Abstract
Mitotic spindle orientation is essential for cell fate decisions, epithelial maintenance, and tissue morphogenesis. In most animal cell types, the dynein motor complex is anchored at the cell cortex and exerts pulling forces on astral microtubules to position the spindle. Early studies identified the evolutionarily conserved Gαi/LGN/NuMA complex as a key regulator that polarizes cortical force generators. In recent years, a combination of genetics, biochemistry, modeling, and live imaging has contributed to decipher the mechanisms of spindle orientation. Here, we highlight the dynamic nature of the assembly of this complex and discuss the molecular regulation of its localization. Remarkably, a number of LGN‐independent mechanisms were described recently, whereas NuMA remains central in most pathways involved in recruiting force generators at the cell cortex. We also describe the emerging role of the actin cortex in spindle orientation and discuss how dynamic astral microtubule formation is involved. We further give an overview on instructive external signals that control spindle orientation in tissues. Finally, we discuss the influence of cell geometry and mechanical forces on spindle orientation.
Keywords: actin cortex, astral microtubules, cell geometry, NuMA, spindle orientation
Subject Categories: Cell Cycle
Glossary
- Antxr2a
anthrax receptor 2a
- APC
adenomatous polyposis coli
- aPKC
atypical protein kinase C
- APs
apical progenitors
- Arp3
actin‐related protein 3
- AurA
Aurora A
- CDK1
cyclin‐dependent kinase 1
- CYK4
Rho family GTPase‐activating protein CYK4/MgcRacGAP
- Dgrip75
Drosophila grip‐motif‐polypeptide 75
- Dlg
Discs large
- Dsh DEP domain
dishevelled/EGL10/pleckstrin domain
- EB1
end binding family member 1
- EB3
end binding family member 3
- ECM
extracellular matrix
- Ed
echinoid
- ERM
Ezrin–radixin–moesin
- EVL
enveloping cell layer
- 4.1G
band 4.1‐like 2 protein/EPB41L2
- 4.1R
band 4.1 protein/EPB41
- Fz–Dsh
frizzled/disheveled
- GAP
GTPase‐activating protein
- GEF
guanine exchange factor
- GOA1
guanine nucleotide‐binding protein G (o) subunit alpha
- GPA16
G protein alpha subunit
- GPR
G protein regulator
- GPR1/2
G protein regulator 1/2
- HTT
huntingtin
- ILK
integrin‐linked kinase
- Insc
inscuteable
- Lgl
lethal giant larvae
- LGN
leucine–glycine–asparagine
- LIN5
spindle apparatus protein lin‐5
- MAP4
microtubule‐associated protein 4
- MDCK
Madin–Darby canine kidney
- MISP
mitotic interactor and substrate of Plk1
- MKLP1
mitotic kinesin‐like protein 1
- MT
microtubule
- Mud
mushroom body defect
- NB
neuroblast
- NRK
normal rat kidney
- NuMA
nuclear and mitotic apparatus
- Par3
partitioning‐defective 3
- Pcnt
pericentrin
- PCP
planar cell polarity
- Pins
partner of inscuteable
- PIP
phosphatidylinositol phosphate
- PIP2
phosphatidylinositol 4,5‐bisphosphate
- PKC‐ζ
protein kinase C‐ζ
- Plk1
polo‐like kinase 1
- Rab11
Ras‐related protein Rab11, recycling endosome GTPase
- Ran
Ras‐related nuclear protein
- SOP
sensory organ precursor
- T2055
threonine 2055
- TCJs
tricellular junctions
- TPR
tetratricopeptide repeats
- Tre1
trapped in endoderm 1
- zDia2
diaphanous‐related formin 2 (in zebrafish)
- γ‐TuRCs
γ‐tubulin ring complexes
Introduction
During division, cells face numerous challenges. First, to avoid genetic aberrations, the genetic material has to be correctly segregated into the daughter cells. This is achieved through the formation of a dedicated bipolar structure, the mitotic spindle, which involves profound remodeling of the microtubule network: Kinetochore microtubules (MTs) attach to chromosomal kinetochores, interpolar and central spindle MTs position the furrow upon cytokinesis, and astral microtubules anchor the furrow to the cell cortex 1, 2. Second, depending on the cell type and the developmental stage, a dividing cell may need to produce daughters of different size and/or fate. Here again, the spindle plays a critical role, since its orientation determines the axis of cell division and thereby decides about symmetry or asymmetry of the division. Correct spindle orientation is important for asymmetric segregation of polarized cell fate determinants in Drosophila neuroblasts thus allowing asymmetric cell divisions 3. Similarly, during the first division of the C. elegans zygote, spindle displacement toward the posterior pole is crucial for the production of two daughter cells of asymmetric size and different fate 4, 5. Third, daughter cells resulting from a division must be correctly positioned in order to maintain tissue structure and/or contribute to tissue morphogenesis in metazoans. In epithelia, planar orientation of divisions is required for the maintenance of daughter cells in the plane of the tissue 6, 7, 8. In addition, polarized orientation of cell divisions within the epithelium plane can contribute to tissue elongation 9, 10. Conversely, spindle orientation along the apico‐basal axis is necessary for asymmetric cell division and epithelial stratification during skin development in the mouse embryo 11. Altogether, spindle orientation and positioning are involved in fundamental developmental processes and in tissue homeostasis, and their deregulation has been correlated with different pathologies, including microcephaly and cancer 12, 13. This underscores the importance of understanding the mechanisms mediating these processes. The multiple roles of oriented cell divisions in animal development and pathologies have been reviewed elsewhere 14, 15, 16, 17, 18, 19. The focus of this review was to provide a comprehensive overview of the mechanisms and regulatory inputs of spindle orientation in metazoans. Of note, spindle positioning mechanisms are also extensively studied during asymmetric division of the budding yeast; however, this model shows important differences to higher eukaryotes and therefore will not be discussed here (see Box 1 for a brief overview).
Box 1: Spindle orientation in budding yeast.
Spindle positioning is well characterized during the asymmetric division of the budding yeast S. cerevisiae 23. In this model system, the spindle is positioned in relation to the bud neck to allow the correct segregation of chromosomes between mother and daughter cells. Spindle orientation in yeast also depends on the interaction of astral microtubules with cortically localized factors; however, cortical factors (e.g., Num1; reviewed in 24) are not homologous to those found in higher eukaryotes. In addition, spindle positioning is achieved by two sequential and clearly distinct pathways 25. In pre‐anaphase, spindle orientation along the mother–bud axis is not linked to dynein‐dependent forces, but instead depends on the displacement of astral microtubules along actin cables. This process depends on the interaction between the MT tip protein Bim1 (homologous to EB1) and the myosin Myo2 via the yeast‐specific adaptor Kar 9 (homologous to APC) 25, 26. In anaphase, spindle displacement into the bud neck is mediated by pulling forces exerted by cortically anchored dynein. The switch between both pathways is linked to the removal of the dynein inhibitor She1 from astral microtubules in the metaphase–anaphase transition 27. In anaphase, dynein (Dyn1) is delivered to the cortex, where it binds to the cortical factor Num1 through a mechanism of “off‐loading” from astral microtubules 28. Thus, pre‐targeting of dynein to microtubule plus‐ends is necessary for spindle positioning in yeast 25, 28. Dynein pre‐targeting depends on Pac1 and Bik1 (LIS1 and CLIP‐170 homologs, respectively) 25. Interestingly, in mammalian interphase cells, dynein localizes to MT plus‐ends in a CLIP170‐ and EB1‐dependent manner 29. However, whether the localization of dynein to MT plus‐ends is important for its delivery to the cortex during vertebrate mitosis remains to be investigated. Notably, both pathways acting in spindle positioning in yeast do not rely on the polarization of cortical anchors as it is seen in higher eukaryotes. Instead, they rely on the asymmetric localization of Kar 9 and Dyn1 to the astral microtubule plus‐ends emanating from the daughter spindle pole 26.
The orientation of the mitotic spindle in animal cells can be influenced by geometric cues, internal cues, and external cues. Hertwig first proposed more than a century ago that cells orient their spindles along the long axis of the cell, arguing for a role of cellular geometry in controlling the plane of division 20. While this rule applies to many situations, orientation of the spindle is also often set by specific polarity cues. At the turn of the century, the realization that the orientation of the axis of division often correlates with fate choices in Drosophila and C. elegans models of asymmetric cell division prompted a series of studies that linked regulators of cell polarity with the molecular control of spindle positioning and orientation. In this context, a role of Gαi subunits of heterotrimeric G proteins and the adaptor molecule LGN (leucine–glycine–asparagine) in spindle orientation was initially identified in Drosophila embryonic neuroblasts 21, 22. Later work revealed the evolutionary conservation of this complex in numerous metazoans, and how it interacts with the NuMA (nuclear and mitotic apparatus) adaptor to recruit the dynein motor complex to the cell cortex in symmetrically and asymmetrically dividing cells. Indeed, in most animal cell types oriented cell divisions involve the transmission of localized pulling forces located at the cell cortex to astral microtubules, resulting in the positioning of the mitotic spindle. As a consequence, the cell cortex, the specific mechanisms that recruit and localize force generators, and the astral microtubule network have emerged as the three essential levels of regulation for spindle orientation.
In this review, we will first briefly review the role of the so‐called LGN complex and discuss recent literature that refines our understanding of the spatial and temporal regulation of the activity of this complex. We will also discuss recently described alternative mechanisms for the recruitment of force generators at the cell cortex. In the second part of the review, we will review the emerging roles of the actin cortex on spindle orientation. In the third part, we will show how mechanisms that regulate astral microtubule nucleation, dynamics, and interaction with the cortex can influence the transmission of cortical forces to the spindle and control the orientation of cell divisions. Finally, we will consider cells in their tissue context and present the extracellular signaling pathways that have been recently demonstrated to modulate spindle orientation in different organisms and tissues. We will also review recent studies that address the influence of geometrical cues and external forces applied at the tissue scale on spindle orientation without the involvement of polarized signaling pathways.
Revisiting an old leader: the Gαi/LGN/NuMA/dynein complex
The historical complex
A number of genetic studies have shown that an evolutionarily conserved molecular complex composed of the heterotrimeric Gα protein Gαi, LGN, and NuMA (respectively Gαi, Pins, and Mud in Drosophila, and GOA1/GPA16, GPR1/2, and LIN5 in C. elegans, and hereafter called the LGN complex for simplicity; see Table 1 and Fig 1) has core functions in spindle orientation and positioning in different tissues both in invertebrate and vertebrate species 6, 7, 11, 21, 22, 30, 31, 32, 33, 34, 35 (reviewed in 19). During mitosis, this complex is localized to a particular subcortical domain and directs the recruitment of the minus‐end‐directed microtubule motor dynein 36, 37, 38 (Fig 1A). The directed movement of cortically anchored dynein along astral microtubules generates pulling forces on the spindle poles leading to the orientation and/or positioning of the spindle. Therefore, the specific localization of the LGN complex determines the site of force concentration and the axis of spindle orientation. Consistently, the apical localization of Pins/Mud or LGN/NuMA directs spindle orientation along the apico‐basal axis in Drosophila neuroblasts (Fig 1B) and mouse skin progenitors, respectively 11, 22, 39, 40, 41, 42. In the C. elegans zygote, enrichment of GPR1/2 at the posterior cortex is necessary for spindle positioning along the antero‐posterior axis 32, 34 (Fig 1B). Furthermore, the lateral localization of the LGN complex regulates planar spindle orientation of progenitors in chick and mouse neuroepithelium 6, 7, 33 as well as during epithelial morphogenesis of Drosophila and mammalian cells 8, 43 (Fig 1B).
Table 1.
Genes mentioned in this review and names of their homologs in model organisms
| Vertebrates | Drosophila | C. elegans |
|---|---|---|
| Gαi 1, 2, 3 | Gαi | GOA1/GPA16 |
| LGN (GPSM2, mPins) | Pins (partner of inscuteable, Rapsynoid) | GPR1/2 |
| NuMA | Mud | Lin‐5 |
| Insc (mInsc) | inscuteable | – |
| Par3 | Bazooka (dPar3) | Par3 |
| Afadin | Canoe | Ce‐AF‐6 |
| RGS14, RGS12 | loco | RGS‐7 |
| Ric8a | dRic8 | Ric8 (synembrin) |
| Dlg1 (SAP97, hDLG) | Dlg | DLG‐1 |
| Dia2 (zDia2) | diaphanous | – |
| ERM (ezrin–radixin–moesin) | moesin | ERM‐1 |
| Dynactin‐1 (DCTN1, p150) | p150‐glued | DNC‐1 |
| GCP4 | Dgrip75 | – |
Figure 1. The LGN complex.
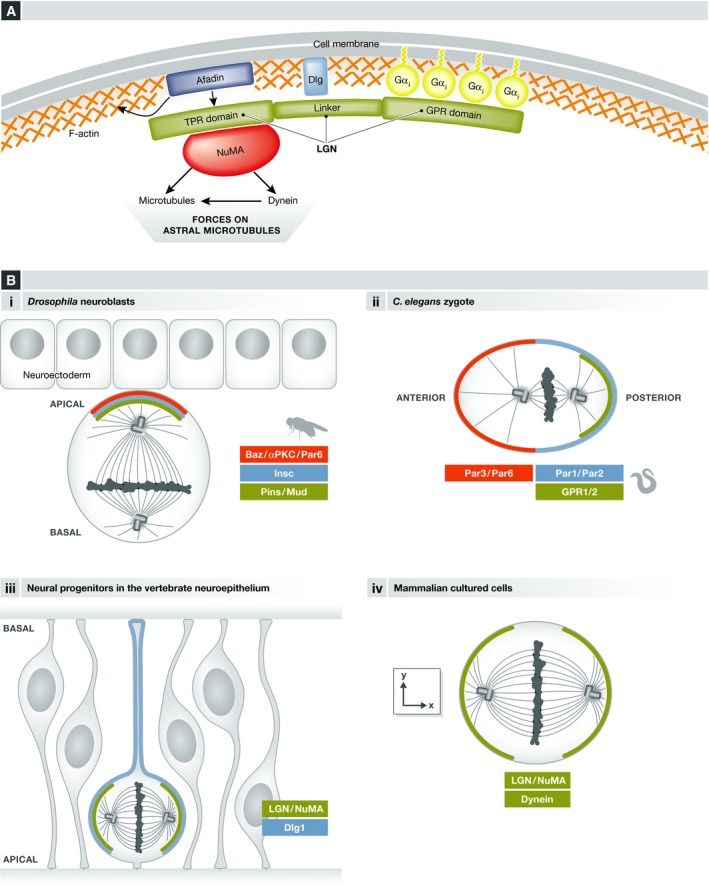
(A) The scheme shows the LGN domains and its interactions with Gαi membrane‐anchored subunits, and with NuMA, as well as the interaction with cortical proteins (Dlg, Afadin) that regulate LGN cortical localization. (B) LGN complex localization in different systems, showing the polarity proteins regulating this specific localization when applicable. (i) Drosophila embryonic neuroblasts, (ii) C. elegans zygote, (iii) neural progenitors in the vertebrate neuroepithelium, and (iv) mammalian cell lines.
LGN
Biochemical and structural analysis defined the different components of the LGN complex. LGN was first identified biochemically as an interaction protein that binds to Gαi subunits 44. LGN is a modular protein composed of three main domains. Its N‐terminal TPR domain contains 7 (6 or 8 depending on the authors) tetratricopeptide repeats, which mediate the interactions with multiple binding partners, including NuMA. Its central “linker” domain does not show any recognizable organization or binding motif, but is crucially required for its function (Fig 1). The C‐terminal GPR (G protein regulator) domain contains four (3 in Drosophila Pins and 1 in C. elegans GPR1/2) GoLoco domains that mediate interaction with Gαi/o subunits. LGN interacts with Gαi only when it is bound to GDP, and has a guanine dissociation inhibitory (GDI) activity 45. The GTPase‐activating protein (GAP) RGS14/Loco/RGS‐7 (in vertebrates, Drosophila and C. elegans, respectively) and the guanine exchange factor (GEF) Ric8a control the interaction between LGN and Gαi, and therefore the stability of the complex, by modulating the GTPase activity of Gαi subunits and thereby the phosphorylation state of bound guanosine 46, 47, 48, 49, 50, 51, 52, 53.
NuMA
NuMA is a coiled‐coil protein that can interact with LGN, dynein as well as with microtubules (Fig 1) 38, 40, 54, 55, 56. NuMA is also present on the spindle, enriched near the poles, and regulates spindle formation and organization. In dividing cells, LGN and NuMA are usually observed as cortical crescents facing one or both spindle poles (Fig 1). It is well established that this specific cortical localization is instructive for spindle orientation in many systems.
Gαi
Gαi subunits localize to the plasma membrane through myristylation, where they serve as an anchor to the complex. By default, Gαi subunits cover the whole cell inner surface and do not contribute to the polarization of LGN and NuMA crescents.
Additional factors, and in particular polarity proteins, must regulate the polarized cortical distribution of LGN and NuMA. Indeed, the LGN homolog Pins (partner of inscuteable) was initially identified in Drosophila in interaction screens with inscuteable 21, 22, an apical protein known for its role in apico‐basal spindle orientation in neuroblasts 3. Apical localization of Insc, and consequently of Pins, requires the polarity protein Bazooka (Drosophila Par3) 22, as well as atypical protein kinase C (aPKC) 57, 58. Similarly, the posterior cortical enrichment of GPR1/2 requires the Par2 and Par3 polarity proteins during the first division of the C. elegans embryo 32.
More recent work has revealed a surprising diversity in the mechanisms that control LGN localization at restricted cortical domains. While Pins/LGN localizes apically in Drosophila neuroblasts, it is found in a ring at the lateral cortex during planar spindle orientation in different epithelial contexts 7, 8, 43. Remarkably, while aPKC is required for the apical recruitment of Pins in neuroblasts 57, it inhibits the apical localization of LGN and favors its lateral enrichment during cystogenesis in MDCK cells 8 (see Box 2). The mechanism mediating this inhibition involves the phosphorylation of LGN by apical aPKC, which increases locally LGN affinity to a 14‐3‐3 protein, competing with the interaction of LGN with Gαi at the apical domain 59, and favoring the planar orientation of the spindle in these cells.
Box 2: Models for studying spindle orientation.
The core components of spindle orientation have been discovered in invertebrate in vivo models of spindle orientation, which continue to be useful for dissecting new regulators and understanding the dynamics of this cellular process. In addition, an induced polarity assay has been developed in Drosophila S2 cells 60. In this model (see the figure below), intracellular fusion to the transmembrane and extracellular domains of the echinoid (Ed) homophilic cell–cell adhesion protein is used to localize a protein of choice to the contacts between clustered cells, in this way generating a polarized distribution in each cell (i). Polarized localization of Pins by using this method results in spindle orientation in the direction of the Ed‐Pins enrichment, constituting a model where the function of molecules in spindle orientation downstream of Pins can be evaluated. Alternatively, by fusing proteins or protein domains to echinoid, their ability to orient the spindle has been evaluated in different studies 60, 61, 62, 63.
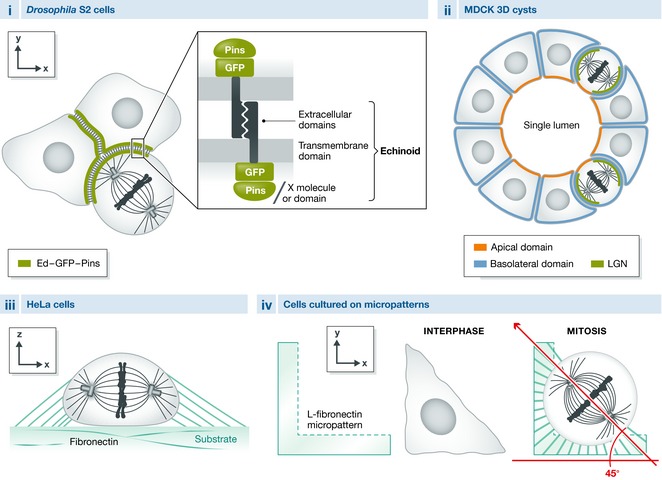
In vertebrate systems, in addition to the in vivo models of spindle orientation (e.g., mouse skin progenitors, mouse and chick neuroepithelial cells, fish epiblast cells), in vitro cultured cells are frequently used to study the molecular details and dynamics of this cellular process. The most frequently used in vitro models are:
MDCK cysts (ii): a 3D model of epithelial morphogenesis. By culturing dog MDCK cells in Matrigel, cysts with a central lumen and defined polarity domains are generated. In this context, spindle orientation occurs in the plane of the epithelium and depends on LGN which localizes to the lateral cell cortex 8. Defective spindle orientation commonly results in cysts with multiple lumina.
HeLa cells cultured on a fibronectin substrate (iii): This human cell line has been shown to orient the mitotic spindle parallel to the substrate, which depends on astral microtubules 64.
Cells cultured on micropatterns (iv; e.g., HeLa cells, MDCK cells, fibroblasts, MCF cells): In this model, single cells are cultured on micro surfaces of defined geometry, which dictates a specific shape and adhesion pattern to the cells. The adhesion pattern can induce a specific spindle orientation in the xy‐plane. This orientation is dependent on the distribution of actin retraction fibers, as well as on astral microtubules 65, 66, 67, 68.
It should be noted that knockdown of LGN or NuMA only shows weak spindle orientation phenotypes in these last two systems, suggesting that the involvement of the LGN complex is only marginal and that it acts in combination with, or as a complement to, additional pathways.
New insights into the molecular regulation of LGN complex recruitment/stability at the cortex
Discs large
In contrast to MDCK cysts, aPKC does not regulate the lateral localization of Pins/LGN, and the resulting planar orientation of divisions, in Drosophila follicular epithelia and chick embryonic neuroepithelium 7, 43. This function relies on the polarity protein Discs large (Dlg), known as a tumor suppressor in Drosophila. The role of Dlg in spindle orientation was identified in Drosophila larval sensory organ precursor (SOP) cells, where it regulates Pins localization to the anterior cell cortex 69. Previous work in fly embryonic neuroblasts had identified a role for Dlg (and other tumor suppressors lgl and scribble) in the asymmetric division of these cells, but not in spindle orientation 70, 71. Later work identified that Dlg is part of a non‐essential microtubule‐based pathway driving cortical localization of LGN in neuroblasts, acting in parallel with the dominant inscuteable recruitment pathway 72 (reviewed in 19). However, both the SOP and NB models relied on a polarized localization of Dlg, which differed from its classical “baso‐lateral” localization, raising the question of its potential role in symmetrically dividing cells in canonical epithelia. Recent studies have shown that depleting Dlg/Dlg1 affects Pins/LGN cortical localization and results in defects in planar spindle orientation in Drosophila epithelia and in chick neuroepithelium 43, 73. Quite remarkably, Dlg/Dlg1 acts differently in each of these tissues. In Drosophila follicular epithelia, Pins becomes localized all around the cortex upon Dlg depletion, indicating that Dlg may act by restricting Pins localization to the lateral cortex 43. In contrast, in the chick neuroepithelium, LGN is lost from the cortex when Dlg1 is depleted, suggesting that Dlg acts to recruit/stabilize LGN at the cortex in this context 73. Similarly, DLG1 depletion in human HeLa cells reduces LGN/NuMA cortical localization and is associated with defects in micropattern‐guided spindle orientation (see Box 2) 73. Importantly, while Dlg/Dlg1 has been described as a polarity protein, acute depletion of Dlg/Dlg1 does not generate obvious defects in tissue polarity in follicular epithelia or in the neuroepithelium, indicating that this protein plays a specific role in spindle orientation independent of its function in cell polarity 43, 73. Direct interaction between Dlg/Dlg1 and Pins/LGN relies on the phosphorylation of a conserved serine residue in the linker domain of LGN 60, 74, 75, 76 (Fig 1A). In Drosophila, Pins is phosphorylated by AurA and regulates the interaction between Dlg and Pins. In contrast, AurA activity is not required for LGN cortical recruitment in HeLa cells 77, suggesting that AurA does not control LGN interaction with DLG1 in these cells.
Afadin
The Drosophila scaffolding protein Canoe (Afadin in mammals) also regulates LGN complex formation and spindle orientation. This role was initially described in Drosophila neuroblasts, where Canoe localizes to the apical cortex and regulates apical–basal spindle orientation 78. The molecular details have been dissected in the S2 cell induced polarity assay (Box 2) 62, where Canoe interacts with Pins and acts specifically in the spindle orientation pathway mediated by PinsTPR/Mud 60. In particular, Canoe is necessary for Mud recruitment to cortical Pins crescents through its interaction with the TPR domains 62, 78. Canoe interaction with RanGTP is necessary for Mud recruitment and spindle orientation, but the mechanisms by which RanGTP regulates these activities remain elusive 62. Canoe's vertebrate homolog Afadin also plays a role in spindle orientation, albeit through a distinct mechanism: Afadin binds simultaneously cortical F‐actin and the TPR region of LGN (Fig 1A). Afadin interaction with LGN is in competition with the NuMA/LGN interaction, although the affinity for Afadin is lower. However, upon Afadin depletion, LGN cortical recruitment is reduced, NuMA and dynein are not recruited, and the spindle is misoriented, both in adherent cells and in 3D cell cultures 79. As NuMA is nuclear in interphase and only released upon nuclear envelope breakdown, one possible interpretation is that Afadin is necessary for the initial recruitment of LGN to the cortex and its interaction with Gαi subunits in early mitosis, before LGN interacts with and recruits NuMA at the cell cortex.
More generally the interplay between Gαi, Afadin, and Dlg1 in LGN cortical localization is not well understood. Gαi appears as an obligate membrane anchor, since LGN is completely absent from the cortex when the Gαi/LGN interaction is disrupted; in contrast cortical levels of LGN are only reduced in the absence of Afadin and Dlg1 73, 79. Whether Afadin and Dlg1 are important for the initial recruitment of LGN by Gαi, or whether they are involved in maintaining LGN at the cortex, remains unclear. In addition, Dlg1 is involved in the polarization of the cortical localization of LGN at least in epithelia.
Huntingtin
A series of recent investigations have focused on the role of huntingtin (HTT) in spindle orientation. This protein, mutated in Huntington's disease, regulates spindle orientation in mouse neural progenitors and basal mammary cells in vivo, as well as in Drosophila neuroblasts 80, 81. The mechanisms of action of HTT have been further evaluated in cultured cells, where it regulates spindle orientation with respect to the substrate 80. HTT depletion leads to a decrease in the cortical levels of LGN, NuMA, and members of the dynactin/dynein complex. Contrary to the LGN interacting proteins Afadin and DLG1, HTT localizes to spindle poles during mitosis. Because HTT plays a role in anterograde vesicular transport in neurons, the proposed hypothesis is that HTT regulates the transport of LGN and dynein complex members via astral MTs from the spindle poles to the cortex. Accordingly, this transport depends on the plus‐end‐directed motor kinesin 1 80.
Phosphorylation of Mud and NuMA
Two recent reports identified the phosphorylation of Mud and NuMA as necessary for their cortical localization. In Drosophila, phosphorylation of a serine residue in the coiled‐coil domain of Mud by the Hippo pathway kinase Warts induces a conformation change that uncovers the Pins binding domain and allows interaction with cortical Pins 82. Warts can also phosphorylate human NuMA in vitro, but the relevance of this modification has not been tested in cells. However, phosphorylation of a distinct serine residue in another domain of NuMA by the mitotic kinase Aurora A (AurA) is also necessary for its cortical recruitment in human cells 77. While the two mechanisms are different, it is remarkable that both Warts and AurA kinases localize to spindle poles in mitotic cells, suggesting that they act there to promote the release of phosphorylated Mud/NuMA from the spindle pole and thereby allow its interaction with cortical Pins/LGN. Accordingly, upon pharmacological inhibition or knockdown of AurA, NuMA is lost from the cortex and its concentration increases at the spindle poles 77.
Temporal and spatial regulation of LGN complex localization
In specific cell types, the centrosome maintains its position during all the cell cycle and the spindle forms directly with its correct orientation 83, 84. However, in many cases, the spindle forms in prometaphase with a random orientation, and the final axis of division observed at anaphase is set through spindle rotation during prometaphase and metaphase 7. The switch from interphase to mitosis, and mitotic progression itself, are accompanied by the sequential activation of numerous signaling pathways and major changes in the organization of cellular structures. This section highlights a number of recent studies that describe the dynamics and molecular regulation of the subcellular recruitment of the LGN complex in relationship to mitotic progression (Fig 2).
Figure 2. Temporal–spatial regulation of LGN complex localization.
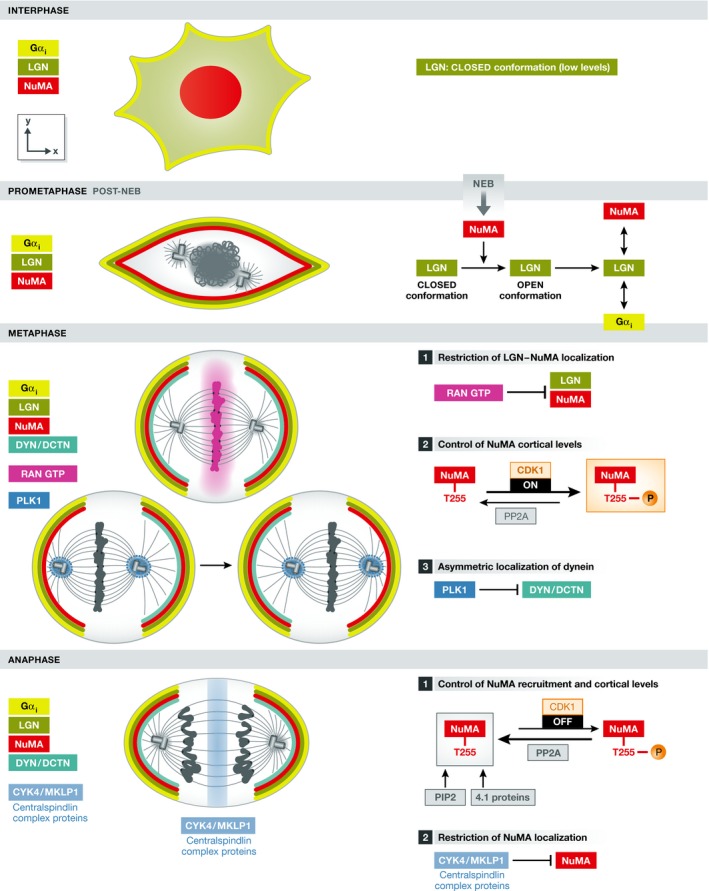
Left: Scheme of interphase and mitotic phases indicating the distribution of LGN, NuMA, and dynein, as well as the localization of specific molecules (Ran‐GTP, PLK1, and centralspindlin proteins) that are involved in controlling this distribution in cultured HeLa cells. Note that NuMA and dynein cortical levels increase in anaphase. Right: Detail of the molecular mechanisms involved in the control of LGN/NuMA localization and of dynein by Ran‐GTP/centralspindlin and PLK1, respectively. The control of NuMA cortical levels by CDK1 activity is also indicated.
Temporal regulation of the LGN complex formation in early mitosis
In the last few years, different labs have studied the temporal and spatial aspects of LGN/dynein complex formation by mainly using mammalian cell lines. In HeLa cells, LGN protein levels increase during mitosis 30, which contributes to restrict LGN complex formation only in mitosis, but the molecular regulation of this increase in unknown. In addition, Du and Macara have also demonstrated that during interphase, LGN exists in a closed conformation and because it interacts poorly with Gαi in this state, it does not localize to the cortex. They proposed that interaction with NuMA is necessary to switch LGN to an open conformation that increases its ability to bind Gαi subunits. Because NuMA localizes to the nucleus during interphase, the formation of the Gαi/LGN/NuMA cortical complex would then be further restricted to mitosis in vertebrate cells 30 (Fig 2). However, whether NuMA is required for the cortical recruitment of LGN is unclear: Knockdown of the NuMA homolog lin‐5 in C. elegans embryos results in the loss of cortical GPR1/2 (LGN) 34, whereas knockdown of NuMA in the chick neuroepithelium does not prevent LGN cortical localization 7. Besides, upon AurA inhibition NuMA is lost from the cortex, but not LGN 77. In contrast to vertebrate cells, Drosophila Mud is not nuclear and shows cortical localization during interphase in neuroblasts and the overlying neuroectoderm 22, 40. This may explain why Pins shows also cortical patterns in interphase in these cells. Similarly, LIN5 in C. elegans is not a nuclear protein.
Spatial regulation of LGN localization in early mitosis
The subcellular localization of the LGN complex is very dynamic throughout mitosis (Fig 2). Using HeLa cells that stably express GFP–LGN cultured on fibronectin, Kiyomitsu and Cheeseman showed that LGN is initially recruited all around the cell cortex during prometaphase but its localization is later restricted to two cortical crescents facing the spindle poles during metaphase and anaphase 85. Using different drugs that affect spindle organization and chromosome alignment, they observed that an abnormal proximity of chromosomes with the cortex inhibited cortical localization of LGN and NuMA, and concluded that chromosome‐derived signals normally exclude LGN/NuMA from cortical sites in proximity to the chromosomal plate. Using a RanT24N dominant‐negative mutant to disrupt the RanGTP chromosomal gradient, they went on to show that this gradient is responsible for cortical exclusion of LGN 85. Therefore, the spatiotemporal restriction of LGN complex localization during mitosis is proposed to rely on a gradient of RanGTP that inhibits the formation of the complex in the vicinity of chromosomes. Once the metaphase plate is formed, LGN and NuMA are excluded from this location and appear enriched as two cortical crescents overlying each spindle pole (Fig 2). Therefore, in this model the localization of the complex is established once spindle orientation is set. This is in marked contrast to models in which the orientation is constrained by polarized molecular cues, such as the asymmetric division of fly neuroblasts. Interestingly, when HeLa cells are cultured on polarized micropatterns (see Box 2), the asymmetric distribution of retraction fibers imposes such a constraint, and both LGN and dynein complexes show a restricted cortical localization before the spindle is oriented along the correct axis 68, 86.
In mitotic HeLa cells, the cortical distribution of dynactin/dynein complexes is dynamic during metaphase. Live imaging revealed redistribution of a polarized crescent that alternates between the cortical domains that face each spindle pole 85. Remarkably, these oscillations are independent of the distribution of LGN and NuMA, which remain localized in two cortical crescents (Fig 2). They are followed by the asymmetric positioning of the spindle, whose poles are alternatively attracted to the dynein enriched cortical domains. Here, the kinase Plk1, localized at the spindle poles, negatively controls the cortical localization of dynein/dynactin. The proximity of a spindle pole to the cortex excludes dynein from this cortical site. Concomitantly, dynein/dynactin accumulates to the side of the cell facing the opposing (and more distant) spindle pole and generates pulling forces, which in turn will reposition the spindle 85. Cortical targeting of dynein/dynactin during this oscillatory phase depends on astral microtubules 86.
A specific spatiotemporal regulation in anaphase
More recently, different labs have described changes in the cortical recruitment of NuMA between metaphase and anaphase 87, 88, 89. In contrast to LGN levels, cortical levels of NuMA increase from metaphase to anaphase. These changes are related to its phosphorylation state at T2055, which is regulated by the balance between the activities of the CDK1 kinase and the PP2CA phosphatase. During metaphase, phosphorylated NuMA is observed at spindle poles, while the cortical protein would correspond to non‐phosphorylated NuMA. At anaphase onset, the decrease in CDK1 activity results in an increase in non‐phosphorylated NuMA, which allows further enrichment of this protein at the cortex (Fig 2). Accordingly, altering NuMA phosphorylation states results in defects in spindle orientation with respect to the substrate 90.
In contrast to metaphase, an absence of LGN or Gαi in anaphase does not result in complete loss of NuMA from the cortex. Besides, LGN cortical levels do not increase in anaphase, indicating that additional molecules contribute to NuMA localization after the metaphase/anaphase transition. Kiyomitsu and Cheeseman showed that cortical 4.1G and 4.1R proteins interact directly with non‐phosphorylated NuMA in anaphase, providing a potential mechanism for this increase 87. However, whereas removal of both LGN and the 4.1 proteins completely deplete cortical NuMA in anaphase, depletion of the 4.1 proteins alone had no effect 87. Moreover, Kotak et al 89 found that a GFP‐tagged version of NuMA lacking the 4.1 protein interaction domain shows the same localization and levels during metaphase and anaphase than wild‐type GFP‐NuMA. As an alternative model, they proposed that an interaction between NuMA and the phosphoinositides PIP/PIP2 is involved in NuMA cortical recruitment in anaphase. Using different approaches to perturb PIP2 levels, they show changes in NuMA and dynein levels in anaphase. The authors found that this PIP2‐mediated recruitment pathway is anaphase‐specific. However, the dependence of the NuMA–PIP2 interaction on NuMA phosphorylation states remains to be elucidated. In the same study, Kotak and colleagues show that NuMA is excluded from the equatorial cortex by the centralspindlin proteins CYK4 and MKLP1 during anaphase 89, therefore maintaining the exclusion initiated in metaphase by the Ran‐GTP signal. Increased cortical levels of NuMA in anaphase are important for spindle elongation and chromosome separation in human cultured cells 88. However, whether this increase is also important for spindle orientation itself is not clear.
Taken together, these recent experiments in cultured symmetrically dividing human cells have revealed a complex regulation of the dynamics of LGN, NuMA, and dynein localization during mitosis by molecules located on chromosomes, centrosomes and at the cortex. Several questions remain. Firstly, are these pathways active and necessary to achieve oriented divisions in vivo? In the neuroepithelium, it is unlikely that the Ran‐GTP mechanism is at play in metaphase. A continuous and homogenous ring of LGN/NuMA is observed at the lateral cortex, and the levels of LGN/NuMA are not lower in the vicinity of the metaphase plate, despite the very small cell size 7. Secondly, how do these pathways integrate with the instructive signals, such as polarized inscuteable, that control spindle orientation in complex tissues? Further investigations in the field will shed light on these aspects.
In this section, we have shown how cell cycle‐regulated changes in the activity of kinases and the assembly of central spindle complexes in anaphase result in a differential regulation of the cortical localization of NuMA. Remarkably, this reveals that NuMA can be recruited to the cortex independently of Gαi and LGN in anaphase. In the next section, we further discuss this notion by describing alternative spindle orientation complexes that converge on NuMA and dynein cortical recruitment, independently of LGN.
Not a monopoly: Gαi/LGN‐independent pathways in spindle orientation
The frizzled/disheveled (Fz/Dsh) PCP pathway regulates spindle orientation in different contexts, including zebrafish gastrulation and asymmetric division of the SOP pI cell (reviewed in 91). pI cells in the fly notum divide along the antero‐posterior axis, and the spindle is slightly tilted relative to the tissue surface. Although Gαi, Pins, and Dlg accumulate at the anterior cell cortex and are involved in the near planar orientation of the spindle, they are not necessary for its antero‐posterior alignment 48. This orientation is regulated by the Fz receptor and its cortical effector Dsh, which are located at the posterior cortex. Using the S2 cell induced polarity assay 60 (see Box 2), Segalen and colleagues identified Mud as the downstream effector of Dsh 63. Accordingly, they showed that Mud recruitment by Dsh at the posterior apical cortex of the pI cell is necessary for spindle orientation along the antero‐posterior axis (Fig 3A). Similarly, during zebrafish gastrulation, dishevelled and NuMA are necessary for spindle orientation along the animal–vegetal axis in epiblast cells 63. This suggests that the dishevelled–NuMA pathway is conserved across different species.
Figure 3. The role of actin in spindle orientation.
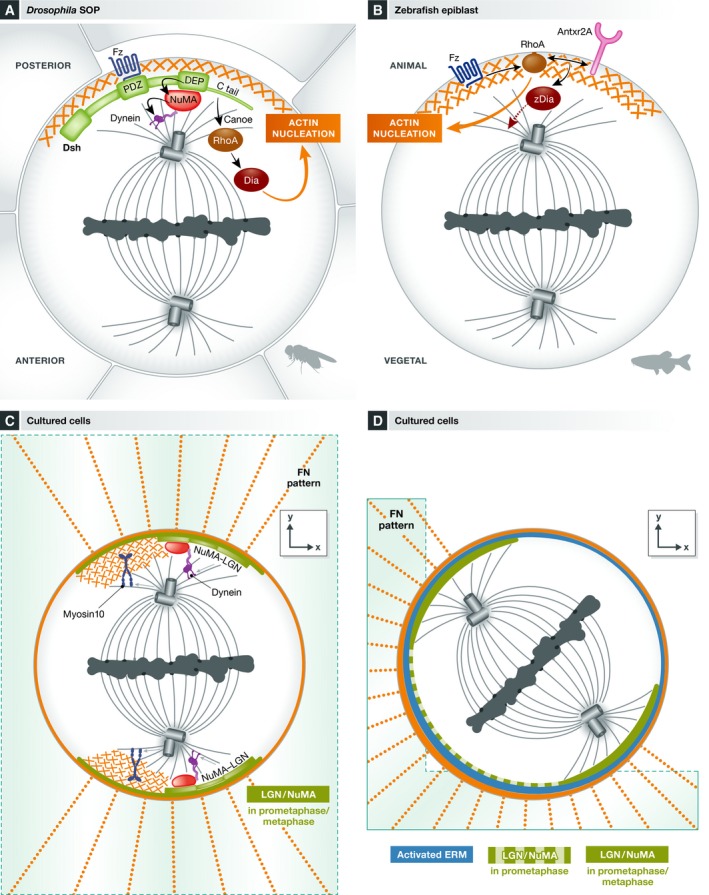
(A) In Drosophila SOP, dishevelled localizes to the posterior cortex activating two parallel pathways required for spindle orientation: i) The recruitment of NuMA via the DEP domain allows dynein enrichment at this site, ii) a molecular cascade involving the tail domain of dishevelled, and the Canoe and RhoA molecules leads to the activation of the actin nucleator diaphanous at this cortical site. (B) RhoA and the anthrax receptor 2a (Antxr2a) orient the spindle along the animal–vegetal axis in zebrafish epiblast. Activation of Fzz promotes RhoA recruitment to the “animal cortex”. In turn, RhoA induces actin nucleation leading to the formation of an actin cap, and together with the anthrax receptor activates the downstream effector zDia. (C, D) Involvement of different actin‐related molecules in xy spindle orientation in single cells cultured on fibronectin micropatterns. In this context, the distribution of actin retraction fibers dictates the orientation of the spindle. (C) Polarized actin subcortical clouds make the link between the distribution of retraction fibers and spindle orientation. Myosin 10 mediates the link between actin and microtubules in this context. The classical LGN/dynein complexes are proposed to act in parallel to this pathway, leading to robust spindle orientation. (D) The ezrin–radixin–moesin proteins are enriched in the adhesive cortex in cells cultured in L patterns. These proteins control the initial distribution of LGN and NuMA, during prometaphase, which favor spindle rotation along the depicted axis.
Mechanistically, the Dsh DEP (dishevelled/EGL10/pleckstrin) domain mediates the recruitment of Mud and dynein. However, Johnston and colleagues found that the Dsh/NuMA pathway does not act alone and uncovered an accessory pathway in Dsh‐mediated spindle orientation 61. Using the induced polarity assay in S2 cells (Box 2), they showed that the DEP domain of Dsh on its own indeed recruits Mud, but surprisingly this was not sufficient to orient the spindle. Robust spindle orientation required the C‐terminal domains of Dsh in addition to the DEP domain (DEP‐CT) and an interaction of the Dsh‐PDZL domain with Canoe. Indeed, RNAi experiments showed that Canoe is necessary for robust spindle orientation in this assay. However, contrary to its previously described role downstream of Pins 62, in this case Canoe does not work through the recruitment of Mud. Instead, it is required for the recruitment of RhoA and the formin diaphanous to the DEP‐CT construct. Both RhoA and the actin nucleation activity of its effector diaphanous are necessary for spindle orientation. Consistently, actin accumulates in the cortical domain where DEP‐CT is localized. The mechanisms that link cortical actin nucleation to spindle rotation, however, remain to be investigated. In support of their in vitro data, the authors demonstrated that diaphanous is indeed necessary for Dsh‐mediated spindle orientation along the antero‐posterior axis in Drosophila SOP cells 61 (Fig 2A).
While NuMA is a central component in several spindle orientation pathways (see “Spindle orientation in context: roles of cell geometry and mechanical forces” for an additional NuMA‐dependent pathway), the molecular details of how it regulates spindle orientation remain to be clarified. Artificial targeting of dynein to the cell membrane independently of its interaction with endogenous NuMA induces excessive spindle rotation 38. This suggests that dynein alone is sufficient to exert forces on astral microtubules and that NuMA may only be a passive anchor for the motor complex. However, artificially high levels of cortical dynein may cause excessive spindle rotations and bypass a regulation of dynein by NuMA that may occur at physiological expression levels. Alternatively, NuMA could itself contribute to force generation, either by regulating the motor activity of dynein, or through its ability to directly interact with microtubules 55. It was proposed that NuMA localization to microtubule ends via its MT binding domain is necessary for spindle orientation 92. Mouse keratinocytes depleted for the NuMA MT binding domain show spindle orientation defects without changes in the localization of the dynein complex. However, the exact mechanisms by which MT end‐localized NuMA contributes to spindle orientation remain to be elucidated.
The emerging role of actin in spindle orientation
In the previous sections, we have used the words “cell cortex” and “cortical recruitment” in an improper (but very widely employed) manner while referring to the inner surface of the cell membrane. The cell cortex is actually defined as a cross‐linked network of actin, myosin, and associated proteins located directly underneath the plasma membrane. In this section, we discuss the emerging role of this network in spindle orientation. The mechanics of the cell cortex are essential in the control of cell deformations that occur during cell division, and contribute to the transmission of forces. Furthermore, as already alluded to in the previous paragraph, several studies describe that specific polarization of the actin cortex controls spindle orientation. Finally, recent work shows that actin cross‐linkers regulate the recruitment of the LGN complex.
Requirement of an intact actin cortex
When a cell enters mitosis, remodeling of its actin cytoskeleton leads to cell rounding and the establishment of a thinner, but stiffer actin/myosin cortex 93 (reviewed in 94). Impairment of the actin cortex by latrunculin A/B or cytochalasin D treatment generates spindle orientation defects in cultured cells and in vivo in the mouse embryonic skin and in Drosophila wing disks 95, 96, 97. In cultured cells and in the developing mouse skin, LGN cortical localization was perturbed by these treatments 68, 95, 97. Hence, an intact cortex is required for the correct localization of the spindle orientation machinery and for the stabilization of force generators. How F‐actin influences force generator localization at the cortex remains elusive. Recent data showing that Afadin can bind simultaneously actin and LGN provide for the first time a direct mechanistic link between cortical actin and the force generator machinery 79. Simultaneously, a sufficiently stiff actin cortex is likely important to prevent membrane deformations and to balance the forces exerted by force generators at the cell surface that pull the spindle, as suggested by experiments in the C. elegans zygote 98. In addition, changes in cellular shape generated by disruption of the actin cortex may be involved in the observed phenotypes, as we will discuss in detail in “Spindle orientation in context: roles of cell geometry and mechanical forces”. While the presence of an intact and stiff actin cortex can be seen as permissive for the correct localization of force generators and mitotic cell rounding, an active (or instructive) role of actin and actin‐related molecules in guiding spindle orientation is becoming more apparent (as discussed below).
Anthrax receptor and actin polarization
In the context of zebrafish gastrulation, Castanon and colleagues described a novel molecular cascade controlling oriented divisions in epiblast cells 99. Interestingly, the authors have observed the formation of an F‐actin cap that co‐localizes with an anthrax receptor (Antxr2a) cap during cell division. Depletion of Antxr2a causes spindle misorientation. By following spindle rotation and cap formation in a series of gene knockdown experiments, the authors dissected the cascade of events that leads to cap formation and spindle rotation. They propose that local activation of RhoA by Wnt leads to the cortical enrichment of actin in an oriented manner. Actin recruits Antxr2a to the actin cap where it contributes to the activation of a diaphanous‐related formin, zDia2, which in turn allows spindle rotation in the direction of the cap (Fig 3B). However, it is not clear whether this pathway acts on dynein, which is also involved in spindle orientation in these cells 100. It is also unknown whether Dsh and NuMA 63 act in parallel to or downstream of the Antxr2a orientation pathway.
Polarized subcortical actin clouds
In addition to cortical actin, Mitsushima and colleagues described the presence of a subcortical cluster of actin during mitosis in cultured cells. This actin cloud undergoes a rotational movement during metaphase and disappears into the contractile ring upon cytokinesis. The formation of this amorphous actin‐rich structure depends on Arp3 101. Further studies support a role for the actin cloud in spindle orientation. When cells are cultured on micropatterned surfaces (Box 2), the adhesion pattern of the cell controls spindle orientation in the planar (xy axis) in an actin‐ and microtubule‐dependent manner 65, 67. The polarized distribution of retraction fibers during mitosis constitutes a memory of the adhesion pattern in interphase and influences the orientation of the spindle, as seen by laser ablation experiments 67. Interestingly, the adhesion pattern and distribution of retraction fibers influenced the polarized distribution and movements of actin clouds, and dynamic analyses suggested that clouds influence the rotation of the mitotic spindle in an astral MT‐dependent manner 67. More recently, the function of these actin clouds in spindle orientation was formally demonstrated by inhibiting the Arp2/3 complex 102. Kwon and colleagues further demonstrated that the unconventional microtubule binding myosin 10, an actin motor involved in spindle formation and integrity 103, regulates spindle orientation with respect to polarized actin clouds in cells cultured on micropatterns. This activity depends on its MT binding domain. Interestingly, myosin 10 localizes to retraction fibers and to dynamic actin clouds but it does not modify their dynamics or assembly. In contrast, depletion of myosin 10 specifically increases astral microtubule dynamics and decreases the interaction of these MT with the cortex, as demonstrated by dynamic analyses of EB3 in metaphase. This suggests that actin‐localized myosin 10 regulates spindle orientation by modulating astral MT dynamics, constituting a link between actin and microtubules in the context of spindle orientation (Fig 3C). Of note, the action of myosin 10 differs from that of dynein, as myosin 10 depletion does not change the frequency of lateral transitions of microtubules in anaphase, in contrast to cells lacking cortical dynein caused by depletion of LGN using RNAi. In addition, depletion of myosin 10 and LGN together results in more dramatic defects on spindle orientation than depleting each protein alone, suggesting that the actin/myosin 10 and LGN/dynein pathways act in parallel to orient the spindle 102. This reinforces the idea that in some cellular contexts, multiple pathways act to promote robust spindle orientation.
ERM proteins
The ezrin–radixin–moesin (ERM) proteins are a family of actin/membrane cross‐linkers which control cortical rigidity and stability 104. Depletion of moesin, the single member of the family in Drosophila, leads to massive cortical instability and blebbing in mitotic S2 cells. This results in exaggerated spindle oscillations and mispositioning 105, 106. Defects in spindle morphology (such as short spindle and asymmetric asters) make it difficult to properly evaluate spindle orientation in this model. In contrast, in the Drosophila larval wing disk, moesin RNAi does not induce massive blebbing during division, but affects cell rounding so that cells are more elongated along the apico‐basal axis. This correlates with a loss of planar spindle orientation 96. ERM proteins have been recently studied for their role in spindle orientation in vertebrate cells. In dividing human cells cultured on L‐shaped micropatterns, activated ERM proteins are asymmetrically distributed, with an enrichment in the cortical domain facing the adhesive surface 66, 68 (Fig 3D). Here, depletion of the three proteins as well as impairment of their activation through depletion of the SLK kinase (which was found to directly activate ERM proteins through phosphorylation) leads to spindle misorientation in the xy axis (see Box 2) 68. This phenotype is associated with the loss of LGN and NuMA cortical localization and with reduced spindle rotation, suggesting that activated ERM proteins are necessary for cortical recruitment of LGN/NuMA or stability in this context. Importantly, in contrast to the effects observed upon depletion of moesin in Drosophila 105, 106, depletion of ERM proteins does not generate obvious alterations in cell shape and spindle morphology in human cells, arguing for a specific role of these proteins in orienting the spindle by the control of LGN/NuMA localization 68. ERM proteins probably act at the level of LGN, since no effect was observed on Gαi localization upon ERM inactivation or depletion. In addition, overactivation of ERM around the cortex leads to ectopic localization of LGN/NuMA and exaggerated spindle rotation, which also results in defects in spindle orientation. Remarkably, perturbing ERM activation in mouse apical neural progenitors in vivo impairs spindle orientation 68. However, whether ERM proteins regulate LGN complex localization also in this context remains to be studied. Intriguingly, activated ERM can also bind microtubules and thus could also influence spindle orientation directly 107. Detailed time‐lapse microscopy indicated that spindles rotate in prometaphase in cells cultured on L‐shaped micropatterns 66, 68. The finding that LGN and NuMA are first localized asymmetrically as a large crescent facing the adhesive matrix 68 likely explains the stereotyped spindle orientation in this system, as anticipated by previous theoretical modeling 65.
In this section we highlighted the role of actin and actin regulators in spindle orientation in different model systems. An important challenge is to understand the crosstalk between actin/dynein and NuMA/dynein pathways. Remarkably, actin‐related pathways both seem to modulate or to act independently of the LGN/NuMA pathways. Indeed, ERM actin cross‐linkers regulate the cortical localization of the LGN complex in cultured cells 68. Whether this regulation goes through modulation of the actin cortex or if alternatively there is a direct molecular link between ERMs and LGN/NuMA, remains to be determined. In contrast, actin subcortical clouds and myosin 10 act in parallel to the LGN/dynein pathway to regulate spindle orientation in cells cultured on micropatterns 102. Similarly, dishevelled controls spindle orientation in Drosophila S2 and SOP cells by activating two parallel cascades: a NuMA/dynein and a RhoA/diaphanous/actin pathway 61. In these cases, it would be interesting to study if parallel pathways act simultaneously or not during spindle orientation. For instance, it could be imagined that one pathway determines the initial orientation of the spindle, having a more instructive role, while the other cascade maintains the orientation once it is set.
The molecular complexes that recruit force generators are located at the plasma membrane. Despite the size of these complexes, due to the thickness of the mitotic cortex (190 nm 93), it is unlikely that force generators stick out beyond the cortex in the cytoplasm, and more probable that astral microtubules reach motor complexes close to the plasma membrane by growing through the actin meshwork. This also provides an additional layer of regulation for the cortical capture of microtubules, which will be addressed in the following section, dedicated to the regulation of astral microtubules. It will be interesting to explore whether the actin regulators described above influence this meshwork.
Modulation of spindle orientation through the specific regulation of astral microtubules
Except for positioning of meiotic spindles, which lack astral microtubules (reviewed in 108), spindle orientation is thought to be achieved by the interaction of astral microtubules with force generators at the cellular cortex (in the broader definition that includes the plasma membrane). Therefore, defects in spindle morphology and/or astral microtubules (MTs) can affect spindle orientation. Shorter spindles may indirectly affect the distance between astral microtubules and the cortex. Alternatively, abnormal astral microtubules may affect the correct transmission of forces necessary to orient the spindle. Indeed, many proteins affecting astral microtubules perturb spindle orientation. We will discuss how modulation of (i) astral MT nucleation/anchoring at the centrosome, (ii) astral MT dynamics and stability, (iii) astral MT cortical capture, (iv) astral MT behavior at the cortex, and (v) astral MT subpopulations impact on spindle orientation (Fig 4).
Figure 4. Modulation of spindle orientation through regulation of astral microtubules.
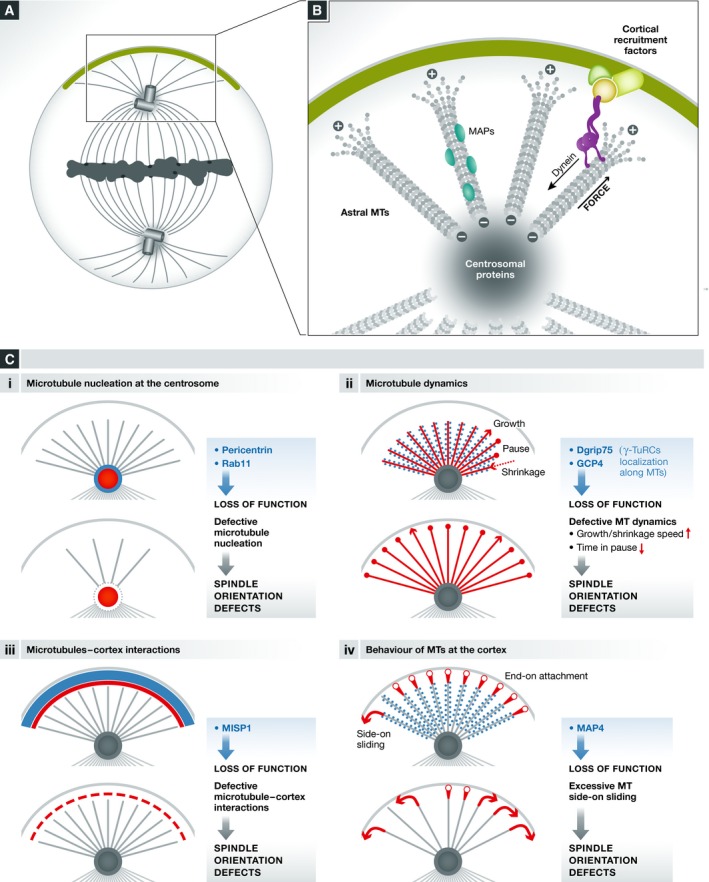
(A, B) Schema illustrating the centrosome and astral microtubules as well as generic proteins localized on these structures. Cortically recruited dynein is believed to walk on the minus‐end direction of astral MT, generating the force that orients the spindle. (C) Regulation of different processes (i–iv) controls the density, length, and behavior of astral microtubules, and thus spindle orientation. The process and cellular structure concerned are indicated in red. Loss of function of specific proteins (in light blue) results in defects in the indicated processes and spindle misorientation. In (iii), MISP acts from the cellular cortex regulating cortex–MT interaction.
Astral microtubules nucleation
The role of the centrosomal protein pericentrin (Pcnt) in spindle orientation has been addressed by using cultured MEFs derived from Pcnt−/− knockout mice 109. In these cells, both astral microtubule length and density (determined by measuring α‐tubulin signal intensity) are decreased, and spindle orientation with respect to the substrate (see Box 2) is impaired. In addition, Pcnt was found necessary for spindle pole localization of a particular set of centrosomal proteins including ninein, centriolin, and Cep215. While the localization of these proteins at the centrosome is required for spindle orientation, it remains to be analyzed how each of them affects astral microtubules. However, the data obtained so far suggest that defective recruitment of centrosomal proteins by Pcnt depletion leads to defects in astral microtubules nucleation at the centrosome and thus induces spindle misorientation. Importantly, the cortical localization of NuMA and the dynactin subunit p150 glued are not affected in Pcnt−/− cells, suggesting that their transport and/or turnover are not affected by the observed defects on astral microtubules. This suggests that defects in centrosomal protein localization and/or astral microtubule density are responsible for the observed defects in spindle orientation. In agreement with the in vitro data, the authors found spindle orientation defects in neural progenitors and in heart septums of Pcnt−/− mice 109.
Related to MT nucleation activity, a novel role of Rab11 recycling endosomes (RE) in spindle orientation has been recently demonstrated in human cells 110. These endosomes associate with the spindle and with spindle poles in a Rab11‐dependent manner. Impairment of Rab11 function generates spindle misorientation with respect to the substrate. The authors proposed that disruption of astral microtubules is related to this phenotype, which could be explained by the fact that Rab11 RE transport microtubule nucleation components like γ‐tubulin and GCP4. These effects may not be astral MT‐specific as the overall spindle microtubule density is affected upon Rab11 depletion 110. Assays of microtubule nucleation from spindle poles in Rab11 depleted vs. control cells demonstrated that Rab11 is indeed important for spindle pole MT nucleation. In conclusion, Rab11 endosomes would be important for the delivery of MT‐nucleating components to the spindle poles, which would affect MT nucleation, spindle morphology and consequently, spindle orientation. However, it should be noted that Rab11 depletion also generates misaligned chromosomes. The proximity of misaligned chromosomes to the cortex could affect the cortical localization of LGN/NuMA in a RanGTP‐mediated manner and thus indirectly affect spindle orientation, as described above 85. Alternatively, recent data show that artificially induced chromosomal misalignments result in kinetochore‐derived Plk1 signaling, whose proximity to the cortex can locally inhibits LGN and NuMA recruitment 111, which may also explain the phenotype of Rab11 depletion. In any case, these results originally link membrane traffic with spindle orientation.
Astral microtubules dynamics and stability
Defects in astral microtubule stability also affect spindle orientation. Toyoshima and Nishida have first shown that depletion of the microtubule plus‐end protein EB1, a regulator of microtubule stability, results in spindle misorientation with respect to the substrate in cultured cells (see Box 2), accompanied by a reduction in spindle length and of astral microtubules 112. More recently, Bouissou and colleagues have shown in Drosophila S2 cells and human HeLa cells that γ‐tubulin ring complexes (γ‐TuRCs) localize to astral microtubules in addition to their well‐known localization at centrosomes and spindle microtubules. Depletion of the γ‐TuRCs component Dgrip75 in Drosophila impairs spindle orientation mediated by Ed‐PinsTPR+Linker in the S2 induced polarity assay (see Box 2) and apico‐basal spindle orientation in neuroblasts 113. Similarly, depletion of GCP4, the human Dgrip75 ortholog, generates defects in spindle orientation with respect to the substrate in cultured human cells. Associated with these defects, spindles show longer astral microtubules in S2 cells. Interestingly, changes in astral microtubules do not result from defects in microtubule nucleation activity, a canonical function assigned to γ‐TuRCs. In contrast, γ‐TuRCs act by regulating astral microtubule dynamics. Indeed, depletion of Dgrip75 increases astral MT dynamics and the time that MTs spend in the growing state, possibly explaining the overall increase in MT length. Importantly, by suppressing MT dynamics using drug‐ and knockdown‐based approaches, the authors were able to rescue spindle orientation defects in S2 cells 113. This suggests that perturbed astral MT dynamics is directly responsible for the spindle orientation phenotypes observed.
While the effects on spindle orientation generated by the absence or shortening of astral MTs can easily be explained by the lack of interactions between the spindle and the force generators, the link between longer and more dynamic astral MTs and defective spindle orientation is less clear. One possibility is that longer astral microtubules establish abnormal interactions with the cortical sites facing the initial axis of spindle orientation, which in consequence could affect the rotation of the spindle to the cortical domains enriched in force generators 65. Alternatively, the interaction of force generators with highly dynamic microtubules may be less effective. Consistently, exaggerated spindle oscillations are seen upon Dgrip75 depletion in S2 cells, which could indicate unstable MT–cortex interactions 113.
Astral MT cortical capture
While microtubule nucleation and dynamics regulate the number of microtubules reaching the cortex, these microtubules need to establish proper contacts with the cortex. The interaction between the cortex and astral MTs can be modified by molecules localized at the cortex. For instance, the actin‐associated protein MISP localizes to the cellular cortex during mitosis and regulates spindle orientation with respect to the substrate in HeLa cells 114. Depletion of MISP results in reduced astral microtubule intensity, which is not caused by defects in microtubule nucleation, as in vitro and in vivo polymerization assays showed. Because MISP does not localize to the spindle but to the cortex, the authors proposed that astral MT attachment to the cortex is impaired in the absence of MISP, resulting in destabilized astral MTs. However, it is noteworthy that MISP depletion generates fragmented centrosomes that are often located at the interior of the spindle, which could also contribute to disrupt astral MTs.
Behavior of astral microtubules at the cortex
Once microtubule plus‐ends contact the cortex by end‐on attachment, two different scenarios have been observed. After a few seconds of cortical dwell, they either undergo catastrophe and shrink or continue to grow along the cell cortex, a process known as side‐on sliding. Samora and colleagues have shown that the microtubule‐associated protein MAP4 regulates spindle orientation and positioning by modifying the behavior of astral MTs at the cellular cortex 115. Dynamic analyses of EB3‐Tomato during metaphase revealed that upon depletion of MAP4, side‐on sliding of astral microtubules at the cortex is increased and leads to spindle pole displacement. Interestingly, these effects are lost upon the impairment of dynein activity, suggesting that MAP4 acts by moderating dynein‐dependent forces that generate abnormal MT–cortex interactions.
Modulation of specific astral MT subpopulations
While most of the studies describing the role of astral MT in spindle orientation have been performed in cultured cells, progress has been made recently to understand their characteristics and in vivo function in apical progenitors (APs) of the mouse neocortex 116. The authors defined two different astral MT subpopulations, which are differentially regulated between proliferating and neurogenic APs. In neurogenic APs, the numbers of apical and basal astral MTs (but not of central MTs), decrease with respect to proliferating APs, in correlation with an increase in the amplitude of spindle oscillation during metaphase. Therefore, the density of apical/basal astral MTs may regulate the stability of spindle orientation. Indeed, specific perturbation of this astral MT subpopulation impacts the amplitude of spindle oscillations observed in proliferating APs. Interestingly, this subpopulation of astral MTs is in part controlled by LGN enrichment in the basal cortex, which is higher in proliferating than in neurogenic progenitors. This suggests that cortical anchoring of apical/basal astral MTs by the LGN complex regulates their stability. While it could be imagined that a broader cortical distribution of the LGN complex would lead to a less stable spindle orientation, the authors propose that it acts in the opposite manner: Basal LGN would favor the stabilization of the spindle by anchoring apical/basal astral MTs. It can be hypothesized that forces exerted on apical/basal astral MTs are smaller than those exerted on central astral MTs. This would allow spindle orientation along the plane of the tissue, which will be further stabilized by the anchoring of astral MTs to the apical/basal domains. Whether specific subpopulations of astral MTs exist in other cellular contexts and how they regulate spindle orientation remains to be investigated.
Finally, it should be pointed out that shortening of astral MTs might differentially impact spindle orientation depending on the spindle size relative to the cell size. In addition, reduced astral MT density can result in different outcomes depending on the available cues for spindle orientation that in turn determine the level of enrichment of force generators at the cortex.
Extracellular stimuli influencing spindle orientation
In a tissue, cells are exposed to a variety of environmental stimuli that can influence their axis of division by mobilizing and polarizing the internal machinery for spindle orientation discussed above. In this section, we will review the increasing diversity in signaling pathways involved in the upstream regulation of spindle orientation (Fig 4).
Tre1 GPCR signaling
The observation that spindle orientation was random in dissociated fly neuroblasts led to the proposal that the axis of division of these cells may be influenced by extrinsic cues from the underlying neuroectoderm 117. More recently, Yoshiura and colleagues have shown that Tre1 GPCR expressed in neuroblasts is the receptor for polarizing signals from the neuroectoderm. Remarkably, signaling downstream of Tre1 recruits Pins to the cell membrane closest to the neuroectoderm, through unconventional (and Drosophila‐specific) Pins binding to activated GTP‐bound Gαo. Pins in turn recruits inscuteable and Par3, thereby aligning the apical–basal polarity axis of the neuroblast relative to the neuroectoderm. In addition, Pins controls the orientation of the spindle along the defined axis through Mud. In Tre1 mutants, the polarity axis of the cell (defined by Par3/Insc localization) and the orientation of the spindle remain coordinated with each other, but they are randomized relative to the neuroectoderm. Therefore, an unidentified signal from the neuroectoderm acts as a positional cue for the polarity axis through Tre1 signaling 118, and Pins plays a dual role in interpreting the external polarity cue and orienting the spindle.
Planar cell polarity (PCP) pathways
The role of planar cell polarity pathways in the modulation of spindle orientation has been extensively documented 19, 91. Both Wnt/Fz and Fat/Dachsous/Four‐jointed (Fat/Ds/Fj) pathways modulate spindle orientation in both asymmetrically and symmetrically dividing cells.
One of the best‐understood models regarding these signaling pathways is the EMS cell division in the C. elegans 4‐cell embryo (Fig 5Ai). In this system, the EMS cell receives a secreted Wnt (MOM2) signal from the neighboring P2 cell, and this leads to the activation of Fz (MOM5). As a consequence, signaling events trigger the accumulation of dynactin at the P2‐EMS contact site, directing spindle rotation in the direction of this site. Similarly, during the asymmetric division of Drosophila SOP, the Fz/Dsh pathway dictates the orientation of the spindle along the anterior–posterior axis. This mechanism involves the localization of Fz and Dsh to the posterior cortex of the SOP cell, which allows the recruitment of NuMA, and possibly dynein 63. In addition, PCP proteins are necessary for the correct localization of Dlg/Pins/Gαi to the anterior lateral cortex, which controls the orientation of the spindle relative to the apico‐basal axis. The Fat/Ds/Fj pathway also controls spindle orientation in some invertebrate and vertebrate tissues. In the context of tissue elongation of Drosophila wing disks, the Fat/Ds/Fj pathway is necessary for spindle orientation along the proximal–distal axis 119. Similarly, this pathway regulates the orientation of cell division in the context of tubule elongation during postnatal development of the murine kidney 120.
Figure 5. Extracellular stimuli influencing spindle orientation.
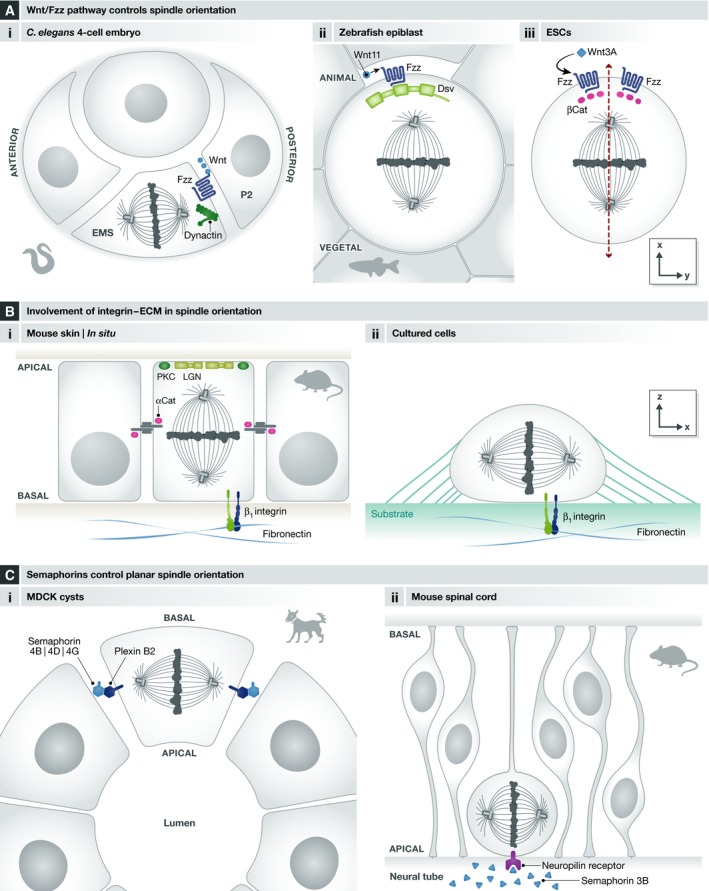
(A) The Wnt/Fz pathway controls spindle orientation in diverse contexts including (i) the 4‐cell C. elegans embryo, (ii) zebrafish epiblast cells, and (iii) embryonic stem cells. (B) Involvement of integrins–ECM in spindle orientation: I) In mouse skin progenitors during stratification, β1 integrin is necessary for correct cell polarity (PKC), LGN apical localization and thus spindle orientation; II) in cultured cells, interaction of β1 integrin with extracellular matrix components is necessary for spindle orientation in parallel to the substrate. (C) Semaphorins control planar spindle orientation (i) in the context of epithelial morphogenesis in MDCK cysts and (ii) in neural progenitors of the mouse spinal cord.
PCP pathways also modulate mitotic spindle orientation during symmetric division in the context of tissue morphogenesis. In particular, Wnt11 and Dsh regulate division orientation along the animal–vegetal axis in epiblast cells during zebrafish gastrulation 121 (Fig 5aii). In addition, the Wnt/Fz and Fat/Ds/Fj pathways regulate spindle orientation in kidney development in the mouse. Likewise, spindle orientation along the proximal–distal axis during limb bud growth in the mouse embryo is under the control of Wnt5A 122.
Finally, localized extracellular Wnt signals have recently been shown to orient the mitotic spindle in cultured embryonic stem cells 123. In this system, asymmetric exposure of ES cells to Wnt3a beads promotes the asymmetric localization of Wnt components such as β‐catenin, APC and frizzled. In this context, the spindle is oriented perpendicularly to the Wnt3A bead (Fig 5Aiii). This leads to the question how Wnt3A orients the spindle in ES cells. One possibility is the involvement of the Fz/Dsh/NuMA pathway 63, considering the asymmetric distribution of Fz‐GFP observed in this context.
Integrins
The influence of signals from the extracellular matrix (ECM) on spindle orientation has first been demonstrated in the context of in vivo mouse skin stratification. Apical recruitment of LGN by inscuteable and Gαi controls the apico‐basal spindle orientation of asymmetrically dividing progenitors during skin stratification 11, 39, 124. In knockout mice for β1 integrin, a protein that is critical for basement membrane integrity, spindle orientation is randomized 11. This phenotype correlates with the loss of PKC‐ζ (a key determinant of the apical domain) from the apical cortex and the randomization of the position of LGN crescents. This suggests that the absence of β1 integrin signaling disrupts cell polarity and correct apical localization of the LGN complex, thus randomizing spindle orientation. Similarly, the loss of the cell–cell adhesion molecule α‐catenin impairs the apical localization of PKC‐ζ and spindle orientation. However, LGN is no longer observed in the cell cortex. Because of the influence of adherens junctions on the actin cytoskeleton, the effects of α‐catenin knockout on LGN complex formation could be due to defects in the actin cortex (see “The emerging role of actin in spindle orientation”). Therefore, both the basement membrane and cell–cell contacts are necessary for spindle orientation during skin stratification. In summary, β1 integrin and α‐catenin may be seen as permissive rather than instructive cues for spindle orientation. By maintaining cell polarity, they allow LGN to be specifically localized to the apical cortex. In addition, α‐catenin would be necessary for LGN recruitment or stability in the cell cortex (Fig 5Bi).
Similarly, Toyoshima and Nishida have shown that spindle orientation with respect to the cell–substrate adhesion plane is influenced by the extracellular matrix (ECM) in cultured cells 112 (Fig 5Bii). Cells cultured on fibronectin and collagen, which are integrin‐binding ECM components, orient their spindles parallel to the substrate more tightly in comparison with cells cultured on poly‐L‐lysine, which does not bind to integrin. In addition, by using blocking antibodies and siRNA approaches, the authors showed that this adhesion‐dependent orientation depends specifically on β1 integrin. Recent work by Morris and colleagues indicates that the direct interaction between integrin‐linked kinase (ILK) and dynactin‐2 (p50) links integrins to the dynein complex and controls the position of force generators 125. It should be noted that in cells cultured on poly‐L‐lysine or in cells in which integrin, ILK, or p50 have been knocked down, spindle orientation remains strongly biased to angles lower than 40°, indicating that additional cues guide spindle orientation in this context (see “Spindle orientation in context: roles of cell geometry and mechanical forces”).
Semaphorins
Semaphorin‐mediated signaling has recently been proposed to regulate spindle orientation in different tissues. Xia and colleagues have demonstrated that cell–cell communication mediated by semaphorin–plexin signaling orients the mitotic spindle in kidney cells 126 (Fig 5Ci). In particular, plexin B2 is localized at the baso‐lateral membrane in different epithelial tissues in vivo and in MDCK cells grown in 3D cultures. Depletion of plexin B2 generates defects in spindle orientation both in MDCK cysts and in renal tubules in the context of tissue repair 126. Based on observations made in knockout mice, the authors suggested that the ligands semaphorin 4B, 4D, and 4G redundantly activate plexin B2. In addition, they demonstrated the involvement of Cdc42 in mediating plexin B2 effects on spindle orientation. Interestingly, upon plexin B2 depletion in MDCK cysts and regenerating renal tubules, spindle orientation is not randomized but appears to be biased toward the apico‐basal axis. Whether this results from the re‐localization of spindle orientation regulators like LGN or from the takeover of an alternative pathway remains to be investigated.
Similarly, Arbeille and colleagues have reported the effects of semaphorin 3B on the orientation of neural progenitor divisions in the mouse spinal cord 127 (Fig 5Cii). In semaphorin 3B knockout mice, the authors observed a small but significant change in the distribution of progenitor division angles at E10.5. Interestingly, these mice do not show defects in apico‐basal polarity and F‐actin organization, indicating that spindle orientation is not indirectly affected by changes in cell polarity. The authors showed that semaphorin 3B released from the ventral floor plate into the central canal binds to neuropilin receptors located at the apical surface of neural progenitors. At the molecular level, GSK3 and the microtubule stabilizing protein CRMP2 have been shown to be involved, but the pathway between signal reception at the apical surface and spindle positioning remains elusive. In particular whether the localization of LGN complex members is affected has not been determined, and whether semaphorin 3B signals during prometaphase/metaphase when the orientation of the spindle is set up is unknown.
Spindle orientation in context: roles of cell geometry and mechanical forces
Mitotic rounding is a common and remarkable feature of most dividing animal cells, whether in adherent cell culture or in intact tissues. Mitotic rounding implies reorganization of the actin cytoskeleton (reviewed in 128), and cell ballooning is achieved through an increase in intracellular osmotic pressure 129. The mitotic actin cortex is thinner, but stiffer than in interphase 93. Mitotic rounding is viewed as a way to generate sufficient intracellular space to accommodate spindle formation and is indeed important for chromosome capture and bipolar spindle maintenance 130 (reviewed in 94). Apart from non‐adherent cells (such as one‐cell zygotes), mitotic rounding in mitosis implies a profound remodeling of cells adhesion with their neighbors and/or the extracellular matrix.
Despite their rounding, mitotic cells are exposed to external forces generated by the contact with neighboring cells and with the substrate. These forces depend on the position of the cell within a tissue and on the changes in the tissue itself, especially during morphogenesis, and reflect a memory of cell shape and adhesion in interphase. In addition, rounding itself is often imperfect and cells retain a slightly elongated shape that corresponds to their shape in interphase and scales with tissue tension. In the following section, we will describe that both the memory of cell shape in interphase and a more direct sensing of cell shape in mitosis can influence spindle orientation.
Intrinsic cell geometry in mitosis impacts on spindle orientation
The empirical century‐old “long axis” or “Hertwig rule”, initially proposed by Oscar Hertwig in the late 19th century, posits that cells usually place their cleavage plane at the center of their mass and perpendicular to their longest axis 20. Hertwig had explored this property through experimental deformation of single cell echinoderm embryos. These cells are normally perfectly spherical, and their first division is symmetric with no preferential orientation. However, by gently squeezing embryos between glass plates, Hertwig observed that the orientation of division could be controlled by the deformation and aligned with the elongated axis. In line with Hertwig's observations, O'Connell and Wang used a similar approach to probe the relationship between cell shape and spindle orientation in cultured mammalian cells 131. Using micromanipulation with glass pipettes, they forced shape deformations in dividing normal rat kidney (NRK) cells, which do not round up during mitosis and keep their interphase shape. In these cells, although the mitotic spindle can sometimes be observed orthogonal to the cell's longest axis in early metaphase, by anaphase it is aligned parallel to the longest axis. They observed that upon experimental deformation of mitotic cells, the spindle constantly reacted to cell shape changes and adapted by moving to the new cell center and realigning with the induced longest axis. They further showed that spindle movements occurred in an astral microtubule‐ and dynein‐dependent manner.
Hertwig's rule was recently revisited in sea urchin embryos by Minc and colleagues, who used microfabricated 3‐D molds to apply specific anisotropic shape deformations 132. While the rule applied to most shapes, some specific shapes did not conform to its predictions. A model in which forces applied to spindle poles scaled with the length of individual astral microtubules predicted much better the orientations observed in all tested shapes. This model is difficult to reconcile with force generators combined at or near the cell cortex, which is the dominant model in other cell types, and it implies a role of force generators in the cytoplasm 132. It should also be noted that single cell zygotes are usually very large and their spindle is comparatively small, with very long astral microtubules. In contrast, in many cell types, the size of the spindle scales with cell size 133, 134; mitotic rounding is indeed essential to allow sufficient space for the formation of the spindle, and artificial confinement results in chromosome missegregation 130.
Recently, Lazaro‐Dieguez and colleagues studied the relationship between spindle orientation and cell shape in the context of imperfect rounding, making use of the natural variability of rounding in adherent MDCK and HeLa cells in mitosis 135. In control conditions, adherent cells divide very precisely in the plane of the substrate, making it difficult to address the question. Disruption of force generation either by knockdown of the LGN pathway or pharmacological removal of astral microtubules disrupts this orientation, but a strong bias toward planar orientation remains. The authors attribute this bias to imperfect cell rounding: They compared the orientation of the spindle in cells treated with low doses of nocodazole (that primarily disrupt astral microtubules and abolish force generation) between perfectly round and more “flat” mitotic cells and found that the bias toward planar orientation was much more pronounced in flat cells (relative to the substrate), while orientation was close to random in cells with a more spherical shape. This indicates that cell shape can directly influence orientation independently of cortical force generators. The authors observed frequent deformation of the metaphase plate in these flat cells, suggesting that the effect on orientation may be a direct consequence of steric hindrance in cells where cytoplasmic volume and cell size are just sufficient to accommodate the size of the metaphase spindle.
This notion can be transposed to in vivo situations, where cell packing imposes constraints on cell shape both in interphase and during mitosis, and where rounding is unlikely to be perfect. In the mouse developing skin, where interphase cells are flat (e.g., with a relatively short apico‐basal length), orientation of the spindle is biphasic: Symmetric divisions orient in the plane of the tissue and asymmetric divisions are perpendicular to this plane in an Insc/Gαi/LGN/NuMA‐dependent manner 11, 124. Remarkably, disruption of force generators via knockdown of NuMA or p150 does not lead to random spindle orientation, but most divisions are planar, according to the main axis of cell elongation (Fig 6A). This suggests that a cell shape sensing mechanism independent of cortical force generators contributes to default planar orientation in this tissue 39.
Figure 6. Role of cell geometry and external forces.
(A) Cells in the basal layer of the developing mouse epidermis adopt a binary orientation: Symmetric divisions occur in the plane of the epithelium, and asymmetric divisions divide along the apico‐basal axis in an Insc/Gαi/LGN/NuMA/dynein manner, with one daughter cell delaminating into the suprabasal cell layer. Upon NuMA or p150 depletion, cortical force generators are not functioning and most divisions now take place in the plane of the epithelium, suggesting that the “default” planar orientation may be dictated by the flat cell shape in this tissue. Green lines: Insc and Gαi3 apical accumulation; orange lines: force generators (dynein). See Williams et al (2011) 39. (B) In single cells cultured on fibronectin micropatterns (light blue), a field of maximal force is associated with polarized retraction fibers (blue lines). Cells cultured on “cross”‐shaped patterns orient their spindle along the long arms of the cross, where maximal forces are observed. Laser ablation of retraction fibers on the long arms induces a 90° spindle rotation and alignment to face the “new” maximal forces. See Fink et al (2011) 67. (C) In the fly notum epithelium, NuMA accumulates at tricellular junctions in the G2 phase. Left panel: A vector corresponding to the cells long axis (gray bar) or to the geometry of tricellular junctions (or Mud accumulation, red dots) can be drawn (blue bar). In elongated cells, both vectors are aligned (top light red cell), whereas they do not always align in cells with an isotropic shape (bottom light red cell). Middle panel: The “Mud accumulation” vector predicts the orientation of cell divisions more accurately than the long axis. Right panel: Position and shape of the daughter cells after division. See Bosveld et al (2016) 140.
Role of surrounding forces in spindle orientation
External forces influence spindle orientation in single cells in vitro
In cultured adherent cells, the distribution of retraction fibers in mitosis reflects the geometry of the adhesion of the cell to its substrate in the previous interphase. As mentioned above, the distribution of retraction fibers dictates, and therefore can indeed be used to predict the orientation of the mitotic spindle within the plane of the substrate 65, 66, 67. Fink and colleagues demonstrated the function of retraction fibers by performing laser ablation of these cellular structures and by analyzing spindle movements (Fig 6B). Importantly, these authors observed changes in cell shape upon retraction fiber ablation, suggesting that retraction fibers exert forces on the cell. Indeed, this was confirmed upon measurement of the forces associated with retraction fibers by using optical tweezers. Remarkably, applying stretch forces to a cell without affecting its shape is sufficient for spindle rotation along the axis of the dominant force field 67. Collectively, these experiments demonstrate that adhesion‐related forces can control spindle orientation in single cells. In these experiments, reorientation was reduced in the presence of low doses of nocodazole that disrupt astral microtubules, indicating that force generators acting on the microtubule network work downstream of retraction fibers. The entire molecular cascade that links external forces from retraction fibers to the recruitment and activation of internal force generators is not completely understood and involves several pathways, as already detailed. On one hand, activation of the actin cloud/myosin 10 pathway provides a direct link with microtubule dynamics 102; on the other hand, forces exerted on the cortex influence local ERM activation 66, 68, which directly or indirectly promotes the asymmetric localization of LGN/NuMA and presumably of dynein to the cortex 68.
While retraction fibers have not been described in tissues, distinct structures may mediate the establishment of forces in an analogous manner through cell–matrix attachment or cell–cell interactions, as described below.
Influence of external forces on spindle orientation in vivo
The influence of external forces on spindle orientation has also been addressed in vivo in developing tissues. During the spreading of the enveloping cell layer (EVL) of the zebrafish gastrula 136, cells orient their spindle along the animal–vegetal axis which coincides with the axis of maximal tension in this tissue. Importantly, artificial induction of local tension in the perpendicular direction induces spindle rotation and reorientation toward the axis of induced tension. Using a computational model, the authors found that the cell shape parameter in interphase can reliably be used to predict the visualized orientation of divisions in the EVL. Mechanistically, they showed that the molecular motor myosin II is involved in cell shape regulation and in mediating the connection between shape and spindle orientation 136.
Wyatt et al 137 used micromanipulation to apply stretch forces to a suspended monolayer of cultured MDCK cells. In response to stretch, cells elongate parallel to this homogeneous field force and divide along their longest axis. This restores cell shape isotropy in the stretched tissue; in short, divisions relieve tension. Similar results were obtained in the developing Drosophila wing disk, where cells divide according to local tension fields, therefore reducing the tension in the tissue 138, 139. During this morphogenetic process, the tension fields themselves are generated by local variations in proliferation rates, showing an interesting feedback loop between proliferation, tissue tension, and oriented cell divisions.
How do cells sense “tension” and translate it into spindle orientation? When a tissue is under tension, cells tend to adopt an elongated shape that generally aligns with the axis of maximal tension. However, a minority of cells do not behave like this. Remarkably, Wyatt et al 137 found that the spindle aligns with the long axis even in the minority of cases when the long axis is not aligned with the stretch applied to the tissue, indicating that cell shape may be a better predictor than global tissue tension itself. However, a recent study by Bosveld and colleagues shows that while cell shape in interphase is a good indicator of spindle orientation when anisotropy is high, it does not predict orientation as efficiently in nearly isotropic cells. Under these conditions the topology of a cell's contacts with its neighbors during interphase is a better parameter 140. In the epithelium of the fly pupal notum, the authors found that tricellular junctions (TCJs; the vertex where three neighboring cells are in contact) localize force generators in a Mud‐dependent manner. Remarkably, Mud starts to accumulate at TCJs during G2 phase. When cells round up for mitosis, the position of cortical patches of Mud reflects the geometry of the cell contacts with its neighbors and dictates where greater forces will be generated. The authors show that a model using the position of the TCJs, and therefore of the Mud patches (“Mud intensity model”), to predict force generation faithfully recapitulates experimental data in this tissue. Remarkably, predictions in this particular tissue are more accurate than with a model that uses cell shape as one of its main parameters 132. Bosveld and colleagues propose that in addition to their function as epithelial barrier structures, TCJs serve as polarity cues promoting geometry and mechanical sensing in epithelial tissues 140 (Fig 6C). Quite remarkably, this new orientation mechanism depends on Mud, dynein, and Dlg, but does not require Gαi or Pins, providing another example of a Pins‐independent, but Mud‐dependent pathway. In contrast to Drosophila Mud, vertebrate NuMA is nuclear in interphase and has not been described at cellular junctions in epithelia; it is therefore unclear whether the mechanism described above reflects a generic property of TCJs. Future experiments in other model systems, either in tissues or in experimentally stretched cell layers (like those described by Wyatt et al), should explore whether TCJs carry a similar geometric information independently of NuMA.
In summary, it appears that mechanisms acting in interphase and during mitosis sense extrinsic tension and intrinsic geometry and contribute to translate these cell shape parameters into an oriented spindle. Despite the increase in osmotic pressure, mitotic rounding is probably never perfect in a tissue where cells are subjected to forces of adhesion and compaction. It is therefore difficult to completely uncouple the factors that depend on external forces from those related to intrinsic shape.
Conclusions
Over the last two decades, our knowledge of the molecular mechanisms that control the orientation of the mitotic spindle has significantly increased. Early findings in a limited number of models uncovered a common set of evolutionarily conserved basic players. However, recent work looking at different species has highlighted that regulatory mechanisms vary a lot, and a number of alternative pathways for force generation have been discovered. Remarkably, within a single organism, there are important tissue‐specific differences concerning the modalities of spindle orientation and which pathway is used.
The actin cortex and astral microtubules are not just passive players that respectively anchor force generators and serve as a substrate for those forces: They are the core of complex regulatory pathways that tune force transmission to finely modulate spindle orientation, and many levels of regulation certainly remain to be uncovered.
Over the last years, advances in live molecular imaging have played a decisive role in this field, highlighting the importance of dynamic studies in the description and understanding of this highly choreographed process. Combinations of theoretical modeling with live observations, genetics, and physical manipulations have helped to refine Hertwig's remarkable observations, bringing forth a number of different interpretations and molecular explanations to his classical rule in different cell and tissue types.
The role of spindle orientation in normal and pathological development and homeostasis has been acknowledged, but is still poorly understood 14, 15, 16, 17, 18, 19. The research performed over the last few years, illustrates the remarkable diversity of strategies and mechanisms used in different systems. Hence, it is increasingly evident that we need to explore, in addition to further research into the details of known pathways in classical models of spindle orientation, the conservation and diversity in spindle orientation pathways in new cell and tissue types (Box 3).
Box 3: In need of answers.
How do different cortical molecules (like Gαi, Afadin, and Dlg1) interplay to regulate the recruitment and maintenance of LGN to the cellular cortex?
In cultured cells the dynamics of LGN, NuMA, and dynein localization during mitosis are regulated by molecules located on chromosomes, centrosomes and at the cortex. Are these pathways active and necessary to achieve oriented divisions in vivo? How do they integrate with the instructive signals, such as polarized inscuteable, that control spindle orientation in complex tissues?
How are LGN/dynein complex members transported to the cellular cortex in higher eukaryotes? Are they transported by astral MT as suggested by the dynein off‐loading model in yeast?
How do actin and NuMA/dynein pathways interact during spindle orientation?
What other alternative force tethering systems exist?
How do geometric and molecular cues integrate in a complex tissue?
Can the recent models and molecular interpretations of the “long axis rule” be transferred in tissues with a different organization?
What is the actual contribution of spindle orientation to development and tissue homeostasis? How does spindle orientation contribute to cancer development and in what specific contexts?
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
Work in the Morin laboratory has received support from Fondation ARC (LIVESPIN 2011), ANR (LIVESPIN 2012), Cancéropôle Ile‐de‐France (2013‐2‐INV‐05), FRM (Equipe FRM DEQ20150331735), and under the program “Investissements d'Avenir ” launched by the French Government and implemented by the ANR, with the references: ANR‐10‐LABX‐54 MEMO LIFE and ANR‐11‐IDEX‐0001‐02 PSL* Research University. FdP was the recipient of doctoral grants from the French Ministry of Higher Education and Research (MESR) and from the FRM. Work in the Echard laboratory was supported by Institut Pasteur, CNRS, FRM (Equipe FRM DEQ20120323707), INCa (2014‐1‐PL BIO‐04‐IP1), ANR (AbCyStem) and IXCORE Foundation.
EMBO Reports (2016) 17: 1106–1130
See the Glossary for abbreviations used in this article.
References
- 1. Tanaka TU (2010) Kinetochore‐microtubule interactions: steps towards bi‐orientation. EMBO J 29: 4070–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glotzer M (2009) The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol 10: 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA (1996) Role of inscuteable in orienting asymmetric cell divisions in Drosophila . Nature 383: 50–55 [DOI] [PubMed] [Google Scholar]
- 4. Cheng NN, Kirby CM, Kemphues KJ (1995) Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par‐2 and par‐3. Genetics 139: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kemphues KJ, Priess JR, Morton DG, Cheng NS (1988) Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320 [DOI] [PubMed] [Google Scholar]
- 6. Morin X, Jaouen F, Durbec P (2007) Control of planar divisions by the G‐protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci 10: 1440–1448 [DOI] [PubMed] [Google Scholar]
- 7. Peyre E, Jaouen F, Saadaoui M, Haren L, Merdes A, Durbec P, Morin X (2011) A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol 193: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng Z, Zhu H, Wan Q, Liu J, Xiao Z, Siderovski DP, Du Q (2010) LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol 189: 275–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aigouy B, Farhadifar R, Staple DB, Sagner A, Roper JC, Julicher F, Eaton S (2010) Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila . Cell 142: 773–786 [DOI] [PubMed] [Google Scholar]
- 10. Baena‐Lopez LA, Baonza A, Garcia‐Bellido A (2005) The orientation of cell divisions determines the shape of Drosophila organs. Curr Biol 15: 1640–1644 [DOI] [PubMed] [Google Scholar]
- 11. Lechler T, Fuchs E (2005) Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB (2006) Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA 103: 10438–10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, Ridgway RA, Sansom OJ, Nathke IS (2010) Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6: 175–181 [DOI] [PubMed] [Google Scholar]
- 14. Bergstralh DT, St Johnston D (2014) Spindle orientation: what if it goes wrong? Semin Cell Dev Biol 34: 140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noatynska A, Gotta M, Meraldi P (2012) Mitotic spindle (DIS)orientation and DISease: cause or consequence? J Cell Biol 199: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Januschke J, Nathke I (2014) Stem cell decisions: a twist of fate or a niche market? Semin Cell Dev Biol 34: 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peyre E, Morin X (2012) An oblique view on the role of spindle orientation in vertebrate neurogenesis. Dev Growth Differ 54: 287–305 [DOI] [PubMed] [Google Scholar]
- 18. Gillies TE, Cabernard C (2011) Cell division orientation in animals. Curr Biol 21: R599–R609 [DOI] [PubMed] [Google Scholar]
- 19. Morin X, Bellaïche Y (2011) Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell 21: 102–119 [DOI] [PubMed] [Google Scholar]
- 20. Hertwig O (1884) Das Problem der Befruchtung und der Isotropie des Eies, eine Theory der Vererbung. Jenaische Zeitschrift fuer Naturwissenschaft 18: 21–23 [Google Scholar]
- 21. Schaefer M, Shevchenko A, Knoblich JA (2000) A protein complex containing Inscuteable and the Galpha‐binding protein Pins orients asymmetric cell divisions in Drosophila . Curr Biol 10: 353–362 [DOI] [PubMed] [Google Scholar]
- 22. Yu F, Morin X, Cai Y, Yang X, Chia W (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100: 399–409 [DOI] [PubMed] [Google Scholar]
- 23. McNally FJ (2013) Mechanisms of spindle positioning. J Cell Biol 200: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore JK, Stuchell‐Brereton MD, Cooper JA (2009) Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton 66: 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Markus SM, Lee WL (2011) Microtubule‐dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture 1: 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Markus SM, Kalutkiewicz KA, Lee WL (2012) Astral microtubule asymmetry provides directional cues for spindle positioning in budding yeast. Exp Cell Res 318: 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woodruff JB, Drubin DG, Barnes G (2009) Dynein‐driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell 20: 3003–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Markus SM, Lee WL (2011) Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell 20: 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lansbergen G, Komarova Y, Modesti M, Wyman C, Hoogenraad CC, Goodson HV, Lemaitre RP, Drechsel DN, van Munster E, Gadella TW Jr et al (2004) Conformational changes in CLIP‐170 regulate its binding to microtubules and dynactin localization. J Cell Biol 166: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du Q, Macara IG (2004) Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119: 503–516 [DOI] [PubMed] [Google Scholar]
- 31. Gotta M, Ahringer J (2001) Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol 3: 297–300 [DOI] [PubMed] [Google Scholar]
- 32. Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J (2003) Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol 13: 1029–1037 [DOI] [PubMed] [Google Scholar]
- 33. Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F (2008) Neuroepithelial progenitors undergo LGN‐dependent planar divisions to maintain self‐renewability during mammalian neurogenesis. Nat Cell Biol 10: 93–101 [DOI] [PubMed] [Google Scholar]
- 34. Srinivasan DG, Fisk RM, Xu H, van den Heuvel S (2003) A complex of LIN‐5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev 17: 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA (2001) Heterotrimeric g proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107: 183–194 [DOI] [PubMed] [Google Scholar]
- 36. Nguyen‐Ngoc T, Afshar K, Gonczy P (2007) Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol 9: 1294–1302 [DOI] [PubMed] [Google Scholar]
- 37. Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M (2007) Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans . J Cell Biol 179: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kotak S, Busso C, Gonczy P (2012) Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol 199: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams SE, Beronja S, Pasolli HA, Fuchs E (2011) Asymmetric cell divisions promote Notch‐dependent epidermal differentiation. Nature 470: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA (2006) The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell 10: 731–742 [DOI] [PubMed] [Google Scholar]
- 41. Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F (2006) Drosophila Pins‐binding protein Mud regulates spindle‐polarity coupling and centrosome organization. Nat Cell Biol 8: 586–593 [DOI] [PubMed] [Google Scholar]
- 42. Siller KH, Cabernard C, Doe CQ (2006) The NuMA‐related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol 8: 594–600 [DOI] [PubMed] [Google Scholar]
- 43. Bergstralh DT, Lovegrove HE, St Johnston D (2013) Discs large links spindle orientation to apical‐basal polarity in Drosophila epithelia. Curr Biol 23: 1707–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mochizuki N, Cho G, Wen B, Insel PA (1996) Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene 181: 39–43 [DOI] [PubMed] [Google Scholar]
- 45. Willard FS, Kimple RJ, Siderovski DP (2004) Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem 73: 925–951 [DOI] [PubMed] [Google Scholar]
- 46. Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gonczy P (2004) RIC‐8 is required for GPR‐1/2‐dependent Galpha function during asymmetric division of C. elegans embryos. Cell 119: 219–230 [DOI] [PubMed] [Google Scholar]
- 47. Couwenbergs C, Spilker AC, Gotta M (2004) Control of embryonic spindle positioning and Galpha activity by C. elegans RIC‐8. Curr Biol 14: 1871–1876 [DOI] [PubMed] [Google Scholar]
- 48. David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaiche Y (2005) Drosophila Ric‐8 regulates Galphai cortical localization to promote Galphai‐dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol 7: 1083–1090 [DOI] [PubMed] [Google Scholar]
- 49. Hampoelz B, Hoeller O, Bowman SK, Dunican D, Knoblich JA (2005) Drosophila Ric‐8 is essential for plasma‐membrane localization of heterotrimeric G proteins. Nat Cell Biol 7: 1099–1105 [DOI] [PubMed] [Google Scholar]
- 50. Wang H, Ng KH, Qian H, Siderovski DP, Chia W, Yu F (2005) Ric‐8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat Cell Biol 7: 1091–1098 [DOI] [PubMed] [Google Scholar]
- 51. Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH (2010) Ric‐8A and Gi{alpha} Recruit LGN, NuMA, and Dynein to the Cell Cortex to Help Orient the Mitotic Spindle. Mol Cell Biol 30: 3519–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hess HA, Roper JC, Grill SW, Koelle MR (2004) RGS‐7 completes a receptor‐independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans . Cell 119: 209–218 [DOI] [PubMed] [Google Scholar]
- 53. Yu F, Wang H, Qian H, Kaushik R, Bownes M, Yang X, Chia W (2005) Locomotion defects, together with Pins, regulates heterotrimeric G‐protein signaling during Drosophila neuroblast asymmetric divisions. Genes Dev 19: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du Q, Stukenberg PT, Macara IG (2001) A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol 3: 1069–1075 [DOI] [PubMed] [Google Scholar]
- 55. Haren L, Merdes A (2002) Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J Cell Sci 115: 1815–1824 [DOI] [PubMed] [Google Scholar]
- 56. Merdes A, Ramyar K, Vechio JD, Cleveland DW (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87: 447–458 [DOI] [PubMed] [Google Scholar]
- 57. Izumi Y, Ohta N, Itoh‐Furuya A, Fuse N, Matsuzaki F (2004) Differential functions of G protein and Baz‐aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol 164: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wodarz A, Ramrath A, Grimm A, Knust E (2000) Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol 150: 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG (2010) Par3 Controls Epithelial Spindle Orientation by aPKC‐Mediated Phosphorylation of Apical Pins. Curr Biol 20: 1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnston CA, Hirono K, Prehoda KE, Doe CQ (2009) Identification of an Aurora‐A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138: 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Johnston CA, Manning L, Lu MS, Golub O, Doe CQ, Prehoda KE (2013) Formin‐mediated actin polymerization cooperates with Mushroom body defect (Mud)‐Dynein during Frizzled‐Dishevelled spindle orientation. J Cell Sci 126: 4436–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wee B, Johnston CA, Prehoda KE, Doe CQ (2011) Canoe binds RanGTP to promote Pins(TPR)/Mud‐mediated spindle orientation. J Cell Biol 195: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Segalen M, Johnston CA, Martin CA, Dumortier JG, Prehoda KE, David NB, Doe CQ, Bellaiche Y (2010) The Fz‐Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev Cell 19: 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E (2007) PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell 13: 796–811 [DOI] [PubMed] [Google Scholar]
- 65. Thery M, Jimenez‐Dalmaroni A, Racine V, Bornens M, Julicher F (2007) Experimental and theoretical study of mitotic spindle orientation. Nature 447: 493–496 [DOI] [PubMed] [Google Scholar]
- 66. Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M (2005) The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol 7: 947–953 [DOI] [PubMed] [Google Scholar]
- 67. Fink J, Carpi N, Betz T, Betard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D et al (2011) External forces control mitotic spindle positioning. Nat Cell Biol 13: 771–778 [DOI] [PubMed] [Google Scholar]
- 68. Machicoane M, de Frutos CA, Fink J, Rocancourt M, Lombardi Y, Garel S, Piel M, Echard A (2014) SLK‐dependent activation of ERMs controls LGN‐NuMA localization and spindle orientation. J Cell Biol 205: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier ML, O'Kane CJ, Bryant PJ, Schweisguth F (2001) The Partner of Inscuteable/Discs‐large complex is required to establish planar polarity during asymmetric cell division in Drosophila . Cell 106: 355–366 [DOI] [PubMed] [Google Scholar]
- 70. Ohshiro T, Yagami T, Zhang C, Matsuzaki F (2000) Role of cortical tumour‐suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408: 593–596 [DOI] [PubMed] [Google Scholar]
- 71. Peng CY, Manning L, Albertson R, Doe CQ (2000) The tumour‐suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408: 596–600 [DOI] [PubMed] [Google Scholar]
- 72. Siegrist SE, Doe CQ (2005) Microtubule‐induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123: 1323–1335 [DOI] [PubMed] [Google Scholar]
- 73. Saadaoui M, Machicoane M, di Pietro F, Etoc F, Echard A, Morin X (2014) Dlg1 controls planar spindle orientation in the neuroepithelium through direct interaction with LGN. J Cell Biol 206: 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johnston CA, Doe CQ, Prehoda KE (2012) Structure of an enzyme‐derived phosphoprotein recognition domain. PLoS ONE 7: e36014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ (2005) mPins modulates PSD‐95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol 7: 1179–1190 [DOI] [PubMed] [Google Scholar]
- 76. Zhu J, Shang Y, Xia C, Wang W, Wen W, Zhang M (2011) Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho‐protein‐binding modules. EMBO J 30: 4986–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gallini S, Carminati M, De Mattia F, Pirovano L, Martini E, Oldani A, Asteriti IA, Guarguaglini G, Mapelli M (2016) NuMA Phosphorylation by Aurora‐A Orchestrates Spindle Orientation. Curr Biol 26: 458–469 [DOI] [PubMed] [Google Scholar]
- 78. Speicher S, Fischer A, Knoblich J, Carmena A (2008) The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol 18: 831–837 [DOI] [PubMed] [Google Scholar]
- 79. Carminati M, Gallini S, Pirovano L, Alfieri A, Bisi S, Mapelli M (2016) Concomitant binding of Afadin to LGN and F‐actin directs planar spindle orientation. Nat Struct Mol Biol 23: 155–163 [DOI] [PubMed] [Google Scholar]
- 80. Elias S, Thion M, Yu H, Moreira Sousa C, Lasgi C, Morin X, Humbert S (2014) Huntingtin Regulates Mammary Stem Cell Division and Differentiation. Stem Cells Reports 2: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Godin JD, Colombo K, Molina‐Calavita M, Keryer G, Zala D, Charrin BC, Dietrich P, Volvert ML, Guillemot F, Dragatsis I et al (2010) Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron 67: 392–406 [DOI] [PubMed] [Google Scholar]
- 82. Dewey EB, Sanchez D, Johnston CA (2015) Warts phosphorylates mud to promote pins‐mediated mitotic spindle orientation in Drosophila, independent of Yorkie. Curr Biol 25: 2751–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rebollo E, Roldan M, Gonzalez C (2009) Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development 136: 3393–3397 [DOI] [PubMed] [Google Scholar]
- 84. Yamashita YM, Jones DL, Fuller MT (2003) Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301: 1547–1550 [DOI] [PubMed] [Google Scholar]
- 85. Kiyomitsu T, Cheeseman IM (2012) Chromosome‐ and spindle‐pole‐derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 14: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tame MA, Raaijmakers JA, van den Broek B, Lindqvist A, Jalink K, Medema RH (2014) Astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning. Cell Cycle 13: 1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kiyomitsu T, Cheeseman IM (2013) Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154: 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kotak S, Busso C, Gonczy P (2013) NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J 32: 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kotak S, Busso C, Gonczy P (2014) NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J 33: 1815–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kotak S, Gonczy P (2014) NuMA phosphorylation dictates dynein‐dependent spindle positioning. Cell Cycle 13: 177–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Segalen M, Bellaiche Y (2009) Cell division orientation and planar cell polarity pathways. Semin Cell Dev Biol 20: 972–977 [DOI] [PubMed] [Google Scholar]
- 92. Seldin L, Muroyama A, Lechler T (2016) NuMA‐microtubule interactions are critical for spindle orientation and the morphogenesis of diverse epidermal structures. eLife 5: e12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Clark AG, Dierkes K, Paluch EK (2013) Monitoring actin cortex thickness in live cells. Biophys J 105: 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cadart C, Zlotek‐Zlotkiewicz E, Le Berre M, Piel M, Matthews HK (2014) Exploring the function of cell shape and size during mitosis. Dev Cell 29: 159–169 [DOI] [PubMed] [Google Scholar]
- 95. Luxenburg C, Pasolli HA, Williams SE, Fuchs E (2011) Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol 13: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nakajima Y, Meyer EJ, Kroesen A, McKinney SA, Gibson MC (2013) Epithelial junctions maintain tissue architecture by directing planar spindle orientation. Nature 500: 359–362 [DOI] [PubMed] [Google Scholar]
- 97. Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q (2013) Evidence for dynein and astral microtubule‐mediated cortical release and transport of Galphai/LGN/NuMA complex in mitotic cells. Mol Biol Cell 24: 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Redemann S, Pecreaux J, Goehring NW, Khairy K, Stelzer EH, Hyman AA, Howard J (2010) Membrane invaginations reveal cortical sites that pull on mitotic spindles in one‐cell C. elegans embryos. PLoS ONE 5: e12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Castanon I, Abrami L, Holtzer L, Heisenberg CP, van der Goot FG, Gonzalez‐Gaitan M (2013) Anthrax toxin receptor 2a controls mitotic spindle positioning. Nat Cell Biol 15: 28–39 [DOI] [PubMed] [Google Scholar]
- 100. Quesada‐Hernandez E, Caneparo L, Schneider S, Winkler S, Liebling M, Fraser SE, Heisenberg CP (2010) Stereotypical cell division orientation controls neural rod midline formation in zebrafish. Curr Biol 20: 1966–1972 [DOI] [PubMed] [Google Scholar]
- 101. Mitsushima M, Aoki K, Ebisuya M, Matsumura S, Yamamoto T, Matsuda M, Toyoshima F, Nishida E (2010) Revolving movement of a dynamic cluster of actin filaments during mitosis. J Cell Biol 191: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kwon M, Bagonis M, Danuser G, Pellman D (2015) Direct Microtubule‐Binding by Myosin‐10 Orients Centrosomes toward Retraction Fibers and Subcortical Actin Clouds. Dev Cell 34: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Woolner S, O'Brien LL, Wiese C, Bement WM (2008) Myosin‐10 and actin filaments are essential for mitotic spindle function. J Cell Biol 182: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fehon RG, McClatchey AI, Bretscher A (2010) Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 11: 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F (2008) Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol 180: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kunda P, Pelling AE, Liu T, Baum B (2008) Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol 18: 91–101 [DOI] [PubMed] [Google Scholar]
- 107. Solinet S, Mahmud K, Stewman SF, Ben El Kadhi K, Decelle B, Talje L, Ma A, Kwok BH, Carreno S (2013) The actin‐binding ERM protein Moesin binds to and stabilizes microtubules at the cell cortex. J Cell Biol 202: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Almonacid M, Terret ME, Verlhac MH (2014) Actin‐based spindle positioning: new insights from female gametes. J Cell Sci 127: 477–483 [DOI] [PubMed] [Google Scholar]
- 109. Chen CT, Hehnly H, Yu Q, Farkas D, Zheng G, Redick SD, Hung HF, Samtani R, Jurczyk A, Akbarian S et al (2014) A unique set of centrosome proteins requires pericentrin for spindle‐pole localization and spindle orientation. Curr Biol 24: 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hehnly H, Doxsey S (2014) Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev Cell 28: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tame MA, Raaijmakers JA, Afanasyev P, Medema RH (2016) Chromosome misalignments induce spindle‐positioning defects. EMBO Rep 17: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Toyoshima F, Nishida E (2007) Integrin‐mediated adhesion orients the spindle parallel to the substratum in an EB1‐ and myosin X‐dependent manner. EMBO J 26: 1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bouissou A, Verollet C, de Forges H, Haren L, Bellaiche Y, Perez F, Merdes A, Raynaud‐Messina B (2014) gamma‐Tubulin Ring Complexes and EB1 play antagonistic roles in microtubule dynamics and spindle positioning. EMBO J 33: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhu M, Settele F, Kotak S, Sanchez‐Pulido L, Ehret L, Ponting CP, Gonczy P, Hoffmann I (2013) MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J Cell Biol 200: 773–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD (2011) MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol 13: 1040–1050 [DOI] [PubMed] [Google Scholar]
- 116. Mora‐Bermudez F, Matsuzaki F, Huttner WB (2014) Specific polar subpopulations of astral microtubules control spindle orientation and symmetric neural stem cell division. eLife 3: e02875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Siegrist SE, Doe CQ (2006) Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133: 529–536 [DOI] [PubMed] [Google Scholar]
- 118. Yoshiura S, Ohta N, Matsuzaki F (2012) Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev Cell 22: 79–91 [DOI] [PubMed] [Google Scholar]
- 119. Mao Y, Tournier AL, Bates PA, Gale JE, Tapon N, Thompson BJ (2011) Planar polarization of the atypical myosin Dachs orients cell divisions in Drosophila . Genes Dev 25: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, Quaggin SE, Harrison R, Mount R, McNeill H (2008) Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet 40: 1010–1015 [DOI] [PubMed] [Google Scholar]
- 121. Gong Y, Mo C, Fraser SE (2004) Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature 430: 689–693 [DOI] [PubMed] [Google Scholar]
- 122. Gros J, Hu JK, Vinegoni C, Feruglio PF, Weissleder R, Tabin CJ (2010) WNT5A/JNK and FGF/MAPK pathways regulate the cellular events shaping the vertebrate limb bud. Curr Biol 20: 1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Habib SJ, Chen BC, Tsai FC, Anastassiadis K, Meyer T, Betzig E, Nusse R (2013) A localized Wnt signal orients asymmetric stem cell division in vitro. Science 339: 1445–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E (2014) Par3‐mInsc and Galphai3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol 16: 758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Morris EJ, Assi K, Salh B, Dedhar S (2015) Integrin‐linked kinase links dynactin‐1/dynactin‐2 with cortical integrin receptors to orient the mitotic spindle relative to the substratum. Sci Rep 5: 8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xia J, Swiercz JM, Banon‐Rodriguez I, Matkovic I, Federico G, Sun T, Franz T, Brakebusch CH, Kumanogoh A, Friedel RH et al (2015) Semaphorin‐Plexin Signaling Controls Mitotic Spindle Orientation during Epithelial Morphogenesis and Repair. Dev Cell 33: 299–313 [DOI] [PubMed] [Google Scholar]
- 127. Arbeille E, Reynaud F, Sanyas I, Bozon M, Kindbeiter K, Causeret F, Pierani A, Falk J, Moret F, Castellani V (2015) Cerebrospinal fluid‐derived Semaphorin3B orients neuroepithelial cell divisions in the apicobasal axis. Nat Commun 6: 6366 [DOI] [PubMed] [Google Scholar]
- 128. Lancaster OM, Baum B (2014) Shaping up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol 34: 109–115 [DOI] [PubMed] [Google Scholar]
- 129. Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA (2011) Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 469: 226–230 [DOI] [PubMed] [Google Scholar]
- 130. Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek‐Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B (2013) Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell 25: 270–283 [DOI] [PubMed] [Google Scholar]
- 131. O'Connell CB, Wang YL (2000) Mammalian spindle orientation and position respond to changes in cell shape in a dynein‐dependent fashion. Mol Biol Cell 11: 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Minc N, Burgess D, Chang F (2011) Influence of cell geometry on division‐plane positioning. Cell 144: 414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R (2013) Cytoplasmic volume modulates spindle size during embryogenesis. Science 342: 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Courtois A, Schuh M, Ellenberg J, Hiiragi T (2012) The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 198: 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lazaro‐Dieguez F, Ispolatov I, Musch A (2015) Cell shape impacts on the positioning of the mitotic spindle with respect to the substratum. Mol Biol Cell 26: 1286–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Campinho P, Behrndt M, Ranft J, Risler T, Minc N, Heisenberg CP (2013) Tension‐oriented cell divisions limit anisotropic tissue tension in epithelial spreading during zebrafish epiboly. Nat Cell Biol 15: 1405–1414 [DOI] [PubMed] [Google Scholar]
- 137. Wyatt TP, Harris AR, Lam M, Cheng Q, Bellis J, Dimitracopoulos A, Kabla AJ, Charras GT, Baum B (2015) Emergence of homeostatic epithelial packing and stress dissipation through divisions oriented along the long cell axis. Proc Natl Acad Sci USA 112: 5726–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Legoff L, Rouault H, Lecuit T (2013) A global pattern of mechanical stress polarizes cell divisions and cell shape in the growing Drosophila wing disc. Development 140: 4051–4059 [DOI] [PubMed] [Google Scholar]
- 139. Mao Y, Tournier AL, Hoppe A, Kester L, Thompson BJ, Tapon N (2013) Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J 32: 2790–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gaugue I, Ainslie A, Christophorou N et al (2016) Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature 530: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]


