Abstract
Background
Cellular respiration is a catabolic pathway for producing adenosine triphosphate (ATP) and is the most efficient process through which cells harvest energy from consumed food. When cells undergo cellular respiration, they require a pathway to keep and transfer electrons (i.e., the electron transport chain). Due to oxidation-reduction reactions, the electron transport chain produces a transmembrane proton electrochemical gradient. In case protons flow back through this membrane, this mechanical energy is converted into chemical energy by ATP synthase. The convert process is involved in producing ATP which provides energy in a lot of cellular processes. In the electron transport chain process, flavin adenine dinucleotide (FAD) is one of the most vital molecules for carrying and transferring electrons. Therefore, predicting FAD binding sites in the electron transport chain is vital for helping biologists understand the electron transport chain process and energy production in cells.
Results
We used an independent data set to evaluate the performance of the proposed method, which had an accuracy of 69.84 %. We compared the performance of the proposed method in analyzing two newly discovered electron transport protein sequences with that of the general FAD binding predictor presented by Mishra and Raghava and determined that the accuracy of the proposed method improved by 9–45 % and its Matthew’s correlation coefficient was 0.14–0.5. Furthermore, the proposed method enabled reducing the number of false positives significantly and can provide useful information for biologists.
Conclusions
We developed a method that is based on PSSM profiles and SAAPs for identifying FAD binding sites in newly discovered electron transport protein sequences. This approach achieved a significant improvement after we added SAAPs to PSSM features to analyze FAD binding proteins in the electron transport chain. The proposed method can serve as an effective tool for predicting FAD binding sites in electron transport proteins and can help biologists understand the functions of the electron transport chain, particularly those of FAD binding sites. We also developed a web server which identifies FAD binding sites in electron transporters available for academics.
Keywords: Electron transport protein, FAD binding site, Transporter, Annotation, Feature selection, Position specific scoring matrix, Significant amino acid pairs
Background
Cellular respiration is the process for producing adenosine triphosphate (ATP) and enables cells to obtain energy from foods. During cellular respiration, cells break down food molecules, such as sugar, and release energy. The objective of cellular respiration is to harvest electrons from organic compounds to create ATP, which is used to provide energy for most cellular reactions. Figure 1 shows the architecture of the cellular respiration process.
Fig. 1.
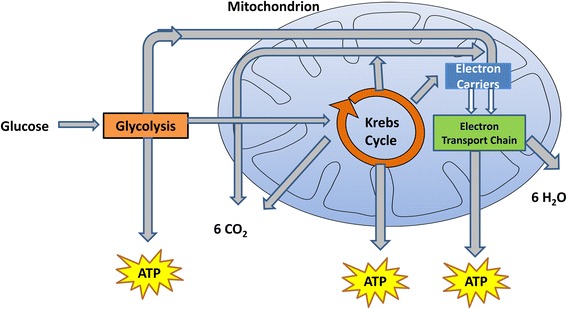
Cellular respiration process
As cells undergo cellular respiration, they require a pathway to store and transport electrons (i.e., the electron transport chain). The electron transport chain components are organized into four complexes (Complex I, Complex II, Complex III, and Complex IV) and ATP synthase (which can be called Complex V). The process of electron transport chain starts from the mitochondrial inner membrane, which electrons transfer from Complex I with nicotinamide adenine dinucleotide (NADH) and succinate (Complex II) to oxygen. In the next step, a carrier (coenzyme Q) that embeds in the cell membrane receives electrons from complex I and pass to Complex III (cytochrome b, c1 complex). Electrons bypass Complex II, the succinate dehydrogenase complex, which is an independent starting stage and is not a component of the NADH pathway. The pathway from Complex III leads to cytochrome c then moves to Complex IV (cytochrome oxidase complex). In the final step, ATP synthase is active by the proton electrochemical to utilize the flow of H+ to generate ATP, which provides energy in numerous cellular processes.
Flavin adenine dinucleotide is one of the most vital molecules in the electron transport chain. It is mainly in Complex II, which is an enzyme complex bound to the inner mitochondrial membrane of mammalian mitochondria and many bacterial cells. Regarding the reaction mechanism of Complex II, succinate is bound and a hydride is transferred to FAD to generate FADH2. After the electrons are derived from succinate oxidation through FAD, they tunnel along the [Fe-S] relay to the [3Fe-4S] cluster. These electrons are subsequently transferred to an awaiting ubiquinone molecule within the active site. The fundamental role of Complex II in the electron transfer chain of mitochondria renders it vital in most organisms, and removing Complex II from the genome has been shown to be lethal at the embryonic stage in mice.
Predicting FAD binding sites in electron transporters is vital for helping biologists clearly understand the operating mechanisms of the electron transport chain and Complex II. In this study, we developed a method that is based on position specific scoring matrix (PSSM) profiles and significant amino acid pairs (SAAPs) for identifying FAD binding residues in electron transport proteins.
FAD binding sites have attracted the interest of numerous researchers because of their relevance in electron transport chains. Prominent studies conducted on FAD binding sites include those by Mishra and Raghava [1] and Fang [2]. Mishra and Raghava [1] used support vector machines to predict FAD binding residues. They also developed a free web server for identifying FAD binding residues in specific sequences. Moreover, Fang [2] used evolutionary information to improve the prediction performance.
Numerous studies have also been conducted on transport proteins. For example, Saier [3] provided a web database containing the sequence, classification, structural, and evolutionary information of transport systems from various living organisms. Furthermore, Ren [4] presented transportDB, which is a comprehensive database of transporters and outer membrane channels. Chen [5] divided electron transport targets into four types of transport proteins to conduct prediction and analysis. After the prediction and analysis, Chen classified the transport proteins and determined the functions of each protein type in the transport protein. Ou [6] attempted to discriminate metal-binding sites in electron transport by using radial basis function networks (RBFNs).
The current study proposes an approach based on PSSM profiles and SAAPs for identifying FAD binding sites in electron transport proteins. We used a set of 55 FAD binding proteins as the training data set and six FAD binding proteins in electron transport proteins as an independent data set. We applied the independent data set to evaluate the performance of the proposed method, which demonstrated an accuracy of 69.84 %. Compared with the general FAD binding predictor developed by Mishra and Raghava, the proposed method exhibited a 9 %–45 % improvement in accuracy and Matthew’s correlation coefficient (MCC) of 0.14–0.5 when applied to two newly discovered electron transport protein sequences. The proposed method also reduces the number of false positives significantly and offers useful information for biologists. The proposed method can serve as an effective tool for predicting FAD binding sites in electron transport proteins and can help biologists understand electron transport chain functions, particularly those of FAD binding sites.
Methods
This study focused on identifying FAD binding sites in electron transport proteins. Figure 2 illustrates a flowchart of the study, which included three subprocesses in each phase: data collection, feature set generation, and model evaluation. According to this flowchart, we developed a novel approach that is based on PSSM profiles and SAAPs for predicting FAD binding sites in electron transport proteins. The details of the proposed approach are described as follows.
Fig. 2.
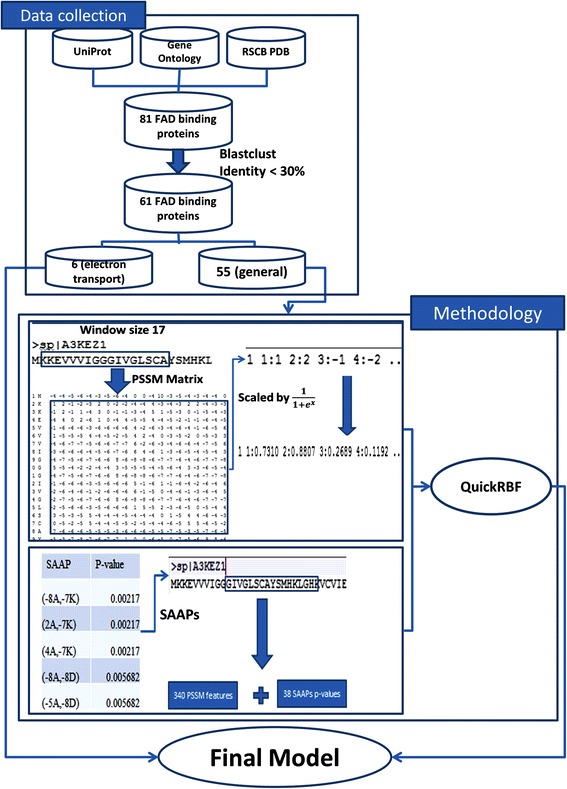
Flowchart of the proposed method for identifying FAD binding sites in electron transport proteins
Data set
First, we collected data about transport proteins and electron transport proteins from the UniProt [7] database. Subsequently, we removed sequences without the annotation “evidence at protein level” or “complete.” After this exclusion, 6694 transport proteins and 889 electron transport proteins remained and were surveyed. Next, we retrieved all FAD binding sites in the electron transport proteins. We collected data on only nine FAD binding proteins. However, creating a precise model requires using a higher number of proteins; thus, we collected data on additional general FAD binding proteins from other sources. We retrieved data from the Gene Ontology (GO) [8] and Protein Data Bank (PDB) [9, 10] databases by using the molecular function of FAD binding. In the GO database, we applied three molecular functions of FAD binding: GO:0050660 (FAD binding), GO:0071949 (FAD binding), and GO:0071950 (FADH2 binding). From these three molecular functions, we obtained data on a total of 42 FAD binding proteins. We applied the same approach to the PDB database and obtained data on a total of 72 FAD binding proteins. We removed duplicated proteins and 81 general FAD binding proteins remained. Next, BLAST [11] was applied to exclude sequences with a sequence identity of more than 40 % from the data set. Finally, 61 FAD binding proteins were used in this study (Table 1).
Table 1.
Statistics of all retrieved FAD binding proteins with FAD and non-FAD binding sites
| Number of proteins | FAD binding sites | Non-FAD binding sites | |
|---|---|---|---|
| FAD binding in electron transport | 6 | 63 | 3030 |
| General FAD binding proteins | 55 | 940 | 26475 |
We divided the collected protein sequences into two data sets: training and independent test data sets. In this phase, the training data set was used for identifying FAD binding sites, and the independent test data set was used for evaluating the performance of the proposed method. We used all six FAD binding proteins in the electron transport chain as the independent data set; thus, the training data set comprised 55 general FAD proteins (containing 863 FAD binding sites and 24408 non-FAD binding sites). Table 2 lists the details of all data sets.
Table 2.
Details of all 61 FAD binding proteins with a UniProt ID in the present study (six FAD binding proteins in electron transport served as an independent data set)
| Independent dataset | Training dataset | ||||
|---|---|---|---|---|---|
| P00455 | O95831 | P21890 | P08165 | Q5SJP8 | Q92947 |
| Q03103 | P00371 | P26440 | Q5SH33 | Q5SK63 | Q945K2 |
| Q96HE7 | P00390 | P37747 | P66004 | Q5UVJ4 | Q96329 |
| Q9YHT1 | P07342 | P38038 | Q0QLF4 | Q709F0 | Q9AL95 |
| P55931 | O53355 | P39662 | Q28943 | Q7SID9 | C6ELC9 |
| A3KEZ1 | O54050 | P41367 | P97275 | Q7WZ62 | D0VWY5 |
| O60341 | P45954 | Q2GBV9 | Q7X2H8 | O52582 | |
| P0A6U3 | P47989 | Q389T8 | Q7ZA32 | Q9RSY7 | |
| P15651 | P49748 | Q47PU3 | Q8DMN3 | Q9UBK8 | |
| P19920 | P55789 | Q52437 | Q8X1D8 | Q9UKU7 | |
| P07872 | P09622 | Q9HJI4 | Q9HKS9 | Q9HTK9 | |
Sequence information
Sequence information is one of the first features set in predicting the secondary structure of proteins [12, 13]. In this feature, each amino acid sequence is represented by a number 0 or 1, creating a binary matrix. From the binary matrix, the value for each amino acid can be calculated. For example, if the sequence of amino acids is ARNDCQEGHILKMFPSWYV and the value for amino acid N must be calculated, the third position is set to 1 and the others are set to 0. In this study, we also used two types of advance sequence information, namely PAM250 and BLOSUM62.
PAM250
A percent accepted mutation (PAM) [14] matrix represents the elements involved in the conversion of amino acids into amino acids within a variable probability of evolutionary distance. A PAM matrix was created in the protein sequence alignment and various phylogenetic trees with the assumption that amino acids are amino acids and that each amino acid is substituted with another amino acid, to establish an acceptable point mutation matrix (accepted point mutation matrix).
A matrix is usually employed to mark aligned peptide sequences in order to identify the similarity of such sequences. By comparing aligned protein sequences with a known homology and determining the “accepted point mutations”, the aforementioned numbers were derived. The frequencies of such mutations were arranged in a table as a “log odds matrix”:
where Mij is the matrix component and Rij is the probability of that substitution, then divided by the standardized frequency of amino acid sequences. Note that all the numbers are rounded to the integer number. The base-10 log is utilized so that the numbers can be added instead of multiplied to decide the score of a practical set of sequences.
BLOSUM62
The block substitution matrix (BLOSUM) [15] is used to assess differences in effectiveness between evolutions of protein sequence alignment methods. They are retrieved from the BLOCKS database, and some of the protein amino acid sequences are retained; the calculated relative amino acid is replaced by the calculated frequency and probability. A BLOSUM62 matrix is commonly collected in a database sequence BLOCKS with 62 % sequence similarity, and the sequence is then deduced from a score matrix.
PSSM profiles
PSSM is a matrix commonly used for representing motifs in biological sequences [16]. It is a matrix of score values and provides a weighted match to any specific substring of fixed length. This matrix has one row for each letter of the alphabet and one column for each position in the pattern.
In recent years, the PSSM has widely been considered an indicator of the properties of protein sequences. The PSSM is used in determining the evolution of sequence information in a specific location as well as the amino acid replacement ratio to identify protein sequences; such sequences represent the original 20 amino acid types in the protein and are used to replace an amino acid with its degree of influence. The PSSM has been extensively used for predicting the secondary structure of proteins as well as subcellular locations and other biological information, and it has been reported to produce favorable results.
We collected all sequence data from BLAST [11] and the non-redundant protein database and used them to establish the sequences in a PSSM. After the PSSM sequences were established, we calculated the optimal protein sequence for each amino acid. We placed 20 types of amino acids in the calculated sequences, leading to the creation of a matrix. If a window size of 17 is used, then the matrix size is 17 * 20 = 340 (because the calculated value for each amino acid was 20). This matrix should be added to predict the properties of the protein sequence. Identical amino acid residues can be replaced with a specific value of amino acids. We used the following numerical normalization formula to convert the values to values between 0 and 1:
F-score
In binary classification analysis, an F-score is a simple parameter applied for measuring the accuracy of a test by using two sets of real numbers [17]. The F-score is defined as follows:
where n+ is the number of positive instances and n− is the number of negative instances. Furthermore, , and are the averages of the ith feature of the entire, positive, and negative data sets, respectively; x(+)k,i is the ith feature of the kth positive instance; and x(−)k,i is the ith feature of the kth negative instance. We calculated all F-score values for all feature sets of FAD binding sites in electron transport proteins. A higher F-score indicates that the corresponding feature has a higher amount of special information. Therefore, we added the F-score values to the PSSM features. In this study, we added the 30 highest F-scores to the PSSM features.
Significant amino acid pairs
We adopted SAAPs to improve the performance of the proposed method in predicting FAD binding sites in electron transport proteins. The SAAPs around the FAD binding sites were identified on the basis of six FAD binding proteins, and the remaining SAAPs were identified on the basis of a statistical distribution measurement. Each amino acid pair surrounding FAD binding sites was calculated using a p-value:
where N denotes the number of sequences in the entire data set, M denotes the number of sequences in the positive data set, and (N-M) denotes the number of sequences in the negative data set; n, x, and n-x denote the number of sequences including a kth SAAP in the entire data set, positive data set, and negative data set. Figure 3 shows the method used for calculating the p-value from FAD binding sites in electron transport chains.
Fig. 3.

Proposed method for calculating initial SAAP values
A p-value less than 0.13 indicates that the amino acid pair surrounding FAD binding sites is significant. That is, numerous special features exist, with some features having a p-value less than 0.13. After we calculated the p-values for all amino acid pairs surrounding FAD binding sites with a window size of 17, we added the ranked SAAPs to the feature set in descending order. Finally, 38 SAAPs were added to the feature set of FAD binding sites in electron transport proteins.
Radial basis function networks
We employed the QuickRBF package [18] to construct RBFN classifiers. Figure 4 shows the architecture of the RBF network. Furthermore, we assigned a constant bandwidth of 5 for each kernel function in the network. We also used all training data as centers. Subsequently, the RBFN classifier was used to identify FAD binding sites according to the output function value. We explained the details of the network structure and design in our previous article [19].
Fig. 4.

Architecture of the RBFN
RBFN-based classifications have been used in several applications in bioinformatics to predict cleavage sites in proteins [20], interresidue contacts [21], and protein disorder [22]; furthermore, they have been applied for discriminating β-barrel proteins [23], classifying transporters [24, 25], identifying O-linked glycosylation sites [26], and identifying ubiquitin conjugation sites [27].
The general mathematical form of output nodes in an RBFN is expressed as follows:
where gj(x) is the function corresponding to the jth output node and is a linear combination of k radial basis functions with center mi and bandwidth si; in addition, wji is the weight associated with the correlation between the jth output node.
Assessment of predictive ability
We measured the predictive performance of the proposed method by using sensitivity, specificity, accuracy, and MCC metrics. TP, FP, TN, and FN represent true positive, false positive, true negative, and false negative, respectively.
Sensitivity
This parameter enables computing the percentage of accurately predicted FAD binding sites.
Specificity
This parameter enables computing the percentage of accurately predicted non-FAD binding sites.
Accuracy
This parameter enables computing the percentage of accurately predicted FAD and non-FAD binding sites.
MCC
This parameter represents the quality of prediction and is used for resolving imbalance in data sets. An MCC value of 1 indicates a perfect prediction.
Results and discussion
Amino acid composition analysis
We analyzed the composition of interacting and non-interacting FAD binding sites by computing the occurrence frequency of amino acids in these sites. Regarding the interacting FAD binding sites, the amino acids G, S, A, and T exhibited the significantly highest occurrence frequency in two interaction instances (general FAD binding proteins and FAD binding proteins in electron transport proteins) (Figs. 5 and 6). We inferred that glycine is vital for the interaction with FAD binding sites. Regarding non-interacting binding sites, the amino acids A, L, and G exhibited the highest occurrence frequency in both instances.
Fig. 5.

Amino acid composition of FAD binding interacting residues and noninteracting residues in 55 general FAD binding proteins
Fig. 6.

Amino acid composition of FAD interacting residues and noninteracting residues in six FAD binding proteins in the electron transport chain
Figure 7 shows a comparison between general FAD binding proteins and FAD binding proteins in electron transport proteins. We observed some differences between the two types of proteins, and the amino acids V, E, and I exhibited considerable differences.
Fig. 7.

Comparison of percentage composition of FAD interacting residues in six FAD binding proteins in the electron transport chain and 55 general FAD binding proteins
Performance in predicting FAD binding sites in electron transport proteins by using various window sizes
We created an FAD binding classifier by using the 61 FAD binding proteins collected. We applied the QuickRBF classifier by using window sizes ranging from 13 to 19 for comparison (Table 3). We measured the predictive performance of the proposed PSSM-based method. As shown in Table 3, changing the window size did not exert considerable effects on the result. The result obtained when the window size was set to 17 was favorable, and the measured sensitivity, specificity, accuracy, and MCC were approximately 80.8 %, 80.2 %, 80.2 %, and 0.27, respectively. Although the MCC was low, all the other performance metrics were approximately 80. We used the experiment with a window size of 17 to create the FAD binding classifier model.
Table 3.
Comparison of performance in identifying FAD binding sites in the electron transport chain with different window sizes
| Window Size | True positive | False positive | True negative | False negative | Sens | Spec | Acc | MCC | |
|---|---|---|---|---|---|---|---|---|---|
| 5-fold | WS13 | 139 | 973 | 3909 | 33 | 80.8 | 80.1 | 80.1 | 0.27 |
| WS15 | 139 | 990 | 3893 | 33 | 80.8 | 79.7 | 79.8 | 0.26 | |
| WS17 | 139 | 966 | 3917 | 33 | 80.8 | 80.2 | 80.2 | 0.27 | |
| WS19 | 138 | 1004 | 3879 | 35 | 79.8 | 79.4 | 79.5 | 0.26 | |
| indept | WS13 | 50 | 1444 | 1586 | 13 | 79.4 | 52.3 | 52.9 | 0.09 |
| WS15 | 50 | 1223 | 1807 | 13 | 79.4 | 59.6 | 60 | 0.11 | |
| WS17 | 51 | 1169 | 1861 | 12 | 81 | 61.4 | 61.8 | 0.12 | |
| WS19 | 51 | 1225 | 1805 | 12 | 81 | 59.6 | 60 | 0.12 |
As shown in Figs. 8 and 9, the sequence frequency logo was generated using a tool provided by the WebLogo server [28]. The window size was set to 17 and used to confirm the FAD binding fragment for comparison. These two figures indicate that some differences exist between the general FAD binding proteins and FAD binding proteins in the electron transport chain. For example, the amino acids T, K, I, and R exhibited clear differences at positions ranging from −4 to −1.
Fig. 8.
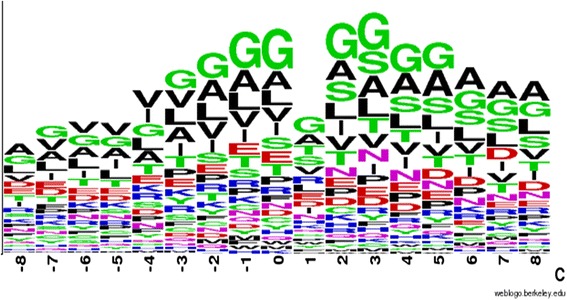
Sequence logo for 55 general FAD binding proteins (generated from WebLogo)
Fig. 9.
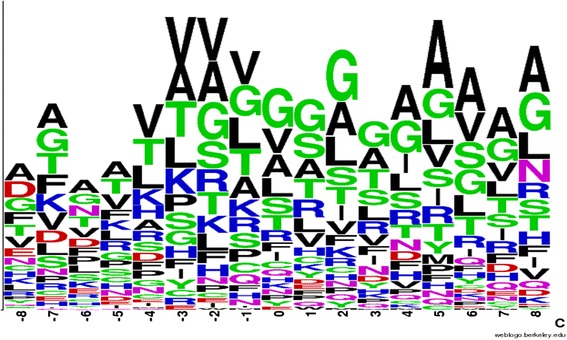
Sequence logo for six FAD binding proteins in the electron transport chain (generated from WebLogo)
Performance in predicting FAD binding sites in electron transport proteins with different feature sets
Table 4 shows the performance assessment results obtained by discriminating FAD binding sites in electron transport chains with different feature sets. We used the established FAD classifier to predict our independent data set (six FAD binding proteins in the electron transport chain) by setting the window size to 17. As shown in Table 4, the predictive performance of the proposed method was more favorable than that of the other methods (i.e., BINARY, BLOSUM62, PAM250, and F-Score). Although the performance of the proposed method was not extremely high (sensitivity = 80.95 %, specificity = 69.6 %, accuracy = 69.84 %, and MCC = 0.15), it was still superior to that of the other methods. We observed that the performance improved when we added SAAPs from FAD binding proteins in the electron transport chain to the PSSM. Thus, the proposed method was effective in predicting FAD binding sites in electron transport proteins.
Table 4.
Comparison of performance in identifying FAD binding sites in the electron transport chain with different feature sets
| Feature set | True positive | False positive | True negative | False negative | Sens | Spec | Acc | MCC |
|---|---|---|---|---|---|---|---|---|
| BINARY | 45 | 972 | 2058 | 18 | 71.43 | 67.92 | 67.99 | 0.12 |
| BLOSUM62 | 41 | 977 | 2053 | 22 | 65.08 | 67.76 | 67.7 | 0.1 |
| PAM250 | 42 | 996 | 2032 | 21 | 66.67 | 67.11 | 67.1 | 0.1 |
| PSSM | 51 | 1169 | 1861 | 12 | 80.95 | 61.42 | 61.82 | 0.12 |
| PSSM + F-score | 51 | 1142 | 1888 | 12 | 80.95 | 62.31 | 62.69 | 0.13 |
| PSSM + SAAPs | 54 | 1074 | 1955 | 9 | 85.71 | 64.54 | 64.97 | 0.15 |
Significance analysis based on the proposed method
Receiver operating characteristic (ROC curve) and area under the curve (AUC) are also used in presenting the accuracy of the test in the presented results [29, 30]. Figure 10 plots the ROC curve based on Sensitivity and Specificity of our prediction results. According to the ROC curve, we calculated the AUC to measure the accuracy. The AUC from this study reached 0.8618325, and therefore we can use this model to identify FAD binding sites in the electron transport chains with good results.
Fig. 10.

ROC Curve for performance of predicting FAD binding sites in electron transport proteins with PSSM and SAAPs
Performance in predicting FAD binding sites in electron transport proteins with different classifiers
Table 5 shows the performance assessment results obtained by discriminating FAD binding sites in electron transport chains with different classifiers. We applied our method in independent dataset with different classifiers, i.e., kNN, RandomForest (in WEKA package [31, 32]) and LibSVM classifiers [33]. The results show in Table 5 can prove that our classifier perform well than the others. Therefore we can use our method to identify FAD binding sites in electron transport proteins with high results.
Table 5.
Comparison of performance in identifying FAD binding sites in the electron transport chain with different classifiers
| Classifier | True positive | False positive | True negative | False negative | Sens | Spec | Acc | MCC |
|---|---|---|---|---|---|---|---|---|
| kNN | 45 | 1448 | 1581 | 9 | 85.71 | 52.2 | 52.88 | 0.11 |
| RandomForest | 31 | 1055 | 1974 | 32 | 49.21 | 65.17 | 64.84 | 0.04 |
| LibSVM | 53 | 1149 | 1880 | 10 | 84.13 | 62.07 | 62.52 | 0.13 |
| QuickRBF | 54 | 1074 | 1955 | 9 | 85.71 | 64.54 | 64.97 | 0.15 |
Leave-one-out analysis with six FAD binding proteins in electron transport chains
Table 6 shows the final results obtained from a leave-one-out analysis of six FAD binding proteins in electron transport chains. Although the number of proteins used in the experiment was not high, we conducted this experiment to obtain a reference for comparison and validate the performance of the proposed method in predicting FAD binding sites in electron transport chains. The analysis results revealed that the proposed method performed well, exhibiting an average sensitivity of 97.37 %, average specificity of 96.36 %, average sensitivity of 96.39 %, and average MCC of 0.66.
Table 6.
Comparison of performance in identifying FAD binding sites in the electron transport chain with PSSM and SAAPs
| Protein | True positive | False positive | True negative | False negative | Sens | Spec | Acc | MCC |
|---|---|---|---|---|---|---|---|---|
| Q9YHT1 | 17 | 223 | 417 | 9 | 65.38 | 65.16 | 65.17 | 0.12 |
| P00455 | 9 | 111 | 247 | 3 | 75 | 68.99 | 69.19 | 0.17 |
| Q03103 | 6 | 1 | 557 | 0 | 100 | 99.82 | 99.82 | 0.92 |
| Q96HE7 | 6 | 68 | 394 | 1 | 85.71 | 85.28 | 85.29 | 0.24 |
| A3KEZ1 | 6 | 5 | 400 | 0 | 100 | 98.77 | 98.78 | 0.73 |
| P55931 | 6 | 0 | 612 | 0 | 100 | 100 | 100 | 1 |
Comparison of the proposed method with another method
We compared the performance of the proposed method with that of the FADPred approach presented by Mishra and Raghava [1]. In this comparison, in addition to the six FAD binding proteins in the electron transport chain, we used two new proteins, namely Q96HE7 and A3KEZ1, which have been demonstrated in experiments conducted after 2010. We subsequently evaluated the results of the proposed method in analyzing these two proteins and compared them with results of the FADPred approach [1]. Table 7 shows the comparison results, indicating that the proposed method demonstrates superior performance relative to the FADPred method [1].
Table 7.
Comparison of performance in identifying FAD binding sites in two newly discovered proteins
Identification of new FAD binding sites in electron transport protein
In this part, we applied our method for prediction of FAD binding sites in electron transport human proteins. The testing dataset retrieved from Swiss-Prot [34], which is a famous protein database. After using BLAST to remove sequence similarity more than 30 %, the rest of dataset contained 100 proteins, which including 21985 amino acids. Then our model can found 1136 FAD binding sites from dataset. Thus our research can help biologists discover some new FAD binding sites in electron transport proteins.
Web server for predicting FAD binding sites in electron transport protein
The web server FAD-ETC.-RBF was built for presenting our method in this study. FAD-ETC.-RBF trained for the identification of FAD binding sites in electron transport proteins by using QuickRBF classification based on PSSM profiles and SAAPs. The web server can be access at http://140.138.155.226/~kahn/Bioinformatics/. We developed friendly web interface including many page menus that users can easily use to retrieve information and submit their sequences. Moreover, the users just wait for the short time to receive the prediction result because the performance of this server is especially fast. In the result page, users can easily check the results because the amino acids predicted were displayed as different colors. According to this web server, biologists can discover new FAD binding sites in electron transport protein to understand clearly the operating mechanism of electron transport chains.
Conclusions
Predicting FAD binding sites in electron transporters is vital in helping biologists clearly understand the operating mechanisms of electron transport chains and Complex II. In this study, we developed a method based on PSSM profiles and SAAPs for identifying FAD binding residues in electron transport proteins. We used the independent data set to evaluate the performance of the proposed method, which achieved an accuracy of 69.84 %. We compared the performance of the proposed method in analyzing two newly discovered electron transport protein sequences with that of the general FADPred approach of Mishra and Raghava. We observed that the accuracy of the proposed method improved by 9 %–45 % and its MCC was 0.14–0.5. The proposed method can serve as an effective tool for predicting FAD binding sites in electron transport proteins and can help biologists understand the functions of the electron transport chain, particularly those FAD binding sites. We also developed a web server for the method described in this paper.
The contributions of this study provide a basis for further research that can enrich the field. However, this study still has some limitations related to the small sample size and limited time. The number of suitable FAD binding proteins in electron transport chains was not sufficient, potentially affecting the performance of the proposed method. To create a more effective model, we must identify additional FAD binding proteins in electron transport proteins. Doing so can enable us to conduct a comparative study and enhance prediction performance.
Abbreviations
ATP, adenosine triphosphate; AUC, area under curve; BLOSUM, block substitution matrix; FAD, flavin adenine dinucleotide; MCC, Matthew’s correlation coefficient; NADH, nicotinamide adenine dinucleotide; PAM, percent accepted mutation; PSSM, position specific scoring matrix; RBFN, radial basis function network; ROC, receiver operating characteristic; SAAPs, significant amino acid pairs
Acknowledgement
We would like to acknowledgement Department of Computer Science and Engineering, Yuan Ze University for supporting good condition to finish this study.
Funding
This research is partially supported by Ministry of Science and Technology, Taiwan, R.O.C. under Grant no. MOST 104-2221-E-155-037 and 105-2221-E-155-065.
Availability of data and material
The data sets supporting the results of this article are included within the article.
Authors’ contributions
Analyzed the data: YYO NQKL. Designed and performed the experiments: YYO NQKL. Wrote the paper: YYO NQKL. Read and approved the final version YYO NQKL.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Contributor Information
Nguyen-Quoc-Khanh Le, Email: khanhlee87@gmail.com.
Yu-Yen Ou, Email: yienou@gmail.com.
References
- 1.Mishra NK, Raghava GP. Prediction of FAD interacting residues in a protein from its primary sequence using evolutionary information. BMC Bioinformatics. 2010;11(1):1. doi: 10.1186/1471-2105-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang C, Noguchi T, Yamana H Prediction of FAD Binding Residues with Combined Features from Primary Sequence. Int Proc Computer Sci Inf Technol. 34;47–153.
- 3.Saier MH, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(suppl 1):D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim K-L, Dawson VL. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16(2):223–32. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 5.Chen S-A, Ou Y-Y, Lee T-Y, Gromiha MM. Prediction of transporter targets using efficient RBF networks with PSSM profiles and biochemical properties. Bioinformatics. 2011;27(15):2062–7. doi: 10.1093/bioinformatics/btr340. [DOI] [PubMed] [Google Scholar]
- 6.Ou Y-Y, Chen S-A, Wu S-C. ETMB-RBF: discrimination of metal-binding sites in electron transporters based on RBF networks with PSSM profiles and significant amino acid pairs. PLoS One. 2013;8(2):e46572. doi: 10.1371/journal.pone.0046572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33(suppl 1):D154–9. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abola EE, Bernstein FC, Koetzle TF. The protein data bank. In neutrons in biology. Springer US; 1984. pp. 441–441.
- 10.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35(suppl 1):D301–3. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(suppl 2):W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rychlewski L, Li W, Jaroszewski L, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9(2):232–41. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–50. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 14.Dayhoff MO, Schwartz RM. A model of evolutionary change in proteins. In Atlas of protein sequence and structure. 1978.
- 15.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci. 1992;89(22):10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y-W, Lin C-J. Combining SVMs with various feature selection strategies. In: Feature extraction. Springer; 2006. p. 315–24.
- 18.Ou Y-Y. QuickRBF: a package for efficient radial basis function networks. 2005. [Google Scholar]
- 19.Ou YY, Oyang YJ, Chen CY. A novel radial basis function network classifier with centers set by hierarchical clustering. In Proceedings. 2005 IEEE International Joint Conference on Neural Networks. 2005;3. pp. 1383–1388. IEEE.
- 20.Yang ZR, Thomson R. Bio-basis function neural network for prediction of protease cleavage sites in proteins. Ieee Transactions on Neural Networks. 2005;16(1):263–74. doi: 10.1109/TNN.2004.836196. [DOI] [PubMed] [Google Scholar]
- 21.Zhang GZ, Huang DS. Prediction of inter-residue contacts map based on genetic algorithm optimized radial basis function neural network and binary input encoding scheme. J Comput Aided Mol Des. 2004;18(12):797–810. doi: 10.1007/s10822-005-0578-7. [DOI] [PubMed] [Google Scholar]
- 22.Su CT, Chen CY, Ou YY. Protein disorder prediction by condensed PSSM considering propensity for order or disorder. BMC Bioinformatics. 2006;7(1):1. doi: 10.1186/1471-2105-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou YY, Gromiha MM, Chen SA, Suwa M. TMBETADISC-RBF: Discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Comput Biol Chem. 2008;32(3):227–31. doi: 10.1016/j.compbiolchem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Ou YY, Chen SA, Gromiha MM. Classification of transporters using efficient radial basis function networks with position‐specific scoring matrices and biochemical properties. Proteins: Structure, Function, and Bioinformatics. 2010;78(7):1789–97. doi: 10.1002/prot.22694. [DOI] [PubMed] [Google Scholar]
- 25.Ou YY, Chen SA. Using efficient RBF networks to classify transport proteins based on PSSM profiles and biochemical properties. In International Work-Conference on Artificial Neural Networks. Springer Berlin Heidelberg; 2009. pp. 869–876.
- 26.Chen SA, Lee TY, Ou YY. Incorporating significant amino acid pairs to identify O-linked glycosylation sites on transmembrane proteins and non-transmembrane proteins. Bmc Bioinformatics. 2010;11(1):1. doi: 10.1155/2010/423589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TY, Chen SA, Hung HY, Ou YY. Incorporating Distant Sequence Features and Radial Basis Function Networks to Identify Ubiquitin Conjugation Sites. PLoS One. 2011;6(3):e17331. doi: 10.1371/journal.pone.0017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn. 1997;30(7):1145–59. doi: 10.1016/S0031-3203(96)00142-2. [DOI] [Google Scholar]
- 30.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 31.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD explorations newsletter. 2009;11(1):10–8. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 32.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20(15):2479–81. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 33.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2(3):27. [Google Scholar]
- 34.Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31(1):365–70. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]


