Abstract
Background
In ventral hernia surgery, mesh implants are used to reduce recurrence. Infection after mesh implantation can be a problem and rates around 6–10% have been reported. Bacterial colonization of mesh implants in patients without clinical signs of infection has not been thoroughly investigated. Molecular techniques have proven effective in demonstrating bacterial diversity in various environments and are able to identify bacteria on a gene-specific level.
Objective
The purpose of this study was to detect bacterial biofilm in mesh implants, analyze its bacterial diversity, and look for possible resemblance with bacterial biofilm from the periodontal pocket.
Methods
Thirty patients referred to our hospital for recurrence after former ventral hernia mesh repair, were examined for periodontitis in advance of new surgical hernia repair. Oral examination included periapical radiographs, periodontal probing, and subgingival plaque collection. A piece of mesh (1×1 cm) from the abdominal wall was harvested during the new surgical hernia repair and analyzed for bacteria by PCR and 16S rRNA gene sequencing. From patients with positive PCR mesh samples, subgingival plaque samples were analyzed with the same techniques.
Results
A great variety of taxa were detected in 20 (66.7%) mesh samples, including typical oral commensals and periodontopathogens, enterics, and skin bacteria. Mesh and periodontal bacteria were further analyzed for similarity in 16S rRNA gene sequences. In 17 sequences, the level of resemblance between mesh and subgingival bacterial colonization was 98–100% suggesting, but not proving, a transfer of oral bacteria to the mesh.
Conclusion
The results show great bacterial diversity on mesh implants from the anterior abdominal wall including oral commensals and periodontopathogens. Mesh can be reached by bacteria in several ways including hematogenous spread from an oral site. However, other sites such as gut and skin may also serve as sources for the mesh biofilm.
Keywords: ventral hernia, mesh, implants, oral bacteria, 16S rRNA, DNA
Surgery to repair abdominal wall hernia is considered one of the most common procedures in general surgery. Ever since the introduction of polyethylene mesh and the concept of tension-free repair, many different mesh types have made progress to the outcome of hernia surgery. There is substantial documentation of late complications after mesh repair such as mesh shrinkage and hernia recurrence, adhesion formation, foreign body reaction, seroma, and infection (1). Infection after mesh implantation can be a serious problem, and rates around 6–10% have been reported (2). Bacteria can colonize the mesh during surgery or postoperatively due to surgical drains, catheters, and tubes, leading to subsequent biofilm formation on the mesh. Even translocation of bacteria through a sick or healthy intestinal wall could occur, but so far the evidence is anecdotal.
Periodontal diseases are chronic infections resulting in variable degrees of connective tissue breakdown and bone loss around the teeth, and they are considered a heterogenous disease group caused by the complex actions and interactions of the subgingival biofilm microbiota and modified by the host immune system. These infections are polymicrobial due to the strong indications of several bacterial species taking part in the initiation and progression of the disease. It is well established that untreated advanced periodontal disease constitutes a chronic source of bacterial dissemination which can result in hematogenous spread to other parts of the body.
Severe forms of periodontitis affect approximately 10–20% of the world's population (3). More than 600 different bacteria can be detected in the oral cavity (4). Studies have shown that bacterial species considered as commensals in the oral cavity may be associated with systemic diseases, for example, endocarditis (5). The subgingival biofilm is dominated by obligate and facultative anaerobic bacteria. Most related to the progression of periodontal disease are the obligate anaerobic, Gram-negative species Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola (red complex bacteria). Fusobacterium nucleatum and Prevotella spp. are also considered important (6). These species are part of the normal oral microbiota and are not considered as exogenous pathogens. In periodontal healthy individuals, there is a predominance of Streptococcus species (7).
The aim of the present study was to find evidence for bacterial biofilm in mesh implants, analyze its bacterial diversity, and look for possible resemblance with biofilm bacteria from the periodontal area.
Materials and methods
Design and participants
The study was conducted at the Akershus University Hospital, University of Oslo, Lørenskog, Norway, in collaboration with the Department of Oral Biology, Faculty of Dentistry, University of Oslo, Oslo, Norway. All participants signed a written informed consent prior to inclusion, and the protocol was approved by the Regional Ethics Committee and the Norwegian Social Science Data Service. From May 2010 to January 2012, 36 patients with painful recurrence after former ventral hernia mesh repair were enrolled for the study with the intention of periodontal examination before new hernia repair surgery. The patients were referred to our hospital by general practitioners. Five patients refused either dental examination or surgery, and in one patient no mesh was detected during surgery. In the final cohort of 30 patients, recurrences were verified by MRI or CT-scan in 25 cases. Information about former hernia surgery was extracted from medical records available. We used ASA score (American Society of Anesthesiologists physical status score) for evaluation of comorbidities (8) (Supplementary Fig. 1). None of the patients had any dental or periodontal treatment between subgingival plaque and mesh sample collection.
Mesh insertion technique
The original hernia mesh repair was either done by laparoscopy or by open technique. The type of surgical approach and mesh selected were based on the surgeon's preferences and experience.
Cephalothin 2 g was given intravenously prior to mesh repair for large hernias. In laparoscopic ventral hernia mesh repair (LVHR), the access to the abdominal cavity was established with open introduction of a 12 mm trocar. Capnoperitoneum was established with a pressure of 12 mm Hg. Two or three additional abdominal trocars, 5 or 10 mm, were positioned on the surgeon's side or on the contralateral side if appropriate. Adhesions were detached with scissors and occasionally with LigaSure® or ultracision. Fatty tissue on the inner abdominal wall was removed. The hernia sac was not routinely removed. The defect was measured. The mesh was introduced through the 12 mm trocar and placed over the defect with a minimum of 5 cm hernia overlap using tacks or transfacial non-absorbable sutures according to the surgeon's preferences. The mesh did not necessarily cover the entire scar with a 5 cm overlap.
In open ventral hernia mesh repair (OVHR), the incision was made over the hernia thus exposing the hernia content. The hernia sac was removed if possible. The peritoneum or posterior rectus sheet was dissected from the rectus muscle. The posterior sheet was not routinely closed with running absorbable sutures. The mesh was anchored in a retromuscular position with running non-resorbable transfacial sutures and seeking to achieve a 5 cm overlap. The anterior rectus sheet was not routinely closed.
Mesh sample collection
A small piece of incorporated mesh (1×1 cm) was collected, either during LVHR (n=18) or OVHR (n=12) for recurrence. The piece was arbitrary excised with scissors where the mesh was most easily accessible. The samples were immediately placed in an empty sterile glass container, transported on ice, and stored at −80°C. In one patient (ID=15), we could not find the implanted mesh and chose to set up a blindfold sample by taking a small piece of mesh directly from the sterile package and stored it at −80°C.
Periodontal examination and microbial sampling
The periodontal examination was conducted by an experienced dentist (JCÅ). Gingivitis was assessed by bleeding on probing (BOP) (9). Periodontitis was defined as the presence of one or more teeth with at least one site with probing depth ≥4 mm and BOP (10). Any severity grading of periodontal disease was beyond the scope of our interest. Periodontal pockets were measured in four sites for each tooth. Subgingival plaque specimens were collected from each pocket ≥4 mm by insertion of several sterile paper points (pooled samples) to the bottom of the pocket for 10 s. In pockets <4 mm, the same procedure was repeated, but only from a representative site of the first molar. If the first molar was missing, the second premolar was chosen, and then the first premolar. The collected plaque samples for each patient were pooled in a 1.5 mL microcentrifuge tube containing 1 mL sterile phosphate-buffered saline and stored at −80°C. The alveolar bone loss was analyzed by periapical digital radiographs taken by an experienced dentist (JCÅ) and analyzed by an experienced periodontist (ME). The distance between the cementum-enamel junction and limbus alveolaris was recorded. Due to lack of a protocol for standardization of radiographic recordings, differential diagnosis of bone loss was not possible. The aim of this assessment was left with the detection of alveolar bone loss indicative of periodontal disease.
DNA extraction and PCR
DNA extractions of samples from mesh and subgingival plaque were performed using the MasterPure DNA isolation kit from Epicentre (MCD85201, Epicentre Biotechnologies, Madison, WI). 16S rRNA gene fragments from bacterial DNA were amplified with PCR using universal eubacterial primers, forward primer 334f (5′- CCAGACTCCTACGGGAGGCAGC-3′), and reverse primer 939r (5′- CTTGTGCGGGCCCCCGTCAATTC-3′) (11) targeting the V3-V5 hypervariable region. PCR reactions were performed with 32 cycles in 25 µL mixture of Accuprime supermix II (Invitrogen, Carlsbad, CA) in an Applied Biosystem (Foster City, CA) PCR cycler.
Cloning and sequencing
PCR products were ligated to the pCR4-TOPO vector and transformed into Escherichia coli DH5a cells using the TOPO-TA cloning kit according to the manufacturer's instructions (Invitrogen). From each sample, 96 clones were picked. The partial sequencing of the clones was performed with BigDye Terminator v1.1 (Applied Biosystem) and M13 forward sequencing primer on ABI 3730. All sequences were trimmed for elimination of vector sequences and adjusted for quality values by using Sequencher 5.0 (Gene Codes Corporation, Ann Arbor, MI).
Identification of 16S rRNA gene sequences
We performed a BLAST search, comparing the consensus sequences with known sequences against the Ribosomal Database Project (RDP, update 10) (12) and the Human Oral Microbiome Database (HOMD) (www.homd.org/). Alignment of the nucleotide sequences was conducted with Clustalw2 with the default program settings (www.ebi.ac.uk/Tools/msa/clustalw2/). A phylogenetic tree was generated by the neighbor-joining method, using the Clustal W 2.0 program. The Molecular Evolutionary Genetics Analysis (MEGA) software (version 5.2) was used to visualize sequence differences and to generate dendrograms (13).
The nucleotide sequences from mesh and plaque analysis have been submitted to NCIB with GenBank accession numbers (Supplementary Table 1).
Statistical analyses
Statistical analyses were performed using SPSS version 20 (SPSS Inc., Chicago, IL, USA). Categorical variables were compared by χ2-test or Fisher's exact test when applicable. Symmetrically distributed continuous variables were compared using the independent samples t-test. The Shannon–Weaver index of diversity (H′) (12, 14) was used to determine the diversity of bacteria present in the subgingival pockets and mesh samples by the following equation:
where s is the number of species (species richness) and p i is the proportion of species in sample i. H′ was compared for subjects by the Mann–Whitney U-test as was other continuous in case of skewed distribution. Variables associated with mesh bacterial diversity at the P<0.1 level in bivariate analyses, were subjected to multivariate regression analysis.
Spearman's rank correlation test (rS) was used for correlation analyses. Principal component analysis was carried out on mesh bacteria and mesh insertion technique. Data are presented as median or mean with range or standard deviation. P values<0.05 were considered statistically significant.
Results
Patient and clinical characteristics with 16S rRNA results are presented in Tables 1 and 2.
Table 1.
Characteristics at the time of index hernia surgery
| Detection of 16S rRNA gene products in mesh samples | |||||
|---|---|---|---|---|---|
| Characteristics | 16S rRNA not detected | 16S rRNA detected | P | ||
| Age, years, mean (±SD) | 54.7 | (18.4) | 46.6 | (13.6) | 0.18 |
| ASA score (±SD) | 2.0 | (0.47) | 1.85 | (0.59) | 0.49 |
| BMI (kg/m2), mean (±SD) | 35.3 | (7.0) | 32.0 | (7.0) | 0.25 |
| Gender: male | 2 | (20.0) | 8 | (40.0) | 0.27 |
| Recurrent hernia | 0.09 | ||||
| First time | 3 | (10.0) | 7 | (23.3) | |
| Second time | 7 | (23.3) | 7 | (23.3) | |
| Third time | 0 | – | 6 | (20.0) | |
| Type of hernia mesh repair | 0.60 | ||||
| LVHR | 5 | (38.5) | 8 | (61.5) | |
| OVHR | 5 | (29.4) | 12 | (70.6) | |
| Periop. antibiotics | 0.63 | ||||
| LVHR | 2 | (28.6) | 5 | (71.4) | |
| OVHR | 4 | (28.6) | 6 | (42.9) | |
| Information not available | 0 | – | 4 | (28.6) | |
| Preoperative complications | |||||
| None | 10 | (33.3) | 14 | (46.7) | |
| Intestinal resection | 0 | – | 2 | (6.7) | |
| Information not available | 0 | – | 4 | (13.3) | |
| Postop. complications | |||||
| None | 4 | (13.3) | 13 | (43.3) | |
| Wound secretion | 3 | (10.0) | 0 | 0 | |
| Wound hematoma | 3 | (10.0) | 0 | 0 | |
| Subcutaneous abscess | 0 | – | 2 | (6.7) | |
| Pneumonia | 1 | (3.3) | 0 | 0 | |
| Ileus | 0 | – | 1 | (3.3) | |
| Information not available | 0 | – | 3 | (10.0) | |
| Postoperative and late complications | 0.06 | ||||
| Yes | 6 | (20.0) | 4 | (13.3) | |
| No | 4 | (13.3) | 13 | (43.3) | |
| Information not available | 3 | (1.0) | |||
Table 2.
Characteristics at the time of periodontal examination and mesh sample collection
| Detection of 16S rRNA gene products in mesh samples | |||||
|---|---|---|---|---|---|
| Characteristics | 16S rRNA not detected | 16S rRNA detected | P | ||
| Age (a), years, mean (±SD) | 57.0 | (18.0) | 51.1 | (11.9) | 0.29 |
| Age (b), years, mean (±SD) | 57.2 | (18.0) | 51.3 | (11.8) | 0.29 |
| BOP, mean (±SD) | 10.2 | (8.4) | 8.4 | (5.7) | 0.48 |
| Pocket depth > 4 mm | 4 | (13.3) | 6 | (20.0) | 0.58 |
| Number of sites | 16 | (34.8) | 30 | (65.2) | 0.30 |
| Mesh implant time, years (±SD) | 2.47 | (1.57) | 4.66 | (3.28) | 0.06 |
| Number of taxa detected, mean (±SD) | – | 7 | (9) | ||
| Type and number of mesh samples extracted | |||||
| PET+collagen/PEG/glycerol (Parietex Comp) | 5 | (16.7) | 8 | (26.7) | |
| Number of taxa detected, mean (±SD) | – | 16 | (11) | ||
| PP Monofil+Ti coating (TiMesh) | 0 | 1 | (3.3) | ||
| PP Monofil+PTFE (Bard Comp) | 0 | 1 | (3.3) | ||
| PP Monofil (Prolene) | 2 | (6.7) | 7 | (23.3) | |
| Number of taxa detected, mean (±SD) | – | 13 | (17) | ||
| Biological (Permacol) | 1 | (3.3) | 2 | (6.7) | |
PP, Polypropylene; PET, Polyester (polyethylene terephthalate); PEG, Polyethylene glycol; PTFE, Polytetrafluoroethylene; Mesh classification after Coda et al. (15).
Periodontal examination.
Mesh sample collection.
Data from periodontal examination
Dental examinations were carried out relatively close to surgery (mean 0.2 years, range 0.6–1.8 years, SD±0.44 year).
Periodontitis was detected in 10 (33.3%) patients. Six (20%) patients with periodontitis were subjected to bacterial analysis of subgingival plaque samples. BOP was seen in all patients except for one. The mean number of sites with BOP was 11.61, SD±17.5. BOP was not correlated with any patient characteristic. Periodontal disease was only correlated with comorbidity (rS=0.426/P=0.019). Periapical radiographs revealed 17 (56.7%) subjects with and 10 (33.3%) subjects without alveolar bone loss compatible with chronic periodontitis. Three x-rays (10.0%) were abandoned due to technical problems.
Surgical data
The time from periodontal examination to mesh sample collection was 2.5 months (SD±5.3). Ten patients (33.3%) presented with their first hernia recurrence, 14 patients (46.7%) with their second, and six patients (20.0%) with their third recurrence. The last recorded ventral hernia mesh repair (index hernia mesh repair) was done by laparoscopy in 13 patients (43.3%) and by open surgery in 17 patients (56.7%). There were two intestinal resections during the index operation, and a biological mesh was therefore selected. One patient (ID 9) also needed reoperation due to postoperative ileus without detection of intestinal injury. The mean age of the eligible group of 30 patients at the time of mesh sample collection, was 53.3 years (range 25.4–78.5 years, SD±14.1 years). The mean time from index operation to mesh sample collection was 3.9 years (range 0.8–14.0 years, SD±3.0 years). Of those mesh samples analyzed, 13 (43.3%) comprised polyester meshes (Parietex Composite), 11 (36.7%) were polypropylene (PP) containing meshes, and three (9.1%) were biological meshes (Permacol) (9.1%). Three mesh samples (9.1%) were of unknown origin (Table 2) (15).
Data from analysis of 16S rRNA gene products
Positive 16S rRNA gene PCR products were obtained from 20 meshes (66.7%). In 70.6% of the meshes implanted by OVHR, 16S rRNA gene products were revealed as compared to 61.5% after LVHR (P=0.60). In a sterile mesh sample, there were no detectable 16S rRNA gene products. There was DNA from detectable bacterial taxa in eight (61.5%) of the polyester meshes and nine (81.8%) of the PP meshes (P=0.28). There were no significant differences in bacterial diversity between the main mesh types. In all six patients with two recurrences, there was bacterial DNA in mesh samples (P=0.07). A sequence similarity threshold of 97% for identification of bacterial sequences in mesh revealed 90 different taxa detected from a total of 357 different sequences (Supplementary Table 2). Of these, 261 were named, 45 were unnamed cultivable taxa, and 51 were unnamed so far uncultivable taxa, that is, phylotypes. The mean number of taxa found in mesh samples was 18.6± SD=12.7 (range 5–56).
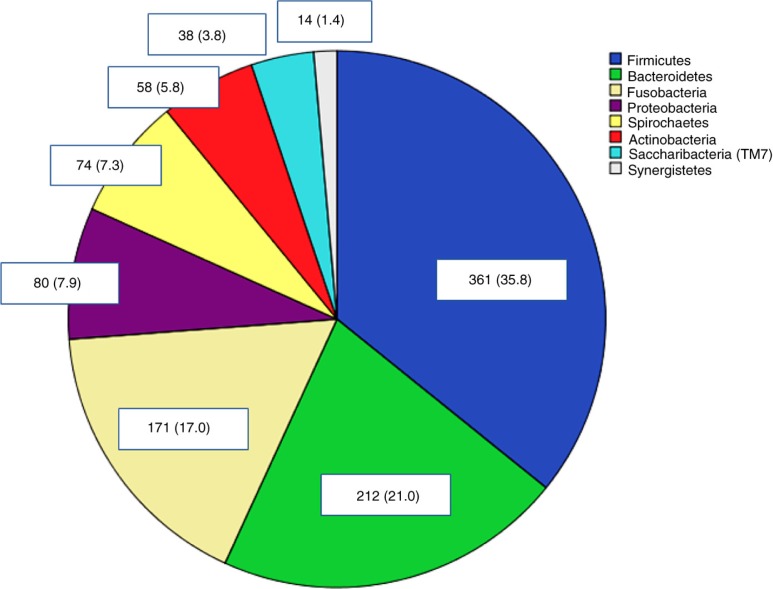
The plaque microbiota was dominated by the phyla Firmicutes (35.8%), Bacteroidetes (21.0%), and Fusobacteria (17%) (Fig. 1).
Fig. 1.
Distribution of phyla in plaque samples. Percentages in brackets.
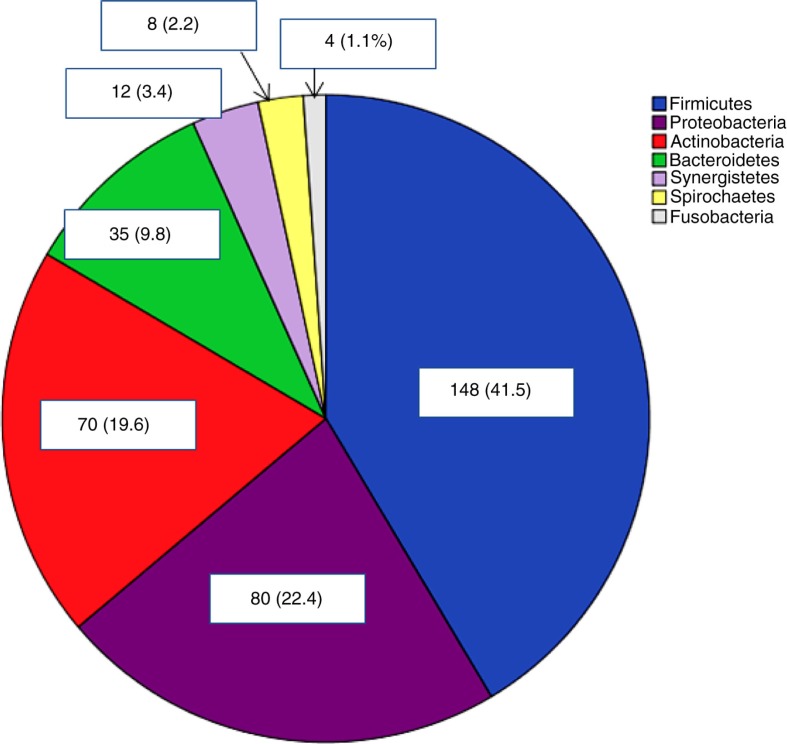
The mesh microbiota was also dominated by Firmicutes (41.5%), but contained significantly higher levels of Proteobacteria (22.4%) and Actinobacteria (19.6%) (Fig. 2).
Fig. 2.
Distribution of phyla in mesh samples. Percentages in brackets.
The plaque microbiota comprised 197 different taxa from a total of 1,008 different sequences. Streptococcaceae, Fusobacteriaceae, Veillonellaceae, and Prevotellaceae accounted for 49.3% of all families. The red complex bacterial species associated with severe periodontitis (16) was found only in four patients. Aggregatibacter actinomycetemcomitans was found with two different strains in another patient. None of these patients were diagnosed with periodontitis.
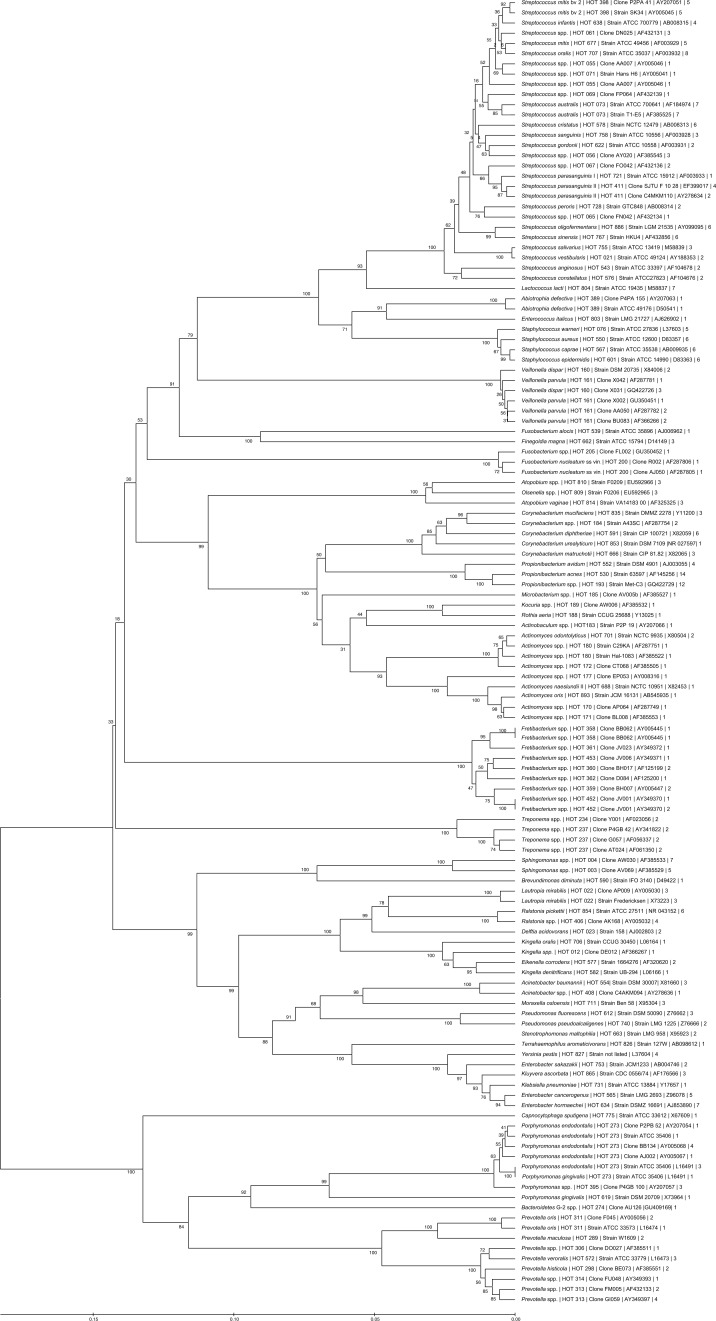
The phylogeny of species and subspecies found in mesh samples is presented in Fig. 3. Examples of phylogenetic trees generated are presented in Figs. 4 and 5. The most abundant species in all mesh samples were Propionibacterium acnes, Streptococcus australis, and Streptococcus spp., which contributed 11.7% of all taxa. Fretibacterium spp., Propionibacterium spp., and Sphingomonas spp. accounted for 10.2% of all taxa and were found in 14 (46.7%) subjects. Typical oral bacterial taxa were more abundant (55.7%) than typical skin taxa (19.9%) and enteric taxa (11.5%).
Fig. 3.
Neighbor-joining tree of sequence alignments of 16S rRNA gene hypervariable segments of most bacterial species detected in mesh samples. Name of taxon is followed by human oral taxon number, clone/strain number, accession number GenBank, and number of strains detected.
Fig. 4.
Dendrogram from patient (ID 6) with corresponding hits. ^99.6% overlap between S. oralis in mesh (M) and plaque (P). ^^99.1% overlap between C. matruchoti in mesh (M) and plaque (P).
Fig. 5.
Dendrogram from patient (ID 22) with corresponding hits. ^100% overlap between P. acnes in mesh (M) and plaque (P).
Putative periodontopathogens found in mesh samples were F. nucleatum, P. gingivalis, Prevotella spp., and Treponema spp., which comprised 9.8% of the taxa.
Among typical skin bacteria detected were Staphylococcus spp. including S. aureus, S. epidermidis, S. caprae, and S. warneri which contributed to 6.4% of all taxa.
Enterobacter spp., Enterococcus spp., E. coli, and Klyvera ascorbate comprised 7.0% of all taxa.
The time from index hernia operation to the study operation reflecting mesh implantation time, was longer in cases with detectable 16S rRNA gene products in mesh samples (P=0.056) (Table 2) and was also longer after OVHR (P=0.054) (Table 3). Mesh implantation time was only correlated with skin bacterial habitants after OVHR (rS 0.56/P=0.018).
Table 3.
Mesh bacteria characteristics relative to perioperative antibiotics and mesh insertion technique
| Mesh insertion technique | |||||
|---|---|---|---|---|---|
| Mesh biocharacteristics | Open | Laparoscopic | P | ||
| Mesh implantation time, years, mean (±SD) | 4.8 | (3.4) | 2.7 | (1.9) | 0.05 |
| Diversity index, mean (±SD) | 0.23 | (0.24) | 0.13 | (0.16) | 0.20 |
| Without perioperative antibiotics | 0.37 | (0.27) | 0.10 | (0.12) | 0.05 |
| Oral bacterial sequences, mean (±SD) | 9.24 | (11.50) | 3.46 | (8.97) | 0.15 |
| Without perioperative antibiotics | 15.43 | (14.19) | 0.67 | (1.63) | 0.03 |
| Enteric bacterial sequences, mean (±SD) | 1.24 | (1.92) | 2.00 | (2.58) | 0.36 |
| Without perioperative antibiotics | 2.29 | (2.63) | 1.50 | (2.81) | 0.61 |
| Skin bacterial sequences, mean (±SD) | 2.82 | (2.86) | 1.77 | (3.19) | 0.35 |
| Without perioperative antibiotics | 3.71 | (2.56) | 3.17 | (4.30) | 0.78 |
There was a high degree of correlation between plaque and mesh bacterial diversity, both after LVHR (rS 0.95/P<0.0001) and after OVHR (rS 0.69/P=0.002). There was also strong correlation between the number of oral sequences and mesh bacterial diversity after OVHR (rS 0.92/P<0.0001) (Table 4).
Table 4.
Mesh insertion technique: Spearman's rank correlation (rS) between mesh implantation time, diversity index in mesh and diversity index in plaque
| Mesh implantation time | Diversity index mesh | Diversity index plaque | Oral bacterial seqa | Enteric bacterial seqa | Skin bacterial seqa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Open mesh insertion | |||||||||||
| Mesh implantation time, rS (P value) | 1.000 | 0.447 | (0.07) | 0.349 | (0.17) | 0.256 | (0.32) | 0.455 | (0.07) | 0.564 | (0.02) |
| Diversity index mesh | 1.000 | 0.685 | (<0.01) | 0.916 | (<0.0001) | 0.466 | (0.06) | 0.740 | (<0.01) | ||
| Diversity index plaque | 0.685 | (<0.01) | 1.000 | 0.574 | (0.02) | 0.468 | (0.06) | 0.565 | (0.02) | ||
| Laparoscopic mesh insertion | |||||||||||
| Mesh implantation time, rS (P value) | 1.000 | 0.209 | (0.49) | 0.288 | (0.34) | 0.078 | (0.80) | 0.020 | (0.94) | 0.429 | (0.14) |
| Diversity index mesh | 1.000 | 0.953 | (<0.0001) | 0.585 | (0.04) | 0.701 | (<0.01) | 0.525 | (0.07) | ||
| Diversity index plaque | 0.953 | (<0.0001) | 1.000 | 0.378 | (0.20) | 0.793 | (<0.01) | 0.619 | (0.02) | ||
Bacterial sequences in mesh further arranged according to typical habitat.
Number of oral, enteric and skin bacterial sequences in mesh samples.
Mesh bacterial diversity was not associated with periodontitis (P=0.57) or gingivitis (P=0.48) and neither was periodontal disease associated with the detection of 16S RNA gene products in mesh samples (P=0.60).
Intraoperative complications were registered during index hernia operation. In one patient that needed intestinal resection (ID 9), a total of 17 taxa were found. They were dominated by oral Streptococcus and Prevotella species (76.5%). The other patient (ID 34) had five taxa in mesh samples only and four taxa (80.0%) were Enterobacter species or E. coli. Permacol mesh was used in both patients. Both patients were also diagnosed with periodontal disease.
Ten (33.3%) patients were registered with postoperative or late complications, while three patients (10.0%) could not be accounted for. Absence of complications was closely associated with detection of bacterial DNA in mesh samples (P=0.058) (Table 1). Mesh bacterial diversity of those cases registered with any postoperative or late complication was also significantly lower than in those without (P=0.011). Skin bacteria such as Pseudomonas spp. and Corynebacterium spp. were however more frequently detected as compared to enteric and oral bacteria in patients with wound complications after index hernia mesh repair. Patient ID 9 and ID 30 had postoperative and late laparotomy due to ileus. Seventeen taxa (ID 9) were detected in the mesh sample, mostly oral bacteria (82.4%) and without detection of typical enteric bacteria. The other patient (ID 30) had only six taxa in the mesh sample, with enteric, skin, and environmental species. Bacterial diversity index was not different between PP and Parietex Composite mesh (P=0.56) or between these meshes and Permacol (P=0.44). The inflammation marker CRP exceeded normal levels in eight (26.7%) patients (>8 mg/L) while leukocyte counts were elevated in four patients (>10.0 10*9/L). There was no association between elevated CRP (P=0.101) or elevated leukocyte count (P=0.951) and mesh bacterial diversity.
Cephalothin 2 g intravenously was given to 12 (40.0%) patients prior to index hernia mesh repair. Six (30.0%) of these patients in addition to one (3.3%) patient, received Cefuroxime 1.5 g×3 iv and Metronidazole 1.5 g postoperatively for 1–3 days. Four (13.3%) patients after OVHR could not be accounted for (Table 1). Perioperative antibiotics in OVHR, were associated with reduction in typical skin bacteria (P=0.031). A reduction in enterics (P=0.058) and bacterial diversity (P=0.068) was also seen only after OVHR (Table 5). Only subgingival plaque bacterial diversity was associated with mesh bacterial diversity in multivariate analysis (P<0.01) (Table 6).
Table 5.
Perioperative antibiotics during index hernia mesh repair
| Perioperative antibiotics | |||||
|---|---|---|---|---|---|
| Mesh biocharacteristics | Yes | No | P | ||
| Diversity index, mean (±SD) | |||||
| Open | 0.11 | (0.17) | 0.37 | (0.27) | 0.07 |
| Laparoscopic | 0.18 | (0.21) | 0.10 | (0.11) | 0.40 |
| Oral bacterial sequences, mean (±SD) | |||||
| Open | 5.5 | (8.7) | 15.4 | (14.2) | 0.17 |
| Laparoscopic | 5.9 | (12.0) | 0.7 | (1.6) | 0.32 |
| Enteric bacterial sequences, mean (±SD) | |||||
| Open | 0 | (0) | 2.3 | (2.6) | 0.06 |
| Laparoscopic | 2.4 | (2.5) | 1.5 | (2.8) | 0.54 |
| Skin bacterial sequences, mean (±SD) | |||||
| Open | 0.8 | (3.1) | 3.7 | (2.6) | 0.03 |
| Laparoscopic | 0.6 | (1.1) | 3.2 | (4.3) | 0.15 |
Table 6.
Predictor variables for mesh bacterial diversity. Univariate analysis
| Variable | B (95%CI) | P |
|---|---|---|
| Subgingival plaque bacterial diversity | 0.64 (0.34; 0.94) | <0.001 |
| Mesh insertion technique | 0.099 (−0.06; 0.26) | 0.20 |
| ASA score | −1.22 (−0.26; 0.02) | 0.08 |
| Age | −0.06 (−0.12; 0.002) | 0.05 |
| Overall complications | −0.04 (−0.13; 0.04) | 0.30 |
| Pocket depth > 4 mm, sites | 0.003 (−0.03; 0.03) | 0.85 |
| Gingival index score > 2 | −0.003 (−0.02; 0.009) | 0.57 |
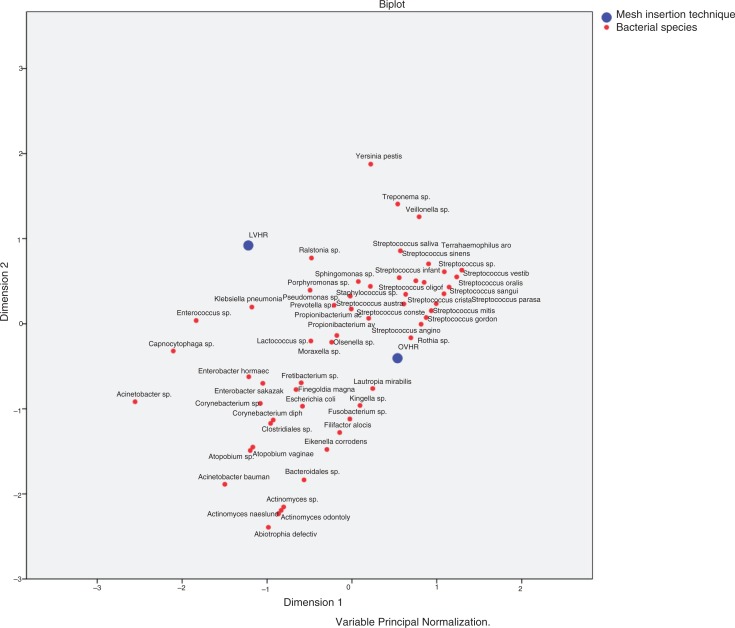
Species diversity according to mesh insertion technique was analyzed by principal component analysis and showed clustering on both components for OVHR rather than for LVHR (Fig. 6). Interestingly, bacteria known to be part of the commensal oral microbiota or opportunistic oral pathogens were found in 13 (43.3%) mesh samples and comprised a total of 58 (16.2%) species within 28 (7.8%) different species identical in HOT numbers with the species from plaque samples (Table 7). There was high correlation between mesh diversity index and HOT number resemblance (rS=0.794/P<0.001).
Fig. 6.
Principal Component Analysis of mesh bacteria in relation to mesh insertion technique.
Table 7.
Corresponding bacterial taxa mesh and plaque
| ID | Bacterial species | Strain number | Resemblancea |
|---|---|---|---|
| 6 | Corynebacterium matruchotii | ∣ HOT_666 ∣ Strain_CIP 81.82 ∣ X82065 ∣ | 99.1/nt=540 |
| 6 | Streptococcus oralis | ∣ HOT_707 ∣ Strain_ATCC 35037 ∣ AF003932 ∣ | 99.6/nt=519 |
| 7 | Fretibacterium spp. | ∣ HOT_452 ∣ Clone_JV001 ∣ AY349370 ∣ | 91.1/nt=697 |
| 7 | Fretibacterium spp. | ∣ HOT_453 ∣ Clone_JV006 ∣ AY349371 ∣ | |
| 9 | Prevotella spp. | ∣ HOT_313 ∣ Clone_FM005 ∣ AF432133 ∣ | 98.0/nt=528 |
| 9 | Streptococcus australis | ∣ HOT_073 ∣ Strain_ATCC 700641 ∣ AF184974 ∣ | NA |
| 9 | Streptococcus australis | ∣ HOT_073 ∣ Strain_T1-E5 ∣ AF385525 ∣ | NA |
| 9 | Streptococcus gordonii | ∣ HOT_622 ∣ Strain_ATCC 10558 ∣ AF003931 ∣ | NA |
| 9 | Streptococcus sinensis | ∣ HOT_767 ∣ Strain_HKU4 ∣ AF432856 ∣ | NA |
| 11 | Treponema spp. | ∣ HOT_234 ∣ Clone__Y001 ∣ AF023056 ∣ | NA |
| 11 | Treponema spp. | ∣ HOT_237 ∣ Clone__G057 ∣ AF056337 ∣ | NA |
| 11 | Treponema spp. | ∣ HOT_237 ∣ Clone_AT024 ∣ AF061350 ∣ | NA |
| 11 | Treponema spp. | ∣ HOT_237 ∣ Clone_P4GB_42 ∣ AY341822 ∣ | NA |
| 13 | Streptococcus oralis b | ∣ HOT_707 ∣ Strain_ATCC 35037 ∣ AF003932 ∣ | 99.5/nt=417 |
| 13 | Streptococcus mitis b | ∣ HOT_677 ∣ Strain_ATCC 49456 ∣ AF003929 | |
| 13 | Streptococcus mitis bv 2 | ∣ HOT_398 ∣ Clone_P2PA_41 ∣ AY207051 ∣ | NA |
| 13 | Streptococcus mitis bv 2 | ∣ HOT_398 ∣ Strain_SK34 ∣ AY005045 ∣ | NA |
| 13 | Streptococcus sinensis | ∣ HOT_767 ∣ Strain_HKU4 ∣ AF432856 ∣ | NA |
| 13 | Veilonella dispar | ∣ HOT_160 ∣ Clone__X031 ∣ GQ422726 ∣ | 98.2/nt=396 |
| 14 | Corynebacterium matruchotii | ∣ HOT_666 ∣ Strain_CIP 81.82 ∣ X82065 ∣ | NA |
| 19 | Porphyromonas endodontalis | ∣ HOT_273 ∣ Clone_BB134 ∣ AY005068 ∣ | |
| 19 | Porphyromonas endodontalis | ∣ HOT_273 ∣ Strain_ATCC 35406 ∣ L16491 ∣ | 99.2/nt=612 |
| 19 | Porphyromonas spp. | ∣ HOT_395 ∣ Clone_P4GB_100 ∣ AY207057 ∣ | 90.6/nt=640 |
| 19 | Prevotella maculosa | ∣ HOT_289 ∣ Strain_W1609 ∣ EF534315 ∣ | 98.7/nt=651 |
| 19 | Prevotella oris | ∣ HOT_311 ∣ Clone__F045 ∣ AY005056 ∣ | 98.7/nt=651 |
| 19 | Prevotella oris | ∣ HOT_311 ∣ Strain_ATCC 33573 ∣ L16474 ∣ | |
| 19 | Streptococcus australis | ∣ HOT_073 ∣ Strain_ATCC 700641 ∣ AF184974 ∣ | NA |
| 19 | Streptococcus cristatus | ∣ HOT_578 ∣ Strain_NCTC 12479 ∣ AB008313 ∣ | 98.7/nt=605 |
| 19 | Streptococcus mitis | ∣ HOT_677 ∣ Strain_ATCC 49456 ∣ AF003929 ∣ | NA |
| 19 | Streptococcus mitis bv 2 | ∣ HOT_398 ∣ Clone_P2PA_41 ∣ AY207051 ∣ | NA |
| 19 | Streptococcus mitis bv 2 | ∣ HOT_398 ∣ Strain_SK34 ∣ AY005045 ∣ | NA |
| 19 | Streptococcus oralis | ∣ HOT_707 ∣ Strain_ATCC 35037 ∣ AF003932 ∣ | 99.8/nt=631 |
| 19 | Streptococcus parasanguinis II | ∣ HOT_411 ∣ Clone_SJTU_F_10_28 ∣ EF399017 ∣ | NA |
| 19 | Streptococcus peroris | ∣ HOT_728 ∣ Strain_GTC848 ∣ AB008314 ∣ | NA |
| 19 | Streptococcus salivarius | ∣ HOT_755 ∣ Strain_ATCC 13419 ∣ M58839 ∣ | NA |
| 19 | Streptococcus sanguinis | ∣ HOT_758 ∣ Strain_ATCC 10556 ∣ AF003928 ∣ | NA |
| 19 | Streptococcus sinensis | ∣ HOT_767 ∣ Strain_HKU4 ∣ AF432856 ∣ | NA |
| 19 | Streptococcus spp. | ∣ HOT_056 ∣ Clone_AY020 ∣ AF385545 ∣ | NA |
| 19 | Streptococcus vestibularis | ∣ HOT_021 ∣ Strain_ATCC 49124 ∣ AY188353 ∣ | 99.7/nt=651 |
| 20 | Prevotella spp. | ∣ HOT_313 ∣ Clone_FM005 ∣ AF432133 ∣ | NA |
| 20 | Prevotella spp. | ∣ HOT_313 ∣ Clone_GI059 ∣ AY349397 ∣ | NA |
| 20 | Streptococcus sinensis | ∣ HOT_767 ∣ Strain_HKU4 ∣ AF432856 ∣ | NA |
| 21 | Streptococcus australis | ∣ HOT_073 ∣ Strain_ATCC 700641 ∣ AF184974 ∣ | NA |
| 21 | Streptococcus australis | ∣ HOT_073 ∣ Strain_T1-E5 ∣ AF385525 ∣ | NA |
| 21 | Streptococcus parasanguinis II | ∣ HOT_411 ∣ Clone_SJTU_F_10_28 ∣ EF399017 ∣ | NA |
| 22 | Propionibacterium acnes | ∣ HOT_530 ∣ Strain_63597 ∣ AF145256 ∣ | 100/nt=661 |
| 22 | Propionibacterium spp. | ∣ HOT_193 ∣ Strain_Met-C3 ∣ GQ422729 ∣ | NA |
| 23 | Propionibacterium acnes | ∣ HOT_530 ∣ Strain_63597 ∣ AF145256 ∣ | 99.7/nt=639 |
| 23 | Staphylococcus aureus | ∣ HOT_550 ∣ Strain_ATCC 12600 ∣ D83357 ∣ | NA |
| 23 | Staphylococcus caprae | ∣ HOT_567 ∣ Strain_ATCC 35538 ∣ AB009935 ∣ | 99.7/nt=675 |
| 23 | Staphylococcus epidermidis | ∣ HOT_601 ∣ Strain_ATCC 14990 ∣ D83363 ∣ | NA |
| 24 | Bacteroidales [G-2] spp. | ∣ HOT_274 ∣ Clone_AU126 ∣ AY005072 ∣ Unnamed | 99.4/nt=672 |
| 24 | Fusobacterium nucleatum ss vin. | ∣ HOT_200 ∣ Clone__R002 ∣ AF287806 ∣ | NA |
| 24 | Fusobacterium nucleatum ss vin. | ∣ HOT_200 ∣ Strain_ATCC 49256 ∣ NZ_AABF02000026 ∣ | 95.1/nt=654 |
| 24 | Streptococcus gordonii | ∣ HOT_622 ∣ Strain_ATCC 10558 ∣ AF003931 ∣ | 98.2/nt=670 |
| 24 | Streptococcus sanguinis | ∣ HOT_758 ∣ Strain_ATCC 10556 ∣ AF003928 ∣ | NA |
| 31 | Streptococcus oralis | ∣ HOT_707 ∣ Strain_ATCC 35037 ∣ AF003932 ∣ | 99.7/nt=696 |
| 31 | Streptococcus sinensis | ∣ HOT_767 ∣ Strain_HKU4 ∣ AF432856 ∣ | NA |
Name of taxon is followed by human oral taxon number, clone/strain number, and listed accession number in GenBank.
Resemblance after pairwise alignment. NA (not applicable).
Pairwise alignment between S. mitis in plaque and S. oralis in mesh.
In eight patients (26.7%), there was a high degree of resemblance (≥99.5%) between certain bacteria in mesh and subgingival plaque samples (Table 7).
Discussion
The prevalence of infection after hernia mesh repair is difficult to estimate, due to the lack of standardized criteria defining infection, the lack and the variability in follow-up examinations, and the effort made to really intervene in those cases having postsurgical symptoms (17). Mesh infection can be subtle with chronic, persistent, or recurrent symptoms and also with skin rubor, abscess formation, or abscess secretion. Bacteria in biofilm can also be dormant giving no sign of infection. Infection is related to the type of mesh, surgical approach, medical conditions, and the strategy to prevent infection (14). Our knowledge of mesh microbiology is mainly from extracted mesh samples due to infection utilizing cultivation methods or microscopy (18). DNA sequencing enabled a more detailed study of bacteria present in biofilm. The methods of DNA sequencing can capture and classify extremely small amounts of bacteria, cultivable as well as non-yet cultivable (19). To our best knowledge, this is the first publication utilizing DNA sequencing to characterize bacterial diversity in mesh implants.
Several bacterial species have been reported from mesh infections such as S. aureus, S. epidermidis, Streptococcus pyogenes, beta-hemolytic streptococci, Enterococcus spp., E. coli, peptostreptococci, Mycobacterium spp., and Acinetobacter baumanii, among others (20).
Mesh characteristics (15, 16), including hydrophobicity, electrostatic charge, number of filaments in yarn, and chemical composition, have influenced the infection rate (17). There are several reports on PP mesh infection as the most common reason for mesh explantation (21). Other reports on PP mesh (19) have demonstrated reduced growth of MRSA compared to multifilament, composite anti-adhesive barrier meshes with hydrophilic polyester (Parietex Composite). Due to increased pore size, this mesh could therefore be relatively resistant to infection (22). Engelsman et al. (20) suggested that both types of meshes have clinical comparable rates of infection. In our series, there was no significant difference in bacterial diversity between the main mesh types. In two of three samples of Permacol, a cross-linked biological mesh, we found 16S rRNA gene products. Several typical oral species were detected such as Prevotella oris and several streptococci together with Enterobacter species among others. The concept that a biological mesh, cross-linked or non-cross-linked, will be resistant to infection has scarce evidence in the literature. Mesh growth of bacteria on biological mesh has been shown both in vivo and in vitro (23). Though 66.7% of all mesh samples harbored bacterial species, this could be an underestimation due to topographic and methodological reasons. One could also argue that a small piece of mesh only reflects a glimpse of the entire biofilm covering all or some parts of the mesh.
Mavros et al. (24) showed acute operation, ASA, length of operation, smoking, and age at the operation to be associated with mesh infection. In our study, none of these variables could explain bacterial diversity. Postoperative and late complications were inversely associated with detection of mesh PCR and mesh bacterial diversity. The reason may be due to perioperative antibiotics. Both mesh implantation time and the number of recurrent repairs were closely associated with mesh PCR detection. The striking association between plaque and mesh bacterial diversity could be coincidental or a reflection of a direct hematogenous route. In univariate analysis, neither periodontal disease nor BOP was associated with mesh bacterial diversity or detection of oral bacteria in mesh. Association statistics covering large numbers of diverse bacteria, often fails in exploring causal relationships with symptoms or disease. Literally, all patients were diagnosed with BOP. The impact of periodontal disease on association statistics was obviously negligible.
Mesh infection is also a risk factor for hernia recurrence (25). Bacterial biofilm without signs of infection could a priori also promote recurrence. For instance, loosening of hip prostheses has been related to bacteria in the synovial fluid without any biochemical or clinical signs of infection (26).
Open mesh repair gives significantly higher rates of surgical site infections than laparoscopic mesh repair (27). In our series, mesh bacterial diversity was slightly higher after open mesh repair (P=0.20). Perioperative antibiotics were associated with a reduction in bacterial diversity, but only after OVHR. This is in accordance with other reports, who have documented lower frequency of surgical site infection after OVHR with preoperative antibiotics (24). A significant reduction in typical skin bacteria, and to some extent enterics, was also seen after administration of perioperative antibiotics. LVHR did not seem to benefit from perioperative antibiotics.
Bacteremia is common after brushing of periodontitis-affected teeth and invasive dental procedures and has led to routine administration of prophylactic antibiotics to those who had earlier endocarditis and prosthetic valves procedures. Bahrani-Mougeot et al. (28) found 98 different species in blood from 290 subjects after tooth extraction and 43 different species after tooth brushing by using 16S rRNA gene sequencing techniques. The most common species detected were Streptococcus spp., Parvimonas micra (Peptostreptococcus micros), Veillonella dispar, or V. parvula. Antibiotic prophylaxis with amoxicillin before single tooth extraction decreased the overall incidence of bacteremia by 61%.
In our series, there was an overall 43.4% incidence reduction of all mesh harboring taxa after perioperative antibiotics. After OVHR and perioperative antibiotics, there was a 65.5% reduction of Streptococcus species and a 100% reduction of Veillonella dispar or V. parvula. Enterobacteriaceae and Staphylococcaceae were also absent after perioperative antibiotics. During open mesh insertion, a relatively large open wound is created where oral bacteria in the blood could escape and attack the mesh construct. Skin bacteria likely contaminate the mesh during intraoperative handling, more during OVHR than LVHR (29). Despite small numbers, antibiotic prophylaxis was shown to eradicate skin and enteric bacteria only after OVHR in this study. Bacteriemia following toothbrushing or periodontal disease could in fact nourish the mesh biofilm by time and explain the abundance of oral bacteria both after OVHR and LVHR.
With multiplex PCR and lactulose breath test, Jun et al. (30) found that nearly 29% of individuals without any bowel or hepatic disease showed evidence of bacterial translocation.
Bacteria are phagocytized at epithelial linings in the distal gut and reach the blood. The macrophages are activated and reach the blood vessel interface where they transform into cholesterol-laden foam cells which in turn contribute to arterosclerotic plaque production (31). A similar mechanism could explain for the construction of mesh biofilm.
Multiple opportunistic pathogens proliferate in subgingival plaque, release proteolytic enzymes that break down host tissues resulting in periodontal inflammation, loss of periodontal attachment, periodontal pocket formation, and alveolar bone destruction (6). Typical proteolytic periodontopathogens such as F. nucleatum, P. gingivalis, Prevotella, and Treponema spp. were found in the mesh samples. These species have not previously been reported from mesh biofilm. Our detection of microbial DNA in mesh samples could, however, originate from dead bacteria as well as engulfed material after phagocytosis (32).
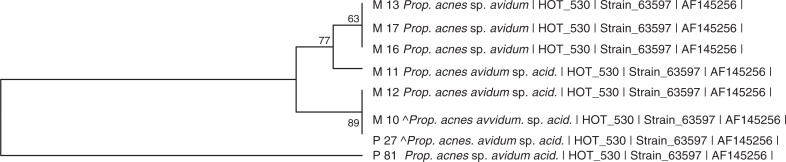
The relatively high frequency of P. acnes found in mesh samples may be due to contamination from skin during surgery. There was, however, no association between laparoscopic and open hernia mesh insertion and detection of P. acnes (P=0.27). Propionibacterium acnes and other Propionibacterium spp. are also commonly detected in carious dentin and root canal infections (33). Two patients (ID 22 and ID 23), both after laparoscopic mesh insertion, had the same strain in mesh and plaque for P. acnes with 100 and 99.7% sequence overlap, respectively. They were not diagnosed with periodontal disease, and no demographic or surgical characteristics could link these subjects together.
Staphylococci can be part of the normal commensal flora. Studies in adults have found oral carriage of S. aureus in 24–36% and different Staphylococcus species in 94–100% (34). One patient (ID 23) had three different strain equivalents in mesh and plaque samples of S. aureus, S. epidermidis, and S. caprae showing 99.7% identity. Whether these bacteria reach the mesh by ingestion or directly from an oral or periodontal site are questions that need further investigation.
Some oral bacteria, especially F. nucleatum, produce FadA adhesin that binds to vascular endothelial cadherin that is essential for the sealant of the endothelial cell junction. In this way, the endothelial cell junctions start to leak and make the blood vessels more permeable. When this happens, neighboring bacteria also leak through the permeabilized vessels (35). Fusobacterium nucleatum can survive, spread hematogenously, and replicate at distant sites from the oral cavity (36).
Our results demonstrated a great diversity of bacteria in the mesh biofilm. Using universal primers, we were able to identify most bacteria at the species level in both plaque and mesh samples. Our eubacterial primer doesn't contain the V2 region necessary for detection of some streptococci. Due to lack of both blood and stool samples for 16S rRNA analysis, we cannot estimate the magnitude of periodontal or intestinal ancestry. The small fraction of the 16S rRNA gene sequences subjected to analysis in our study only suggest a role of periodontitis as a pathogenic factor explaining mesh biofilm constituents.
However, the results of this study clearly show that the oral cavity is an important source for the development of hernia mesh biofilm.
Conclusion
The results show great bacterial diversity of mesh implants from the anterior abdominal wall including typical oral commensals and periodontopathogens, enterics, and skin bacteria. Mesh can be reached by bacteria in several ways including hematogenous spread from an oral site. However, other sites such as gut and skin may also be the sources of dissemination.
Supplementary Material
Acknowledgements
OL wants to acknowledge funding through Akershus University Hospital, Faculty of Medicine, University of Oslo, Norway. IO wants to thank the European Commission (FP7-HEALTH-306029 ‘TRIGGER’) for funding. Jens Christian Årving is thanked for his contribution in dental examination of the cohort.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Sanchez VM, Abi-Haidar YE, Itani KM. Mesh infection in ventral incisional hernia repair: incidence, contributing factors, and treatment. Surg Infect. 2011;12:205–10. doi: 10.1089/sur.2011.033. [DOI] [PubMed] [Google Scholar]
- 2.Hawn MT, Snyder CW, Graham LA, Gray SH, Finan KR, Vick CC. Long-term follow-up of technical outcomes for incisional hernia repair. J Am Coll Surg. 2010;210:648–55. doi: 10.1016/j.jamcollsurg.2009.12.038. [DOI] [PubMed] [Google Scholar]
- 3.Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–53. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feder HM, Jr, Roberts JC, Salazar J, Leopold HB, Toro-Salazar O. HACEK endocarditis in infants and children: two cases and a literature review. Pediatr Infect Dis J. 2003;22:557–62. doi: 10.1097/01.inf.0000069795.12338.cf. [DOI] [PubMed] [Google Scholar]
- 6.Filoche S, Wong L, Sissons CH. Oral biofilms: emerging concepts in microbial ecology. J Dent Res. 2010;89:8–18. doi: 10.1177/0022034509351812. [DOI] [PubMed] [Google Scholar]
- 7.Abiko Y, Sato T, Mayanagi G, Takahashi N. Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. J Periodontal Res. 2010;45:389–95. doi: 10.1111/j.1600-0765.2009.01250.x. [DOI] [PubMed] [Google Scholar]
- 8.Owens WD, Felts JA, Spitznagel EL., Jr ASA physical status classifications: a study of consistency of ratings. Anesthesiology. 1978;49:239–43. doi: 10.1097/00000542-197810000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Lang NP, Adler R, Joss A, Nyman S. Absence of bleeding on probing. An indicator of periodontal stability. J Clin Periodontol. 1990;17:714–21. doi: 10.1111/j.1600-051x.1990.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 10.WHO. Geneva, Switzerland: World Health Organization; 1997. Oral health surveys: basic methods. [Google Scholar]
- 11.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–55. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobb WS, Carbonell AM, Kalbaugh CL, Jones Y, Lokey JS. Infection risk of open placement of intraperitoneal composite mesh. Am Surg. 2009;75:762–7. discussion 67–8. [PubMed] [Google Scholar]
- 15.Coda A, Lamberti R, Martorana S. Classification of prosthetics used in hernia repair based on weight and biomaterial. Hernia. 2012;16:9–20. doi: 10.1007/s10029-011-0868-z. [DOI] [PubMed] [Google Scholar]
- 16.da Silva-Boghossian CM, do Souto RM, Luiz RR, Colombo AP. Association of red complex, A. actinomycetemcomitans and non-oral bacteria with periodontal diseases. Arch Oral Biol. 2011;56:899–906. doi: 10.1016/j.archoralbio.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Collage RD, Rosengart MR. Abdominal wall infections with in situ mesh. Surg Infect. 2010;11:311–18. doi: 10.1089/sur.2010.029. [DOI] [PubMed] [Google Scholar]
- 18.Sjollema J, Sharma PK, Dijkstra RJ, van Dam GM, van der Mei HC, Engelsman AF, et al. The potential for bio-optical imaging of biomaterial-associated infection in vivo . Biomaterials. 2010;31:1984–95. doi: 10.1016/j.biomaterials.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 19.Horz HP, Scheer S, Huenger F, Vianna ME, Conrads G. Selective isolation of bacterial DNA from human clinical specimens. J Microbiol Methods. 2008;72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Engelsman AF, van der Mei HC, Ploeg RJ, Busscher HJ. The phenomenon of infection with abdominal wall reconstruction. Biomaterials. 2007;28:2314–27. doi: 10.1016/j.biomaterials.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Cevasco M, Itani KM. Ventral hernia repair with synthetic, composite, and biologic mesh: characteristics, indications, and infection profile. Surg Infect. 2012;13:209–15. doi: 10.1089/sur.2012.123. [DOI] [PubMed] [Google Scholar]
- 22.Blatnik JA, Krpata DM, Jacobs MR, Gao Y, Novitsky YW, Rosen MJ. In vivo analysis of the morphologic characteristics of synthetic mesh to resist MRSA adherence. J Gastrointest Surg. 2012;16:2139–44. doi: 10.1007/s11605-012-1992-5. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Pumarino R, Pascual G, Rodriguez M, Perez-Kohler B, Bellon JM. Do collagen meshes offer any benefits over preclude® ePTFE implants in contaminated surgical fields? A comparative in vitro and in vivo study. J Biomed Mater Res B Appl Biomater. 2014;102:366–75. doi: 10.1002/jbm.b.33015. [DOI] [PubMed] [Google Scholar]
- 24.Mavros MN, Athanasiou S, Alexiou VG, Mitsikostas PK, Peppas G, Falagas ME. Risk factors for mesh-related infections after hernia repair surgery: a meta-analysis of cohort studies. World J Surg. 2011;35:2389–98. doi: 10.1007/s00268-011-1266-5. [DOI] [PubMed] [Google Scholar]
- 25.Awaiz A, Rahman F, Hossain MB, Yunus RM, Khan S, Memon B, et al. Meta-analysis and systematic review of laparoscopic versus open mesh repair for elective incisional hernia. Hernia. 2015;19:449–63. doi: 10.1007/s10029-015-1351-z. [DOI] [PubMed] [Google Scholar]
- 26.Bereza PL, Ekiel A, Augusciak-Duma A, Aptekorz M, Wilk I, Kusz DJ, et al. Identification of silent prosthetic joint infection: preliminary report of a prospective controlled study. Int Orthop. 2013;37:2037–43. doi: 10.1007/s00264-013-1955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvilla SA, Thusu S, Panesar SS. Analysing the benefits of laparoscopic hernia repair compared to open repair: a meta-analysis of observational studies. J Minim Access Surg. 2012;8:111–17. doi: 10.4103/0972-9941.103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahrani-Mougeot FK, Paster BJ, Coleman S, Ashar J, Barbuto S, Lockhart PB. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008;46:2129–32. doi: 10.1128/JCM.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown RH, Subramanian A, Hwang CS, Chang S, Awad SS. Comparison of infectious complications with synthetic mesh in ventral hernia repair. Am J Surg. 2013;205:182–7. doi: 10.1016/j.amjsurg.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Jun DW, Kim KT, Lee OY, Chae JD, Son BK, Kim SH, et al. Association between small intestinal bacterial overgrowth and peripheral bacterial DNA in cirrhotic patients. Dig Dis Sci. 2010;55:1465–71. doi: 10.1007/s10620-009-0870-9. [DOI] [PubMed] [Google Scholar]
- 31.Campbell LA, Kuo CC. Chlamydia pneumoniae – an infectious risk factor for atherosclerosis? Nat Rev Microbiol. 2004;2:23–32. doi: 10.1038/nrmicro796. [DOI] [PubMed] [Google Scholar]
- 32.Kane TD, Johnson SR, Alexander JW, Babcock GF, Ogle CK. Detection of intestinal bacterial translocation using PCR. J Surg Res. 1996;63:59–63. doi: 10.1006/jsre.1996.0223. [DOI] [PubMed] [Google Scholar]
- 33.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson MS, Bagg J, Gupta MN, Sturrock RD. Oral carriage of staphylococci in patients with rheumatoid arthritis. Rheumatology. 1999;38:572–5. doi: 10.1093/rheumatology/38.6.572. [DOI] [PubMed] [Google Scholar]
- 35.Fardini Y, Wang X, Temoin S, Nithianantham S, Lee D, Shoham M, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468–80. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velsko IM, Chukkapalli SS, Rivera-Kweh MF, Chen H, Zheng D, Bhattacharyya I, et al. Fusobacterium nucleatum alters atherosclerosis risk factors and enhances inflammatory markers with an atheroprotective immune response in ApoE(null) Mice. PLoS One. 2015;10:e0129795. doi: 10.1371/journal.pone.0129795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.