Abstract
Human prostate cancer is a complex heterogeneous disease that mainly affects elder male population of the western world with a high rate of mortality. Acquisitions of diverse sets of hallmark capabilities along with an aberrant functioning of androgen receptor signaling are the central driving forces behind prostatic tumorigenesis and its transition into metastatic castration resistant disease. These hallmark capabilities arise due to an intense orchestration of several crucial factors, including deregulation of vital cell physiological processes, inactivation of tumor suppressive activity and disruption of prostate gland specific cellular homeostasis. The molecular complexity and redundancy of oncoproteins signaling in prostate cancer demands for concurrent inhibition of multiple hallmark associated pathways. By an extensive manual curation of the published biomedical literature, we have developed Human Prostate Cancer Hallmarks Map (HPCHM), an onco-functional atlas of human prostate cancer associated signaling and events. It explores molecular architecture of prostate cancer signaling at various levels, namely key protein components, molecular connectivity map, oncogenic signaling pathway map, pathway based functional connectivity map etc. Here, we briefly represent the systems level understanding of the molecular mechanisms associated with prostate tumorigenesis by considering each and individual molecular and cell biological events of this disease process.
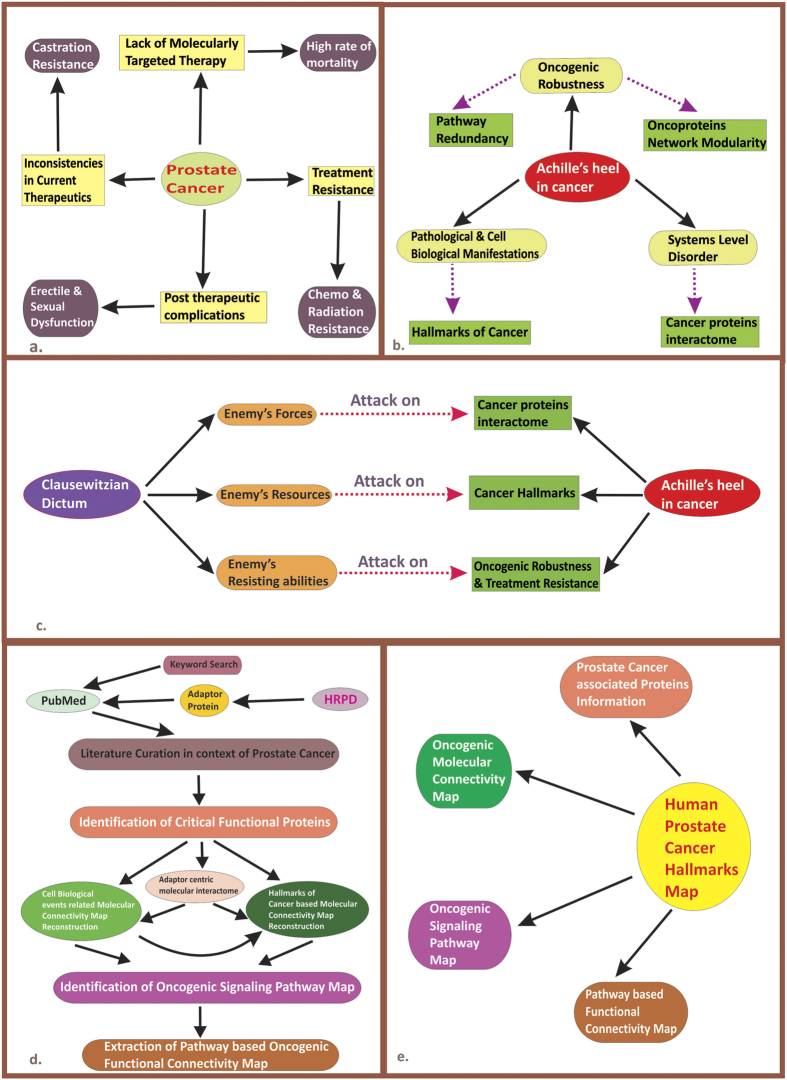
It is generally believed that cancer is a complex array of heteropathological disease manifestation united by some common hallmarks1,2,3. Prostate cancer is not an exception to this point4, and it is one of the most common malignancies responsible for male cancer deaths in the western world4,5,7,8. The heterogeneous5, multifocal5,6 nature of prostate cancer along with the late appearance of castration resistant7,8 disease form and metastatic bone invasion7,8 features makes the scenario frustratingly complex and untreatable (Fig. 1a).
Figure 1. Human Prostate Cancer Hallmarks Map- A Hypothesis Driven Project.
(a) Prostate cancer at a glance- represents some basic facts around current prostate cancer treatment approach, its limitations and its associated post therapeutic complications. (b) Achilles’ heel in cancer- depicts three important factors and their underlying features that most fundamentally challenges and dampens current molecularly targeted therapies in cancer. (c) ‘Clausewitzian Dictum’ in cancer biology- Here we propose for a multi target strategy, ‘Clausewitzian Dictum’ that essentially calls for a three level targeting- attack at the network level, attack at the cancer hallmark level, and for functional modular level. On the other hand, these three factors-cancer protein interactome network, cancer hallmark functional ability and its modular signaling network architecture together comprises the Achilles’ heel in cancer. (d) Our Bottom Up Approach- Adaptor protein comprises our initial seed protein. Binary protein interaction database [Human Protein Reference Database (HRPD)] provides information for adaptor protein and its physical interacting partner. For accessing the role of adaptor protein and its binary interacting partner in the context of prostate cancer, we manually curated the literature for each of adaptor protein and its corresponding interacting partner. Based on these information, we reconstruct adaptor centric molecular interactome in human prostate cancer. For gaining a functional organizational view, we then extracts various oncogenic signaling pathways maps, subnetworks and modules associated with diverse cell biological events and hallmark related phenomenon with other literature mined information’s (detailed in the material and methods section). (e) Human Prostate Cancer Hallmarks Map Features- It provides a comprehensive resource for revealing molecular architecture of human prostate cancer at four specific levels namely, prostate cancer hallmark & its underlying feature based phenomenon, prostate cancer hallmark based and cell biological function based molecular connectivity map, hallmark based and its underlying events related oncogenic signaling pathway map, hallmark and inter hallmark related pathway based functional connectivity map.
The principal challenges of current cancer therapy inclusive of prostate cancer can be fundamentally grouped into three pivotal factors that together constitute an ‘Achilles heel’ in the context of molecularly targeted therapies in cancer (Fig. 1b). Its inherent nature implies that as cancer as a systems level disease3,9, there is an urgent need for intervening the cancer network at multiple hallmark level1,2. Here in this article we propose for a systems medicine based strategy (Fig. 1c) that essentially aims for a simultaneous targeting at three level- first, attack the key oncocogenic network, second, attack at the hallmark level and attack in order to break the systems modularity or oncogenic robustness. In an analogical manner, it is quiet comparable to ‘Clausewitzian dictum’, an earlier multi targeted war strategy invented and implemented by general Carl von Clausewitz during Napoleonic wartime10. In his most insightful scholastic masterpiece, ‘Vom Kriege’(On War)10 general Clausewitz, essentially pointed out that, in the context of multi target attacking, strategy should simultaneously focus both at enemy’s forces and his resources and should most specifically try to dampen enemy’s resisting ability10,11. In a parallel logical connotation, here we envision that enemy’s forces should indicate cancer protein interactome as signaling proteins in cancer is supposed to behave as a network of multiple nested networks with modular interconnections2,12. Enemy’s resources most specifically address the multifunctional role of cancer proteins as cancer is supposed to emerge from the complex interplay of different onco-functional features1,2,3. In another way, it could be assumed that an intertwined relationship between various cancer hallmarks most intrinsically strengthen the enemy’s resources1,2. The classic cancer hallmark concept most crucially tries to link molecular signaling events to pathological outcomes1,2,3. To this line, protein-protein interactions provide the most fundamental unit in oncogenic network and essentially play a vital role in the acquisition of various cancer hallmark abilities13. The enemy’s resisting ability here should be understood in terms of oncogenic robustness which arises as a typical consequence of functional redundancies and due to presence of alternative compensatory signaling circuitries as cancer has been previously depicted as a disease of pathways3,9. The emergence of treatment resistance in terms of chemo/radiation resistance14,15 and castration resistance (CRPC)7,8 additionally imparts the flexibilities of enemy’s resisting ability.
In support of our hypothetical proposal, here we attempt to represent a comprehensive molecular framework for unraveling the complexities of human prostate cancer associated molecular mechanisms by structuring the existing knowledge. For concept of our work, please visit at- http://www.bioinformatics.org/canjovdb/concept.php
Results
Human Prostate Cancer Hallmarks Map Aims & Features
Human Prostate Cancer Hallmarks Map (HPCHM) is a manually constructed onco-functional atlas of human prostate tumorigenesis and is inherently based on classic cancer hallmarks concepts. It covers molecular signaling (both connectivity & pathway based) maps of most of the major cellular and pathogenic mechanisms implicated in prostate cancer progression, including 13 cancer hallmarks (10 classical & 3 prostate cancer unique hallmarks) and 11 cell biological feature based phenomenon (Table 1). It provides a comprehensive resource for revealing molecular architecture of human prostate cancer associated signaling at five specific level-first key molecular components of prostate cancer hallmarks & their underlying features (Table 2), second prostate cancer hallmark and cell biological function based molecular connectivity map (Table 1), third hallmark based and its underlying events related oncogenic signaling pathway map (Table 3), fourth hallmark and underlying cell biological feature related pathway based functional connectivity map (Tables 4 and 5), fifth inter hallmark & cell biological feature based functional connectivity map (Tables 6 and 7). Human Prostate Cancer Hallmark Map (Fig. 1e) aims to represent the systems level understanding of prostate tumorigenesis, which can be accessed at- http://www.bioinformatics.org/canjovdb.
Table 1. Content of Human Prostate Cancer Hallmarks Map.
| Cancer Hallmarks/ Cell Bilogical Features | Molecular Components | Network Statistics |
Oncogenic Signaling Pathway | Pathway based Functional Connectivity Map | |
|---|---|---|---|---|---|
| Classical Cancer Hallmarks | Proteins | Nodes | Edges | Proteins | No of Connectivity Maps |
| Evasion of growth suppression | 62 | 88 | 110 | 90 | 14 |
| Sustaning proliferative signaling | 100 | 163 | 468 | 91 | 21 |
| Tumour promoting inflammation | 12 | 23 | 63 | 29 | 16 |
| Genome instability & Mutation | 56 | 56 | 125 | — | 19 |
| Cell death resistance | 36 | 92 | 90 | 89 | 13 |
| Angiogenesis | 23 | 43 | 41 | 42 | 15 |
| Metastasis | 106 | 157 | 427 | 101 | 14 |
| Metabolic reprogramming | 23 | — | — | 78 | — |
| Avoidance of Immune Destruction | 22 | — | — | 43 | — |
| Replicative Immortalization | 29 | — | — | 38 | — |
| Tumour Microenvironment | 30 | — | — | 74 | — |
| Prostate Cancer Unique Hallmarks | |||||
| AR mediated signaling | 17 | 116 | 115 | 58 | — |
| Androgen Independence | 29 | — | — | 143 | — |
| Castration Resistance | 49 | — | — | 130 | — |
| Cell Bilogical Features | |||||
| Cell Cycle | 27 | — | — | 75 | — |
| Tumour Suppression | 39 | — | — | 93 | — |
| Cell Growth | 59 | 95 | 103 | 90 | 18 |
| Cell Survival | 35 | 91 | 95 | 91 | 9 |
| Cell Proliferation | 56 | 91 | 101 | 90 | 15 |
| Chemoresistance | 7 | 37 | 32 | 37 | — |
| Cell Migration | 46 | 82 | 93 | 82 | 18 |
| Cell Motility | 33 | 68 | 89 | 66 | 18 |
| Cell Adhesion | 21 | 41 | 42 | 40 | 4 |
| Cell Invasion | 53 | 71 | 73 | 71 | 16 |
| Bone Metastasis | 13 | 13 | 24 | — | 13 |
| Epithelial Mesenchymal Transition (EMT) | 22 | 39 | 34 | 38 | — |
Node represents number of proteins and edge indicates number of binary (physical) interactions between the proteins present in network.
Table 2. Molecular components of Human Prostate Cancer Hallmarks Map.
| Cancer Hallmarks/Cell Bilogical Features | Total Proteins(Intrinsic Components) | Adaptors | Receptors | Kinases | Enzymes | Transcription Factors & Regulators | Other Signaling Proteins | Last Update |
|---|---|---|---|---|---|---|---|---|
| Classical Cancer Hallmarks | ||||||||
| Evasion of growth suppression | 62 | 9 | 8 | 4 | 4 | 16 | 21 | 2015 |
| Sustaning proliferative signaling | 100 | 23 | 25 | 18 | 6 | 14 | 14 | 2015 |
| Tumour promoting inflammation | 12 | 2 | 5 | — | — | 3 | 2 | 2015 |
| Genome instability & Mutation | 56 | 16 | 14 | 5 | — | 8 | 13 | 2015 |
| Cell death resistance | 36 | 14 | 8 | 5 | 2 | 4 | 3 | 2015 |
| Angiogenesis | 23 | 1 | 8 | 6 | — | 4 | 4 | 2015 |
| Metastasis | 106 | 18 | 20 | 14 | 5 | 22 | 27 | 2015 |
| Metabolic reprogramming | 23 | 4 | 2 | 3 | 3 | 2 | 9 | 2015 |
| Avoidance of Immune Destruction | 22 | — | 4 | — | 2 | 3 | 13 | 2015 |
| Replicative Immortalization | 29 | 1 | 4 | 3 | 3 | 9 | 9 | 2015 |
| Tumour Microenvironment | 30 | 1 | 8 | 1 | 5 | 3 | 12 | 2015 |
| Prostate Cancer Unique Hallmarks | ||||||||
| AR mediated signaling | 17 | 1 | 4 | 4 | 1 | 5 | 2 | 2015 |
| Androgen Independence | 29 | 3 | 7 | 3 | 3 | 4 | 9 | 2015 |
| Castration Resistance | 49 | 5 | 7 | 9 | 5 | 9 | 14 | 2015 |
| Cell Biological Features | ||||||||
| Cell Cycle | 27 | 5 | 2 | 3 | 1 | 4 | 12 | 2015 |
| Tumour Suppression | 39 | 5 | 5 | 1 | 3 | 12 | 13 | 2015 |
| Cell Growth | 59 | 10 | 17 | 14 | 3 | 7 | 8 | 2015 |
| Cell Survival | 35 | 9 | 11 | 7 | 1 | 3 | 4 | 2015 |
| Cell Proliferation | 56 | 12 | 11 | 6 | 3 | 8 | 16 | 2015 |
| Chemoresistance | 7 | 3 | 1 | 1 | — | 1 | 1 | 2015 |
| Cell Migration | 46 | 10 | 12 | 5 | 1 | 10 | 8 | 2015 |
| Cell Motility | 33 | 8 | 5 | 7 | 1 | 4 | 8 | 2015 |
| Cell Adhesion | 21 | 5 | 6 | 1 | — | 1 | 8 | 2015 |
| Cell Invasion | 53 | 10 | 11 | 7 | 2 | 10 | 13 | 2015 |
| Epithelial Mesenchymal Transition (EMT) | 22 | — | 5 | 2 | 2 | 7 | 6 | 2015 |
| Bone Metastasis | 13 | — | 7 | 2 | — | 2 | 2 | 2015 |
Table 3. Oncogenic signaling pathway map statistics.
| Cancer Hallmarks/Cell Bilogical Features | Total Proteins | Adaptors | Receptors | Transcription Factors & Regulators | OtherSignaling Proteins | Oncogenes | TumourSuppressor Proteins | Modulators | Gene expressions | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Classical Cancer Hallmarks | ||||||||||
| Evasion of growth suppression | 90 | 10 | 14 | — | 66 | 14 | 19 | 6 | — | 24 |
| Sustaning proliferative signaling | 91 | 6 | 8 | 12 | 65 | 17 | 3 | 14 | 30 | 36 |
| Tumour promoting inflammation | 29 | 3 | 5 | 3 | 18 | 5 | 2 | 3 | 1 | 9 |
| Cell death resistance | 89 | 13 | 11 | 11 | 54 | 15 | 7 | 5 | 16 | 24 |
| Angiogenesis | 42 | 4 | 10 | 5 | 23 | 7 | 3 | 2 | 7 | 18 |
| Metastasis | 101 | 11 | 24 | 17 | 49 | 16 | 5 | 5 | 27 | 45 |
| Metabolic reprogramming | 78 | 3 | 16 | 5 | 54 | 15 | 8 | 17 | 12 | 47 |
| Avoidance of Immune Destruction | 43 | 1 | 9 | 3 | 30 | 6 | 1 | 2 | 17 | 21 |
| Replicative Immortalization | 38 | 2 | 3 | 9 | 24 | 7 | 7 | 7 | 3 | 27 |
| Tumour Microenvironment | 73 | 3 | 19 | 9 | 42 | 9 | 1 | 6 | 12 | 37 |
| Prostate Cancer Unique Hallmarks | ||||||||||
| AR mediated signaling | 58 | 6 | 4 | 7 | 41 | 9 | 2 | 7 | 16 | 23 |
| Androgen Independence | 143 | 8 | 18 | 27 | 90 | 25 | 10 | 23 | 22 | 54 |
| Castration Resistance | 130 | 7 | 12 | 21 | 90 | 18 | 4 | 11 | 29 | 63 |
| Cell Biological Features | ||||||||||
| Cell Cycle | 75 | 4 | 9 | 14 | 48 | 14 | 6 | 2 | 13 | 36 |
| Tumour Suppression | 93 | 4 | 9 | 0 | 80 | 14 | 56 | 3 | 0 | 45 |
| Cell Growth | 90 | 8 | 12 | 11 | 59 | 22 | 3 | 6 | 21 | 47 |
| Cell Survival | 91 | 5 | 20 | 12 | 54 | 19 | 4 | 6 | 23 | 47 |
| Cell Proliferation | 90 | 10 | 15 | 9 | 56 | 17 | 7 | 5 | 18 | 58 |
| Chemoresistance | 37 | 2 | 9 | 7 | 19 | 14 | 3 | 4 | 12 | 19 |
| Cell Migration | 82 | 13 | 18 | 11 | 40 | 14 | 7 | 5 | 12 | 53 |
| Cell Motility | 66 | 9 | 16 | 6 | 35 | 18 | 3 | 3 | 12 | 28 |
| Cell Adhesion | 40 | 10 | 10 | 2 | 18 | 7 | 2 | 6 | 6 | 20 |
| Cell Invasion | 71 | 11 | 17 | 11 | 32 | 19 | 7 | 9 | 15 | 50 |
| Epithelial Mesenchymal Transition (EMT) | 38 | 3 | 5 | 9 | 21 | 11 | 2 | 8 | 7 | 16 |
Table 4. Hallmarks related oncogenic functional connectivity map (pathway based) statistics.
| Name of Pathway | Network statistics |
||||||
|---|---|---|---|---|---|---|---|
| EVGS | SPS | TPI | GIM | ANG | MET | CDR | |
| Actin cytoskeletal regulation | N-11, E-14 | ||||||
| Adherens junction | N-4, E-5 | N-12, E-19 | N-4, E-2 | N-13, E-21 | |||
| Alpha6 Beta4 mediated signaling | N-15, E-32 | N-8, E-12 | N-4, E-4 | ||||
| Androgen receptor mediated signaling | N-4, E-5 | N-16, E-35 | N-11, E-19 | N-6, E-9 | N-13, E-30 | ||
| AP-1 transcription factor associated signaling | N-10, E-15 | N-21, E-55 | |||||
| ATM dependent DNA damage response | N-3, E-2 | ||||||
| Bladder cancer associated pathogenesis | N-10, E-9 | ||||||
| CDC42 mediated signaling | N-21, E-55 | ||||||
| Chronic myeloid leukemia pathogenesis | N-6, E-5 | N-12, E-14 | N-7, E-8 | N-15, E-22 | N-9, E-11 | ||
| Class I PI3K mediated signaling | N-48, E-110 | N-13, E-20 | N-20, E-27 | ||||
| Colorectal cancer pathogenesis | N-5, E-7 | N-17, E-29 | N-8, E-13 | N-12, E-18 | N-4, E-3 | N-10, E-14 | |
| DNA damage response | N-14, E-17 | N-9, E-12 | |||||
| EGFR mediated signaling | N-19, E-44 | N-12, E-14 | |||||
| Endometrial cancer associated pathogenesis | N-9, E-8 | ||||||
| ERBB mediated signaling | N-13, E-21 | N-9, E-16 | N-4, E-4 | ||||
| Focal Adhesion | N-16, E-32 | N-10, E-20 | N-7, E-7 | N-14, E-27 | N-5, E-6 | ||
| Glioma associated pathogenesis | N-9, E-9 | N-8, E-12 | N-14, E-27 | N-6, E-5 | N-14, E-19 | N-6, E-7 | |
| IGF1 mediated signaling | N-46, E-134 | ||||||
| IFN-gamma signaling | N-26, E-63 | ||||||
| IL-2 mediated signaling | N-15, E-24 | N-9, E-11 | |||||
| IL-3 mediated signaling | N-19, E-40 | ||||||
| IL-5 mediated signaling | N-14, E-23 | ||||||
| IL-6 mediated signaling | N-17, E-39 | N-7, E-7 | |||||
| IL-7 mediated signaling | N-7, E-7 | ||||||
| IL-11 mediated signaling | N-8, E-9 | ||||||
| Insulin mediated signaling | N-48, E-110 | N-20, E-27 | |||||
| Integrin-linked kinase associated signaling | N-21, E-55 | ||||||
| JAK-STAT signaling | N-6, E-5 | ||||||
| Kit receptor associated signaling | N-5, E-4 | N-4, E-3 | |||||
| Leptin signaling | N-11, E-25 | N-5, E-5 | |||||
| MAPK signaling | N-5, E-3 | N-13, E-15 | N-4, E-3 | ||||
| Melanoma pathogenesis | N-9, E-8 | N-13, E-15 | N-7, E-5 | ||||
| Microtubule cytoskeletal regulation | N-5, E-6 | ||||||
| Neurotrophin signaling | N-4, E-3 | ||||||
| Non small lung cancer associated pathogenesis | N-10, E-11 | ||||||
| Oncostatin M signaling | N-12, E-19 | N-10, E-9 | N-4, E-3 | ||||
| Pancreatic cancer pathogenesis | N-5, E-7 | N-11, E-13 | N-8, E-10 | N-13, E-13 | N-6, E-5 | N-20, E-30 | N-7, E-8 |
| PAR1 mediated thrombin signaling | N-10, E-15 | ||||||
| PDGFR-beta associated signaling | N-23, E-63 | N-13, E-20 | |||||
| Prolactin signaling | N-10, E-17 | N-11, E-12 | N-14, E-21 | ||||
| Senescence & autophagy associated signaling | N-5, E-3 | N-10, E-11 | N-5, E-4 | ||||
| TGF beta associated signaling | N-4, E-5 | N-20, E-42 | N-11, E-21 | N-13, E-19 | |||
| TRAIL signaling | N-23, E-29 | ||||||
| VEGF signaling | N-4, E-4 | ||||||
Abbreviations: EVGS-Evasion of growth suppression, SPS-sustaining proliferative signaling, TPI-Tumor promoting inflammation, GIM-Genomic instability & mutation, ANG-Angiogenesis, MET-Metastasis, CDR-Cell death resistance, N- Node, number of protein present in the corresponding network, E-Edge, number of binary (physical) interactions between the proteins present in the corresponding network
Table 5. Cell biological feature related oncogenic functional connectivity map (pathway based) statistics.
| Name of Pathway | Network statistics |
|||||||
|---|---|---|---|---|---|---|---|---|
| CPN | CGH | CSV | CMG | CML | CIV | CAH | OVP | |
| Actin cytoskeletal regulation | N-7, E-7 | |||||||
| Adherens junction | N-8, E-9 | N-8, E-12 | N-9, E-10 | N-4, E-3 | N-8, E-13 | |||
| Alpha6 Beta4 mediated signaling | N-11, E-21 | N-6, E-6 | N-7, E-10 | |||||
| Androgen receptor mediated signaling | N-11, E-21 | N-12, E-24 | N-9, E-16 | N-7, E-8 | N-9, E-17 | N-3, E-2 | ||
| ARF6 trafficking event | N-16, E-31 | |||||||
| ATM dependent DNA damage response | N-8, E-9 | N-6, E-5 | ||||||
| Chemokine mediated signaling | N-11, E-18 | N-8, E-6 | N-8, E-8 | |||||
| Chronic myeloid leukemia pathogenesis | N-8, E-8 | N-6, E-5 | ||||||
| Class I PI3K mediated signaling | N-30, E-56 | N-21, E-28 | ||||||
| Colorectal cancer pathogenesis | N-8, E-9 | N-10, E-15 | N-4, E-3 | N-8, E-9 | ||||
| EGFR mediated signaling | N-8, E-7 | N-13, E-28 | N-3, E-2 | N-11, E-15 | ||||
| Estrogen receptor mediated signaling | N-31, E-59 | N-8, E-9 | N-10, E-10 | |||||
| Endometrial cancer associated pathogenesis | N-7, E-7 | |||||||
| ERBB mediated signaling | N-8, E-7 | N-11, E-18 | N-6, E-7 | |||||
| Focal Adhesion | N-9, E-11 | N-14, E-31 | N-8, E-8 | N-8, E-10 | N-9, E-13 | N-8, E-11 | N-4, E-4 | N-7, E-7 |
| Glioma associated pathogenesis | N-7, E-7 | |||||||
| HGF mediated signaling | N-8, E-10 | |||||||
| IGF1 mediated signaling | N-7, E-6 | |||||||
| IFN-gamma signaling | N-17, E-35 | N-10, E-14 | ||||||
| IL-2 mediated signaling | N-4, E-3 | |||||||
| IL-3 mediated signaling | N-11, E-21 | N-6, E-5 | N-9, E-11 | |||||
| IL-5 mediated signaling | N-10, E-15 | N-5, E-4 | ||||||
| IL-6 mediated signaling | N-13, E-27 | N-4, E-3 | N-6, E-6 | |||||
| IL-11 mediated signaling | N-6, E-5 | |||||||
| Insulin mediated signaling | N-6, E-5 | N-8, E-7 | ||||||
| Integrin mediated cell adhesion | N-7, E-10 | |||||||
| Kit receptor associated signaling | N-9, E-18 | N-5, E-4 | ||||||
| Leptin signaling | N-8, E-6 | N-9, E-11 | ||||||
| MAPK signaling | N-7, E-5 | N-6, E-5 | ||||||
| Melanoma pathogenesis | N-7, E-6 | |||||||
| Neurotrophin signaling | N-8, E-7 | N-5, E-5 | ||||||
| Oncostatin M signaling | N-7, E-8 | |||||||
| Pancreatic cancer pathogenesis | N-12, E-13 | N-6, E-5 | N-7, E-5 | N-5, E-3 | ||||
| p75(NTR) mediated signaling | N-8, E-7 | N-7, E-5 | ||||||
| Prolactin signaling | N-12, E-12 | N-10, E-15 | N-3, E-2 | |||||
| TGF beta associated signaling | N-9, E-8 | N-12, E-20 | N-8, E-6 | N-9, E-12 | N-8, E-9 | |||
| TRAIL signaling | N-9, E-9 | |||||||
| VEGF signaling | N-5, E-4 | N-6, E-5 | ||||||
| Wnt signaling | N-10, E-10 | N-8, E-8 | ||||||
| uPAR mediated signaling | N-21, E-28 | |||||||
Abbreviations: CPN-Cell proliferation, CGH-Cell growth, CSV-Cell survival, CMG-Cell migration, CML-Cell motility, CIV-Cell invasion, CAH-Cell adhesion, OVP-Overexpression, N-Node, number of protein present in the corresponding network, E-Edge, number of binary [physical] interactions between the proteins present in the corresponding network.
Table 6. A summary of inter-hallmark functional connectivity map.
| EVGS | SPS | TPI | GIM | CDR | ANG | MET | |
|---|---|---|---|---|---|---|---|
| EVGS | 6 | — | 9 | 3 | — | 6 | |
| SPS | 17 | 34 | 17 | 13 | 44 | ||
| TPI | 12 | 4 | 7 | 13 | |||
| GIM | 16 | 7 | 23 | ||||
| CDR | 19 | 12 | |||||
| ANG | 13 | ||||||
| MET |
Abbreviations: EVGS-Evasion of growth suppression, SPS-sustaining proliferative signaling, TPI-Tumor promoting inflammation, GIM-Genomic instability & mutation, ANG-Angiogenesis, MET-Metastasis, CDR-Cell death resistance. Number indicates common protein between two corresponding hallmark.
Table 7. A summary of inter-cell biological feature based functional connectivity map.
| CGH | CSV | CPN | EMT | CMG | CML | CAH | CIV | CMR | BMT | OVP | TSP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CGH | 10 | 17 | 20 | 17 | 9 | 19 | 8 | 16 | — | 10 | — | |
| CSV | 12 | 16 | — | 29 | 14 | 15 | 17 | 6 | — | — | ||
| CPN | 18 | 13 | 32 | 21 | 12 | 15 | 6 | 7 | 5 | |||
| EMT | 19 | 16 | 9 | 15 | 19 | — | — | — | ||||
| CMG | 35 | 24 | 23 | 26 | 10 | 9 | — | |||||
| CML | 20 | — | 35 | 5 | — | — | ||||||
| CAH | 23 | 8 | — | — | — | |||||||
| CIV | 39 | 9 | 10 | — | ||||||||
| CMR | — | — | — | |||||||||
| BMT | 4 | — | ||||||||||
| OVP | — | |||||||||||
| TSP |
Abbreviations: CGH-Cell growth, CSV-Cell survival, CPN-Cell proliferation, EMT-Epithelial mesenchymal transition, CMG-Cell migration, CML-Cell motility, CAH-Cell adhesion, CIV-Cell invasion, CMR-Chemoresistance, MBT-Bone metastasis, OVP-Overexpression, TSP-Tumour suppressor protein. Number indicates common proteins between two corresponding cell biological features.
By taking cellular signaling scaffolds or adaptor protein16 as seed protein, we have reconstructed the corresponding adapter centric protein-protein binary interactome map in human prostate cancer with literature mined published experimental reports (Fig. 1d). This adapter protein centric molecular interactome provides the crucial ‘scaffold’ for gaining the holistic overview of prostate tumorigenesis in the context of its various hallmarks functioning. Apart from this, we have also incorporated information by key word searching (detailed in the material and methods section). For doing so, we have manually screened over 40,000 research articles and extracted the information from 955 scientific literatures (Supplementary File 1). Our manual curation effort led to the identification of 770 molecules (Supplementary File 2) involved in various aspects of prostate cancer progression and pathogenesis. For components of adaptor centric molecular interactome map in human prostate cancer, please visit- http://www.bioinformatics.org/canjovdb/components.php.
HPCHM content and depiction of prostate cancer allied molecular mechanisms
Here, we attempt to focus on key salient features of prostate cancer associated hallmarks in light of Human Prostate Cancer Hallmarks Map (HPCHM).
Hallmark- Insensitivity to antigrowth signals: prostate cancer cells become unresponsive to growth restraining signals
One of the fundamental features underlying the prostate tumorigenesis involves acquisition of insensitivity to growth inhibitory signals or evasion of growth suppression. The alterations in the functional behavior of diverse sets of tumor suppressor genes by various unfavorable circumstances such as deletion, mutation, loss of expression, rearrangement, promoter methylation and haplo-insufficiency greatly contributes in this acquisition process1,17. Apart from the actions of tumor suppressor, many aberrantly working cell proliferation negative regulators, deregulated cell cycle proteins, neutralization of pro-apoptotic stimulations and extracellular ATP also plays the key leading role in the establishment of insensitivity to antigrowth signaling during the course of tumorigenesis1,18. In HPCHM, systems medicine based view of insensitivity to antigrowth signaling hallmark consists of a core network of 88 proteins with 110 physical interactions, 14 pathway based functional modules and a signaling pathway map of 90 proteins, supported by information from 24 articles. The insensitivity to antigrowth signaling hallmark pathway map depicts 3 major signaling circuitries associated with cell growth & proliferation regulation, cell cycle regulation and suppression of apoptosis1,4. For holistic map of human prostate cancer specific insensitivity to anti growth signaling, visit - http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H1.
Cell Biological Feature- Tumour Suppression
Suppression of tumor suppressive activity most critically aids in these acquisition process as progression of prostate tumorigenesis is closely associated with chromosomal rearrangement and alterations of multiple number of chromosomal regions including 1p, 6q, 7p, 7q, 8p, 10q, 13q, 16q, 17p, 17q and 18q, harboring the sites for many important tumor suppressors19,20. As a consequence of this loss of expression or functional inactivation of tumor suppressors, prostate cancer cell activates an intricate signaling circuitry that operates across diverse oncofunctional features associated signaling pathways. In HPCHM, systems medicine based view of tumor suppression contains a signaling pathway map of 93 proteins, supported by information from 45 articles. For holistic map of human prostate cancer associated tumor suppression, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F1
Cell Biological Feature- Cell Cycle
Cell cycle deregulation plays an essential leading role in the suppression of anti-proliferative signaling as majorities of anti-proliferative signals are channeled through the proteins associated with cell cycle regulatory activities and G1 to S phase transition. All of the major cell cycle regulators in prostate cancer, which also responds to anti proliferative signaling, including p21Cip1, p27Kip1, Rb, p53 are deregulated by multiple ways and are subjected to loss of function21. Mutational inactivation that occurs during prostate tumorigenesis have been reported for p14, p16, p27, p53 and Rb genes21. In HPCHM, systems medicine based view of cell cycle contains a signaling pathway map of 75 proteins, supported by information from 36 articles. For holistic map of human prostate cancer specific cell cycle, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?featurety=P
Hallmark- Self-sufficiency in growth signaling: prostate cancer cells produce their own growth signals to trigger cell growth, proliferation and survival
Three principal factors can be accounted for achieving the state of self-sufficiency in growth signaling or sustaining proliferative signaling in prostate cancer. The factors are, first, aberrant functioning of androgen receptor22,23,24, second, modulation of growth factor receptor activity25 and third, modulation of an alternative intracellular signaling circuitries associated with cell growth, survival and proliferation6,8,26. Most strikingly, these factors also significantly contribute in the acquisition of androgen independent phenotypes and consequent emergence of castration resistant prostate cancer27. Androgen receptor mediated signaling, androgen independence and castration resistance can be considered as prostate cancer unique hallmarks7,8,22,23. In HPCHM, systems medicine based view of self sufficiency in growth signaling hallmark comprises a core network of 163 proteins with 468 physical interactions, 21 pathway based functional modules and a signaling pathway map of 91 proteins, supported by information from 36 articles. The sustaining proliferative signaling hallmark pathway map represents three major signaling circuitries related to prostate cancer cell growth & tumorigenesis, cell proliferation and cell viability1,4. For holistic map, please visit- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H2
Prostate Cancer Unique Hallmark- Androgen Receptor (AR) mediated signaling: a nuclear receptor signaling axis that plays the most pivotal role in each and every steps of prostate tumorigenesis
Numerous molecular mechanisms have been proposed for shifting of AR activation from the balanced mode to promiscuous mode in castration resistant prostate cancer (CRPC). These molecular mechanisms can be basically grouped into three major categories-First, hyper activation of androgen receptor(AR)22,23 as a result of mutation, amplification, increase in local synthesis of androgen, proteolysis, ligand independent activation and PTEN loss mediated nuclear exclusion of FOXO1 etc. Second, promiscuous activation of AR by overexpression of AR coactivators and down regulation of its corepressors23. Third, multiple alternative activation pathways mediated by transactivation24 of AR. It has been proposed that a multitude of cellular signaling pathways including PI3K/AKT, ERK, SRC etc acts in concert and converge on AR for enhancing AR mediated non-genomic response and thereby impacting self-sustenance in growth signaling22,24. For holistic view of androgen receptor mediated signaling, please visit- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H12
Prostate Cancer Unique Hallmark- Androgen Independence: prostate cancer cells develop a variety of mechanisms to survive and flourish in an androgen deprived environment
The emergence of androgen independent phenotype is a typical characteristic of human prostate cancer development and is largely responsible for castration resistant disease progression22. The growth of androgen independent castration resistant prostate tumor is intensified by up regulation of both autocrine and paracrine growth factors as a consequence of an adaptive response to androgen ablation and it essentially also fuels self sufficiency in growth signaling25. Apart from androgen ablation therapy, various factors and a series of oncogenic signaling events contributed for the acquisition of prostate cancer androgen independence22. In HPCHM, systems medicine based view of androgen independence contains a signaling pathway map of 143 proteins, supported by information from 54 articles. The androgen independence map is composed of six major pathway modules devoted to the mechanisms of prostate cancer androgen independence. The modules are associated with hypersensitive pathway, outlaw pathway, bypass pathway, co-activation pathway, promiscuous pathway and lurker cell pathway22,25. For holistic map of human prostate cancer androgen independence, please visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H13.
Prostate Cancer Unique Hallmark- Castration Resistance: a late stage aggressive manifestation that results from re-growth of prostate tumor cells that has accustomed to the hormone deprived environment of the prostate
90% of prostate cancer patients finally develop castration resistant prostate cancer (CRPC), an incurable form of disease and is largely accounted for prostate cancer associated deaths with a mean survival time of only 16–18 months7,8. Modulation of an extensively rewired pathway is supposed to act for the acquisition of both self sustenance in growth signaling and the late appearance of castration resistance disease features7,8,26. These alternative growth and survival pathways performs normal cellular signaling processes, but become oncogenic as a consequence of two distinct means: First due to an early adaptive response to androgen ablation therapy8, Second and most importantly as a result of functional inactivation of many tumor suppressors, cell growth regulators and negative feed-back regulators. In HPCHM, systems medicine based view of castration resistance contains a signaling pathway map of 130 proteins, supported by information from 63 articles. The castration resistance map depicts the global view of signaling events that underlie the pathogenesis of castration resistance4. For holistic map of human prostate cancer castration resistance (CRPC) associated signaling pathways, please visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H14
Cell Biological Feature- Cell proliferation
Deregulated cellular proliferation is an often considered hallmark feature of prostate tumorigenesis. Although the normal prostate epithelium is generally characterized by its low rate of cell proliferation, the prostatic intraepithelial neoplasia (PIN) and early invasive prostate carcinoma exhibits seven to ten fold increase in cell proliferation rate28. In HPCHM, systems medicine based view of cell proliferation comprises a core network of 91 proteins with 101 physical interactions, 15 pathway based functional modules and a signaling pathway map of 90 proteins, supported by information from 58 articles. For holistic map of prostate cancer cell proliferation, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F7
Cell Biological Feature- Cell survival
A complex set of molecular mechanisms involving actions of many growth factors, intricate interplay between pro apoptotic and anti apoptotic proteins along with enhanced cell proliferation contributes together towards the enhancement of prostate cancer cell survival8,26,27. In HPCHM, systems medicine based view of cell proliferation comprises a core network of 91 proteins with 95 physical interactions, 9 pathway based functional modules and a signaling pathway map of 91 proteins, supported by information from 47 articles. For holistic map, visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F3
Cell Biological Feature- Cell growth
The normal human prostatic epithelium cell growth rate critically balances its death rate but several causes including dysregulation of AR, inactivation of tumor suppressors, evasion of apoptotic pathway and an extensively altered metabolic activity critically mediates disruption of growth regulation, resulting in enhancement of prostate tumorigenesis8,26,27. In HPCHM, systems medicine based view of cell growth comprises a core network of 95 proteins with 103 physical interactions, 18 pathway based functional modules and a signaling pathway map of 90 proteins, supported by information from 47 articles. For holistic map of cell growth, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F4
Hallmark- Cell death resistance: prostate cancer cells resist and modulate apoptotic program
Inhibition or evasion of apoptosis in comparison with enhanced cellular proliferation behaves as a critical pathophysiological factor for initiation and progression of majority of prostate adenocarcinomas28,29. In HPCHM, systems medicine based view of cell death resistance hallmark comprises a core network of 92 proteins with 90 physical interactions, 13 pathway based functional modules and a signaling pathway map of 89 proteins, supported by information from 24 articles. Mechanisms represented on cell death resistance map are involved in the deregulation of extrinsic & intrinsic apoptotic pathways, modulation of apoptosis resistance, inhibition of apoptotic programs, inhibition of autophagy and various androgen independent & dependent signaling events. For holistic map of prostate cancer cell death resistance, visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H7
Pathophenomic Feature- Chemoresistance & Radioresistance
A majority of the metastatic prostate cancer manifests their resistance against available cytotoxic chemotherapeutic drugs including docetaxel and paclitaxel14,15. In HPCHM, systems medicine based view of chemoresistance comprises a core network of 37 proteins with 32 physical interactions and a signaling pathway map of 37 proteins, supported by information from 19 articles. The chemoresistance map characterize the various underlying factors of prostate cancer associated chemo & radiation resistance, comprising docetaxel & paclitaxel resistance, radiation resistance, androgen receptor mediated signaling, neuroendocrine behavior and the role of transcription factors. For holistic map of human prostate cancer associated chemo-resistance & radiation resistance event, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F34
Hallmark- Angiogenesis: prostate cancer cells trigger the formation of new blood vessels
Angiogenesis is needed for each and every steps in prostate tumorigenesis, ranging from tumor outgrowth to its metastatic dissemination30,31. The microvessel density in invasive tumors of human prostatic adenoma follows a direct correlation with tumors state, grade and its clinical courses31. In HPCHM, systems medicine based view of angiogenesis hallmark comprises a core network of 43 proteins with 41 physical interactions, 15 pathway based functional modules and a signaling pathway map of 42 proteins, supported by information from 18 articles. The angiogenic pathway map presents the detail signaling circuitries of prostate cancer associated angiogenesis, including the activities of four distinct angiogenic switches (VEGF, HIF1A, HO-1 and CEACAM1), growth factor receptor signaling and the role of transcription factors. For holistic map of prostate cancer angiogenesis, please visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H5
Hallmark- Metastasis: the ability of prostate tumor to escape from primary organ by evading the neighboring cells and disseminate to new sites
The underlying molecular mechanism of prostate cancer metastasis is highly complex multi-step process and involves interplay of various interrelated cell biological events. Although human prostate cancer most typically follows an exceptionally slow progression pattern, but still metastatic dissemination comprises the principal cause for prostate cancer associated mortalities7,8,32. The development of prostate cancer metastasis mainly depends on an intense orchestration of various coordinated key cell biological events, namely cell migration, invasion, cell adhesion, cell motility and epithelial mesenchymal transition33. In HPCHM, systems medicine based view of metastasis hallmark comprises a core network of 157 proteins with 427 physical interactions, 14 pathway based functional modules and a signaling pathway map of 101 proteins, supported by information from 45 articles. The metastasis pathway map shows crucial cell signaling cascades underlying multifaceted metastasis hallmark, including bone metastasis1,4. For holistic map of human prostate cancer specific metastasis, visit- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H6
Cell Biological Feature- Epithelial mesenchymal transition (EMT)
The epithelial mesenchymal transition in prostate cancer plays a leading role in the emergence of metastatic castration resistant prostate cancer (mCRPC) and functions as crucial determinants of prostate cancer metastatic dissemination33. In HPCHM, systems medicine based view of EMT contains a core network of 39 proteins with 34 physical interactions and a signaling pathway map of 38 proteins, supported by information from x articles. For holistic map of prostate cancer specific epithelial mesenchymal transition (EMT), please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F27
Cell Biological Feature- Cell adhesion
During prostate tumorigenesis, the cell adhesion molecules33 provide a framework for mediating prostate cancer cell-cell interactions and an aberrant interaction between prostate glandular epithelial cells and the extracellular matrix. In HPCHM, systems medicine based view of cell adhesion comprises a core network of 41 proteins with 42 physical interactions, 4 pathway based functional modules and a signaling pathway map of 40 proteins, supported by information from 20 articles. For holistic map of human prostate cancer specific cell adhesion event, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F28
Cell Biological Feature- Cell Invasion
The earliest step of prostate cancer metastasis is the local invasion33 which involves entry of cancer cells from the location of primary prostate tumour into the surrounding extracellular matrix and tumor associated stromal cell layer. In HPCHM, systems medicine based view of cell invasion comprises a core network of 71 proteins with 73 physical interactions, 16 pathway based functional modules and a signaling pathway map of 71 proteins, supported by information from 50 articles. For holistic map of human prostate cancer cell invasion, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F25
Cell Biological Feature- Cell motility
Cell motility plays a very important role during the metastatic dissemination of prostate cancer33. In HPCHM, systems medicine based view of cell motility comprises a core network of 68 proteins with 89 physical interactions, 18 pathway based functional modules and a signaling pathway map of 66 proteins, supported by information from 28 articles. For holistic map of cell motility, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F26
Cell Biological Feature- Cell Migration
Cell migration plays a crucial role in prostate cancer metastatic progression33. In HPCHM, systems medicine based view of cell migration comprises a core network of 82 proteins with 93 protein-protein interactions, 18 pathway based functional modules and a signaling pathway map of 82 proteins, supported by information from 53 articles. For details, please visit at- http://www.bioinformatics.org/canjovdb/feature.php?feature=F29
Hallmark- Inflammation: promotion of prostate tumor through enhancement of inflammatory response
Prostatic inflammation accelerates the aggressiveness of prostate tumorigenesis and its subsequent transformation into castration resistant metastatic pathological forms34. Prostate epithelial cells play an important role in initiating and amplifying the prostatic inflammatory processes through simultaneous activation of NF-κB pathway and by locally producing diverse pro-inflammatory cytokines that supports the recruitment and activation of additional immune cells, such as macrophage, to the prostate35. In HPCHM, system medicine based view of inflammation hallmark comprises a core network of 23 proteins with 63 physical interactions, 16 pathway based functional modules and a signaling pathway map of 29 proteins, supported by information from 9 articles. The inflammation signaling pathway map represents four important signaling mechanisms that critically drives prostate tumour inflammation. The signaling mechanisms include modulation of NF-kB activation, modulation of inflammatory response, inflammasome activation and immune cell activation. For details, please visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H3
Hallmark- Metabolic Reprogramming: prostate cancer cells deregulate metabolic program and energetics to support tumor formation
Although normal human prostatic epithelium cell growth rate critically balances its death rate but several factors including multivariate metabolic regulation by androgen receptor36, inactivation of tumor suppressors37 and metabolic reprogramming38 crucially fuels for the disruption of normal metabolic regulation, leading to an enhancement of prostate tumorigenesis. In HPCHM, systems medicine based view of metabolic reprogramming contains a signaling pathway map of 78 proteins, supported by information from 47 articles. The metabolic reprogramming oncogenic signaling map contains 12 critical metabolic switches that most significantly promotes prostate cancer cell metabolism. The metabolic switches are associated with PTEN inactivation, AMPK regulation, Warburg effect, HIF1A regulation, De novo fatty acid synthesis, De novo cholesterol synthesis, Cholesterol accumulation, Intratumoral steroidogenesis, D-Lactate metabolism, Restoration of TCA cycle, Decrease in zinc uptake and Reverse Warburg Effect. For holistic map of human prostate cancer specific metabolic reprogramming, please visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H8
Hallmark- Enabling Replicative Immortality: prostate cancer cells have unlimited replicative potential
Telomere dysfunction and reactivation of the telomerase reverse transcriptase activity, which is typically associated with telomere maintenance, plays the profound significant roles in imparting limitless replicative potential of the prostate tumor cells39,40. Telomerase activity has not been detected in both normal prostate and benign prostatic hyperplastic tissue, but over 90% of prostate cancer exhibits significantly higher level of telomerase activity40. Although replicative senescence is assumed a significant impediment to cellular immortalization and transformation1, but the deregulation of a broad spectrum of senescence regulators greatly contributes in surpassing classical senescence program that eventually enhances prostate tumorigenesis. In HPCHM, systems medicine based view of replicative immortalization contains a signaling pathway map of 38 proteins, supported by information from 27 articles. The replicative immortalization signaling map represents detail molecular mechanisms associated with expression, modulation & enhancement of telomerase (hTERT) activity, down-regulation of replicative senescence and the role of androgen deprivation therapy in modulation of prostate tumors senescence program. For holistic map of prostate cancer replicative immortalization, visit- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H10
Hallmark- Avoidance of Immune Destruction: prostate cancer cells evade the monitoring capability of immune system
Tumor immune evasion is currently recognized as an emerging hallmark of cancer1 and the current research strongly suggests that evasion of immune destruction plays a crucial role in the development of prostate cancer along with its pathogenic progression41. A significant alteration in cell mediated immune response is particularly observed during the course of prostate tumorigenesis and it involves multiple potential immune suppressive mechanisms41,42. In HPCHM, the avoidance of immune destruction holistic map represents an integrated view of signaling events between prostate cancer cell and various immune cells, which can be accessed at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H9
Hallmark- Tumor Microenvironment: the surroundings of prostate tumor which critically fuels other hallmark manifestations
The tumor microenvironment in prostate cancer, which involves complex reciprocal interactions between prostate cancer epithelial cell and surrounding ‘reactive stroma’, plays the most influential role in survival, angiogenesis, inflammation and its metastatic dissemination43. Prostate cancer ‘reactive stroma’ is typically characterized by the presence of cancer associated fibroblastic (CAF) and myofibroblastic cell populations along with a consequential loss of smooth muscle cells and is centrally responsible for the genesis of intratumoural heterogeneity43,44. Additionally, adipocytes play a very essential role in the modulation of prostate tumour microenvironment by secreting multiple growth factors (IGF1, TGF-beta, TNF-alpha, IL-6) and inflammatory adipokines45. In HPCHM, the tumor microenvironmental holistic map depicts the detail paracrine signaling interactions between prostate cancer cell and its surrounding ‘reactive stroma’ components including CAF, myofibroblast and adipocytes1,43. For details of prostate cancer associated tumor microenvironment, visit at- http://www.bioinformatics.org/canjovdb/hallmark.php?hall=H11
Discussion
Currently there is no effective molecularly targeted treatment strategy available for metastatic castration resistant prostate cancer7,8. Consequently, there is an actual need for identification of potential molecular targets and global molecular signaling map in the context of human prostate tumorigenesis, progression and treatment resistance7,26. But several challenging factors make the scenario frustratingly complex and practically infeasible. First, progression of prostate cancer involves a number of molecular and cellular processes at the same time46, second, prostate tumor’s heterogeneous and multifocal nature5,6, third, prostate cancer’s transition from androgen dependent to androgen unresponsive castration resistant form7,8. In addition, integrating information from multifarious sources of information across diverse systems is also a formidable challenge because of limitation of the available computational tools2,47. As a consequence, our understanding of the pathway signaling map and their modular interplay at prostate cancer hallmark level is largely lacking4. This is due to three principal reasons: first, although the classic cancer hallmark concept1 alone provides the most fundamental organizing principles of neoplastic diseases, it remains descriptive2 and no prostate cancer specific views are available yet4,7,26. Second, comprehensive information for most of cancer hallmark network in a generalized and global basis is largely unavailable there by restricting any computational based prediction2. Third, currently available pathway databases mainly offer the ‘canonical pathway’ based depiction, predominantly ignoring the facts that the multi stage process of tumorigenesis involves concomitantly multiple cellular processes and their signaling events48. For addressing all these issues, we have taken an initiative from a system medicine perspective for systems level exploration of prostate cancer associated molecular process and signaling cascades.
Although the concept of system medicine offer an unique opportunity for simplification of complex systems (by transforming medicine into a network based platform in a context specific fashion)47,49, but it is assumed that the currently available interactome maps (generated by high-throughput methods) are more than 80% incomplete50,47. From this current standpoint, compilation of already available published experimental report in the literature provides a paramount opportunity for reconstruction of interactomes thereby inferring as realistic as possible views of molecular signaling maps underlying diseases51. To this line, literature mined binary protein interactome maps plays a potential role in attaining hypothesis about focused biological mechanisms13,51. In the current study, we have followed a bottom up approach, which is essentially based on network medicine47,51 (Fig. 1d). Our effort reveals HPCHM, a web based contrivance, which provides the basic blue print of human prostate cancer specific ‘hallmarkosme’, an assembly of cancer hallmarks and their underlying cell biological events. This ‘hallmarkosome’ represents a set of comprehensive molecular signaling map of each and individual cellular and molecular processes related with human prostate cancer pathogenesis. For comprehensive map of human prostate cancer associated hallmarks, please visit at the following link- http://www.bioinformatics.org/canjovdb/comprehensive.php
To our knowledge, Human Prostate Cancer Hallmarks Map (HPCHM) provides the first time depiction of global molecular signaling events that are underlying human prostate tumorigenesis. Several key features make the holistic maps unique in its representation: (1) It shows most of the crucial molecular signaling cascades, their crosstalks and regulatory molecular processes and also, ‘a disease of pathway’ based view that are usually deregulated during prostate cancer development and progression; (2) It covers all major human prostate tumour specific features including androgen dependent & independent signaling views, androgen receptor (AR) mediated signaling and their regulation, the role of prostate tumour specific modulators etc; (3) It presents detailed functional view of various onco & tumour suppressor proteins, whose collaborative action functions as one of the key driver of human prostate tumorigenesis; (4) It offers an inter hallmark connectivity map, as it has been currently hypothesized that cancer hallmarks are closely intertwined2.
In summary, here we represent a new frame work for gaining a comprehensive understanding of cellular and molecular events underlying of human prostate cancer pathogenesis by accumulating the existing knowledge. Most importantly, Human Prostate Cancer Hallmarks Map (HPCHM) provides a major step towards systems level mechanistic representation of prostate tumorigenesis that will surely give a new space for the development of novel therapeutic approaches.
Methods
Initial seed gene listing
Adapter protein comprises our initial seed gene. Human Protein Reference Database (HRPD)52 provides information for different aspects of human proteins ranging from protein-protein interactions to molecular class information, post–translational modification to disease associations. From HRPD molecule class we got the list of 339 human adapter genes under the name of ‘Adaptor molecule’.
Gene name normalization
A gene might appear under different synonyms. For example TICAM2 indicates toll- like receptor adaptor molecule 2 gene might shows TIRP, TRAM, TIRAP3, MyD 88-4, TICAM-2, different synonyms in biological textual article. For these reason we used the gene name provided by the HUGO Gene Nomenclature Committee GGNC database53.
Literature Database searching
We manually curated the text for each of 339 adapter protein for their corresponding role in human prostate cancer. For literature search we used the NCBI PubMed database, for instance, by typing BCL2 in prostate cancer yields 159 different article containing experimentally verified information for the role of BCL2 adapter protein in prostate cancer. We manually curated each & individual of these articles for accessing the role of adapter proteins in prostate cancer.
Additionally, an extensive literature survey was followed using NCBI Pubmed for retrieving articles related to human prostate cancer associated cell biological events by key word searching, for example using search terms ‘prostate cancer cell proliferation’ etc (corresponding cell biological events). The full text articles were manually screened for cell signaling, molecular interactions, oncogenic activation & tumor suppression processes, nuclear translocation events, transcriptional gene regulation and the hyperlinked PubMed identifier for the corresponding publication. For enhancing the accuracy of this textual curation, each of the information has been simultaneously cross verified three times by different researchers.
Binary Interaction partner selection
Adaptor proteins physical interacting partners were selected from the Human Protein Reference Database (HRPD)52. Each of the binary interacting partners is subjected for pubmed search in previously mentioned ways for identifying their role in human prostate cancer. In this manner, we constructed adaptor centric interactome map in prostate cancer based on proteins binary interaction along with literature curetted experimentally verified information.
Oncogenic Signaling Map extraction
We manually constructed the comprehensive pathway maps [Molecular Connectivity Map & Oncogenic Signaling Pathway Map] for prostate cancer hallmarks, their underlying cell biological events and its associated pathophenomic features (Androgen Independence, Castration Resistance etc). These maps are intrinsically based on the molecular interactions documented in adapter centric molecular interactome. In addition, we have also incorporated information from current existing publication on molecular events associated with pathogenesis and progression of human prostate cancer.
Pathway based Oncogenic Functional Connectivity Map reconstruction
For pathway based information, we incorporated data from various sources, ranging from literature mined information to pathway databases, including KEGG54, Pathway Commons55 and WikiPathways56 through both manual curation and by using WEB-based GEne SeT AnaLysis Toolkit57. The subnetworks were derived by using Pajek58 and represented in Pajek58 and Cytoscape59 based visualization platform.
Additional Information
How to cite this article: Datta, D. et al. Human Prostate Cancer Hallmarks Map. Sci. Rep. 6, 30691; doi: 10.1038/srep30691 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Rahul Shaw at the Yale-NUS College, Singapore for technical and computational help. We also thank Samiran Kundu Roy at Department of Physics, Asutosh College, Kolkata, India for valuable assistance.
Footnotes
Author Contributions D.D. conceived the idea, collected all information by literature curation, reconstructed all connectivity maps & associated signaling pathways and oversaw all aspects of the project, D.K.G. helped D.D. for reconstruction of molecular connectivity maps, Md.A. constructed database, S.R. and P.S. provided critical insights. D.D. wrote the manuscripts with contributions from all authors.
References
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell. 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- Wang E. et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 30, 4–12 (2015). [DOI] [PubMed] [Google Scholar]
- Kreeger P. K. & Lauffenburger D. A. Cancer systems biology: a network modeling perspective. Carcinogenesis. 31, 2–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei N. Review of hallmarks of prostate cancer (PCa). Health Professional Student Journal. 2, 1 (2015). [Google Scholar]
- Mazaris E. & Tsiotras A. Molecular pathways in prostate cancer. Nephrourol Mon. 5, 792–800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algaba F. & Montironi R. Impact of prostate cancer multifocality on its biology and treatment. J Endourol. 24, 799–804 (2010). [DOI] [PubMed] [Google Scholar]
- Katsogiannou M. et al. The hallmarks of castration-resistant prostate cancers. Cancer Treat Rev. 41, 588–597 (2015). [DOI] [PubMed] [Google Scholar]
- Karantanos T., Corn P. G. & Thompson T. C. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 32, 5501–5511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol A., Mosca R., Farrés J. & Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol Sci. 31, 115–123 (2010). [DOI] [PubMed] [Google Scholar]
- von Clausewitz C. On War. Ed./trans. Howard M. & Paret P., Knopf Alfred. A."Everyman's Library" edition, New York, NY, 1993. [Google Scholar]
- Csermely P., Agoston V. & Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 26, 178–182 (2005). [DOI] [PubMed] [Google Scholar]
- Haura E. B. From modules to medicine: How modular domains and their associated networks can enable personalized medicine. FEBS Lett. 586, 2580–2585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A. A., Khuri F. R. & Fu H. Targeting protein-protein interactions as an anti cancer strategy. Trends Pharmacol Sci. 34, 393–400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M., Kashiwagi E., Yokomizo A., Takeuchi A. & Dejima T. Interaction between docitaxel resistance and castration resistance in prostate cancer: implications of Twist1, YB-1, and androgen receptor. Prostate. 73, 1336–1344 (2013). [DOI] [PubMed] [Google Scholar]
- Fitzgerald T. J. et al. Prostate carcinoma and radiation therapy: therapeutic treatment resistance and strategies for therapeutic targeted therapeutic intervention. Expert Rev Anticancer Ther. 8, 967–974 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reebye V., Frilling A., Habib N. A. & Mintz P. J. Intracellular adaptor molecules and AR signaling in the tumour microenvironment. Cell Signal. 23, 1017–1021 (2011). [DOI] [PubMed] [Google Scholar]
- Karan D., Lin M. F., Johansson S. L. & Batra S. K. Current status of the molecular genetics of human prostatic adenocarcinomas. Int J Cancer. 103, 285–293 (2003). [DOI] [PubMed] [Google Scholar]
- Prevarskaya N., Skryma R. & Shuba Y. Ion channels and the hallmarks of cancer. Trends Mol Med. 16, 107–121 (2010). [DOI] [PubMed] [Google Scholar]
- Rubin M. A. & De Marzo A. M. Molecular genetics of human prostate cancer. Mod Pathol. 17, 380–388 (2004). [DOI] [PubMed] [Google Scholar]
- Isaacs W. & Kainu T. Oncogenes and tumour suppressor genes in prostate cancer. Epidemiol Rev. 23, 36 (2001). [DOI] [PubMed] [Google Scholar]
- Lee J. T. et al. Targeting prostate cancer based on signal transduction and cell cycle pathways. Cell Cycle 7, 1745–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B. J. & Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer 1, 34–45 (2001). [DOI] [PubMed] [Google Scholar]
- Devlin H. L. & Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Lett. 274, 177–186 (2009). [DOI] [PubMed] [Google Scholar]
- Lamont K. R. & Tindall D. J. Minireview: Alternative activation pathways for the androgen receptor in prostate cancer. Mol Endocrinol. 25, 897–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. J., Bennett S. & Stricker P. Growth factor involvement in the progression of prostate cancer. Clin Chem. 44, 705–723 (1998). [PubMed] [Google Scholar]
- Inoue T. & Ogawa O. Role of signaling transduction pathways in development of castration-resistant prostate cancer. Prostate Cancer 2011, 647987 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parray A., Siddique H. R., Nanda S., Konety B. R. & Saleem M. Castration-resistant prostate cancer: potential targets and therapies. Biologics. 6, 267–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate-Shen C. & Shen M. M. Molecular genetics of prostate cancer. Genes Dev. 14, 2410–2434 (2000). [DOI] [PubMed] [Google Scholar]
- Tu H., Jacobs S. C., Borkowski A. & Kyprianou N. Incidence of apoptosis and cell proliferation in prostate cancer: relationship with TGF-beta1 and bcl-2 expression. Int J Cancer 69, 357–363 (1996). [DOI] [PubMed] [Google Scholar]
- Lu S. et al. Smad3 is overexpressed in advanced human prostate cancer and necessary for progressive growth in prostate cancer cells in nude mice. Clin Cancer Res. 13, 5692–5702 (2007). [DOI] [PubMed] [Google Scholar]
- Strohmeyer D. et al. Tumour angiogenesis is associated with progression after radial prostatectomy in pT2/ pT3 prostate cancer. Prostate. 42, 26–33 (2000). [DOI] [PubMed] [Google Scholar]
- Yu H. S., Lin T. H. & Tang C. H. Bradykinin enhances cell migration in human prostate cancer cells through B2 receptor/ PKCδ/ c-Src dependent signaling pathway. Prostate. 73, 89–100 (2013). [DOI] [PubMed] [Google Scholar]
- Clarke N. W., Hart C. A. & Brown M. D. Molecular mechanisms of metastasis in prostate cancer. Asian J Androl. 11, 57–67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M. & Batra S. K. Development of animal models underlining mechanistic connections between prostate inflammation and cancer. World J Clin Oncol. 4, 4–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. P., Bray T. M. & Ho E. Induction of proinflammatory response in prostate cancer epithelial cells by activated macrophages. Cancer Lett. 276, 38–46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfeld S. J., Itkonen H. M., Urbanucci A. & Mills I. G. Androgen-regulated metabolism and biosynthesis in prostate cancer. Endocr Relat Cancer. 21, T57–T66 (2014). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 8, 1461–1474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin R., Zadra G. & Loda M. Metabolic alterations and targeted therapies in prostate cancer. J Pathol. 223, 283–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z. et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148, 896–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biroccio A. & Leonetti C. Telomerase as a new target as a new target for the treatment of hormone-refractory prostate cancer. Endocr Relat Cancer 11, 407–421 (2004). [DOI] [PubMed] [Google Scholar]
- Silvestri I., Cattarino S., Aglianò A. M., Collalti G. & Sciarra A. Beyond the Immune Suppression: The Immunotherapy in Prostate Cancer. Biomed Res Int. 2015, 794968, doi: 10.1155/2015/794968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarubendra N., Lawrentschuk N., Bolton D. M., Klotz L. & Davis I. D. Prostate cancer immunology-an update for Urologists. BJU Int. 107, 1046–1051 (2011). [DOI] [PubMed] [Google Scholar]
- Barron D. A. & Rowley D. R. The reactive stroma microenvironment and prostate cancer progression. Endocr Relat Cancer 19, R187–R204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser C. J. Targeting prostatic tumor micro-environment to address therapy resistance. Transl Cancer Res. 2, 62–63 (2013). [Google Scholar]
- Toren P. & Venkateswaran V. Periprostatic adipose tissue and prostate cancer progression: new insights into the tumor microenvironment. Clin Genitourin Cancer 12, 21–26 (2014). [DOI] [PubMed] [Google Scholar]
- Arnold J. T. & Isaacs J. T. Mechanisms involved in the progression of androgen-independent prostate cancers: it is not only the cancer cell’s fault. Endocr Relat Cancer 9, 61–73 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A. L., Gulbahce N. & Loscalzo J. Network medicine: a network based approach to human disease. Nat Rev Genet. 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperstein I. et al. Atlas of Cancer Signalling Network: a systems biology resource for integrative analysis of cancer data with google maps. Oncogenesis. 4, e160, doi: 10.1038/oncsis.2015.19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. & Linding R. Network Medicine. FEBS Lett. 582, 1266–1270 (2008). [DOI] [PubMed] [Google Scholar]
- Menche J. et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601, doi: 10.1126/science.1257601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Cusick M. E. & Barabási A. L. Interactome networks and human disease. Cell. 144, 986–998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad T. S. et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 37, D767–72, doi: 10.1093/nar/gkn892 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain H. M., Lush M. J., Ducluzeau F., Khodiyar V. K. & Povey S. Genew: the Human Gene Nomenclature Database, 2004 updates. Nucleic Acids Res. 32, D255–D257 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. The KEGG database. Novartis Found Symp. 247, 91–101 (2002). [PubMed] [Google Scholar]
- Cerami E. G. et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–D690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico A. R. et al. WikiPathways: pathway editing for the people. PLoS Biol. 6, e184, doi: 10.1371/journal.pbio.0060184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kirov S. & Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagelj V. & Mrvar A. Pajek: Program for large network analysis. Connections. 21, 47–57 (1998). [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 11, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.