by RNA polymerase II (RNA pol II), capped, spliced, and polyadenylated [28, 34]. In addition, they can be transcribed from both sense and antisense strands of the genome [35]. LncRNAs can be classified by their genomic position relative to protein-coding genes and include the long intergenic ncRNA (lincRNA), intronic lncRNA, antisense lncRNA, transcribed pseudogene lncRNAs, and enhancer RNA [36].
Cellular functions
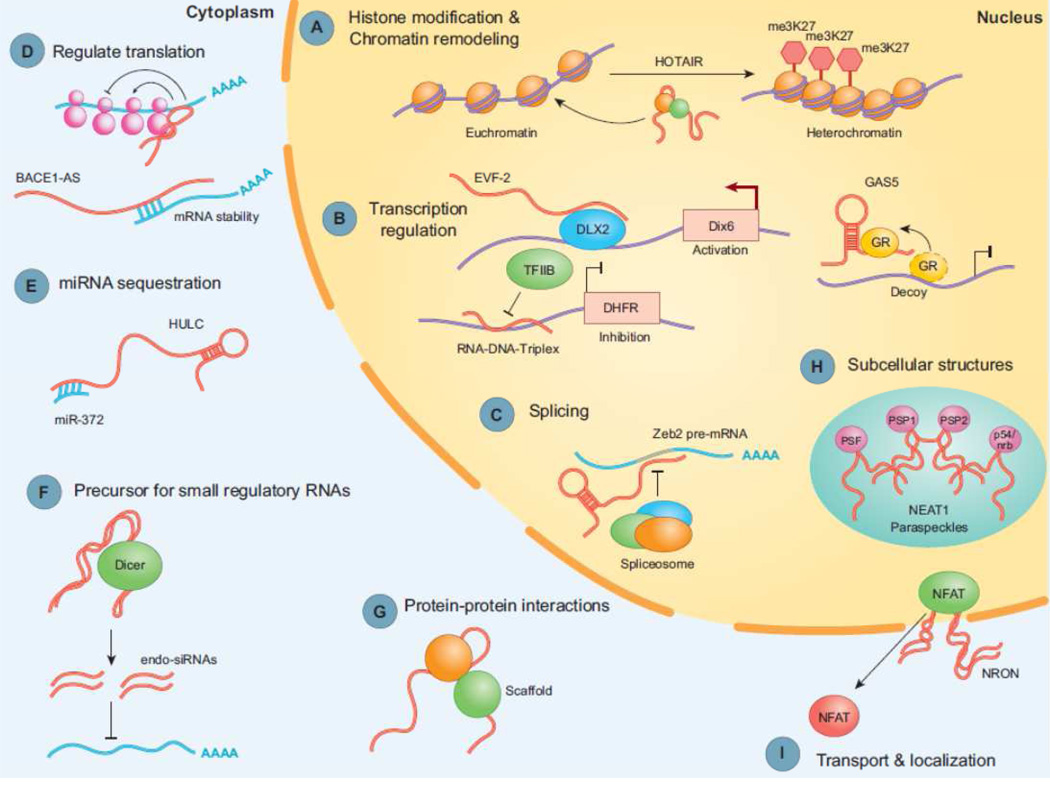
The intrinsic nucleic acid nature of lncRNAs gives them the dual ability to function as ligands for proteins and to mediate base-pairing interactions that guide lncRNA-containing complexes to specific RNA or DNA target sites, explaining their involvement in many regulatory functions [37–39]. Figure 1 shows examples of lncRNA cellular functions. In contrast to small ncRNAs, lncRNAs can fold into complex secondary and higher order structures, increasing the potential for both protein and target recognition [37–40]. LncRNAs can inhibit or facilitate the recruitment of RNA pol II, transcription factors and/or cofactors to gene promoters, thereby controlling transcription of target genes. They can also regulate alternative splicing of pre-mRNAs [41]. Moreover, their flexible scaffold nature enables lncRNAs to join multiple protein factors that would not interact or functionally cooperate if they relied solely on protein–protein interactions [37, 39, 42]. The scaffold function is also important for protein activity and localization as well as subcellular structures [43, 44]. Additionally, lncRNAs can guide specific chromatin remodeling complexes (e.g. Polycomb Repressive Complex 2; PRC2) to the correct chromosomal locations in cis or in trans providing target specificity to these complexes. Through these interactions lncRNAs can control local or global chromatin packing and the balance between transcriptionally active euchromatin and silent heterochromatin [28, 45]. Furthermore, lncRNAs can base-pair with mRNA molecules and affect their stability or translation as well as compete for miRNA binding and thereby preventing their function [46, 47]. Many lncRNAs may be processed into short ncRNAs such as siRNAs that can downregulate gene expression by degrading the mRNA transcripts [48–52]. Some lncRNAs may actually code for peptides and small proteins [53, 54].
Figure 1.
Example of lncRNA cellular functions. LncRNAs can bind to DNA, RNA, and proteins and act in diverse ways within the cell. LncRNAs regulate gene expression by multiple mechanisms. They can guide chromatin remodeling complexes to the correct chromosomal locations controlling the balance between transcriptionally active euchromatin and silent heterochromatin both locally and globally (A). Furthermore, lncRNAs can inhibit or facilitate the recruitment of RNA pol II, transcription factors and/or cofactors to gene promoters, thereby controlling transcription of target genes (B). They can regulate alternative splicing of pre-mRNAs and thereby contribute to the transcriptome complexity (C). Moreover, they can affect the stability and translation of mRNA by base-pairing with mRNA molecules (D). LncRNAs can compete for miRNA binding and thereby preventing their function and influencing the expression of miRNA target gene expression (E). They can also be processed into small, single- or double-stranded RNAs that act as siRNAs and target other RNAs, which subsequently could result in target degradation (F). Their flexible scaffold nature enables lncRNAs to join multiple protein factors that would not interact or functionally cooperate if they only relied on protein–protein interactions (G). The scaffold function is also important for protein activity and localization as well as subcellular structures (H, I). (Adapted from: Gutschner T, Diederichs S: The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012 Jun;9(6):703–19) [59].
lncRNAs, cancer and other disease
There is accumulating evidence that lncRNAs are important regulators of physiological and pathological responses [55–57]. Table 1 shows examples of lncRNAs that have been associated with human disease. The function of lncRNAs has mostly been studied in relation to tumorigenesis where they play important roles in the regulatory mechanisms of oxidative stress, inflammation, apoptosis, cell growth, and viability [23, 58–61]. For example, the HOX transcript antisense RNA (HOTAIR) plays a key role in gene regulation and chromatin dynamics and is misregulated in a variety of cancers [60, 62]. HOTAIR functions as a molecular scaffold that links the two histone modification complexes PRC2 and Lysine-specific histone demethylase 1 (LSD1) and modifies chromatin structure in trans to induce gene silencing and promote cancer cell proliferation, progression, and metastasis (Figure 1A) [60, 62]. HULC (highly upregulated in liver cancer) is another lncRNA that is associated with cancer. It can sequester and inhibit the function of several miRNAs, including the tumor suppressor miR-372 (Figure 1E)[47, 63] However, dysregulation of lncRNA is not only associated with several types of cancers but a variety of human diseases including: cardiovascular disease [64–66], diabetes [67], membranous nephropathy [68], neural pathogenesis such as autism spectrum disorder and Alzheimer’s disease [57, 69, 70], and alterations of both the innate and adaptive immune system [71, 72]. Moreover, recent studies have revealed that lncRNAs also are localized within extracellular vesicles (EVs) and may be important for long distant intercellular communication and function as biomarkers [73–76].
Table 1.
LncRNAS: Examples of biological functions and associations with human disease
| Disease | lncRNA | Status1 | Molecular mechanisms/role in disease | Ref. |
|---|---|---|---|---|
| Colorectal cancer (CRC) |
PINT | ↓ | PINT acts as a tumor suppressor that reduces cell proliferation by regulating the expression of genes involved in p53 signaling via a PRC2-dependent mechanism. |
[130] |
| Liver tumor | HULC | ↑ | HULC act as a molecular sponge that can bind and inhibit the function a number of miRNA, including the tumor suppressor miR-372. |
[47, 63] |
| Breast, uterus, ovary tumors |
SRA | ↑ | SRA forms ribonucleoprotein complexes with a number of nuclear receptors generally acting to stimulate transcriptional activation. SRA is a potential biomarker of steroid-dependent tumors |
[131, 132] |
| Breast, colorectal tumors, prostate cancer etc |
HOTAIR | ↑ | HOTAIR function as a molecular scaffold to link and target PRC2 and LSD1, leading to chromatin remodeling via H3K27 methylation and H3K4 demethylation and silencing genes implicated in inhibiting cancer progression/metastasis. |
[29, 62, 133] |
| Breast tumor, type 2 diabetes |
GAS5 | ↓ | GAS5 act as a decoy and competes for binding to the DNA binding domain of the glucocorticoid receptor. GAS5 expression induces growth arrest and apoptosis. Decreased serum levels of GAS5 has been associated with diabetes |
[67, 89, 91] |
| Cancer, type 2 diabetes, coronary artery disease, myocardial infarction |
ANRIL | - | Several SNPs in the ANRIL locus on chromosome 9p are involved in coronary artery disease, diabetes and cancer. ANRIL binds PRC1/PRC2 and regulate the tumor suppressors CDKN2A/B. However, the clear role in the pathogenesis of these conditions is yet to be understood |
[97, 134– 138] |
| Myocardial infarction, diabetic retinopathy, schizophrenia |
MIAT or GOMAFU |
- | MIAT is involved in pathological angiogenesis and is suggested as a predictor of myocardial infarction. MIAT forms ribonucleoprotein complex with three splicing proteins, SRSF1, SF-1, and QKI. Downregulation of MIAT leads to alternative splicing, suggesting a lncRNA-driven mode of splicing-defect pathogenesis. |
[57, 66, 139– 141] |
| Alzheimer’s disease |
BACE1-AS | ↑ | BACE1-AS increases BACE1 mRNA stability leading to accelerated amyloid β42 accumulation |
[69] |
| Autism spectrum disorder |
MSNP1AS | ↑ | MSNP1AS regulates the moesin protein, regulator of synapse development and function, by stabilizing moesin mRNA. This mechanism may causally connect SNP variants in the MSNP1AS locus to autism spectrum disorder pathogenesis. |
[70, 142] |
↓ downregulated, ↑ upregulated
LncRNAs are important regulators of physiological and pathological responses. Their role and functions have been mostly studied in tumorigenesis but dysregulation of lncRNAs is not only associated with several types of cancers but a variety of human diseases.
Abbreviations: PINT (p53-induced non-coding transcript), HULC (highly upregulated in liver cancer), SRA (steroid receptor RNA activator), HOTAIR (HOX transcript antisense RNA), GAS5 (growth arrest-specific 5), ANRIL (antisense non-coding RNA in the INK4 locus), MIAT (myocardial infarction associated transcript), GOMAFU (spotted pattern” in Japanese), BACE1 (beta-site APP-cleaving enzyme 1) BACE1-AS (BACE1 antisense RNA), MSNP1AS (moesin pseudogene 1 antisense RNA).
Immune function
LncRNAs are emerging as important regulators of immune cell differentiation and activation [36, 77]. Several studies have reported aberrant expression of lncRNAs in various inflammatory conditions, including autoimmune and allergic disease [78–84]. For instance, a recent study discovered the novel C5T1lncRNA, in the rheumatoid arthritis risk locus TRAF1-C5, which influences transcript levels of C5 that are important for the pathogenesis [85]. Psoriasis is another autoimmune disorder generally thought to be a genetic disease triggered by environmental factors. PRINS (psoriasis susceptibility-related RNA gene induced by stress) is a lncRNA that may contribute to the psoriasis susceptibility [86, 87].
Metabolic disease
The regulation of metabolism and glucose homeostasis is a complex interplay of tissues/organs and includes several mechanistically important lncRNAs [88]. For example, the lncRNA growth-arrest specific transcript 5 (GAS5) regulates cell growth and is induced under conditions of nutrient deprivation and cellular stress [89–91]. Functionally, GAS5 is both a precursor for small RNA [51] and a glucocorticoid receptor (GR) decoy (Figure 1B) [89]. By competing with GR DNA-binding sequences, GAS5 suppresses transactivation of GR-dependent gene promoters [89]. GAS5 also appears to repress the effects of other steroid hormone receptors [89] and may have a role in saving energy resources as an adaptive response to starvation by restricting the expression of steroid-responsive genes and has been reported to have proapoptotic functions [89–91]. Significant efforts have been made to reach a better understanding of the causes of diabetes at the molecular level [92]. Interestingly, Carter and coworkers have demonstrated that decreased serum levels of GAS5 were associated with type 2 diabetes in a cohort of U.S. military veterans [67].
Cardiovascular disease
Recent studies also suggest critical roles of lncRNAs in modulating the initiation and progression of cardiovascular disease [64, 93, 94]. LncRNAs such as Bvrt, Fendrr, ANRIL, MIAT, and MyHeart (Mhrt) play important roles in cardiovascular development and heart disease, including myocardial infarction, cardiomyopathy, heart failure, and atherosclerosis [66, 95–99]. For instance, Mhrt prevents myopathy in mice by binding to the helicase domain of Brg1, inhibiting chromatin targeting and aberrant gene regulation otherwise induced by this stress triggered chromatin-remodeling factor [95].
Brain disease
A significant part of all sequenced lncRNAs are expressed specifically in the brain, where they show strictly regulated temporal and spatial expression patterns [57, 100]. Genome-wide association studies and comparative transcriptomic studies have associated lncRNA expression with neurological disorders including schizophrenia, bipolar disorder, depression, autism spectrum disorder, Asperger’s syndrome, attention deficit hyperactivity disorder, neuropathic pain, epilepsy, and neurodegenerative disorders such as amyotrophic lateral sclerosis, Alzheimer’s and Parkinson’s disease [57, 70, 101–103]. For example: the lncRNA BACE1-AS is transcribed antisense to the BACE1 gene and expressed at 2- to 6-fold higher levels in the brains of Alzheimer’s disease patients compared to controls [69]. The BACE1 gene encodes a transmembrane beta-secretase protein that drives overproduction of pathogenic Aβ-42 peptides in Alzheimer’s disease [69]. Functionally, BACE1-AS positively regulates BACE1 by binding to and stabilizing the BACE1 mRNA (Figure 1D). Experimental data suggest a positive feedback loop, in which BACE1-AS drives overproduction of toxic Aβ-42 peptides, which then further induce BACE1-AS overexpression and accelerating amyloid accumulation [69].
lncRNAs and environmental exposures
The role of miRNA in environmental health has been previously reviewed [6, 104–106]. Numerous studies demonstrate that miRNAs functionally interact with a variety of environmental factors including environmental chemicals, drugs, alcohol, cigarette smoking, viruses, and bacterial pathogens. It is therefore likely that lncRNAs are also important for driving exposure-disease associations or function as novel biomarkers of environmental exposure. However, while lncRNAs have been found to be dysregulated in a variety of human disease that are known to include environmental factors in the etiology, little is currently known about lncRNA interactions with environmental exposures. Most of the available data is derived from cell studies and no population-based studies have yet been published.
Cell studies
To adapt to environmental changes and survive different damages, eukaryotic cells have evolved networks of different protection mechanisms, including the heat shock response, which detect and controls diverse forms of stress. Heat shock proteins (HSPs) are produced by cells in response to exposure to environmental stressors, such as heat shock or various chemicals [107]. HSPs, also referred to as stress proteins, perform a chaperone function by stabilizing new proteins to ensure correct folding or by helping to refold proteins that were damaged by the stress [108]. The upregulation of HSPs is induced primarily by the heat shock transcription factor 1 (HSF1) and requires the lncRNA HSR1 [109, 110]. HSR1 undergoes a structural conformational change in response to heat shock, and forms a complex with the elongation factor eEF1A, stimulating the trimerization of HSF1, and thereby promoting activation of heat shock response genes [110]. More recently cell studies have shown that several lncRNAs respond to numerous environmental stressors such as genotoxic agents, ultraviolet (UV)-C irradiation, oxidative stress, cigarette smoke extracts, heavy metals, and endocrine disrupting chemicals [111–119]. For example, serum starvation or exposure to translation inhibitors results in an increase in GAS5 that functions as a riborepressor for GR by binding to its DNA-binding domain leading to altered expression of several glucocorticoid-responsive genes. GAS5 has been reported to be proapoptotic and sensitize human cells to cell death by environmental stressors [111, 120]. Another example is PRINS, whose expression levels are increased by many stressors such as UV-B irradiation, viral infection, translational inhibition, and which may contribute to psoriasis susceptibility [86, 87].
Animal experiments
Increasing evidence suggests that lncRNAs take part in gene regulation, from the single gene to chromosome level, and cell studies have demonstrated that lncRNAs are involved in responses to external stimuli such as chemical exposure. However, to date, the in vivo effects of chemical toxicants on lncRNAs are not well studied. Martinez-Guitarte and coworkers analyzed lncRNAs levels after pollutant exposure of the aquatic reference organism Chironomus riparius [121]. Three lncRNA sequences were studied: telomeric repeats, Cla repetitive elements, and the SINE CTRT1, after 24-hour exposure to bisphenol A (BPA), benzyl butyl phthalate (BBP) and the heavy metal cadmium (Cd). BPA exposure upregulated both telomeric and Cla transcripts, whereas Cd and BBP did not significantly affect of any of the sequences [121]. More recently, Bhan and coworkers investigated if the lncRNA HOTAIR, which is transcriptionally regulated by estradiol and a key player in breast cancer, is misregulated by BPA and diethylstilbestrol (DES) exposure [118]. The results revealed that HOTAIR expression is induced after exposure to nanomolar concentrations of BPA and DES in breast cancer cells (MCF7) as well as in the mammary glands of ovariectomized rats. Exposure to these endocrine disrupting chemicals leads to recruitment of estrogen-receptors (ERs) and ER-coregulators at the HOTAIR promoter, chromatin modification, resulting in an increased HOTAIR expression [118]. As HOTAIR is important for gene silencing and highly expressed in variety of cancers including breast tumors a chemically-induced increase in HOTAIR expression could potentially lead to adverse health effects [29, 42, 118].
Human exposure studies
Tobacco smoke is a complex chemical mixture containing thousands of compounds, several known to be carcinogens, cocarcinogens, and/or mutagens. Smoking is a common risk factor for the development of diseases such as cardiovascular disease, lung cancer, and chronic obstructive pulmonary disease (COPD). Genome-wide lncRNA expression in lung tissue resections from three non-smokers without COPD, five smokers without COPD and five smokers with COPD, showed that smoking alters the expression of lncRNAs [122]. Hundreds of differentially expressed lncRNAs (≥2-fold change) were found when comparing smokers and non-smokers with RNA44121|UCSC-2000-3182 and RNA43510|UCSC-1260-3754 being the most over- and under-expressed, respectively. Gene ontology and pathway analysis revealed that these lncRNAs are associated with changes in key pathogenic processes of COPD caused by smoking [122]. Although the number of subjects was low this study indicates that lncRNAs may play a role in the pathological changes generated by cigarette smoking. This is supported by cell studies reporting that cigarette smoke extracts increased levels of HOTAIR in human bronchial epithelial (HBE) cells. HOTAIR then mediates epithelial-mesenchymal transition, a process involved in the malignant transformation of cells caused by cigarette smoke extracts [119]. Knockdown of HOTAIR by siRNA reversed the induced epithelial-mesenchymal transition, formation of cancer stem cells, and malignant transformation [119]. In addition, HOTAIR epigenetic silencing of p21 via enhancer of zeste homolog 2 (EZH2) mediated tri-methylation of Lys 27 of histone H3 contributes to changes in the cell cycle induced by cigarette smoke extracts [123]. Similar studies indicate that the lncRNAs MALAT1 also is involved in cigarette smoke extract induced epithelial-mesenchymal transition and malignant transformation of HBE cells [124].
Future perspectives
The developments in genomics and bioinformatics have resulted in a rapidly growing number of identified lncRNAs. Although an increasing number of lncRNAs are emerging as important disease players, surprisingly little is known about lncRNA’s roles in environmental health. Studies are warranted to clarify whether and how lncRNAs may drive exposure-disease associations or function as novel biomarkers of environmental exposure. LncRNA interactions with a variety of environmental factors such as environmental chemicals, cigarette smoking and air pollution are likely. For instance, the health effects of ambient air pollution include allergic, respiratory, and cardiovascular diseases [125–127]. Numerous studies from the last several years have demonstrated that epigenetic modifications are susceptible to air pollution exposure and may be important for the biological mechanisms mediating these disease associations [128]. LncRNAs may be critical for the biological responses to air pollutants as they regulate numerous cellular processes suggested to drive these exposure-disease associations [129]. Furthermore, as lncRNAs have crucial roles in the control of developmental processes, they are also of particular interest in the context of the developmental origins of health and disease paradigm (DOHaD). Not only are population-based studies needed to establish lncRNA roles in environmental health, but experimental studies to clearly establish causality and detailed mechanisms are also warranted. However, as lncRNAs have a low evolutionary conservation integration of animal and human studies can be challenging.
Conclusion
Although only a small selection of all known lncRNAs have been functionally characterized to date, they have been demonstrated to control every layer of gene regulation including transcriptional gene silencing via regulation of the chromatin structure, and DNA methylation. Research shows that epigenetic and nonepigenetic mechanisms are involved in how the lncRNAs regulate various biological functions. Several lncRNAs have been reported to play an important role in the pathogenesis of human diseases, and their interactions with environmental exposure are exciting, but many questions remain unanswered. Further experimental and population-based epidemiological studies are warranted to clarify lncRNAs role in the interaction of genetic and environmental factors underpinning the development of many common diseases. Characterization of the specific roles of lncRNAs in epigenetic regulation and patterns of dysregulation in the lncRNAs-environmental factor interactions can help to understand the mechanisms of diseases and provide insights into disease pathogenesis.
Footnotes
Conflict of Interest
Oskar Karlsson and Andrea A. Baccarelli declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
- 1.Jorde L. Genes, environment, lifestyle and common diseases. In: McCance K, Huether S, editors. Pathophysiology: The Biologic Basis for Disease in Adults and Children. Elsevier Health Sciences; 2015. pp. 164–182. [Google Scholar]
- 2.Wu S, et al. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2015 doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto AM, Sonnenschein C. Environmental causes of cancer: endocrine disruptors as carcinogens. Nat Rev Endocrinol. 2010;6(7):363–370. doi: 10.1038/nrendo.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das UN. Obesity: genes, brain gut environment. Nutrition. 2010;26(5):459–473. doi: 10.1016/j.nut.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson O, Lindquist NG. Melanin affinity and its possible role in neurodegeneration. Journal of neural transmission. 2013 doi: 10.1007/s00702-013-1062-5. [DOI] [PubMed] [Google Scholar]
- 6.Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015;123(5):399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HS. Impact of Maternal Diet on the Epigenome during In Utero Life and the Developmental Programming of Diseases in Childhood and Adulthood. Nutrients. 2015;7(11):9492–9507. doi: 10.3390/nu7115467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Kevin C, Chang Howard Y. Molecular Mechanisms of Long Noncoding RNAs. Molecular Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19(14):1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 11.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 12.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 13.Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790(9):936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist pre-empts X inactivation choice without RNA stabilization. Mol Cell. 2006;21(5):617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Mohammad F, et al. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 16.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 18.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium EP, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 21.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-proteincoding RNA. Biochim Biophys Acta. 2010;1799(9):597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brosius J. Waste not, want not--transcript excess in multicellular eukaryotes. Trends Genet. 2005;21(5):287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 26.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 27.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 28.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 31.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29(3):288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 32.Ravasi T, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercer TR, et al. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2012;2(6):189–199. doi: 10.4161/bioa.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, et al. CREB up-regulates long non-coding RNA HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453(7196):798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pang KC, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182(12):7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 51.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol Cell. 2008;31(3):309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9(1):59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92(4):337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 56.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briggs JA, et al. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 58.Thai P, et al. Characterization of a novel long noncoding RNA SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunner AL, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13(8):R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35(10):9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 63.Panzitt K, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 64.Archer K, et al. Long Non-Coding RNAs as Master Regulators in Cardiovascular Diseases. Int J Mol Sci. 2015;16(10):23651–23667. doi: 10.3390/ijms161023651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishii N, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 67.Carter G, et al. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015;4:102–107. doi: 10.1016/j.bbacli.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang YS, et al. Urinary Xist is a potential biomarker for membranous nephropathy. Biochem Biophys Res Commun. 2014;452(3):415–421. doi: 10.1016/j.bbrc.2014.08.077. [DOI] [PubMed] [Google Scholar]
- 69.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerin T, et al. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4(128):128ra40. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- 71.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Balkom BW, et al. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J Extracell Vesicles. 2015;4:26760. doi: 10.3402/jev.v4.26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gezer U, et al. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38(9):1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- 76.Song J, et al. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. 2015;15(1):121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 77.Sigdel KR, et al. The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J Immunol Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Z, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsitsiou E, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129(1):95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol. 2013;132(4):1005–1008. doi: 10.1016/j.jaci.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 81.Stuhlmuller B, et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol. 2003;163(3):901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li B, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134(7):1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Q, et al. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66(4):969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 84.Muller N, et al. Interleukin-6 and tumour necrosis factor-alpha differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine. 2014;68(1):65–68. doi: 10.1016/j.cyto.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 85.Messemaker TC, et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2015 doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 86.Sonkoly E, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280(25):24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 87.Szegedi K, et al. Expression and functional studies on the noncoding RNA, PRINS. Int J Mol Sci. 2012;14(1):205–225. doi: 10.3390/ijms14010205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kino T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barthel A, Schmoll D. Novel concepts in insulin regulation of hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2003;285(4):E685–E692. doi: 10.1152/ajpendo.00253.2003. [DOI] [PubMed] [Google Scholar]
- 91.Mourtada-Maarabouni M, et al. GAS5, a non-protein-coding RNA controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 92.Kameswaran V, Kaestner KH. The Missing lnc(RNA) between the pancreatic beta-cell and diabetes. Front Genet. 2014;5:200. doi: 10.3389/fgene.2014.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 94.Wang P, et al. Differential lncRNAmRNA coexpression network analysis revealing the potential regulatory roles of lncRNAs in myocardial infarction. Mol Med Rep. 2015 doi: 10.3892/mmr.2015.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han P, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Broadbent HM, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17(6):806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 98.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu C, Arora P. Long noncoding Mhrt RNA: molecular crowbar unravel insights into heart failure treatment. Circ Cardiovasc Genet. 2015;8(1):213–215. doi: 10.1161/CIRCGENETICS.115.001019. [DOI] [PubMed] [Google Scholar]
- 100.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Syrbe S, et al. De novo loss- or gain-of-function mutations in KCNA2 cause epileptic encephalopathy. Nat Genet. 2015;47(4):393–399. doi: 10.1038/ng.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Talkowski ME, et al. Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. Am J Hum Genet. 2012;91(6):1128–1134. doi: 10.1016/j.ajhg.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao X, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16(8):1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu C, Chen G, Cui Q. Towards the understanding of microRNA and environmental factor interactions and their relationships to human diseases. Sci Rep. 2012;2:318. doi: 10.1038/srep00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang J, Cui Q. Specific Roles of MicroRNAs in Their Interactions with Environmental Factors. Journal of Nucleic Acids. 2012;2012:10. doi: 10.1155/2012/978384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutat Res. 2011;714(1–2):105–112. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santoro MG. Heat shock factors and the control of the stress response. Biochem Pharmacol. 2000;59(1):55–63. doi: 10.1016/s0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 108.De Maio A. Heat shock proteins: facts, thoughts, and dreams. Shock. 1999;11(1):1–12. doi: 10.1097/00024382-199901000-00001. [DOI] [PubMed] [Google Scholar]
- 109.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 110.Shamovsky I, et al. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440(7083):556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 111.Tani H, Torimura M. Development of cytotoxicity-sensitive human cells using overexpression of long non-coding RNAs. J Biosci Bioeng. 2015;119(5):604–608. doi: 10.1016/j.jbiosc.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 112.Zhou Z, et al. Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep. 2015;5:15293. doi: 10.1038/srep15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jacob MD, et al. Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol Biol Cell. 2013;24(18):2943–2953. doi: 10.1091/mbc.E13-04-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tani H, Torimura M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem Biophys Res Commun. 2013;439(4):547–551. doi: 10.1016/j.bbrc.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 115.Tani H, et al. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS One. 2014;9(8):e106282. doi: 10.1371/journal.pone.0106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mizutani R, et al. Identification and characterization of novel genotoxic stress-inducible nuclear long noncoding RNAs in mammalian cells. PLoS One. 2012;7(4):e34949. doi: 10.1371/journal.pone.0034949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bhan A, et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. This paper demonstrate that BPA and DES exposure alters the epigenetic programming of the HOTAIR promoter leading to its endocrine disruptionin vitro and in vivo.
- 119.Liu Y, et al. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;282(1):9–19. doi: 10.1016/j.taap.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 120.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8(1):e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Martinez-Guitarte JL, Planello R, Morcillo G. Overexpression of long non-coding RNAs following exposure to xenobiotics in the aquatic midge Chironomus riparius. Aquat Toxicol. 2012;110–111:84–90. doi: 10.1016/j.aquatox.2011.12.013. This paper demonstrated for the first time the ability of BPA to increase lncRNA expression
- 122. Bi H, et al. Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm Res. 2015;64(2):119–126. doi: 10.1007/s00011-014-0790-9. This paper indicates that smoking may alter the expression of lncRNAs in human lung tissue
- 123.Liu Y, et al. Epigenetic silencing of p21 by long non-coding RNA HOTAIR is involved in the cell cycle disorder induced by cigarette smoke extract. Toxicol Lett. 2016;240(1):60–67. doi: 10.1016/j.toxlet.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 124.Lu L, et al. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;289(2):276–285. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 125.Franchini M, Mannucci PM. Air pollution and cardiovascular disease. Thromb Res. 2012;129(3):230–234. doi: 10.1016/j.thromres.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 126.Kelly FJ, Fussell JC. Air pollution and airway disease. Clin Exp Allergy. 2011;41(8):1059–1071. doi: 10.1111/j.1365-2222.2011.03776.x. [DOI] [PubMed] [Google Scholar]
- 127.Takizawa H. Impact of air pollution on allergic diseases. Korean J Intern Med. 2011;26(3):262–273. doi: 10.3904/kjim.2011.26.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Breton C, Marutani A. Air Pollution and Epigenetics: Recent Findings. Current Environmental Health Reports. 2014;1(1):35–45. [Google Scholar]
- 129.Stanek LW, et al. Air pollution toxicology--a brief review of the role of the science in shaping the current understanding of air pollution health risks. Toxicol Sci. 2011;120(Suppl 1):S8–S27. doi: 10.1093/toxsci/kfq367. [DOI] [PubMed] [Google Scholar]
- 130.Marin-Bejar O, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hatchell EC, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22(5):657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 132.Lanz RB, et al. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23(20):7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kogo R, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 134.Cunnington MS, et al. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6(4):e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Holdt LM, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30(3):620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 136.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pasmant E, et al. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 138.Congrains A, et al. ANRIL: molecular mechanisms and implications in human health. Int J Mol Sci. 2013;14(1):1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yan B, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 140.Liao J, et al. LncRNA MIAT: Myocardial infarction associated and more. Gene. 2016;578(2):158–161. doi: 10.1016/j.gene.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 141.Barry G, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 142.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459(7246):528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]