Abstract
Cartilage is a mechanosensitive tissue, which can means that it can perceive and respond to biomechnical signals. Despite the known importance of biomechanical signals in the etiopathogenesis of arthritic diseases, and their effectiveness in joint restoration, little is understood about their actions at the cellular level. Recent molecular approaches have revealed that specific biomechanical stimuli and cell interactions generate intracellular signals that are powerful inducers or suppressors of proinflammatory and reparative genes in chondrocytes. Biomechanical signals are perceived by cartilage in magnitude-, frequency-, and time-dependent manners. Static and dynamic biomechanical forces of high magnitudes induce proinflammatory genes and inhibit matrix synthesis. Contrarily, dynamic biomechanical signals of low/physiologic magnitudes are potent antiinflammatory signals that inhibit interleukin-1β (IL-1β)–induced proinflammatory gene transcription and abrogate IL-1β/tumor necrosis factor-α–induced inhibition of matrix synthesis. Recent studies have identified nuclear factor-κB (NF-κB) transcription factors as key regulators of biomechanical signal–mediated proinflammatory and antiinflammatory actions. These signals intercept multiple steps in the NF-κB signaling cascade to regulate cytokine gene expression. Taken together, these findings provide insight into how biomechanical signals regulate inflammatory and reparative gene transcription, underscoring their potential in enhancing the ability of chondrocytes to curb inflammation in diseased joints.
Keywords: biomechanical signals, chondrocytes, inflammation
I. INTRODUCTION
Biomechanical signals play a major role in the development and homeostasis of the musculoskeletal system. Cytoskeletal rearrangments provide the morphogenic signals necessary for embryonic cell movement and force generation. Morphogenesis is intrinsically a cascade of biomechanical signal transductions that direct the molecular events and genetic cues that promote embryonic pattern formation and tissue differentiation. Biomechanical signals provide the bridge between gross morphologic signals and molecular gene expression. Compressive, tensile, and shear forces interplay to orchestrate a complex and tightly regulated series of events that ultimately direct gene expression within the cell. Although nearly all cell types examined demonstrate significant responses to biomechanical stimuli (osteoblasts, chondrocytes, myoblasts, and mesechymal, epithelial, endothelial, and fibroblastic cells), the precise patterns of gene expression that manifest are less well characterized. The functional responses to biophysical forces, on the other hand, have been well documented in the literature.1
This review focuses on some of the recent stimulating findings centered around the specific transcriptional responses of cartilage and chondrocytes to external biomechanical stimuli. The link among mechanical signals, mechanotransduction, and gene expression has only recently started being fully experimentally explored. It is clear that biomechanical signal transduction progresses from the single-cell intracellular environment to apposed cells and extracellular matrices (ECMs) to the tissue microenvironment to the organ macroenvironment to eventually being functionally manifested at the organism level.
There are three types of cartilagenous tissues that perceive biomechanical stimuli: hyaline articular cartilage, fibrocartilage, and elastic cartilage. Hyaline articular cartilage is an avascular tissue present at articulating surfaces that absorbs loads and dissipates the frictional forces realized at these joints. Fibrocartilage and elastic cartilage are both vascularized. Whereas elastic cartilage provides flexibility to the tissues, fibrocartilage takes part in absorbing loads and facilitating smooth joint movements.
II. CARTILAGE, CHONDROCYTES, AND BIOMECHANICAL FORCES
Under physiologic conditions, articular cartilage is often simultaneously exposed to axial compressive loading (normal to articular surface), lateral radial and circumferential tensile strain, and fluid shear forces. This complex modeling may be reproduced to varying degrees in vitro using cartilage explants, chondrocyte-seeded tissue–engineered scaffoldings, and monolayered chondrocyte cell culture systems. The physiologic recapitulation of the explant models makes them attractive systems for mimicking the in situ complexity of joint movement kinematics. However, the reductionistic approach of isolated chondrocytes in three-dimensional scaffolding environments or monolayers on silastic stretchable membranes permits a simplified, tightly controlled, and molecularly dissectable model system. It is clearly a combination of these different models that will ultimately allow investigators to delineate the complicated signaling pathways activated in response to physiologic mechanical forces in the in situ tissue microenvironent.
The structure of hyaline (articular) cartilage allows the tissue to respond to mechanical forces in unique ways. The ECM of cartilage, which can be considered a viscoelastic material with a rigid organic component of collagen and aggregated proteoglycans (PGs) integrated with a hydrated interstitial fluid component, presents as a structured porous matrix (Fig. 1). The combination of fluid–flow-generated signals coupled to matrix mechanotransduction generates a complex series of signaling cascades and ultimately a biomechanical signal–dependent transcriptional response. It is estimated that only 5% of the volume of articular cartilage can be accounted for by the resident cell population of chondrocytes, leaving the vast majority of the cartilage volume occupied by the ECM and interstitial fluids. It is this network of fibrillar collagen, especially collagen type II (COL2) and negatively charged aggrecans (ACANs), that defines the signal transduction matrix essential for chondrocyte stimulation. This charged fluidic matrix of cross-linked PGs and electrolytes permits the electrostatic osmotic pressure regulation of the dynamic stiffness, as well as resilience to frictional forces, of the articular cartilage under physiologic conditions.2
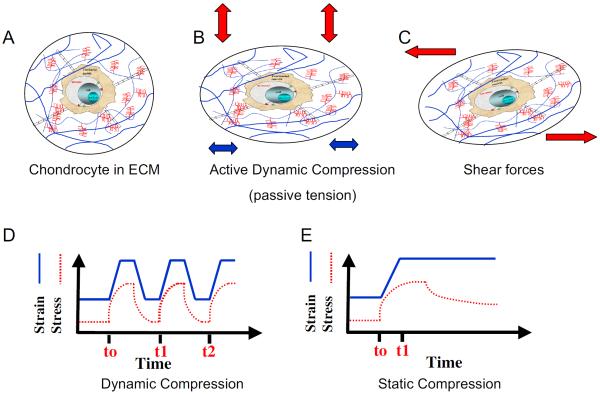
FIGURE 1.
Schematic representation of types of biomechanical forces exerted on chondrocytes. (A) A normal chondrocyte in a lacunae surrounded by extracellular matrix (ECM). (B) Deformation of chondrocytes and ECM during active compression that consequently resulted in passive tensile forces, radial fluid flow, and increased nutrient transport. (C) Deformation of chondrocytes and ECM in response to shear forces that resulted in minimal hydrostatic pressure, fluid flow, and nutrient transport. (D) Schematic representation of dynamic compressive forces that can be of equal time intervals or of varying time intervals (frequencies). Dynamic compression leads to cyclic matrix and chondrocyte deformation, changes in hydrostatic pressure, and enhanced flow of nutrients. (E) Static compression showing ramp- and hold-type effects that increase hydrostatic pressure and induce matrix deformation that is followed by static conditions and minimal transport of nutrients.2,4
The biomechanical signals generated during joint movement are essential components of the cells' and tissue's abilities to repair and recover following physiologic insults, as well as to maintain homeostasis. Chondrocytes are positioned as primary responders to biomechanical signals as they are transduced through the viscoelastic ECM. It is well documented that chondrocytes in cartilage are mechanosensitive cells and can perceive mechanical signals and respond to them by converting physical signals into biochemical events.
A. Compressive Forces Regulate Cartilage Damage and Repair
The normal physiologic movement of joints exerts compressive loading on articular cartilage with either intermittent or cyclic durations. This dynamic compression of the ECM results in synovial fluid exudation and reabsorption that facilitate the diffusion of nutrients and oxygen into avascular cartilage tissue. Additionally, the biomechanical signals received by the viscoelastic ECM are transduced and propagated to the resident chondrocytes. The molecular effects of compressive forces on gene expression in chondrocytes has been studied using several different models, largely ex vivo, including cartilage explants3–8 and in vitro systems such as agarose,9–11 alginate,5 and other polymeric scaffolds.12–14
Although the cartilage explant tissue models provide exceptional physiologic simulation of in vivo biomechanics, the tissue-level environment and signaling complexity creates challenges for dissecting the individual molecular responses to the applied compressive forces. The gene expression in chondrocytes under compression is dependent on the magnitude,3,8,12 frequency,10 and duration5,6,8,10,12,13,15 of applied compressive forces. Dynamic compression of cartilage explant cultures at low magnitudes (3%–5% strain), whether intermittent or periodic, leads to cyclic changes in pressure, deformation, and fluid flow in the cartilage. At the molecular level, the gene expression of cartilage oligomeric matrix protein (COMP) in chondrocytes under dynamic compression has revealed that direction (uniaxial or multiaxial), orientation (rotation about cylindric axis), and periodicity (oscillation of scaffold) of the applied forces, as well as culture period, impact the transcriptional profile of chondrocytic gene expression.14 Dynamic compression has been shown to upregulate expression of anabolic genes such as ACAN, COL2 α1 (COL2A1), and tissue inhibitor of metalloproteinase 3 (TIMP3)15 while downregulating specific genes of the matrix metalloproteinase (MMP) family.4,9,15,16 More importantly, compressive forces at low magnitudes have been shown to be antiinflammatory in nature. This is evidenced by the findings that when chondrocytes seeded into agarose hydrogel scaffolds were stimulated by exogenous interleukin-1β (IL-1β), sinusoidal form compression at 15% strain suppressed aggrecanase 1 (a disintegrin-like and metalloprotease domain (reprolysin-type) with thrombospondin type I motifs 4 [ADAMTS4]) and aggrecanase 2 (ADAMTS5) but not MMP3 gene expression. The compression also prevented downregulation of ACAN in the presence of inflammation.3,6 Similarly, compressive forces inhibited IL-1β–inducible nitric oxide synthase (INOS/NOS2A) and cyclooxygenase 2 (COX2)/prostaglandin G/H synthase 2 (PTGS2) expression17,18 and upregulated PG synthesis and cell division in the presence or absence of IL-1β (Fig. 2).19,20
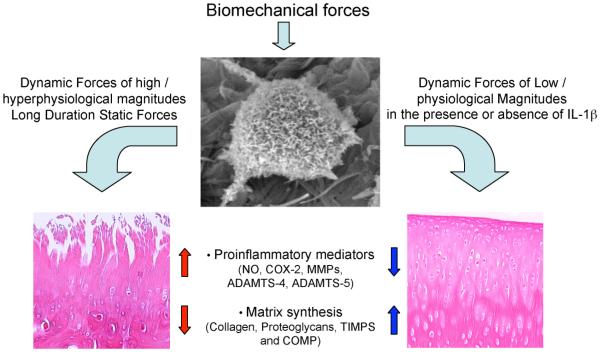
FIGURE 2.
Transcriptional regulation of proinflammatory and reparative genes in response to various types of mechanical forces. Dynamic compressive and tensile forces of high magnitudes and static forces induce expression of proinflammatory genes that are associated with matrix destruction such as inducible nitric oxide synthase, cyclooxygenase 2, matrix metalloproteinases, a disintegrin-like and metalloprotease domain (reprolysin-type) with thrombospondin type I motifs 4 (ADAMTS4), and ADAMTS5. In parallel, these forces inhibit expression of matrix-associated molecules such as aggrecan, collagen type II (COL2), tissue inhibitor of metalloproteinases (TIMPs), and cartilage oligomeric matrix protein (COMP). On the other hand, dynamic compressive and tensile forces of low magnitudes upregulate synthesis of matrix-associated proteins such as proteoglycans, COL2, aggrecan, TIMPs, and COMP. In addition, biomechanical signals of low magnitudes inhibit interleukin-1β–induced expression of proinflammatory genes as well as abrogate cytokine-mediated inhibition of synthesis of matrix-associated proteins.
Interestingly, exposure of chondrocytes to dynamic compression of higher magnitudes (2 MPa) for as little as 1 hour can be traumatic, inducing generation of NO, COX2/PTGS2, prostaglandin E2 (PGE2), and MMP1 and inhibiting PG and decorin mRNA expression.21–23 Similarly, long-term (24–48 hours) compressive strain of 25% to 50% is catabolic and substantially downregulates expression of ACAN and COL2A1 and upregulates MMP3, MMP9, MMP13, and ADAMTS4 expression to induce cartilage destruction.16,24
Contrary to dynamic loading, static compression induces an initial accumulation of interstitial hydrostatic pressure within the cartilage; however, this pressure reaches an equilibrium stasis due to stress relaxation of the tissue.3 Static compression invariably downregulates anabolic gene expression4,12,16 and upregulates catabolic (MMP3, MMP9, MMP13, and ADAMTS4)4 and inflammatory (tumor necrosis factor-α [TNF-α; TNFA], COX2/PTGS2, iNOS/NOS2A).4,6 gene expression. Murata et al.6 have shown that static compression activates the IL-1 signaling pathway by upregulating IL-1α, IL-1β, and NOS2A gene expression in the presence and absence of IL-1 receptor antagonist (Fig. 2).6
At the cellular level, the presence of a developed pericellular matrix appears to be essential for the appropriate mechanotransduction of biomechanical forces into gene expression in chondrocytes.25 Giannoni et al.5 showed the upregulation of COMP gene expression in either alginate or cartilage explant cultures under long-term dynamic compression. However, the mechanosensitivity required for COMP activation was lost by inhibition of the availability of the transmembrane receptor protein β1-integrin, possibly by the disabling of fibronectin-dependent signaling.26 These findings emphasize the requirement of operative cell–matrix signaling to achieve chondrocyte transcriptional activation.
Currently there is little known about the mechanisms by which compressive forces are converted into intracellular biochemical events. β-integrins (also called fibronectin receptors and CD29) are the mechanoreceptors responsible for transducing many biomechanical signals to the cytoplasm, and the biomimetic peptide–amphiphile, GRGDSP, abolishes the integrin-mediated compression-induced chondrocytic responses to compressive forces.19,27,28 Recently investigators showed that the biomechanical signaling is processed at the level of nuclear envelope deformation.29 Both static and dynamic compressive forces induce intracellular actin and vimentin reorganization; however, the significance of this reorganization has yet to be fully defined.30
B. Regulation of Inflammation and Repair in Chondrocytes by Tensile Strain
During joint movement, in addition to compressive forces, chondrocytes in cartilage are exposed to tensile and shear forces. Examination of the tensile and compressive properties of cartilage has shown that fluid-flow–dependent viscoelasticity dominates the compressive response of cartilage, whereas intrinsic solid matrix viscoelasticity dominates the tensile response. Furthermore, the dynamic compressive modulus of cartilage is critically dependent upon elevated values of the dynamic tensile modulus, suggesting that tensile forces are generated during joint movement.2
Early investigations of the responses of chondrocytes to tensile forces applied in vitro showed that similar to compressive forces, chondrocytes perceive and respond to tensile forces in a magnitude-dependent manner. At low magnitudes, tensile forces act as potent antiinflammatory signals and inhibit IL-1β–, TNF-α–, and lipopolysaccharide-induced proinflammatory gene transcription.2,31,32 However, at high/hyperphysiologic magnitudes, these signals act as traumatic signals and induce production of proinflammatory mediators such as NOS2A, COX2, MMPs, and NO, irrespective of the presence of an inflammatory stimulus.21 One of the pathways regulated in chondrocytes following mechanical stimulation is the signaling cascade involved in inflammatory responses. Dynamic tensile strains of low magnitudes (2.5%–7.5% elongation) significantly suppress IL-1β– and TNF-α– dependent INOS/NOS2A, COX2/PTGS2, MMP13, and MMP1 expression, as well as PGE2 and NO production in articular chondrocytes.32–36 In parallel, cyclic tensile strain also inhibits cartilage degradation by upregulating the mRNA expression of TIMP2 and COL2A1 following their IL-1β–dependent suppression.37 Furthermore, cyclic tensile strain could augment cartilage repair by facilitating the induction of ACAN mRNA and attenuation of IL-1β–induced suppression of PG synthesis (Fig. 2).
In subsequent reports, elaboration of the findings on the antiinflammatory effects of dynamic tensile strain in fibrochondrocytes of the temporomandibular joint (TMJ) and knee meniscus has also shown inhibition of IL-1β– and TNF-α–induced INOS/NOS2A, COX2/PTGS2, and MMP1 mRNA expression, NO and PGE2 production, and MMP1 protein synthesis. Extensive in vivo evidence has shown that PG synthesis is an important regulator of cartilage repair and has demonstrated that the application of exogenous PGs can prevent cartilage loss in arthritic joints. In parallel to inhibiting proinflammatory gene transcription, cyclic tensile strain also abrogates IL-1β–induced downregulation of PG, COL2 α1, and TIMP2 synthesis.38,39 Thus, tensile forces of low magnitudes are not only antiinflammatory but are also reparative in their actions.
Continuing investigations on the role of cyclic tensile strain on the expression and regulation of MMPs in fibrochondrocytes of the TMJ demonstrated a significant increase in mRNA expression and synthesis of MMP3, MMP7, MMP8, MMP9, MMP13, MMP16, and MMP17 in response to IL-1β. This IL-1β–induced upregulation of MMPs was significantly inhibited by the application of dynamic tensile strain. However, MMP2, MMP11, MMP14, TIMP1, TIMP2, and TIMP3 mRNA expression was not affected by either IL-1β–induced inflammation or the application of tensile strain.40 Interestingly, studies on the expression of proinflammatory mediators in fibrochondrocytes from the TMJ or knee meniscus showed that 5% to 20% magnitude of tensile strain did not produce any proinflammatory effects, whereas 15% tensile strain was optimal for more than 90% suppression of the above-mentioned proinflammatory mediators. This suggested that mechanical signals inhibit proinflammatory gene expression in both chondrocytes and fibrochondrocytes and that the threshold of their sensitivities differs with the cell type.
Mechanical signals also regulate expression of TNF superfamily member 11 (TNFSF11/receptor activator of NF-κB ligand [RANKL]), TNF receptor superfamily member 11A (TNFRSF11A/RANK), and osteoprotegerin (OPG/TNFRSF11B) in fibrochondrocytes of the rat meniscus. Whereas IL-1β increases the expression of RANKL/TNFSF11 and RANK/TNFRSF11A, cyclic tensile strain suppresses this expression.39 Using the same biomechanical and inflammatory model, TNFRSF11B/OPG expression was unaffected by the application of cyclic tensile strain. These effects of cyclic tensile strain were also found to be both magnitude and frequency dependent.39
1. Dynamic Tensile Forces Inhibit Inflammation in a Sustained Manner
The findings that the application of dynamic tensile strain can cause a rapid inhibition of the actions of IL-1β and TNF-α in chondrocytes in vitro led to the inquiry of how long the actions of mechanical signals last in suppressing proinflammatory gene induction. In this model of biomechanical strain, chondrocytes were subjected to cyclic tensile forces for the initial 1.5, 4, or 8 hours of the experimental design while being exposed continuously to an IL-1β inflammatory environment over a 24-hour time course. These results showed that the suppressive effects of biomechanical strain were sustained over various time intervals of application during a 24-hour experimental time course, and as short as 90 minutes of tensile forces were sufficient to inhibit IL-1β–induced COX2/PTGS2, INOS/NOS2A, MMP9, and MMP13 expression for the ensuing 8 hours, despite the continued presence of a proinflammatory environment.32,40 Similarly, an 8-hour exposure of dynamic tensile strain of low magnitudes was sufficient to inhibit proinflammatory gene induction for the ensuing 16 hours but was not sufficient to inhibit INOS/NOS2A expression for an additional 28 to 40 hours. These observations suggested that the antiinflammatory cascade initiated by the application of dynamic tensile forces persisted despite the cessation of the biomechanical stimuli.
2. Dynamic Tensile Strain Regulates the NF-κB Pathway to Induce or Inhibit Proinflammatory Gene Transcription
The preceding findings that dynamic tensile strain can cause rapid induction or inhibition of proinflammatory genes in magnitude- and frequency-dependent manners provided the essential foundation for further exploration of pathways through which the actions of mechanical signals are converted into functional responses. To extend these observations, attention soon turned to the NF-κB signaling pathway as a possible link between tensile loading and chondrocytic responses to proinflammatory cytokines. First, the NF-κB signaling cascade is the major pathway that controls proinflammatory gene transcription. Second, both low and high magnitudes of tensile strain regulate proinflammatory gene transcription. Finally, the effects of tensile forces are not mediated by the immediate downregulation of IL-1β or TNF-α receptors on chondrocytes. Together, these findings suggest that chondrocytic responses to mechanical loading occur largely independently of traditional cytokine receptors and that biomechnical activation of the NF-κB signaling pathway is essential for the propagation of these actions.
To identify the key target molecule(s) that are regulated by dynamic tensile forces, it is essential to understand the many roles of NF-κB and its complex web of intracellular regulation. This knowledge provides the fundamental rationale for focusing on IL-1β–induced NF-κB nuclear translocation and the prerequisite upstream events involved in its activation in response to dynamic biomechanical stimuli. In chondrocytes, biomechanical signals are perceived in magnitude- and frequency-dependent manners to promote or attenuate proinflammatory gene transcription.37 Biomechanical signals are transduced to cells by surface molecules such as β-integrins and focal adhesion kinases/protein-tyrosine 2 kinases).41 The proinflammatory response exhibited by articular chondrocytes subjected to tensile strain of higher magnitudes is paralleled by an increase in NF-κB nuclear import, providing support for a central role of NF-κB in the proinflammatory signals generated by tensile loading.38 Conversely, at lower magnitudes, biomechanical signals inhibit nuclear translocation of NF-κB transcription factors and act as potent inhibitors of IL-1β– and TNF-α–dependent proinflammatory gene transcription.37,42,43
Multiple cytokine-induced proinflammatory pathways converge at the signalosome composed of inhibitor of κ light polypeptide gene enhancer in B cells kinase, IKKα (IKKA/IKK1/conserved helix-loop-helix ubiquitous kinase); IKKβ (IKKB/IKK2/IKBKB); and IKKγ (NF-κB essential modifier/IKBKG) to activate downstream events in the NF-κB cascade. Upon phosphorylation by IKKs, inhibitor of κ light polypeptide gene enhancer in B cells protein, I-κB, is ubiquinated and marked for proteosomal degradation. The liberation of NF-κB from I-κB complexes is followed by its phosphorylation at multiple sites in a stimulant-dependent manner and eventual translocation into the nucleus. The binding of NF-κB to its consensus sequences leads to transcription of a variety of genes including proinflammatory cytokines and mediators, as well as several of the molecules required for the activation of the NF-κB signaling cascade. This classical model of NF-κB activation by TNF-α or IL-1β is well documented, and its complexity evolved from its regulation at multiple intracellular levels in cell- and stimulus-dependent manners (Fig. 3).
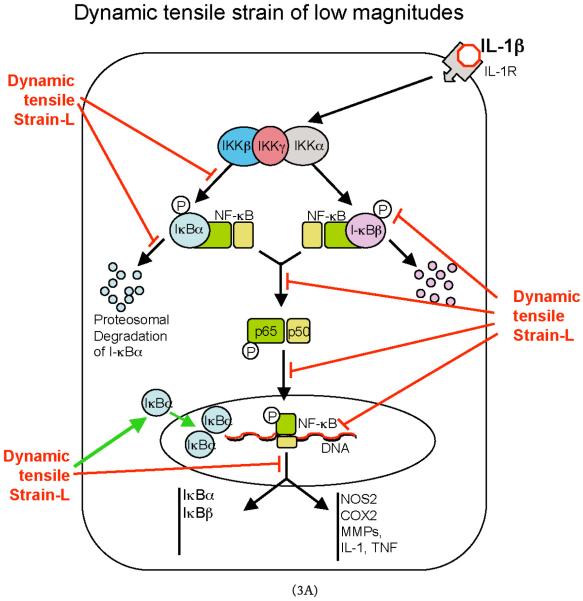
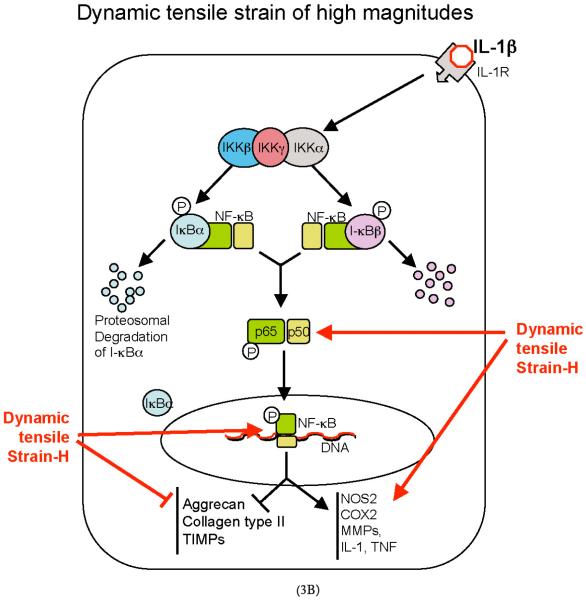
FIGURE 3.
Schematic representation of the mechanisms of intracellular actions of dynamic tensile strain (DTS). (A) DTS of low magnitudes (DTS-L) suppresses interleukin-1β (IL-1β)–induced proinflammatory gene induction by intercepting salient steps in the nuclear factor-κB (NF-κB) signaling cascade to inhibit its transcriptional activity. (1) DTS suppresses IL-1β–induced inhibitor of κ light polypeptide gene enhancer in B cells kinase activation and thus phosphorylation and proteosomal degradation of I-κBα and I-κBβ. This leads to the inhibition of NF-κB nuclear translocation. (2) During the initial stages of IL-1β–mediated activation of cells, DTS upregulates I-κBβ nuclear translocation to prevent NF-κB binding to the DNA and to facilitate export of nuclear NF-κB, which may enter the nucleus. (3) DTS represses IL-1β–induced I-κBα and I-κBβ mRNA expression. Collectively, these actions of DTS inhibit proinflammatory gene induction as well as expression of multiple molecules involved in regulation of the NF-κB signaling cascade to suppress IL-1β–induced inflammation. (B) DTS of high magnitudes (DTS-H) upregulates proinflammatory gene transcription by inducing I-κBα and I-κBβ degradation and subsequent nuclear translocation of NF-κB. This results in the transcriptional activation of proinflammatory mediators including nitric oxide synthase 2, cyclooxygenase 2, matrix metalloproteinases, IL-1β, and tumor necrosis factor-α (TNF-α) and inhibition of the expression of matrix-associated proteins aggrecan, collagen type II, and tissue inhibitor of metalloproteinases. Black arrows indicate IL-1β–mediated activation of NF-κB transcription factors. Red arrows indicate steps in the NF-κB cascade that are inhibited by DTS-L, and the green arrows indicate points that are upregulated by DTS-L.
Characterization of antiinflammatory actions of dynamic tensile strain has shown that it intercepts multiple steps along the NF-κB signaling cascade to block IL-1β–induced proinflammatory gene transcription. As an initial step in regulating NF-κB signaling, dynamic tensile strain markedly abrogates IL-1β–dependent IKK activation, leading to drastic reductions in the phosphorylation and subsequent degradation of I-κBα (NFKBIA) and I-κBβ (NFKBIB). Consequently, NF-κB remains sequestered in the cytoplasm by I-κBα and I-κBβ. This results in the suppression of NF-κB nuclear translocation and repression of transcriptional activation of proinflammatory genes. Additionally, signals generated by tensile strain abrogate the IL-1β–induced increase in NFKBIA and NFKBIB gene expression to unstimulated control levels. Whereas NFKBIA expression is controlled by NF-κB transcriptional activity, NFKBIB has not been shown to be under the transcriptional control of NF-κB.44 Because the pathways that culminate in I-κBβ resynthesis after its degradation are incompletely characterized, the main conclusion of these findings is that the signals generated by cyclic tensile strain likely interact with proteins other than those controlled by NF-κB to regulate proinflammatory gene induction. Finally, dynamic tensile strain induces a rapid upregulation of I-κBα shuttling from the cytoplasm into the nucleus to faciliate the export of any translocating NF-κB to prevent/terminate its transcriptional activity. Thus, the collective actions of cyclic tensile strain at multiple regulatory levels within the NF-κB signal transduction pathway prevent the activation of NF-κB transcription factors and ultimately result in the inhibition of proinflammatory gene transcription (Fig. 3).45
3. Shear Forces, Chondrocyte Metabolism, and Gene Expression
Studies have shown that fluid-induced shear stress regulates chondrocyte metabolism. Articular chondrocytes exhibit dose- and time-dependent responses to shear stress that result in the release of soluble mediators and ECM macromolecules.46 Additionally, Lee et al.47 showed that exposure to shear stress induces NO release in both dose- and time-dependent manners. Shear stress–induced NOS2A mRNA expression and NO production have been shown to be associated with decreased mRNA expressions for the cartilage matrix proteins ACAN and COL2 α1.46,47 Similarly, fluid shear forces have been shown to induce COX2/PTGS2 mRNA expression and suppress phosphoinositide-3 kinase activity, which in turn is associated with modulation of apoptotic and antioxidant pathways.48 On the contrary, Fitzgerald et al.49 showed an upregulation of the transcription of PGs and COL2A1 by cyclic shear stress. These findings suggest that the biomechanical signals introduced by the fluid shear deformation of chondrocytes and cartilage is an additional important component of cartilage homeostasis.
III. SUMMARY
Many recent investigations have demonstrated, in vitro and in vivo, that both exercise and passive motion can exert reparative effects on inflamed joints, whereas inappropriate or excessive mechanical forces initiate cartilage destruction such as is observed in osteoarthritic joints. However, the specific intracellular signaling cascades that transduce mechanical signals into the biochemical events responsible for cartilage destruction or repair remain somewhat elusive, sometimes paradoxic, and clearly complex. This review summarized how the signals generated by biomechanical stress may initiate either the repair or destruction of cartilage, depending upon their magnitude, frequency, and duration.
Although the use of subjectively “appropriate” joint movement has been suggested for centuries to be therapeutic for arthritic joints, the specific molecular mechanisms and defined biomechanical signals that regulate inflammation and promote cartilage repair remain an enigma. Examination of the transcriptional regulation of proinflammatory and matrix-associated genes has provided a framework for examining how mechanical signals regulate cartilage metabolism. Chondrocytes respond to all three major types of biomechanical forces in magnitude-, frequency-, and duration-dependent manners. However, it appears that dynamic forces provide the initiating stimulus capable of suppressing proinflammatory gene induction, whereas static forces invariably induce proinflammatory gene expression. Dynamic biomechanical signals of high magnitudes activate proinflammatory genes via the activation of the NF-κB family of transcription factors. Conversely, at lower/physiologic magnitudes, these signals attenuate proinflammatory gene induction. The biomechanical signals generated by appropriate magnitudes of cyclic tensile strain are able to attenuate IL-1β–dependent activation of IKK and result in the inhibition of NF-κB transcriptional activity. These signals act at multiple steps within the NF-κB signaling cascade to inhibit the transcriptional activity of NF-κB itself by preventing its nuclear import and inhibiting the activation and gene expression of its inhibitors, I-κBα and I-κBβ. Biomechanical signal–enforced dynamic shuttling of NF-κB/I-κBα complexes may represent yet another level of regulation, which could inhibit the activity of NF-κB and the subsequent downstream signaling events involved in proinflammatory gene induction (Fig. 3). Compressive, tensile, and shear forces of appropriate low/physiologic magnitudes also promote the upregulation of PG and collagen synthesis that is drastically inhibited in inflamed joints. Thus, the beneficial effects of physiologic levels of biomechanical signals or exercise may be explained by their abilities to suppress the signal transduction pathways of proinflammatory/catabolic mediators while simultaneously stimulating the anabolic pathways. Whether these anabolic signals are a consequence of the inhibition of NF-κB signaling or are mediated via distinct anabolic pathways has yet to be elucidated. Regardless, it is this complex interplay of magnitude-and frequency-dependent reparative signals within the inflammatory microenvironment of the cartilage that ultimately allows the remarkable and beneficial effects of dynamic tensile forces to be realized.
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health grants AR04878, DE15399, AT00646, and HD40939.
REFERENCES
- 1.Grodzinsky AJ, Levenston ME, Jin M, Frank EH. Cartilage tissue remodeling in response to mechanical forces. Annu Rev Biomed Eng. 2000;2:691–713. doi: 10.1146/annurev.bioeng.2.1.691. [DOI] [PubMed] [Google Scholar]
- 2.Park S, Hung CT, Ateshian GA. Mechanical response of bovine articular cartilage under dynamic unconfined compression loading at physiological stress levels. Osteoarthritis Cartilage. 2004;12(1):65–73. doi: 10.1016/j.joca.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Fehrenbacher A, Steck E, Rickert M, Roth W, Richter W. Rapid regulation of collagen but not metalloproteinase 1, 3, 13, 14 and tissue inhibitor of metalloproteinase 1, 2, 3 expression in response to mechanical loading of cartilage explants in vitro. Arch Biochem Biophys. 2003;410(1):39–47. doi: 10.1016/s0003-9861(02)00658-6. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald JB, Jin M, Grodzinsky AJ. Shear and compression differentially regulate clusters of functionally related temporal transcription patterns in cartilage tissue. J Biol Chem. 2006;281(34):24095–103. doi: 10.1074/jbc.M510858200. [DOI] [PubMed] [Google Scholar]
- 5.Giannoni P, Siegrist M, Hunziker EB, Wong M. The mechanosensitivity of cartilage oligomeric matrix protein (COMP) Biorheology. 2003;40(1–3):101–9. [PubMed] [Google Scholar]
- 6.Murata M, Bonassar LJ, Wright M, Mankin HJ, Towle CA. A role for the interleukin-1 receptor in the pathway linking static mechanical compression to decreased proteoglycan synthesis in surface articular cartilage. Arch Biochem Biophys. 2003;413(2):229–35. doi: 10.1016/s0003-9861(03)00129-2. [DOI] [PubMed] [Google Scholar]
- 7.Valhmu WB, Raia FJ. Myo-inositol 1,4,5-trisphosphate and Ca2+/calmodulin-dependent factors mediate transduction of compression-induced signals in bovine articular chondrocytes. Biochem J. 2002;361(Pt 3):689–96. doi: 10.1042/0264-6021:3610689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valhmu WB, Stazzone EJ, Bachrach NM, Saed-Nejad F, Fischer SG, Mow VC, Ratcliffe A. Load-controlled compression of articular cartilage induces a transient stimulation of aggrecan gene expression. Arch Biochem Biophys. 1998;353(1):29–36. doi: 10.1006/abbi.1998.0633. [DOI] [PubMed] [Google Scholar]
- 9.Mio K, Saito S, Tomatsu T, Toyama Y. Intermittent compressive strain may reduce aggrecanase expression in cartilage: a study of chondrocytes in agarose gel. Clin Orthop Relat Res. 2005;433:225–32. doi: 10.1097/01.blo.0000150466.30696.c6. [DOI] [PubMed] [Google Scholar]
- 10.Mauck RL, Byers BA, Yuan X, Tuan RS. Regulation of cartilaginous ECM gene transcription by chondrocytes and MSCs in 3D culture in response to dynamic loading. Biomech Model Mechanobiol. 2007;6(1–2):113–25. doi: 10.1007/s10237-006-0042-1. [DOI] [PubMed] [Google Scholar]
- 11.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99(15):9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter CJ, Imler SM, Malaviya P, Nerem RM, Levenston ME. Mechanical compression alters gene expression and extracellular matrix synthesis by chondrocytes cultured in collagen I gels. Biomaterials. 2002;23(4):1249–59. doi: 10.1016/s0142-9612(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 13.Stoddart MJ, Ettinger L, Hauselmann HJ. Enhanced matrix synthesis in de novo, scaffold free cartilage-like tissue subjected to compression and shear. Biotechnol Bioeng. 2006;95(6):1043–51. doi: 10.1002/bit.21052. [DOI] [PubMed] [Google Scholar]
- 14.Grad S, Gogolewski S, Alini M, Wimmer MA. Effects of simple and complex motion patterns on gene expression of chondrocytes seeded in 3D scaffolds. Tissue Eng. 2006;12(11):3171–9. doi: 10.1089/ten.2006.12.3171. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald JB, Jin M, Dean D, Wood DJ, Zheng MH, Grodzinsky AJ. Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem. 2004;279(19):19502–11. doi: 10.1074/jbc.M400437200. [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Fitzgerald JB, Dimicco MA, Grodzinsky AJ. Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis Rheum. 2005;52(8):2386–95. doi: 10.1002/art.21215. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury TT, Bader DL, Lee DA. Dynamic compression counteracts IL-1β induced iNOS and COX-2 activity by human chondrocytes cultured in agarose constructs. Biorheology. 2006;43(3–4):413–29. [PubMed] [Google Scholar]
- 18.Chowdhury TT, Bader DL, Lee DA. Dynamic compression counteracts IL-1β-induced release of nitric oxide and PGE2 by superficial zone chondrocytes cultured in agarose constructs. Osteoarthritis Cartilage. 2003;11(9):688–96. doi: 10.1016/s1063-4584(03)00149-3. [DOI] [PubMed] [Google Scholar]
- 19.Chowdhury TT, Appleby RN, Salter DM, Bader DA, Lee DA. Integrin-mediated mechanotransduction in IL-1β stimulated chondrocytes. Biomech Model Mechanobiol. 2006;5(2–3):192–201. doi: 10.1007/s10237-006-0032-3. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhury TT, Bader DL, Lee DA. Anti-inflammatory effects of IL-4 and dynamic compression in IL-1β stimulated chondrocytes. Biochem Biophys Res Commun. 2006;339(1):241–7. doi: 10.1016/j.bbrc.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Guilak F, Fermor B, Keefe FJ, Kraus VB, Olson SA, Pisetsky DS, Setton LA, Weinberg JB. The role of biomechanics and inflammation in cartilage injury and repair. Clin Orthop Relat Res. 2004;423:17–26. doi: 10.1097/01.blo.0000131233.83640.91. [DOI] [PubMed] [Google Scholar]
- 22.Upton ML, Chen J, Guilak F, Setton LA. Differential effects of static and dynamic compression on meniscal cell gene expression. J Orthop Res. 2003;21(6):963–9. doi: 10.1016/S0736-0266(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 23.Tomiyama T, Fukuda K, Yamazaki K, Hashimoto K, Ueda H, Mori S, Hamanishi C. Cyclic compression loaded on cartilage explants enhances the production of reactive oxygen species. J Rheumatol. 2007;34(3):556–62. [PubMed] [Google Scholar]
- 24.Griffin TM, Guilak F. The role of mechanical loading in the onset and progression of osteoarthritis. Exerc Sport Sci Rev. 2005;33(4):195–200. doi: 10.1097/00003677-200510000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Alexopoulos LG, Setton LA, Guilak F. The biomechanical role of the chondrocyte pericellular matrix in articular cartilage. Acta Biomater. 2005;1(3):317–25. doi: 10.1016/j.actbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Lucchinetti E, Bhargava MM, Torzilli PA. The effect of mechanical load on integrin subunits α5 and β1 in chondrocytes from mature and immature cartilage explants. Cell Tissue Res. 2004;315(3):385–91. doi: 10.1007/s00441-003-0836-8. [DOI] [PubMed] [Google Scholar]
- 27.Orazizadeh M, Cartlidge C, Wright MO, Millward-Sadler SJ, Nieman J, Halliday BP, Lee HS, Salter DM. Mechanical responses and integrin associated protein expression by human ankle chondrocytes. Biorheology. 2006;43(3–4):249–58. [PubMed] [Google Scholar]
- 28.Chowdhury TT, Salter DM, Bader DL, Lee DA. Integrin-mediated mechanotransduction processes in TGFβ-stimulated monolayer-expanded chondrocytes. Biochem Biophys Res Commun. 2004;318(4):873–81. doi: 10.1016/j.bbrc.2004.04.107. [DOI] [PubMed] [Google Scholar]
- 29.Guilak F. The deformation behavior and viscoelastic properties of chondrocytes in articular cartilage. Biorheology. 2000;37(1–2):27–44. [PubMed] [Google Scholar]
- 30.Knight MM, Toyoda T, Lee DA, Bader DL. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. J Biomech. 2006;39(8):1547–51. doi: 10.1016/j.jbiomech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Long P, Gassner R, Agarwal S. Tumor necrosis factor α-dependent proinflammatory gene induction is inhibited by cyclic tensile strain in articular chondrocytes in vitro. Arthritis Rheum. 2001;44(10):2311–9. doi: 10.1002/1529-0131(200110)44:10<2311::aid-art393>3.0.co;2-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madhavan S, Anghelina M, Rath-Deschner B, Wypasek E, John A, Deschner J, Piesco N, Agarwal S. Biomechanical signals exert sustained attenuation of proinflammatory gene induction in articular chondrocytes. Osteoarthritis Cartilage. 2006;14(10):1023–32. doi: 10.1016/j.joca.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deschner J, Hofman CR, Piesco NP, Agarwal S. Signal transduction by mechanical strain in chondrocytes. Curr Opin Clin Nutr Metab Care. 2003;6(3):289–93. doi: 10.1097/01.mco.0000068964.34812.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gassner R, Buckley MJ, Piesco N, Evans C, Agarwal S. Cytokine-induced nitric oxide production of joint cartilage cells in continuous passive movement. Anti-inflammatory effect of continuous passive movement on chondrocytes: in vitro study. Mund Kiefer Gesichtschir. 2000;(Suppl 2):S479–84. doi: 10.1007/PL00012696. [DOI] [PubMed] [Google Scholar]
- 35.Gassner R, Buckley MJ, Georgescu H, Studer R, Stefanovich-Racic M, Piesco NP, Evans CH, Agarwal S. Cyclic tensile stress exerts antiinflammatory actions on chondrocytes by inhibiting inducible nitric oxide synthase. J Immunol. 1999;163(4):2187–92. [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Buckley MJ, Evans CH, Agarwal S. Cyclic tensile strain acts as an antagonist of IL-1β actions in chondrocytes. J Immunol. 2000;165(1):453–60. doi: 10.4049/jimmunol.165.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferretti M, Srinivasan A, Deschner J, Gassner R, Baliko F, Piesco N, Salter R, Agarwal S. Anti-inflammatory effects of continuous passive motion on meniscal fibrocartilage. J Orthop Res. 2005;23(5):1165–71. doi: 10.1016/j.orthres.2005.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal S, Deschner J, Long P, Verma A, Hofman C, Evans CH, Piesco N. Role of NF-κB transcription factors in antiinflammatory and proinflammatory actions of mechanical signals. Arthritis Rheum. 2004;50(11):3541–8. doi: 10.1002/art.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deschner J, Wypasek E, Ferretti M, Rath B, Anghelina M, Agarwal S. Regulation of RANKL by biomechanical loading in fibrochondrocytes of meniscus. J Biomech. 2006;39(10):1796–803. doi: 10.1016/j.jbiomech.2005.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deschner J, Rath-Deschner B, Agarwal S. Regulation of matrix metalloproteinase expression by dynamic tensile strain in rat fibrochondrocytes. Osteoarthritis Cartilage. 2006;14(3):264–72. doi: 10.1016/j.joca.2005.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angele P, Yoo JU, Smith C, Mansour J, Jepsen KJ, Nerlich M, Johnstone B. Cyclic hydrostatic pressure enhances the chondrogenic phenotype of human mesenchymal progenitor cells differentiated in vitro. J Orthop Res. 2003;21(3):451–7. doi: 10.1016/S0736-0266(02)00230-9. [DOI] [PubMed] [Google Scholar]
- 42.Angele P, Schumann D, Angele M, Kinner B, Englert C, Hente R, Füchtmeier B, Nerlich M, Neumann C, Kujat Cyclic, mechanical compression enhances chondrogenesis of mesenchymal progenitor cells in tissue engineering scaffolds. Biorheology. 2004;41(3–4):335–46. [PubMed] [Google Scholar]
- 43.Trindade MC, Shida J, Ikenoue T, Lee MS, Lin EY, Yaszay B, Yerby S, Goodman SB, Schurman DJ, Smith RL. Intermittent hydrostatic pressure inhibits matrix metalloproteinase and pro-inflammatory mediator release from human osteoarthritic chondrocytes in vitro. Osteoarthritis Cartilage. 2004;12(9):729–35. doi: 10.1016/j.joca.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Griffin BD, Moynagh PN. In vivo binding of NF-κB to the IkBβ promoter is insufficient for transcriptional activation. Biochem J. 2006;400(1):115–25. doi: 10.1042/BJ20060786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anghelina M, Dossumbekova A, Madhavan S, He L, Quan N, Knobloch T, Agarwal S. Biomechanical signals inhibit IKK activity to attenuate NF-κB transcriptional activity in inflamed chondrocytes. Arthritis Rheum. 2007 doi: 10.1002/art.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane Smith R, Trindade MC, Ikenoue T, Mohtai M, Das P, Carter DR, Goodman SB, Schurman DJ. Effects of shear stress on articular chondrocyte metabolism. Biorheology. 2000;37(1–2):95–107. [PubMed] [Google Scholar]
- 47.Lee MS, Trindade MC, Ikenoue T, Goodman SB, Schurman DJ, Smith RL. Regulation of nitric oxide and bcl-2 expression by shear stress in human osteoarthritic chondrocytes in vitro. J Cell Biochem. 2003;90(1):80–6. doi: 10.1002/jcb.10611. [DOI] [PubMed] [Google Scholar]
- 48.Healy ZR, Lee NH, Gao X, Goldring MB, Talalay P, Kensler TW, Konstantopoulos K. Divergent responses of chondrocytes and endothelial cells to shear stress: cross-talk among COX-2, the phase 2 response, and apoptosis. Proc Natl Acad Sci U S A. 2005;102:14010–5. doi: 10.1073/pnas.0506620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald JB, Jin M, Grodzinsky AJ. Shear and compression differentially regulate clusters of functionally related temporal transcription patterns in cartilage tissue. J Biol Chem. 2006;281(34):24095–103. doi: 10.1074/jbc.M510858200. [DOI] [PubMed] [Google Scholar]