Abstract
The human genome contains millions of fragments from retrotransposons—highly repetitive DNA sequences that were once able to “copy and paste” themselves to other regions in the genome. However, the majority of retrotransposons have lost this capacity through acquisition of mutations or through endogenous silencing mechanisms. Without this imminent threat of transposition, retrotransposons have the potential to act as a major source of genomic innovation. Indeed, large numbers of retrotransposons have been found to be active in specific contexts: as gene regulatory elements and promoters for protein‐coding genes or long noncoding RNAs, among others. In this review, we summarise recent findings about retrotransposons, with implications in gene expression regulation, the expansion of gene isoform diversity and the generation of long noncoding RNAs. We highlight key examples that demonstrate their role in cellular identity and their versatility as markers of cell states, and we discuss how their dysregulation may contribute to the formation of and possibly therapeutic response in human cancers.
Keywords: endogenous retrovirus, lncRNA, regulation, retrotransposon, transcription
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Transcription
Glossary
- BANCR
BRAF‐activated non‐protein‐coding RNA
- Cas9
CRISPR‐associated protein 9
- Cdx2
caudal type homeobox 2
- ChIP
chromatin immunoprecipitation
- ChIP‐Seq
chromatin immunoprecipitation followed by DNA sequencing
- CRISPR
clustered regularly interspaced short palindromic repeats
- CSF1R
colony‐stimulating factor 1 receptor
- DNase‐Seq
DNase I hypersensitive sites sequencing
- DNMTI
DNA methyltransferase inhibitor
- Elf5
E74‐like factor 5
- Env
envelope
- Eomes
eomesodermin
- ERK2
extracellular signal‐regulated kinase 2
- ERV
endogenous retrovirus/endogenous retroviral element
- ESC
embryonic stem cell
- Gag
group‐specific antigen
- H3K27ac
histone 3 lysine 27 acetylation
- H3K4me3
histone 3 lysine 4 trimethylation
- H3K9ac
histone 3 lysine 9 acetylation
- HBV
hepatitis B virus
- HERV
human endogenous retrovirus
- HPAT5
human pluripotency‐associated transcript 5
- KAP1
KRAB‐associated protein‐1
- LBP9
lipid‐binding protein 9
- linc‐RoR
long intergenic non‐protein‐coding RNA, regulator of reprogramming
- LINE
long interspersed nuclear elements
- lncRNA
long noncoding RNA
- LTR
long terminal repeat
- OCT4
octamer‐binding transcription factor 4
- ORF
open reading frame
- Pol
polymerase
- RNAi
RNA interference
- RNA‐Seq
RNA sequencing
- SAMMSON
survival‐associated mitochondrial melanoma‐specific oncogenic noncoding RNA
- shRNA
small hairpin RNA
- SINE
short interspersed nuclear elements
- siRNA
small interfering RNA
- SVA
SINE‐R, VNTR and Alu
- TE
transposable element
- TF
transcription factor
- TRIM28
tripartite motif‐containing 28
- TSS
transcription start site
- UCA1
urothelial cancer‐associated 1
Introduction
The human genome consists of 3 billion nucleotides, the sequence that provides the blueprint for human life. The genetic information is organised in regulatory networks, consisting of trans‐acting protein‐coding or noncoding genes, and cis‐acting regulatory elements that control expression patterns. The interplay of these elements facilitates the establishment of cellular diversity during embryogenesis, and the subsequent development of tissues and organs. However, only a subset of the human genome actively contributes to these regulatory networks. High‐throughput genomics technology has been used to map the active elements in the human genome sequence, through initiatives such as ENCODE 1 and the Roadmap Epigenomics Project 2. These efforts have used ChIP‐Seq to generate genomewide profiles of transcription factor binding sites and landscapes of histone modifications 1, 2; DNase‐Seq to identify regions of accessible chromatin 3; and RNA‐Seq to measure transcription 4, providing in‐depth information about the regulatory networks encoded in the space of the human genome sequence.
About 50% of the human genome consists of repetitive elements—DNA sequences that occur multiple times in almost‐identical copies 5. The largest fraction of repetitive DNA is contributed by retrotransposons, a family of transposable elements (TEs) that are able to “copy and paste” their own DNA in the host genome. There are three major classes of retrotransposons: long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs) and endogenous retroviral elements (ERVs) 6, 7, 8, 9, 10. Today, almost all retrotransposons have lost their capacity for transposition. However, the remaining fragments contain the major ingredients of regulatory networks: cis‐regulatory sequences, transcription start sites, and even genes that can be transcribed. Indeed, retrotransposons are frequently detected across all genomic assays, suggesting that they can contribute to both regulation and transcription in the human genome.
Compared to protein‐coding genes, retrotransposon sequences are much less conserved, and they can differ substantially between species 11, 12, 13, 14, 15, 16. While this may suggest that they are not essential for human life, they nevertheless show some biochemical activity 3. The presence of biochemical activity amidst low evolutionary conservation highlights one of the key challenges in retrotransposon research: how to identify elements that are biologically relevant (“functional” for simplicity) among all active retrotransposons (Box 1).
In this review, we discuss recent evidence showing that retrotransposons make large‐scale contributions to gene regulatory elements, noncoding genes and protein‐coding genes. In particular, we highlight studies that demonstrate a phenotype attributed to retrotransposon activation or describe a mechanism by which this occurs, and we summarise recent evidence implicating retrotransposon activation in cancer. This review mainly focuses on the ERV class of retrotransposons; however, it also highlights common themes among ERVs, LINEs and SINEs that have emerged from genomewide studies of their regulation and contribution to the transcriptome.
Box 1: Active versus functional.
Large‐scale genomics assays are powerful tools to map specific characteristics such as transcription factor binding, histone modifications, chromatin accessibility, chromatin interactions, methylation, transcription or even transcript structure to the human genome. The vast majority of the genome is detected in at least one such assay; however, not all of that is evolutionarily conserved, as shown by comparisons of genome sequences from different species. In this review, we will use the terms active and functional to discriminate the observation of biochemical activity at a genomic locus (active) from the demonstration that this activity has a consequence (functional). In this sense, functional does not imply that an element is evolutionarily conserved or essential for human life, but in the case of retrotransposons distinguishes possible noise from elements whose activity may contribute to early embryonic development, innate immunity or human diseases.
LINEs, SINEs and ERVs/LTRs and their contribution to the human genome sequence
The three classes of retrotransposons (ERV, LINE, SINE) can be distinguished based on their retrotransposition mechanisms; however, the naming of individual elements follows a similar hierarchical system (Fig 1; numbers were calculated from the GRCh38 RepeatMasker annotations). Within each class of retrotransposons, there are two additional layers that group the individual elements based on their similarity 17, 18, 19, 20, 21, 22, 23, 24. The first layer reflects the family of retrotransposons, such as Alu (SINE), L1 (LINE) or ERV1 (ERVs) (Fig 1B). The second layer captures the subfamily, such as AluSx (family: Alu), L1M5 (family: L1) or HERVH (family: ERV1) (Fig 1C). Elements belonging to the same subfamily can show a very high sequence similarity, and often hundreds or thousands of almost‐identical copies can be found in the human genome (Fig 1D and E).
Figure 1. Retrotransposon classes, naming and genomic distribution.
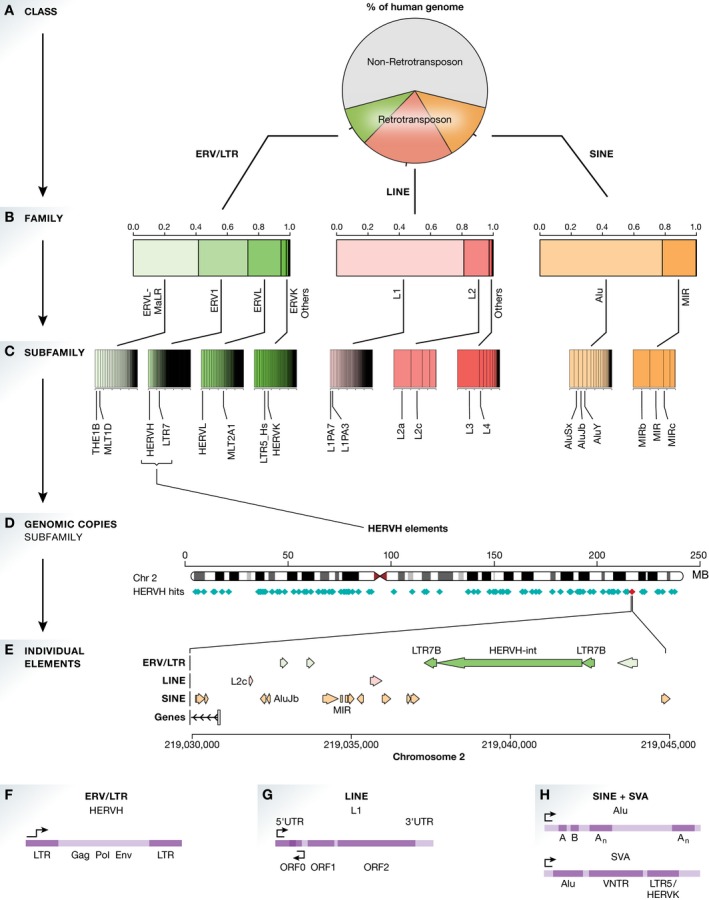
(A) The contribution of the three major classes of retrotransposons to the human genome sequence. (B) Each retrotransposon class contains several families; shown is the relative contribution of each family to the respective retrotransposon class. (C) Each family contains several subfamilies; the relative contribution to the families is shown, and selected examples are highlighted. (D) Shown are all HERVH fragments on chromosome 2. (E) Visualisation of a LTR7B‐HERVH retrotransposon on chromosome 2 consisting of the two flanking LTR elements and the internal HERVH element. (F) Model for ERVs. Three genes (Gag, Pol, Env) are surrounded by two long terminal repeats (LTRs) that contain a promoter. (G) Model for LINEs. Human LINE 1 elements contain 2 (sometimes 3) open reading frames (ORFs), which are regulated by two promoters. (H) Model of SINEs and SVAs. Unlike the other retrotransposons, SINEs do not contain protein‐coding genes. SVAs are a combination of SINEs and ERVs.
ERVs and LTRs
ERVs are genomic elements that resemble retroviruses—viruses which multiply their DNA by inserting it into the genome of the host cell. If retroviruses infect cells of the germline or cells that give rise to the germline, their DNA can be passed to the next generation, giving rise to endogenous retroviruses 25. A complete endogenous retrovirus consists of a set of genes (Gag, Pol, Env) that facilitate the retrotransposition, and two identical long terminal repeats (LTRs) that flank these genes and contain the promoter element (Fig 1F) 26, 27, 28, 29, 30. A complete ERV spans several kilobases. However, this structure is only preserved in a subset of genomic elements, as most ERVs are fragments or solitary LTRs. Estimated to contribute to 9% of the human genome, ERVs are the smallest retrotransposon family. Compared with the more repetitive retrotransposon classes, ERVs contribute a larger diversity of potential regulatory elements to the human genome, making them particularly interesting in the context of transcriptional regulation 3, 31, 32, 33.
LINEs, SINEs and SINE‐related nonautonomous retrotransposons
Similar to ERVs, LINEs contain all of the ingredients required for retrotransposition 34. Complete LINE elements can be more than 6 kilobases long. LINEs contain two open reading frames (ORFs) encoding an RNA‐binding protein, and an endonuclease and reverse transcriptase 35, 36, 37 (Fig 1G). In primates, a short third open reading frame was described for a subset of LINE elements 38. Unlike LTR promoters, LINE promoters transcribe themselves, and are considered weaker, sometimes requiring additional regulatory input to activate cell‐type‐specific expression 39. Some LINEs contain an additional antisense promoter that can drive transcription into adjacent genes 39, 40, 41. Many times, LINEs are only fragments, and due to 5′ truncation, it is estimated that only a small fraction of all LINEs contains the promoter element 42, 43. LINEs contribute the largest fraction (21%) of retrotransposon‐derived DNA to the human genome.
SINEs are shorter in length than ERVs or LINEs (usually fewer than 500 bp), and unlike these autonomous retrotransposons, SINEs do not contain ORFs (Fig 1H) 44. SINEs originate from small functional RNAs 45, 46, and instead of encoding their own proteins, they require the machinery from LINE elements for retrotransposition 39, 47, 48. SINEs also contribute to the small class of SVA retrotransposons, a family of nonautonomous retrotransposons that also contain LTR sequences 49, 50, 51 (Fig 1G). Thirteen percent of the human genome resembles SINE elements. With more than 1 million copies, the Alu family of SINEs is the most frequent retrotransposon in the human genome.
Silencing of retrotransposons
Due to their potential to disrupt DNA sequences and impair genome stability, most ERVs, LINEs and SINEs are silenced. Silencing is orchestrated by a combination of sequence‐specific transcription factors and epigenetic modifiers that affect histone modifications or DNA methylation.
The transcription factor (TF) family that extensively binds to retrotransposons is named Kruppel‐associated box‐zinc finger (KRAB‐ZFP) proteins 29, 52, 53, 54, 55, 56. These KRAB‐ZFP proteins are among the fastest evolving group of genes in the human genome 57, and it is estimated that this diversity enables their ability to recognise a large number of genomic retroelements 52. One of the first KRAB‐ZFP TFs that were found to contribute to retrotransposon silencing was ZFP809 58, 59. ZFP809 recognises and binds a sequence element in the promoter of ERVs and then recruits the epigenetic silencing machinery, at the core of which is TRIM28 59. KRAB‐ZFPs also silence other retrotransposons, in addition to ERVs. For example, the primate‐specific TFs ZNF91/93 evolved in response to expansion of the LINE L1PA3 subfamily and SINE‐related SVA elements in what has been called an evolutionary arms race 52. Further, the binding of KRAB‐ZNF proteins to a broad range of retrotransposons was confirmed on a larger scale through the analysis of genomewide binding profiles of 18 KRAB‐ZFP TFs 55. The majority (16 out of 18) of KRAB‐ZFPs were shown to bind to retrotransposons from LINE, ERV and SVA families.
Binding of sequence‐specific TFs to retrotransposons is the first step in the cascade that facilitates their epigenetic silencing. Sequence‐specific TFs recruit TRIM28 (also known as KAP1), which plays a pivotal role in the silencing cascade by facilitating histone tail methylation and DNA methylation. This ensures transposon silencing during early embryogenesis, and even in adult tissues 53, 54, 58, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71. Most retrotransposons are well under control through these sequence‐specific and epigenetic silencing mechanisms and not able to undergo transposition. Noteworthy, deletion of Trim28 in the maternal germline results in embryonic lethality 72, demonstrating the essentiality of this dynamic and robust defence system that ensures genomic and epigenetic stability.
Retrotransposons as a source of regulatory elements, noncoding genes and alternative gene isoforms
Retrotransposons in the human genome are identified based on their sequence similarity to retrotransposons, rather than their ability to copy and paste their DNA. In fact, only a very small subset of retrotransposons in the human genome can transpose their DNA, suggesting that the majority do not function as retrotransposons. Instead, these elements can acquire novel functions, either through a change in their retrotransposon identity over a long period of time or through the more rapid process of exaptation, where retrotransposon sequences are partially preserved but gain functionality in a different context. Such retrotransposons not only alter their own function, many times they confer novel or altered functions to the host.
Broadly, exaptation of retrotransposons can be classified according to the mechanism through which they influence the transcriptome. Firstly, retrotransposons can be co‐opted as enhancers (Fig 2A), influencing the expression of nearby genes without activating the retrotransposon itself. Secondly, retrotransposons can act as promoters that initiate transcription at the retroelement. Such elements can increase gene isoform diversity and introduce novel cell‐type specificity for existing protein‐coding genes (Fig 2B). In addition, they may act as their own promoter, driving expression of retrotransposon‐derived RNAs (Fig 2C and D). Thirdly, retrotransposons can directly be integrated into existing genes, increasing gene isoform diversity and influencing posttranscriptional regulation 73, 74. There are additional examples of retrotransposon exaptation that impact on small RNA pathways, and chromatin architecture and accessibility (see 6, 75, 76). In the following sections, we provide an overview of how exaptation of the regulatory elements of retrotransposons and specifically ERVs integrates them into existing regulatory networks as enhancers, promoters or as a source of novel noncoding and protein‐coding genes.
Figure 2. ERVs regulate and expand the transcriptome.
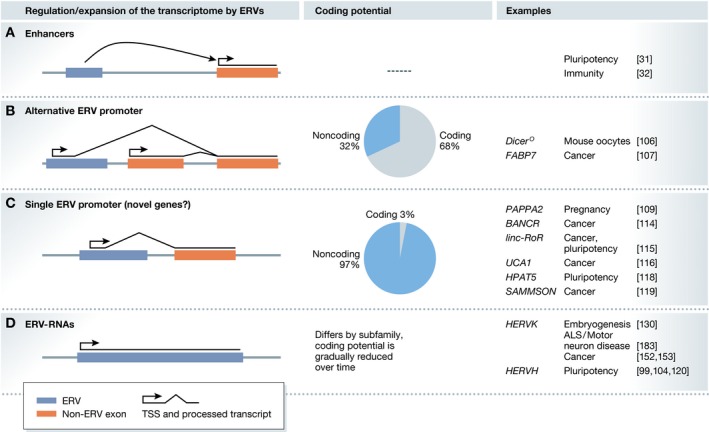
(A) ERVs can act as enhancers, regulating genes in the proximity. (B) ERVs can act as alternative promoters for protein‐coding and noncoding genes. (C) ERVs can provide the only promoter for a gene; such ERV‐derived genes are largely noncoding. (D) ERVs can be transcribed over their full length. Transcribed ERVs can generate proteins and peptides, but they can also generate noncoding RNAs.
Retrotransposons as enhancers
Despite the existence of multiple layers of repression targeted at the regulatory sequences of retrotransposons, many have been implicated in gene expression activation 77. Comparative genomics studies have shown that retrotransposons provide a rich source of regulatory innovation 78, 79; a number of these are under purifying selection, suggesting that exaptation of retrotransposon as regulatory elements occurs frequently 79, 80, 81, 82, 83, 84, 85. Indeed, many retrotransposons are unmethylated, bound by transcription factors and marked by H3K4me3, H3K27ac and H3K9ac, indicating an epigenetic state reminiscent of active enhancers and promoters (Fig 2A) 86, 87, 88, 89.
In an attempt to systematically identify active elements in the human genome, Thurman et al 3 analysed DNase‐Seq data from a large variety of human cell lines. DNase‐Seq captures regions of open chromatin, many of which were found to include transposable elements, among which ERVs were the most enriched class. The authors showed that ERVs were frequently cell‐type‐specific and active as enhancers 3. Among the primate‐specific open chromatin regions, transposable elements contribute up to 63% 90. While not all of these active chromatin regions contribute to regulatory networks, these numbers are a striking illustration of the potential of transposable elements to impact on the genomic regulatory landscape.
Specific examples of regulatory networks that have been systematically altered through retrotransposon‐derived enhancers can be found in embryonic stem cells (ESCs), germ cells and cells from organs related to sexual reproduction. Successful expansion of retrotransposon requires integration events in cells that pass their DNA to the offspring, and all of the above cell types provide such a window of opportunity 91. In ESCs, up to 25% of binding sites for the key pluripotency transcription factors OCT4 and NANOG were shown to originate from transposable elements 31. These binding sites are often primate‐specific and can integrate new genes into existing regulatory networks, demonstrating that they indeed act as regulatory elements. Besides these embryonic cells, the organs involved in pregnancy such as the placenta or endometrium have been found to employ a large number of retrotransposon‐derived enhancers 33, 92, 93, 94. By genomewide profiling of epigenetic marks and TF binding sites in mouse and rat trophoblast stem cells, Chuong et al 33 identified a specific class of ERVs, RLTR13D5, which was significantly enriched in enhancers that are active in the placenta. These ERVs contained binding sites for Eomes, Cdx2 and Elf5, transcription factors that are central to the trophoblast regulatory network; and genomewide binding site profiling by ChIP‐Seq has confirmed the binding of these TF to the sites in cultured cells. Through a similar genomics profiling approach, Lynch et al 94 identified a large number of retrotransposons that contribute regulatory elements to drive the endometrium expression profile in humans. Even though these retrotransposon‐derived regulatory elements are not able to copy and paste their DNA, these studies show that retrotransposon‐derived enhancer activity is frequently associated with tissues that are linked to embryogenesis, probably because these tissues are most likely to transmit new retrotransposon copies.
Large‐scale genomics surveys provide a genomewide overview, and transcriptomics data link retrotransposons to proximal gene expression; however, revealing the functions of individual elements is challenging. Chuong et al 32 used CRISPR–Cas9 to delete specific ERVs that are bound by transcription factors downstream of the interferon signalling pathway. Without these ERVs, expression of nearby genes was uncoupled from interferon signalling, impairing the inflammatory response to infection. Important binding sites tend to be well preserved between species 95; however, retrotransposon‐derived enhancers can contribute to interspecies differences by rewiring regulatory networks. These studies on retrotransposon enhancers demonstrate that this can affect a variety of cell types and cellular functions: from pluripotency in early embryos to the immune response in adults.
Retrotransposons as promoters: retrotransposon‐derived RNAs
The original purpose of retrotransposon regulatory sequences was to drive expression not of distal, but of the proximal DNA of the retrotransposon genes. Analysis of large‐scale transcriptome data sets has shown that many retrotransposons, and in particular ERVs, still act as transcription start sites (TSS) in different cell types and scenarios (Fig 2B–D). ERV regulatory elements act as promoters and TSS in early embryos 96, 97, 98, embryonic stem cells 99, 100 and adult tissues 90, 98, 101, 102, 103. Global estimates suggest that more than 30% of TSS overlap with retrotransposons 98. Many ERV‐derived promoters are highly cell‐type‐specific 96, 98, 104, and thereby increase transcriptome complexity.
The promoter and TSS of ERVs reside within the LTR elements (Fig 1D and E). Accordingly, most of the cell‐type specificity originates from these LTRs. The LTR sequence—and particularly splice sites within or downstream of the LTR—determines the impact on the transcriptome. LTR families that are devoid of splice sites initiate transcription of the adjacent, often proviral DNA. In contrast, LTR families with splice sites often generate “fusion transcripts” containing an ERV fragment at the 3′ end, and other, often non‐ERV exons in the remaining transcript 76, 96, 97, 101, 105.
LTRs with splicing elements as cell‐type‐specific alternative promoters
LTR elements that contain a splice donor site and are transcribed can be spliced to a splice acceptor site further downstream. If such an LTR promoter resides upstream of genes, the splice acceptor site can be provided by the genes' exons (Fig 2B). In this way, ERVs are a source of alternative promoters, generating transcript isoforms that are active in a specific cell type or that encode a truncated protein or transcript. One such example is Dicer, a gene that is central to the microRNA and RNAi pathways. In mice, Dicer has an oocyte‐specific transcript that is initiated in a rodent‐specific LTR element 106, and gives rise to a truncated Dicer isoform that more efficiently processes siRNAs. Deletion of the LTR element causes sterility in females with meiotic spindle defects and increases the expression of endogenous siRNA targets and of retrotransposons. Hence, this demonstrates that retrotransposons can become essential as cell‐type‐specific alternative promoters and first exons. Additional examples of tissue‐specific regulation of gene expression by retrotransposons exist in humans (107, 108, see 109 for an overview). Among all alternative TSS that overlap with an ERV element, 68% belong to protein‐coding genes (Fig 2B, estimated using RepeatMasker and Ensembl annotations for GRCh38). The large number of ERVs in promoters suggests that other examples like Dicer exist, where the alternative promoter initiated in an ERV has become essential for human biology.
Retrotransposons as a source of long noncoding RNAs
For many genes, the TSS in the retrotransposon is the only TSS. For transcription of these single‐TSS genes, the retrotransposons is essential as it provides the only promoter. The vast majority (97%) of such genes are long noncoding RNAs (lncRNAs; Fig 2C, estimated using RepeatMasker and Ensembl annotations for GRCh38).
LncRNAs show higher cell‐type specificity compared with protein‐coding genes, and their expression levels are generally lower 110. Thousands of lncRNAs have been discovered, and many are involved in human diseases such as cancer 111, 112, 113. Strikingly, 75–83% of lncRNAs contain transposable elements (TEs), a considerably higher percentage compared with protein‐coding genes 101, 104. Nineteen percent of lncRNAs consist of more than 50% TE sequence 101, suggesting that exaptation of TEs and evolution of lncRNAs are closely related. In fact, many lncRNAs were discovered without the knowledge that they substantially overlap with retrotransposons 114, 115, 116. TEs which are part of lncRNA exons show higher conservation in primates. This is indicative of purifying selection, reinforcing the idea that retrotransposons are a key element of lncRNAs 100, 101.
Most ERV‐initiated lncRNAs have promoters and TSS of retroviral origin. However, sometimes only a small part of the final transcript contains sequences from ERVs. As a result, their cellular function is often very specific to the individual lncRNA, and a large diversity of different ERV‐derived lncRNAs has been described. Among the lncRNAs that are initiated in retrotransposons and were shown to be functional are: linc‐RoR, a ncRNA that is required for pluripotency and influences p53 levels in response to DNA damage 115, 117; HPAT5, a lncRNA that plays a role in human preimplantation development and reprogramming, possibly through interaction with the let7 miRNA family 118; and the lncRNAs BANCR, SAMMSON and UCA1 that were discovered in cancer cells 114, 116, 119. The diversity of functions attributed to these lncRNAs—all initiated through an ERV promoter—demonstrates that retrotransposons contribute novel functions also by expanding the noncoding transcriptome.
Transcribed ERVs contribute to the coding and noncoding transcriptome
Some LTR families have a highly cell‐type‐specific promoter but are not spliced, and one example is LTR7 96, 99, 104, 120. LTR7 is bound by key transcription factors such as NANOG, LBP9 or the kinase ERK2 and shows high levels of H3K4me3, which is associated with promoter activity 96, 121, 122. LTR7 is the LTR of the human endogenous retrovirus h (HERVH), which provides one of the most striking examples for exaptation of retrotransposons in the human genome 99, 123, 124, 125. Unlike other LTRs that act as alternative promoters of non‐ERV exons, LTR7 primarily drives transcription of HERVH, generating hundreds of transcripts across the genome that are dominated by retroviral genes (Fig 2D). HERVH expression is highly specific to the pluripotent state of hESCs 99, 104, 120, 123. In contrast to LTRs which only provide a similar promoter and TSS to otherwise diverse genes, LTR7‐HERVH elements provide both promoter and a large part of the transcript sequence itself, suggesting that they form a class that shares some functionality. To investigate this hypothesis, Lu et al 99 generated multiple shRNAs against different parts of the HERVH transcripts, all of which led to differentiation of hESCs. These HERVH transcripts have lost their protein‐coding potential, suggesting that HERVH forms a class of lncRNAs that are essential for the maintenance of pluripotency in humans. HERVH transcripts are localised in the nucleus where they interact with proteins, potentially working together as noncoding regulators of gene expression 99. Notably, not all HERVH elements are fully transcribed, and they also contribute a number of lncRNAs through splicing 115, 118. Due to the repetitive nature of retrotransposons, it is rather difficult to identify the essential copies; an in‐depth understanding of the role of HERVH as a class and of individual HERVH‐derived transcripts in human pluripotency requires additional research efforts. While HERVH is the most prominent family, other retrotransposons also show transcription of the retroviral genes during early embryogenesis 96. These examples suggest that retrotransposons contribute to novel noncoding transcripts in the human genome and that exaptation of retroviral genes as noncoding RNAs may constitute a general mechanism.
Since retrotransposons frequently occur as fragments or mutated copies, most transcribed retrotransposons do not have strong protein‐coding potential. However, there are a few notable exceptions. Among the ERVs, the HERVK family is the most recently integrated family in the human genome 126. Proteins and viral particles transcribed from HERVK have been reported in germ cell tumours and human embryonal carcinoma cells 127, 128, 129. Recently, proteins from the HERVK genes have been identified in human blastocysts, indicating that human embryogenesis tolerates, or possibly benefits from HERVK expression 130. One of the HERVK‐encoded proteins was shown to interact with cellular RNAs, indicating that HERVK may be integrated into the cellular pathways 130. HERVK expression in early embryos is associated with an antiviral response mechanism, possibly protecting the embryo from exogenous viruses 130. However, among the ERV families which are transcribed in early embryos, only HERVK shows coding potential comparable to mRNAs 96, suggesting that the majority of ERVs in early embryos generate noncoding transcripts.
ERVs as markers of cell identity and cell potency
The unifying theme for ERV exaptation is that their highly specific regulatory elements contribute to genome regulation and transcription as cell‐type‐specific enhancers and promoters. Unlike protein‐coding genes that are most often used as markers of cellular identity, there are hundreds of ERV copies in the human genome. Further, ERV promoters are robustly activated and very specific for the cell type of interest. ERVs and ERV promoters therefore provide a unique resource as markers of cellular identity.
The notion of cellular identity is particularly relevant for work with pluripotent cells. ESCs are derived from the blastocyst and correspond to the pluripotent preimplantation epiblast cells within the inner cell mass 131, 132, 133. However, there are many differences between the in vitro expression profiles of ESCs and the in vivo expression profile of cells from the human blastocyst 134. These differences have been attributed to ESCs being in a developmentally more advanced, “primed” state, compared to the “naïve” state of pluripotency found in mouse ESCs and in the cells from human blastocysts 135. By using specific culture conditions, it is possible to alter the cell state of hESCs in a way that they more closely resemble naïve pluripotent cells 136, 137, 138, 139, 140. Strikingly, ERVs were found to be a key indicator for these alternative states of pluripotency (Fig 3A). In humans, the HERVK‐associated LTR elements (LTR5_Hs) are expressed specifically in naïve pluripotent cells and embryonic carcinoma cells 96, 129, 130, the HERVH‐associated LTR7Y elements have been used as a reporter for naïve pluripotency 96, and the HERVH‐associated LTR7 was used to isolate naïve human ESCs 122. The ability to demarcate specific cell states reflects the highly specific ERV expression in the distinct stages of early embryonic development: LTR7Y is specific for the blastocyst stage and only weakly expressed in primed ESCs, LTR7 is specific for the pluripotent cells from the blastocyst, and LTR5_Hs is expressed in the blastocyst and morula stage 96. LTR families that are specifically expressed in earlier stages such as the morula (LTR7B) and the 8‐cell stage (MLT2A1) have also been identified using single‐cell RNA‐Seq data from human embryos 96, 134, 141. In mice, a two‐cell specific ERV family (MuERVL) has been used to identify a subpopulation within mESCs that resemble properties of the totipotent two‐cell stage (Fig 3B) 142, 143, 144. The highly specific regulatory elements within the ERV promoters provide a powerful tool to identify novel cell states and evaluate cell potency even beyond currently available cell models.
Figure 3. Specific ERVs mark the different cellular identities in early embryonic development.
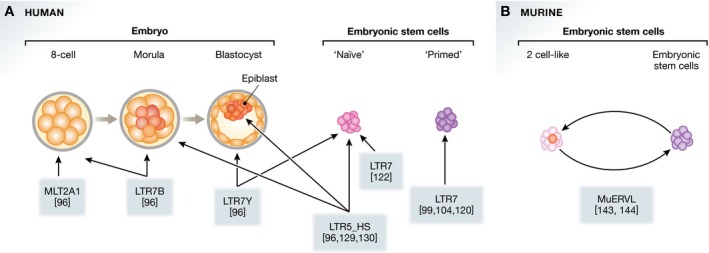
(A) Specific ERV families are expressed in the early human embryo, and in naïve and primed human embryonic stem cells (ESCs). (B) In mouse, it was found that ERVs are specifically activated in the two‐cell stage. These ERVs are spontaneously expressed in cells which show features of two‐cell‐like totipotent cells.
Retrotransposons in cancer: oncogenic lncRNAs, genomic instability and a link to immunotherapy
Many insights into the regulation, transcription and function of retrotransposons have been obtained from models of early embryonic development. However, retrotransposon expression can be observed across adult tissues 90, 98, 102, 103. This extensive contribution of retrotransposons to the transcriptome of adult tissues and nonembryonic cell types highlights the possibility that their dysregulation may be a factor in diseases such as cancer.
By comparing the expression profiles of tumours with the corresponding healthy tissue, it has been found that ERVs are among the dysregulated genes across a number of cancer types 145, 146, 147, 148, 149, 150, 151. The spectrum of possible functions of ERVs in cancer is as wide as their diversity in healthy tissues. In B‐cell‐derived Hodgkin lymphomas, ERVs from the THE1B family are systematically activated, leading—among others—to transcription of an alternative promoter of the oncogene CSF1R 151. Transcripts from the HERVK family were shown to be translated, generating proteins and peptides that are absent in normal tissues 26, 152, 153. In hepatocellular carcinomas, upregulation of ERV elements was specifically associated with a subtype of tumours that are HBV‐positive 146. The expression of these upregulated ERVs was driven by their promoter sequence, and many of the ERV‐derived RNAs were noncoding. While some cancers show concerted upregulation of ERVs, individual ERV‐derived lncRNAs contribute to the disease in other cases. The lncRNA SAMMSON is specifically induced in melanomas where it contributes to cancer cell‐specific mitochondrial functions 119. Inhibition of SAMMSON led to reduced tumour growth, and its cancer cell specificity makes it a candidate therapeutic target. The TSS of SAMMSON originates from an LTR1 retrotransposon; therefore, the specific upregulation may be related to its retroviral origin. Other ERV‐derived lncRNAs that appear to play a role in cancer are: BANCR—a lncRNA involved in melanoma cell migration 114; UCA1—a lncRNA that promotes cell growth and invasion in bladder cancer 116; and the aforementioned linc‐RoR that was found to contribute to breast cancer 154, pancreatic cancer 155 and hepatocellular carcinoma 156. Additional lncRNAs that are central to cancer have been discovered, reinforcing their important role in the disease 111, 112, 113. Many of these lncRNAs are likely to consist partly of retrotransposons, highlighting the potential impact that ERV‐derived transcripts might have in cancer.
While these studies allude to a possible oncogenic function for ERVs, other studies show that this does not always have to be the case. ERVs play an intriguing role in cancer drug responses, with applications in immunotherapy 157. DNA methyltransferase inhibitors (DNMTIs) can reduce DNA methylation levels, thereby activating genes that are silenced in tumours, some of which may be tumour suppressors. Genomic demethylation by DNMTIs also results in upregulation of ERVs and the generation of double‐stranded RNAs. These retroviral RNAs are associated with activation of antiviral response genes, indicating that ERV expression may potentially be clinically relevant in the context of immunotherapy 157, 158. Expression of ERVs was also associated with immune cytolytic activity 159, and neoantigens critical for successful immunotherapy 160, 161. Thus, ERVs may have opposing roles: as oncogenic drivers, but also possibly contributing to successful cancer treatment.
In contrast to ERVs—where a disease association is attributed to their cis‐regulatory potential or to their RNA product in trans—the research on LINE elements in cancer has been directed at their function as retrotransposon. The L1 family of LINEs is able to retrotranspose both in the germline and in somatic cells 162, 163, 164, 165; hence, this family is particularly interesting as it has potential (yet to some degree uncertain) roles in genomic instability and mutagenesis in cancer 166, 167. A systematic analysis of somatic retrotransposon events in five cancer types using whole‐genome sequencing of the tumour sample and matched blood samples confirmed that L1 elements are the most active family also in cancer (183 L1, 10 Alu and 1 ERV out of 194 in total) 168. L1 insertions are enriched in hypomethylated regions and associated with a change in gene expression 168. An interesting property of L1 elements is that they can copy and paste nonrepetitive sequences downstream of the element itself in the genome. This so‐called transduction was used by Tubio et al 169 to map somatic retrotransposition events back to their original loci. Ninety‐five percent of transduction events originated from only 72 L1 elements, which is similar to estimates for the number of retrotransposition‐competent elements found in the human population 164, suggesting that the majority of LINE instances are well under control or incapable of transpositions even in de‐regulated cancer genomes. For these somatically active instances, promoter hypomethylation indicated a potential loss of silencing that has been reported in other studies as well 170. Even though some of these transposition events disrupted exons and genes, the majority did not seem to affect gene expression, contrasting Lee et al 168, and suggesting that L1 retrotranspositions are most often harmless by‐products of a potentially important mutational process. These large pan‐cancer studies have the power to detect general mechanisms across cancer types; however, the occurrence of L1 retrotransposition events is often cancer‐type‐specific 168, 169. Additional insights into the frequency and relevance of L1 retrotransposition events have been obtained from studies focused on single cancer types, such as hepatocellular carcinoma 147, pancreatic ductal adenocarcinoma 171, oesophageal carcinoma 172, colorectal cancer 173 and gastric cancer 174, among others. Although the extent to which the somatic or germline retrotransposition contributes to cancer is still debatable, the phenomenon is frequently observed 175, and in specific cases has been directly linked to cancer initiation 176. This suggests that further exploration of both ERV and non‐ERV retrotransposons in cancer has the potential to uncover novel aspects of tumour biology.
Summary
Millions of fragments resembling retrotransposons exist in the human genome. Large‐scale genomics technologies have enabled researchers to obtain a genomewide view on retrotransposon activation, their regulation and their contribution to the transcriptome. Besides their contribution to innovation of genome biology, their contribution to human diseases has become a focus of research 175. Apart from cancer, expression of retrotransposons has been reported to be associated with multiple sclerosis 177, 178, 179, 180, schizophrenia 181 and amyotrophic lateral sclerosis 182, 183. Following these data‐driven discoveries of their activation, the key challenge is to understand the regulation of retrotransposons, identify essential elements among the large number of copies and ultimately assign a function for retrotransposon‐derived enhancers and transcripts in vitro and in vivo (see Box 2).
Overall, the pervasiveness of retrotransposons in the human genome and transcriptional landscape shows that there is a clear need to identify and assign functions to retrotransposons. However, this presents many challenges: the technical difficulties in analysing highly repetitive elements, combined with the huge diversity of possible mechanisms outlined in this review, demand new analytical approaches that deviate from methods applied to nonrepetitive genes. Despite these difficulties, there are many examples of retrotransposons that have acquired functions as enhancers, promoters or lncRNAs, suggesting that there are many additional cases yet to be discovered. In addition, retrotransposons are functionally important, with roles in development, cellular identity and disease. Technological developments that bring longer reads for sequencing, insights into genome or transcriptome structure, single‐cell resolution for transcriptomics and regulatory genomics, and almost base pair precision will be instrumental in interrogating the repetitive part of the human genome in more detail. Together with precise genome editing technology and integration of additional big data, research on retrotransposons promises to yield many new insights. This presents an exciting opportunity to advance our understanding of some of the most complex genomic elements that contribute to almost half of the human genome.
Box 2: In need of answers.
Among the many retrotransposons that are transcribed or which show regulatory activity, which elements are biologically relevant? Precise genome editing technology will enable the investigation of individual loci.
What is the function of individual elements and transcripts generated from retrotransposons? What are the pathways and interaction partners for retrotransposon‐derived RNAs?
Which retrotransposons are translated, and which retrotransposon‐derived RNAs are noncoding? Which retrotransposons are further processed into small RNAs?
Does co‐expression of similar retrotransposons such as HERVH indicate a common function? Only LTR7‐HERVH elements are expressed in ESCs, what is the difference to the LTR7Y and LTR7B subclasses of HERVH?
What are the precise sequences of retrotransposon‐derived RNAs? Will long read sequencing technology help overcome current limitations?
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Chloe Yap, Kevin Gonzales and Iwona Szczerbinska for discussion and comments on the manuscript and Engin Cukuroglu for help with data visualisation. This work was supported by the Agency for Science, Technology and Research (A∗STAR) of Singapore.
EMBO Reports (2016) 17: 1131–1144
See the Glossary for abbreviations used in this article.
References
- 1. Consortium TEP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi‐Moussavi A, Kheradpour P, Zhang Z, Wang J et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B et al (2012) The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F et al (2012) Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- 6. Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- 8. Goodier JL, Kazazian HH (2008) Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35 [DOI] [PubMed] [Google Scholar]
- 9. Smit AF (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9: 657–663 [DOI] [PubMed] [Google Scholar]
- 10. Smit AF (1996) The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6: 743–748 [DOI] [PubMed] [Google Scholar]
- 11. Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E (2013) On the immortality of television sets: “function” in the human genome according to the evolution‐free gospel of ENCODE. Genome Biol Evol 5: 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Souza FS, Franchini LF, Rubinstein M (2013) Exaptation of transposable elements into novel cis‐regulatory elements: is the evidence always strong? Mol Biol Evol 30: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niu D‐K, Jiang L (2013) Can ENCODE tell us how much junk DNA we carry in our genome? Biochem Biophys Res Commun 430: 1340–1343 [DOI] [PubMed] [Google Scholar]
- 14. Doolittle WF (2013) Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci USA 110: 5294–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Medstrand P, Mager DL (1998) Human‐specific integrations of the HERV‐K endogenous retrovirus family. J Virol 72: 9782–9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD et al (2013) Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seberg O, Petersen G (2009) A unified classification system for eukaryotic transposable elements should reflect their phylogeny. Nat Rev Genet 10: 276 [DOI] [PubMed] [Google Scholar]
- 18. Wicker T, Sabot F, Hua‐Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982 [DOI] [PubMed] [Google Scholar]
- 19. Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- 20. Bao W, Kojima KK, Kohany O (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smit A, Hubley R (2010) RepeatMasker. http://www.repeatmasker.org
- 22. Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS et al (2016) The UCSC genome browser database: 2016 update. Nucleic Acids Res 44: D717–D725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AF, Wheeler TJ (2016) The Dfam database of repetitive DNA families. Nucleic Acids Res 44: D81–D89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wheeler TJ, Clements J, Eddy SR, Hubley R, Jones TA, Jurka J, Smit AF, Finn RD (2013) Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res 41: D70–D82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rowe HM, Trono D (2011) Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- 26. Lower R, Lower J, Kurth R (1996) The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA 93: 5177–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Ann Rev Genet 42: 709–732 [DOI] [PubMed] [Google Scholar]
- 28. Weiss RA (2006) The discovery of endogenous retroviruses. Retrovirology 3: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13: 283–296 [DOI] [PubMed] [Google Scholar]
- 30. Mager DL, Henthorn PS (1984) Identification of a retrovirus‐like repetitive element in human DNA. Proc Natl Acad Sci USA 81: 7510–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G (2010) Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- 32. Chuong EB, Elde NC, Feschotte C (2016) Regulatory evolution of innate immunity through co‐option of endogenous retroviruses. Science 351: 1083–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chuong EB, Rumi MA, Soares MJ, Baker JC (2013) Endogenous retroviruses function as species‐specific enhancer elements in the placenta. Nat Genet 45: 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kazazian HH Jr, Goodier JL (2002) LINE drive. Retrotransposition and genome instability. Cell 110: 277–280 [DOI] [PubMed] [Google Scholar]
- 35. Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916 [DOI] [PubMed] [Google Scholar]
- 36. Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810 [DOI] [PubMed] [Google Scholar]
- 37. Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L (1987) Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1: 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ et al (2015) Primate‐specific ORF0 contributes to retrotransposon‐mediated diversity. Cell 163: 583–593 [DOI] [PubMed] [Google Scholar]
- 39. Deininger PL, Batzer MA (2002) Mammalian retroelements. Genome Res 12: 1455–1465 [DOI] [PubMed] [Google Scholar]
- 40. Speek M (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swergold GD (1990) Identification, characterization, and cell specificity of a human LINE‐1 promoter. Mol Cell Biol 10: 6718–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macia A, Munoz‐Lopez M, Cortes JL, Hastings RK, Morell S, Lucena‐Aguilar G, Marchal JA, Badge RM, Garcia‐Perez JL (2011) Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol 31: 300–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beck CR, Garcia‐Perez JL, Badge RM, Moran JV (2011) LINE‐1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12: 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- 45. Ullu E, Tschudi C (1984) Alu sequences are processed 7SL RNA genes. Nature 312: 171–172 [DOI] [PubMed] [Google Scholar]
- 46. Houck CM, Rinehart FP, Schmid CW (1979) A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol 132: 289–306 [DOI] [PubMed] [Google Scholar]
- 47. Okada N, Hamada M, Ogiwara I, Ohshima K (1997) SINEs and LINEs share common 3′ sequences: a review. Gene 205: 229–243 [DOI] [PubMed] [Google Scholar]
- 48. Dewannieux M, Esnault C, Heidmann T (2003) LINE‐mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- 49. Ostertag EM, Goodier JL, Zhang Y, Kazazian HH (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73: 1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY (1994) Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon‐intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem 269: 8466–8476 [PubMed] [Google Scholar]
- 51. Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA (2005) SVA elements: a hominid‐specific retroposon family. J Mol Biol 354: 994–1007 [DOI] [PubMed] [Google Scholar]
- 52. Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D (2014) An evolutionary arms race between KRAB zinc‐finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard‐Liesching H, Verp S, Marquis J et al (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- 54. Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D (2012) The KRAB‐ZFP/KAP1 system contributes to the early embryonic establishment of site‐specific DNA methylation patterns maintained during development. Cell Rep 2: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM et al (2015) C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol 33: 555–562 [DOI] [PubMed] [Google Scholar]
- 56. Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D (2013) De novo DNA methylation of endogenous retroviruses is shaped by KRAB‐ZFPs/KAP1 and ESET. Development 140: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nowick K, Hamilton AT, Zhang H, Stubbs L (2010) Rapid sequence and expression divergence suggest selection for novel function in primate‐specific KRAB‐ZNF genes. Mol Biol Evol 27: 2606–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS (2015) The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev 29: 538–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolf D, Goff SP (2009) Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turelli P, Castro‐Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D (2014) Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 24: 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pezic D, Manakov SA, Sachidanandam R, Aravin AA (2014) piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28: 1410–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- 63. Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat‐associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP et al (2007) Genome‐wide maps of chromatin state in pluripotent and lineage‐committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fadloun A, Le Gras S, Jost B, Ziegler‐Birling C, Takahashi H, Gorab E, Carninci P, Torres‐Padilla ME (2013) Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE‐1 by RNA. Nat Struct Mol Biol 20: 332–338 [DOI] [PubMed] [Google Scholar]
- 66. Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP et al (2015) Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Castro‐Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D (2014) Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Walsh CP, Chaillet JR, Bestor TH (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- 69. Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511: 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J et al (2014) The DNA methylation landscape of human early embryos. Nature 511: 606–610 [DOI] [PubMed] [Google Scholar]
- 71. Friedli M, Trono D (2015) The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 31: 429–451 [DOI] [PubMed] [Google Scholar]
- 72. Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson‐Smith A, Knowles BB (2012) Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335: 1499–1502 [DOI] [PubMed] [Google Scholar]
- 73. Zemojtel T, Penzkofer T, Schultz J, Dandekar T, Badge R, Vingron M (2007) Exonization of active mouse L1s: a driver of transcriptome evolution? BMC Genom 8: 392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sorek R, Ast G, Graur D (2002) Alu‐containing exons are alternatively spliced. Genome Res 12: 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Elbarbary RA, Lucas BA, Maquat LE (2016) Retrotransposons as regulators of gene expression. Science 351: aac7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cowley M, Oakey RJ (2013) Transposable elements re‐wire and fine‐tune the transcriptome. PLoS Genet 9: e1003234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D (2013) TRIM28 repression of retrotransposon‐based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Polak P, Domany E (2006) Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genom 7: 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lowe CB, Haussler D (2012) 29 mammalian genomes reveal novel exaptations of mobile elements for likely regulatory functions in the human genome. PLoS ONE 7: e43128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D (2006) A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441: 87–90 [DOI] [PubMed] [Google Scholar]
- 81. del Rosario RC, Rayan NA, Prabhakar S (2014) Noncoding origins of anthropoid traits and a new null model of transposon functionalization. Genome Res 24: 1469–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lowe CB, Bejerano G, Haussler D (2007) Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci USA 104: 8005–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thompson PJ, Macfarlan TS, Lorincz MC (2016) Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62: 766–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gerdes P, Richardson SR, Mager DL, Faulkner GJ (2016) Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol 17: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rebollo R, Romanish MT, Mager DL (2012) Transposable elements: an abundant and natural source of regulatory sequences for host genes. Ann Rev Genet 46: 21–42 [DOI] [PubMed] [Google Scholar]
- 86. Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH et al (2008) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Marino‐Ramirez L, Jordan IK (2006) Transposable element derived DNaseI‐hypersensitive sites in the human genome. Biol Direct 1: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gifford WD, Pfaff SL, Macfarlan TS (2013) Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 23: 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, Tapscott SJ (2013) DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet 9: e1003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jacques PE, Jeyakani J, Bourque G (2013) The majority of primate‐specific regulatory sequences are derived from transposable elements. PLoS Genet 9: e1003504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Levin HL, Moran JV (2011) Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lynch VJ, Leclerc RD, May G, Wagner GP (2011) Transposon‐mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159 [DOI] [PubMed] [Google Scholar]
- 93. Johansen T, Holm T, Bjorklid E (1989) Members of the RTVL‐H family of human endogenous retrovirus‐like elements are expressed in placenta. Gene 79: 259–267 [DOI] [PubMed] [Google Scholar]
- 94. Lynch VJ, Nnamani MC, Kapusta A, Brayer K, Plaza SL, Mazur EC, Emera D, Sheikh SZ, Grutzner F, Bauersachs S et al (2015) Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep 10: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Göke J, Jung M, Behrens S, Chavez L, O'Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M (2011) Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput Biol 7: e1002304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I (2015) Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16: 135–141 [DOI] [PubMed] [Google Scholar]
- 97. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- 98. Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T et al (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- 99. Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH (2014) The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425 [DOI] [PubMed] [Google Scholar]
- 100. Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A et al (2014) Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 46: 558–566 [DOI] [PubMed] [Google Scholar]
- 101. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, Trowbridge CA, Maller JB, Tukiainen T, Lek M (2015) The Genotype‐Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ et al (2015) Human genomics. The human transcriptome across tissues and individuals. Science 348: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kelley D, Rinn J (2012) Transposable elements reveal a stem cell‐specific class of long noncoding RNAs. Genome Biol 13: R107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Medstrand P, Landry J‐R, Mager DL (2001) Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein CI genes in humans. J Biol Chem 276: 1896–1903 [DOI] [PubMed] [Google Scholar]
- 106. Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P (2013) A retrotransposon‐driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155: 807–816 [DOI] [PubMed] [Google Scholar]
- 107. Lock FE, Rebollo R, Miceli‐Royer K, Gagnier L, Kuah S, Babaian A, Sistiaga‐Poveda M, Lai CB, Nemirovsky O, Serrano I et al (2014) Distinct isoform of FABP7 revealed by screening for retroelement‐activated genes in diffuse large B‐cell lymphoma. Proc Natl Acad Sci USA 111: E3534–E3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. van de Lagemaat LN, Landry JR, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530–536 [DOI] [PubMed] [Google Scholar]
- 109. Cohen CJ, Lock WM, Mager DL (2009) Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114 [DOI] [PubMed] [Google Scholar]
- 110. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q et al (2015) Comprehensive genomic characterization of long non‐coding RNAs across human cancers. Cancer Cell 28: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21: 1253–1261 [DOI] [PubMed] [Google Scholar]
- 114. Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA (2012) BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res 22: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S et al (2010) Large intergenic non‐coding RNA‐RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang F, Li X, Xie X, Zhao L, Chen W (2008) UCA1, a non‐protein‐coding RNA up‐regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 582: 1919–1927 [DOI] [PubMed] [Google Scholar]
- 117. Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, Lu Z, Bai C, Watabe K, Mo YY (2013) The human long non‐coding RNA‐RoR is a p53 repressor in response to DNA damage. Cell Res 23: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Durruthy‐Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, Davila J, Mall M, Wong WH, Wysocka J et al (2016) The primate‐specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet 48: 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K et al (2016) Melanoma addiction to the long non‐coding RNA SAMMSON. Nature 531: 518–522 [DOI] [PubMed] [Google Scholar]
- 120. Santoni FA, Guerra J, Luban J (2012) HERV‐H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9: 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Goke J, Chan YS, Yan J, Vingron M, Ng HH (2013) Genome‐wide kinase‐chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol Cell 50: 844–855 [DOI] [PubMed] [Google Scholar]
- 122. Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV et al (2014) Primate‐specific endogenous retrovirus‐driven transcription defines naive‐like stem cells. Nature 516: 405–409 [DOI] [PubMed] [Google Scholar]
- 123. Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A et al (2014) Dynamic regulation of human endogenous retroviruses mediates factor‐induced reprogramming and differentiation potential. Proc Natl Acad Sci USA 111: 12426–12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Robbez‐Masson L, Rowe HM (2015) Retrotransposons shape species‐specific embryonic stem cell gene expression. Retrovirology 12: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Izsvak Z, Wang J, Singh M, Mager DL, Hurst LD (2016) Pluripotency and the endogenous retrovirus HERVH: conflict or serendipity? BioEssays 38: 109–117 [DOI] [PubMed] [Google Scholar]
- 126. Mager DL, Medstrand P (2005) Retroviral repeat sequences. eLS doi: 10.1038/npg.els.0005062
- 127. Tonjes RR, Lower R, Boller K, Denner J, Hasenmaier B, Kirsch H, Konig H, Korbmacher C, Limbach C, Lugert R et al (1996) HERV‐K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol 13(Suppl. 1): S261–S267 [DOI] [PubMed] [Google Scholar]
- 128. Boller K, Konig H, Sauter M, Mueller‐Lantzsch N, Lower R, Lower J, Kurth R (1993) Evidence that HERV‐K is the endogenous retrovirus sequence that codes for the human teratocarcinoma‐derived retrovirus HTDV. Virology 196: 349–353 [DOI] [PubMed] [Google Scholar]
- 129. Fuchs NV, Loewer S, Daley GQ, Izsvák Z, Lower J, Lower R (2013) Human endogenous retrovirus K (HML‐2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 10: 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY et al (2015) Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Thomson JA, Itskovitz‐Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- 132. Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- 133. Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J et al (2013) Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139 [DOI] [PubMed] [Google Scholar]
- 135. Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4: 487–492 [DOI] [PubMed] [Google Scholar]
- 136. Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L et al (2014) Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez‐Caliani AJ, Deng X, Cavanaugh C, Cook S et al (2014) Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA 111: 4484–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH (2013) Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13: 663–675 [DOI] [PubMed] [Google Scholar]
- 139. Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben‐Yosef D, Kalma Y, Viukov S, Maza I, Zviran A et al (2013) Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286 [DOI] [PubMed] [Google Scholar]
- 140. Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W et al (2014) Resetting transcription factor control circuitry toward ground‐state pluripotency in human. Cell 158: 1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE et al (2013) Genetic programs in human and mouse early embryos revealed by single‐cell RNA sequencing. Nature 500: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ishiuchi T, Enriquez‐Gasca R, Mizutani E, Boskovic A, Ziegler‐Birling C, Rodriguez‐Terrones D, Wakayama T, Vaquerizas JM, Torres‐Padilla ME (2015) Early embryonic‐like cells are induced by downregulating replication‐dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671 [DOI] [PubMed] [Google Scholar]
- 143. Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kigami D, Minami N, Takayama H, Imai H (2003) MuERV‐L is one of the earliest transcribed genes in mouse one‐cell embryos. Biol Reprod 68: 651–654 [DOI] [PubMed] [Google Scholar]
- 145. Wang‐Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL (2003) Quantitation of HERV‐K env gene expression and splicing in human breast cancer. Oncogene 22: 1528–1535 [DOI] [PubMed] [Google Scholar]
- 146. Hashimoto K, Suzuki AM, Dos Santos A, Desterke C, Collino A, Ghisletti S, Braun E, Bonetti A, Fort A, Qin XY et al (2015) CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus‐induced tumors. Genome Res 25: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shukla R, Upton KR, Munoz‐Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S et al (2013) Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kassiotis G (2014) Endogenous retroviruses and the development of cancer. J Immunol 192: 1343–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller‐Lantzsch N (2008) Endogenous retroviruses and cancer. Cell Mol Life Sci 65: 3366–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yi J‐M, Kim H‐M, Kim H‐S (2004) Expression of the human endogenous retrovirus HERV‐W family in various human tissues and cancer cells. J Gen Virol 85: 1203–1210 [DOI] [PubMed] [Google Scholar]
- 151. Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E et al (2010) Derepression of an endogenous long terminal repeat activates the CSF1R proto‐oncogene in human lymphoma. Nat Med 16: 571–579 [DOI] [PubMed] [Google Scholar]
- 152. Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J (1995) Identification of a Rev‐related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV‐K. J Virol 69: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Downey RF, Sullivan FJ, Wang‐Johanning F, Ambs S, Giles FJ, Glynn SA (2015) Human endogenous retrovirus K and cancer: innocent bystander or tumorigenic accomplice? Int J Cancer 137: 1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J (2014) LincRNA‐ROR induces epithelial‐to‐mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5: e1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zhan H‐X, Wang Y, Li C, Xu J‐W, Zhou B, Zhu J‐K, Han H‐F, Wang L, Wang Y‐S, Hu S‐Y (2016) LincRNA‐ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett 374: 261–271 [DOI] [PubMed] [Google Scholar]
- 156. Takahashi K, Yan IK, Kogure T, Haga H, Patel T (2014) Extracellular vesicle‐mediated transfer of long non‐coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 4: 458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A et al (2015) Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ et al (2015) DNA‐demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kassiotis G, Stoye JP (2016) Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 16: 207–219 [DOI] [PubMed] [Google Scholar]
- 161. Cherkasova E, Scrivani C, Doh S, Weisman Q, Takahashi Y, Harashima N, Yokoyama H, Srinivasan R, Linehan WM, Lerman MI et al (2016) Detection of an immunogenic HERV‐E envelope with selective expression in clear cell kidney cancer. Cancer Res 76: 2177–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE (2010) A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV (2010) LINE‐1 retrotransposition activity in human genomes. Cell 141: 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Brouha B, Schustak J, Badge RM, Lutz‐Prigge S, Farley AH, Moran JV, Kazazian HH Jr (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 100: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH (2010) LINE‐1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci 33: 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE (2010) Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y (1992) Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52: 643–645 [PubMed] [Google Scholar]
- 168. Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ III, Lohr JG, Harris CC, Ding L, Wilson RK et al (2012) Landscape of somatic retrotransposition in human cancers. Science 337: 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K et al (2014) Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong‐ngam D, Voravud N, Sriuranpong V, Mutirangura A (2004) Distinctive pattern of LINE‐1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23: 8841–8846 [DOI] [PubMed] [Google Scholar]
- 171. Rodić N, Steranka JP, Makohon‐Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P (2015) Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med 21: 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Doucet‐O'Hare TT, Rodic N, Sharma R, Darbari I, Abril G, Choi JA, Young Ahn J, Cheng Y, Anders RA, Burns KH et al (2015) LINE‐1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc Natl Acad Sci USA 112: E4894–E4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B et al (2012) Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon‐Moore A et al (2015) Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hancks DC, Kazazian HH Jr (2016) Roles for retrotransposon insertions in human disease. Mob DNA 7: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE (2016) A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res 26: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C (2011) Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim Biophys Acta 1812: 162–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Perron H, Garson JA, Bedin F, Beseme F, Paranhos‐Baccala G, Komurian‐Pradel F, Mallet F, Tuke PW, Voisset C, Blond JL et al (1997) Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc Natl Acad Sci USA 94: 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Perron H, Lalande B, Gratacap B, Laurent A, Genoulaz O, Geny C, Mallaret M, Schuller E, Stoebner P, Seigneurin JM (1991) Isolation of retrovirus from patients with multiple sclerosis. Lancet 337: 862–863 [DOI] [PubMed] [Google Scholar]
- 180. Perron H, Geny C, Laurent A, Mouriquand C, Pellat J, Perret J, Seigneurin JM (1989) Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particles. Res Virol 140: 551–561 [DOI] [PubMed] [Google Scholar]
- 181. Karlsson H, Schröder J, Bachmann S, Bottmer C, Yolken R (2004) HERV‐W‐related RNA detected in plasma from individuals with recent‐onset schizophrenia or schizoaffective disorder. Mol Psychiatry 9: 12–13 [DOI] [PubMed] [Google Scholar]
- 182. Manghera M, Ferguson J, Douville R (2014) Endogenous retrovirus‐K and nervous system diseases. Curr Neurol Neurosci Rep 14: 488 [DOI] [PubMed] [Google Scholar]
- 183. Li W, Lee MH, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K et al (2015) Human endogenous retrovirus‐K contributes to motor neuron disease. Sci Transl Med 7: 307ra153 [DOI] [PMC free article] [PubMed] [Google Scholar]


