Abstract
The objective of this manuscript is to introduce a catalogue of salivary proteins that are altered secondary to carcinoma of the breast. The catalogue of salivary proteins is a compilation of twenty years of research by the authors and consists of 233 high and low abundant proteins which have been identified by LC-MS/MS mass spectrometry, 2D-gel analysis and by enzyme-linked immunosorbent assay. The body of research suggests that saliva is a fluid suffused with solubilized by-products of oncogenic expression and that these proteins may be useful in the study of breast cancer progress, treatment efficacy and the tailoring of individualized patient care.
Despite the numerous advances made in breast cancer research, carcinoma of the breast is still the most common, disfiguring and deadliest cancer among women1. Screening for breast cancer has led to earlier detection; however, the age old dilemma of “watch-and-wait” or to aggressively treat breast lesions haunt clinicians1,2. The understanding of BRCA1 and BRCA2 gene mutations has saved lives, but these findings account for only 5 to 10% of breast cancer cases1. Additional paradigms, i.e. biological models, may be needed in order to increase early detection and treatment efficacy1,2.
Biological models, such as those for breast cancer, simulate the simultaneous operations and interactions of multiple processes and molecular networks, in an attempt to re-create and predict the appearance of complex phenomena such as breast cancer progression3. A model that can reflect the presence and progression of malignancy among women would enhance our knowledge of breast cancer, and serve as an enabling system for biomarker discovery and credentialing of candidate markers so that the biological causality of any analyte(s) can be assessed. With respect to the study of breast cancer, there are currently three major methods for creating models for studying breast cancer progression. The three methods utilize either breast cancer tumor cell lines (BCTCL), xenografts of cell lines, and the third method uses animals – in this case genetically engineered mice for creating various models for studying breast cancer1. All three models have generated useful insight into cancer progression; however, despite their utility no individual model recapitulates all aspects of cancer progression4.
In the case of BCTCL research, the main question is - are the cell lines representative of human breast cancer and how well do they capture breast cancer tumorigenesis in the context of the unique tumor-stroma microenvironment? This question is debatable with strong evidence supporting both sides of the discussion. The fact is that no single cell line is representative of the disease process. To date, there are 51 BCTCL used to detect genetic abnormalities associated with breast cancer. However, the major concern is that multiple variants of the same cell line may exist and display distinct phenotypes. This suggests that they have acquired genetic changes that might preclude comparison between different studies1,2,3. Moreover, the ability of ex vivo cell line experiments to recapitulate the tumor microenvironment is in question, with stoma and extracellular matrix interactions, immune cell involvement, vasculature, and the complex milieu of the blood itself all contributing to the tumor cell biology. Additionally, cell lines are prone to genotypic and phenotypic drift during their continual culture and there is always the problem of contamination1,2,3,4.
Xenografts models are also very useful in studying breast cancer and have certainly helped understand genetic pathways associated with breast cancers but they are also problematic. Many of the problems that plague cell line studies also affect xenograft-driven modeling; however, a major problem is that xenografts must be established in immunocompromised mice4,5,6. The absence of an intact immune system may profoundly affect tumor development and progression as this microenvironment is entirely artificial. In fact, there is increasing evidence of roles for the immune system in both early stage breast cancer and metastasis1,4,5,6,7. There are also stoma differences between mouse and human breast tissues the must be taken into account. However, the major problem with xenograft models has to deal with breast cancer metastasis because metastatic cells preferentially colonize the lungs in mice, (instead of developing brain and liver metastasis) contain less fibrosis and inflammation then human tumors, nearly all are hormone independent as opposed to approximately half of human breast cancers that are hormone dependent, Another major draw-back is the lack of histological concordance between tumors from genetically engineered mice and the common types of human breast cancers. Finally, most tumors from mice do not resemble the most common subtypes of breast cancer. Taken together, clinicians and researchers may benefit by having an additional model system for assessing breast cancer tumorigenesis and in particular a predictive model for treatment response4.
One question emanating from the aforementioned paragraphs is what is the alternative to those models? The answer we are proposing is the use of the salivary proteome to study and assess cancer progression. The basic secretory units of the salivary glands are phenotypically similar to those of the mammary glands8,9,10,11,12. With the submandibular gland being the exception; they both have origins from the ectodermal germ layer. Both tissues are compound exocrine glands composed of specialized glandular epithelia. The tissues are characterized with two epithelial cell types i.e., ductal and acinar cells along with myoepithelial cells which contract to move fluid from the acinar lumen to the ducts. The ductal epithelial cells (terminal ducts) adjacent to the acinar units are cuboidal8,9,10,11,12.
From an immunohistological perspective there are a number of similarities between the mammary and salivary gland tissues. Both tissues have HER2/neu receptors on their ductal epithelial cells which can be overexpressed in malignant transformation8,9,10,11,12. Additionally, epithelial cells of both tissues have estrogen, progesterone, and androgen receptors that can be overexpressed8,9,10,11,12. p53 tumor suppressor gene is often mutated with overexpression of ineffective, mutated protein in carcinomas of the breast or the salivary glands. More interesting is that these proteins can be found in mammary ductal fluids and saliva8,9,10,11,12. As a consequence, the purpose of this manuscript is to provide evidence for the use of salivary protein profiles as an in vivo adjunct model for studying breast cancer progression.
Results and Discussions
The results of the proteomic analysis yielded 233 proteins that were either up or down regulated secondary to the presence of carcinoma of the breast. The 233 proteins represent 46% of the total number of proteins (505) identified among normal individuals assayed in this report. Table 1 lists the 233 proteins. Of the 233 proteins, 142 were up-regulated and 91 were down regulated. The down regulated proteins are emboldened in Table 1. In addition, the profile consists of both high and low abundant salivary proteins assayed by both mass spectrometry and enzyme-linked immunosorbent assay (ELISA) respectively.
Table 1. A Catalogue of Salivary Proteins Altered Secondary to Breast Cancer.
| UniProt | Gene | Protein Name | Function | Stage |
|---|---|---|---|---|
| Genomic Integrity Related Proteins (10, 4% of total) | ||||
| P16403 | H12 | Histone H1.2 | Transcription, DNA repair, replication | IIa, H+ |
| Q8IUE6 | H2A2B | Histone H2A | Transcription, DNA repair, replication | IIb |
| P16104 | H2AX | Histone H2A.x (H2a/x) | Transcription, DNA repair, replication | IIa, H+ |
| Q99880 | H2B1L | Histone H2B type 1-L | Transcription, DNA repair, replication | IIb, H+ |
| Q99877 | H2B1N | Histone H2B type 1-N | Transcription, DNA repair, replication | 0 |
| Q16778 | H2B2E | Histone H2B type 2-E | Transcription, DNA repair, replication | IIa, H+ |
| Q71DI3 | H32 | Histone H3.2 | Transcription, DNA repair, replication | IIa, H+ |
| P62805 | H4 | Histone H4 | Transcription, DNA repair, replication | IIa, H+ |
| P12004 | PCNA | Prolif. cell nuclear antigen | DNA repair | IIa, H+ |
| P35637 | TLS | TLS Oncogene | Maintenance of genomic integrity | IIa, H+ |
| Molecular Chaperones/Heat Shock Proteins (14, 6% of total) | ||||
| Q99933 | BAG-1 | BAG chaperone reg. 1 | Inhibits chaperone activity HSP70/HSC70 | 0, I, IIa, IIb, H+ |
| Q6P5S2 | CF058 | C6orf58 | Chromosome 6 open reading frame 58 | IIa, H+ |
| P14211 | CRP55 | Calreticulin | Major endoplasmic reticular activity | 0, I, IIa, IIb, H+ |
| P11021 | GRP78 | Glucose-regulated protein | Assembly multimeric prot. complexes | IIa, H+ |
| P11142 | HSP10 | Heat Shock 10 protein | Repressor of transcriptional activation | IIb |
| Q12988 | HSP27 | Heat Shock 27 protein | Inhibitor of actin polymerization | I, IIa |
| P25685 | Hsp40 | Heat shock protein 40 | Interacts with HSP70 | IIa, H+ |
| P08107 | HSP70 | Heat Shock 70 protein | Stabilizes preexistent proteins - aggregation | 0, I, IIa, IIb, H+ |
| P20585 | MSH3 | DNA repair protein | Post-replicative DNA mismatch repair sys | IIa, H+ |
| P05307 | PDI | Protein disulfide isomerase; | Catalyzes the rearrangement of -S-S- bonds | IIa, H+ |
| P07237 | PDIA1 | Protein disulfide-isomerase | Catalyzes rearrangement of -S-S- bonds | IIb |
| P62937 | PPIA | Peptidyl-prolyl isomerase A | PPIases accelerate the folding of proteins | IIa, IIb, H+ |
| P23284 | PPIB | Peptidyl-prolyl isomerase B | PPIases accelerate the folding of proteins | IIa, IIb, H+ |
| Q13546 | RIP | Recept.-interacting protein 1 | DNA damage repair | IIa, H+ |
| Cell Growth Related Proteins (17, 7% of total) | ||||
| P63104 | 1433Z | 14-3-3 protein zeta/delta | Protein kinase C inhibitor | IIb |
| P31749 | AKT-1 | α- serine/threonine-prot. kinase | Cell survival, growth & angiogenesis | IIa, H+ |
| P04040 | CATA | Catalase | Promotes growth of cells | IIa, IIb |
| P32577 | Csk | Tyrosine-protein kinase CSK | Regulates cell growth | IIa, H+ |
| P01133 | EGF | epidermal growth factor | Stimulates growth of epid. & epith. tissues | 0, I, IIa, IIb, H+ |
| P00533 | EGFR | epidermal growth factor recep. | EGF receptor | 0, I, IIa, IIb, H+ |
| P09038 | FGF | Fibroblastic growth factor | Growth factor | IIa, H+ |
| P15692 | VEGF | Vasc. Endothel. Growth Factor | Plays an important role in angiogenesis | IIa, H+ |
| P29354 | GRB2 | Growth factor recept. protein 2 | Links cell surface growth factor receptor | IIa, H+ |
| P04626 | HER2/neu | Epid. growth factor receptor 2 | EGF receptor | 0, I, IIa, IIb, H+ |
| P15531 | NDKA | Nucleoside diphosphate kin. A | Cell proliferation, differentiation | IIa, H+ |
| P29474 | NOS3 | Nitric oxide synthase | Mediates vascular angiogenesis | IIa, H+ |
| P29476 | NOSI | NOS type I | Nitric oxide producer | IIa, H+ |
| P20936 | Ras-GAP | Ras GTPase-act. prot. 1 | Inhibitory reg. Ras-cyclic AMP pathway | IIa, H+ |
| P25815 | S100P | S100-P | Stimulate cell proliferation | 0 |
| Q9UKW4 | VAV3 | VAV 3 oncogene | Plays an important role in angiogenesis | IIa, H+ |
| P01135 | TGF-α | Transforming growth factor α | Potent mitogenic polypeptide | 0, I, IIa, IIb, H+ |
| Apoptosis Related Proteins (13, 5% of total) | ||||
| P31947 | 1433S | 14-3-3 protein sigma | p53 regulated inhibitor | IIa, IIb |
| O14727 | Apaf-1 | Apop. prot. activity factor-1 | Apoptosis activity | IIa, H+ |
| P99999 | CYC | Cytochrome c | Apoptosis activity | IIa, H+ |
| P10909 | CLU | Clusterin | Apoptosis activity | 0, I, IIa, H+ |
| P80188 | NGAL | Oncogene 24p3 | Involved apoptosis, innate immunity | IIa, IIb |
| P38936 | p21 | WAF-1 | Inhibitor of cellular proliferation | IIa, IIb, H+ |
| P04637 | p53 | protein 53 | Apoptosis | IIa, IIb, H+ |
| P19525 | PKR | p68 kinase | Apoptosis, cell proliferation | IIa, H+ |
| P62988 | UBIQ | Ubiquitin | Apoptosis | IIa, IIb, H+ |
| P98170 | XIAP | E3 ubiquitin-protein ligase XIAP | Apoptosis | IIa, H+ |
| P04156 | PRIP | Protein prion | Possible apoptotic activity | IIa |
| O43550 | CDC25B | CDC25B | Tyrosine phosphatase activity | IIa, H+ |
| P01375 | TNF-α | Tumor necrosis factor alpha | Induces cell death. | 0, I, IIa, IIb |
| Immunity Related Proteins (52, 22% of total) | ||||
| P02763 | A1AG1 | Alpha-1 acid glycoprotein 1 | Anti-inflammatory activity | IIa, IIb, H+ |
| P61769 | B2MG | Beta-2 microglobulin precursor | Component of the MHC I complex | IIa, |
| Q8N4F0 | BPIL1 | Palate, lung & nasal epith. | Involved in the innate immune response | 0, I, IIa, H+ |
| P01024 | C3 | Complement C3 precursor | Effector of innate and adaptive immunity | 0, I, IIa, IIb, H+ |
| P0C0L4 | C4 | Complement 4A | Of the classical complement pathway | 0, I, IIa, IIb, H+ |
| P28907 | CD38 | ADP-ribosyl cyclase 1 | Receptor in cells of the immune system | IIa, H+ |
| P53618 | COPB | Coatomer subunit beta | Degradation of CD4 & MHC I antigens | IIa |
| P54108 | CRIS3 | Cysteine-rich secret. prot. 3 | Ligand of alpha1B-glycoprotein in plasma | 0, IIa, |
| Q9UGM3 | DMBT1 | GP 300 | Interaction of tumor cells & immune system | IIa, H+ |
| P08246 | ELNE | Leukocyte elastase | Modifier of monocytes & granulocytes | IIa, H+ |
| P06241 | FYN | Fyn-Tyrosine-protein kinase | Regulates cell growth | IIa, H+ |
| P01762 | HV301 | Ig heavy chain V-III region TRO | Fc-epsilon receptor signaling pathway | IIb |
| P01777 | HV316 | Ig heavy chain V-III region TEI | Fc-epsilon receptor signaling pathway | IIa, H+ |
| P01781 | HV320 | Ig heavy chain VIII region GAL | Complement activation | IIa, IIb |
| P01579 | IFN-γ | Interferon gamma | Potent activator of macrophages | 0, I, IIa, IIb, H+ |
| P01876 | IGHA1 | Ig alpha1 chain C region | Defends against local infection | IIa, IIb |
| P01877 | IGHA2 | Ig alpha-2 chain C region | Defends against local infection | IIb, H+ |
| P01857 | IGHG1 | Ig gamma-1 chain C region | Complement activation | 0, I, IIa, IIb, H+ |
| P01859 | IGHG2 | Ig gamma-2 chain C region | Complement activation | IIb, H+ |
| P01591 | IGJ | Immunoglobulin J chain | Links two monomer units of IgM or IgA | IIa, IIb |
| P05112 | IL 4 | Interleukin - 4 | Activates B-cell and T-cell proliferation | 0, I, IIa, IIb |
| P05231 | IL 6 | Interleukin - 6 | Inducer of the acute phase response | 0, I, IIa, IIb |
| P10145 | IL 8 | Interleukin - 8 | A chemotatic factor | 0, I, IIa, IIb |
| P22301 | IL-10 | Interleukin - 10 | Cytokine Inhibitor (IFN-γ, IL-2, IL-3, TNF) | 0, I, IIa, IIb |
| P18510 | IL1RA | Interleukin-1 recept. antagonist | Antagonist to IL-1 alpha, beta | IIa, H+ |
| Q9BY25 | IL1β | IL-1beta | Inhibits neutrophil apoptosis | 0, I, IIa, IIb |
| P30740 | ILEU | Leukocyte elastase inhibitor | Regulates neutrophil proteases | IIb |
| P01834 | KAC | Ig kappa chain C region | immunoglobulin κ chains | IIa, IIb, H+ |
| P06870 | KLK1 | Kallikrein-1 precursor | Cleaves Met-Lys and Arg-Ser bonds | 0, I, IIa, IIb |
| P06309 | KV205 | Ig kappa chain V-II region | Fc-epsilon receptor signaling pathway | IIa, H+ |
| P18135 | KV312 | Ig kappa chain VIII region HAH | Surface immunoglobulin M autoantibody | IIa, IIb |
| P01842 | LAC | Ig lambda chain C regions | Complement activation | IIa, H+ |
| P06239 | LCK | Lck-Tyrosine-protein kinase | Selection & maturation of T-cells in thymus | IIa, H+ |
| P31025 | LCN1 | Lipocalin1 precursor | Regulates activity of neutrophil proteases | 0, I, IIa, IIb, H+ |
| P13500 | MCP-1 | Monocyte chemotactic prot.-1 | Chemotactic factor attracts monocytes | 0, I, IIa, IIb |
| O88888 | Mint3 | Minit-3 | Activates macrophages | IIa, H+ |
| P22894 | MMP8 | Matrix metalloproteinase-8 | Degrades fibrillar type I, II & III collagens | IIa, H+ |
| P14780 | MMP9 | Matrix metalloproteinase-9 | Basement membrane dissolution | IIa, H+ |
| P01871 | MUC | Ig mu chain C region | Role in primary defense mechanisms | I, IIa, IIb, H+ |
| O43240 | NES1 | Kallikrein 10 | Tumor suppressor in breast cancer | IIa, H+ |
| P22079 | PERL | Lactoperoxidase | Airway host defense against infection | 0, IIa, IIb, H+ |
| P05164 | PERM | Myeloperoxidase | Host defense system of leukocytes | IIa, IIb, H+ |
| P01833 | PIGR | Poly-IG receptor protein | Binds IgA & IgM at basolateral surface | IIa, IIb, H+ |
| P12273 | PIP | Prolactin-inducible protein | Pathological conditions mammary gland | I, IIa, IIb |
| P13796 | PLSL | Plastin-2 | Modulates cell surface expression IL2RA/CD25 | IIa, H+ |
| Q9NP55 | PLUNC | BPI fold-containing family A1 | Associated with tumor progression | 0, I, IIa, IIb |
| P13501 | RANTES | RANTES | Chemokine (CCL5) | 0, I, IIa, IIb |
| P13405 | Rb | Retinoblastoma-assoc. prot. | Acts as a tumor suppressor | IIa, H+ |
| P29508 | SPB3 | Serpin B3 | Immune response against tumor cells | IIa, H+ |
| P42224 | Stat1 | Transcription factor ISGF-3 | Mediates responses to cytokines | IIa, H+ |
| P01135 | TGF-α | Transforming growth factor α | Potent mitogenic polypeptide | 0, I, IIa, IIb |
| P01375 | TNF-α | Tumor necrosis factor alpha | Secreted to induce cell death | 0, I, IIa, IIb |
| Cytoskeleton Related Proteins (40, 17% of total) | ||||
| P63261 | ACTG | Actin, cytoplasmic 2 | Cytoskeleton | IIa, H+ |
| Q01518 | CAP1 | Adenylyl cyclase | Actin cytoskeleton organization | IIa, H+ |
| P12830 | CDH1 | Cadherin-1 | Epithelial Adherens junction protein | IIa, H+ |
| P06731 | CEA | Carcin-embryonic antigen | Cell adhesion and in intracellular signaling | 0, I, IIa, IIb, H+ |
| P23528 | COF1 | Cofilin, non-muscle isoform | Cytoskeleton | IIa, H+ |
| Q02487 | DSC2 | Desmocollin-2 precursor | Intercellular desmosome junctions | IIb |
| P15311 | EZRI | Ezrin | Cytoskeleton | 0, I, IIa, IIb |
| P02671 | FIBA | Fibrinogen α chain precursor | Involved in cell adhesion, cell motility | IIa, |
| P02675 | FIBB | Fibrinogen beta chain precursor | Involved in cell adhesion, cell motility | 0, I, IIa, IIb |
| P06396 | GELS | Gelsolin | Assembly of monomers into filaments | IIa, H+ |
| Q15151 | JUP | g-Catenin | Associated with desmosome junctions | IIa, H+ |
| P13646 | K1C13 | Cytokeratin-13 | Cytoskeleton protein | 0, IIa, IIb, H+ |
| P08779 | K1C16 | Cytokeratin-16 | Cytoskeleton protein | 0, I, IIa, IIb |
| P35527 | K1C9 | Cytokeratin-9 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P04264 | K2C1 | Cytokeratin 1 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P19013 | K2C4 | Cytokeratin 4 | Cytoskeleton protein | IIa, H+ |
| P13647 | K2C5 | Cytokeratin-5 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P02538 | K2C6A | Cytokeratin-6A | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P48666 | K2C6C | Cytokeratin 6C | Cytoskeleton protein | 0, |
| P13645 | KRT10 | Cytokeratin-10 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P02533 | KRT14 | Cytokeratin-14 | Cytoskeleton protein | 0, I, IIb, |
| P19012 | KRT15 | Cytokeratin-15 | Cytoskeleton protein | I, IIa, |
| Q04695 | KRT17 | Cytokeratin-17 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| P08727 | KRT19 | Cytokeratin-19 | Cytoskeleton protein | IIa |
| P35908 | KRT2 | Cytokeratin 2 | Cytoskeleton protein | 0, I, IIa, IIb, H+ |
| Q7Z3Z0 | KRT25 | Cytokeratin-25 | Cytoskeleton protein | IIa, H+ |
| P08729 | KRT7 | Cytokeratin-7 | Cytoskeleton protein | 0, I, IIa, IIb |
| P22894 | MMP8 | Matrix metalloproteinase-8 | Degrades fibrillar type I, II & III collagens | IIa, H+ |
| P14780 | MMP9 | Matrix metalloproteinase-9 | Basement membrane dissolution | IIa, H+ |
| P26038 | MOES | Moesin | Involved in major cytoskeletal structures | IIa, H+ |
| P49024 | Paxillin | Paxillin | Involved in actin-membrane attachment | IIa, H+ |
| P07737 | PROF1 | Profilin-1 | Binds to actin & affects the cytoskeleton | 0, IIa, IIb, H+ |
| P03749 | Rho | Rho | Regulates intracellular actin dynamics | IIa, H+ |
| P31949 | S10AB | S100-A11 | Cornification of keratinocytes | IIa, |
| P35321 | SPR1A | Cornifin-A | Keratinization activity | I, IIa, IIb, H+ |
| P22528 | SPR1B | Cornifin-B | Keratinization activity | IIb, H+ |
| Q9UBC9 | SPRR3 | Cornifin beta | Envelope protein of keratinocytes | 0, IIa, H+ |
| P10636 | TAU | Microtubule-associated prot. - τ | Promotes microtubule assembly and stability | IIa, H+ |
| P62328 | TYB4 | Thymosin beta 4 | Role in the organization of the cytoskeleton | IIa, |
| P08670 | VIME | Vimentin | Class-III intermediate filaments | IIa, IIb, H+ |
| Metabolism Related Proteins (56, 24% of total) | ||||
| P22303 | ACTH | Acetylcholinesterase | Signal transduction at NMJ | IIa, H+ |
| P07108 | ACBP | AcylCoA binding protein | Binds medium & long-chain acyl-CoA | IIa, H+ |
| Q15848 | ADIPO | Adiponectin | Adipokine involved in fat metabolism | 0, I, IIa, IIb, H+ |
| Q9DCT1 | AK1E1 | Aldo-keto reductase | Catalyst | 0, |
| P04217 | A1BG | Alpha-1B-glycoprotein | Functions as transport protein in blood | IIa, H+ |
| P01023 | A2MG | Alpha-2 macroglobulin | Proteinase inhibitor | IIa, IIb, H+ |
| Q92746 | ST8SIA2 | Alpha-2,8-sialyltransferase 8B | Protein modification; protein glycosylation | IIa, H+ |
| P19961 | AMYC | Alpha-amylase 2B precursor | Carbohydrate metabolism | 0, I, IIa, IIb |
| P06733 | ENOA | Alpha-enolase | Enzyme that has a role in glycolysis | IIa, IIb, H+ |
| P03973 | ALK1 | Antileukoproteinase 1 precursor | Proteinase inhibitor | IIa, H+ |
| P02647 | APOA1 | Apolipoprotein A-I | Promotes efflux of cholesterol from cell | IIb, H+ |
| P02649 | APO-E | Apolipoprotein -E | Catabolizes lipoproteins | IIa, H+ |
| P06576 | ATPB | ATP synthase subunit beta | ATP synthesis | 0, IIa, IIb |
| Q8WZ76 | BCRP | Breast cancer resistance prot. | ATP hydrolysis-dependent efflux transport | I |
| P00915 | CAH1 | Carbonic anhydrase 1 | Reversible hydration of carbon dioxide | IIb, |
| P23280 | CAH6 | Carbonic anhydrase 6 | Reversible hydration of carbon dioxide | 0, I, IIa, IIb |
| P07339 | CATD | Cathepsin D | Pathogenesis of breast cancer | 0, I, IIa, IIb, H+ |
| P01040 | CYTA | Cystatin A | Protease inhibitor | 0, I, IIa, H+ |
| P04080 | CYTB | Cystatin B | Intracellular thiol proteinase inhibitor | IIa, IIb, H+ |
| P01034 | CYTC | Cystatin C | Local regulator of this enzyme activity | 0, I, IIa, IIb, H+ |
| P28325 | CYTD | Cystatin-D | Proteinase inhibitor | 0, I, IIa, IIb, H+ |
| P01036 | CYTS | Cystatin-S precursor | Protein inhibits papain and ficin | 0, I, IIa, IIb |
| P09228 | CYTT | Cystatin-SA precursor | Thiol protease inhibitor | IIa, IIb, H+ |
| P01037 | CYTN | Cystatin-SN precursor | Cysteine proteinase inhibitors | IIa, IIb |
| P20813 | CYP2B6 | Cytochrome p450 | NADPH-dependent electron transport | 0, I, IIa, IIb, H+ |
| Q01469 | FABPE | Fatty acid binding protein | Involved in keratinocyte differentiation | IIa, H+ |
| P80303 | NUCB2 | Gastric cancer antigen Zg4 | Calcium-binding protein | 0, IIa, IIb, H+ |
| P06744 | G6PI | Glucose-6-phosphate isom. | Glycolytic enzyme | IIb |
| P09211 | GSTP1 | Glutathione S-transferase P | Regulates negatively CDK5 activity | IIa, IIb, H+ |
| P00738 | HPT | Haptoglobin precursor | Captures & combines with hemoglobin | 0, IIa, IIb, H+ |
| Q9Y5Z4 | HEBP2 | Heme-binding protein 2 | Promotes mitochondrial permeability transition | IIa |
| P69905 | HBA | Hemoglobin subunit alpha | Oxygen transport from lung | IIb, H+ |
| P68871 | HBB | Hemoglobin subunit beta | Oxygen transport from lung | IIb, H+ |
| P02790 | HEMO | Hemopexin precursor | Binds heme and transports to liver | IIb |
| P04075 | ALDOA | Fructose biphosphate aldol. | Role in glycolysis & gluconeogenesis | IIa, IIb |
| Q14764 | LRP | Lung resistance-related protein | Role in nucleo-cytoplasmic transport | I |
| P40926 | MDHM | Malate dehydrogenase | Catalytic activity | 0, IIa, IIb, H+ |
| O15438 | MRDP | Multidrug resistance protein | Inducible transporter of organic anions | I |
| Q06830 | PRDX1 | Peroxiredoxin-1 | Redox regulation of the cell | IIa, IIb, H+ |
| P30041 | PRDX6 | Peroxiredoxin-6 | Redox regulation of the cell | IIa, |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | Glycolytic enzyme | IIa, IIb, H+ |
| Q96PX9 | PKH4B | Pleckstrin family G member 4B | PH domains bind various proteins | IIa, |
| P20742 | PZP | Pregnancy zone protein | Inhibits all four classes of proteinases | 0, I, IIa, IIb, H+ |
| P08129 | PP1 | Protein phosphatase 1 | Regulation of glycogen metabolism | IIa, H+ |
| Q08188 | TGM3 | Transglutaminase E | Formation of isopeptide cross-links | IIa, H+ |
| P14618 | PKM2 | Pyruvate kinase PKM2 | Caspase independent cell death of tumor cells | IIa, IIb, H+ |
| P02768 | ALBU | Serum albumin precursor | Regulation of colloidal osmotic pressure | 0, I, IIa, IIb |
| P10599 | THIO | Thioredoxin | Cytoplasmic antioxidant | IIb |
| P37837 | TALDO | Transaldolase | Balances metabolites | IIb, H+ |
| P20061 | TCO1 | Transcobalamin1 precursor | Vitamin B12-binding protein | IIa, IIb |
| P02787 | TRFE | Transferrin | Iron transporter | 0, IIa, IIb, H+ |
| P29401 | TKT | Transketolase | Transfers ketol group to aldose acceptor | IIa, |
| P60174 | TPIS | Triosephosphate isomerase | Catalytic activity | IIa, H+ |
| P02774 | VTDB | Vitamin D-binding protein | Transport | IIb |
| P25311 | ZA2G | Zinc-alpha-2-glycoprotein | Stimulates lipid degradation in adipocytes | 0, |
| Q96DA0 | U773 | Zymogen granule protein | Protein trafficking | IIa, H+ |
| Membrane and Calcium Binding Related Proteins (17, 7% of total) | ||||
| P04083 | ANXA1 | Annexin A1 | Membrane fusion & exocytosis | 0, I, IIa, IIb, H+ |
| P46193 | ANXA1 | Annexin I | Membrane fusion & exocytosis | 0, I, IIa, IIb, H+ |
| P07355 | ANXA2 | Annexin II | Calcium-regulated membrane-binding prot. | 0, I, IIa, IIb, H+ |
| P12429 | ANXA3 | Annexin A3 | Inhibitor of phospholipase A2 | IIb, |
| P02765 | FETUA | Alpha-2-Z-globulin | Promotes endocytosis | IIa, IIb |
| P52566 | GDIS | Rho GDP-dissociation inhib. 2 | Regulates GDP/GTP exchange reaction | IIa, IIb |
| P20810 | ICAL | Calpastatin | Specific inhibition of calpain | IIa, |
| P97799 | p24 | Neurensin-1 | Role in neural organelle transport | IIa, H+ |
| P15154 | Rac1 | p21 Rac-1 | Plasma membrane-associated small GTPase | IIa, H+ |
| P11233 | Ral A | Ras-related protein Ral-A | GTPase involved in cellular processes | IIa, H+ |
| P62158 | CALM | Calmodulin | Control of a large number of enzymes | IIb |
| P26447 | S10A4 | S100-A4 | Regulation of I-kappaB kinase/NF-kappaB | IIa, |
| P06703 | S10A6 | S100-A6 | Modulator of cellular calcium signaling | IIa, |
| P31151 | S100P | S100-A7 | Calcium binding protein | 0, I, IIa, IIb, H+ |
| P05109 | S10A8 | S100-A8 | Inflammatory processes, immune response | 0, I, IIa, IIb, H+ |
| P06702 | S10A9 | S100-A9 | Inflammatory processes, immune response | 0, I, IIa |
| P80511 | S10AC | S100-A12 | Binding protein, immune response | 0, I, IIa, IIb, H+ |
| Oral Anti-Microbial Related Proteins (14, 6% of total) | ||||
| P08311 | CATG | Cathepsin G | Antibacterial to Gram-negative bact. | 0, IIb |
| P59666 | DEF3 | Neutrophil defensin 3 | Antimicrobial to Gram-negative/positive bact. | I, IIa, IIb |
| P15515 | HIS1 | Histatin-1 precursor | Exhibit antibacterial & antifungal activities | IIa, IIb |
| P61626 | LYSC | Lysozyme C precursor | Bacteriolytic function | IIa, IIb |
| Q9HC84 | MUC5B | Mucin-5B precursor | Clearance of bacteria in the oral cavity | 0, I, IIa, IIb, H+ |
| Q8TAX7 | MUC7 | Mucin-7 | Clearance of bacteria in the oral cavity | 0, I, IIa, H+ |
| P02812 | PRB2 | Salivary prol-rich prot. 2 | Oral antimicrobial activity | IIa, H+ |
| Q16378 | PROL4 | Proline-rich protein 4 | Antimicrobial activity | IIa, H+ |
| P02814 | SMR3B | Submax. gland androgen-reg. 3 | Salivary PRP | 0, I, IIa, H+ |
| Q96DR5 | SPLC2 | Parotid secretory protein | Innate immune response | 0, I, IIa, IIb, H+ |
| P02788 | TRFL | Lactotransferrin | Antimicrobial activity | 0, IIa, IIb, H+ |
| P02810 | PRPC | Acidic proline-rich prot. | Inhibitors of crystal growth | IIa, IIb |
| P15941 | MUC-1 | Cancer antigen 15-3 | Adhesion and an anti-adhesion protein | 0, I, IIa, IIb, H+ |
| Q02817 | MUC-2 | Mucin 2 | Mucosal protection | IIa, IIb |
Note: The UniProt identification number, gene identification protein name and protein function are from UniProt database (www.uniprot.org). The proteins are classified according to molecular function. Additionally all down-regulated proteins are bold; otherwise, the remaining proteins are up-regulated.
Staging Abbreviations: 0 = Stage 0, I = Stage I, IIa = Stage IIa, IIb = Stage IIb, H+ = Her2/neu positive.
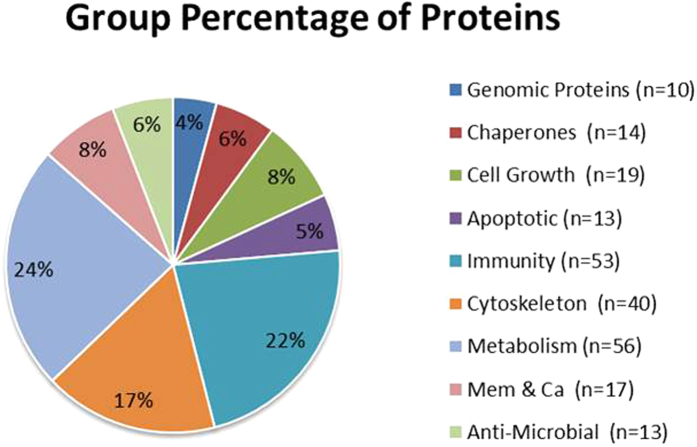
In order to compare the results to published proteomic cancer cell analysis, the proteins were categorized into ten groups of cellular activity13,14. The groups are illustrated in Fig. 1 and are as follows: 1) Genomic proteins; 2) molecular chaperones; 3) cell growth; 4) apoptotic proteins; 5) anti-inflammatory and immunoresponse proteins; 6) cytoskeletal proteins; 7) metabolic proteins; 8) membrane associated proteins and 9) antimicrobial proteins13,14.
Figure 1. The figure represents the number and percentage of proteins as classified by function.
As illustrated in Fig. 1 and Table 1, the metabolic protein category reflected 24% of the 236 proteins while the inflammatory/immunoresponse and the cytoskeletal categories exhibited 23% and 17% of the total number of proteins respectively. Eighty four (36%) of the proteins were detected in the early stage carcinomas (Stage 0 & Stage I).
Table 2 demonstrates how each protein was assayed and which proteins were present in varying cancer cell lines cited in the literature15,16,17,18. Fifty one of the 233 proteins (22%) could be identified in the SKBR3 cell line, 34 (14%) in the MCF7, 4 (2%) in the T47D, 29 (12%) in the MB-MDA-231, 26 (11%) in the 8701-BC and 43 (18%) in malignant tumor tissues. Twenty four (11%) were common to three or more of the cell lines.
Table 2. Salivary Protein Presence in Breast Cancer Cell Lines and Exosomes.
| UniProt | Protein Name | Method of Identification | Presence in Cell Lines | Exosomes In Saliva | Exosomes in T.T. | Ref. |
|---|---|---|---|---|---|---|
| Genomic Integrity Related Proteins | ||||||
| P16403 | Histone H1.2 | MS | TT | 25 | ||
| Q8IUE6 | Histone H2A | MS | TT | 25 | ||
| Q99880 | Histone H2B type 1-L (H2B.c) | MS | TT | 25 | ||
| Q71DI3 | Histone H3.2 | MS | TT | 28 | ||
| P62805 | Histone H4 | MS | Yes | 73 | ||
| P12004 | Prolif. cell nuclear antigen | AA | T, TT | 8, 20, 77 | ||
| Molecular Chaperones/Heat Shock Proteins | ||||||
| Q6P5S2 | C6orf58 | 2D, MS | Yes | 85 | ||
| P27797 | Calreticulin | MS, AA | S, B, M, MD, TT | Yes | 8, 15, 16, 17, | |
| P11021 | Glucose-regulated protein | AA | S, B, M, MD, TT | Yes | Yes | 8, 15, 16, 17, 74, 85 |
| P11142 | Heat Shock 10 protein | MS | Yes | Yes | 74, 85 | |
| Q12988 | Heat Shock 27 protein | MS | B, M, MD, TT | 14, 15, 16 | ||
| P08107 | Heat Shock 70 protein | MS | B, M, T | Yes | Yes | 15, 16, 74, 85 |
| P05307 | Protein disulfide isomerase; p55 | AA | Yes | 8, 15, 16, 17, 85 | ||
| P07237 | Protein disulfide-isomerase | MS | B, M, MD, TT | 14, 15, 16 | ||
| P62937 | Peptidyl-prolyl cis-trans isom. A | MS | S, B, M, MD | Yes | Yes | 15, 16, 17, 74, 85 |
| Cell Growth Related Proteins | ||||||
| P63104 | 14-3-3 protein zeta/delta | AA | B, M, MD, TT | Yes | 8, 15, 18, 85 | |
| P00533 | epidermal growth factor receptor | E | S, M | Yes | Yes | 10, 17, 74, 85 |
| P29354 | Growth factor receptor protein 2 | AA | S, M, MD | Yes | 8, 17, 85 | |
| P04626 | epidermal growth factor receptor2 | WB, E | S | Yes | Yes | 10, 17, 74, 85 |
| P15531 | Metastatic process-associ. prot. | AA | M | 8 | ||
| Apoptosis Related Proteins | ||||||
| P31947 | 14-3-3 protein sigma | MS | TT | 18, 37 | ||
| P38936 | WAF-1 | AA, E | S | 8, 17, 39 | ||
| P04637 | protein 53 | E | TT | 8, 36 | ||
| P62988 | Ubiquitin | MS, WB | S, B | Yes | Yes | 15, 16, 17, 85 |
| Immunity Related Proteins | ||||||
| P61769 | Beta-2 microglobulin precursor | MS | S, B | Yes | Yes | 15, 16, 17, 74, 85 |
| Q8N4F0 | Long palate, lung and nasal epith. | MS | Yes | Yes | 74, 85 | |
| Q9UGM3 | GP 300 | MS | Yes | 85 | ||
| P01579 | Interferon gamma | AA | TT | 8, 57 | ||
| P01876 | Ig alpha1 chain C region | MS | Yes | 85 | ||
| P01877 | Ig alpha-2 chain C region | MS | Yes | 85 | ||
| P01857 | Ig gamma-1 chain C region | MS | Yes | 15, 85 | ||
| P01859 | Ig gamma-2 chain C region | MS | Yes | 85 | ||
| P01591 | Immunoglobulin J chain | MS | Yes | 85 | ||
| P05112 | Interleukin - 4 | AA, E | TT | 8, 10, 87 | ||
| P05231 | Interleukin - 6 | AA | TT | 8, 46, 87 | ||
| P10145 | Interleukin - 8 | AA | TT | 8, 46, 87 | ||
| P22301 | Interleukin - 10 | AA | M, TT | 8, 46, 87 | ||
| Q9BY25 | IL-1beta | AA | TT | 8, 46, 87 | ||
| P30740 | Leukocyte elastase inhibitor | MS | M, MD | 85 | ||
| P01834 | Ig kappa chain C region | MS | Yes | 85 | ||
| P01842 | Ig lambda chain C regions | MS | Yes | 85 | ||
| P31025 | Lipocalin1 precursor | MS | TT | 85 | ||
| P01871 | Ig mu chain C region | MS | Yes | 85 | ||
| P22079 | Lactoperoxidase | MS | Yes | 85 | ||
| P05164 | Myeloperoxidase | MS | Yes | 85 | ||
| P01833 | Poly-IG receptor protein | MS | TT | 85 | ||
| P12273 | Prolactin-inducible protein | MS | Yes | 85 | ||
| P26447 | S100-A4 | MS | S | 17 | ||
| P05109 | S100-A8 | MS, WB | S, TT | Yes | 18, 85 | |
| P06702 | S100-A9 | MS | S | Yes | 17 | |
| P29508 | Serpin B3 | MS | M, MD | Yes | 14, 85 | |
| P42224 | STAT-1 | MS, AA | S, M | 17, 80 | ||
| P01135 | Transforming growth factor alpha | AA | S | 8, 17 | ||
| P01375 | Tumor necrosis factor alpha | AA | TT | 8, 17 | ||
| Cytoskeleton Related Proteins | ||||||
| P63151 | Actin, cytoplasmic 2 | MS | S, B | 15, 16, 17 | ||
| Q01518 | Adenylyl cyclase | MS | S | 17 | ||
| P12830 | Cadherin-1 | MS, AA | MD, TT | 8, 15, 16, 17 | ||
| P06731 | Carcin-embryonic antigen | MS | Yes | Yes | 85 | |
| P23528 | Cofilin, non-muscle isoform | MS | S, B, M, MD, TT | Yes | 14, 18 | |
| P15311 | Ezrin | MS | S, B | Yes | Yes | 17, 80, 85 |
| P06396 | Gelsolin | MS | S | Yes | Yes | 17, 18, 85 |
| P13646 | Cytokeratin-13 | MS | S | 17 | ||
| P08779 | Cytokeratin-16 | MS | Yes | 85 | ||
| P35527 | Cytokeratin-9 | MS | S, B | Yes | 15 | |
| P35908 | Keratin, type II | MS | S | 17 | ||
| P04264 | Cytokeratin 1 | MS | S, M, MD | Yes | Yes | 17 |
| P19013 | Cytokeratin 4 | MS | Yes | 85 | ||
| P13647 | Cytokeratin-5 | MS | S | 28 | ||
| P02538 | Cytokeratin-6A | MS | Yes | 80 | ||
| P13645 | Cytokeratin-10 | MS | S | 17, 87 | ||
| P02533 | Cytokeratin-14 | MS | TT | Yes | 85 | |
| P19012 | Cytokeratin-15 | MS | M, MD, TT | Yes | 85 | |
| Q04695 | Cytokeratin-17 | MS | TT | 45 | ||
| P08727 | Cytokeratin-19 | MS | S, TT | Yes | 17, 87 | |
| P08729 | Cytokeratin-7 | MS | S, TT | 17, 80, 85 | ||
| P26038 | Moesin | MS | M, MD | Yes | Yes | 80 |
| P07737 | Profilin-1 | MS, WB, E | S, B, M, MD, T | Yes | Yes | 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 85 |
| P03749 | Rho | AA | S | 8, 15, 17 | ||
| P31949 | S100-A11 | MS | TT | Yes | Yes | 85 |
| P35321 | Cornifin-A | MS | Yes | Yes | 85 | |
| P08670 | Vimentin | MS | S, B, M, MD | Yes | 15, 16, 17 | |
| Metabolism Related Proteins | ||||||
| P07108 | AcylCoA binding protein | AA | B | Yes | 15, 16 | |
| Q9DCT1 | Aldo-keto reductase | MS | TT | 14 | ||
| P06733 | Alpha-enolase | MS, WB, E | S, B, M, MD, TT | Yes | Yes | 14, 15, 16, 17, 18, 85 |
| P03973 | Antileukoproteinase 1 precursor | MS | Yes | 85 | ||
| P02647 | Apolipoprotein A-I | MS | S, TT | Yes | Yes | 15, 17 |
| P02649 | Apolipoprotein -E | AA | Yes | 8, 15, 85 | ||
| P06576 | ATP synthase subunit beta | MS | M, MD | 80 | ||
| P00915 | Carbonic anhydrase 1 | MS | TT | Yes | 14, 18 | |
| P23280 | Carbonic anhydrase 6 | MS | Yes | 85 | ||
| P07339 | Cathepsin D | MS, AA, E | M, MD, T, S | 8, 14, 17 | ||
| P04080 | Cystatin B | MS | S | Yes | 17, 85 | |
| P01034 | Cystatin C | MS | B | 16 | ||
| P28325 | Cystatin-D | MS | Yes | 85 | ||
| P01036 | Cystatin-S precursor | MS | Yes | 85 | ||
| P09228 | Cystatin-SA precursor | MS | Yes | 85 | ||
| P01037 | Cystatin-SN precursor | MS | Yes | 85 | ||
| Q01469 | Fatty acid binding protein | MS | S, TT | Yes | Yes | 8, 18, 85 |
| P06744 | Glucose-6-phosphate isomerase | MS | S, M, MD | Yes | 14, 17 | |
| P09211 | Glutathione S-transferase P | MS | S, B, M, MD, TT | 14, 15, 16, 17, 18 | ||
| P04075 | Fructose biphosphate aldolase | MS | M, MD | 14 | ||
| P40926 | Malate dehydrogenase | MS | S, M, MD, T | 17 | ||
| Q06830 | Peroxiredoxin-1 | MS | S, B, TT | Yes | 15, 16, 17, 85 | |
| P30041 | Peroxiredoxin-6 | MS | B, M, MD, TT | 14, 15, 16, | ||
| P00558 | Phosphoglycerate kinase 1 | MS | S | Yes | Yes | 17, 85 |
| P14618 | Pyruvate kinase PKM2 | AA | B, M, MD | Yes | 14, 15, 16, 18, 85 | |
| P10599 | Thioredoxin | MS | S, B, TT | Yes | Yes | 15, 16, 17, 85 |
| P37837 | Transaldolase | MS | S | 17 | ||
| P29401 | TranSetolase | MS | S | 17 | ||
| P60174 | Triosephosphate isomerase | MS | B, M | Yes | 14, 15, 16 | |
| P25311 | Zinc-alpha-2-glycoprotein | MS, 2DMS, E | S, MC | Yes | 17 | |
| Q96DA0 | Zymogen granule protein | MS | Yes | 85 | ||
| Membrane Related Proteins | ||||||
| P04083 | Annexin A1 | AA | S, B, M, MD, TT | Yes | Yes | 15, 16, 17, 74, 85 |
| P46193 | Annexin I | MS, WB | S, B, M, MD | 15, 16, 17 | ||
| P07355 | Annexin II | MS, WB | S, B, M, MD, TT | Yes | Yes | 14, 18, 74, 85 |
| P12429 | Annexin A3 | MS | Yes | 14, 85 | ||
| P15154 | p21 Rac-1 | AA | S | 8, 17 | ||
| P11233 | Ras-related protein Ral-A | AA | TT | 8, 18 | ||
| Calcium Binding Related Proteins | ||||||
| P62158 | Calmodulin | MS | B, M, MD | Yes | Yes | 15, 16, 74, 85 |
| P26447 | S100-A4 | MS | S, B | 15, 16, 17 | ||
| P06703 | S100-A6 | MS | S | Yes | Yes | 17, 74, 85 |
| P31151 | S100-A7 | MS | S, TT | 16, 17 | ||
| Anti-Microbial Related Proteins | ||||||
| P59666 | Neutrophil defensin 3 | MS | Yes | 85, 87 | ||
| P15515 | Histatin-1 precursor | MS | Yes | 85 | ||
| Q9HC84 | Mucin-5B precursor | MS | Yes | 85 | ||
| Q8TAX7 | Mucin-7 | MS | Yes | 85 | ||
| P02814 | Submax. gland androgen-reg. prot. | MS, 2DMS, E | S, M, MB | 17 | ||
| P02788 | Lactotransferrin | MS | Yes | 85 | ||
| P15941 | Cancer antigen 15-3 | E | Yes | Yes | 29, 85 | |
Abbreviations: The UniProt identification number and protein name are from UniProt database (www.uniprot.org). Method of Identification: MS = Mass spectrometry, 2DMS = 2D-Gel Spot Mass Spectrometry, E = ELISA, WB = Western blot, AA = Antibody Array. Cell Lines: S = SKBR3, M = MCF7, T = T47D, MD = MB-MDA-231, B = 8701-BC, TT = Tumor Tissue. Ref. = References.
Additionally, information is provided as to which proteins are contain within both salivary and breast tissue exosomes. Seventy one (30%) proteins from the panel were found to be contained within salivary exosomes and 35 (15%) proteins were within breast cancer tissue exosomes. There were twenty seven (11%) proteins that were common to both the salivary and the breast tissue exosomes.
Table 3 displays the results from the GO and AmiGO analyses. It illustrates the overlapping functional diversity of the panel with respect to their associated molecular processes.
Table 3. Altered Salivary Proteins According to Molecular Function.
| Go Function | Go ID | Gene Frequency & % |
|---|---|---|
| Anatomical Structure Development | GO:0048856 | 94 of 204 genes, 46% |
| Biosynthetic Process | GO:0009058 | 63 of 204 genes, 46% |
| Carbohydrate Metabolic Process | GO:0005975 | 21 of 204 genes, 10% |
| Catabolic Process | GO:0009056 | 54 of 204 genes, 46% |
| Cell Adhesion | GO:0007155 | 26 of 204 genes, 13% |
| Cell Cycle | GO:0007049 | 22 of 204 genes, 11% |
| Cell to Cell Signaling | GO:0007267 | 19 of 204 genes, 9% |
| Cell Death | GO:0008219 | 65 of 204 genes, 46% |
| Cell Differentiation | GO:0030154 | 62 of 204 genes, 46% |
| Cell Proliferation | GO:0008283 | 50 of 204 genes, 25% |
| Cellular Component Assembly | GO:0022607 | 45 of 204 genes, 22% |
| Cellular Nitrogen Metabolic Process | GO:0034641 | 73 of 204 genes, 46% |
| Cellular Protein Modification Process | GO:0006950 | 53 of 204 genes, 46% |
| Chromosome Organization | GO:0051276 | 14 of 204 genes, 7% |
| Cytoskeleton | GO:0007010 | 25 of 204 genes, 12% |
| DNA Metabolic Process | GO:0006259 | 27 of 204 genes, 13% |
| Generation of metabolites & Energy | GO:0006091 | 14 of 204 genes, 7% |
| Growth | GO:0040007 | 26 of 204 genes, 13% |
| Homeostatic Process | GO:0042592 | 53 of 204 genes, 46% |
| Immune System | GO:0002376 | 83 of 204 genes, 42% |
| Lipid Metabolic Process | GO:0006629 | 24 of 204 genes, 12% |
| Membrane Organization | GO:0061024 | 18 of 204 genes, 9% |
| Morphogenesis | GO:0006950 | 25 of 204 genes, 12% |
| Response to Stress | GO:0006950 | 107 of 204 genes, 52% |
| Signal Transduction | GO:0007165 | 86 of 204 genes, 46% |
| Small Molecule Metabolic Process | GO:0044281 | 50 of 204 genes, 25% |
| Transmembrane Transport | GO:0055085 | 11 of 204 genes, 5% |
| Transport | GO:0006810 | 89 of 204 genes, 52% |
| Vesicle-mediated Transport | GO:0016192 | 49 of 204 genes, 24% |
Table 4 represents a sampling of some of the significant pathways calculated by the National Cancer Institute’s Pathway Interaction Database. The signaling events mediated by HDAC Class III was highly significant at the p < 0.01 × 10−16 level as was the glucocorticoid receptor regulatory network at the p < 0.01 × 10−6 level.
Table 4. National Cancer Institute’s Pathway Interaction Database Analysis.
| Pathway | Proteins | p value |
|---|---|---|
| Signaling events mediated by HDAC Class III | CDKN1A, HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, HIST2H4B, HIST4H4, TP53 | 6.22E-18 |
| Glucocorticoid receptor regulatory network | AKT1, CDKN1A, IFNG, IL4, IL6, IL8, KRT14, KRT17, KRT5, SFN, STAT1, TP53 | 6.73E-08 |
| VEGFR1 specific signals | AKT1, CALM1, CALM2, CALM3, NOS3, RASA1, VEGFA | 1.52E-06 |
| AP-1 transcription factor network | BAG1, CCL2, IFNG, IL10, IL4, IL6, IL8, MMP9, TP53 | 9.53E-06 |
| Caspase Cascade in Apoptosis | AKT1, APAF1, ARHGDIB, GSN, RIPK1, TNF, VIM, XIAP | 1.43E-05 |
| Insulin-mediated glucose transport | AKT1, CALM1, CALM2, CALM3, SFN, YWHAZ | 1.84E-05 |
| SHP2 signaling | EGF, EGFR, IFNG, IL6, LCK, NOS3, STAT1, VEGFA | 1.85E-05 |
| a6b1 and a6b4 Integrin signaling | AKT1, CDH1, EGF, EGFR, ERBB2, SFN, YWHAZ | 2.94E-05 |
| Angiopoietin receptor Tie2-mediated signaling | AKT1, CDKN1A, FGF2, FYN, NOS3, RASA1, TNF | 4.45E-05 |
| ErbB1 downstream signaling | AKT1, CALM1, CALM2, CALM3, EGF, EGFR, RALA, SFN, STAT1, YWHAZ | 5.51E-05 |
| IFN-gamma pathway | AKT1, CALM1, CALM2, CALM3, IFNG, STAT1 | 1.80E-04 |
| ErbB receptor signaling network | EGF, EGFR, ERBB2, TGFA | 2.31E-04 |
| HIF-1-alpha transcription factor network | AKT1, ALDOA, ENO1, PGK1, PKM, TF, VEGFA | 3.18E-04 |
| Calcium signaling in the CD4+ TCR pathway | CALM1, CALM2, CALM3, IFNG, IL4 | 3.20E-04 |
| LKB1 signaling events | CTSD, EZR, MAPT, SFN, TP53, YWHAZ | 3.29E-04 |
| Ceramide signaling pathway | AKT1, CTSD, EGF, EIF2AK2, RIPK1, TNF | 3.68E-04 |
| Direct p53 effectors | APAF1, CDKN1A, CTSD, EGFR, HSPA1A, HSPA1B, PCNA, SFN, TGFA, TP53 | 3.82E-04 |
| Signaling events mediated by PTP1B | AKT1, EGF, EGFR, FYN, LCK, TXN | 5.05E-04 |
| Signaling events mediated by VEGFR1 and VEGFR2 | AKT1, CALM1, CALM2, CALM3, FYN, NOS3, VEGFA | 5.30E-04 |
| EGF receptor (ErbB1) signaling pathway | EGF, EGFR, GSN, RASA1, STAT1 | 5.66E-04 |
The analyses for nodal & Her2/neu receptor status were also performed and published prior to this manuscript10,11. Briefly, the results yielded approximately 174 differentially expressed proteins in the saliva specimens lymph node status. There were 55 proteins that were common to both cancer stages in comparison to each other and healthy controls. In contrast, there were there were 20 proteins unique to Stage IIa and 28 proteins that were unique to Stage IIb10. The results Her2/neu receptor status yielded approximately 71 differentially expressed proteins in the saliva specimens. There were 34 up-regulated proteins and 37 down regulated proteins11.
The results of the Harvard Partners Center for Genetics and Genomics, Cambridge, MA., proteomic analyses confirmed our findings and provided us with additional markers for this manuscript12.
The ensuing paragraphs further detail each protein category and how the proteins of each category relate to carcinogenesis of the breast.
Genomic Integrity Related Proteins (10, 4%)
Ten salivary proteins related to genomic integrity were found to be variant (over-expressed) in the presence of carcinoma of the breast. Eight of the ten proteins were histones while the remaining two proteins were associated with genomic maintenance. Of the two aforementioned proteins, the TLS oncogene maintains genomic integrity and mRNA/microRNA processing, while the other protein, Proliferating Cell Nuclear Antigen, is implicated with DNA repair. Both proteins are involved in breast cancer progression19,20.
The remaining eight belong to the histone family of proteins. Histones are a group of basic proteins that are involved with nuclear DNA and help condense it into chromatin21. Histones are basic proteins, and their positive charges allow them to associate with DNA, which is negatively charged. Some histones function as molecular reels for the thread-like DNA to wrap around. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. The chain of nucleosomes is then wrapped into a 30 nm spiral called a solenoid, where additional H1 histone proteins are associated with each nucleosome to maintain the chromosome structure19,20,21,22,23,24,25.
Of particular interest within the group of histones is the H2A family. This group in particular is epigenetically associated with carcinoma of the breast26. The H2AX variant for example (Table 1), functions as a sensor of DNA damage and responds by defining the cellular response for DNA repair or apoptosis. It is also an indicator of tumor radio-sensitivity and is associated with BRCA1 and E-Cahedrin1 activity. E-Cahedrin1 was found to be down regulated in saliva25,26,27.
Histone 3.2 is also present in saliva in the protein profile and is associated with invasive ductal cell carcinoma28. It is routinely used in tumor cell immunohistochemistry to determine the grade of the tumor. The presence of the over-expressed H3, H2AX and p300 proteins may possibly explain the upregulated presence of p21(Waf−1) and CA 15-3 in saliva29.
Molecular Chaperones/Heat Shock proteins (14, 6%)
Heat shock proteins (HSPs), also referred to as stress proteins, are a group of cellular proteins that respond to extreme temperature changes, infection, inflammation and oxygen deprivation. The proteins function as molecular chaperones and assist in the folding and maintenance of newly translated proteins, the refolding of denatured proteins and the further unfolding of misfolded or destabilized proteins to assist in their degradation30. Alterations in HSP expression may also increase due to other sources of cellular stress, including osmotic stress and the unfolded protein response, mediated by the ATF family of transcription factors. HSP expression and function can be deregulated during pathophysiological processes such as breast carcinogenesis30.
Table 1 exhibit fourteen proteins that function as molecular chaperones. Included among these are the overexpressed proteins Hsp27, Hsp40 and Hsp70. Hsp70 is especially important as this chaperone can block the programmed cell death that often accompanies malignant transformation. Additionally, Hsp70 may be involved in mitotic spindle formation and cell proliferation31.
In addition to the heat shock proteins, there were a number of proteins associated with protein folding and the catalyzation of –S–S– bonds. Of importance is the protein disulfide isomerase (PDI), which has shown to be upregulated in breast cancer tissues30,31.
Growth Factors and Their Receptors (19, 7%)
Growth factors are polypeptides that stimulate cell proliferation by binding to membrane receptors; EGF for example has been cited as a salivary protein for nearly four decades and is recognized as being up-regulated in the presence of carcinoma of the breast along with the solubilized receptors EGFR1 and Her2/neu32,33. TGFα, another ligand of the EGF/Her2 signaling pathway, was found to over-expressed. Within the EGF/Her2 signaling pathway, the following downstream proteins were overexpressed: AKT1, CALM, CDH1, CALM3, EGF, EGFR, GRB2, MUC1, PAX, RAC1, STAT1, and UB34.
VEGF, a growth factor implicated in angiogenesis was also upregulated as were its associated downstream proteins AKT1, FGF2, FYN, NOS1, NOS3, p21, TNFα and VAV335.
Apoptotic Related Proteins (13, 5%)
There were thirteen proteins related to apoptosis that are presented in Table 1. Among the thirteen proteins there were five prominent proteins of the intrinsic apoptotic pathway: p53, Apaf-1, 14-3-3δ, Xiap and p21WAF−1. p53 and Apaf-1 were down regulated while14-3-3δ, Xiap and p21WAF−1 were up-regulated. This pattern appears to be consistent with cancer progression as cellular proliferation is one the hallmarks of this disease36. For example XIAP is a protein that impedes apoptotic cell death. XIAP is a member of the inhibitor of apoptosis family of proteins and is the most potent human IAP protein currently identified. In addition, there is the up-regulation of 14-3-3δ among this group of proteins. 14-3-3δ is a p53 inhibitor via Mdm2 inactivation. Additionally, when bound to KRT17, it regulates protein synthesis and cell growth by stimulating the Akt/mTOR pathway36,37,38.
Finally, the up-regulated presence of the p21WAF−1 protein suggests a possible anti-apoptotic role by suppressing pro-apoptotic genes. Phosphorylated p21WAF1 expression, for example in breast cancer, may be associated with an inability of p21WAF1 to inhibit cell cycle progression38,39,40. Additionally, p21WAF1, when phosphorylated via PI3K pathway may bind with PCNA and facilitate the inactivation of capase-3 which is essential in the apoptotic process. It can also bind with PCNA inhibiting DNA repair. The p21WAF1 may also be unable to regulate Cdc25C binding with PCNA which is necessary for cell cycle arrest the G2/M checkpoint41,42. Collectively, the aforementioned panel of proteins may have utility in assessing anti-apoptotic activity in cancer progression36,37,38,39,40,41,42,43.
Chronic Inflammatory/Immunoresponse Proteins (53, 23%)
Fifty three (23%) of the salivary proteins identified in Table 1 were associated with the presence of a Chronic Inflammatory Response. Of the 53 proteins of this group, 10 (19%) were Th1/Th2 cytokines. The presence of VEGF, IL-10 and IL-6 may be a consequence of mutations in the serine/threonine-protein kinase B-Raf pathway (BRAF)44,45. Additionally, mutations in the BRAF oncogene also promotes the secretion of IL-1β, an innate inflammatory cytokine mediator which can drive neoplastic cells to up-regulate molecules that inhibit the function of anti-tumor lymphocytes. This may also be the reason for increased salivary presence of IL-1RA among cancer patients44,45.
Additionally, AKT1, CDKN1A, IFNγ, IL4, IL6, IL8, KRT14, KRT17, KRT5, SFN, STAT1 and p53 are associated with numerous overlapping pro-inflammatory, immunological pathways such as the glucocorticoid receptor regulatory network (GR) and the IFN-γ/Stat1 pathway44,45,46,47. The activated GR complex up-regulates the expression of anti-inflammatory proteins in the nucleus or represses the expression of pro-inflammatory proteins in the cytosol by preventing the translocation of other transcription factors from the cytosol into the nucleus. This coupled with the up-regulated activities of the IFN-γ/Stat1 pathway and the increase concentrations of TNF-α and IFN-γ may be significant as this suggests possible T cell exhaustion from constant exposure to tumor antigens or the absence of the HLA-A2 allele44,45,46,47.
Cytoskeletal (40, 17%)
The cytoskeleton is present in all eukaryotic cells and provides the cell with structure, shape, mobility and by excluding macromolecules from some of the cytosol. It is also involved in cell migration and is believed to be involved in tumor dissemination48. Cytoskeleton protein complexes are also important regulators of migration, angiogenesis, cell polarity, cell morphology, intracellular trafficking and signal transduction. Many cytoskeleton proteins associated with cancer and are currently being used as histopathological biomarkers48,49,50.
The most abundant group in Table 1 is the cytokeratins. There are 16 cytokeratins among the cytoskeletal proteins. Nine of the cytokeratins are acidic Type 1 cytokeratins while seven are neutral Type II cytokeratins. The wide range of cytokeratin expression may result from the different types of salivary tissues contributing to the composition of whole saliva i.d., serous, mixed and mucinous secretions. However, similar to salivary gland ducts, normal breast ducts contain at least 3 types of epithelial cells: luminal (glandular) cells, basal/myoepithelial cells and stem cells50. Myoepithelial and luminal epithelia can be distinguished by their different cytokeratin expression patterns. Myoepithelial cells typically express cytokeratin 5/6 and cytokeratin 17, while luminal cells typically express cytokeratins 8 and 18. A small fraction of breast cancers express CK5 together with its major partners CK14 and CK17. Of particular interest is the presence of Cytokeratins 5, 14 and 17 as they are generally associated with poor prognosis and short disease-free survival48,49,50,51.
Within the salivary group of cytoskeletal proteins, of particular interest, are the proteins E-cadherin, γ-catenin, gesolin and vimentin which were down regulated in saliva. Together, these proteins are associated with the noncanonical planar cell polarity pathway which regulates the cytoskeleton that is responsible for the shape of the cell. E-cadherin to regulate b-catenin signaling in the canonical Wnt pathway; its potential to inhibit mitogenic signaling through growth factor receptors and the possible links between cadherins and the molecular determinants of epithelial polarity52,53. Each of these potential mechanisms provides insights into the complexity that is likely responsible for the tumor-suppressive action of E-cadherin52,53.
Metabolic Proteins (56, 24%)
The metabolic proteins represent the largest of the nine categories of salivary proteins that were changed as a consequence of the presence of breast cancer. Table 1 shows that the 56 metabolic protein composed of 20 enzymes, 10 enzyme inhibitors, 10 transport proteins, 4 detoxification and redox proteins and 14 proteins of miscellaneous metabolic functions.
One interesting fact concerning the array of metabolic proteins is that alpha enolase, glucose-6-phosphate isomerase, fructose biphosphate aldolase, malate dehydrogenase, phosphoglycerate kinase 1, and triophosphate isomerase, are all associated with the glycolytic anaerobic pathway54,55,56,57,58,59. However, it is the presence of pyruvate kinase (PKM2) which suggests the occurrence of the Warburg effect54,55. The Warburg effect, suggested by Otto Warburg in 1927, suggests that among cancer cells there is a shift from ATP generation through oxidative phosphorylation to ATP generation through glycolysis even in the presence of normal oxygen concentrations54,55,56,57,58,59,60. The presence of the PKM2 protein suggests the occurrence of a glycolytic shift which is the metabolic hallmark of proliferating cells54,55,56,57,58,59,60,61,62.
A pathway analysis was performed on the list of these proteins and it revealed that ALDOA, ENOA, PGK1 and PKM2 are members of the Hypoxia-Inducible transcription Factor-1 alpha network (HIF-1α). The HIF-1α pathway deregulation can produce consequences in disease settings with a chronic inflammatory component. It has also been shown that chronic inflammation is self-perpetuating and that it distorts the microenvironment as a result of aberrantly active transcription factors. As a consequence, alterations in growth factor, chemokine, cytokine, and ROS balance occur within the cellular milieu that in turn provide the axis of growth and survival needed for de novo development of cancer and metastasis. It promotes angiogenesis and is consistent with the hypoxic events associated with the Warburg effect54,55,56,57,58,59,60,61,62,63.
Membrane and Calcium Binding Related Proteins (18, 7%)
The most numerous proteins within this category are the annexins and the S100 family of proteins. The annexins are a large family of proteins which can be both intra and extracellular in presence having a wide variety of physiological functions64. They can be associated with membrane scaffolding, which is relevant to changes in the cell’s shape and have been shown to be involved in trafficking and organization of vesicles exocytosis. They are also associated with calcium ion channel formation. More importantly, annexins are associated with inflammation and apoptosis. With respect to carcinoma of the breast, the over expression of ANXA1, ANXA2, ANXA3 have been implicated with this malignancy64,65,66. ANXA1, expression, for example, was significantly associated with disease progression and metastases. ANXA2 and ANXA3 have also been related to poor prognosis and may have potential as prognostic indicators67,68.
S100 proteins, similar to the annexins, are implicated in a wide variety of intracellular and extracellular functions. For example, they are involved in regulation of protein phosphorylation, transcription factors, cytoskeleton dynamics, cell growth/differentiation, and the inflammatory response69,70,71,72.
A comprehensive study concerning the relationship between S100 proteins and breast cancer was conducted by Cancemi et al. in 201072. The study identified the up-regulation of S100 protein expression in breast cancer tissues. Specifically, they observed S100A2, S100A4, S100A6, S100A7, S100A8, S10011 and S10013 as being disparate in the presence of breast cancer. Similarly, the proteins S100A4, S100A6, S100A7, S100A8 and S10011 were over expressed in saliva72.
Anti-Microbial Proteins (14, 6%)
Table 1 illustrates 14 proteins that were not similar due to the presence of carcinoma of the breast. As illustrated, there are three mucins, which in health protect the integrity of the epithelia provide a barrier against microbial invasion. The balance of the panel consists of proline-rich proteins, lysozymes and histatins which have been documented in the dental literature as having antibacterial properties73,74.
The reason for their alteration in expression is unknown and may be due to changes in the oral microbiota75,76. As to whether the oral microbiome was altered as a consequence of tumor development or was an etiological cause for tumorigenesis to begin with, is also unknown. Our microbiota might be considered unknown variable at present, but they are likely to become more familiar considering the accelerated pace of research in this area75,76,77,78.
Study Limitations
The manuscript presents a “proof of concept”, but has its limitations. Further study is required to address the phenotypic diversity breast cancer tumors. Tumors that are of different histological types, pathological grade and molecular subtypes should be proteomically assessed. Late stage tumors should also be studied and proteomically characterized according to the metastatic site of the tumor. High-throughput analysis is necessary in order to assay numerous individual specimens, where pooled specimens were used here to limit expense. The final task would be to combine the genomic, proteomic and other omic profiles together in an attempt to obtain a broader vision of how a carcinoma progresses.
Conclusions
The authors of this manuscript have presented a catalogue of salivary proteins which are altered in concentration as a consequence of the presence of ductal carcinoma of the breast. These findings are supported by other investigators employing proteomic analysis of breast cancer cell lines, breast cancer tissues, tissue microenvironment and serum79,80,81. Additionally, nearly 29% of the panel of proteins has been technically validated by either western blot or by ELISA. A breakdown of these proteins has also been analyzed according staging and Her2/neu receptor status12,82. The investigators have also found that the protein concentrations can be modulated while undergoing cancer treatment and respond differently according pathological cell type9.
The main question arising from this line of research is how can tumors that are remote from the oral cavity influence salivary protein profiles? The prominent hypothesis to explain the aforementioned phenomenon is that exosomes which are shed and escape extracellular breakdown diffuse throughout the body and appear in biological fluids such as blood, urine, saliva semen breast milk, nipple aspirates and malignant effusions83. In doing so, they initiate exosome-mediated cell-to-cell communication by fusing with membranes of their cellular targets84,85. Upon entering the cellular he cytoplasm, their contents are released and activate downstream events in recipient cells73. It is also possible that extracellular proteases degrade the exosomes and their contents become soluble ligands binding to cell surface receptors. Experimentally, it has been demonstrated by Lau et al. that breast cancer-derived exosomes communicated and activated transcription within salivary gland cells and alter the proteomic composition of salivary gland cell-derived exosomes86.
Table 2 illustrates the presence of proteins carried by both salivary gland cell-derived exosomes and breast cancer-derived exosomes with 20 of the proteins being common to both cell types. The finding is circumstantial at this point in time as exosome research is in its infancy; however, as research continues in this field one could expect further support for the proposed mechanism for salivary protein alterations73,74,83,84,85.
In summary, the manuscript proposes a novel method for studying breast cancer progression which includes the tumor microenvironment and inflammatory progress87. Clinically, many of the markers have shown utility for monitoring treatment efficacy and tumor recurrence. Several proteins such as Lung Resistance Protein may be useful as a prognostic indicator. Additionally, key pathway markers such as EGFR, E-Cadherin, p53, Apaf-1, 14-3-3δ, Xiap, p21WAF−1 and others could be placed in a microarray assay and render a probability (risk assessment) of having carcinoma of the breast rather than a “yes or no” response which is subject to many false positive and negative assessments.
Further research is required in order determine the effects of tumor and tumor receptor phenotypes and the alteration of the panel as the tumor progresses from one stage to the next. It is also important to know which proteins from the profile are associated with tumor dormancy and tumor resistance to therapy. Considering that the salivary protein profile was modified secondary to the presence of a tumor remote from the oral cavity, it may provide information on the metastatic process which is the main cause of death for breast cancer patients.
Methods and Materials
Proteomic Design
The investigators analyzed pooled, stimulated whole saliva specimens. Each pooled specimen within a cohort consisted of ten individual patient saliva specimens from a bank of control and cancer specimens frozen at −80 °C. One pooled saliva specimen consisted of ten specimens from ten healthy volunteers, another specimen was a pooled saliva specimen from ten subjects diagnosed with ductal carcinoma in situ (DCIS)10. Similarly assembled pooled specimens came from Stage I, Stage IIa, Stage IIb, Her2/neu positive and Her2/negative ductal carcinoma volunteers11,12. The cancer cohort, internally, was estrogen and progesterone receptor status negative as determined by the pathology report. All subjects were closely matched for age and race and were non-tobacco users.
The study was conducted in accordance with the Declaration of Helsinki, and the University of Texas Health Science Center Institutional Review Board approved the protocol and informed consent form prior to study initiation. All participating volunteers were explained their participation rights and signed an IRB consent form. The saliva specimens and related patient data are non-linked and bar coded in order to protect patient confidentiality. This study was performed under the UTHSC IRB approved protocol# HSC-DB-05-0394.
To increase accuracy and assure reproducibility, a duplicate set of specimens were sent to the Harvard Partners Center for Genetics and Genomics, Cambridge, MA., and were proteomically analyzed using both bottom-up and gel based approaches. A Thermo Finnigan LTQ FT ICR Hybrid Mass Spectrometer was used for the proteome analysis.
In order to present a complete catalogue of salivary cancer related proteins, proteins from previous studies were added to this analysis. These proteins were determined by either antibody array and/or by ELISA8,9,10. The reason for adding these antibody based protein determinations was to provide information concerning the presence of some of the low abundance proteins that were changed secondary to carcinoma of the breast.
Saliva Collection and Sample Preparation
Stimulated whole salivary gland secretion is a reflex response occurring during the mastication of a bolus of food. Usually, a standardized bolus (1 gram) of paraffin or a gum base (generously provided by the Wrigley Co., Peoria, IL) is given to the subject to chew at a regular rate. The individual, upon sufficient accumulation of saliva in the oral cavity, expectorates periodically into a preweighed disposable plastic cup between the hours of 8:00 a.m. and 5:00 p.m. As this is a reflexive collection, circadian rhythms are not a factor on salivary flow rates10. This procedure is continued for a period of five minutes. The volume and flow rate is then recorded along with a brief description of the specimen’s physical appearance11. Saliva specimens tainted with the presence of blood were not used in the study. The cup with the saliva specimen is reweighed and the flow rate determined gravimetrically. The specimens were placed on ice and immediately transported from the clinic to the laboratory for processing. Specimens were collected by one calibrated individual working in the same location.
The specimens were aliquoted and centrifuged in an Eppendorf centrifuge 5415R with temperature control (4 °C) for five minutes in order to remove debris and any unwanted particulates. The supernants were removed and a protease cocktail inhibitor (trypsin, calpain, papain, cathepsin B, chymotrypsin, kallikrein, human leukocyte elastase and aminopeptidases) from Sigma Co (St. Louis, MI, USA) was added along with enough dithiothreitol from a 1 M stock solution to bring its concentration 1 mM. The 1 ml aliquots were frozen at −80 °C11.
Bottom-Up Mass Spectrometry Using iTRAQ Labeling
Briefly, the saliva samples were thawed and immediately centrifuged to remove insoluble materials10,11,12. The supernatants were assayed for protein using the Bio-Rad protein assay (Hercules, CA, USA) and an aliquot containing 100 μg of each specimen was precipitated with 6 volumes of −20 °C acetone. Specimens were normalized for analysis by using total protein concentrations. The precipitate was resuspended and treated according to the manufacturer’s instructions. Protein digestion and reaction with iTRAQ labels were carried out as previously described and according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). Briefly, the acetone precipitable proteins were centrifuged in a table top centrifuge at 15,000 × g for 20 minutes. The acetone supernatants were removed and their pellets resuspended in 20 ul dissolution buffer. The soluble fractions were denatured and the disulfides reduced by incubation in the presence of 0.1% SDS and 5 mM TCEP (tris-(2-carboxyethyl)phosphine)) at 60 °C for one hour. Cysteine residues were blocked by incubation at room temperature for 10 minutes with MMTS (methyl methane-thiosulfonate). Trypsin was added to the mixture to a protein: trypsin ratio of 10:1. The mixtures were incubated overnight at 37 °C. The protein digests were labeled by mixing with the appropriate iTRAQ reagent and incubating at room temperature for one hour. On completion of the labeling reaction, the four separate iTRAQ reaction mixtures were combined. Since there are a number of components that can interfere with the LC-MS/MS analysis, the labeled peptides were partially purified by a combination of strong cation exchange followed by reverse phase chromatography on preparative columns. The combined peptide mixtures were diluted 10 fold with loading buffer (10 mM KH2PO4 in 25% acetonitrile at pH 3.0) and applied by syringe to an ICAT Cartridge-Cation Exchange column (Applied Biosystems, Foster City, CA) column that has been equilibrated with the same buffer.
The column is washed with 1 ml loading buffer to remove contaminants. To improve the resolution of peptides during LC-MS/MS analysis, the peptide mixtures were partially purified by elution from the cation exchange column in 3 fractions. Stepwise elution from the column was achieved with sequential 0.5 ml aliquots of 10 mM KH2PO4 at pH 3.0 in 25% acetonitrile containing 116 mM, 233 mM and 350 mM KCl respectively. The fractions were evaporated Speed Vac to about 30% of their volume to remove the acetonitrile and then slowly applied to an Opti-Lynx Trap C18 100 ul reverse phase column (Alltech, Deerfield, IL) with a syringe. The column was washed with 1 ml of 2% acetonitrile in 0.1% formic acid and eluted in one fraction with 0.3 ml of 30% acetonitrile in 0.1% formic acid. The fractions were dried by lyophilization and resuspended in 10 ul 0.1% formic acid in 20% acetonitrile. Each of the three fractions was analyzed by reverse phase LC-MS/MS.
Reverse Phase LC-MS/MS
The desalted and concentrated peptide mixtures were quantified and identified by nano-LC-MS/MS on an API QSTAR XL mass spectrometer (ABS Sciex Instruments) operating in positive ion mode. The chromatographic system consists of an UltiMate nano-HPLC and FAMOS autosampler (Dionex LC Packings). Peptides were loaded on a 75 μm × 10 cm, 3 μm fused silica C18 capillary column, followed by mobile phase elution: buffer (A) 0.1% formic acid in 2% acetonitrile/98% Milli-Q water and buffer (B): 0.1% formic acid in 98% acetonitrile/2% Milli-Q water. The peptides were eluted from 2% buffer B to 30% buffer B over 180 minutes at a flow rate 220 nL/min. The LC eluent was directed to a NanoES source for ESI/MS/MS analysis. Using information-dependent acquisition, peptides were selected for collision induced dissociation (CID) by alternating between an MS (1 sec) survey scan and MS/MS (3 sec) scans. The mass spectrometer automatically chooses the top two ions for fragmentation with a 60 s dynamic exclusion time. The IDA collision energies parameters were optimized based upon the charge state and mass value of the precursor ions.
Random control and cancer specimens from the specimen bank were selected and blindly sent for proteomic analysis to ascertain quantification repeatability and to address issues of variability, proteomic inconsistency and issues (pooled variance) surrounding the use of pooled specimens. Additionally, western blots were performed on both pooled and individual specimens for technical validation88,89,90,91.
Bioinformatics and Statistical Methods
The accumulated LC-MS/MS spectra were analyzed by ProQuant and ProGroup software packages (Applied Biosystems) using the SwissProt database for protein identification. The ProQuant analyses were carried out with a 75% confidence cutoff with a mass deviation of 0.15 Da for the precursor and 0.1 Da for the fragment ions. The ProGroup reports were generated with a 95% confidence level for protein identification.
The Swiss-Prot database was employed for protein identification while the PathwayStudio® bioinformatics software package was used to determine Venn diagrams were also constructed using the NIH software program (http://ncrr.pnl.gov). Pathways were retrieved from three databases: DAVID, KEGG, BioCarta, and the NCI’s Protein Interaction Database (PID)86,92,93,94,95. Gene ontologies were determined by employing the GO and AmiGO databases95,96.
Routine statistical evaluations were performed using the IBM SPSS Statistics 23 software. These evaluations include frequency, cross-tabulations and descriptive statistics. Mean comparisons were performed using parametric statistical analysis.
Additional Information
How to cite this article: Streckfus, C. F. and Bigler, L. A Catalogue of Altered Salivary Proteins Secondary to Invasive Ductal Carcinoma: A Novel In Vivo Paradigm to Assess Breast Cancer Progression. Sci. Rep. 6, 30800; doi: 10.1038/srep30800 (2016).
Acknowledgments
The research presented in this manuscript was supported by the Avon Breast Cancer Foundation (#07-2007-071), Komen Foundation (KG080928), Gillson-Longenbaugh Foundation and the Texas Ignition Fund. The authors would also like to thank Dr. William Dubinsky of the UT School of Medicine and Dr. David Sarracino of the Harvard Partners Center for Genetics and Genomics, Cambridge, MA., for the LC-MS/MS salivary mass spectrometry analyses.
Footnotes
Author Contributions C.F.S. and L.B. have been colleagues regarding salivary biomarker research for twenty years and shared efforts in the production of this manuscript. Both have contributed to its writing and editing.
References
- Kiberstis P. & Roberts L. A race still unfinished. Science 343, 1451 (2014). [DOI] [PubMed] [Google Scholar]
- Eccles S. A. et al. Clinical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. R92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni Y. M., Suarez V. & Klinke D. J. Inferring predominant pathways in cellular models of breast cancer using limited sample proteomic profiling. BMC Cancer 10, 291–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo-Gogola T. & Rosen J. M. Modelling breast cancer: one size does not fit all. Nature Reviews Cancer 7, 659–672 (2007). [DOI] [PubMed] [Google Scholar]
- Tordai A. et al. Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast Cancer Res. 10, R37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L. Mouse models for breast cancer. Breast Cancer Res. 2(1), 2–7 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia G., Cruz-Manos W., Man S., Xu P. & Kerbel R. S. Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat. Rev. Cancer 11, 135–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigler L. G. & Streckfus C. F. A unique protein screening analysis of stimulated whole saliva from normal and breast cancer patients. Preclinica 2(1), 52–56 (2004). [Google Scholar]
- Streckfus C. F. & Bigler L. The Use of soluble, salivary c-erbB-2 for the detection and post-operative follow-up of breast cancer in women: The results of a five year translational study. J. Adv. Dental Res. 18, 17–22 (2005). [DOI] [PubMed] [Google Scholar]
- Streckfus C. F. et al. Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Invest. 26(2), 159–167 (2008). [DOI] [PubMed] [Google Scholar]
- Streckfus C. F., Bigler L., Storthz K. & Dubinsky W. P. A comparison of the oncoproteomic profiles in pooled saliva specimens from individuals diagnosed with Stage IIa and Stage IIb ductal carcinoma of the breast and healthy controls. J. Oncology 1–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streckfus C. F., Arreola D., Edwards C. & Bigler L. A comparison of salivary protein profiles between her2/neu receptor positive and negative breast cancer patients: support for using salivary protein profiles for modeling breast cancer progression. J. Oncology, Article ID 413256, 9 pages, http://dx.doi.org/10.1155/2012/413256 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski M. & Anderson N. L. A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights 2, 1–48 (2006). [PMC free article] [PubMed] [Google Scholar]
- Chou H. C. & Chan H. L. Proteomic analysis of potential breast cancer biomarkers in Breast Cancer - Recent Advances in Biology, Imaging and Therapeutics (ed. Done S. J.) (InTech, 2011). [Google Scholar]
- Minafra I. P. et al. Expanding the protein catalogue in the proteome reference map of human breast cancer cells. Proteomics 6, 2609–2625 (2006). [DOI] [PubMed] [Google Scholar]
- Minafra I. P. et al. Proteomic profiling of 13 paired ductal infiltrating breast carcinomas and non-tumoral adjacent counterparts. Proteomics Clin. Appl. 1, 118–129 (2007). [DOI] [PubMed] [Google Scholar]
- Wu S., Hancock W., Goodrich G. & Kunitake S. An approach to the proteomic analysis of a breast cancer cell line (SKBR-3). Proteomics 3, 1037–1046 (2003). [DOI] [PubMed] [Google Scholar]
- Somiari R. I. et al. High – throughput proteomic analysis of infiltrating ductal carcinoma of the breast. Proteomics 3, 1863–1873 (2003). [DOI] [PubMed] [Google Scholar]
- Bernstein C., Nfonsam V., Prasad A. & Bernstein H. Epigenetic field defects in progression to cancer. World J. Gastrointest. Oncol. 5(3), 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoimenov I. & Helleday T. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37(3), 605–613 (2009). [DOI] [PubMed] [Google Scholar]
- Munot K. et al. Pattern of expression of genes linked to epigenetic silencing in human breast cancer. Human Pathol. 37, 989–999 (2006). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. & Ohm J. E. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 6, 107–116 (2006). [DOI] [PubMed] [Google Scholar]
- Herman J. G. & Baylin S. B. Gene silencing in cancer in association with promoter hypermethylation. New Engl. J. Med. 349, 2042–2054 (2003). [DOI] [PubMed] [Google Scholar]
- Elsheikh S. et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 69(9), 3802–3809 (2009). [DOI] [PubMed] [Google Scholar]
- Monteiro A., Zhang S., Phelan C. & Narod S. Absence of constitutional H2AX gene mutations in 101 hereditary breast cancer families. J. Med. Genet. 40, e51 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Nayak S., Jankowitz J., Davidson N. & Oesterreich S. Epigenetics in breast cancer. Breast Cancer Res. 13(6), 225–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M. & Castilho R. Histones: controlling tumor signaling circuitry. J. Carcinogene Mutagene S5, 001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. et al. p21-actvated kinase-1 interacts with and phosphorylates H3 in breast cancer cells. EMBO Reports 3(8), 767–773 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood S., Khaleque A., Sawyer D. & Ciocca D. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends in Biochemical Sciences 31(3), 164–172 (2006). [DOI] [PubMed] [Google Scholar]
- Vargas-Roig L. M. et al. Heat shock proteins and cell proliferation in human breast cancer biopsy samples. Cancer Detect. Prev. 21(5), 441–451 (1997). [PubMed] [Google Scholar]
- Updike M. S. et al. Primary cultured human breast epithelial cells up-regulate protein disulfide isomerase in response to zeranol. Anticancer Res. 27(1A), 407–410 (2007). [PubMed] [Google Scholar]
- Streckfus C. F., Bigler L., Tucci M. & Thigpen J. T. The presence of CA 15-3, c-erbB-2, EGFR, Cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest. 18(2), 101–109 (2000). [DOI] [PubMed] [Google Scholar]
- Navarro M. A. et al. Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Can. Res. Treat. 42, 83–86 (1997). [DOI] [PubMed] [Google Scholar]
- Skálová A. et al. Mammary analogue secretory carcinoma of salivary glands with high-grade transformation: report of 3 cases with the ETV6-NTRK3 gene fusion and analysis of TP53, β-catenin, EGFR, and CCND1 genes. Am. J. Surg. Pathol. 38(1), 23–33 (2014). [DOI] [PubMed] [Google Scholar]
- Brooks M. N. et al. Salivary protein factors are elevated in breast cancer patients. Mol. Med. Rep. 1(3), 375–8 (2008). [PMC free article] [PubMed] [Google Scholar]
- Adams R., Dellinger T., Kuhn M., Streckfus C. & Bigler L. p53 as a Diagnostic tool for the detection of cancer. J. Miss. Acad. Sci. 46(4), 163–167 (2001). [Google Scholar]
- Yang W., Dicker D. T., Chen J. & El-Deiry W. S. ARPs enhance p53 turnover by degrading 14-3-3 sigma and stabilizing MDM2. Cell Cycle 7, 670–682 (2008). [DOI] [PubMed] [Google Scholar]
- Mhawech P. 14-3-3 proteins-an update. Cell Research 15(4), 228–236 (2005). [DOI] [PubMed] [Google Scholar]
- Ando T. et al. Involvement of the interaction between p21 and Proliferating Cell Nuclear Antigen for the maintenance of G2/M arrest after DNA damage. J. Biol. Chem. 276(46), 42971–42977 (2001). [DOI] [PubMed] [Google Scholar]
- Piccolo M. & Crispi S. The dual role played by p21 may influence the apoptotic or anti-apoptotic fate in cancer. J. Cancer Res Updates 1, 189–202 (2012). [Google Scholar]
- Albert H. et al. Differential expression of CDC25 phosphatases splice variants in human breast cancer cells. Clin. Chem. Lab. Med. 49(10), 1707–1714 (2011). [DOI] [PubMed] [Google Scholar]
- Kristjansdottir K. & Rudolph J. Cdc25 Phosphatases and cancer. Chem. & Biol. 11, 1043–1051 (2004). [DOI] [PubMed] [Google Scholar]
- Harper J. & Elledge S. The DNA damage response: ten years after. Molecular Cell 28, 739–745 (2007). [DOI] [PubMed] [Google Scholar]
- Harris T. J. & Drake C. G. Primer on tumor immunology and cancer immunotherapy. J. Immunotherapy Cancer 1(12), 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Banuch A. Inflammatory cells, cytokines and chemokines in breast cancer progression: reciprocal tumor-microenvironment interactions. Breast Cancer Res. 5, 31–36 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Liu S. & Wicha M. S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Invest. 121(10), 3804–3809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allinen M. et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6, 17–32 (2004). [DOI] [PubMed] [Google Scholar]
- Moll R. Cytokeratins in the histological diagnosis of malignant tumors. Int. J. Biol. Markers 9, 63–69 (1994). [DOI] [PubMed] [Google Scholar]
- Moll R., Krepler R. & Franke W. W. Complex cytokeratin polypeptide patterns observed in certain human carcinomas. Differentiation 23(3), 256–269 (1983). [DOI] [PubMed] [Google Scholar]
- Abd El-Rehim D. M. et al. Expression of luminal and basal cytokeratins in human breast carcinoma. J. Pathol. 203(2), 661–671 (2004). [DOI] [PubMed] [Google Scholar]
- Gusterson B. A., Ross D. T., Heath V. J. & Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 7(4), 143–148 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes A., Gottardi C. & Yap A. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol J. P., Neil J. R., Schiemann B. J. & Schiemann W. P. The use of cystatin C to inhibit epithelial–mesenchymal transitionand morphological transformation stimulated by transforming growth factor-β. Breast Cancer Res. 7, R844–R853 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- Warburg O., Franz-Wind F. & Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 8(6), 519–530 (1927). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci-Minafra I., Fontana S., Cancemi P., Alaimo G. & Minafra S. Proteomic patterns of cultured breast cancer cells and epithelial mammary cells. Ann. N.Y. Acad. Sci. 963, 122–139 (2002). [DOI] [PubMed] [Google Scholar]
- Koukourakis M. I., Giatromanolaki A., Harris A. L. & Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 66(2), 632–637 (2006). [DOI] [PubMed] [Google Scholar]
- Dang C. V. Links between metabolism and cancer. Genes Dev. 26, 877–890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns R. A., Harris I. A. & Mak T. W. Regulation of cancer cell metabolism. Cancer Res. 66(2), 632–637 (2006). [DOI] [PubMed] [Google Scholar]
- Mazurek S., Boschek C. B., Hugo F. & Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 15(4), 300–308 (2005). [DOI] [PubMed] [Google Scholar]
- Christofk H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M. & Cantley L. C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 (2008). [DOI] [PubMed] [Google Scholar]
- Porporato P. E., Dhup S., Dadhich R. K., Copetti T. & Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Frontiers in Pharmacology: Pharmacology of Anti-Cancer Drugs 2(49), 1–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V. & Moss S. E. Annexins: from structure to function. Physiol. Rev. 82(2), 331–371 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu S., Guo C., Zong J. & Sun M. Z. The association of annexin A2 and cancers. Clin. Transl. Oncol. 14(9), 634–640 (2012). [DOI] [PubMed] [Google Scholar]
- Wang L. P. et al. Annexin A1 expression and its prognostic significance in human breast cancer. Neoplasma 57(3), 253–259 (2010). [DOI] [PubMed] [Google Scholar]
- Zeng C. et al. Annexin A3 is associated with a poor prognosis in breast cancer and participates in the modulation of apoptosis in vitro by affecting the Bcl-2/Bax balance. Exp. Mol. Path. 95(1), 23–31 (2013). [DOI] [PubMed] [Google Scholar]
- de Graauw M. et al. Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc. Natl. Acad. Sci. USA 107(14), 6340–6345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- West N. R. & Watson P. H. S100A7 (psoriasin) is induced by the proinflammatory cytokines oncostatin-M and interleukin-6 in human breast cancer. Oncogene 29(14), 2083–2092 (2010). [DOI] [PubMed] [Google Scholar]
- Arai K. et al. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr. Cancer Drug Targets 8(4), 243–252 (2008). [DOI] [PubMed] [Google Scholar]
- Jiang W. G., Watkins G., Douglas-Jones A. & Mansel R. E. Psoriasin is aberrantly expressed in human breast cancer and is related to clinical outcomes. Int. J. Oncol. 25(1), 81–85 (2004). [PubMed] [Google Scholar]
- Cancemi P. et al. Large-scale proteomic identification of S100 proteins in breast cancer tissues. BMC Cancer 10, 476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. & Wong D. Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell derived exosome-like microvesicles in vitro. PLoS ONE 7(3), e33037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger S. et al. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 14(44), 1–10, www.biomedcenter.com/1471-2407/14/44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe R. F. & Jobin C. The microbiome and cancer. Nature Reviews Cancer 13, 800–812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. & Tribble G. Chapter Five: Salivary diagnostics and the oral microbiome in Advances in Salivary Diagnostics (ed. Streckfus C. F.) 83–119 (Springer Press, 2015). [Google Scholar]
- Xuan C. et al. Microbial dysbiosis is associated with human breast cancer. PloS ONE 9(1), e83744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S. J. Emerging roles of the microbiome in cancer. Carcinogenesis 35(2), 249–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfkuhle J. D. et al. Proteomics of human breast carcinoma in situ. Cancer Res. 62, 6740–6749 (2002). [PubMed] [Google Scholar]
- Alldridge L. et al. Proteome profiling of breast tumors by gel electrophoresis and nanoscale electrospray ionization mass spectrometry. J. Proteome Res. 7(4), 1458–1469 (2008). [DOI] [PubMed] [Google Scholar]
- Hudelist G. et al. Proteomic analysis in human breast cancer: Identification of a characteristic protein expression profile of malignant breast epithelium. Proteomics 6, 1989–1902 (2006). [DOI] [PubMed] [Google Scholar]
- Schaefer J. S., Arreola D., Rendon M., Bigler L. & Streckfus C. F. Salivary cytokine alterations secondary to breast carcinoma. J. Dent. Res., 86(A), 1651 (2007). [Google Scholar]
- Lässer C. et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Translat. Med. 9, 9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzolo G. et al. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer. Res., 32(3), 847–860 (2012). [PubMed] [Google Scholar]
- Ogawa Y. et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol. Pharm. Bull. 34(1), 13–23 (2011). [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 4(1), 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Celis J. E. et al. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics 3(4), 327–344 (2004). [DOI] [PubMed] [Google Scholar]
- Bell A. W. et al. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nature Methods 6(6), 423–430, doi: 10.1038/nmeth.1333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg A. L. & Vitek O. Statistical design of quantitative mass spectrometry-based proteomic experiments. J. Proteome Res. 8(5), 2144–2156, doi: 10.1021/pr8010099 (2009). [DOI] [PubMed] [Google Scholar]
- Bignert A., Eriksson U., Nyberg E., Miller A. & Danielsson S. Consequences of using pooled versus individual samples for designing environmental monitoring sampling strategies. Chemosphere 94, 177–182, doi: 10.1016/j.chemosphere.2013.09.096 (2014). [DOI] [PubMed] [Google Scholar]
- Ting L. et al. Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol. Cell Proteomics 8(10), 2227–2242, doi: 10.1074/mcp.M800462-MCP200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37(1), 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura D. BioCarta. Biotech Software & Internet Report 2(3), 117–120 (2001). [Google Scholar]
- Kanehisa M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer C. et al. PID: The pathway interaction database. Nucleic Acids Res. 37, D674–679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]