Abstract
Quantitative methods for detection of biological molecules are needed more than ever before in the emerging age of “omics” and “big data.” Here, we provide an integrated approach for systematic analysis of the “lipidome” in tissue. To test our approach in a biological context, we utilized brain tissue selectively deficient for the transcription factor Specificity Protein 2 (Sp2). Conditional deletion of Sp2 in the mouse cerebral cortex results in developmental deficiencies including disruption of lipid metabolism. Silver (Ag) cationization was implemented for infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) to enhance the ion abundances for olefinic lipids, as these have been linked to regulation by Sp2. Combining Ag-doped and conventional IR-MALDESI imaging, this approach was extended to IR-MALDESI imaging of embryonic mouse brains. Further, our imaging technique was combined with bottom-up shotgun proteomic LC-MS/MS analysis and western blot for comparing Sp2 conditional knockout (Sp2-cKO) and wild-type (WT) cortices of tissue sections. This provided an integrated omics dataset which revealed many specific changes to fundamental cellular processes and biosynthetic pathways. In particular, step-specific altered abundances of nucleotides, lipids, and associated proteins were observed in the cerebral cortices of Sp2-cKO embryos.
Keywords: IR-MALDESI, Proteomics, Lipidomics, Mass spectrometry imaging, Embryonic brain
Introduction
In the age of biological “omics” and the consequent “big data,” there is an immediate need for development of analytical methods to systematically elucidate molecular mechanisms that underlie various loss-of-function and gain-of-function biological perturbations in living animal models. Current mass spectrometry-based techniques combined with new bioinformatics tools allow collection of large datasets and contextual interpretation of biological information. Here, we report the development and evaluation of a new method for mass spectrometric tissue imaging and its application in forebrain tissues from mouse embryos. We illustrate the integration of our method with LC-MS-based proteomics discovery for quantitative assessment of lipids and their associated proteins in a genetic mouse model in which lipid metabolism is disrupted selectively in the developing cerebral cortex through conditional deletion of the transcription factor Specificity Protein 2 (Sp2) in the cerebral cortex. Sp2-dependent transcription regulates a variety of housekeeping, tissue-specific, developmental, and fundamental cell cycle processes [1, 2]. In a past study, we showed that Sp2 expression is critical for cortical development through regulation of the cell cycle in neural progenitors [3], in part through presumptive regulation of cholesterol metabolism [2].
Infrared matrix-assisted electrospray ionization (IR-MALDESI) is a mass spectrometry-based imaging technique developed in our lab [4–6] which employs an IR laser tuned to excite water molecules deposited evenly in the form of an ice matrix coating the tissue sample. The excitation ablates the tissue, allowing molecules to partition into an electrospray droplet. This approach provides many advantages over conventional MS-based imaging techniques by avoiding sample preparation that involves organic matrices. Moreover, IR-MALDESI is performed under ambient pressure and utilizes a softer method of ionization than MALDI, providing sensitive detection and high spatial resolution of molecules within a tissue. Due to their high abundance and hydrophobic nature, lipids are ideal targets for IR-MALDESI. While lipids have been analyzed by chromatographic techniques [7–11], many of these studies have focused on specific classes of lipids due to their diverse chemical properties. Additionally, many lipids are not soluble or easily extracted in LC-MS-compatible solvents. Furthermore, nano-LC analyses of lipids present a greater difficulty due to carryover [12]. Direct shotgun analysis has removed the challenges of separation techniques [13]; however, none of these approaches provide spatial information regarding these molecules. Additionally, our recently published results on the direct analysis of vitamin D in human serum demonstrated an enhancement of sensitivity and selectivity in IR-MALDESI direct analysis of unsaturated lipids by silver cationization [14].
Tissue-specific LC-MS-based shotgun proteomics provides an additional dimension in which reverse-phase nanoflow liquid chromatography can be used to analyze nanogram quantities of sample obtained from tissues, separating peptides by hydrophobicity using a gradient elution. Moreover, high-resolving-power tandem mass spectrometry, and speed of data-dependent acquisition provided by the Orbitrap technology, has the capability to confidently identify thousands of peptides and proteins within a complex biological mixture in a single run [15–18]. Complementary to MS imaging of lipids which provides spatial information regarding the lipidome, LC-MS-based proteomics utilizes tandem mass spectrometry that enables extraction of sequence-specific information regarding the proteome. Additionally, both approaches have the capacity to provide quantitative information regarding the lipidome/proteome. Here, we report that combining Ag-doped and conventional IR-MALDESI imaging with LC-MS-based proteomics allows for a systems approach to studying changes to fundamental cellular and molecular pathways involved in lipid biosynthesis under the control of Sp2-dependent mechanism in the mouse embryonic cerebral cortex.
Experimental procedures
Materials
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade solvents were purchased from Honeywell Burdick & Jackson (Muskegon, MI), and silver nitrate (99.85 % p.a.) was purchased from Fisher Scientific (Nazareth, PA, USA). All materials were used as purchased.
Tissue samples
Sp2-floxed mice were generated and genotyped as described previously [3] and successively crossed to Emx1cre mice (Jackson Laboratory, Bar Harbor, ME, USA; catalogue #005628) to generate Emx1cre:Sp2F/F mice (Sp2-cKO). Cre-negative embryos were used as littermate controls (WT). Genotyping for cre was performed using the primers generic cre F: 5′ CACCCTGTTACGTATAGCCG 3′ and generic cre R: 5′ GAGTCATCCTTAGCGCCGTA 3′ using standard polymerase chain reaction (PCR) conditions. Pregnant dams were sacrificed on embryonic day 13.5 (E13.5) by cervical dislocation, prior to immediate removal of embryos from the uterus. Harvested embryos were rapidly decapitated on ice in 0.1 M phosphate-buffered saline (PBS), heads fresh frozen over isopentane cooled in a dry ice/methanol bath and stored at −80 °C until sectioning. Ten-micrometer-thick coronal sections were prepared at −20 °C using a CM1950 cryostat (Leica, Buffalo Grove, IL, USA). The tissue sections were thaw mounted on either standard microscope slides (VWR) for IR-MALDESI imaging or PEN membrane slides for laser capture microdissection and stored at −80 °C until needed.
Histochemistry
Adjacent 10-μm-thick tissue sections from embryonic mouse brains were collected for immuno-histology. Thaw-mounted tissue sections were fixed in 4 % paraformaldehyde in 0.1 M PBS for 5 min. Following a washing step in 0.1 M PBS for 2 min, nucleic bases were stained in a 1-μg/mL solution of the fluorescent probe 4′,6-diamidino-2-phenylindole (DAPI) in 0.1 M PBS for 1 min. The tissue sections were then washed twice in 0.1 M PBS, embedded in mounting medium, and covered with a glass coverslip. Fluorescence imaging was conducted using an Olympus Fluoview FV1000 (Center Valley, PA, USA).
Laser capture microdissection
Fresh frozen 10-μm mouse brain tissue sections on individual PEN membrane slides were allowed to warm up to room temperature for 20 s and processed immediately. The tissue sections were fixed in 70 % ethanol (anhydrous KOPTEC USP, VWR, Radnor, PA) at −20 °C for 30 s. After the slides were washed in cold H2O for 10 s, the tissues were stained with Mayer’s hematoxylin for 10 s and then washed again in H2O for 10 s. Each slide was subsequently rinsed with 70 % ethanol for 10 s, 95 % ethanol for 10 s, and 100 % ethanol for 30 s. The slide was then placed into xylene for approximately 5 min for complete dehydration of the tissue section. Rostral forebrain cortex tissue sections (see Electronic Supplementary Material, ESM) were excised utilizing a laser microdissection system (LMD 7000, Leica Microsystems, Buffalo Grove, IL).
Sample preparation for LC-MS/MS
Mircodissected tissue sections were lysed and reduced in 3 μL of 100 mM DTT, 4 % SDS, and 0.1 M Tris/HCl at pH 8.0 per 10 nL (106×10 μm3) tissue for 5 min at 95 °C (mixed for 1 min before incubation). Tissue lysate was transferred to a 30-kDa MWCO-filter unit (Vivaproducts, Littleton, MA) and diluted with 200 μL 8 M urea in 0.1 M Tris/HCl pH 8.0. Samples were centrifuged at 14,000×g for 10 min at 20 °C, washed with 200 μL of 8 M urea in 0.1 M Tris/HCl pH 8.0, and centrifuged again at 14,000×g for 10 min. Samples were then alkylated on the filter with 100 μL of 0.05 M iodoacetamide and 8 M urea in 0.1 M Tris/HCl pH 8.0, mixed and incubated for 20 min in the dark. Samples were then centrifuged at 14,000×g for 10 min, washed twice with 100 μL 8 M urea in 0.1 M Tris/HCl pH 8.0, and centrifuged at 14,000×g for 10 min. Filter units were then equilibrated with 3×100 μL of 50 mM Tris/HCl at pH 7.0 and centrifuged at 14,000×g for 10 min. The collection tube was changed, and 50 μL of modified porcine trypsin (reconstituted in 50 mM Tris/HCl pH 7.0) was added at an enzyme to protein ratio of 1:50, mixed for 1 min (assumed approx. 1 μg protein/sample). Parafilm was wrapped around the filters to prevent evaporation of sample during digestion. Digestion was performed overnight in an incubation chamber at 37 °C. Digestion was quenched with 50 μL of 1 % formic acid (v/v) and 0.001 % Zwittergent 3–16 (Calbiochem, La Jolla, CA), and the sample was centrifuged at 14,000×g for 10 min. Samples were then rinsed with 400 μL of 1 % formic acid (v/v) and 0.001 % Zwittergent 3–16 and centrifuged at 14,000×g for 15 min. The eluate was frozen, dried (Speed Vac), and stored at −80 °C prior to LC-MS analysis.
IR-MALDESI imaging
Detailed information on the construction and operation of the IR-MALDESI imaging source has been published elsewhere [5, 6]. In brief, the sample is placed on a cooled Peltier stage at −10 °C in an enclosed housing. The enclosed housing allows controlling the inner atmosphere in a way that a defined ice layer can be deposited on the tissue section and the number of ambient ions is reduced during MS analysis. For IR-MALDESI imaging of the embryonic mouse brain sections, the region of interest was rasterized at a spot-to-spot distance of 100 μm with a tunable mid-IR laser (IR-Opolette 2371, Opotek, Carlsbad, CA, USA) to thermally desorb neutrals, which were post-ionized in charged droplets of an electrospray plume. The electrospray solvent was 50 % MeOH/50 % H2O (v/v) doped with either 0.2 % formic acid (referred to as conventional IR-MALDESI) or 100 μmol/L AgNO3 (Ag-doped IR-MALDESI) as optimized previously [14].
The IR-MALDESI source was coupled to a QExactive mass spectrometer (Thermo Scientific, Bremen, Germany), which was operated in positive ion mode and fully synchronized with the IR laser to acquire one scan per voxel [19]. The ion injection time was fixed at 150 ms, allowing trapping of ions from two consecutive laser pulses at 20 Hz prior to a single Orbitrap measurement at a resolving power of 140, 000 at m/z 200. Tissue samples were analyzed at m/z 300–1200 (Ag-doped IR-MALDESI) or m/z 250–1000 (conventional IR-MALDESI). To increase the mass measurement accuracy, the acquired mass spectra were automatically recalibrated by the Thermo acquisition software using two ambient ions (protonated and sodiated diisooctyl phthalate at m/z 391.2843 and m/z 413.2662) as lock-masses [20].
IR-MALDESI data analysis
The QExactive. RAW data files were first converted into either the mzXML or the imzML format (for concatenated heat maps) as described previously [14] and further processed in the MSiReader software environment, which allows exporting ion heat maps, to extract unique peaks from a user-defined region of interest (ROI) as well as to normalize heat maps pixel per pixel. In our recent study about Ag MALDESI, we found that the distinctive ion abundance ratio and mass difference of [M+107Ag+]+ and [M+109Ag+]+ can be applied as effective peak filter criteria and thus increase the confidence in mass spectral peak assignments [14]. Here, we extend the previously developed methodology to tissue imaging using the in-house software GlycoHunter and MatLab R2012b (MathWorks, Natick, MA, USA) to identify peak pairs and to investigate covariance of the two isotopic peaks.
For a statistical comparison of the IR-MALDESI datasets from wild-type and Sp2-cKO embryonic mouse brains, two-dimensional ion abundance matrices were exported from a user-defined ROI or the whole imaging dataset. The matrix entries corresponded either to the maximum ion abundance in a defined bin width or, if exported for defined m/z values associated with a peak list, the maximum ion abundance within 5 ppm tolerance. JMP Pro 11.0.0 (SAS Institute, Cary, NC, USA) “multivariate analysis of variance” (MANOVA) and “response screening” functionalities were applied to perform univariate statistical tests and false discovery rate (FDR) corrections.
For the histology-independent analysis, a peak list was first generated from the ROI to extract ion abundance matrices from the whole imaging dataset. This strategy strictly excluded non-tissue-related background ions as well as low abundant species from the computational analysis without, however, compromising the mass resolution. Principal component analysis on the extracted ion abundance matrices was performed using the MatLab R2012b built-in princomp function in “econ” mode without modification. Principal component score maps were compiled from the score matrix, and corresponding loading plots were generated from the coefficient matrix.
Peaks of interest were tentatively assigned to molecular species based on accurate mass data and METLIN database search results with a maximum mass tolerance of ±10 ppm. For conventional IR-MALDESI, the search comprised [M+H+]+, [M+H+−H2O]+, [M+Na+]+, and [M+K+]+ species; for silver cationized species, the search was performed with calculated neutral masses for [M+Ag+]+ and [M+Ag+−H2O]+.
Histology-defined analysis of IR-MALDESI imaging data
For the histology-driven strategy, peaks unique to the cerebral cortex were extracted accordingly. In total, six embryos (n = 3 per genotype) were analyzed in duplicates via IR-MALDESI and the data from each hemisphere were examined separately (N = 24). To obtain aligned spectra across all samples, a concatenated peak list was generated by combining extracted peak lists from each embryo. The original list contained a total of 288 unique m/z values of ions that were coincident with the embryonic brain outlines. Absolute ion abundances associated with the peak list were subsequently extracted for each sample from each voxel in the dorsolateral cortex within a 5-ppm mass measurement accuracy window. To account for phenotypical differences in the thickness of the cortex between Sp2-cKO and WT [3], the ion abundances were individually normalized by the number of voxels and low abundant m/z values were excluded from further analysis by introduction of an average ion abundance threshold of 100 a.u. Eight m/z values with more than 15 % of the observations below the threshold value were thus removed from the dataset. For the remaining 280 m/z values, ion abundances below the threshold (0.4 %) were set to 100 a.u.
The reduced dataset was subsequently normalized to ensure the comparability between the samples and to minimize experimental biases. Several normalization strategies have been proposed in the scientific discussion and applied to MSI datasets [21, 22]. The most common normalization strategy is normalization to summed intensities of all detected m/z values [21, 22]. This approach is straightforward to implement and sufficiently corrects for global effects, e.g., fluctuations in the ionization process. However, we reasoned that alterations in the abundance of ambient ions in IR-MALDESI might mask variation in the lower abundant tissue-related ions. To overcome this limitation, only a small subset of the total ion current (280 tissue-related m/z values) was selected for normalization in the present study (“TIC of informative peaks”) [22]. No significant differences in the summed and TIC normalized abundances of these ions were observed. The reduced and normalized dataset was subsequently grouped into WT and Sp2-cKO samples.
Applying the ANOVA [23] functionalities of JMP Pro 11, each m/z value was independently tested against the null hypothesis that the corresponding mean values from Sp2-cKOs and WTs were derived from the same sample population [24]. The t test revealed 66 m/z values with significantly changed means (p value <0.05). A Benjamini-Hochberg FDR adjustment was subsequently applied to correct for multiple comparisons [25]. FDR correction refined the list to 25 unique m/z values that exhibited significantly different relative abundances in the dorsolateral cortex of WT and Sp2-cKO mouse embryos. Six m/z values were additionally removed as they were assigned to 13C isotopes by manual revision of the raw mass spectra. The list of the remaining 19 m/z values was subjected to a METLIN database search [26].
The workflow developed above was subsequently applied to the Ag-doped IR-MALDESI imaging dataset of adjacent tissue sections from the same embryos. To avoid redundancy with the previous section, the analysis was exclusively limited to potential Ag+ adducts. In total, 133 unique peak pairs were extracted from the six brain sections, whereas only the lower m/z value ([M+107Ag+]+) was considered for further analysis. Ion abundances were extracted from the cerebral cortices for each m/z value and normalized to the sample area. Nine m/z values were removed from the dataset as more than 15 % of the averaged ion abundances were below the threshold of 100 a.u. For the remaining 124 m/z values, observations below the threshold (0.8 %) were set to 100 a.u. as described above. Prior to performing ANOVA, the dataset was normalized to the sum of all extracted ion abundances and grouped into WT and Sp2-cKO cohorts. ANOVA yielded nine distinct m/z values with p values <0.05. FDR correction and manual inspection of the raw mass spectra refined this list to only one significant hit, m/z 491.2457.
LC-MS/MS analysis
Samples were reconstituted in 30 μL of 0.001 % Zwittergent 3–16. Eight microliters of tryptic digest was subjected to online desalting and reverse-phase nano-LC separation using a direct inject column configuration on a Thermo Easy nano-LC 1000 system coupled to a QExactive High Field mass spectrometer (Thermo Scientific, Bremen, Germany). Analytical columns were created using 75 μm×20 cm PicoFrit columns (New Objective, Woburn, MA) which were self-packed with Kinetex C18 2.6-μm particles (Phenomenex, Torrance, CA, USA). The sample was loaded using a 12-μL injection volume of mobile phase A (98 % water, 2 % acetonitrile, and 0.2 % formic acid) with a flow rate achieved at a maximum pressure of 500 bar. A 120-min gradient from 5 to 30 % mobile phase B (98 % acetonitrile, 2 % water, 0.2 % formic acid) was performed at a flow rate of 300 nL/min. A full-MS top 20 data-dependent MS/MS (dd-MS/MS) analysis was performed with the following parameters: a spray voltage of +2000 V, a capillary temperature of 300 °C, an S-lens voltage of 55 V, a 300–1600 m/z scan range, 120k MS resolving power, 3E6 MS AGC target, 50 ms MS fill time, 15k MS/MS resolving power, a 1E5 MS/MS AGC target, 30 ms MS/MS fill time, 2.0 m/z isolation window, 27 normalized collision energy, a 1 % underfill ratio, and a dynamic exclusion of 20 s. Unassigned and +1 charges were excluded from selection for MS/MS and peptide match was set to “preferred.”
LC-MS/MS data analysis
Raw data were searched using Sequest HT and Proteome Discoverer with Percolator to filter at a 1 % FDR peptide confidence, along with precursor ions area detector for label-free quantification. Spectra were filtered at 350–5000 Da and a signal-to-noise threshold of 1.5 with a minimum peptide length of six amino acids. Data was searched using the Mus musculus Uniprot database containing 16,718 reviewed sequences. The enzyme used was trypsin, allowing two missed cleavages. Precursor mass tolerance was set to 5 ppm and fragment mass tolerance was 0.02 Da. Dynamic modifications included deamidation (NQ) and oxidation (M) with a maximum of four modifications per peptide. Carbamidomethyl (C) was set as a static modification. Strict parsimony principle was applied to the dataset which requires protein groups to have at least one unique peptide in order to label it as a distinct protein group. Peak area used to quantify protein groups was calculated based on an average of up to the three most abundant peptides for a given protein group. Peak area data was summed for each injection and each sample then normalized to the run with the greatest total peak area. Spectral counts were normalized to the total spectral counts for each sample and filtered prior to analysis, removing any protein groups below an average of five normalized spectral counts (NSpC) in both WT and Sp2-cKO samples. Peak area and normalized spectral counts were compared using Student’s t test for equal variances. Protein groups determined to be significantly different (p < 0.05) by peak area and spectral counts were then functionally analyzed by the DAVID algorithm to obtain biological process and molecular function information [27]. Fold change in peak area and normalized spectral counts was calculated with respect to the wild type.
Western blot confirmation of identified lipid metabolic pathways
Pregnant dams were sacrificed at E13.5, embryos were removed from the uterus, and cortices were microdissected in 1×Hank’s balanced salt solution (HBSS, Gibco) on ice. Cortices were collected in radioimmunoprecipitation assay (RIPA) buffer containing 1 % Triton X-100, protease inhibitors (Roche Diagnostics Corporation, Indianapolis, IN), and phosphatase inhibitors (Thermo Scientific, Rockford, IL). Samples were homogenized in a bullet blender at setting 8 for 3 min then continued lysis on a rocker for 1 h. Supernatant was collected after samples were centrifuged at 13,000 rpm for 20 min at 4 °C. Pierce BCA analysis (Thermofisher Scientific, Rockford, IL) was performed to determine protein concentration. Samples were resolved in an SDS-PAGE gel, transferred to a PVDF membrane, and blotted with the indicated antibodies. Antibodies used in this study are rabbit anti-LSS (1:1000, Abcam, Cambridge, MA), rabbit anti-GPDH (1:1000, Abcam, Cambridge, MA), rabbit anti-AACS (1:1000, Novus Biologicals, Littleton, CO), rabbit anti-Sp2 (1:750, Sigma-Aldrich, St. Louis, MO), and Actin-HRP (1:2000, Santa Cruz Biotechnologies, Dallas, TX). Binding of anti-rabbit HRP-conjugated secondary antibodies was detected using the chemiluminescent HRP substrate Clarity (BioRad, Hercules, CA). Images were captured by film (Kodak, Rochester, NY).
Results
Examination of lipid species in the developing mouse forebrain via IR-MALDESI
Based on our past results from direct analysis of serum samples [14], we reasoned that combining IR-MALDESI and Agdoped IR-MALDESI may significantly enhance the lipid coverage in tissue analyses as well. Since Sp2 is suggested to have a role in cholesterol metabolism [2], we specifically aimed to increase sensitivity toward detection of olefins, such as sterols and unsaturated fatty acid esters, via Ag cationization.
In an initial experiment, we examined the mass spectra acquired from the forebrain of embryonic mouse brains via IR-MALDESI as well as Ag-doped IR-MALDESI in detail (ESM Fig. S2). To facilitate mass spectral interpretation and highlight tissue-related ions, off-tissue electrospray background was subtracted using Thermo Excalibur software tools. Applying conventional IR-MALDESI conditions (i.e., 50:50 MeOH/H2O + 0.2 % FA as electrospray solvent) yielded a complex mass spectrum dominated by the cholesterol [M+H+−H2O]+ peak at m/z 369.3517 (ESM Fig. S2A). This finding concurs the biological abundance of cholesterol in the brain, which is the most cholesterol-rich organ and contains almost 25 % of total cholesterol present in the body [28, 29]. A precise regulation of cholesterol homeostasis is crucial to maintain the physicochemical properties of membranes in both neurons and glia [4, 30, 31]. In particular, this is pivotal during early embryonic development, when a remarkable cellular expansion takes place [32, 33].
The second major constituent of neuronal and glial membranes is phospholipids [30], which yielded an abundant mass spectral pattern of sodiated, potassiated, and protonated molecules between m/z 700 and 900. The most abundant signals in this regime corresponded to phosphatidic acids (PA), phosphatidylethanolamines (PE), phosphatidylcholines (PC), and phosphatidylserines (PS). The high resolving power of the Orbitrap mass analyzer assisted in peak assignment and distinguished different species at the same nominal m/z (for instance PE(38:6) at m/z 764.5225 and PS(34:0) at m/z 764.5436). However, structural isomers that exhibit the same molecular formula remained unresolved. In particular, PC (n:m) and PE (n+3:m) were indistinguishable. A differentiation between the two isobars would require tandem mass spectrometry, which is, as for MSI, limited to a relatively small number of pre-selected ions [34], requires re-interrogation of the sample [35], or comes at the expense of spatial information [36]. These restrictions contradict the untargeted and unbiased study design proposed in here as well as the demand for high spatial resolution. However, previous tandem MS experiments from rat brain tissue indicated a prevalent detection of PCs in the CNS as compared with PEs [37]. Based on this information, m/z 732.5548 was assigned to PC(32:1) while 760.5859, 782.5684, and 798.5420 were tentatively assigned to PC(34:1) [M+H+]+, [M+Na+]+, and [M+K+]+ respectively.
In total, 288 individual ions that coincided with the tissue outline were extracted using the MSiReader software. A simple METLIN database search based on accurate mass yielded tentative identifications for 205 m/z values within 5-ppm mass measurement accuracy (ESM Table S1). Identified species included small molecules with critical cellular functions, such as glutathione (m/z 308.0912) or intermediates of fatty acid metabolism, e.g., acylcarnitines (m/z 400.3421, 450.3574), as well as several lipid species. The list of lipids comprised phospholipids, as well as ceramides, sulfatides, and acylglycerides. These results are in excellent accordance with previous studies from adult mouse brains, e.g., [38, 39], as well as previous IR-MALDESI studies from different tissue types [5, 40].
To evaluate the capabilities of Ag-doped IR-MALDESI, adjacent tissue sections were analyzed using previously optimized electrospray conditions (100 μM AgNO3 in MeOH/H2O) [14] (ESM Fig. S2B). In agreement with previously described results by conventional IR-MALDESI, the base peak at m/z 369.3521 corresponded to [M+H+−H2O]+ of cholesterol. Additionally, a large number of Ag cationized species were detected, as confirmed by the characteristic isotopic pattern. Cholesterol yielded the most abundant silver adduct at m/z 493.2598 and 495.2582 ([M+107Ag+]+ and [M+109Ag+]+) at a relative abundance of approximately 50 %. Interestingly, abundant silver adducts of squalene and lanosterol were observed (m/z 517.2970, [M+107Ag+]+ and 533.2916, [M+107Ag+]+). Squalene is a triterpene and a biosynthetic precursor of the sterol family. In the course of cholesterol biosynthesis, squalene is oxidized to epoxysqualene, which subsequently undergoes isomerization into lanosterol [41]. With respect to the proposed influence of Sp2-dependent transcription on sterol biosynthesis, these two key intermediates were of special interest for the present study [2].
Additional peak doublets in the 700–900 m/z region corresponded to the following silver cationized lipid species: PA(36:2) at m/z 807.4115, PC(32:1) at m/z 838.4542, PC(34:1) at m/z 866.4858, and PC(36:2) at m/z 892.5011 ([M+107Ag+]+ for each). In total, 133 unique peak pairs ([M+107Ag+]+ and [M+109Ag+]+) were extracted from the brain region by applying the previously developed peak picking algorithm based on the exact mass difference of 1.99965 Da and the isotopic abundance ratio of 0.93 [14]. A METLIN database search yielded tentative identifications for 65 of the calculated neutral masses (ESM Table S2). In line with the results obtained under conventional IR-MALDESI conditions, the list comprised acylglycerides, phosphatidylcholines, phosphatidic acids as well as ceramides. As expected from the distinct affinity of silver cations toward olefins, unsaturated compounds were exclusively observed. Forty-four compounds were exclusively silver cationized (22 if only considering [M+Ag+]+), whereas 52 (36 if only considering [M+Ag+]+) compounds were equally detected by both approaches. Compounds that were exclusively detected via Ag-doped IR-MALDESI comprised eight compounds for which the calculated m/z values were below the lower limit of the m/z range for conventional IR-MALDESI, thus limiting comparability. Remarkably, cholesteryl esters (CE) and other cholesterol derivatives were exclusively detected as silver adducts. This is an enticing result with regard to the regulatory effect of Sp2 on cholesterol metabolism.
The mass spectrum between m/z 700 and 900 was strongly interfered by protonated and, to a lesser extent, sodiated and potassiated lipid molecules (ESM Fig. S2B). These signals matched those observed in the absence of silver cations, for instance, [PC(30:0)+H+]+ at m/z 706.5405 and [PC(32:1)+ H+]+ at m/z 732.5564. It was therefore concluded that the gas phase equilibrium of silver cationized vs. protonated molecules was not entirely shifted toward the metal adduct even though Ag+ was added in large excess. This was particularly pronounced for compounds which were easily protonated on the one hand, and exhibited mediocre affinities toward silver cations on the other. This included lipid species with only one double bond, e.g., cholesterol or PC(34:1), the latter of which was predominantly detected in its protonated form at m/z 760.5873. In contrast, the corresponding silver adduct ([M+107Ag+]+, m/z 866.4858) was observed at approximately 40 % relative abundance of the protonated species.
Molecular imaging of embryonic mouse rostral forebrain
A simple comparison of the spatial dimensions of postnatal and embryonic mouse brains highlights the need for high spatial resolution to adequately address anatomical features (Fig. 1). The typical width of a coronal section of an adult mouse brain is approximately 9 mm [42]. In contrast, the width of embryonic sections examined here typically ranged from 2 to 3 mm. To examine molecular differences in the cerebral cortex of developing mouse brains in which Sp2 was conditionally deleted [3], a sufficient resolution of physiological features in the embryonic forebrain was thus prerequisite. Focus was placed on base-line characterization of rostral forebrain sections for this part of the study as a prelude to comparison of control and Sp2-cKO cortices in these sections. Representative examples for IR-MALDESI images of coronal sections of an embryonic mouse brain at E13.5 are depicted in Fig. 2. As evident from these results, the distinct localization of tissue-specific ions correlated well with the histochemical annotation of major anatomical features. Both hemispheres of the rostral forebrain yielded unique lipid profiles and were readily distinguished from adjacent areas of the tissue section. Importantly, the lateral ventricles as well as the thin dorsolateral and medial cortices were well resolved at the examined spot-to-spot distance of 100 μm. One example for the highly abundant class of glycerophospholipids is the ion at m/z 766.5384 corresponding to the [M+H+]+ ion of PC(35:5) and/or PE(38:5) (Fig. 2a). According to the generic biological role of glycerophospholipids as membrane lipids [30], the signal was observed across the tissue section in high frequency and with high signal-to-noise ratios. However, the lipid appeared slightly more abundant in the brain regions of the sliced tissue as compared with the remainder of the body, with abundant signal exhibited in the ventricular zones of the cortex and the lateral ganglionic eminence in the ventral telencephalon.
Fig. 1.
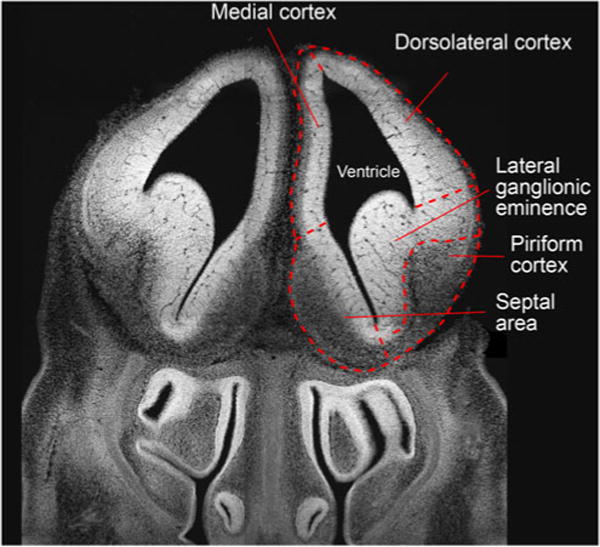
Fluorescence (DAPI, white) image of a coronal section of an E13.5 embryonic mouse brain at the level of the rostral forebrain. Major anatomical features are outlined with dashed lines and annotated accordingly
Fig. 2.
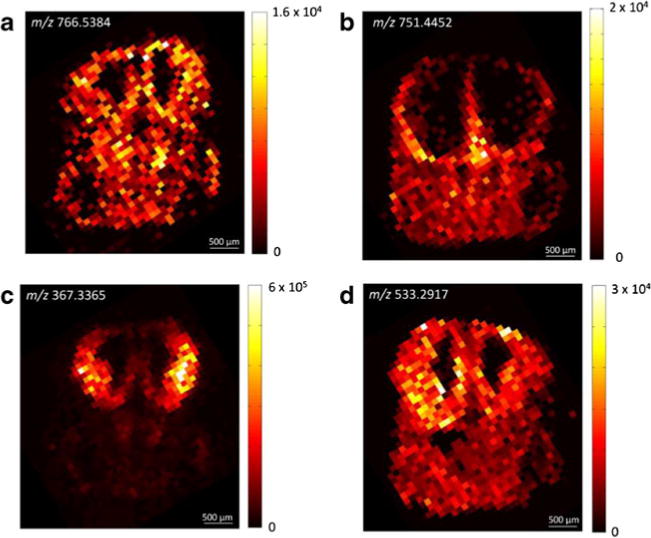
IR-MALDESI imaging of coronal sections of an E13.5 embryonic mouse brain. The ion maps show the spatial distribution of a [PC(35:5)+H+]+, b [DAG(38:4)+107Ag+]+, c [desmosterol+H+−H2O]+, and d [lanosterol+107Ag+]+ (cf. text for details on the peak assignment)
A more distinct spatial distribution was obtained for the silver adduct of diacylglyceride (DAG) 38:4 at m/z 751.4452 (Fig. 2b). DAGs are well established intracellular second messengers and are known to regulate protein kinase C [43–45]. As such, DAGs are also thought to play important roles in neural development [46]. DAG(38:4) appeared specifically localized in the marginal zones of several ventral areas. Most strikingly, the signal was not observed in the ventricular zones of the entire brain, whereas it was abundant in outer superficial regions of the developing forebrain.
The lateral ganglionic eminence was predominantly characterized by a highly abundant ion at m/z 367.3365 (Fig. 2c), which was tentatively identified as [MH−H2O]+ of a cholesterol precursor (7-dehydrocholesterol, zymosterol, or desmosterol). Previous studies of cholesterol metabolism in early central nervous system development of mice found up to 30 % of total sterol content comprised of desmosterol for the first 2 weeks of life [47]. Interestingly, a second biosynthetic precursor of cholesterol, epoxysqualene (or lanosterol, cf. above), exhibited a very similar spatial distribution. The silver cationized molecule was detected in high abundance at m/z 533.2917 and 535.2914 in the lateral ganglionic eminence (Fig. 2d). In addition, abundant signals were observed in the ventricular zones of the medial cortex and, to a lesser extent, the dorsolateral cortex. However, the ion signal was not exclusively limited to the forebrain, and was moderately abundant throughout the entire tissue section.
Mapping unsaturated lipids via specific silver adduct formation
Based on the applicability of both Ag-IR-MALDESI and conventional IR-MALDESI for a comprehensive analysis of lipids in embryonic mouse rostral forebrain, we further hypothesized that the distinct ionization characteristics and mass spectral features observed in Ag-doped IR-MALDESI will permit MSI of unsaturated lipid species, which would otherwise fail to be detected due to low gas phase basicity and low binding affinities to alkali cations. Moreover, we postulated that Ag-doped IR-MALDESI would facilitate the assignment of MS signals due to the soft ionization and the distinctive natural abundance of silver isotopes. Three examples for unsaturated lipids that are involved in metabolic pathways of cholesterol are shown in Fig. 3. Squalene as well as CE(16:1) and CE(18:2) formed abundant silver adducts, which was confirmed throughout the tissue section by examination of the distinctive isotopic ion abundance ratio (107Ag/109Ag 1:0.93) as well as the high spatial correlation of the isotopic peaks indicated by Pearson correlation coefficients >0.95. In the absence of silver cations, the protonated molecule of squalene was detected, whereas both cholesteryl esters were predominantly potassiated.
Fig. 3.
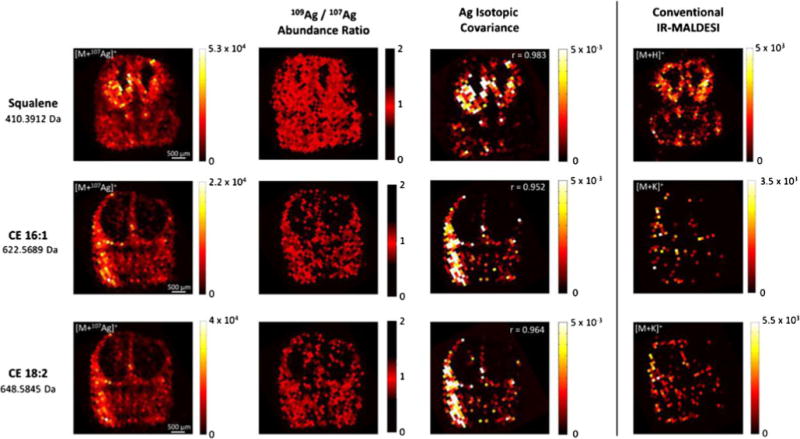
Comparison of silver cationization with ion formation under conventional IR-MALDESI imaging conditions. Ion maps of [M+107Ag+]+ species show the spatial distribution of squalene, CE(16:1), and CE(18:2) in coronal embryonic brain sections. Ion abundance ratios close to the theoretical abundance ratio of 0.9291 (NIST Atomic Weights and Isotopic Compositions, version 3.0) and the high spatial correlation of the detected silver isotopes (Pearson correlation coefficients r > 0.95) confirm the formation of Ag+ adducts. Ion maps on the right refer to the most abundant species that were formed using an electrospray solvent composition of MeOH/H2O (1:1)+0.2 % FA
The Ag+ adduct of squalene was observed throughout the tissue section with high signal-to-noise ratios. Squalene was particularly abundant in the brain as compared with different areas of the tissue section (Fig. 3, top panel). Strikingly, squalene was highly abundant in the ventricular zones of the lateral ganglionic eminence and the medial cortex. This spatial distribution was far less apparent under conventional IR-MALDESI conditions. In highly abundant areas, the spatial distribution of the protonated molecule and the silver adduct was coincidental. However, squalene was less frequently detected in areas with low abundant squalene and the signal-to-noise ratio of the protonated molecule was generally lower as compared with the silver adduct. These results indicated a decreased limit of detection through silver cationization and highlight the specific benefits for mapping less abundant unsaturated lipid species.
Similar observations were made for both cholesteryl esters. Mapping of the silver adducts indicated an accumulation of cholesteryl esters in the marginal zones of the ventral telencephalon (Fig. 3, middle and bottom panel). Interestingly, the spatial distribution of the two cholesteryl esters appeared virtually congruent, which is in accord with their complementary biological function. Subtle differences were observed in the absolute ion abundance, as CE(16:1) appeared slightly less abundant. In contrast, the biological abundance of cholesteryl esters appeared close to the LOD of the conventional IR-MALDESI approach. The potassium adduct of CE(16:1) was merely detected in a diminishing number of voxels, which precluded a proper assignment of the spatial distribution to anatomical features. CE(18:2) was detected in higher frequency; however, the spatial information was still diminishing. This example greatly illustrates the increased sensitivity through silver adduct formation which ultimately enabled the mass spectral imaging of otherwise not accessible compounds.
Resolving conflicts in peak assignment by Ag cationization
In addition to elevated ionization efficiencies, silver cationization facilitated the mass spectral peak assignment since these ions are readily identified by their characteristic isotopic ratio and are less susceptible to in-source decay [14]. As discussed above, peak assignment and compound identification in the present study is limited to accurate mass data. This approach is thus limited to the elucidation of molecular compositions, which a priori precludes the identification of structural isomers. However, uncertainties in the assignment of molecular formulae arise when the formation of various adducts is taken into account. Phospholipids, for instance, were readily detected as protonated, sodiated, or potassiated species applying conventional IR-MALDESI conditions. However, the equilibrium between these species is not at a fixed ratio and may vary throughout an imaging experiment in response to altered salt concentrations in the examined tissue areas [48, 49]. Consequently, all three possibilities have to be considered when searching databases for potential compound IDs based on accurate mass.
For example, m/z 768.5537 was observed in high abundance throughout the embryonic tissue section. According to a METLIN database search within 5 ppm mass measurement accuracy, this signal could be traced back to [M+H+]+ of PC(35:4)/PE(38:4) and/or [M+Na+]+ of PC(33:1)/PE(36:1) (ESM Fig. S3). A further discrimination between both species was not possible based on accurate mass alone. However, Ag+ adduct formation circumvented this conflict, as Ag+ cationized species were readily identified based on the characteristic ion abundance ratio and mass difference, with both species being detected. Strikingly, both species were colocalized, whereas [PC35:4/PE(38:4)+107Ag+]+ yielded the more abundant signal as compared with [PC33:1/PE(36:1)+107Ag+]+. Assuming that protonation and Na+ adduct formation occurred simultaneously in the absence of Ag+, m/z 768.5537, the data likely represents the sum of both lipids. In contrast, the corresponding Ag+ adducts were readily separated on the m/z scale. The colocalization is in accord with the structural and functional similarities of both lipids, confirmed by the spatial distribution of the respective [M+H+]+ ion of PC(33:1)/PE(36:1), which was observed colocalized but less abundant.
Our previous study indicated that Ag+ cationization in IR-MALDESI preserves the intact molecule, whereas insource decays were frequently observed under conventional conditions [14]. Therefore, special care must be taken to avoid false peak assignments. Close inspection of m/z 409.3830 and 411.3983 as detected using 50:50 MeOH/H2O+0.2 % FA as electrospray solvent revealed a conflict that can result from in-source decay (ESM Fig. S4). A metabolite database search for m/z 409.3830 yielded two possible hits: [M+H+]+ of C30H48 and [MH−H2O]+ of C30H50O. Similarly, m/z 411.3983 corresponded to either [M+H+]+ of C30H50 or [MH−H2O]+ of C30H52O. Note that in the first case C30H48 could have been tentatively excluded from the list, as the corresponding compound (dehydrosqualene) does not occur in mammalian metabolism; however, the molecular formulae C30H50 and C30 H52O match those from squalene and dihydrolanosterol, both of which are key metabolites in the cholesterol biosynthesis [41] and thus likely to be detected in embryonic mouse brains. A further differentiation based on accurate mass was unfeasible as the corresponding accurate masses differ by less than 1 ppm.
Ag+ adduct formation readily resolved this conflict. Out of the four different candidates, C30H52O (m/z 533.2916) and C30H50 (m/z 517.2970) were detected as [M+107Ag+]+. However, there was no detectable presence of silver cationization of C30H48 or C30H52O. Note that dehydrosqualene and dihydrolanosterol are both unsaturated compounds, which is why Ag cationization would have been expected. Strikingly, the detected silver adducts were coincident with the ions at the nominal m/z 409.3830 and 411.3983. Based on these results, m/z 409.3830 and 411.3983 were assigned to [MH−H2O]+ of lanosterol (C30H52O) and [M+H+]+ of squalene (C30H50), respectively.
Comparison of Sp2-cKO and WT mouse embryonic cortices via IR-MALDESI imaging and histology-dependent analysis
Our application of IR-MALDESI for MSI of embryonic mouse forebrain revealed several key intermediates of the cholesterol biosynthesis pathway. Here, the developed IR-MALDESI imaging approaches were applied to elucidate differences between Sp2-cKO and WT embryonic mouse cerebral cortices in rostral forebrain sections at the molecular level.
Stringent histology-defined and histology-independent (unsupervised) data analysis strategies were applied autonomously to investigate the effect Sp2 deletion on lipid metabolism in the cortex. Consecutive coronal sections of Sp2-cKO and WT mouse brains were collected and subsequently analyzed via conventional and Ag-doped IR-MALDESI. To guarantee the inter-embryo comparability, the sections were collected from equivalent regions on the rostral-to-caudal axis for each embryo. The correct spatial orientation was confirmed via histochemical analysis of adjacent tissue sections using DAPI staining. Univariate and multivariate statistical tests were subsequently employed to analyze the MSI dataset as described in “Materials.”
In total, eight ions were significantly upregulated between two- and sevenfold in Sp2-cKO embryos as compared with the corresponding control samples (Table 1). The ion at m/z 309.0498 was tentatively identified as 2, 4-dioxotetrahydropyrimidine D-ribonucleotide as the other possible matches for the m/z value were drugs and thus not expected to be abundant in the samples. Interestingly, Terrados et al. reported that genes involved in nucleotide metabolism were repressed in Sp2-deficient mouse embryonic fibroblasts (MEFs) [2], which may be linked to the accumulation of a precursor molecule observed in our data. The dicarboxylic fatty acid hexacosanedioic acid and unsaturated lipid heneicosadiene were also upregulated. Other dicarboxylic fatty acid structures have been identified by previous studies [50] and while less abundant, odd carbon long-chain lipids have been associated with disease states in mammals and can be obtained from dietary sources as well as de novo lipid biosynthesis and alpha oxidation pathways [51, 52]. Other upregulated ions observed corresponded to small lipid molecules and phosphatidylglycerol 36:0 (m/z 779.5789).
Table 1.
Results of the histology-defined comparison of the dorsolateral cortices of Sp2-cKO and WT mouse embryos via conventional IR-MALDESI imaging. Ions with significant FDR-corrected p values <0.05 were tentatively identified by a METLIN database search based on accurate mass
| m/z | Fold Change | p-value* | Adduct | Experimental Neutral Mass | Formula | MMA**(ppm) | Name |
|---|---|---|---|---|---|---|---|
| 309.0498 | 2.04 | 0.016 | [M+H]+ | 308.0412 | C15H17BrO2 | 4 | Aplysinal |
| [M+H−H2O]+ | 326.0515 | C9H15N2O9P | 3 | 2,4-Diaxotetrahydropyrimidine D-ribonucleotide | |||
| [M+K]+ | 270.0852 | C11H14N2O6 | 4 | Clitidine | |||
| [M+Na]+ | 286.0606 | C12H15O6P | 0 | 2-Methacryloyloxyethyl phenyl phosphate | |||
| 315.3009 | 2.22 | 0.047 | [M+Na]+ | 292.313 | C21H40 | 4 | Heneicosadiene |
| 427.3787 | 4.30 | 0.032 | [M+H]+ | 426.3709 | C26H50O4 | 1 | Hexacosanedioic acid |
| 683.405 | −0.65 | 0.011 | [M+K]+ | 644.4417 | C35H65O8P | 0 | PA 32:2 |
| 709.421 | −0.69 | 0.034 | [M+K]+ | 670.4574 | C37H67O8P | 0 | PA 34:3 |
| 711.4363 | −0.71 | 0.014 | [M+K]+ | 672.473 | C37H69O8P | 0 | PA 34:2 |
| 737.452 | −0.68 | 0.013 | [M+H]+ | 736.4411 | C41H60N4O8 | 4 | Antanapeptin A |
| [M+H−H20]+ | 754.4549 | C37H72O11P2 | 0 | PPA 34:1 | |||
| [M+K]+ | 698.4887 | C39H71O8P | 0 | PA 36:3 | |||
| 739.4675 | −0.70 | 0.014 | [M+H]+ | 738.4588 | C37H70O12S | 1 | 1,2-Di-O-myristoyl-3-O-(6-sulfoquinovopyranosyl)glycerol |
| [M+K]+ | 700.5043 | C39H73O8P | 0 | PA 36:2 | |||
| 761.4518 | −0.60 | 0.038 | [M+K]+ | 722.4887 | C41H71O8P | 0 | PA 38:5 |
| [M+Na]+ | 738.4588 | C37H70O12S | 4 | 1,2-Di-O-myristoyl-3-O-(6-sulfoquinovopyranosyl)glycerol | |||
| 765.4833 | −0.64 | 0.011 | [M+K]+ | 726.5200 | C41H75O8P | 0 | PA 38:3 |
| 767.4999 | −0.54 | 0.003 | [M+K]+ | 728.5356 | C41H77O8P | 1 | PA 38:2 |
| 779.5789 | 2.12 | 0.014 | [M+H]+ | 778.5723 | C42H83O10P | 0 | PG 36:0 |
| 789.4843 | −0.51 | 0.013 | [M+K]+ | 750.5200 | C43H75O8P | 1 | PA 40:5 |
| 806.5101 | −0.63 | [M+K]+ | 767.5465 | C43H78NO8P | 0 | PE 38:4 | |
| 826.5736 | −0.62 | 0.032 | [M+K]+ | 787.6091 | C44H86NO8P | 1 | PC 36:1 |
| [M+Na]+ | 803.5829 | C47H82NO7P | 1 | PE 42:6 |
MMA mass measurement accuracy
FDR corrected
Eleven compounds were significantly reduced in Sp2-cKO cortices. The fold changes ranged from 0.5 to 0.7, and the corresponding ions were identified as phosphatidic acids (PA) and phosphatidylcholines (PC). PAs are involved in the biosynthesis of triacylglycerides and phosphatidylethanolamines. Transcriptome data from Terrados et al. [2] indicated that the biosynthesis of lipids is elevated in the absence of Sp2, which would be consistent with a lower abundance of PAs as intermediates in these pathways. On the contrary, the ions at m/z 806.5101 and 826.5736 were also found to be downregulated and identified as alkali metal adducts of PCs and/or PEs. However, the corresponding (more abundant) protonated species did not yield significant alterations in the Sp2-cKO cortices. The statistical analysis Ag-doped IR-MALDESI data revealed only one significantly altered ion at m/z 491.2457 (FDR corrected), which corresponded to [M+107Ag+]+ of desmosterol or other cholesterol precursor. The Ag+ adduct formation was confirmed by plotting the 109Ag/107Ag ion abundance ratio and determination of the Pearson correlation coefficient of the two isotopes (>0.99). The statistical significance of the observed 0.7-fold change of desmosterol or other cholesterol precursor (Fig. 4) was exclusively determined for the silver cationized molecule. For the corresponding [MH −H2O]+ ion under conventional IR-MALDESI conditions, a FDR corrected p value of 0.058 was calculated.
Fig. 4.
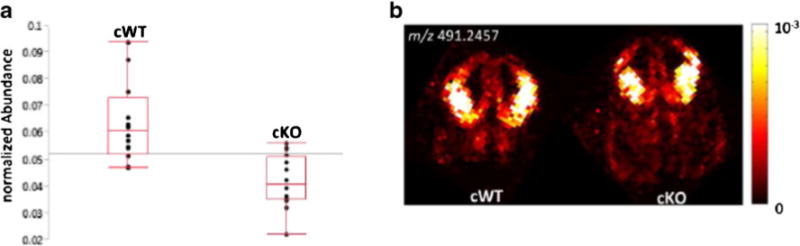
Comparison of WT and Sp2-cKO mouse embryos via Ag-doped IR-MALDESI imaging. a Box plot of averaged and normalized ion abundances of m/z 491.2457 (desmosterol or other cholesterol precursor) from dorsolateral cortex (N = 24). b IR-MALDESI imaging of 491.2457 m/z ion in coronal brain sections from WT and Sp2-cKO embryos
Histology-independent strategy: principal component analysis
The analysis described above was strictly guided by histology. In contrast, the ‘histology-independent’ strategy reveals latent regions within a tissue section that exhibit correlated MS spectra, without the requirement for prior histological information. In particular, rodent brain sections have been successfully subjected to such strategies due to their intrinsic and defined compartmentalization [53, 54]. Here, we applied principal component analysis (PCA) to an IR-MALDESI imaging dataset from an embryonic mouse rostral forebrain preparation (ESM Fig. S5). Each of the three principal components highlighted distinct regions of the dataset that exhibited correlated mass spectra. The first principal component differentiated the tissue from off-tissue background areas, which was in agreement with the large number of ions generated from the tissue. The subsequent principal components highlighted distinct areas within the tissue. The lateral ganglionic eminence, for instance, was clearly differentiated from adjacent forebrain regions as well as from non-brain tissue by principal component 3. Principal component 7 highlighted the medial and dorsolateral frontal cortices. Given that the highlighted regions exhibit distinct mass spectral features, we searched for potential marker ions responsible for the observed mass spectral correlation by reconstructing loading plots of principal components 3 and 7 (ESM Fig. S6). High absolute loadings indicate a high contribution to the principal component and thus to the differentiation visualized by the principal component. The largest contribution to the principal component 3 was given by m/z 367.3366. This was in excellent agreement with the distinct localization of desmosterol or other cholesterol precursor in the lateral ganglionic eminence (LGE). Further contributions to this principal component were seen from lipids detected at the nominal m/z 732, 734, 739, and 760. These ions corresponded to the protonated species of PC(32:1), PC(32:0), PA(36:2), and PC(34:1), which were found to be predominantly abundant in the brain by our initial study of wild-type tissue.
The dorsolateral and medial frontal cortices were largely characterized by PC(32:1) ([M+H+]+, m/z 732.5545) as indicated by principal component 7, but also exhibited contributions from potassium adducts of PC(34:1) (m/z 798.5416) and PC(32:1) (m/z 770.5100). This lipid profile differentiated the frontal cortex from the LGE and other areas of the tissue section, as the latter exhibited distinct contributions from different lipid species, namely PC(32:0) (m/z 734.5709), PC(36:1) (m/z 788.6173), and PC (38:4) (m/z 810.6011). These results indicated that the unsupervised PCA workflow developed here readily highlights correlated tissue areas and provides a rapid readout of the corresponding mass spectral features. We thus aimed to extend this workflow to differentiate WT and Sp2-cKO sample cohorts efficiently. Therefore, two imaging datasets from a WT and a Sp2-cKO embryo were concatenated prior to exporting a total ion current (TIC) normalized ion abundance matrix.
Two principal components in particular highlighted shared features and differences between both brain sections (Fig. 5). Upon interpretation of the loading plots, it was found that the highly abundant signal for m/z 367.3367 in the LGE accounted the most for the similarity between both samples. This finding was in accord with the observations reported above. In addition, both samples shared the distinct abundance of a testosterone derivate at m/z 465.3342 and PC(32:1) at m/z 732.5546. With respect to the ultimate goal of the study to elucidate molecular differences between Sp2-cKO and WT cortices, it was tempting to search a principal component that discriminated against one phenotype. However, there was no clear differentiation between both imaging datasets found. Principal component 7 showed some differentiation between both embryos, as the signals at m/z 465.3342, 732.5546, and 760.5856 appeared specifically associated with the Sp2-cKO brain. However, this result was ambiguous and not clearly limited to the cortex, where the largest variance was expected based on the biological model. In addition, the histology-dependent strategy did not confirm these results, as no significant changes in the ion abundance were observed for the highlighted species.
Fig. 5.
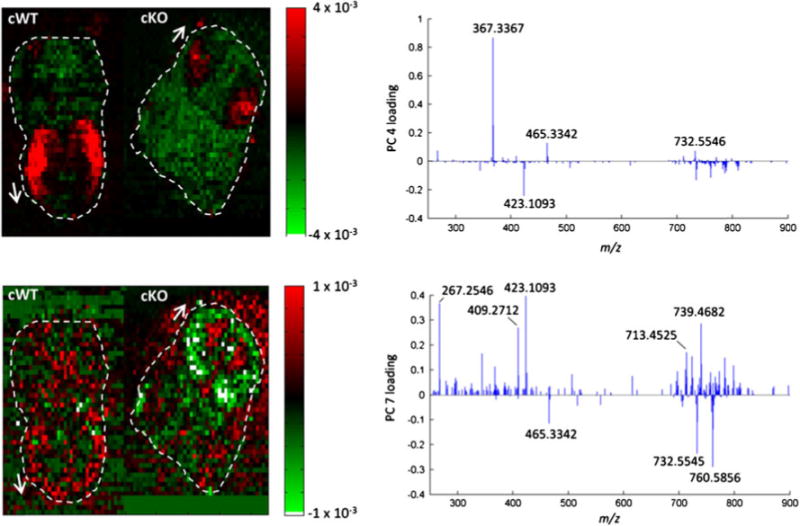
Principal component analysis of a concatenated IR-MALDESI imaging dataset of Sp2 WT and cKO embryonic brain sections. Score plots of principal components 4 (top) and 7 (bottom) indicate mass spectral correlation and anti-correlation between WT and cKO samples. Loading plots on the right visualize the contribution of MS signals to the respective principal component. Dashed lines and arrows indicate the outline and orientation of the tissue sections
PCA was likewise applied to the Ag+ doped IR-MALDESI imaging dataset. To facilitate the interpretation, exclusively, the low m/z values from detected peak pairs were considered. Similar to the previous results, the LGE and the cortex exhibited strongly correlated mass spectra as visualized by principal components 2 and 8 (ESM Fig. S7). In accordance with the previous results, the LGE (principal component 2) was predominantly characterized by the signal associated to desmosterol or other cholesterol precursor (m/z 491.2457). In addition, the silver cationized species of squalene (m/z 517.2970), lanosterol (m/z 533.2917), and distinct lipids contributed to the principal component. The cortical areas (principal component 8) were predominantly characterized by lanosterol and PC(32:1) (m/z 838.4542), PC(34:1) (m/z 866.4858) as well as PA(36:2) (m/z 807.4115). In general, these results correlated well with the manual analysis of the Ag-doped IR-MALDESI dataset and the multivariate analysis of the conventional IR-MALDESI dataset.
Ultimately, we aimed to compare Sp2-cKO and WT species via PCA (Fig. 6). In agreement with the previous observations, the lateral ganglionic eminences of both species largely contributed to the similarity of both datasets. The loadings for the second principal component thus did not differ from those obtained for a single tissue section. Interestingly, a better differentiation of both forebrains was obtained from the Ag-doped approach as compared with the conventional one. Principal component 6 differentiated the ventricular zones of the WT from the Sp2-cKO brain. In particular, squalene and lanosterol contributed to the WT, whereas PA(36:2) (m/z 807.4115) was associated to the Sp2-cKO.
Fig. 6.
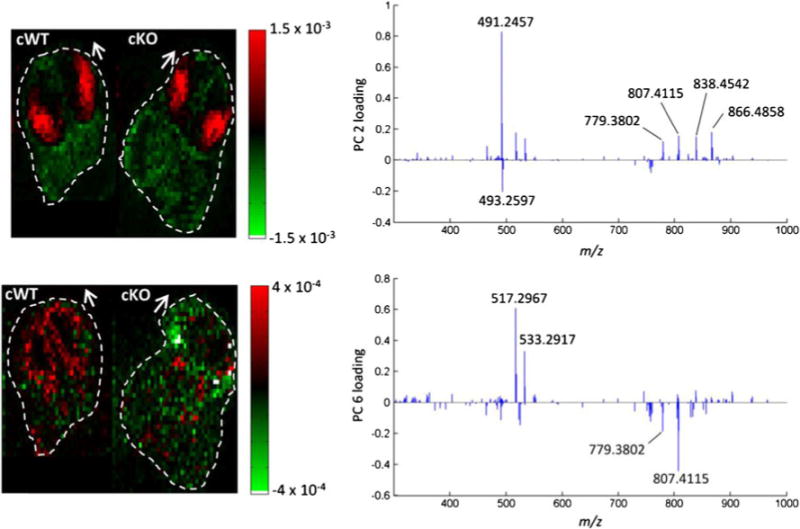
Principal component analysis of a concatenated Ag-doped IR-MALDESI imaging dataset of Sp2 cWT and cKO embryonic brain sections. Score plots of principal components 2 (top) and 6 (bottom) indicate mass spectral correlation and anti-correlation between WT and cKO samples. Loading plots on the right visualize the contribution of MS signals to the respective principal components. Dashed lines and arrows indicate the outline and orientation of the tissue sections
Comparative proteomic profiling of Sp2-cKO and WT cortices via label-free quantification by LC-MS/MS and western blot
Proteins identified in Sp2-cKO and WT cortical samples were compared by label-free quantification of normalized spectral counts and peak area. Based on the normalization factor, Sp2-cKO biological replicate “3” was determined to be an outlier and removed from further analysis. Between two remaining biological replicates, 3350 proteins were quantifiable by peak area and 1450 proteins were shared by both WT and Sp2-cKO samples (see ESM). In comparing proteins by normalized spectral counts (NSpC), 33 protein groups were significantly different (p < 0.05) based on t test; 17 upregulated, 16 downregulated in Sp2-cKO) and normalized peak area indicated, and 147 protein groups were significantly different (p < 0.05; 63 upregulated, 84 downregulated). Between the two methods, NSpC and normalized peak area shared 7 protein groups which were significantly different (p <0.05). In searching the Reactome pathway browser [55], 10 of these proteins were involved with lipid biosynthesis (Table 2). Additionally, several signaling pathways were enriched for including EGFR, FGFR, P13/AKT, ERBB2, PDGF, VEGF, and NGF signaling pathways.
Table 2.
Proteins involved in lipid biosynthesis which were significantly different (p < 0.1) identified by shotgun LC-MS analysis of Sp2-cKO and WT tissue sections
| Protein targets involved in lipid biosynthesis
| |||
|---|---|---|---|
| Fold change in Sp2-c KO | Accession | Protein | p value |
| −0.6 | Q3TFD2 | Lysophosphatidylcholine acyltransferase 1 | 0.025 |
| −0.4 | O70503 | Very-long-chain 3-oxoacyl-CoA reductase/estradiol 17-beta-dehydrogenase 12 | 0.007 |
| +2.0 | Q9CY27 | Trans-2,3-enoyl-CoA reductase | 0.025 |
| −0.7 | Q9D2RO | Acetoacetyl-CoA synthetase | 0.004 |
| +1.1 | Q91V92 | ATP-citrate synthase | 0.027 |
| +1.6 | O54734 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase | 0.041 |
| +7.9 | Q8BLN5 | Lanosterol synthase | 0.055 |
| +2.1 | Q64521 | Glycerol-3-phosphate dehydrogenase | 0.056 |
| −0.4 | Q8KOC4 | Lanosterol 14-alpha demethylase | 0.096 |
| −0.6 | Q99JY8 | Lipid phosphate phosphohydrolase | 0.055 |
Proteins selected for western blot analysis are italicized
Annotation information for molecular function obtained from the DAVID algorithm indicated that ~83 % of the downregulated proteins and 74 % of those upregulated in Sp2-cKO cortices were nucleotide binding (Table 3). With respect to biological processes, proteins involved in macromolecule biosynthesis comprised 39 % of those upregulated in the Sp2-cKO cortices. Of the proteins downregulated in Sp2-cKO cortices, the most highly represented biological processes included: cellular biosynthetic processes, macromolecule metabolic processes, transport, nitrogen compound metabolic processes, nucleobase metabolic processes, and gene expression (Table 4). Further analysis of KEGG pathways (ESM Table S3) revealed the following pathways were upregulated in Sp2-cKO cortices: ubiquitin-mediated proteolysis, glycolysis/gluconeogenesis, and spliceosome and cell cycle pathways. The following KEGG disease pathways were also found to be downregulated in Sp2-cKO mice: Alzheimer’s disease, oxidative phosphorylation, Parkinson’s disease, proteasome, Huntington’s disease, prion diseases, and ribosomal related.
Table 3.
Molecular functions upregulated and downregulated as derived from DAVID algorithm based on proteins significantly differing in abundance by spectral counts and/or peak area in Sp2-cKO mice
| Molecular functions altered in Sp2-cKO
| |||||
|---|---|---|---|---|---|
| Pathway | Genes | p value | Fold change in Sp2-cKO | Fisher exact | Direction in KO |
| Ribonucleotide binding | 16 | 2.70E−03 | +2.2 | 1.20E−03 | Upregulated |
| Purine nucleotide binding | 16 | 4.00E−03 | +2.2 | 1.80E−03 | Upregulated |
| RNA binding | 9 | 4.10E−03 | +3.4 | 1.20E−03 | Upregulated |
| Purine nucleoside binding | 12 | 3.30E−02 | +2 | 1.60E−02 | Upregulated |
| Ligase activity, forming carbon-nitrogen bonds | 4 | 3.90E−02 | +5.2 | 7.10E−03 | Upregulated |
| Hydrolase activity, acting on acid anhydrides | 7 | 4.00E−02 | +2.7 | 1.40E−02 | Upregulated |
| Oxygen transporter activity | 3 | 1.40E−03 | −52.8 | 2.10E−05 | Downregulated |
| Purine nucleoside binding | 17 | 4.10E−03 | −2.1 | 1.80E−03 | Downregulated |
| Protein transporter activity | 4 | 4.60E−03 | −11.7 | 3.70E−04 | Downregulated |
| Purine nucleotide binding | 19 | 4.80E−03 | −2 | 2.30E−03 | Downregulated |
| Heme binding | 5 | 6.20E−03 | −6.7 | 8.70E−04 | Downregulated |
| Ribonucleotide binding | 18 | 7.30E−03 | −1.9 | 3.60E−03 | Downregulated |
| Hydrolase activity, acting on acid anhydrides | 9 | 1.70E−02 | −2.7 | 6.20E−03 | Downregulated |
| Ligase activity, forming phosphoric ester bonds | 2 | 2.00E−02 | −96.9 | 1.60E−04 | Downregulated |
| RNA binding | 9 | 2.00E−02 | −2.6 | 7.40E−03 | Downregulated |
| Phosphoprotein binding | 2 | 8.80E−02 | −21.5 | 3.80E−03 | Downregulated |
Table 4.
Biological processes upregulated and downregulated as derived from DAVID algorithm based on proteins significantly differing in abundance by spectral counts and/or peak area in Sp2-cKO mice
| Biological processes altered in Sp2-cKO
| |||||
|---|---|---|---|---|---|
| Pathway | Genes | p value | Fold enrichment | Fisher exact | Direction in KO |
| Mitotic cell cycle | 7 | 8.10E−04 | +6.2 | 1.20E−04 | Upregulated |
| Organelle fission | 6 | 2.00E−03 | +6.6 | 2.80E−04 | Upregulated |
| Cell cycle phase | 7 | 3.60E−03 | +4.6 | 7.50E−04 | Upregulated |
| Cell cycle process | 7 | 8.60E−03 | +3.9 | 2.10E−03 | Upregulated |
| Microtubule-based movement | 4 | 1.10E−02 | +8.6 | 1.20E−03 | Upregulated |
| Alcohol catabolic process | 3 | 3.50E−02 | +10 | 3.30E−03 | Upregulated |
| Carbohydrate catabolic process | 3 | 5.20E−02 | +8 | 6.20E−03 | Upregulated |
| Cellular carbohydrate metabolic process | 5 | 6.10E−02 | +3.3 | 1.70E−02 | Upregulated |
| Cellular macromolecule metabolic process | 27 | 9.50E−02 | +1.3 | 7.00E−02 | Upregulated |
| Electron transport chain | 7 | 3.40E−05 | −11.2 | 2.90E−06 | Downregulated |
| Generation of precursor metabolites and energy | 9 | 9.00E−03 | −6.2 | 1.40E−05 | Downregulated |
| Gas transport | 3 | 2.20E−03 | −41.4 | 4.60E−05 | Downregulated |
| Cellular macromolecule metabolic process | 38 | 2.90E−03 | −1.5 | 1.90E−03 | Downregulated |
| Ribosome biogenesis | 5 | 3.30E−03 | −8 | 4.00E−04 | Downregulated |
| Transport | 23 | 6.20E−03 | −1.8 | 3.40E−03 | Downregulated |
| Cellular catabolic process | 12 | 6.40E−03 | −2.5 | 2.40E−03 | Downregulated |
| Response to dsRNA | 3 | 6.40E−03 | −24.5 | 2.40E−04 | Downregulated |
| Protein localization | 11 | 7.90E−03 | −2.6 | 2.90E−03 | Downregulated |
| Protein transport | 10 | 9.20E−03 | −2.8 | 3.20E−03 | Downregulated |
| Establishment of protein localization | 10 | 9.60E−03 | −2.7 | 3.40E−03 | Downregulated |
| Cellular nitrogen compound metabolic process | 27 | 1.20E−02 | −1.6 | 7.70E−03 | Downregulated |
| Nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 25 | 1.80E−02 | −1.6 | 1.10E−02 | Downregulated |
| Once-carbon metabolic process | 4 | 2.70E−02 | −6.1 | 4.10E−03 | Downregulated |
| Cell morphogenesis | 6 | 2.80E−02 | −3.5 | 7.50E−03 | Downregulated |
| Embryonic development ending in birth or egg hatching | 7 | 2.90E−02 | −3 | 9.40E−03 | Downregulated |
| Embryonic development | 9 | 3.00E−02 | −2.4 | 1.20E−02 | Downregulated |
| Gene expression | 22 | 3.40E−02 | −1.5 | 2.10E−02 | Downregulated |
| Blastocyst development | 3 | 3.60E−02 | −10 | 3.40E−03 | Downregulated |
| Cellular component morphogenesis | 6 | 4.40E−02 | −3.1 | 1.30E−02 | Downregulated |
| Macromolecule catabolic process | 8 | 6.80E−02 | −2.2 | 2.90E−02 | Downregulated |
| Chromosome organization | 6 | 7.10E−02 | −2.7 | 2.50E−02 | Downregulated |
| Cellular response to stress | 6 | 7.10E−02 | −2.7 | 2.50E−02 | Downregulated |
| Cellular biosynthetic process | 23 | 7.90E−02 | −1.4 | 5.40E−02 | Downregulated |
| Cellular macromolecule localization | 5 | 8.40E−02 | −3 | 2.60E−02 | Downregulated |
| Regulation of binding | 3 | 8.80E−02 | −6 | 1.40E−02 | Downregulated |
| Intracellular transport | 6 | 8.80E−02 | −2.5 | 3.30E−02 | Downregulated |
Integrating omics datasets for pathway analysis
LC-MS/MS proteomic and IR-MALDESI metabolomics datasets were integrated to perform lipid pathway analysis. Several proteins and metabolites involved in steroid and steroid hormone biosynthesis were significantly different between WT and Sp2-cKO cortices (Fig. 7). Lanosterol synthase (LSS) was upregulated nearly eightfold in Sp2-cKO cortices. In addition, the enzyme lanosterol demethylase (LDM) which converts LSS into intermediates of the cholesterol pathway was downregulated 0.4-fold. Eight steps following LDM, the cholesterol precursor (demosterol or other) was downregulated 0.7-fold in Sp2-cKO forebrains by IR-MALDESI. Cholesterol then acts as a substrate for steroid hormone biosynthesis which includes reversible formation of estradiol-17β/estriol and irreversible formation of 11-β-hydroxytestosterone by estradiol dehydrogenase (EDH); which was found downregulated 0.4-fold in Sp2-cKO cortices by LC-MS/MS.
Fig. 7.
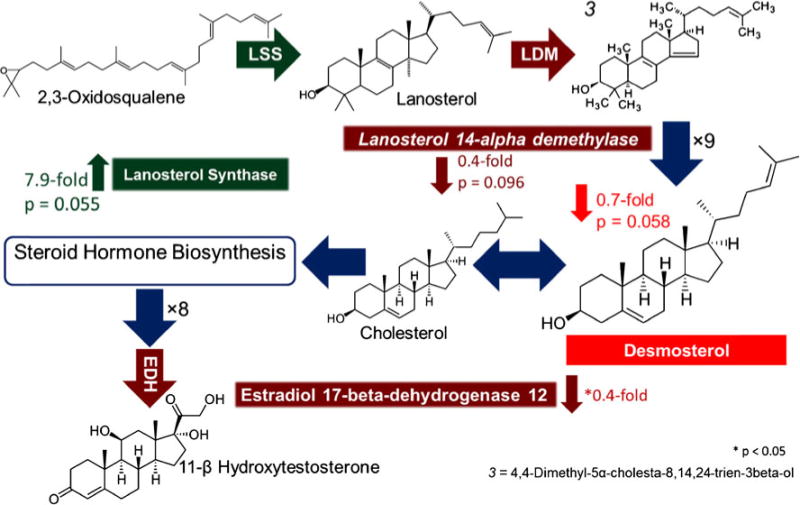
Integrated data analysis of steroid and steroid hormone biosynthesis. LC-MS proteomic data revealed an upregulation of lanosterol synthetase and downregulation of lanosterol 14-alpha demethylase and estradiol 17-beta-dehydrogenase in Sp2-cKO mice while imaging MS data found a decrease in the cholesterol precursor (desmosterol or other). Steroid biosynthesis KEGG pathway (http://www.genome.jp/kegg/pathway/map/map00100.html) and steroid hormone biosynthesis KEGG pathway (http://www.genome.jp/keggbin/show_pathway?map00140)
Several proteins and metabolites belonging to the glycerophospholipid pathway were identified as differing significantly between WT and Sp2-cKO cortices (Fig. 8). One that directly emerged from correlations between metabolomics and proteomic data sets was lysophosphatidylcholine acyltransferase 1 (LPCAT1), found to be downregulated 0.6-fold in Sp2-cKO cortices. The products of this enzyme, phosphocholine and phosphatidic acid, were both downregulated 0.6-fold in Sp2-cKO cortices. Lipid phosphate phosphohydrolase (PPAP2) which converts phosphatidic acid to 1,2-diacyl-sn-glycerol was also found to be downregulated 0.6-fold. Contrastingly, phosphatidylglycerol, an end product of the glycerolphospholipid pathway, was upregulated approximately twofold in Sp2-cKO cortices.
Fig. 8.
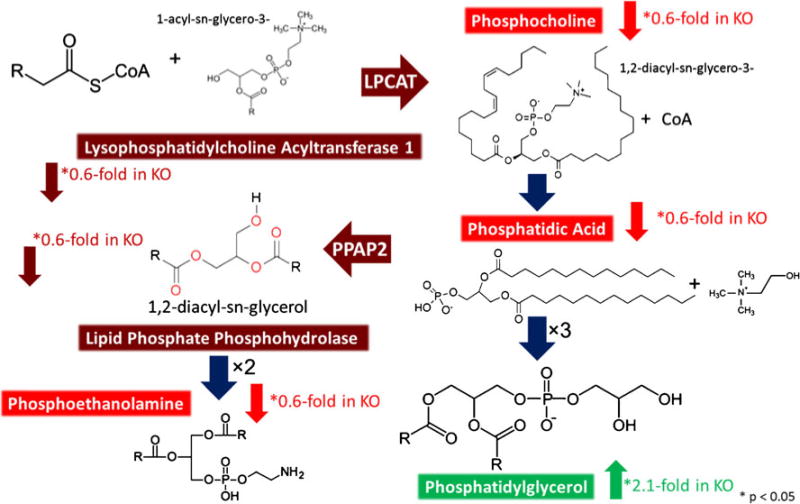
Integrated analysis of glycerophospholipid biosynthesis. LC-MS proteomic data revealed downregulation of lysophosphatidylcholine acyltransferase (LPCAT) and lipid phosphate phosphohydrolase in Sp2-cKO mice. Imaging MS data demonstrated a direct correlation with proteomic data with a downregulation of LPCAT products phosphocholine and phosphatidic acid while the subsequent metabolite, phosphatidylglycerol, was found to be upregulated in Sp2-cKO mice. KEGG glycerophospholipid metabolism (http://www.genome.jp/kegg/pathway/map/map00564.html)
Proteins and metabolites involved in long-chain fatty acid biosynthesis were also found to differ significantly (p <0.05 unless otherwise stated) between WT and Sp2-cKO cortices (Fig. 9). Very-long CoA reductase (synonymous with EDH), which catalyzes the second step in the pathway, was found to be downregulated 0.4-fold in Sp2-cKO cortices. In contrast, trans-2,3-enoyl-CoA reductase (TECR) which catalyzes the fourth step in this pathway, was found to be upregulated twofold in Sp2-cKO cortices. Accompanying these findings, IR-MALDESI found the long-chain dicarboxylic fatty acid hexacosanedioic acid was upregulated fourfold in Sp2-cKO cortices. The fatty acid precursor hexacosanoic acid is produced by single enzymatic step following trans-2,3-enoyl-CoA reductase in which the acyl-CoA is hydrolyzed to the free fatty acid form. Hexacosanedioic acid could be a product of omega oxidation of hexacosanoic acid. This could suggest a defective β-oxidation since this is typically the primary pathway of degradation or a possible response to the accumulation of fatty acids.
Fig. 9.
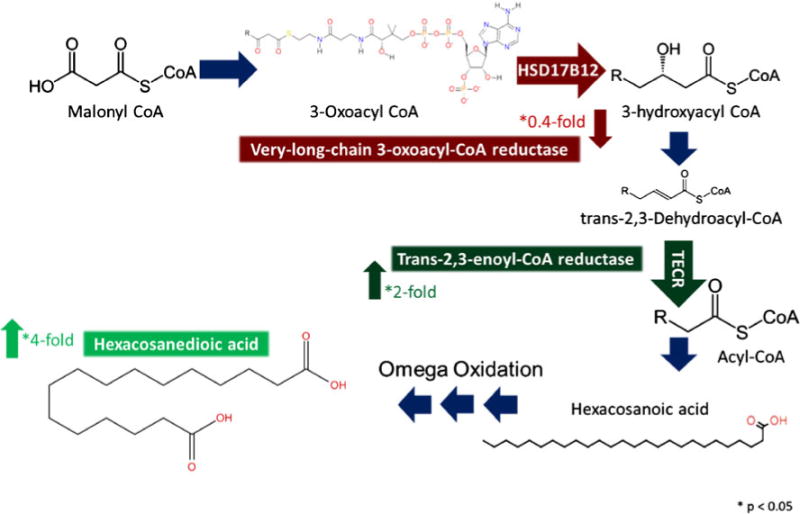
Integrated analysis of fatty acid elongation pathway. LC-MS proteomic data indicated a significant decrease (p < 0.05) in very-long-chain 3-oxoacyl-CoA reductase and a significant increase (p < 0.05) in the subsequent protein in the pathway, trans-2,3-enoyl-CoA reductase, while imaging MS data showed an increase (p < 0.05) in the long-chain dicarboxylic fatty acid, hexacosanedioic acid, in Sp2-cKO mice. KEGG pathway long-chain fatty acid biosynthesis (http://www.genome.jp/keggbin/show_pathway?map00062)
To validate these findings, three of the proteins involved in fatty acid biosynthesis found to differ significantly by label-free quantification were selected for comparative analysis by western blotting (Fig. 10). Lanosterol synthase, the enzyme responsible for formation of lanosterol (a steroid precursor) which was found to be upregulated nearly eightfold (p= 0.055) in Sp2-cKO forebrains, also displayed a twofold increase in abundance by western blot chemiluminescence. Glycerol-3-phosphate dehydrogenase, the enzyme responsible for the reversible hydroxylation of glycerone phosphate (a glycerophospholipid precursor) which was found to be significantly increased twofold (p=0.056) by label-free quantification, was also upregulated twofold by western blotting. Acetoacetyl-CoA synthetase, the enzyme responsible for the generation of coenzyme-A for de novo fatty acid biosynthesis, was found to be downregulated 0.7-fold by label-free quantification and also found to be downregulated twofold by western blot analysis.
Fig. 10.
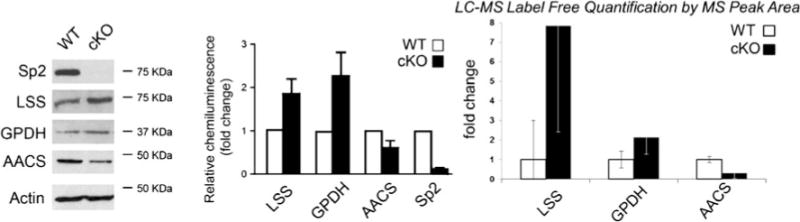
Analysis of three proteins involved in lipid biosynthesis by LC-MS label-free quantification using peak area and western blot showed agreement with an increase in lanosterol synthase (LSS) and glycerol-3-phosphate dehydrogenase (GPDH) abundance and a decrease in acetyl-CoA synthetase (AACS) in Sp2-cKO mice. Sp2 was confirmed to be approximately 10-fold lower in Sp2-cKO mice by western blot
Discussion
Integrated LC-MS proteomic and IR-MALDESI lipidomic imaging data from comparative analysis of WT and Sp2-cKO mice revealed many pathways which were altered by the absence or presence of Sp2. While proteomic analysis discovered changes in many vital pathways (e.g., macromolecule biosynthesis, nucleotide biosynthesis, signaling, and transcription), imaging data revealed alterations to the abundance of 20 lipid species. Correlation of MS and western blot data lends support to the hypothesis that these proteins are differentially regulated in Sp2 cKO and WT samples. Furthermore, the proteins identified within the long-chain fatty acid, steroid/steroid hormone, and glycerophospholipid biosynthesis pathways are also supported as being Sp2 targets by previous transcript studies by Terrados et al. [2]. The direction and magnitude of proteomic changes between WT and Sp2-cKO cortices varied from −0.7-fold to +8-fold. With the exception of the enzyme LSS involved in steroid biosynthesis and TECR involved in glycerophospholipid biosynthesis, precursor and intermediates were primarily downregulated (phosphatidylcholine, phosphatidic acid, and desmosterol/cholesterol precursor) in Sp2-cKO mice while pathway end products were found to be primarily upregulated (phosphatidylglycerol, heneicosadiene, and hexacosanedioic acid). These findings support Sp2 as a direct activator of many lipid biosynthetic enzymes and a repressor of LSS and TECR. Additionally, analysis of changes within the lipidome and proteome highlight the importance of collecting multiple omics datasets for effective elucidation of metabolic steps which are dynamic under physiologic conditions.
Using a novel in-tissue mass spectrometric imaging platform, we successfully elucidated the molecular impact of Sp2-cKO on lipid biosynthesis in developing mouse cortex. Imaging revealed the spatial distribution of these molecules within the rostral forebrain. Further, comparative analysis of Sp2-cKO and WT mice yielded a list of metabolites which differed in abundance within this particular region. Of particular interest, sterol compounds were implicated to be under Sp2 control based on previously reported alterations at the transcriptome level. Even though it is well established that more than 95 % of the brain’s cholesterol is synthesized de novo in the brain [29, 31], the outstanding accumulation of the precursor molecules in the lateral ganglionic eminence of developing mouse brains has not been reported previously. We speculate that this region-specific abundance is linked to distinct proliferation and migratory activity in the LGE which generates distinct classes of neurons for the olfactory bulbs and the ventral forebrain [56–58]. As the generation and gestation of neurons require cholesterol to build plasma membranes [59, 60], this could explain the high abundance of the corresponding molecular precursors within the LGE. It will be interesting to determine whether the same abundance is observed in the medial ganglionic eminence (MGE) in which interneurons destined for the cerebral cortices and several subcortical structures are generated.
Ag-doped IR-MALDESI together with advanced data processing tools were implemented for MSI of embryonic mouse forebrains. Oversampling at a spot-to-spot distance of 100 μm sufficiently resolved major anatomical features of the mouse rostral forebrain without losing spectral sensitivity. Both conventional IR-MALDESI and Ag-doped IR-MALDESI yielded numerous tissue-related ions that were coincident with the physiological fine structure of the tissue sections. By doping trace amounts of AgNO3 into the electrospray solvents, 133 unique peak pairs were detected from the tissue sections, as identified by the peak filtering methods developed above. In accordance with results from conventional IR-MALDESI as well as previous MSI studies of rodent brains, phospholipids were predominantly detected. Most strikingly, Ag dopants decreased the LOD for unsaturated compounds, e.g., cholesteryl esters, and thus enabled imaging of these species. Moreover, as silver adducts are less susceptible to in-source decays, the assignment of mass spectral peaks was clearly facilitated. In this way, multiple biosynthetic precursors of cholesterol, including squalene, lanosterol and desmosterol or other cholesterol precursor, were identified. Interestingly, close inspection of the corresponding ion maps revealed the specific accumulation of these compounds in the lateral ganglionic eminence. This enticing finding will be the subject of further biological studies.
Ultimately, Ag-doped and conventional IR-MALDESI imaging were applied for molecular comparison of Sp2-cKO and WT mouse cortices in sections obtained from the rostral forebrain. Therefore, two complementary data analysis strategies were elaborated. Virtual microdissection of the dorsolateral frontal cortices indicated the regulation of distinct nucleotides and small lipids, which was tentatively correlated with changes in the transcriptome of Sp2 deficient cells. Most strikingly, Ag-doped IR-MALDESI revealed the downregulation of desmosterol or other cholesterol precursor in the dorsolateral frontal cortex of Sp2-cKO mouse embryos, which was presumably linked to alterations in the cholesterol biosynthesis. A second, unsupervised data analysis strategy was applied to reveal latent features in the molecular histology of the tissue sections. PCA readily indicated regions that exhibited distinct mass spectral correlations and provided a rapid readout for those ions that contributed the most to intra-tissue variance.
In conclusion, two complementary data analysis strategies were applied above to determine statistically sound differences between Sp2-cKO and WT sample cohorts. The histology-defined virtual microdissection of the dorsolateral rostral cortices yielded a rich list of compounds that were potentially affected by the conditional deletion of Sp2. This strategy was readily implemented and yielded statistically significant correlations. Histology-independent strategy efficiently reduced the dimensionality of the dataset and yielded a rapid readout for distinct marker ions. Tissue regions that were indicated by principal components correlated well with the histological tissue annotation as well as with manual inspection of the imaging dataset.
Supplementing the molecular information afforded by IR-MALDESI, LC-MS proteomic data revealed many pathways that were significantly different in Sp2-cKO mice by label-free peak area and spectral counting. In agreement with previous research by Terrados et al., nucleoside, RNA, and transcriptional processes in these data also appear to be downregulated in Sp2-cKO mice. Another common finding was that metabolic biosynthetic processes also appeared to be upregulated in Sp2-cKO mice. Previous findings of disrupted cell proliferation phenotypes in Sp2-cKO mice are consistent with the finding that mitotic and cell cycle processes were found to be upregulated and chromosomal organization, gene expression, and cell morphogenesis were downregulated [2]. Moreover, many other fundamental pathways and processes previously identified by Terrados et al. as Sp2 targets were also altered significantly in Sp2-cKO mice including downregulation of protein transport and intracellular transport. Further, other pathways not previously identified as being associated with Sp2 regulation were identified including upregulation of ubiquitin-mediated proteolysis, carbohydrate catabolic processes, spliceosome and downregulation of dsRNA response, proteasome and oxidative phosphorylation. Furthermore, proteins differing in abundance between groups were enriched for cell proliferation and disease-associated signaling, suggesting Sp2 may play a regulatory role in these pathways.
Closer examination of integrated LC-MS proteomic and IR-MALDESI metabolomics datasets revealed that while many pathways were found to be upregulated/downregulated, within these pathways, a more detailed view highlights the complexity of these changes as they are not unidirectional. While 50 % of all 10 lipid biosynthetic proteins and 40 % of the 5 associated metabolites were found to be upregulated in Sp2-cKO mice, 50 % of the proteins and several metabolites were also found to be downregulated. This suggests that Sp2 role in regulating lipid biosynthesis may not be as a global repressor or activator but more pathway and enzyme specific. Several proteins and metabolites involved in steroid biosynthesis differed in abundance in Sp2-cKO mice. Lanosterol synthase (LSS) which was found to be upregulated by proteomic and western blot analysis is a precursor to cholesterol associated with increased oxidative stress [61]. This suggests Sp2 is a direct repressor of LSS which is in agreement with previously published transcript data [2]. Subsequent steps in the pathway including lanosterol 14-alpha demethylase (LDM), estradiol 17-beta-dehydrogenase (HSD17B12), and desmosterol or other cholesterol precursor were all found to be downregulated. These findings could be a result of upregulated LSS. Additionally, the observed downregulation of cholesterol biosynthetic enzymes in Sp2-cKO mice contrasts previous transcript data which suggests Sp2 is a repressor of cholesterol biosynthetic genes (specifically LDM and HSD17B12) in mouse embryonic fibroblasts [2]. This stresses the importance of obtaining measurements of the proteome and lipidome as transcript information alone does not always directly correlate with what is occurring at subsequent levels.
Protein and metabolites involved in the glycerolphospholipid pathway were also found to be altered in abundance. Downregulation of pathway intermediates lysophosphatidylcholine acyltransferase (LPCAT) and its product, phosphatidic acid was the most direct connection between imaging MS and LC-MS proteomic data. Furthermore, subsequent metabolite phosphocholine and the enzyme lipid phosphate phosphohydrolase (PPAP2) were downregulated as well. Downregulation of PPAP2 has been associated with accumulation of lysophosphatidic acid (LPA), proliferation of cells, and cancer [62]. Contrasting this, imaging MS data also found one of the end products of the pathway, phosphatidylglycerol (PG), to be upregulated. PG has been shown to be an activator of nuclear protein kinase C which is necessary for G2 progression of mitosis (a process also upregulated in Sp2-cKO mice) [63]. These observations also suggest a possible shift in lipid class preference in the absence of Sp2 toward PGs. In addition, glycerol-3-phosphate dehydrogenase was determined to be upregulated by proteomic data and western blot. This suggests that the pathway not entirely downregulated but that the flux utilized within this pathway is altered.
An enzyme involved in steroid hormone and long-chain fatty acid biosynthesis, very-long-chain CoA reductase (HSD17B12 or EDH), was found to be downregulated. This suggests Sp2 could be an activator of HSD17B12, an enzyme necessary for normal embryonic development [64]. In HSD17B12KO mice, the absence of this enzyme has been shown to result in lethal effects [65]. Successive steps of long-chain fatty acid biosynthesis also revealed an upregulated trans-2,3-enoyl-CoA reductase (TECR) and hexacosanedioic acid. This suggests Sp2 may also act as a repressor of long-chain saturated fatty acid biosynthesis and/or fatty acid oxidation. Additionally, Sp2 could provide a protective effect by inhibiting saturated fatty acid production which has been associated with stimulation of cytokines and inflammatory pathways [66]. Finally, acetyl-CoA synthetase (AACS), an enzyme responsible for de novo lipid biosynthesis, was also confirmed by MS and western blot to be downregulated in Sp2-cKO mice. Sp2 may activate AACS which has been shown to be developmentally regulated and important during embryogenesis [67]. This finding could also reflect global histone acetylation, a process important in preventing transcriptional errors [68].
Conclusion
The development of a powerful Ag-doped and conventional IR-MALDESI combined analytical tool, allowed for global imaging and label-free quantification of biologically important metabolites, while LC-MS-based proteomics allowed for the identification and quantification of thousands of proteins within specific brain tissue. Integration of data demonstrated the capacity of our approach to provide complementary information, allowing for a better global understanding of complex dynamic changes inherent to metabolic pathways. This analysis revealed fundamental cellular processes altered in Sp2-cKO cortices in proteomics, metabolomics, and lipidomics contexts. Thus, our study represents a new foundation toward solving complex biological problems from a systems perspective, utilizing multiple omics datasets to improve methods for the discovery of important biological pathway targets.
Supplementary Material
Acknowledgments
This work was supported by NIH R01NS089795 (HTG), NIH R01GM087964 (DCM), and the NIH/NCSU Molecular Biotechnology Training Grant 5T32GM00-8776-08 (PL). FM acknowledges travel funding from DAAD (German Academic Exchange Service).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00216-016-9421-3) contains supplementary material, which is available to authorized users.
The authors declare that they have no conflicts of interest.
Compliance with ethical standards Mice used in this study were bred and housed in the College of Veterinary Medicine vivarium according to Institutional Animal Care and Use Committee (IACUC), North Carolina State University regulations, and Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Contributor Information
H. Troy Ghashghaei, Email: Troy_Ghashghaei@ncsu.edu.
David C. Muddiman, Email: dcmuddim@ncsu.edu.
References
- 1.Volkel S, Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. Zinc finger independent genome-wide binding of Sp2 potentiates recruitment of histone-fold protein Nf-y distinguishing it from Sp1 and Sp3. PLoS Genet. 2015;3:e1005102. doi: 10.1371/journal.pgen.1005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terrados G, Finkernagel F, Stielow B, Sadic D, Neubert J, Herdt O, et al. Genome-wide localization and expression profiling establish Sp2 as a sequence-specific transcription factor regulating vitally important genes. Nucleic Acids Res. 2012;16:7844–57. doi: 10.1093/nar/gks544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang H, Xiao G, Yin H, Hippenmeyer S, Horowitz JM, Ghashghaei HT. Neural development is dependent on the function of specificity protein 2 in cell cycle progression. Development. 2013;3:552–61. doi: 10.1242/dev.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom. 2013;1:92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom. 2014;3:319–28. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry JA, Muddiman DC. Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments. Rapid Commun Mass Spectrom. 2011;23:3527–36. doi: 10.1002/rcm.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;5:654–66. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- 8.Shoji N, Nakagawa K, Asai A, Fujita I, Hashiura A, Nakajima Y, et al. LC-MS/MS analysis of carboxymethylated and carboxyethylated phosphatidylethanolamines in human erythrocytes and blood plasma. J Lipid Res. 2010;8:2445–53. doi: 10.1194/jlr.D004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers DS, Ivanova PT, Milne SB, Brown HA. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim Biophys Acta Mol Cell Biol Lipids. 2011;11:748–57. doi: 10.1016/j.bbalip.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ecker J. Profiling eicosanoids and phospholipids using LC-MS/MS: principles and recent applications. J Sep Sci. 2012:10–11. 1227–35. doi: 10.1002/jssc.201200056. [DOI] [PubMed] [Google Scholar]
- 11.Ogiso H, Suzuki T, Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal Biochem. 2008;1:124–31. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Roberg-Larsen H, Vesterdal C, Wilson SR, Lundanes E. Underivatized oxysterols and nanoLC-ESI-MS: a mismatch. Steroids. 2015;99:125–30. doi: 10.1016/j.steroids.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Fhaner CJ, Liu SC, Ji H, Simpson RJ, Reid GE. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;21:8917–26. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier F, Garrard KP, Muddiman DC. Silver dopants for targeted and untargeted direct analysis of unsaturated lipids via infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) Rapid Commun Mass Spectrom. 2014;22:2461–70. doi: 10.1002/rcm.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gokce E, Franck WL, Oh Y, Dean RA, Muddiman DC. In-depth analysis of the Magnaporthe oryzae conidial proteome. J Proteome Res. 2012;12:5827–35. doi: 10.1021/pr300604s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loziuk PL, Parker J, Li W, Lin CY, Wang JP, Li Q, et al. Elucidation of xylem-specific transcription factors and absolute quantification of enzymes regulating cellulose biosynthesis in Populus trichocarpa. J Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00233. [DOI] [PubMed] [Google Scholar]
- 17.Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, et al. The Q exactive HF, a benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteomics. 2014;12:3698–708. doi: 10.1074/mcp.M114.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelstrup CD, Jersie-Christensen RR, Batth TS, Arrey TN, Kuehn A, Kellmann M, et al. Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J Proteome Res. 2014;12:6187–95. doi: 10.1021/pr500985w. [DOI] [PubMed] [Google Scholar]
- 19.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;4:1017–31. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 20.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;12:2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J Proteome. 2012;16:4962–89. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, et al. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal Chem. 2012;3:1310–9. doi: 10.1021/ac201767g. [DOI] [PubMed] [Google Scholar]
- 23.Bray JH, Scott E. Multivariate analysis of variance. Newbury park, CA: Sage; 1985. [Google Scholar]
- 24.Krzywinski M, Altman N. Significance, P values and t-tests. Nat Methods. 2013;11:1041–2. doi: 10.1038/nmeth.2698. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001:1–2. 279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;6:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;9:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietschy JM, Turley SD. Thematic review series: brain lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;8:1375–97. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;5:806–15. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 30.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;2:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orth M, Bellosta S. Cholesterol: its regulation and role in central nervous system disorders. Cholesterol. 2012:292598. doi: 10.1155/2012/292598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herz J, Farese RV., Jr The LDL receptor gene family, apolipoprotein B and cholesterol in embryonic development. J Nutr. 1999;2S(Suppl):473S–5S. doi: 10.1093/jn/129.2.473S. [DOI] [PubMed] [Google Scholar]
- 33.Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, et al. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;4:468–75. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- 34.Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, et al. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2013;20:9596–603. doi: 10.1021/ac401760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landgraf RR, Prieto Conaway MC, Garrett TJ, Stacpoole PW, Yost RA. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal Chem. 2009;20:8488–95. doi: 10.1021/ac901387u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perdian DC, Lee YJ. Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2010;22:9393–400. doi: 10.1021/ac102017q. [DOI] [PubMed] [Google Scholar]
- 37.Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2010;3:982–8. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev. 2011;10:6491–512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gode D, Volmer DA. Lipid imaging by mass spectrometry—a review. Analyst. 2013;5:1289–315. doi: 10.1039/c2an36337b. [DOI] [PubMed] [Google Scholar]
- 40.Nazari M, Muddiman DC. Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) Analyst. 2015 doi: 10.1039/c5an01513h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laboratories K. Steroid biosynthesis, KEGG pathway 00100, version 11/29/13. Available: http://www.genome.jp/kegg-bin/show_pathway?map00100.
- 42.Matusch A, Fenn LS, Depboylu C, Klietz M, Strohmer S, McLean JA, et al. Combined elemental and biomolecular mass spectrometry imaging for probing the inventory of tissue at a micrometer scale. Anal Chem. 2012;7:3170–8. doi: 10.1021/ac203112c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berridge MJ. Inositol trisphosphate and diacylglycerol as second messengers. Biochem J. 1984;2:345–60. doi: 10.1042/bj2200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu Rev Biochem. 1987;56:159–93. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- 45.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;7:484–96. [PubMed] [Google Scholar]
- 46.Adachi N, Oyasu M, Taniguchi T, Yamaguchi Y, Takenaka R, Shirai Y, et al. Immunocytochemical localization of a neuron-specific diacylglycerol kinase beta and gamma in the developing rat brain. Brain Res Mol Brain Res. 2005;2:288–99. doi: 10.1016/j.molbrainres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Quan G, Xie C, Dietschy JM, Turley SD. Ontogenesis and regulation of cholesterol metabolism in the central nervous system of the mouse. Brain Res Dev Brain Res. 2003:1–2. 87–98. doi: 10.1016/j.devbrainres.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Hankin JA, Farias SE, Barkley RM, Heidenreich K, Frey LC, Hamazaki K, et al. MALDI mass spectrometric imaging of lipids in rat brain injury models. J Am Soc Mass Spectrom. 2011;6:1014–21. doi: 10.1007/s13361-011-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiura Y, Konishi Y, Zaima N, Kajihara S, Nakanishi H, Taguchi R, et al. Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res. 2009;9:1776–88. doi: 10.1194/jlr.M900047-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Qin XZ, Chen HQ, Qiu L, Guo YM, Liu H, et al. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2013;1:24–34. doi: 10.1002/rcm.6420. [DOI] [PubMed] [Google Scholar]
- 51.Tucci S, Flogel U, Spiekerkoetter U. Sexual dimorphism of lipid metabolism in very long-chain acyl-CoA dehydrogenase deficient (VLCAD(−/−)) mice in response to medium-chain triglycerides (MCT) Biochim BiophysActa (BBA) - Mol Basis Dis. 2015;7:1442–50. doi: 10.1016/j.bbadis.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Jenkins B, West JA, Koulman A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic acid (C15:0) and heptadecanoic acid (C17:0) in health and disease. Molecules. 2015;2:2425–44. doi: 10.3390/molecules20022425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov T, Becker M, Deininger SO, Ernst G, Wehder L, Grasmair M, et al. Spatial segmentation of imaging mass spectrometry data with edge-preserving image denoising and clustering. J Proteome Res. 2010;12:6535–46. doi: 10.1021/pr100734z. [DOI] [PubMed] [Google Scholar]
- 54.Alexandrov T, Kobarg JH. Efficient spatial segmentation of large imaging mass spectrometry datasets with spatially aware clustering. Bioinformatics. 2011;13:i230–8. doi: 10.1093/bioinformatics/btr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–32. doi: 10.1093/nar/gki072. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deacon TW, Pakzaban P, Isacson O. The lateral ganglionic eminence is the origin of cells committed to striatal phenotypes: neural transplantation and developmental evidence. Brain Res. 1994:1–2. 211–9. doi: 10.1016/0006-8993(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 57.de Carlos JA, Lopez-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;19:6146–56. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;5:461–6. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- 59.van Meer G. Cellular lipidomics. EMBO J. 2005;18:3159–65. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito K, Dubreuil V, Arai Y, Wilsch-Brauninger M, Schwudke D, Saher G, et al. Ablation of cholesterol biosynthesis in neural stem cells increases their VEGF expression and angiogenesis but causes neuron apoptosis. Proc Natl Acad Sci U S A. 2009;20:8350–5. doi: 10.1073/pnas.0903541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;9:2785–97. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanyi JL, Hasegawa Y, Lapushin R, Morris AJ, Wolf JK, Berchuck A, et al. Role of decreased levels of lipid phosphate phosphatase-1 in accumulation of lysophosphatidic acid in ovarian cancer. Clin Cancer Res. 2003;10(Pt 1):3534–45. [PubMed] [Google Scholar]
- 63.Murray NR, Fields AP. Phosphatidylglycerol is a physiologic activator of nuclear protein kinase C. J Biol Chem. 1998;19:11514–20. doi: 10.1074/jbc.273.19.11514. [DOI] [PubMed] [Google Scholar]
- 64.Jokela H, Rantakari P, Lamminen T, Strauss L, Ola R, Mutka AL, et al. Hydroxysteroid (17beta) dehydrogenase 7 activity is essential for fetal de novo cholesterol synthesis and for neuroectodermal survival and cardiovascular differentiation in early mouse embryos. Endocrinology. 2010;4:1884–92. doi: 10.1210/en.2009-0928. [DOI] [PubMed] [Google Scholar]
- 65.Rantakari P, Lagerbohm H, Kaimainen M, Suomela JP, Strauss L, Sainio K, et al. Hydroxysteroid (17{beta}) dehydrogenase 12 is essential for mouse organogenesis and embryonic survival. Endocrinology. 2010;4:1893–901. doi: 10.1210/en.2009-0929. [DOI] [PubMed] [Google Scholar]
- 66.Gupta S, Knight AG, Keller JN, Bruce-Keller AJ. Saturated long-chain fatty acids activate inflammatory signaling in astrocytes. J Neurochem. 2012;6:1060–71. doi: 10.1111/j.1471-4159.2012.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loikkanen I, Haghighi S, Vainio S, Pajunen A. Expression of cytosolic acetyl-CoA synthetase gene is developmentally regulated. Mech Dev. 2002:1–2. 139–41. doi: 10.1016/s0925-4773(02)00097-7. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;2:207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


