Summary
Influenza virus remains a threat because of its ability to evade vaccine-induced immune responses due to antigenic drift. Here, we describe the isolation, evolution, and structure of a broad-spectrum human monoclonal antibody (mAb), MEDI8852, effectively reacting with all influenza A hemagglutinin (HA) subtypes. MEDI8852 uses the heavy-chain VH6-1 gene and has higher potency and breadth when compared to other anti-stem antibodies. MEDI8852 is effective in mice and ferrets with a therapeutic window superior to that of oseltamivir. Crystallographic analysis of Fab alone or in complex with H5 or H7 HA proteins reveals that MEDI8852 binds through a coordinated movement of CDRs to a highly conserved epitope encompassing a hydrophobic groove in the fusion domain and a large portion of the fusion peptide, distinguishing it from other structurally characterized cross-reactive antibodies. The unprecedented breadth and potency of neutralization by MEDI8852 support its development as immunotherapy for influenza virus-infected humans.
Graphical Abstract
Highlights
-
•
Binding to all influenza A subtypes neutralizing seasonal and pandemic strains
-
•
Utilizes a rare VH (VH6-1) and carries a low level of somatic mutations
-
•
Highly conserved epitope encompassing fusion peptide and hydrophobic groove
-
•
Superior therapeutic window compared to oseltamivir in animals
Identification of a human monoclonal antibody that reacts effectively with all influenza A hemagglutinin subtypes paves the way for developing immunotherapy for people infected with the flu virus.
Introduction
Influenza virus infection remains a serious threat to global health and the world economy. Annual epidemics result in a high number of hospitalizations, with an estimated 3–5 million cases of severe disease and 250,000–500,000 deaths globally, and higher mortality rates are possible during pandemics (Wright et al., 2007). Given the emergence of anti-viral drug-resistance, short treatment windows for antivirals and the lack of cross-protective vaccines, there is an unmet medical need for new therapeutic options that can effectively treat influenza infection.
There are three types of influenza viruses, A, B, and C causing disease in humans, and influenza A and B are responsible for frequent seasonal epidemics. However, influenza A infections account for the majority of hospitalizations and are the only type to cause pandemics (Wright et al., 2007). Influenza A is subtyped by its two major surface proteins, hemagglutinin (HA) and neuraminidase (NA). HA is the main target of neutralizing antibodies that are induced by infection or vaccination. The globular HA head domain mediates binding to the sialic acid receptor, while the HA stem mediates the subsequent fusion between the viral and cellular membranes that is triggered in endosomes by the low pH (Skehel and Wiley, 2000). Genetically, there are 16 influenza A subtypes of HA, which form two structurally and antigenically distinct groups (Nobusawa et al., 1991, Russell et al., 2004). In addition, two new HA analogs recovered from bats, H17 and H18, have been included in this classification (Tong et al., 2012, Tong et al., 2013). Currently, H1 and H3 HA subtypes are associated with human disease and viruses containing H5, H7, H9, and H10 HAs are associated with sporadic human infections due to direct transmission from avian species.
The majority of influenza virus neutralizing antibodies elicited by vaccination or infection bind to the globular head of HA and recognize homologous strains within a given subtype (Russell et al., 2008). These antibodies neutralize virus infectivity by blocking sialic acid receptor binding either directly (Knossow and Skehel, 2006, Schmidt et al., 2013) by interacting with the receptor binding site at the tip of the molecule or indirectly, by projecting over the binding site thereby rendering it inaccessible (Fleury et al., 1999, Xiong et al., 2015). These antibodies are involved in the selection of viruses with variant HAs in the process of antigenic drift, necessitating the annual re-development of influenza vaccines.
In the past 8 years, several laboratories have described a new class of influenza-neutralizing antibodies that target conserved sites in the HA stem that showed different levels of cross-reactivity toward group 1 (Corti et al., 2010, Sui et al., 2009, Throsby et al., 2008, Wrammert et al., 2011), group 2 (Dunand et al., 2015, Ekiert et al., 2011, Friesen et al., 2014, Tan et al., 2014) and groups 1 and 2 viruses (Corti et al., 2011, Dreyfus et al., 2012, Nakamura et al., 2013, Wu et al., 2015). Anti-stem antibodies are less potent at direct viral neutralization as compared to anti-head antibodies, but were shown to induce potent antibody-dependent cellular cytotoxicity (ADCC) of infected cells in vitro and in vivo (Corti et al., 2011, Dilillo et al., 2016, DiLillo et al., 2014), while anti-head antibodies were not or less effective at mediating ADCC. In general, the human antibody response to the HA stem region is more frequent against group 1 as compared to group 2 HAs and is dominated by VH1-69 antibodies (Pappas et al., 2014, Sui et al., 2009, Wrammert et al., 2011). Although subdominant, the group 1 stem response was shown to be recalled after heterologous boosts by the new pandemic H1N1 virus in 2009 (Corti et al., 2011, Wrammert et al., 2011). The antibody response to the HA stem region of group 2 HAs is less frequent, possibly due to the presence of a conserved glycan bound to N38 in HA1 that may shield the access to the most conserved sites in the HA stem and to the lack of exposure to heterologous group 2 viruses (i.e., H7) or to new pandemic H3N2 viruses. Finally, antibodies capable of reacting with the HA stem region of both group 1 and 2 subtypes are extremely rare and usually do not show complete coverage of all subtypes. It has been hypothesized that such broadly cross-reactive antibodies might have potential as therapeutic agents and studies on their mechanism of action, epitope specificity, and ontogeny could also inform the design of cross-protective influenza virus vaccines (Corti and Lanzavecchia, 2013, Yewdell, 2013).
A problem related to the development of anti-stem antibodies as immunotherapeutics is their variable neutralizing potency against viruses belonging to different subtypes and the existence of natural escape mutants. In view of the limitations of group 1 and group 2 antibodies isolated so far, we searched for an antibody capable of potently neutralizing group 1 and 2 influenza A viruses within a narrow range of antibody concentrations. In this study, we isolated and optimized an antibody, named MEDI8852, that exhibited unprecedented breadth and potency, being able to neutralize a diverse panel of representative viruses spanning >80 years of antigenic evolution. Unlike other broadly neutralizing stem-reactive antibodies, MEDI8852 is unique in that it uses a rare VH (VH6-1) and carries a low level of somatic mutations. Crystallographic analysis of the Fab alone or in complex with H5 and H7 HA proteins reveals that MEDI8852 binds a highly conserved epitope on H5 and H7 that is markedly different from other structurally characterized stem-reactive neutralizing antibodies. The characterization of this unique epitope and the breadth and potency of neutralization exhibited by MEDI8852 support its development for immunotherapy in influenza virus-infected humans.
Results
Isolation, Genetic Description, and Optimization of MEDI8852
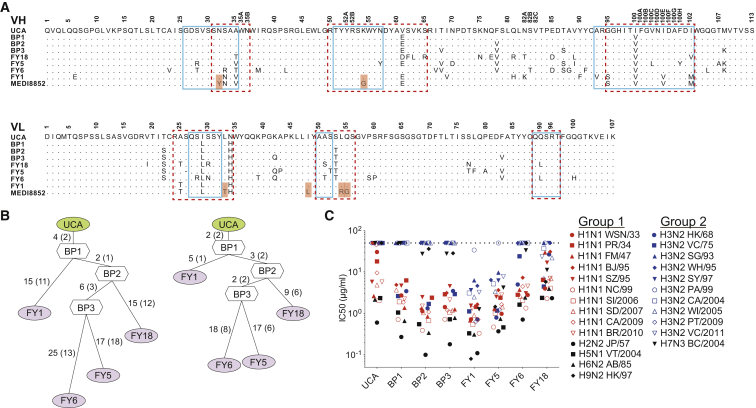
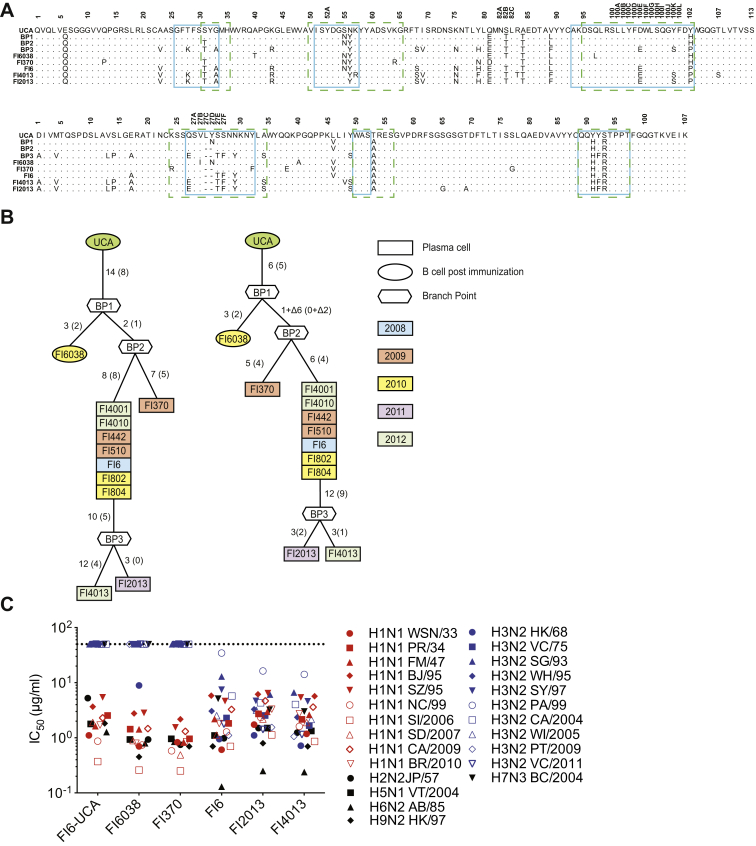
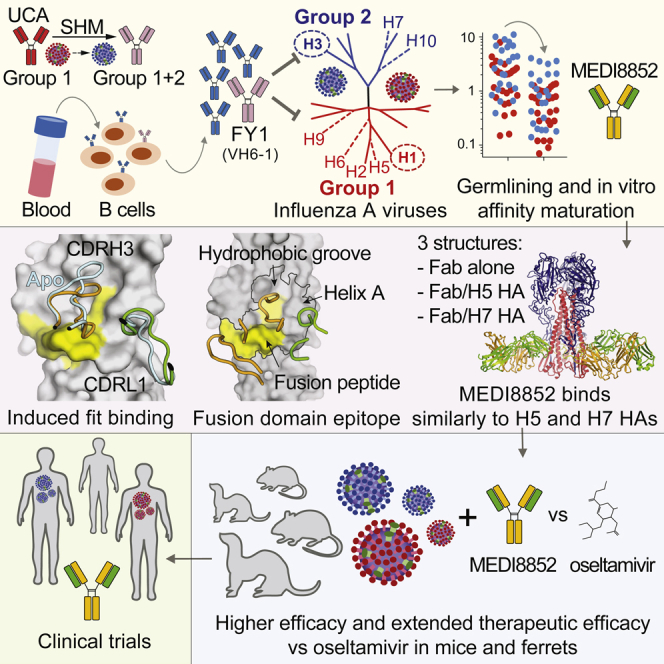
Four broadly reactive antibodies were isolated from the memory B cells of a selected donor based on influenza A HA protein cross-reactivity as previously reported (Traggiai et al., 2004, Pappas et al., 2014). These antibodies (FY1, FY5, FY6, and FY18) belong to the same lineage carrying VH6-1∗01/D3-3∗01/JH3∗02 and VK1-39∗01/JK1∗01 gene segments (Figure 1A). We reconstructed the genealogical trees of this lineage and produced the unmutated common ancestor (UCA), the four clonally derived antibodies, and three antibodies representing the evolutionary branching points (BP) of the lineage (Figure 1B). Purified antibodies were tested for neutralizing activity against multiple viruses of different group 1 and 2 subtypes (Figure 1C). Interestingly, the UCA antibody exhibited neutralizing activity against group 1 viruses, but not group 2 viruses, albeit with lower potency as compared to some of the mutated antibodies. Of note, the first BP (BP1) gained neutralization activity toward early group 2 H3N2 viruses (HK/68 and VC/75). Two antibodies (i.e., FY1 and FY5) of this lineage acquired neutralization activity against group 2 viruses through two independent pathways of somatic mutations. The same analysis was extended to the lineage of FI6, a previously described antibody cross-neutralizing group 1 and group 2 viruses (Corti et al., 2011). Isolation of five additional antibodies from this lineage allowed the reconstruction of a complex genealogy tree (Figure S1). Similarly to what was observed for the FY1 lineage, the FI6-UCA antibody exhibited neutralizing activity against group 1 viruses only and evolved through two independent pathways of somatic mutations that led to the group 1-specific FI370 and FI6038 antibodies and to the group 1 and 2 cross-reactive antibodies FI6, FI2013 and FI4013. Taken together, these findings suggest that in both lineages, the UCA was initially selected by a group 1 virus and developed to a branching point characterized by cross-reactivity toward a limited number of group 2 viruses. From this point, the final antibody may have been selected further for binding to group 1 only (e.g., FY6 and FI370) or group 2 HAs (e.g., FY1 and FI6). These results are consistent with a model in which the development of cross-reactive group 1 and 2 antibodies is started by group 1 HAs and then further selected through boosts by group 2 HAs.
Figure 1.
Developmental Pathway of the MEDI8852 Lineage
(A) Alignment of VH and VL amino acid sequences of four mutated antibodies with their UCA and branchpoint (BP) configurations and MEDI8852. Amino acid substitutions are highlighted in red. Residue positions are according to Kabat numbering. Dots indicate identical residues. Boxes indicate CDR borders according to IMGT (solid line) and Kabat (dashed line).
(B) Genealogy trees of VH (left) and VL (right) nucleotide sequences generated using dnaml. The number of mutations is indicated on the branches with amino acid substitutions in parentheses.
(C) Neutralization of influenza A viruses. IC50 values were determined against a panel of 25 influenza A isolates. Values above 50 μg/ml were scored as negative (dashed line). Average IC50 values were obtained from at least two independent experiments. Full viral strains designations are listed in Table S1.
See also Figure S1.
Figure S1.
Affinity Maturation Pathway of the FI6 Lineage, Related to Figure 1
(A) Alignment of VH and VL amino-acid sequences of the FI6 lineage mutated antibodies with their UCA and branchpoint (BP) configurations. Residue positions are according to Kabat numbering. Dots indicate identical residues. Boxes indicate CDR borders according to IMGT (solid line) and Kabat (dashed line).
(B) Genealogy tree of FI6 lineage VH and VL nucleotide sequences generated using dnaml. The number of mutations is indicated on the branches with amino-acid substitutions in parentheses. Background color and shape identify the origin and the year of isolation.
(C) Neutralization of influenza A viruses. Neutralization IC50 values were determined against a panel of 25 seasonal and non-seasonal influenza A isolates. Values above 50 μg/ml were scored as negative (dashed line). Average IC50 values were obtained from at least two independent experiments.
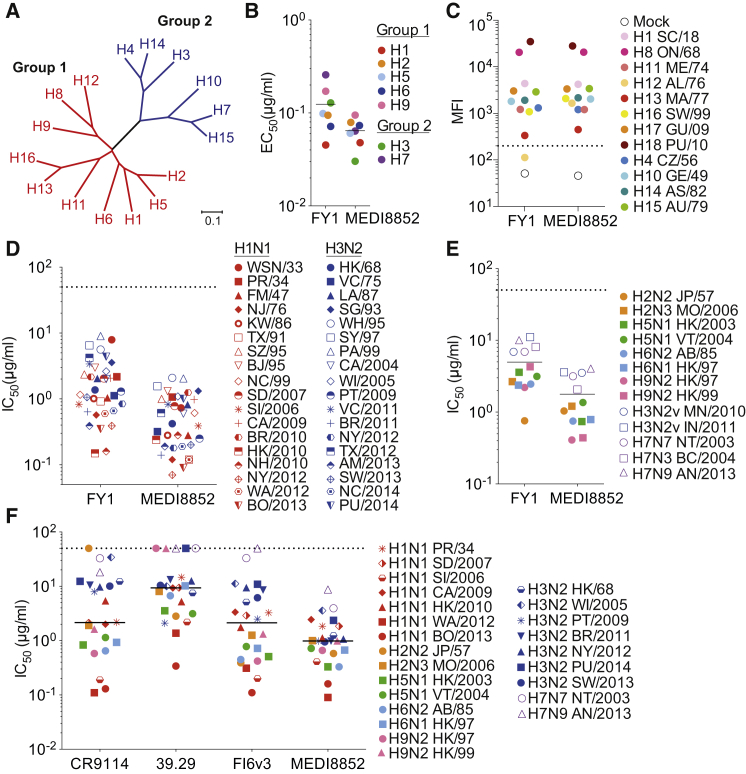
The FY1 antibody was chosen as the lead, based on its potency, breadth, and low somatic mutations for further in vitro optimization through parsimonious mutagenesis of the complementarity determining regions (CDRs) combined with reversion of unnecessary somatic mutations in the frameworks. The optimization focusing on affinity binding resulted in a 14-fold and 5-fold improved Fab affinity to H3 HA and H1 HA proteins determined by surface plasmon resonance, respectively (Table S2). The resulting antibody was named MEDI8852 (VH and VL sequences shown in Figure 1A) and was compared side by side with the parental FY1 antibody for binding and neutralization of a large panel of influenza A viruses. MEDI8852 showed higher binding activity as compared to FY1 against the group 1 HA proteins of H1, H2, H5, H6, and H9 subtypes and group 2 HA proteins of H3 and H7 subtypes, with a mean half-maximal effective concentration (EC50) of 0.064 μg/ml versus 0.124 μg/ml for MEDI8852 and FY1, respectively (Figure 2B; Table S2). In addition, we investigated the binding of FY1 and MEDI8852 to the remaining HAs including the 1918 H1N1 pandemic strain and two recently identified HA analogs recovered from bats (H17 and H18) (Tong et al., 2012, Tong et al., 2013), by flow cytometric analysis of cell-surface expressed HAs (Figure 2C). Of note, MEDI8852 bound to all HAs and gained reactivity against H12 HA over the parental FY1 antibody.
Figure 2.
MEDI8852 Binds to All Influenza A HA Subtypes and Exhibits Neutralization of Influenza A Seasonal and Non-seasonal Viral Strains
(A) Phylogenetic tree of influenza A HAs. Group 1 and group 2 colored in red and blue are further subdivided into 3 clades (H8, H9, and H12; H1, H2, H5, and H6; H11, H13, and H16) and 2 clades (H3, H4, and H14; H7, H10, and H15), respectively.
(B) ELISA binding average EC50 values of FY1 and MEDI8852 to purified recombinant HA proteins.
(C) Binding of FY1 and MEDI8852 to surface-expressed HA proteins as determined by flow cytometry. Shown are MFI values.
(D and E) FY1 and MEDI8852 neutralization IC50 values were determined against a panel of 36 seasonal influenza A isolates (D) and 13 non-seasonal influenza viruses (E).
(F) Neutralization average IC50 values of MEDI8852, 39.29, FI6v3, and CR9114 were determined from at least two independent experiments using a panel of 24 seasonal and non-seasonal influenza viruses and plotted as a single symbol. Full viral strains designations are listed in Tables S1 and S2.
To examine if the higher potency and breadth of binding activity of MEDI8852 as compared to FY1 translated into potent and broad antiviral activity, we measured neutralizing activity of both antibody variants in MDCK cells against a diverse panel of seasonal H1N1 and H3N2 viruses and emerging, potentially epidemic viruses, isolated over a period spanning >80 years (1933–2014). All seasonal influenza viruses tested were neutralized by FY1 and MEDI8852 with median IC50 values of 1.33 μg/ml and 0.51 μg/ml, respectively, resulting in nearly a 3-fold increase in overall potency (Figure 2D). However, both antibodies exhibited comparable activity in neutralizing group 1 and group 2 viruses with similar IC50 values (1.03 and 2.02 μg/ml for FY1 and 0.34 and 0.61 μg/ml for MEDI8852 against 18 H1N1 and 18 H3N2 viruses, respectively) (Figure 2D). The increase in overall activity of MEDI8852 compared to FY1 was also apparent when tested against 13 non-seasonal influenza viruses including H5 and H7 viruses isolated from recent human infections. MEDI8852 neutralized these viruses having an overall median IC50 of 1.21 μg/ml (range 4.05–0.41 μg/ml) versus FY1 with a median IC50 of 3.59 μg/ml (range 11.05–0.76 μg/ml) (Figure 2E). These results indicate that the optimization of MEDI8852 resulted in a 3-fold increase in the potency of neutralization and the ability to bind to all HA subtypes of influenza A viruses.
To extend the evaluation, we directly compared the in vitro neutralization activity and breadth of MEDI8852 with the previously published cross-group neutralizing mAbs FI6v3, CR9114, and 39.29 (Corti et al., 2011, Dreyfus et al., 2012, Nakamura et al., 2013), using a diverse panel of seasonal and non-seasonal influenza strains from group 1 and group 2 (Figure 2F). As reported previously, these antibodies neutralized group 1 and group 2 viruses although they exhibited distinct differences in both potency and breadth. Among all the antibodies tested, MEDI8852 is the only one that demonstrated neutralizing activity against all the viruses tested with a median IC50 0.99 μg/ml (range 8.75–0.09 μg/ml). CR9114 failed to neutralize the human H2N2 A/Japan/57 virus. Both FI6v3 and 39.29 were unable to neutralize the contemporary human isolate H7N9 A/Anhui/2013, and 39.29 was incapable of neutralizing the A/Netherlands/2003 H7N7 virus, both H9N2 viruses, and a contemporary H3N2 virus, A/Palau/2014, at the highest concentration tested (50 μg/ml). In addition to better overall breadth of neutralization, MEDI8852 exhibits equal or greater neutralization potency than the other cross-reactive monoclonal antibodies with a median IC50 of 0.99 μg/ml, compared to 2.13, 7.57, and 1.76 μg/ml for CR9114, 39.29, and FI6v3, respectively, when the non-neutralized viruses are excluded from the analysis.
MEDI8852 Mechanisms of Antiviral Activity
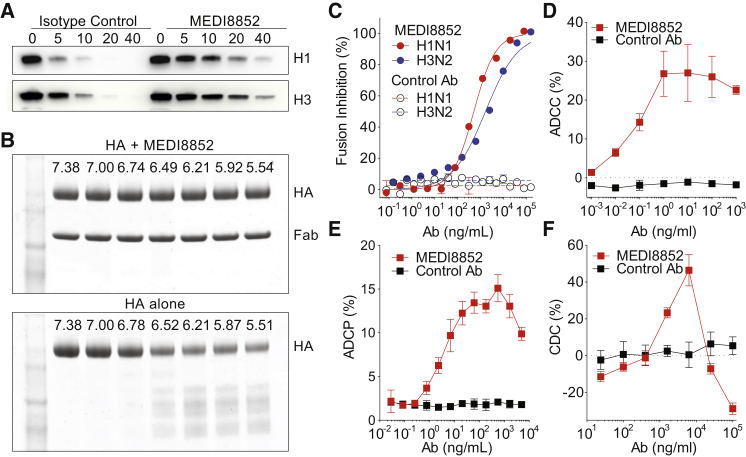
The cross-subtype neutralizing antibodies reported to date inhibit HA-mediated membrane fusion activity in vitro (Corti et al., 2011, Dreyfus et al., 2012, Nakamura et al., 2013). Activation of fusion requires cleavage of the precursor, HA0, and exposure of the cleaved HA to the low pH of endosomes. In assays of these two processes, we have shown that MEDI8852 inhibits the host cell protease cleavage of both H1 (group 1) and H3 (group 2) HA0 that would prevent membrane fusion (Figure 3A), and MEDI8852 binding to cleaved HA also prevents its low pH-induced conformational change, which is required for membrane fusion by stabilizing the pre-fusion conformation (Figures 3B and 3C).
Figure 3.
MEDI8852’s Antiviral Mechanisms of Action
(A) HA cleavage inhibition assay of uncleaved HA0 recombinant proteins of A/New Caledonia/20/99 (H1N1) or A/Hong Kong/8/68 (H3N2), pre-treated with MEDI8852 or a non-relevant isotype control antibody, MPE8v3, after digestion with TPCK-trypsin for 0, 5, 10, 20, or 40 min.
(B) Inhibition of low pH-activated conformational change in HA showing SDS PAGE gels of H5 HA with and without MEDI8852, incubated at decreasing pH values and neutralized after digestion with TPCK- trypsin.
(C) Fusion inhibition assay using MEDI8852 (solid) or MPE8v3 (open) incubated with A/Puerto Rico/8/34 (H1N1) virus (red) or A/Aichi/2/68 (H3N2) virus (blue) and human red blood cells and exposed to low pH to induce viral fusion. Percent fusion inhibition was calculated based on the amount of hemoglobin present in the supernatant.
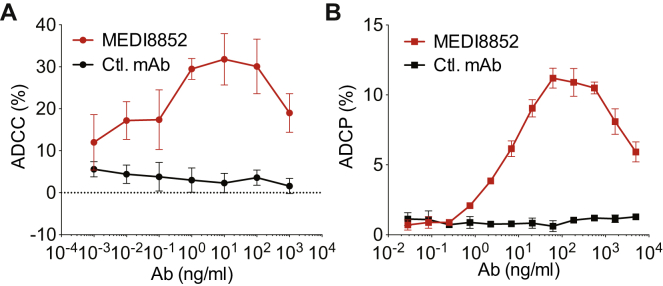
(D) ADCC activity on A549 cells infected with A/Puerto Rico/8/34 (H1N1) and incubated with MEDI8852 (red) or MPE8v3 (black) antibody in the presence of human NK cells, antibody-dependent killing was measured in quadruplicate by LDH release.
(E) ADCP activity on MDCK cells expressing H1 HA from A/South Dakota/06/2007 that were labeled CFSE and incubated with MEDI8852 (red), or an irrelevant control, R347 (black) antibody in the presence of violet-labeled human macrophages in duplicate. Percent phagocytosis was determined by the amount of total macrophages that were labeled with violet and CFSE.
(F) CDC activity on MDCK cells infected with A/Puerto Rico/8/34 (H1N1) and incubated with a serial dilution of MEDI8852 (red) or MPE8v3 (black) antibody in the presence of rabbit complement. Antibody-dependent killing was measured in triplicate by LDH release. Error bars represent two times the SD at each antibody concentration.
See also Figure S2.
In addition, we have shown that MEDI8852 mediates the lysis of infected cells by human primary NK cells (ADCC), the antibody-dependent cellular phagocytosis (ADCP) of MDCK cells expressing H1 or H3 HAs by human monocyte-derived macrophages and the complement-dependent cytotoxicity (CDC) of influenza-infected MDCK cells in the presence of complement (Figures 3D, 3E, 3F, and S2).
Figure S2.
MEDI8852 Fc-Effector Function Activity against H3N3 Viruses, Related to Figure 3
(A) ADCC activity on A549 cells infected with with A/Honk Kong/8/68 (H3N2) and incubated with MEDI8852 (red solid dot) or an irrelevant control, MPE8v3 (black solid dot) antibody in the presence of human NK cells, antibody dependent killing was measured in quadruplicate by LDH release.
(B) ADCP activity on MDCK cells expressing H3 from A/Hong Kong/8/68 that were labeled with CSFE and incubated with MEDI8852 (red solid square), or an irrelevant control, R347 (black solid square) antibody in the presence of violet labeled human macrophages in duplicate. Percent phagocytosis was determined by the amount of total macrophages that were labeled with violet and CSFE. Error bars represent two times the standard deviation at each antibody concentration.
Prophylactic and Therapeutic Efficacy of MEDI8852 in Mice and Ferrets
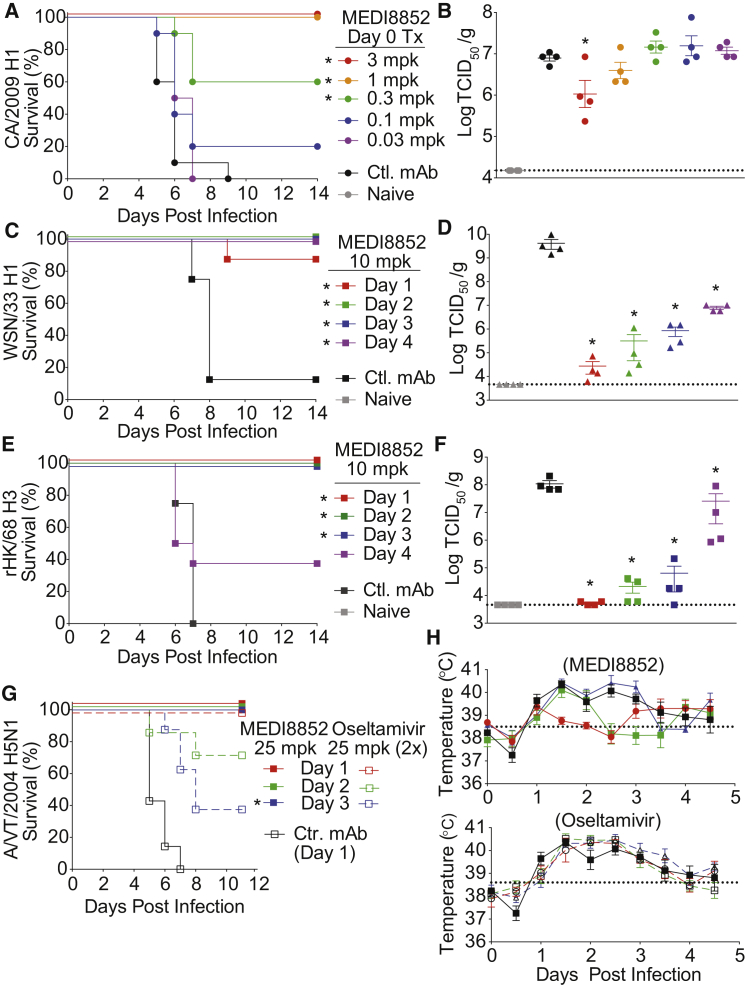
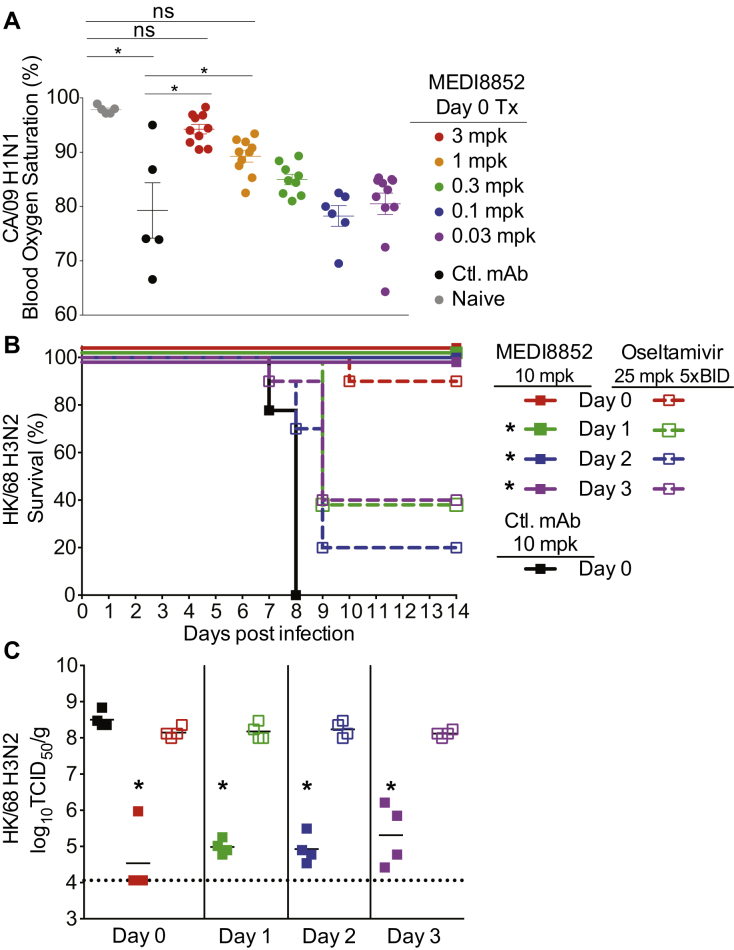
We evaluated the antiviral activity of MEDI8852 in mice challenged with a lethal dose of three different influenza A viruses, A/California/7/2009 H1N1 (CA/2009 H1), A/Wilson Smith N/33 H1N1 (WSN/33 H1), and a reassortant A/Hong Kong/8/68 H3N1 (rHK/68 H3). A dose-ranging study was conducted in which MEDI8852 was administered at the time of virus challenge. Mice (100%) receiving MEDI8852 at 3 or 1 mg/kg survived challenge with CA/2009 H1, while 60% and 20% of mice survived that received 0.3 and 0.1 mg/kg of MEDI8852, respectively (Figure 4A). Consistent with the survival data, lung viral titers were significantly reduced compared to control antibody-treated animals when MEDI8852 was administered at 3 mg/kg (Figure 4B). In addition, we observed that the two highest doses of MEDI8852 significantly protected lung function in mice compared to the control antibody as measured by pulse oximetry (Figure S3A).
Figure 4.
MEDI8852 Provides Dose-Dependent Protection from Lethal Influenza Infection in Mice and Ferrets Even When Treatment Was Delayed
(A) Kaplan-Meier survival curves.
(B) Lung viral titers on day 5 post infection determined by TCID50 assay after mice were treated with MEDI8852 at 3, 1, 0.3, 0.1, and 0.03 mg/kg (single i.p. dose) and then infected with CA/2009 H1.
(C) Kaplan-Meier survival curves.
(D) Lung viral titers on day 5 post infection in mice infected with WSN/33 H1 virus, on study day 0, then treated with MEDI8852 or irrelevant control antibody, R347 (single i.p. dose) at 10 mg/kg at various days post infection.
(E) Kaplan-Meier survival curves.
(F) Lung viral titers on day 5 post infection in mice infected with rHK/68 H3 virus, on study day 0, then treated with MEDI8852 at 10 mg/kg or R347 (single i.p. dose) at various days post infection.
(G) Kaplan-Meier survival curves of ferrets infected with 1LD90 of A/Vietnam/1203/2004 H5N1 virus on study day 0. Treatment with MEDI8852 at 25 mg/kg (closed symbols solid line), oseltamivir at 25 mg/kg (open symbol dashed line), or R347 (open symbol solid line) was initiated at the indicated day post infection.
(H) Temperature of ferrets treated with MEDI8852, or oseltamivir at various days post infection. Dotted line designates the average normal temperature of a ferret at 38.5°C. Error bars represent the SE of the mean for each determination. ∗For murine studies, significance was determined compared to control antibody treatment with p < 0.005 for survival (log-rank test) and p < 0.05 for lung viral titers (Student’s t test); for ferret survival studies, significance was determined by comparing to oseltamivir on the indicated initiation day with p < 0.05 for survival (log-rank test).
Figure S3.
MEDI8852 Provides Dose-Dependent Protection from Lethal Influenza Infection in Mice Even When Treatment Was Delayed, Related to Figure 4
(A) Lung function was measured by pulse oximetry on Day 6 post-infection with CA/09 H1 in mice treated with mice were treated with a single IP dose of MEDI8852 at 3, 1, 0.3, 0.1, and 0.03 mg/kg, 3 mg/kg irrelevant control antibody, or non-treated non-infected animals (naive). Error bars represent two times the standard error of the mean at each antibody concentration. ∗p < 0.05 significance determined by ANOVA with multiple comparison’s test.
(B) Kaplan-Meier survival curves.
(C) Lung viral titers on Day 5 post infection determined by TCID50 assay after mice were infected with rHK/68 H3 virus on study Day 0, then treatment with MEDI8852 at 10 mg/kg (closed symbols with solid line), Oseltamivir at 25 mg/kg BID for 5 days (open symbol with dashed line, or irrelevant control antibody, R347 (black solid) was initiated at the indicated Day post infection. Error bars represent the standard error of the mean for each determination. ∗, For survival studies p < 0.005 significance compared to control antibody treatment by Log-Rank test; for lung titers, p < 0.05 significance compared to control antibody treatment by student’s t test.
To evaluate the therapeutic utility of MEDI8852, we administered MEDI8852 at different time points following infection with WSN/33 H1 or rHK/68 H3 virus. At a dose of 10 mg/kg, survival rates of 90%–100% were achieved even when treatment was delayed until day 4 post infection with WSN/33 H1, or day 3 post infection with rHK/68 H3 (Figures 4C and 4E). Significant survival benefits were also seen with 1 and 3 mg/kg doses when administered on days 1, 2, or 3 post infection, albeit lower survival rates than the 10 mg/kg (Table S3). MEDI8852 treatment of 10 mg/kg at all times post infection resulted in significantly decreased viral titers, compared to control antibody treated and untreated animals, with a clear trend for greater reductions with earlier treatment (Figures 4D and 4F).
To further investigate MEDI8852’s therapeutic potential, we determined the therapeutic window for treating ferrets infected with the highly pathogenic avian influenza virus, A/Vietnam/1204/2004 H5N1 (VT/2004 H5N1). In these studies, ferrets were infected intranasally with 1 LD90 of VT/2004 and then treated with a single intravenous (i.v.) dose of 25 mg/kg of MEDI8852 at 1, 2, or 3 days post infection. We also used as a comparator, the anti-influenza drug oseltamivir at 25 mg/kg twice a day (b.i.d.) (Figure 4G). As expected, all control animals showed signs of infection including fever peaking from days 1–3 post infection and 100% mortality by day 7 post infection (Figures 4G and 4H). In comparison, ferrets treated with MEDI8852 or oseltamivir on day 1 post infection were completely protected. When treatment was delayed until 2 or 3 days post infection, MEDI8852 provided complete protection with 100% survival while oseltamivir only partially protected animals with survival rates of 71% and 29%, respectively. In addition, MEDI8852 treatment resulted in a period of fever reduction following administration, which was not observed in oseltamivir or control antibody-treated animals (Figure 4H). Similar efficacy and therapeutic window results were seen when MEDI8852 and oseltamivir treatments were compared in a lethal murine model (Figures S3B and S3C).
The Structures of Complexes Formed between MEDI8852 Fab and H5, Group 1, and H7, Group 2, HAs
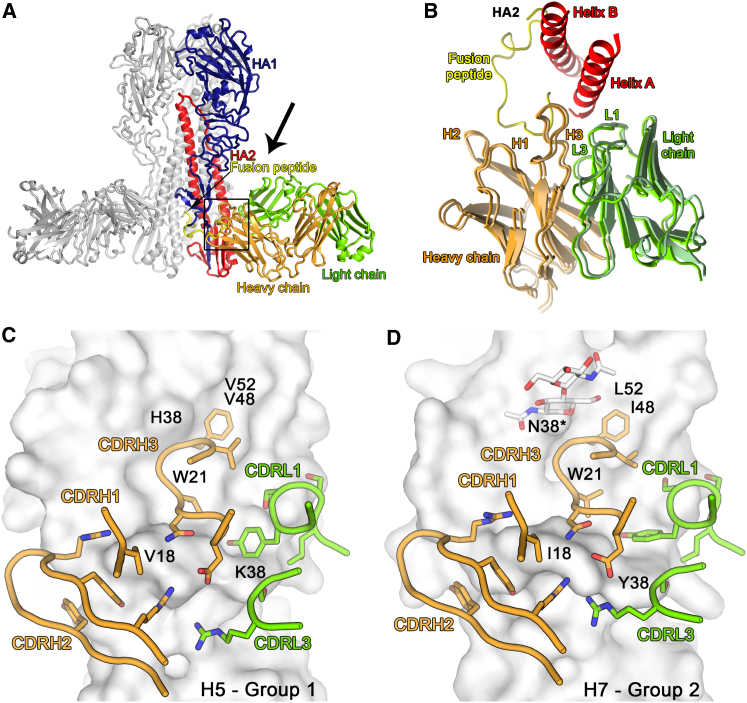
To provide insight into the structural basis for MEDI8852 breadth and potency, we have determined the structures of the MEDI8852 Fab fragment at 1.9 Å and of its complexes with H5 and H7 HA proteins at 3.7 Å and 3.75 Å resolution, respectively (Figures 5A and S4A). The structures of the HA proteins in both complexes are similar to those of the apo structures determined before (Russell et al., 2004, Xiong et al., 2015). MEDI8852 makes similar contacts with both H5 and H7 HAs, by binding in a very similar orientation to both HAs (Figures 5B–5D), each Fab interacting with just one protomer of the HA trimer. Overall, the interactions bury 1,750 and 1,646 Å2 from solvent for the H5 and H7 complexes, respectively, consistent with their high binding affinity (Table S2). MEDI8852 contacts the fusion domain of HA and interacts with three regions of HA2, a central hydrophobic groove, the fusion peptide and helix A, and with specific residues in the HA1 component of the fusion domain (Figure 6). The location of MEDI8852 in the complex with the HA proteins is consistent with the in vitro assays showing that MEDI8852 stabilized the pre-fusion conformation inhibiting fusion as well as blocking the proteolytic cleavage of the HA precursor on the neighboring subunit (Figure 3).
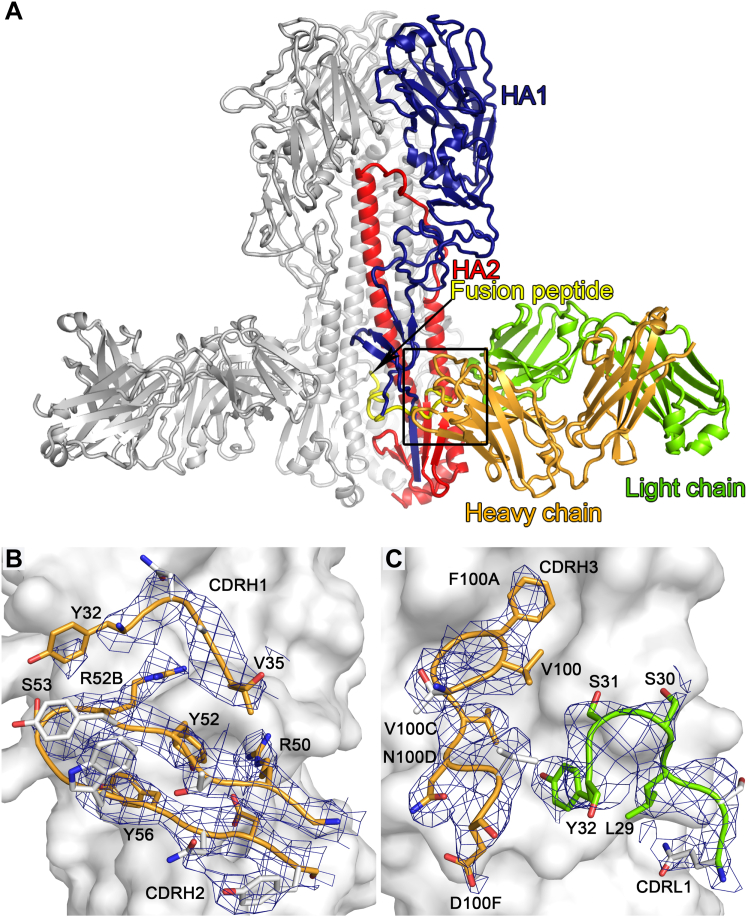
Figure 5.
MEDI8852 Binds to a Unique Site within the H5 and H7 HA Proteins
(A) Overview of MEDI8852 in complex with H5 hemagglutinin. One HA protomer and the cognate MEDI8852 Fab fragment are highlighted in color, the other two copies in the trimer are colored gray. The HA1 polypeptide is colored blue, the HA2 polypeptide is colored red, with the fusion peptide at the N terminus of HA2 highlighted in yellow. The heavy chain of the MEDI8852 Fab is colored orange, the light chain is colored green.
(B) Overlay of MEDI8852 bound to group 1 (H5) and group 2 (H7) HAs. The antibodies are shown in cartoon representation together with Helices A and B of the HA. The components are colored according to (A) and the view orientation is approximately that shown by the black arrow in (A).
(C and D) H5 (C) and H7 (D) HAs are shown in surface representation. Only the HA residues in the MEDI8852 binding epitope that differ between H5 and H7 are labeled. The CDR loops of MEDI8852 that are in contact with HA are shown in cartoon and stick representation and colored by element.
See also Figures S4, S7, and Table S4.
Figure S4.
Overview of MEDI8852 in Complex with H7 Haemagglutinin and Unbiased Electron Density Maps, Related to Figure 5
(A) One H7 HA monomer and the cognate MEDI8852 Fab fragment are highlighted in color, the other two copies in the trimer are colored gray. The HA1 polypeptide is colored blue, the HA2 polypeptide is colored red, with the fusion peptide at the N terminus of HA2 highlighted in yellow. The heavy chain of the MEDI8852 Fab is colored orange, the light chain is colored green.
(B and C) The MEDI8852 CDR loops H1 and H2 (B) and H3 and L1 (C) are shown in complex with H5 HA. All six instances of the variable domain of the heavy or light chains were omitted from the model after rigid-body refinement and omit maps were calculated. Figures of merit were determined in Sigmaa and six-fold NCS averaging was performed in DM. The maps were sharpened by a negative B-factor of 75 Å2. The resulting maps are shown at a σ level of 1.0 within 2.0 Å distance of MEDI8852 atoms. H5 HA is shown in surface representation and colored gray, MEDI8852 is shown in cartoon and stick representation and colored orange for the heavy chain and green for the light chain. MEDI8852 side chains that are not in contact with HA are colored gray.
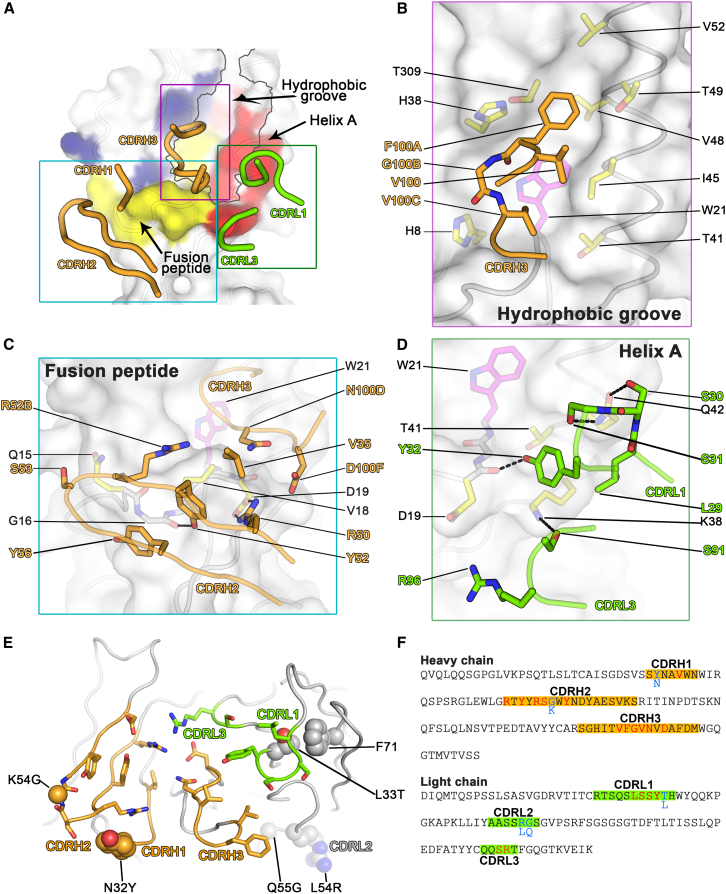
Figure 6.
Binding Epitope of MEDI8852 on H5 HA
(A) HA is shown in surface representation and residues that are contacted by MEDI8852 are highlighted in color (blue for HA1, red for HA2 and yellow for fusion peptide residues). Secondary structure elements of HA are shown in cartoon representation. The hydrophobic groove on HA is outlined in gray. The CDR loops of MEDI8852 that are in contact with HA are shown in cartoon representation and colored orange and green for the heavy and light chains, respectively. The colored boxes indicate the three parts of the binding epitope that are shown in more detail in (B), (C), and (D).
(B) Interactions of MEDI8852 with the hydrophobic groove of H5 HA. HA is drawn in surface representation, with the main chain shown in cartoon representation and amino acids that are in contact with MEDI8852 shown in stick representation. W21, which adopts different rotamers in group1 and group 2 influenza viruses, is colored magenta. MEDI8852 is also shown in cartoon representation, with contact residues shown in stick representation. Hydrogen bonds and salt bridges are indicated by dashed lines.
(C) Interactions of MEDI8852 with the fusion peptide of H5 HA. Shown in the same style as in (B).
(D) Interactions of MEDI8852 with the base of helix A of H5 HA. Shown in the same style as in (B).
(E) Location of mutations found during affinity maturation of FY1 to MEDI8852. The variable domains of MEDI8852 are shown in cartoon representation, viewed from the direction of HA. Regions of the heavy and light chains in contact with HA are colored orange and green, respectively. Interacting sidechains are shown in stick representation. Residues that differ between the parental and affinity-maturated antibody are shown in sphere representation.
(F) Sequences of MEDI8852 variable region framework and CDR residues. CDRs (according to Kabat) are highlighted in orange and green for the heavy and light chains respectively. Residues in contact with HA are colored red and residues changed during affinity maturation from FY1 are colored cyan, with corresponding residues of FY1 indicated.
See also Figure S6.
Although the structures of the complexes were determined at intermediate resolution, the interfaces between the HAs and MEDI8852 Fab are well-ordered and the electron density maps in these areas are among the clearest of the overall complexes (unbiased omit electron density maps are shown in Figures S4B and S4C). We can, therefore, have confidence in our description of the inter-molecular contacts: hydrogen bonds described in the text should be regarded as potential interactions, however, given the limitations of defining the exact geometry of these interactions. The principal contact areas involve three CDRs, CDRH3, CDRH2, and CDRL1, with minor interactions with CDRH1 and CDRL3 (Figure 6A). CDRH3 makes extensive contacts with the bottom of a hydrophobic groove between helix A of HA2 and the fusion domain component of HA1 (Figures 6B and 6C). Phe100A(CDRH3) inserts into this groove made by HA2 residues Ile45, Val48, Thr49, and Val52 of helix A, Trp21 of the fusion peptide, and Thr309 of HA1. Val100C(CDRH3) binds in a lower position in the same groove and interacts with the main chain of HA2 residues 19–21 of the fusion peptide as well as with the side chains of Trp21 and of HA1 His8 (Figure 6B). Asn100D(CDRH3) also makes hydrophobic contact with Val18 in the fusion peptide (Figure 6C). The CDRH2 loop interacts, through VH germline-encoded residues, with HA2 residues 15–19 of the fusion peptide (Figure 6C). In particular, HA2 Val18 is almost completely protected from solvent by Tyr56(CDRH2) and Arg52B(CDRH2). There is also van der Waals interaction between the Cα of HA2 Gly16 and Tyr52(CDRH2). Tyr52(CDRH2) is positioned within hydrogen bonding distance of Gly16. Arg50(CDRH2) forms a salt bridge with HA2 Asp19. Arg50(CDRH2) also contributes to a polar patch on the antibody surface that includes Arg96(CDRL3) and Asp58(CDRH2) and Asp100F(CDRH3). Finally, CDRL1 interacts with the N-terminal region of helix A of HA2 (Figure 6D). Ser30(CDRL1) and Ser31(CDRL1) are in hydrogen bonding distance of HA2 Gln42, while Tyr32(CDRL1) and Leu29(CDRL1) make hydrophobic contacts with the aliphatic moiety of HA2 Lys38 of helix A. The side chain of Tyr32(CDRL1) stacks against HA2 Gln42 of helix A and its hydroxyl group is in hydrogen bonding distance of the main chain of HA2 Asp19 of the fusion peptide.
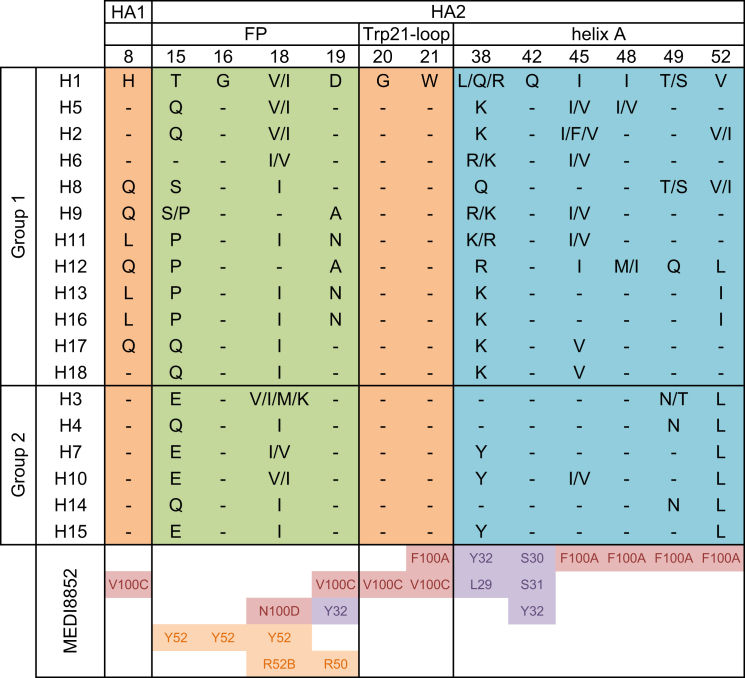
The epitope recognized by MEDI8852 is highly conserved between both HA proteins, consistent with the antibody’s broad activity against group 1 and group 2 influenza viruses (Figures 5C, 5D, and S5). However, the membrane proximal fusion domains of both group 1 and group 2 HAs have a few distinct structural differences such as glycosylation status of HA1 position 38, which could potentially affect the binding of some antibodies (Corti et al., 2011, Ekiert et al., 2009, Sui et al., 2009). In the H7 complex with MEDI8852, the bulky carbohydrate chain attached to HA1 Asn38 changes its orientation to allow antibody binding, as observed before in the FI6 Fab-H3 HA complex (Corti et al., 2011). Another notable feature of the MEDI8852 complex with H7 HA is the involvement of HA2 Tyr38 in an aromatic stacking interaction with Tyr32(CDRL1). Interestingly, the tyrosine at position 38 is not conserved across all HA subtypes only in H7, H10, and H15. The arginine in H6, H9, H11, and H12 could engage in a similar stacking interaction although the leucine, lysine, and glutamine found in the remaining HA proteins could not interact in the same way. Thus, the high and similar affinities with which MEDI8852 reacts with these HAs, suggests that the differences in glycosylation at HA1 38 and the side chain at HA2 38 make a minor contribution to the overall energetics of binding (Figure 2B; Table S2).
Figure S5.
Sequence Comparison of MEDI8852 Epitope among 18 HA Subtypes and Corresponding Contact Residues in the MEDI8852 Paratope, Related to Figures 5 and 6
The occurrence of amino acid identities was analyzed at different HA residues among each HA subtype, only amino acids with a frequency higher than 1% are shown. In total, 12′472 H1, 404 H2, 12′263 H3, 1’005 H4, 694 H5, 1’152 H6, 119 H7, 119 H8, 1’838 H9, 590 H10, 487 H11, 153 H12, 67 H13, 16 H14, 10 H15, 7 H16, 2 H17 and 1 H18 isolates were analyzed, respectively.
Conformational Rearrangements in MEDI8852 on Complex Formation
The availability of a high-resolution structure of MEDI8852 Fab and of well-ordered interfaces in the structures of the complexes formed with H5 and H7 HAs enables us to analyze conformational changes in the Fab upon HA binding, particularly in the CDRH3 and CDRL1 loops. The loop formed by residues 97–100F of CDRH3 undergoes a largely rigid-body rotation, pivoted around Gly96(CDRH3) and Ala100G(CDRH3) (Figure 7A) to facilitate interactions with HA. As a consequence, the side chain of Phe100A(CDRH3) moves by ∼5 Å, to insert into the hydrophobic groove of the epitope, near HA2 48. In addition, residues 27–32(CDRL1) are restructured, with an average displacement of ∼10 Å between apo and bound forms. The reorientation of the side chains of Tyr32(CDRL1) and Leu29(CDRL1) enables them to interact with HA2 Tyr38 in the H7 complex. Ser30(CDRL1) and Ser31(CDRL1) in the complex form a helical structure that places them in hydrogen bonding distance with Gln42 of HA2.
Figure 7.
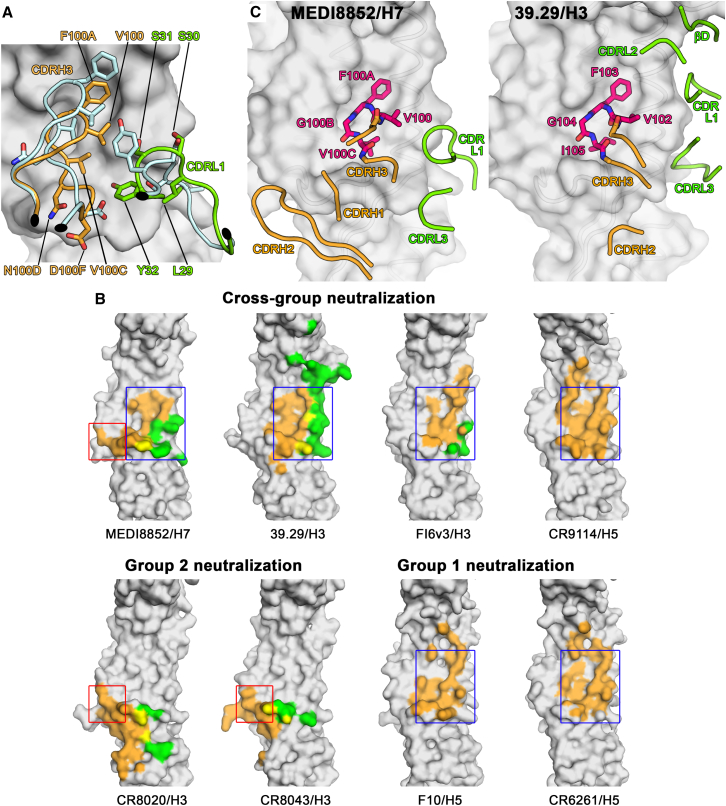
MEDI8852 Binds to a Unique Site within the H5 and H7 HA Proteins through CDR-H3 and CDR-L1 Conformational Rearrangements upon Complex Formation
(A) Conformational rearrangements in MEDI8852 on complex formation. Conformational change of the CDRH3 and CDR-L1 loops upon HA engagement. The apo structure of MEDI8852 is shown in blue, the bound structure is shown in orange and green for the heavy and light chains, respectively. The beginning and end of the moving regions are indicated with black ovals. HA (H7) is shown as a gray surface. The apo structure does not make interactions with HA and does not fit into its surface features—the conformational change is necessary for productive HA engagement.
(B) Epitopes of different broadly neutralizing antibodies on the HA surface. Residues of HA that are in contact with the heavy chain are colored orange, residues that are in contact with the light chain are colored green, and residues that are in contact with both chains are colored yellow. The blue box encases the part of the MEDI8852 epitope (helix A and hydrophobic groove) that can also be found in other broadly neutralizing antibodies as well as group 1 specific ones. The red box encases the part of the MEDI8852 epitope that can also be found in group 2 specific antibodies (middle of fusion peptide).
(C) Comparison of the structures of the conserved CDRH3 tetra-peptide in the complexes between MEDI8852 and H7 HA (left panel) and 39.29 and H3 HA (right panel). In both cases the tetra-peptide is shown in stick representation with other loops of the antibody shown as coil, colored as in panel A. The HAs are shown in surface representation.
See also Figure S7.
There are five mutations found in the vitro optimization of FY1 to MEDI8852 that are not in direct contact with HA, but are contained within CDR loops (Figures 6E and 6F). The mutated residues are seen to stabilize the conformations that the loop regions adopt in complex with HA while the parental residues (FY1) do not appear to be able to make similar stabilizing interactions (Figure S6). Thus, the optimization process of FY1 to MEDI8852 results in the selection of amino acid substitutions that stabilize the induced fit conformation that the CDR loops adopt in complex with HA.
Figure S6.
Mutations Associated with Maturation of FY1 to MEDI8852, Related to Figure 6
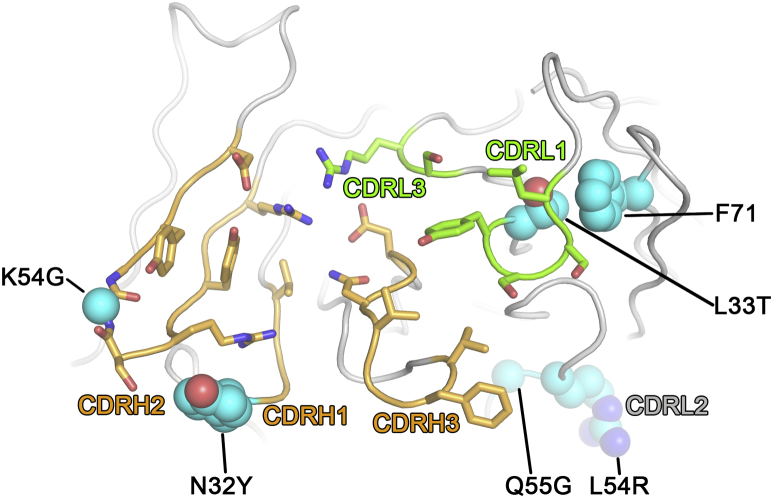
Heavy chain: Y32 (MEDI8852) instead of N32 (parental): The Y32 sidechain is within hydrogen bonding distance with HA and makes a stabilizing π stacking interaction with R52B, which is in direct contact with V18 in the fusion peptide of HA. G54 (MEDI8852) instead of K54 (parental): This amino acid is located in the middle of the CDRH2 loop in an unusual turn between the two beta strands of CDRH2. The backbone at G54 adopts a conformation that is only possible with a glycine sidechain. The parental K54 would lead to a different backbone conformation. Light chain: T33 (MEDI8852) instead of L33 (parental): This residue is next to CDRL1 and especially Y32, which undergo a conformational change upon binding. The T33 sidechain stacks against the sidechain of F71 in unbound and bound conformations, anchoring the entire CDRL1 region during the conformational change. The leucine in the parental antibody could make a similar stacking interaction, but would anchor CDRL1 in a different conformation due to the additional methyl group of the leucine. R54 and G55 instead of L54 and Q55 (parental): R54 and G55 are in CDRL2 and connected with CDRL1 through backbone hydrogen bonds. The sidechain of Q55 in the parental antibody would sterically clash with L46 and completely alter the backbone and sidechain conformation the LCDR1 and LCDR2 loops. G55 has no sidechain and thus does not clash. Therefore, R54 and G55 enable and stablilize the backbone conformation observed in the complex, which might form a stable platform for the conformational change in CDRL1 necessary for binding.
Comparison of the Epitope of MEDI8852 with Those of Other Broadly Neutralizing Antibodies
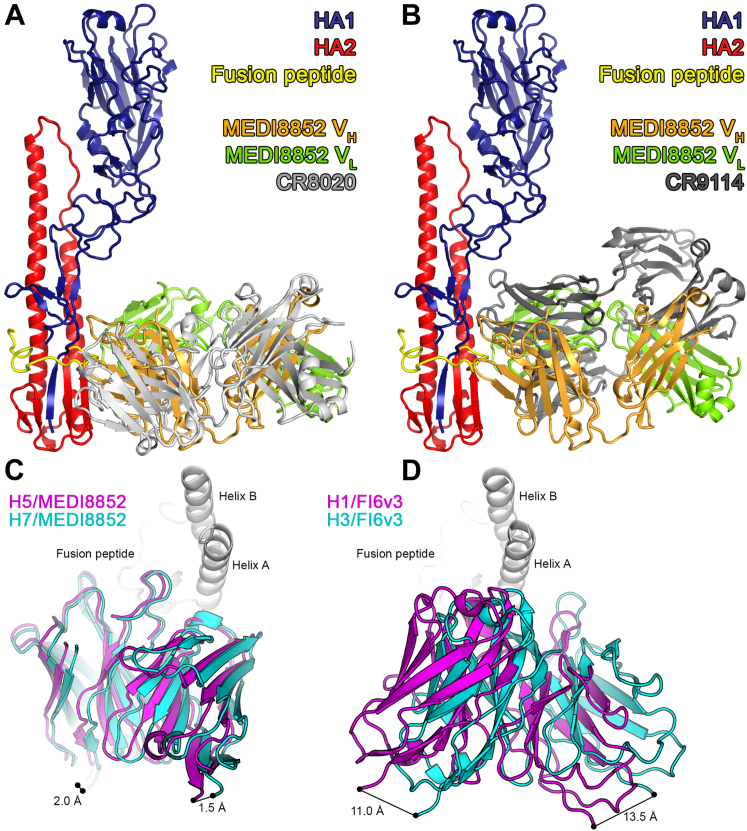
We have compared the mode of interaction of MEDI8852 with other broadly neutralizing antibodies that recognize the membrane proximal fusion domain of HA. The cross-group neutralizing antibodies 39.29, FI6v3, and CR9114 (Corti et al., 2011, Dreyfus et al., 2012, Nakamura et al., 2013), as well as the group 1-neutralizing antibodies F10 and CR6261 (Ekiert et al., 2009, Sui et al., 2009), all recognize helix A of HA2 and the adjacent hydrophobic groove (Figure 7B, blue box). In contrast, the group 2-neutralizing antibodies CR8020 and CR8043 (Ekiert et al., 2011, Friesen et al., 2014) recognize a different region of the fusion peptide and a small β sheet below it in the fusion domain (Figure 7B, red box). The epitope of MEDI8852, uniquely among the cross-group neutralizing antibodies reported to date, represents a combination of both regions. A structural comparison of HA-bound MEDI8852 overlapped with the antibodies CR8020 and CR9114 is shown in Figures S7A and S7B, which reveals that the overall orientation of MEDI8852 is such that it sits slightly higher on the HA than the CR8020 antibody and lower than CR9114. The nearest paratope residue of MEDI8852 is ∼20 Ǻ from the membrane proximal end of the HA.
Figure S7.
Comparison between MEDI8852, CR8020, CR9114, and FI6, Related to Figure 7
(A and B) Antibody-HA complex structures were overlayed by aligning their HA2 polypeptides. For clarity, only the HA of the MEDI8852/H5 complex is shown. All proteins are shown in cartoon representation, with HA1 colored blue, HA2 colored red and the fusion peptide colored yellow. The CR8020 (A) and CR9114 (B) Fab fragments are colored light and dark gray, respectively, and the MEDI8852 Fab fragment is colored orange (heavy chain) and green (light chain).
(C and D) The antibodies are shown in cartoon representation with group 1 bound antibody colored in magenta and group 2 colored in cyan from the N terminus of the heavy chain to the C terminus of the light chain. Helix A and helix B of HA2 are shown in cartoon representation. FI6v3 is rotated between group 1 and group 2 leading to a 11-13.5 Å distance between the ends of the variable domains. By contrast, for MEDI8852, there is a much smaller rotation of about 1 degree and only 2-1.5 Å displacement between the ends of the variable domains in the complexes with H5 HA (group 1) and H7 HA (group 2).
MEDI8852 and 39.29 antibodies bind similarly to residues in the hydrophobic groove and adjacent helix A in the fusion domain. Both antibodies contain the four amino acid sequence ValPheGlyVal/Ile in their otherwise dissimilar CDRH3 loops. These tetrapeptides superpose in the complexes with an all-atom root-mean-square deviation (RMSD) of 0.7 Å, indicating that they interact with their cognate HAs in a similar way (Figure 7C). However, other contacts made by these antibodies with HA are quite different between MEDI8852 and 39.29, reflecting the fact that the two antibodies are not particularly similar in sequence and are derived from different germline sequences (VH6-1∗01 and VK 1-39∗01 for MEDI8852 versus VH3-30∗01 and VK3-15∗01 for 39.29). MEDI8852 contacts the base of helix A and the fusion peptide with its CDRL1 and CDRH2 loops, respectively (Figure 6). By contrast, contacts made by the heavy chain of 39.29 Fab mainly involve CDRH3. 39.29 appears to bury helix A using all three light chain CDR loops. As a consequence, the 39.29-HA interaction buries a total of 2,287 Å2, while MEDI8852 achieves similar affinity with a smaller buried surface of 1,646 Å2.
MEDI8852 is the second cross-group neutralizing antibody for which structures of complexes with both group1 and group 2 HAs have been reported. The first was FI6v3, which, although it recognizes both group 1 and group 2 HAs, has higher in vivo neutralizing activity against group 1 viruses (Corti et al., 2011). The cross-group binding of FI6v3 has been attributed to its long and flexible HCDR3, which can accommodate the differences in conformation and environment of HA2 Trp21 observed between group 1 and group 2 viruses. There is a much more significant rearrangement between FI6v3 bound to H1 HA (group 1) versus H3 HA (group 2) than there is between MEDI8852 in its complexes with H5 HA (group 1) and H7 HA (group 2) which overlap very closely (Figures 5B and S7C). MEDI8852, therefore, binds in a very similar way to HAs of both groups. This greater structural conservation of the binding interface is likely responsible for its broader neutralizing ability.
Discussion
There is an unmet medical need for effective treatments against severe influenza. The potential of broadly infectivity-neutralizing antibodies used therapeutically to address this need has provided a stimulus for their isolation and characterization. Among the antibodies considered to date, the anti-HA human monoclonal antibody MEDI8852 has demonstrated significant breadth of its infectivity-neutralizing capacity. MEDI8852 reacts with HAs of all influenza antigenic subtypes, potently neutralizes diverse virus strains with numerous HA subtypes, and can block infection and lethality caused by influenza viruses when administered up to 4 days after challenge with the virus in mice and up to 3 days post challenge in ferrets with the highly pathogenic H5N1 virus. This potential ability to overcome the unpredictable characteristics of influenza, namely the antigenic shift, that results in disease during pandemic periods, and the antigenic drift, that occurs with the emergence of antigenically novel viruses, is a major advantage for a candidate anti-influenza therapeutic antibody.
The mechanisms of MEDI8852-mediated neutralization of infection involve processes at the beginning and the end of the infection cycle. Binding of the antibody to HAs on the infecting virus inhibits HA-mediated membrane fusion that is required for the initiation of infection. At the end of infection, antibody binding to precursor HA0 can block its cleavage and prevent the formation and spread of newly made infectious virus. Additionally, binding of MEDI8852 to HAs displayed on the surfaces of infected cells results in their recognition and lysis by other components of the immune system: NK cells, macrophages, and complement. These multiple mechanisms exhibited by MEDI8852 presumably combine to ensure the observed effectiveness of antibody treatments in infected mice and ferrets.
The epitope recognized by MEDI8852 is consistent with the ways it blocks HA function in membrane fusion and with the locations of epitopes that have been described previously for influenza group-specific cross-reactive antibodies and for more broadly reactive antibodies. However, the regions of HA that interact with MEDI8852 are a combination of those previously assigned to group 1 specificity (primarily a hydrophobic groove and the adjacent helix A of HA) (Ekiert et al., 2009, Sui et al., 2009) or group 2 specificity (a separate part of the fusion peptide, near its N terminus) (Ekiert et al., 2011, Friesen et al., 2014). The structural characterization of MEDI8852 bound and unbound structures also highlight the coordinated movement of the CDRH3 and the CDRL1 to insert into the hydrophobic grove of the HA, as well as the rearrangement of the orientation of the glycan attached to Asn38 of the H7 virus to allow antibody binding. Importantly, structures of the complexes formed by MEDI8852 with H5 and H7 HAs indicate that the locations and orientations of the bound antibodies are very similar, and this structurally conserved ability to interact with both regions of HA presumably results in effective cross-reactivity.
The comparison of the structures of the complexes formed by MEDI8852 with H5 and H7 HAs with the previously reported complex formed between the cross-reactive monoclonal antibody 39.29 and H3 HA (Nakamura et al., 2013) indicated that the two antibodies had the amino acid sequences, V-F-G-V- MEDI8852 and V-F-G-I- 39.29, in their HCDR3 loops that occupied equivalent positions in the complexes. Conceivably, the structure of this shared component of the antibodies might be used in the preparation of immunogens or to select candidate molecules on the basis of their affinity for the tetra-peptides. Of note, a recent paper described the development of a computationally designed protein binding to Helix A in the stem region of group 1 HAs that showed in vitro and in vivo antiviral efficacy (Koday et al., 2016).
The reconstruction of the developmental pathway of MEDI8852, as well as of FI6, suggests that the generation of such broadly reactive antibodies may require a stepwise stimulation by group 1 HAs, followed by the selection of mutated variants by group 2 HAs. Indeed, the MEDI8852 donor was born in the 1950s and it is possible that this lineage was primed by H2N2 and further matured through multiple H3N2 exposures. This hypothesis is further strengthened by the observation that the UCA mAb neutralized with high potency the strain H2N2 JP/57 (i.e., IC50 = 0.6 μg/ml). The UCA and mutated antibodies of the MEDI8852 and FI6 lineages represent useful tools to design stem-based immunogens that can be used in a heterologous prime-boost mode to prime the group 1 reactive naive B cells and selectively expand those that also cross-react with group 2 HAs.
Based on the results reported, MEDI8852 is currently being evaluated for safety and efficacy in adults with uncomplicated influenza infection in an outpatient setting (https://.clinicaltrials.gov: NCT02603952) prior to conducting studies in patients hospitalized with influenza caused by type A strains.
Experimental Procedures
Monoclonal Antibody Isolation and Ex Vivo Affinity Maturation
Monoclonal antibodies were isolated from memory B cells, as previously described (Pappas et al., 2014, Traggiai et al., 2004) from blood donors who had given written informed consent, following approval by the Cantonal Ethical Committee of Cantone Ticino, Switzerland. FY1 antibody was further modified to revert the non-germline framework amino acid changes and its affinity was improved through parsimonious mutagenesis of CDRs (Supplemental Experimental Procedures).
Recombinant HA Protein and Binding Assays
Recombinant HA proteins were expressed and purified as previously described (Benjamin et al., 2014). The binding of antibodies to HAs was measured by ELISA or by staining HA transfected cells using flow cytometry (Supplemental Experimental Procedures).
Viruses and Microneutralization Assay
Wild-type influenza strains and cold-adapted (ca) live-attenuated influenza vaccine viruses (complete viral strain designations shown in Table S1) were propagated in embryonated chicken eggs, titered, and used to infect MDCK cells to determine neutralizing activity as described in the Supplemental Experimental Procedures.
In Vitro Fusion and HA Cleavage Assays
Antibody-mediated fusion inhibition was tested using a low pH-induced red blood cell fusion model adapted from protocol described in Wang et al. (2010). The ability of MEDI8852 to inhibit the low pH-activated conformational change in trypsin-digested H5 HA was analyzed by SDS/PAGE. The ability of antibody to block the HA0 cleavage by TPCK-treated trypsin was measured by western blot analysis (Supplemental Experimental Procedures).
Measurement of Fc-Effector Function
ADCC activity was measured with the LDH release assay using primary human NK cells as effector cells and H1N1 or H3N2 influenza virus infected A549 cells as a target. ADCP activity was measured by flow cytometry using fluorescently labeled monocyte-derived macrophages and H1 or H3 HA-expressing MDCK as target cells. CDC activity was measured with the LDH release assay using rabbit complement on influenza H1N1-infected MDCK cells (Supplemental Experimental Procedures).
Therapeutic Efficacy Studies in Mice and Ferrets
All animal studies were approved and conducted in accordance with MedImmune’s Institutional Animal Care and Use Committee (murine studies) and Southern Research Institute’s Institutional Animal Care and Use Committee (ferret studies) and performed in Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-certified facilities.
MEDI8852 or R347 control mAb was administered as a single intraperitoneal (i.p.) dose at various days post infection, depending on the virus strain. For oseltamivir comparison studies, mice were administered 25 mg/kg oseltamivir by mouth (PO) b.i.d. for 5 days, or a single 10 mg/kg dose i.v. of MEDI8852 with vehicle PO b.i.d. for 5 days. Viral loads in the lungs were measured by TCID50 assay on day 5 post infection. Five- to six-month-old ferrets were challenged intranasally with A/Vietnam/1203/04 (H5N1) virus and treated with a single 25 mg/kg i.v. dose of MEDI8852 (or R347 control) or oseltamivir at 25 mg/kg BID for 5 days initiated at different days post infection. Bio-metric data systems chip was used for temperature monitoring. (Supplemental Experimental Procedures).
HA-MEDI8852 Complex Preparation, Crystallization, and Structure Determination
H5 and H7 HAs were purified from the virus membrane and mixed with purified MEDI8852 antibody Fab fragments and incubated overnight at 4°C for complex formation. Complexes were further purified by size-exclusion chromatography and concentrated for crystallization. Crystals were frozen by direct immersion in liquid nitrogen and diffraction datasets were collected at 100 K at the IO2 and IO4 beamlines at the diamond light source (Harwell). Structures were solved by molecular replacement and refined using standard protocols. Macromolecular structures have been deposited under the accession numbers PDB: 5JW5 (apo MEDI8852), PDB: 5JW4 (H5 complex), and PDB: 5JW3 (H7 complex). Crystallographic statistics are summarized in Table S4 (Supplemental Experimental Procedures).
Author Contributions
B.F.-R. carried out PCR of immunoglobulin sequences from B cells. G.A. produced and purified antibodies. M.F. analyzed immunoglobulin genetic elements, produced figures, and carried out bioinformatic analysis. D.P. and C.S. carried out donor’s selection and screenings for the identification of cross-reactive antibodies. F.V. and A.F. carried out cloning HAs and testing antibodies for binding in cytofluorimetry. S.B. performed biochemical and cellular assays to test the fusion and HA maturation inhibiting activities of isolated antibodies. B.G. and A.D.M. carried out ADCC and CDC studies. F.S. wrote the paper. A.L. and D.C. directed the B cell isolation studies, analyzed data, and wrote the paper. J.M.M. and E.B. carried out in vivo studies. L.W.-R. carried out ADCP studies. F.J.P.-H. carried out the ELISA binding studies. N.L.K., Q.Z., L.W.-R., and F.J.P.-H. carried out the neutralization studies. A.Q.Y. lead the antibody optimization. J.A.S. edited the paper and provided supervision. N.L.K. and Q.Z. directed antibody optimization, in vitro and in vivo characterization, analyzed the data, and wrote the paper. P.J.C., U.N., P.A.W., M.K.V., R.W.O., S.R.M., S.J.G., and J.J.S. designed and performed structural research, contributed new reagents and analytical tools, analyzed data, and wrote the paper.
Conflicts of Interest
A.L. is the scientific founder of Humabs BioMed SA. A.L. holds shares in Humabs BioMed. B.G., A.D.M., G.A., F.V., and D.C. are employees of Humabs Biomed. This work was funded by MedImmune, LLC, a wholly owned subsidiary of AstraZeneca Pharmaceuticals. N.L.K., J.M.M., E.B., L.W.-R., F.J.P.-H., A.Q.Y., J.A.S., and Q.Z. were employed by MedImmune, LLC when work was executed and may currently hold AstraZeneca stock or stock options.
Acknowledgments
We thank Jose Martinez and the MedImmune Laboratory Animal Resource staff for in vivo study assistance; Robert Woods for additional affinity characterization; and Sandrina Phipps, Arnita Barnes, and Kannaki Senthil for generating HA reagents. This work was supported by the European Research Council (grant 670955 BROADimmune) and the Swiss National Science Foundation (grant 160279). A.L. is supported by the Helmut Horten Foundation. We thank the staff at the Diamond Light Source Synchrotron for assistance and beam-line access under Diamond Light Source Proposal 9826. This work was funded by the Francis Crick Institute, London. U.N. was also funded by a Marie Curie Actions Intra-European Fellowship (grant 629829).
Published: July 21, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.073.
Contributor Information
Qing Zhu, Email: zhuq@medimmune.com.
John J. Skehel, Email: john.skehel@crick.ac.uk.
Accession Numbers
The accession number for the coordinates and structure factors reported in this paper is PDB: 5JW5 (apo MEDI8852, 5JW4 (H5 complex), and 5JW3 (H7 complex). The accession number for the sequences for all of the antibodies reported in this paper is GenBank: KX398429-KX398468.
Supplemental Information
References
- Benjamin E., Wang W., McAuliffe J.M., Palmer-Hill F.J., Kallewaard N.L., Chen Z., Suzich J.A., Blair W.S., Jin H., Zhu Q. A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 hemagglutinin globular head. J. Virol. 2014;88:6743–6750. doi: 10.1128/JVI.03562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Corti D., Suguitan A.L., Jr., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C.J.H., Leon P.E., Kaur K., Tan G.S., Zheng N.-Y., Andrews S., Huang M., Qu X., Huang Y., Salgado-Ferrer M. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J. Clin. Invest. 2015;125:1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Bhabha G., Elsliger M.-A., Friesen R.H.E., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert D.C., Friesen R.H.E., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H.J.W.M., Brandenburg B. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D., Barrère B., Bizebard T., Daniels R.S., Skehel J.J., Knossow M. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat. Struct. Biol. 1999;6:530–534. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- Friesen R.H.E., Lee P.S., Stoop E.J.M., Hoffman R.M.B., Ekiert D.C., Bhabha G., Yu W., Juraszek J., Koudstaal W., Jongeneelen M. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. USA. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M., Skehel J.J. Variation and infectivity neutralization in influenza. Immunology. 2006;119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koday M.T., Nelson J., Chevalier A., Koday M., Kalinoski H., Stewart L., Carter L., Nieusma T., Lee P.S., Ward A.B. A Computationally Designed Hemagglutinin Stem-Binding Protein Provides In Vivo Protection from Influenza Independent of a Host Immune Response. PLoS Pathog. 2016;12:e1005409. doi: 10.1371/journal.ppat.1005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura G., Chai N., Park S., Chiang N., Lin Z., Chiu H., Fong R., Yan D., Kim J., Zhang J. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Nobusawa E., Aoyama T., Kato H., Suzuki Y., Tateno Y., Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- Pappas L., Foglierini M., Piccoli L., Kallewaard N.L., Turrini F., Silacci C., Fernandez-Rodriguez B., Agatic G., Giacchetto-Sasselli I., Pellicciotta G. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Russell R.J., Gamblin S.J., Haire L.F., Stevens D.J., Xiao B., Ha Y., Skehel J.J. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J., Hurt A.C. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- Schmidt A.G., Xu H., Khan A.R., O’Donnell T., Khurana S., King L.R., Manischewitz J., Golding H., Suphaphiphat P., Carfi A. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. USA. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.-M., Santelli E., Stec B., Cadwell G., Ali M. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.S., Lee P.S., Hoffman R.M.B., Mazel-Sanchez B., Krammer F., Leon P.E., Ward A.B., Wilson I.A., Palese P. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J. Virol. 2014;88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M., van den Brink E., Jongeneelen M., Poon L.L.M., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A.A., Chen L.-M., Recuenco S., Ellison J.A., Davis C.T., York I.A. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Tan G.S., Hai R., Pica N., Petersen E., Moran T.M., Palese P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Koutsonanos D., Li G.-M., Edupuganti S., Sui J., Morrissey M., McCausland M., Skountzou I., Hornig M., Lipkin W.I. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P., Neumann G., Kawaoka Y. Fields Virology; 2007. Orthomyxoviruses. [Google Scholar]
- Wu Y., Cho M., Shore D., Song M., Choi J., Jiang T., Deng Y.-Q., Bourgeois M., Almli L., Yang H. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 2015;6:7708. doi: 10.1038/ncomms8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Corti D., Liu J., Pinna D., Foglierini M., Calder L.J., Martin S.R., Lin Y.P., Walker P.A., Collins P.J. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc. Natl. Acad. Sci. USA. 2015;112:9430–9435. doi: 10.1073/pnas.1510816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J.W. To dream the impossible dream: universal influenza vaccination. Curr. Opin. Virol. 2013;3:316–321. doi: 10.1016/j.coviro.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.