Abstract
Recurrent deletions of the long arm of chromosome 5 were detected in 23/200 cases of T-cell acute lymphoblastic leukemia. Genomic studies identified two types of deletions: interstitial and terminal. Interstitial 5q deletions, found in five cases, were present in both adults and children with a female predominance (chi-square, P=0.012). Interestingly, these cases resembled immature/early T-cell precursor acute lymphoblastic leukemia showing significant down-regulation of five out of the ten top differentially expressed genes in this leukemia group, including TCF7 which maps within the 5q31 common deleted region. Mutations of genes known to be associated with immature/early T-cell precursor acute lymphoblastic leukemia, i.e. WT1, ETV6, JAK1, JAK3, and RUNX1, were present, while CDKN2A/B deletions/mutations were never detected. All patients had relapsed/resistant disease and blasts showed an early differentiation arrest with expression of myeloid markers. Terminal 5q deletions, found in 18 of patients, were more prevalent in adults (chi-square, P=0.010) and defined a subgroup of HOXA-positive T-cell acute lymphoblastic leukemia characterized by 130 up- and 197 down-regulated genes. Down-regulated genes included TRIM41, ZFP62, MAPK9, MGAT1, and CNOT6, all mapping within the 1.4 Mb common deleted region at 5q35.3. Of interest, besides CNOT6 down-regulation, these cases also showed low BTG1 expression and a high incidence of CNOT3 mutations, suggesting that the CCR4-NOT complex plays a crucial role in the pathogenesis of HOXA-positive T-cell acute lymphoblastic leukemia with terminal 5q deletions. In conclusion, interstitial and terminal 5q deletions are recurrent genomic losses identifying distinct subtypes of T-cell acute lymphoblastic leukemia.
Introduction
Deletion of the long arm of chromosome 5, del(5q), is the most frequent genomic loss in myeloid diseases.1 Two distinct common deleted regions (CDR) were identified in myelodysplastic syndromes and acute myeloid leukemia. Del(5q), as the sole cytogenetic abnormality, occurs in 10–15% of myelodysplastic syndromes and is known as “the 5q- syndrome”.1 It is characterized by a 1.5 Mb CDR (5q32–q33) where the putative oncosuppressor RPS14 is mapped.2 Del(5q) associated with other cytogenetic changes, often within a complex karyotype, is prevalent in acute myeloid leukemia and in high-risk myelodysplastic syndromes.3 Here it is characterized by a ~1 Mb CDR (5q31) and haploinsufficiency of the oncosuppressor EGR1.4 Conversely, del(5q) has been rarely reported in B-and T- acute lymphoblastic leukemia (ALL).5–8
T-lineage ALL (T-ALL), a heterogeneous group of leukemias, is characterized by co-occurrence of multiple genetic lesions.9 Interestingly, cooperative genetic defects have been described in T-ALL, suggesting that perturbation of specific cell processes are needed for the development of overt leukemia.10,11 Translocations causing TAL/LMO, TLX1, TLX3, MYB, MEF2C, NKX2-1/2, or HOXA over-expression define distinct gene expression signatures and are known as “type A” abnormalities.6,12–14 These rearrangements co-occur with multiple mutations and imbalances, named “type B” abnormalities, which activate oncogenic signaling cascades, including JAK/STAT, PI3K/AKT, and RAS/MEK/ERK. The most prevalent type B abnormalities are loss-of-function mutations and genomic losses, indicating that tumor suppressor genes, such as CDKN2A/B, PHF6, LEF1, PTEN, WT1, ETV6, and PTPN2, play pivotal roles in the initiation of T-ALL.9,15
To provide insights into the prevalence and specific features of del(5q), in T-ALL, we screened a series of 200 TALL cases by fluorescence in situ hybridization (FISH). We found two distinct types of recurrent 5q deletions: interstitial (I-5q) and terminal (T-5q). I-5q was identified in 2.5% of cases with a genomic profile closely resembling immature/early T-cell precursor (ETP) ALL. These findings indicate that I-5q is a recurrent cytogenetic event in T-ALL with early differentiation arrest of blasts. T-5q, identified in 9% of cases, clustered within the HOXA category and was characterized by a high incidence of abnormalities involving the CCR4-NOT complex and its closely related transcription factor BTG1. These observations indicate that HOXA over-expression, haplo-insufficiency of genes at T-5q, and deregulation of the CCR4-NOT complex are characteristic of this subgroup of leukemia.
Methods
We investigated the incidence of del(5q) in a cohort of 66 adult (≥18 years) and 134 pediatric T-ALL patients from the Italian (GIMEMA LAL-0496 and LAL-0904 and AIEOP LLA-2000) and UK clinical trials (MRC, ALL2003 and ALL97) (Online Supplementary Table S1, Online Supplementary Information).10,16,17 All patients or their parents/guardians gave informed consent to sample collection and molecular analyses, in agreement with the Declaration of Helsinki. The study was approved by the local bio-ethical committee (research project 2014-025).
Combined interphase FISH identified T-ALL-associated genomic rearrangements which classified cases into defined subgroups (Online Supplementary Table S2).10,16,17 Deletions of 5q were detected by FISH with LSI EGR1/D5S23, D5S721 Dual Colour probe, RP11-182E4/RP11-453D13 and CTB-31E20/RP11-266N12 for rearrangements of TLX3, RP11-117L6 for NPM1 abnormalities, and RP1-240G13 for deletions of the subtelomeric 5q region. Cases with del(5q) were further characterized with clones for 5p13-qter (Online Supplementary Table S3). Single nucleotide polymorphism array, denaturing high performance liquid chromatography, Sanger sequencing, haloplex polymerase chain reaction,11 transcriptome sequencing,18 and gene expression profiling were performed in cases with available material (Online Supplementary Information).
Results and Discussion
Incidence and distribution of del(5q) in T-cell acute lymphoblastic leukemia
FISH and single nucleotide polymorphism array revealed two types of recurrent del(5q) in 23/200 T-ALL patients: I-5q (5 cases) (Table 1) and T-5q (18 cases) (Table 2). In 11/18 cases with T-5q, the deletion was very large and included the CDR of I-5q cases (Figure 1A). Overall, del(5q) was mainly associated with the HOXA category (16/23; 69.5%). It was found at diagnosis in 22 cases while in the remaining case it was detected only during disease progression (13 months after diagnosis); it belonged to the major abnormal clone in all cases but one. Cytogenetically, both types of del(5q) were always associated with additional chromosome abnormalities and had significantly more DNA copy number abnormalities than cases without del(5q) [median 7 (range, 3–28) versus 3.5 (1–14), P=0.003] (Online Supplementary Table S4).
Table 1.
Clinical, hematologic and molecular-cytogenetic features of 5 cases of T-ALL with interstitial 5q deletion.
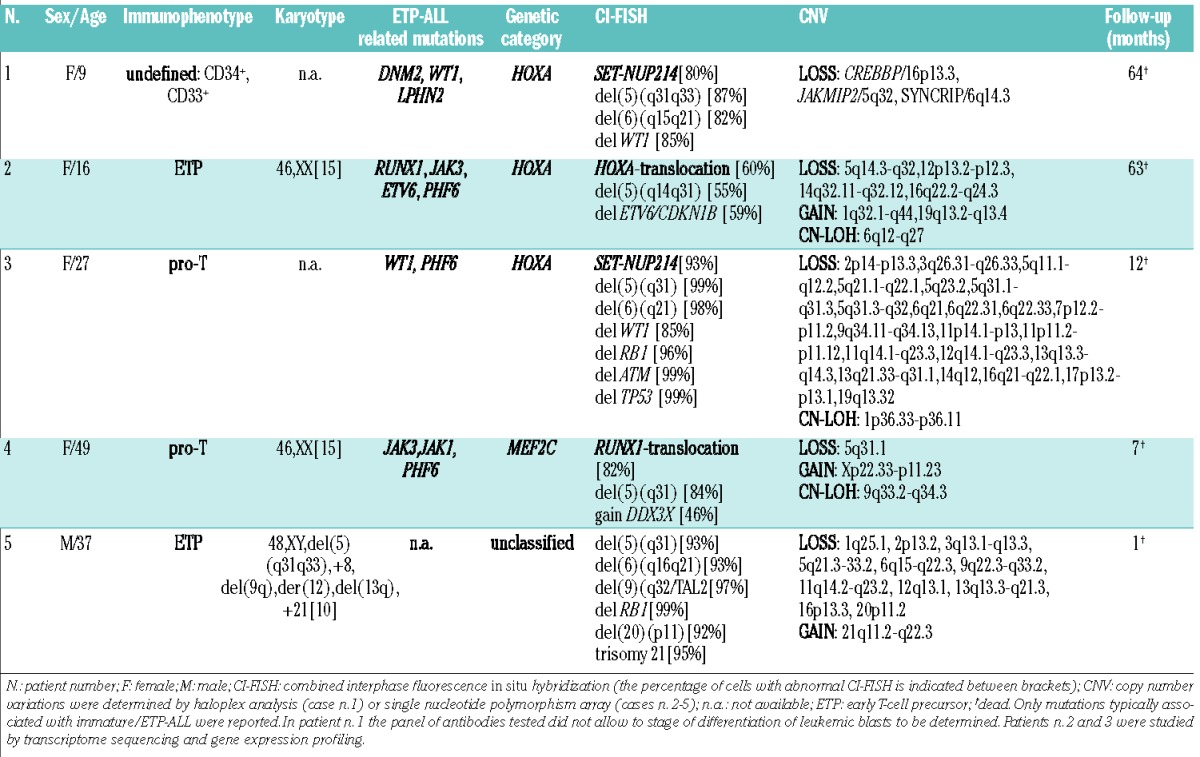
Table 2.
Clinical, hematologic, and molecular-cytogenetic features of 18 cases of T-ALL with terminal 5q deletion

Figure 1.
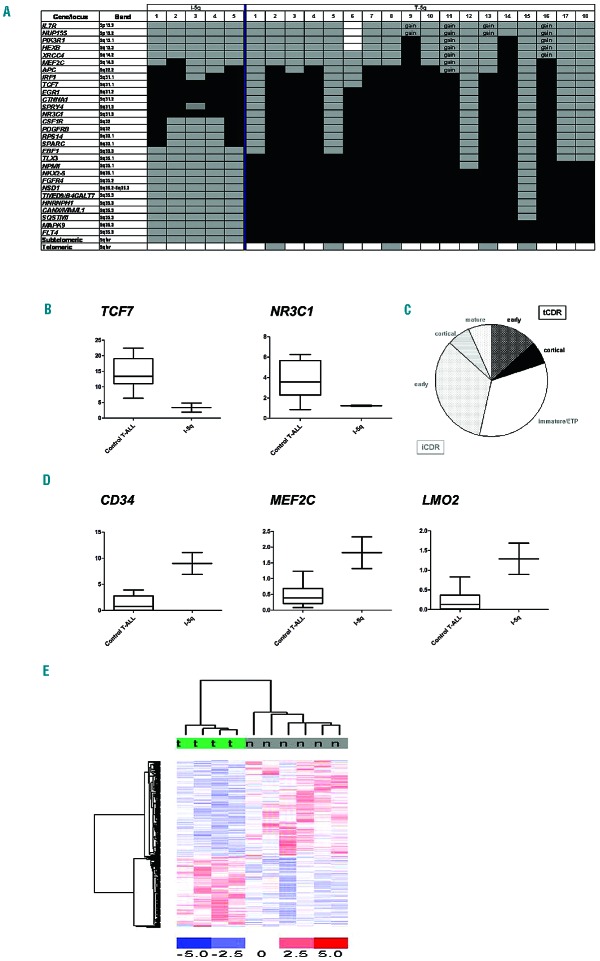
Genomic characteristics of I-5q and T-5q T-ALL. (A) Schematic representation of 5q deletions in our 23 T-ALL cases. Patients’ numbers refer to Table 1 for cases with I-5q, and to Table 2 for cases with T-5q. Black boxes indicate monoallelic deletion; gray boxes, diploidy; white boxes, not tested; gain, presence of three copies; (B) TCF7 and NR3C1 expression in two T-ALL with I-5q vs. control [7 cases without del(5q) and 3 cases with T-5q]; (C) Distribution of terminal CDR (tCDR) (black) (3 cases) and interstitial CDR (iCDR) (gray) (3 cases with I-5q and 9 cases with large T-5q including the I-5q CDR) within the HOXA-category: all five cases with an immature/ETP phenotype as well as five of seven cases of early T-ALL had I-5q or large T-5q including the iCDR; (D) Expression of CD34, MEF2C, and LMO2 in two cases with I-5q vs. ten controls [7 cases without del(5q) and 3 cases with T-5q]; (E) Supervised gene expression profiling analysis of four HOXA positive cases with T-5q (t) and six without (n), identified 327 differentially expressed genes.
Deletions of 5q define two independent subgroups of T-cell acute lymphoblastic leukemia
We have data to show that I-5q and T-5q are cytogenetic markers of two subgroups of T-ALL, with different age and gender distributions and distinct genomic backgrounds.
Interstitial 5q T-cell acute lymphoblastic leukemia
I-5q was detected in five patients (2.5%) (Table 1). Four of these five patients were females (Pearson χ2 test, P=0.012) but there was no distinctive age distribution. Cytogenetically, three cases carried HOXA-activating rearrangements (2 carried SET-NUP214 and 1 carried TCRB-HOXA); patient n. 4 had a translocation involving RUNX1, suggesting likely membership of the MEF2C category;14 patient n. 5 was unclassified (Online Supplementary Information and Online Supplementary Figure S1). The cases of I-5q T-ALL had a higher incidence of genomic losses than cases of T-ALL without del(5q) [mean 9.5 (range, 1–22) versus 3.1 (1–11)], a difference which did not, however, reach statistical significance likely due to the small sample size (Online Supplementary Table S4). They were significantly associated with WT1 deletions (n=2) (χ2, p=0.002) and del(6q) (n=3) (χ2, P=0.002); these del(6q) shared a CDR at band 6q21, encompassing the putative onco-suppressor genes SEC63 and FOXO3 (Online Supplementary Table S5).7,19
All I-5q had a CDR at band 5q31 (Figure 1A), encompassing the 1 Mb CDR of high-risk myelodysplastic syndrome/acute myeloid leukemia del(5q).3,4,20 When we compared I-5q- positive cases with T-5q and T-ALL without del(5q), we found that the expression levels of IRF1, EGR1, CTNNA1, HNRNPAO, TIFAB, and CXXC5, known putative onco-suppressors in in vitro and/or in vivo models20–24 mapping within the CDR, did not differ between the three groups; while the presence of I-5q was associated with significant down-regulation of NR3C1 and TCF7 genes (Figure 1B, Online Supplementary Information and Online Supplementary Figures S2 and S3).
NR3C1 belongs to the nuclear hormone receptor super-family that includes the mineral-corticoid and estrogen receptors. Once NR3C1 binds to steroid hormones, it acts as a direct transcriptional regulator.25 NR3C1 deletion has been associated with relapse in pediatric and adult B-cell ALL and predicted corticosteroid resistance in T-ALL, thus influencing response to treatment.26,27 TCF7 is an essential transcriptional regulator of T-cell specification, commitment, and lineage determination.28 In mouse models, Tcf7−/− induced a T-cell malignancy that was similar to human ETP-ALL as 117 deregulated genes were common to both.29 In vivo studies suggested that TCF7 haplo-insufficiency rendered pre-malignant thymocytes susceptible to later lesions which subsequently transformed them into leukemic blasts. In fact, low Tcf7 expression predisposed murine thymocytes to acquire Notch1-activating mutations which were invariably found in Tcf7−/− lymphomas.29,30 Interestingly, NOTCH1 mutations were present in four out of five cases of T-ALL with I-5q. Together these data suggest strong similarities between mouse Tcf7−/− T-cell malignancies and human T-ALL with I-5q assigning a role to TCF7 haplo-insufficiency in this subgroup of leukemia.
Finally, among HOXA-positive T-ALL, ten of 12 cases with immature/ETP or early phenotype lost the I-5q CDR (Online Supplementary Table S6 and Figure 1C). Among them five cases had the typical features of the high-risk subgroup recently named HOXA-positive ETP ALL.31,32 As far as we know, loss of genes at the I-5q CDR is the first recurrent cytogenetic change so far described in this subgroup.
Terminal 5q T-cell acute lymphoblastic leukemia
T-5q T-ALL was detected in 18 patients (9%), who were mainly adults (χ2, P=0.010) but there was no association with gender (12 males; 6 females), stage of blast differentiation (7 pre-T, 5 cortical, 2 ETP, 1 mature, and 3 undefined), or white blood cell count (Table 2). Cytogenetically, 13 cases belonged to the HOXA category [with rearrangements of: HOXA (n=5), NUP98 (n=3), NUP214 (n=2), MLLT10 (n=2), and MLL (n=1)], two cases to the TLX3 category (Online Supplementary Information and Online Supplementary Figure S4), while three cases remained unclassified. T-5q ALL were significantly associated with del(6)(q14q15) (χ2, P=0.002), and genomic gains (P=0.0038) (Online Supplementary Table S4), of which the most frequent was gain of chromosome 5p arm, found in four out of 17 (23.5%) fully characterized cases. We also found a high incidence of NF1 deletions (4 cases)(χ2, P=0.002) and recurrent N/KRAS mutations (3 cases), consistent with RAS/MEK pathway involvement in ~23% of cases (Online Supplementary Information).33–35
Although T-5q deletions varied greatly in size, they all had a common 1.4 Mb CDR (chr5:179257527-180719789, GRCh37) telomeric of SQSTM1 (Figure 1A). The T-5q CDR contained one LIN gene (long intergenic non-protein coding), five olfactory receptor genes, three microRNA, eight LOC non-coding RNA, and 37 genes. Supervised gene expression profiling analysis showed that only eight out of the 26 genes with probe-sets available at the CDR, i.e. MAPK9, TBC1D9B, RFP130, TRIM52, TRIM52-AS1, HEIH, ZFP62, and CNOT6, were significantly down-regulated in six cases with T-5q T-ALL compared to 22 TALL without.
Genetic profile links interstitial 5q with immature/early T-cell precursor acute lymphoblastic leukemia
ETP-ALL, a distinct subgroup of T-ALL, is defined by a typical immunophenotype which is negative for CD1a and CD8, negative or weakly positive for CD5, and positive for at least one of the following markers: CD34, CD117, HLA-DR, CD13, CD33, CD11b, and CD65.36 ETP-ALL also shows a distinct genetic profile with high expression of two bHLH transcription factors, LYL1 and LMO2, a high incidence of mutations typically associated with the pathogenesis of acute myeloid leukemia, and a low frequency of typical T-ALL lesions, such as CDKN2A/B deletions and NOTCH1 mutations.37 ETP-ALL has been associated with a dismal outcome due to poor response to chemotherapy and a high rate of resistance/early relapse, namely in cases with genomic rearrangements associated with HOXA deregulation.31,32,38 On the other hand, a recent clinical trial demonstrated a high rate of continuous complete remission in children.39
In our study, all I-5q T-ALL were characterized by early differentiation arrest of leukemic blasts with expression of at least one stem cell/myeloid antigen (Table 1). In fact patients n. 2 and n. 5 satisfied all diagnostic criteria for ETP-ALL.36 RB1/13q14 deletions co-occurred in two of five cases. A critical analysis of previous studies showed that del(5q) had already been found in immature/ETP ALL.6,34 Indeed, del(5q) together with del(13q) were the two cytogenetic changes most frequently associated with immature/ETP ALL as they were both detected in 23% of cases (4/17), and co-occurred in 11% (2/17).6,36 Additional evidence that I-5q T-ALL are closely related to the immature/ETP subtype of T-ALL came from identification of PHF6, JAK3, JAK1, DNM2, WT1, ETV6, and/or RUNX1 mutations and lack of CDKN2AB deletion/mutation in all analyzed cases.37,40,41
It is noteworthy that in addition to TCF7, other significantly down-regulated genes in I-5q T-ALL were TDRKH, PCGF5, HDAC4, and MTA3, so that our I-5q patients carried five out ten of the most differentially expressed (down-regulated) genes seen in human ETP-ALL (Online Supplementary Table S7 and Online Supplementary Figures S2, S3, S5-8).29,36 Moreover, among the 22 genes which have been reported to be significantly over-expressed in ETP-ALL,6,14,36,37,40–42 MEF2C, LMO2, and CD34 were significantly up-regulated in I-5q (Figure 1D). Overall, our data highlight two informative aspects of I-5q. First, genomic profiles link I-5q and immature/ETP ALL; furthermore all five cases with I-5q were poor responders to standard therapy. In fact, patient n. 1 was a late remitter despite being assigned to the most intensive arm of the ALL2003 MRC trial. She received a bone marrow transplant from an unrelated donor in second remission, relapsed shortly afterwards and died. Patient n. 2 had an early relapse and also received a bone marrow transplant from an unrelated donor while she was in second remission. She had a second relapse after the transplant and died of her disease. The other three patients had resistant disease and died within 1 year. Although there were too few patients in the present series to draw any definitive conclusions, I-5q marks a particularly high-risk subgroup of immature/ETP ALL for which alternative targeted therapies should be developed. Among them, JAK/STAT inhibitors, which are highly effective in ETP-ALL xenograft models,43 and the BCL2 inhibitor ABT-737, which restores the sensitivity to steroids in cell lines with high MEF2C expression,44,45 might be considered in the treatment of these refractory leukemias.
Terminal 5q is a HOXA-positive cytogenetic subgroup
T-5q was found in 27% of patients belonging to the HOXA group (13/48 cases) (χ2; P<0.001) in which it behaved as a type B event (Table 2). Supervised gene expression profiling analysis compared four HOXA-positive cases with T-5q and six without to determine whether T-5q defined specific pathways within the HOXA category: t-test analysis (P≤0.005) identified 327 genes (Figure 1E). Of the 130 over-expressed genes in T-5q cases, functional annotation analysis revealed enrichment of genes involved in nuclear lumen, DNA replication and mRNA metabolic processes, such as CDC45, CDC5L, CHEK1, E2F3, and FANCD2, suggesting specific deregulation of these pathways. Among the down-regulated genes, FYN and LCK tyrosine kinases, IL7R, ZAP70, ADD3, and the adaptor protein, BTG1, are known oncogenes/tumor suppressors in ALL.
Within the T-5q CDR we observed down-regulation of TRIM41, ZFP62, MAPK9, MGAT1, and interestingly, CNOT6. This is the first report of CNOT6 involvement in human cancer and its down-regulation is consistent with it being a putative onco-suppressor gene as indicated by in vitro data.46 Besides CNOT6 down-regulation, our T-5q TALL cases were associated with a high rate of CNOT3 mutations (18%). Notably, CNOT3 loss-of-function mutations were found in ~7% of adult T-ALL but no specific association with any major molecular subgroups has been reported.47 Both CNOT6 and CNOT3 encode for members of the CCR4-NOT complex, which consists of two major modules: the deadenylase module composed of two subunits with deadenylation enzymatic activity (CNOT6 or CNOT6L and CNOT7 or CNOT8) and the NOT module (CNOT1, CNOT2, and CNOT3). The CCR4-NOT complex is involved in chromatin modification, cellular response to DNA-damage, transcription elongation, RNA export, nuclear RNA surveillance, and miRNA-mediated deadenylation of mRNA.48–50 Although, CCR4-NOT involvement in human tumors has been rarely reported, a recurrent hotspot P131L mutation of RQCD1 (formerly known as CNOT9) has recently been identified in ~4% of cutaneous melanomas.51 Our findings of a high rate of deletion/loss-of-function mutations and down-regulation of members of the CCR4-NOT complex as well as low expression of BTG1, a directly interacting adaptor protein of CCR4-NOT, suggest that this complex plays a role in the subset of HOXA-positive T-ALL with T-5q.
In conclusion, the present study has identified distinct recurrent 5q deletions in T-ALL, corresponding to different genomic landscapes and defining two cytogenetic subgroups. I-5q identified a subgroup of immature T-ALL, found predominantly in females, with an ETP-like genetic profile and poor response to current treatments. T-5q designated a subgroup of HOXA-positive T-ALL, mainly found in adults and associated with a high rate of CCR4-NOT complex abnormalities. In I-5q, two putative onco-suppressors, NR3C1 and TCF7, mapping to the 5q31 CDR, were down-regulated. In T-5q CNOT6, a member of the CCR4-NOT complex, mapping to the 5q35 CDR, was significantly down-regulated. Due to the heterogeneity of treatment and age of our 18 patients, the clinical impact of T-5q could not be established, while the unresponsive T-ALL with I-5q should be considered for new experimental therapies.
Acknowledgments
Associazione Italiana per la Ricerca sul Cancro (AIRC, IG-15525), Fondo per gli Investimenti della Ricerca di Base (FIRB 2011 RBAP11TF7Z_005), Programmi di Ricerca scientifica di rilevante Interesse Nazionale (PRIN Cod. 2010NYKNS7_003), Associazione Sergio Luciani, Fabriano, Italy, and AIRC 5×1000; Bloodwise, formerly Leukaemia and Lymphoma Research.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/8/951
References
- 1.Van Den Berghe H, Michaux L. 5q-, twenty-five years later: a synopsis. Cancer Genet Cytogenet. 1997;94(1):1–7. [DOI] [PubMed] [Google Scholar]
- 2.Boultwood J, Pellagatti A, McKenzie ANJ, Wainscoat JS. Advances in the 5q- syndrome. Blood. 2010;116(26):5803–5811. [DOI] [PubMed] [Google Scholar]
- 3.Lai F, Godley LA, Joslin J, et al. Transcript map and comparative analysis of the 1.5Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q). Genomics. 2001;71(2):235–245. [DOI] [PubMed] [Google Scholar]
- 4.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110(2): 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faderl S, Gidel C, Kantarjian HM, Manshouri T, Keating M, Albitar M. Loss of heterozygosity on chromosome 5 in adults with acute lymphoblastic leukemia. Leuk Res. 2001;25(1):39–43. [DOI] [PubMed] [Google Scholar]
- 6.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. [DOI] [PubMed] [Google Scholar]
- 7.Grossmann V, Haferlach C, Weissmann S, et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer. 2013;52(4):410–422. [DOI] [PubMed] [Google Scholar]
- 8.Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122(1):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durinck K, Goossens S, Peirs S, et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp Hematol. 2015;43(8):625–639. [DOI] [PubMed] [Google Scholar]
- 10.La Starza R, Borga C, Barba G, et al. Genetic profile of T-cell acute lymphoblastic leukemias with MYC translocation. Blood. 2014;124(24):3577–3582. [DOI] [PubMed] [Google Scholar]
- 11.Vicente C, Schwab C, Broux M, et al. Targeted sequencing identifies association between IL7R/JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100(11): 1373–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 2005;106(1): 274–286. [DOI] [PubMed] [Google Scholar]
- 13.Clappier E, Cuccuini W, Kalota A, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110(4):1251–1261. [DOI] [PubMed] [Google Scholar]
- 14.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19(4):484–497. [DOI] [PubMed] [Google Scholar]
- 15.Van Vlieberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorello P, La Starza R, Varasano E, et al. Combined interphase fluorescence in situ hybridization elucidates T-cell acute lymphoblastic leukemia genetic heterogeneity in adults. Haematologica. 2010;95(1):79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Starza R, Lettieri A, Pierini V, et al. Linking genomic lesions with minimal residual disease improves prognostic stratification in children with T-cell acute lymphoblastic leukaemia. Leuk Res. 2013;37(8):928–935. [DOI] [PubMed] [Google Scholar]
- 18.Atak ZK, Gianfelici V, Hulselmans G, et al. Comprehensive analisis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013;9(12):e1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karube K, Nacagawa M, Tsuzuki S, et al. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK-cell neoplasms by genomic and functional analyses. Blood. 2011;118(12):3195–3204. [DOI] [PubMed] [Google Scholar]
- 20.Varney ME, Niederkorn M, Konno H, et al. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J Exp Med. 2015;212(11):1967–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Testa U, Stellacci E, Pelosi E, et al. Impaired myelopiesis in mice devoid of interferon regulatory factor 1. Leukemia. 2004;18(11): 1864–1871. [DOI] [PubMed] [Google Scholar]
- 22.Liu TX, Becker MW, Jelinek J, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13(1):78–83. [DOI] [PubMed] [Google Scholar]
- 23.Young DJ, Stoddart A, Nakitandwe J, et al. Knockdown of Hnrnpa0, a del(5q) gene, alters myeloid cell fate in murine cells through regulation of AU-rich transcripts. Haematologica. 2014;99(6):1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kühnl A, Valk PJ, Sanders MA, et al. Downregulation of the Wnt inhibitor CXXC5 predicts a better prognosis in acute myeloid leukemia. Blood. 2015;125(19): 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray PJ, Cotton RG. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003;21(6): 557–568. [DOI] [PubMed] [Google Scholar]
- 26.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322(5906):1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuster L, Grausenburger R, Fuka G, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signalling. Blood. 2011;117(9):2658–2667. [DOI] [PubMed] [Google Scholar]
- 28.Weber BN, Chi AW, Chavez A, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Science. 2011;476(7358): 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu S, Zhou X, Steinke FC, et al. The TCF-1 and LEF-1 Transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;13(5):813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiemessen MM, Baert MR, Schonewille T, et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10(11):e1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matlawska-Wasowska K, Kang H, Devidas M, et al. Mixed lineage leukemia rearrangements (MLL-R) are determinants of high risk disease in homeobox A (HOXA)-deregulated T-lineage acute lymphoblastic leukemia: a Children’s Oncology Group Study. Blood (ASH Annual Meeting Abstracts). 2015; 126(23):694. [Google Scholar]
- 32.Bond J, Marchand T, Touzart A, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-ALL: a GRAALL study. Blood (ASH Annual Meeting Abstracts). 2015;126(23):808. [Google Scholar]
- 33.Jenkinson S, Kirkwood AA, Goulden N, Vora A, Linch DC, Gale RE. Impact of PTEN abnormalities on outcome in pediatric patients with T-cell acute lymphoblastic leukemia treated on the MRC UKALL2003 trial. Leukemia. 2016;30(1):39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beldjord K, Chevret S, Asnafi V, et al. Oncogenetics and minimal residual disease are independent outcome predictors in adult patients with acute lymphoblastic leukemia Blood. 2014;123(24):3739–3749. [DOI] [PubMed] [Google Scholar]
- 35.Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol. 2013;31(34):4333–4342. [DOI] [PubMed] [Google Scholar]
- 36.Coustan-Smith E, Mullighan CG, Onciu MM, et al. Early T-cell precursor leukemia: a subtype of very risk acute lymphoblastic leukemia identified in two independent cohorts. Lancet Oncol. 2009;10(2):147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukemia. Nature. 2012;481(7380):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain N, Lamb AE, O’Brien S, et al. Early T-cell precursor acute lymphoblastic leukemia/lymphoma (ETP-ALL/LBL) in adolescents and adults: a high-risk subtype. Blood. 2016;127(15):1863–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conter V, Valsecchi MG, Buldini B, et al. Favourable outcome for children with early T-cell precursor acute lymphoblastic leukemia treated in AIEOP centers on the AIEOP-BFM ALL 2009 contemporary protocol. Lancet Hematol. 2016. January 25 [Epub ahead of print]. [Google Scholar]
- 40.Neumann M, Heesch S, Gökbuget N, et al. Clinical and molecular characterization of early T-cell precursor leukemia: a high-risk subgroup in adult T-ALL with a high frequency of FLT3 mutations. Blood Cancer J. 2012;2(1):e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann M, Heesch S, Schlee C, et al. Whole exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121(23):4749–4752. [DOI] [PubMed] [Google Scholar]
- 42.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maude SL, Dolai S, Delgado-Martin C, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015;125(11):1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chonghaile TN, Roderick JE, Glenfield C, et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 2014;4(9): 1074–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawashima-Goto S, Imamura T, Tomoyasu C, et al. BCL2 Inhibitor (ABT-737): a restorer of prednisolone sensitivity in early T-cell precursor-acute lymphoblastic leukemia with high MEF2C expression? PLoS One. 2015;10(7):e0132926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Perez I, Manguan-Garcia C, Menacho-Marquez M, Murguía JR, Perona R. hCCR4/cNOT6 targets DNA-damage response proteins. Cancer Lett. 2009;273(2): 281–291. [DOI] [PubMed] [Google Scholar]
- 47.De Keersmaeker K, Atak ZK, Li N, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45(2):186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collart M, Panasenko OO. The Ccr4-Not complex. Gene. 2012;492(1):42–53. [DOI] [PubMed] [Google Scholar]
- 49.Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol. 2012;47(4):315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta. 2013;1829(6–7):561–570. [DOI] [PubMed] [Google Scholar]
- 51.Wong SQ, Behren A, Mar VJ, et al. Whole exome sequencing identifies a recurrent RQCD1 P131L mutation in cutaneous melanoma. Oncotarget. 2015;6(2):1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]