Abstract
Perivascular, meningeal and choroid plexus macrophages are non-parenchymal macrophages that mediate immune responses at brain boundaries. Although the origin of parenchymal microglia has recently been elucidated, much less is known about the precursors, the underlying transcriptional program and the dynamics of the other macrophages in the central nervous system (CNS). It has been assumed that they have a high turnover with blood-borne monocytes. However, large scale single-cell RNA-sequencing reveals a striking molecular overlap between perivascular macrophages and microglia but not monocytes. Using several fate mapping approaches and parabiosis we demonstrate that CNS macrophages arise from yolk sac precursors during embryonic development and remain a stable population. Notably, the generation of CNS macrophages relies on the transcription factor Pu.1 whereas myb, Batf3 and Nr4a1 are not required. Upon autoimmune inflammation, macrophages undergo extensive self-renewal by local proliferation. Our data provide challenging new insights into brains innate immune system.
Keywords: EAE, inflammation, multiple sclerosis, CX3CR1
INTRODUCTION
Under steady-state conditions, the central nervous system (CNS) hosts a heterogeneous population of myeloid cells, including parenchymal microglia and non-parenchymal perivascular, meningeal and choroid plexus macrophages1–3. The latter myeloid cells are critical effectors and regulators of immune responses at CNS borders during virtually all neuroinflammatory, neurodegenerative and neurooncological diseases.
Unlike microglia, which are derived from early yolk sac precursors prior to birth4–7, all other CNS macrophages found in the perivascular (Robin-Virchow) spaces, meninges, and choroid plexus were believed to originate from short-living blood monocytes after birth that are quickly replaced by bone marrow (BM)- derived cells3, 8. These assumptions were made on the basis of cell transplantation experiments in rodents starting in the 1980s9, 10. Just recently, chemotherapeutical conditioning additionally suggested that perivascular macrophages were present in the CNS parenchyma after BM transplantation11, 12. However, all of these studies used either irradiation or chemotherapy as conditioning regimens thereby inducing an artificial influx of injected BM-derived cells due to damage of the blood-brain barrier and local induction of chemoattractants in the host CNS13–15.
Therefore, it has been believed for decades that microglia and non-parenchymal macrophages in the meninges, perivascular spaces and choroid plexus represent two ontogenetically and genetically distinct myeloid populations. By combining large scale single cell RNA-sequencing with multiple approaches of fate mapping, parabiosis and in vivo imaging we challenged the traditional view and provide new insights into the transcriptional networks, ancestry and turnover of non-parenchymal macrophages at CNS boundaries during health and disease. We further show that CNS macrophages are closely related to microglia but still represent a distinct specialized population of tissue macrophages.
RESULTS
Gene expression profiling of CNS macrophages
Myeloid cells in the brain are a diverse group of cells localized at strategic places of the CNS. The parenchyma is filled with microglia, whereas tissue borders host Iba-1+ macrophages that can be found in the meninges (mMΦ), perivascular spaces (pvMΦ) and choroid plexus (cpMΦ) (Fig. 1a).
Figure 1. Molecular census of non-parenchymal macrophages, microglia and monocytes.
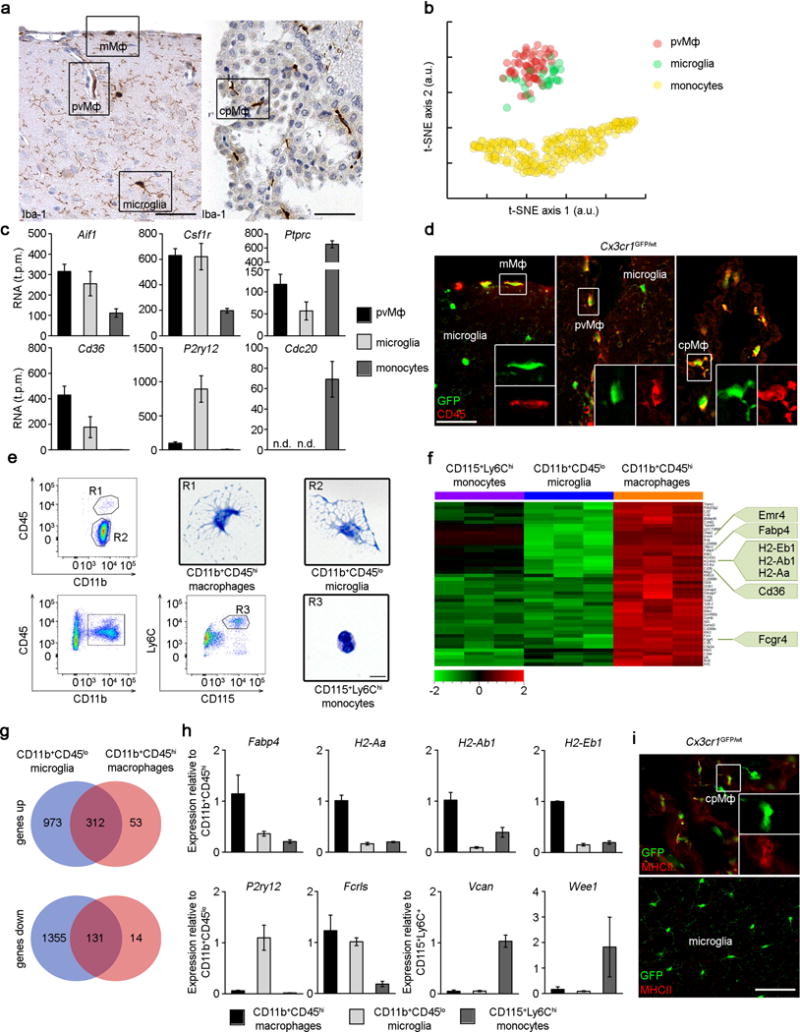
a) CNS histology of the brain cortex (left) and choroid plexus (right) that were subjected to immunohistochemistry for Iba-1 to detect meningeal (mMΦ), perivascular (pvMΦ), choroid plexus (cpMΦ) macrophages and microglia. Scale bars: cortex = 50 μm, choroid plexus = 50 μm. Representative pictures of four examined mice are displayed.
b) Cluster analysis of individual pvMΦ, cortical microglia and monocytes measured by single-cell RNA-sequencing and biclustering.
c) Bar graphs for selected markers for individual pvMΦ (black), cortical microglia (grey) and monocytes (dark grey) evaluated by single cell RNA-seq. Bars represent means ± s.e.m. of single cells.
d) Immunfluorescence microscopy for GFP (green) and CD45 (red). Overview and magnification are shown. Scale bar: 25 μm overview. At least three mice per group were analysed.
e) Flow cytometry for to identify CNS macrophages (R1), microglia (R2) and monocytes (R3), respectively. Cytospins of isolated and May-Grünwald Giemsa-stained cells. Scale bar: 5 μm. Data are representative of two independent experiments.
f) Affymetrix gene chip array-based heat map (standardized and scaled to log2 expression) of significantly induced transcripts in CD11b+CD45hi CNS macrophages compared to microglia and monocytes. Typical genes are highlighted.
g) Venn diagram depicting the different regulated and overlapping between CD45hi macrophages and CD45lo microglia compared to Ly-6Chi monocytes.
h) Quantitative RT-PCR of differentially regulated genes. Data are expressed as ratio of the mRNA expression compared to endogenous Gapdh relative to CD11b+CD45hi (top row), CD11b+CD45lo (bottom row, left) and Ly-6Chi monocytes (bottom row, right). Bars show mean ± s.e.m. At three to five samples per group were analysed.
i) Immunfluorescence for MHC class II (red) on cpMΦ but not on microglia in Cx3cr1GFP/wt mice (green). Overview and magnification are shown. Scale bar: 25 μm (overview). At least three mice per group were analysed.
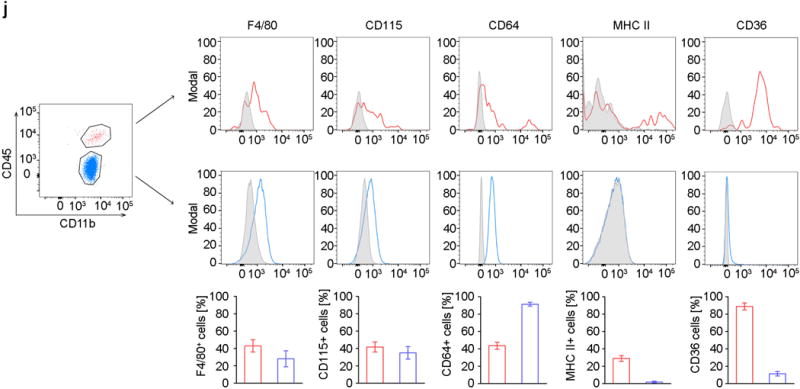
j) Flow cytometric analysis of CNS macrophages and microglia. Characteristic histograms are depicted. Grey areas represent isotype controls. Five to eight independent experiments were performed. Bars represent the mean ± s.e.m of CD11b+CD45hi macrophages (red) and CD11b+CD45lo microglia (blue).
CNS macrophages have historically been classified based on their localization, morphology and expression of selected molecular makers1. Here, we used unbiased quantitative single-cell RNA-sequencing to perform a molecular census of microglia, pvMΦ and their proposed precursors, the monocytes (Fig. 1b,c, Suppl. Fig. 1). Individual RNA molecules were counted using unique molecular identifiers (UMIs) as described previously16, which greatly reduces PCR amplification bias. Dimensionality reduction using t-distributed stochastic network embedding (t-SNE)17, revealed that pvMΦ and microglia were transcriptionally closely related whereas monocytes had a distinct RNA profile. All populations expressed the myeloid markers Cx3cr1, Csf1r and Aif (gene for Iba-1), but pvMΦ were distinguishable based on their expression of Mrc1and Cd36, microglia based on their expression of P2ry12 and monocytes based on their expression of Cdc20. Notably, RNA levels of Ptprc (encoding CD45) were higher in pvMΦ compared to microglia which was confirmed on protein levels for all non-parenchymal macrophages in situ but not in microglia (Fig. 1d). Consequently, cytometry-based distinction of CD45hi macrophages, CD45lo microglia in the CNS is feasible (Fig. 1e) as described before18. Comparison of the gene expression profiles revealed that the 46 most highly expressed genes in CD45hi macrophages were largely immunologically-related transcripts encoding molecules such as antigen presentation (H2 molecules), pattern of recognition/activation (Cd36, Fcgr4) (Fig. 1f, g) which was confirmed by qRT-PCR (Fig. 1h), immunofluorescence (Fig. 1i) and flow cytometry (Fig. 1h). Taken together, these data show a close transcriptional relationship between non-parenchymal macrophages and microglia albeit with some cell type-specific differences.
Prenatal yolk sac origin of CNS macrophages
The current concept that macrophages at CNS borders are derived from blood-borne myeloid cells during adulthood largely depends on previous work on BM chimeras using irradiation of the recipient19, 20 a finding that we could confirm by transplanting BM from actin-GFP mice into irradiated hosts leading to a vigorous engraftment of donor-derived GFP+Iba-1+ macrophages in the meninges, perivascular spaces and choroid plexus whereas microglial exchange was limited (Suppl. Fig. 2).
However, in several organs, resident macrophages arise from embryonic precursors in the yolk sac (YS) that seed the tissue before birth5–7, 21, 22 (Ensan et al. 2015, in revision). In accordance with these findings we already observed Cx3cr1GFP+ macrophages in the meninges at E9.5 and shortly thereafter in the choroid plexus and the perivascular spaces (Suppl. Fig. 3) suggesting prenatal seeding of tissue macrophages in these compartments. To test whether YS progenitors indeed contribute to the pool of CNS macrophages we adapted our recently developed Cx3cr1CreER mouse system23, 24. Female Cx3cr1CreER mice – which upon exposure to tamoxifen (TAM) express Cre recombinase under the control of the CX3CR1 promoter – were crossed to Rosa26-yfp reporter mice and pregnant animals were injected with a single intraperitoneal dose of TAM at E9 to pulse Cx3cr1-expressing progenitor cells in the YS (Fig. 2a). This approach induced irreversible expression of the yfp promoter in YS-derived CX3CR1 expressing cells and their progeny. 55.4 ± 3.3 % Iba-1+ mMΦ, 51.3 ± 11.5 % Iba-1+ cpMΦ and 36.2 ± 7.5 % Iba-1+microglia were yfp+ at E16.0 indicating robust labeling efficiency of YS progenitors (Fig. 2b,c). Considering that microglia are entirely of YS origin3, 5 we can also assume that pvMΦ, mMΦ and cpMΦ have their major source in the YS. Not only microglia but also pvMΦ and mMΦ retained the yfp label in 8–9 weeks old mice, demonstrating that YS labelling persists into adulthood (Fig. 2d, e). In contrast, cpMΦ dramatically lost their yfp label indicating that YS cells were replaced in the choroid plexus. Expression of yfp in pvMΦ in the perivascular space was confirmed by immunoelectron microscopy (Fig. 2f). Several transcription factors have been shown to be important for lineage commitment in myeloid cells25, 26. To determine which transcription factors are required for macrophage development we investigated the presence of CNS macrophages in mutants lacking Sfpi (encoding Pu1.), Irf8, myb and batf3 (Fig. 2g–i, Suppl. Fig.4). Mice lacking Pu.1 were devoid of any pvMΦ, mMΦ and cpMΦ. Notably, we found a reduction of mMΦ but not cpMΦ in Irf8−/− mice, whereas absence of the master transcription factor for stem cell development in the BM, myb did not impair mMΦ and cpMΦ development similar to its redundant role for YS-derived macrophages such as microglia6. Batf3-defiency did not impair any macrophage numbers. In sum, mMΦ, pvMΦ, cpMΦ and microglia have their prenatal origin in the YS and largely depend on similar transcription factors for proper development.
Figure 2. Ontogeny of brain macrophages at brain interfaces.
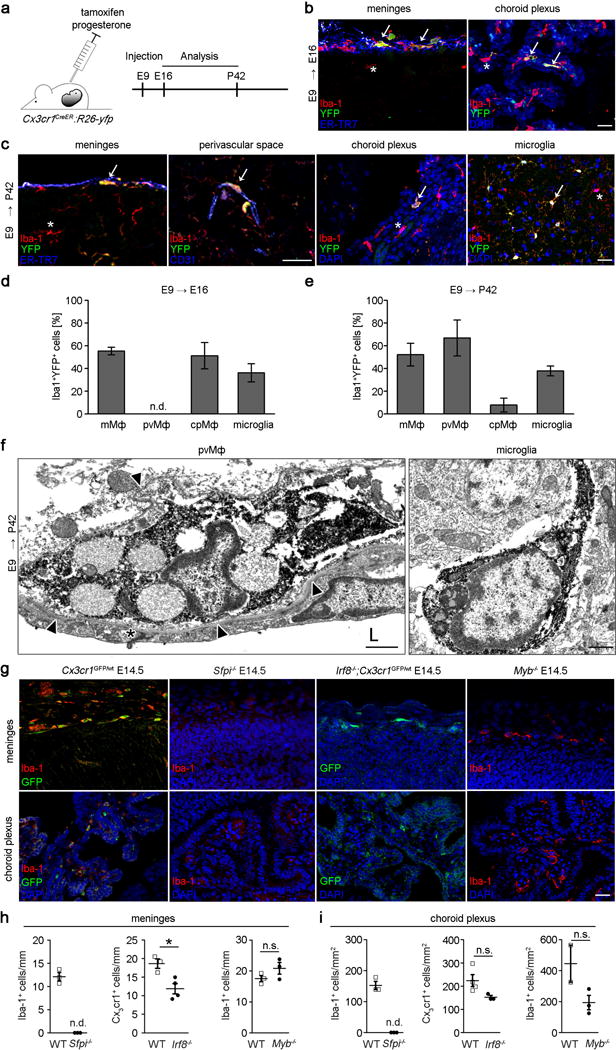
a) Scheme for the induction of recombination (injection of tamoxifen [TAM]) and subsequent analysis in Cx3cr1CreER:R26-yfp animals.
b,c) Representative sections of meninges, choroid plexus and perivascular spaces at defined time points in TAM-induced Cx3cr1CreER:R26-yfp animals using immunofluoresence for YFP (green), the microglia marker Iba-1 (red), 4′,6-diamidino-2-phenylindole (DAPI, blue), CD31 (blue) or the fibroblast marker ER-TR7 (blue), respectively. Arrows highlight YFP+Iba-1+double positive cells. Scale bar = 25 μm.
d, e) Quantitative analysis of regional yfp expression in Iba-1+ macrophages in TAM-induced Cx3cr1CreER:R26-yfp mice at indicated time points. Data are expressed as mean ± s.e.m. At least three mice per group were analysed. N.d.= not detectable.
f) Immunolectron microscopy for yfp in pvMΦ and microglia at postnatal day (P) 60 in E9 TAM-induced Cx3cr1CreER:R26-yfp mice. Arrow heads indicate endothelial basal lamina surrounding the pvMΦ. Asterisk specifies endothelial tight junction. L = vessel lumen. Scale bar = 1 μm. Representative pictures of three mice examined are displayed.
g) Iba-1 immunofluorescence (red) of macrophages in the absence of the respective transcription factors. Alternatively, GFP+ macrophages (green) on the Cx3cr1GFP/wt back ground are shown. DAPI (blue) Scale bar = 25 μm.
h, i) Quantitative examination of Iba-1+ or Cx3cr1GFP/wt macrophages at embryonic day E14.5. Each symbol represents the mean measurement of one mouse. Three sections from at least two mice were examined for each group. N.s. = not significant, *P < 0.05.
Maintenance of pvMΦ, mMΦ and cpMΦ in adulthood
Once established in the CNS, microglia persist throughout the entire life of the organism without any significant input from circulating blood cells due to their longevity and their capacity of self-renewal13, 27. We therefore sought to use this unique feature of microglia to compare the kinetics of persistence with mMΦ, pvMΦ, cpMΦ and blood cells such as Ly-6Clo monocytes. For this purpose we challenged Cx3cr1CreER:R26-yfp animals23, 24 with TAM and determined mMΦ, pvMΦ, cpMΦ labeling at several time points after application (Fig. 3a). FACS analysis demonstrated long-term labeling of yfp+CD45hiCD11b+ macrophages even 30 weeks after TAM treatment (Fig. 3b). Histological examination confirmed the FACS data and revealed a high percentage of Iba-1-labeled mMΦ, pvMΦ, cpMΦ that co-expressed yfp (Fig. 3c). Quantitative examination on histological slices revealed constant high expression of the reporter gene in mMΦ and pvMΦ comparable to microglia (Fig. 3d). These data indicate that mMΦ and pvMΦ are stable populations which do not undergo significant exchange with blood cells within 43 weeks. In contrast, yfp-labeling continuously dropped in Iba-1+ cpMΦ suggesting a slow continuous exchange with blood cells. Due to their ephemerality Cx3cr1-targeted Ly-6Clo monocytes in the blood lost their yfp mark very rapidly and were quickly replaced by the non-targeted progeny. Thorough confocal analysis confirmed the long-term persistence of pvMΦ in perivascular spaces of cerebral vessels (laminin+) at 8 weeks after TAM (Fig. 3e). At this time point, microglia were still labelled but not blood monocytes (Fig. 3f). Notably, yfp-expressing mMΦ and cpMΦ were detected by Immuno-EM 30 weeks after recombination (Fig. 3g). Labelled cpMΦ were found in close proximity to the microvilli of the choroid plexus epithelium indicating that these cells represent Kolmer’s epiplexus cells. To further analyse whether the integration of these long lived myeloid cells into distinct CNS microenvironments shapes their dynamic behaviour we imaged fluorescently labelled mMΦ, pvMΦ and microglia in the intact spinal cord of living Cx3cr1CreER:R26-tomato mice at 8 weeks after TAM by confocal and 2-photon microscopy. Time-lapse imaging revealed that these myeloid cells populations that derived from a common origin can be differentiated in vivo not only based on their anatomical location but also based on their characteristic morphology and distinct dynamic behavior (Fig. 3h–j and Supplementary Videos 1 and 2). While mMΦ present with amoeboid morphology and movement characteristics, microglial cells show a ramified morphology with highly dynamic processes and a rather stable cell body. These cell types can be further differentiated from pvMΦ, which are elongated and highly oriented cells that follow the blood vessel outline and extend and retract their protrusions along the blood vessel wall.
Figure 3. Maintenance of non-parenchymal macrophages in adulthood.
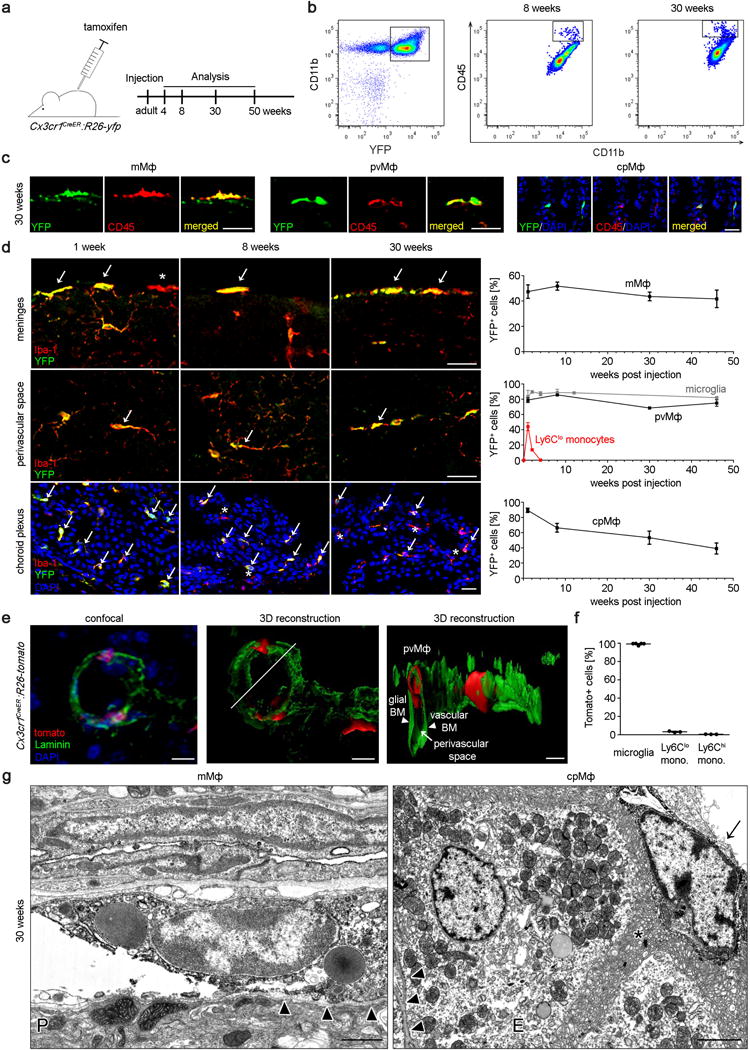
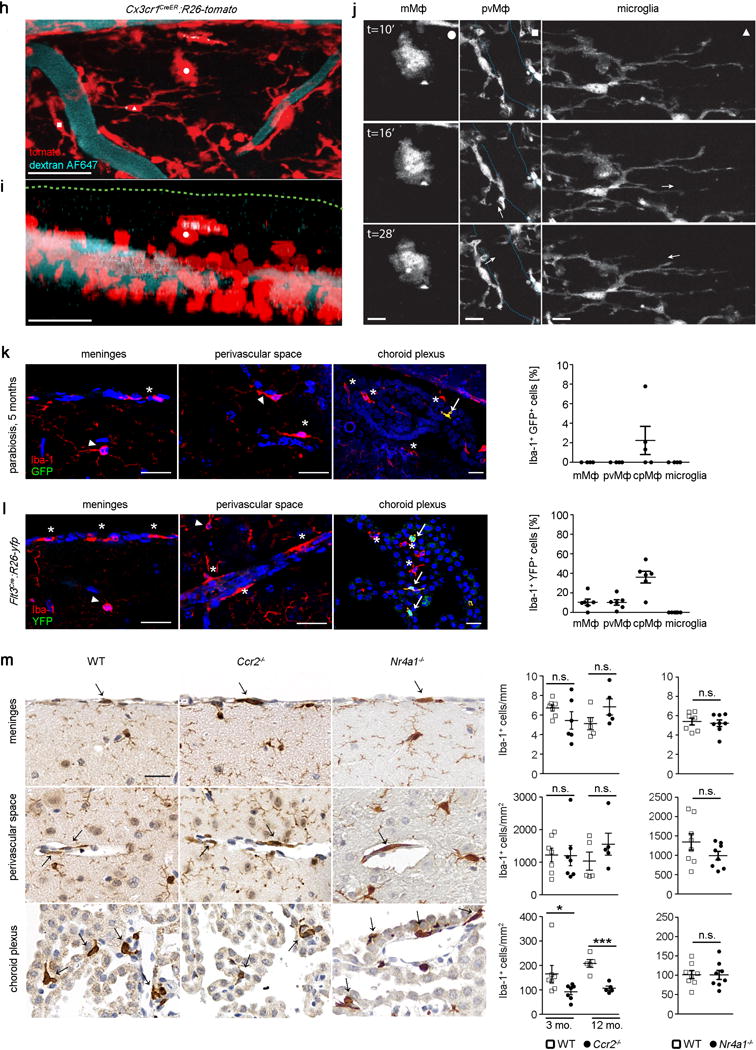
a) Scheme and time line for labelling and analyses of pvMΦ, mMΦ and cpMΦ in adulthood using TAM injection in adult Cx3cr1CreER:R26-yfp animals.
b) Persistence of labelled yfp+CD11b+CD45hi macrophages in adult Cx3cr1CreER:R26-yfp mice. Representative flow cytometric images of five investigated mice are displayed.
c) Representative immunofluorescence for yfp (green), CD45 (red) and DAPI (blue) in pvMΦ, mMΦ and cpMΦ in adult Cx3cr1CreER:R26-yfp mice 30 weeks after TAM application. Scale bars = 25 μm. At least three mice per group were analysed.
d) Kinetics of yfp labelling in Iba-1+ pvMΦ, mMΦ and cpMΦ in adult Cx3cr1CreER:R26-yfp animals upon TAM application. Left: Characteristic brain sections are shown. Scale bars = 25 μm. Right: Quantification thereof. Asterisks indicate single positive cells, arrows label double positive cells. Data represent mean ± s.e.m. of at least three mice per group.
e) Localization of tomato+pvMΦ (red) in the vascular compartment using confocal microscopy (left) and 3D-reconstruction (middle, right) in adult Cx3cr1CreER:R26-tomato animals 8 weeks after TAM injection using laminin (green) to indicate the basal lamina and nuclear staining (DAPI, blue). Arrow heads and arrow point the respective structures. Scale bars = 10 μm, (overview), 5 μm (zoom). Three mice were investigated and typical pictures are shown.
f) Quantification of tomato labelling 8 weeks after TAM application. Data represent mean ± s.e.m. of at least three mice per group.
g) Immuno-EM for yfp in Cx3cr1CreER:R26-yfp animals 30 weeks after TAM application reveals positively labelled mMΦ and cpMΦ. Arrow heads point the basal lamina. Arrow indicates Kolmer’s epiplexus cell. Asterisk designates microvilli of the choroid plexus epithelium. E = epithelium. Scale bar1 = 1 μm (left) 2 μm (right).
h) Confocal projection of the dorsal spinal surface of a Cx3cr1CreER:R26-tomato mice (tomato, red) at 8 weeks after TAM and injected with dextran-AF647 (blue) to reveal the vasculature. An example of a pvMΦ was marked by a white square, a microglial cell by a white triangle, and a mMΦ by a white circle. Scale bar = 25 μm.
i) Z-projection of the confocal image stack shown in (h) that illustrates the localization of the different myeloid cell populations. Green dotted line represents upper limit of the dura membrane as inferred by in vivo staining with Nuclear-ID blue dye (not shown). Scale bar = 25 μm.
j) In vivo 2-photon time-lapse of the myeloid cells marked in illustrating the dynamic behaviour of a mMΦ (left panel), a pvMΦ (middle panel) and a microglia cell (right panels). White arrows indicate examples and direction of dynamic changes in the middle and left panels. The cyan dotted line in the middle panel indicates the outline of the vasculature. Scale bars = 1 μm, left panels; 1.2 μm, middle panels and 2.4 μm, right panels.
k) Negligible exchange of pvMΦ, mMΦ and cpMΦ in a wild-type parabiont after 5 months of parabiosis with an ActinGFP/wt mouse. Left: representative immunofluorescence pictures. Scale bar = 25 μm. Asterisks indicate single positive cells arrows label double positive cells. Right: quantification thereof. Arrow heads point to microglia. One symbol represents one mouse with quantification of a minimum of three tissue sections. Data represent means ± s.e.m. of at least three animals per group.
l) Little Flt3 expression as marker of definitive hematopoiesis in pvMΦ, mMΦ and cpMΦ in adult Flt3Cre:R26-yfp mice. Left: representative immunofluorescence pictures. Asterisks indicate single positive cells arrows label double positive cells. Arrow heads point to microglia. Scale bar = 25 μm. Right: quantification thereof. Each symbol represents one mouse with quantification of a minimum of three tissue sections. Data represent means ± s.e.m. of at least three animals per group.
m) Localization and presence of pvMΦ, mMΦ and cpMΦ in adult wild-type (WT), Ccr2−/− and Nr4a1−/− mice evaluated using Iba-1 immunohistochemistry. Representative figure are presented (left) and quantification (right). Each symbol represents one mouse with quantification of a minimum of three tissue sections. Data represent means ± s.e.m. of at least five animals per group. N.s. = not significant. Significant differences were with asterisks (*P < 0.05, ***P < 0.001).
To evaluate a possible contribution of blood cells to the pool of macrophages at CNS boundaries during non-diseased conditions, we needed to develop a method for tracking circulating cells without affecting the CNS environment or resident cells. We therefore induced peripheral blood chimerism by surgically joining two syngeneic mice, one of which ubiquitously expressed GFP. These parabiotic mice established a rich anastomotic circulation, which quickly led to efficient blood chimerism as described before28. Remarkably, the extent of circulatory exchange of mMΦ and pvMΦ was not measurable after 5 months of parabiosis, whereas cpMΦ had a detectable rate of donor-derived blood cells (Fig. 3k). Further, low gene recombination of mMΦ and pvMΦ in Flt3Cre:R26-yfp animals suggested that their development occurs largely independently of Flt3+ multipotent hematopoietic precursors in the BM (Fig. 3l). Again, only cpMΦ displayed a higher recombination of this gene suggesting a role for HSCs in the development of cpMΦ. Furthermore, we provide genetic evidence that neither Ly-6Chi nor Ly-6Clo monocytes are essential for mMΦ and pvMΦ development because mice lacking Ccr2 or Nr4a1 presented normal amounts of these macrophages subsets (Fig. 3m). Only cpMΦ were reduced in Ccr2-deficient mice indicating that Ly-6Chi monocytes contribute to their homeostasis.
Self-renewal of pvMΦ during autoimmune inflammation
Accumulation of myeloid cells during inflammation can occur through either local proliferation of tissue macrophages or recruitment of peripheral Ly-6Chi monocytes from the blood. It has been shown that engrafted Ly-6Chi monocytes rapidly die during autoimmune inflammation whereas the microglia pool speedily expands due to local self-renewal28. However, the kinetics of macrophages at CNS interfaces during inflammation is only poorly understood. To establish the spatiotemporal relationship between infiltrating monocytes, pvMΦ activation and microgliosis we took advantage of the Cx3cr1CreER:R26-tomato system that allows distinction of CX3CR1+ CNS resident cells (pvMΦ and microglia are tomato+) from incoming CX3CR1+ myeloid cells (Ly-6Chi and Ly-6Clo monocytes are tomato−)24.
Cx3cr1CreER:R26-tomato mice were actively immunized with MOG35–55 and immunohistochemical evaluation of spinal cord sections at the acute (15 dpi) and chronic (30 dpi) phase revealed many cells that were positive for the myelomonocytic marker Iba-1 (Fig. 4a). Recruitment of monocytes (Iba-1+, tomato−) was first observed when animals reached the acute phase and remained visible until the chronic stage (Fig. 4b). Notably, Iba-1+ tomato− cells were initially found in proximity to the meninges, suggesting that the entry of monocytes into the CNS occurs via this compartment. In addition to infiltrating monocytes, also Iba-1+tomato+ myeloid cells dramatically increased during inflammation which could be distinguished between pvMΦ and microglia based on their relationship to vascular basal membrane (Fig. 4c,d). Ki67+ pvMΦ (tomato+, close to laminin+ structures) and microglia (tomato+, far from laminin+ structures) were already evident at the acute phase, suggesting that activation of pvMΦ and microglia is an early event in EAE pathogenesis. Notably, a robust decrease in the frequency of proliferating microglia was observed in the chronic phase whereas pvMΦ proliferation remained high (Fig. 4e). Taken together, these data strongly suggest that during autoimmune inflammation pvMΦ expand by local self-renewal rather than by recruitment of peripheral myeloid cells.
Figure 4. Self-renewal of pvMΦ during autoimmune inflammation.
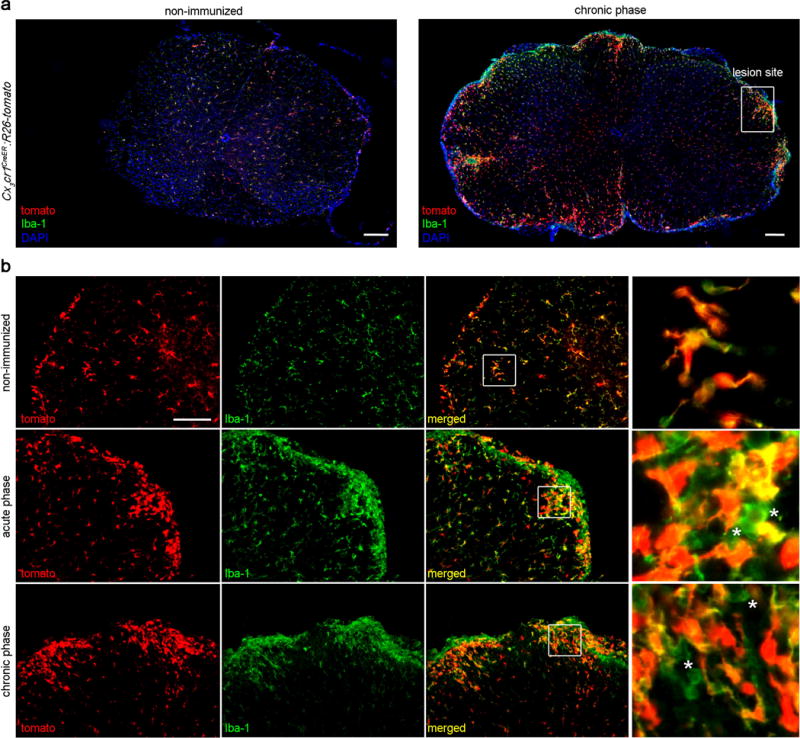
a) Representative images of the spinal cord from Cx3cr1CreER:R26-tomato mice that were either not immunized with MOG35–55 (left) or were immunized (right, chronic phase, 30 days post immunization [dpi]). Spinal cord sections were immunreactive for the mature macrophage and microglia marker Iba-1 (green) and nuclear maker DAPI (blue). Tomato, which identifies long-living CX3CR1+ pvMΦ and microglia, is shown in red. Scale bar =100 μm.
b) Magnification thereof. Acute phase sections represent mice around 16 dpi during EAE. Note that no infiltration of Iba-1-single positive cell was detected in healthy spinal cord. In contrast, during EAE the amount of Iba-1+ myeloid cells strongly increased containing Iba-1+tomato− infiltrating monocytes or Iba-1+tomato+ pvMΦ and microglia. Scale bars = 100 μm (overview) and 10 μm (insert).
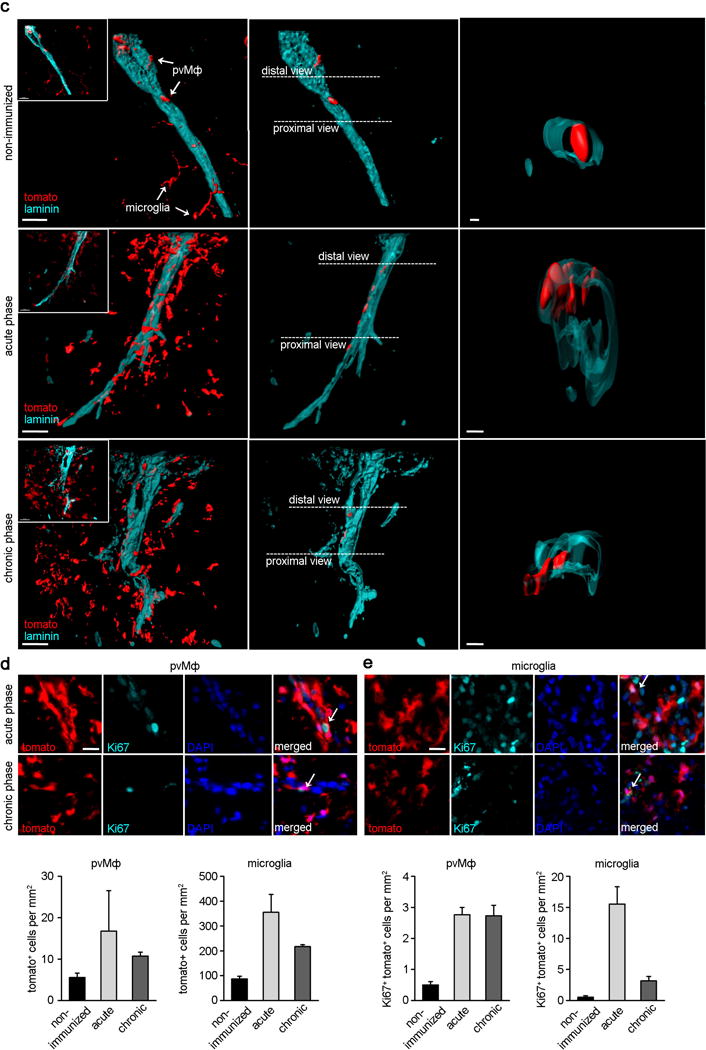
c) Kinetics of pvMΦ and microglia expansion in Cx3cr1CreER:R26-tomato mice that were healthy (upper row) or subjected to EAE (lower rows). Representative high-magnification confocal images of spinal cord sections immunoreactive with laminin to visualize the basal membranes are shown. Note the overall increase of all tomato+ myeloid cells during EAE (left column) and just of tomato+ pvMΦ around the vessels (middle column). Right column: high magnification depicting tomato+ pvMΦ (red) and laminin (turquoise). Scale bars = 30 μm (overview) and 5 μm (insert).
d) Representative high-magnification images of spinal cord sections from diseased Cx3cr1CreER:R26-tomato mice immunoreactive for Ki67 (turquoise) and the nuclear marker DAPI (blue). Tomato+ pvMΦ (red) around the vessels were positive for the proliferation marker Ki67 (arrow, left images) as well as parenchymal microglia (arrow, right images). Scale bar = 10 μm.
e) Left: Numbers of resident pvMΦ (tomato+ cells found within the perivascular space) and microglia (tomato+ cells found in the parenchyma) in the spinal cord at different time points of disease. Right: Kinetics of Ki67-positive pvMΦ and microglia during EAE. Bars represent mean ± s.e.m. of at least two mice per group.
DISCUSSION
This study described the molecular signatures of tissue macrophages at CNS boundaries, their developmental pathways and key mechanisms to ensure their homeostasis and response to inflammation (Suppl. Fig. 5). By using large-scale single cell RNA-sequencing we were able to dissect the individual transcriptional profiles of non-parenchymal macrophages and parenchymal myeloid cells (microglia) in the CNS and found a close relationship between these cells that was not shared by circulating monocytes. Our unbiased high-throughput methodology allows to define extensive functional specialization between cell classes16. In fact, we found that the marker Mrc1 was specific for macrophages at CNS interfaces whereas P2ry12 was expressed only by microglia which confirms previous data that used expression profiling of several thousands of cells in the CNS29, 30. However, our finding that mMΦ, pvMΦ, cpMΦ have only limited relation to circulating myeloid cells represents a major change in the field. Since the 1980s it has been assumed that macrophages at CNS interfaces are solely blood-derived based on BM chimera data using whole-body irradiation9. The involvement of the CNS during the irradiation procedure clearly leads to substantial local priming with concomitant induction of myeloattracting and myelopromoting factors and damage of the BBB7, 13, 14. Consequently, all previous studies on mMΦ, pvMΦ and cpMΦ using these techniques to establish BM chimeras have to be interpreted with caution.
Tissue macrophages arise from two distinct developmental programs; early YS-derived erythromyeloid progenitors (EMPs) that give rise to macrophages without monocyte intermediates and fetal monocytes that derive from myb+ EMPs generated in the YS7, 21, 31. These pathways contribute variously to macrophage development in several tissues including the brain, skin, heart, liver and lung7, 21, 32, 33. Consistent with these findings we found Cx3cr1+ mMΦ, pvMΦ and cpMΦ already during early embryogenesis and fate mapping approaches confirmed the persistence of YS-derived macrophages in embryonic as well adult mice. Development and homeostasis of CNS macrophages is not uniform. Similar to microglia, the maintenance of mMΦ and pvMΦ does not dependent on circulating monocytes and non-parenchymal CNS macrophages persist over a very long period of time. This is in contrast to cpMΦ that show features of relative ephemerality (compared to other CNS macrophages) and some replenishment by blood cells (Suppl. Fig. 5). Neurological diseases in which the choroid plexus (CP) is crucially involved as one of the interfaces between periphery and CNS are abundant34. Several studies suggested a fundamental involvement of the CP in the development and progression of multiple sclerosis (MS) and its animal model, EAE. Firstly, CP inflammation with immune cell activation and infiltration has been described preceding parenchymal inflammation and clinical disease onset in humans and mice35. Secondly, autoaggressive sentinel T cells are thought to enter the CSF through the CP during incipient stages of MS and EAE, travelling to the subarachnoidal space where they are reactivated by local APC presenting cognate antigen36. All findings argue for the dual role of the CP as an immune cell modulatory site as well as an entry gate for peripheral immune cells to the CNS during neuroinflammation. To fulfil these highly specialized tasks during diseases it is very advantageous for the CP to host cpMΦ that are also derived from short-lived circulating cells.
During inflammation pvMΦ perform local self-renewal by proliferation. Notably, our genetic labelling strategy in Cx3cr1CreER:R26-tomato mice does not allow to distinguish between pvMΦ and microglia by marker expression and we therefore decided to use their relative localization to the vascular basal membrane as a discriminating feature. However, we cannot exclude that during inflammation pvMΦ may also migrate to the CNS parenchyma. Further studies are needed to address this issue.
Flt3 is expressed only by definitive hematopoietic progenitors in the embryo and during adulthood37. It can therefore be used to distinguish YS-derived versus fetal liver and bone marrow-derived macrophages38. This approach helped us to define cpMΦ as a distinct population of CNS macrophages that is different from mMΦ, pvMΦ and microglia that have no input from Flt3-dependent definitive hematopoiesis. CpMΦ development from circulating monocytes was confirmed by our results from the parabiosis experiments and in Ccr2-deficient animals. In contrast, genetic labelling using the TAM-inducible Cx3cr1CreER:R26-yfp system revealed that mMΦ and pvMΦ remained remarkably stable when compared between embryonic and adult animals arguing against a significant contribution of fetal liver to these macrophages population which is in contrast to other tissue macrophage populations such as in the liver and gut21.
Strategically positioned at the CNS barriers, mMΦ, pvMΦ, cpMΦ may modulate immune cell entry and phenotype. The myeloid cells in the CNS-adjoining tissues have thus been implicated in various immunopathological processes, including antigen presentation to circulating lymphocytes39, 40 (Brendecke 2015, in press). As presumed guardians of tissue homeostasis, the CNS-surrounding myeloid cell network appears to be crucially involved in the development, progression and resolution of neuroinflammatory, neurodegenerative and neurooncological diseases. Our results help to unravel the regulatory program that controls mMΦ, pvMΦ and cpMΦ function in vivo, and to identify new means to manipulate these cells for the treatment of neural diseases.
MATERIAL AND METHODS
Mice
In this study, C57BL/6 and CD-1 mice were used as WT mice. All transgenic lines (ActinGFP/+, Batf3−/−, Ccr2−/−, Cx3cr1GFP/WT, Myb−/−, Nr4a1−/−, Irf8−/−:Cx3cr1GFP/WT, Sfpi1−/−) have a C57BL/6 background. Mice were bred in-house under pathogen-free conditions. Cx3cr1CreER were crossed to either R26-yfp or R26-tomato mice. Flt3Cre were backcrossed to R26-yfp. All animal experiments were approved by local administration and were performed in accordance to the respective national, federal and institutional regulations.
Tamoxifen treatment
For induction of the Cre recombinase in adult animals, six to eight week-old Cx3cr1CreER mice were treated twice with 4 mg Tamoxifen (TAM, Sigma) solved in 200 μl corn oil (Sigma), injected subcutaneously at two time points 48 hrs apart. For pulse-labelling experiments, the Cre recombinase was induced in Cx3cr1CreER:R26-yfp embryos with 200 μl of 20 mg/ml TAM and 10 mg/ml Progesterone dissolved in corn oil by i.p. injections into pregnant females at 9 days post coitum.
Bone-marrow transplantation
Eight week old WT recipient mice lethally were irradiated and reconstituted with 5×106 bone marrow cells derived from femur and tibia of adult ActinGFP/+ mice, injected into the tail vein of recipients. Mice received whole-body irradiation (11Gy) 24 h prior to bone marrow reconstitution with an RS 2000 Biologica x-Ray irradiator.
Parabiosis
Pairs of WT and ActinGFP/+ were surgically connected for five months as previously described27. Blood chimerism was verified using FACS-Analysis.
Induction of experimental autoimmune encephalitis
Mice from each group were immunized subcutaneously eight weeks after TAM injection with 200 μg of MOG35-55 peptide emulsified in CFA containing 1 mg of Mycobacterium tuberculosis (H37RA; Difco Laboratories, Detroit, Michigan, USA). The mice received intraperitoneal injections with 250 ng pertussis toxin (Sigma-Aldrich, Deisenhofen, Germany) at the time of immunization and 48 hrs later. Mice were analyzed at the acute phase (between day 15–17) and at the chronic phase (day 30) of EAE.
In vivo imaging and image processing
Animals were anesthetized with ketamine and xylazine (ketamine 87 mg/kg, xylazine 13 mg/kg), placed on a heating pad, and then tracheotomized and intubated. The dorsal spinal cord was surgically exposed as previously described41 and the opening constantly superfused with artificial cerebrospinal fluid (aCSF). For the imaging session, the vertebral column was fixed using a spinal clamping device (Narishige STS-a) and the spinal opening surrounded by a 4% agarose well. Mice were injected i.p. with 200 μg of Dextran-AF647 (Life Technologies) to reveal the vasculature and the spinal cord was incubated with Nuclear-ID Blue dye (dilution 1:250, Enzo life sciences) for 15′ at RT to reveal the meningeal surface as previously established42. In vivo imaging was performed using confocal or two-photon laser excitation on a Olympus FV1200 MPE setup equipped with a 25×/1.25 water immersion objective (Olympus) at 1024 ×1024 pixel resolution. For confocal imaging, the Nuclear-ID Blue dye, tdtomato and Dextran-AF647 were sequentially excited using 405 nm, 568 nm, and 647 nm lasers, respectively. For 2-photon time-lapse imaging of microglia/macrophage dynamics, the IR laser was tuned to 950 nm and fluorescence was collected using a standard green/red filter set (BA575–630). For image representation both confocal and 2-photon images have been gamma-adjusted and processed with a despeckling filter using Photoshop software (Adobe). The z-projection and 3D-rendering of the confocal image stack shown in Fig. 3i and Supplementary Movie 1 were performed using Imaris software (Bitplane). Supplementary Movie 2 was assembled using Windows Movie Maker (Windows).
Histology
Histology was performed as described recently 43. Briefly, mice were transcardially perfused with phosphate-buffered saline (PBS), brains were dissected and postfixed in 4% PFA for 24 h. Brain samples were embedded in paraffin and stained with Iba-1. Sections were evaluated using the cell-P software (Olympus).
Cytochemistry
Cells were sorted into chamber slides and incubated o/n in medium. After washing with PBS, cells were fixed and incubated with freshly filtered May-Grünwald solution followed by Giemsa solution.
Fluorescence microscopy
After transcardial perfusion with phosphate-buffered saline (PBS), brains were fixed for in 4% PFA, dehydrated in 30% sucrose and embedded. Cryosections were obtained as described previously (Goldmann2013, 2015). Sections were then blocked with PBS containing 5 % bovine serum albumin and permeabilized with 0.1% Triton-X 100 in blocking solution. Primary antibodies were added over night at a dilution of 1:500 for Iba-1(019-19741, WACO, Japan), 1:1000 for GFP (600-106-215, Rockland Immunochemicals Inc., Gilbertsville, USA), 1:100 for MHC class II (ab23990, Abcam), 1:400 CD45 (BD Pharmingen), 1:1000 ER-TR7 (Abcam ab51824), 1:100 CD31 (BD Pharmingen), 1:500 Laminin (L9393, Sigma-Aldrich), 1:500 Ki67 (ab15580, Abcam) at 4°C. Secondary antibodies were added as follows: Alexa Flour® 488 1:500, Alexa Flour® 555 1:500 and Alexa Fluor® 568 1:500 for 2h at RT. Nuclei were counterstained with DAPI. Images were taken using a conventional fluorescence microscope (Olympus BX-61, Keyence) and the confocal pictures were taken with Fluoview FV 1000 (Olympus).
3-D reconstruction
Free floating 40 μm cryo sections from adult brain tissue were stained overnight with anti-laminin, followed by Alexa Fluor 488–conjugated secondary antibody staining, which was added at a dilution of 1:500 for 4 h at 20–25 °C. Imaging was performed on an Olympus Fluoview 1000 confocal laser scanning microscope using a 20 × 0.95 NA objective. Z stacks were done with 1.1-μm steps in z direction, 1,024 × 1,024 pixel resolution were recorded and analyzed using IMARIS software (Bitplane).
Electron microscopy
Animals were sacrificed and transcardially perfused with PBS followed by 4 % PFA and 0.1% of glutaraldehyde in PBS. Upon removal of the brain, the tissue was kept in the same fixative for 3–6 hours followed by preparation of 60μm consecutive sections using a vibrating microtome (Leica Microsystems, Wetzlar, Germany). The sections were blocked with 3% of bovine serum albumin in Tris-buffered saline (TBS-BSA) followed by incubation of the primary antibody (goat anti GFP, 1:200, Acris antibodies, Herford, Germany). After thorough rinsing, sections were incubated with biotinylated secondary antibodies (rabbit anti goat, 1:250, Sigma-Aldrich, Deisenhofen, Germany). After further rinsing, the sections were incubated with ExtrAvidin (1:100, Sigma-Aldrich) and were finally stained with diaminobenzidine (Sigma-Aldrich) to achieve an electron dense precipitate allowing detection at the level of light as well as electron microscopy. Sections were further stained with osmium tetroxide and uranyl acetate, dehydrated and embedded between coated microscope slides and cover glasses using Durcupan (Sigma-Aldrich, Steinheim, Germany) followed by polymerization at 56°C for 48 hours. After identification of the respective cells by light microscopy the sections were trimmed and transferred on blocks of resin for ultra-thin sectioning using an ultra-microtome (Leica Microsystems). Ultra-thin sections of 55 nm thickness were transferred on formvar-coated copper grids, stained with lead-citrate and analyzed using a Zeiss Sigma electron microscope (Zeiss NTS, Oberkochen, Germany).
Flow cytometry
CNS macrophages were isolated using Percoll gradients from homogenized brain tissue. Monocytes were isolated from bone marrow. Cells stained with primary antibodies directed against CD11b, CD36, CD45, CD115, F4/80(eBioscience, San Diego, USA), CD64,Ly6C (BD Biosciences, Heidelberg, Germany) and MHC II (BioLegend, San Diego, USA) at 4°C for 15 min. Cells were washed and analyzed using a FACSCanto II or sorted using a Cell Sorter FACS Aria Fusion. Viable cells were gated by forward and side scatter pattern. Data were acquired with FACSdiva software (Becton Dickinson). Postacquisition analysis was performed using FlowJo software (Tree Star, Inc.).
Single cell RNAseq
Monocytes were isolated from bone marrow FACS sorting for CD45, CD11b and CD115 and then subjected to single-cell RNA-seq using the C1 AutoPrep instrument (Fluidigm) and STRT/C1 protocol, as previously described16. Each single cell was imaged and manually curated, and only single healthy-looking cells without debris were used for the analyses. Expression profiles were obtained as absolute cDNA molecule counts, and normalized to “transcripts per million” to compensate for differences in total transcriptome size between cell types. For t-SNE, we used the first 25 principal components, with perplexity 50 and theta = 0.5. However, the qualitative results were insensitive to the exact parameter choices and always clearly separated monocytes from the two other cell types.
Microarray analysis
Cells were sorted into 60 μl extraction buffer and RNA was extracted for microarray and qPCR using the Arcturus®PicoPure®RNA Isolation Kit (Life Technologies) according to the manufacturer’s protocol. RNA quantity and quality was assessed using Agilent 2100 Bioanalyzer. Affymetrix GeneChips were used for genomewide expression analysis. Total RNA was processed using the GeneChip Expression 3′ Amplification One-Cycle Target Labeling Kit according to the manufacturer’s instruction. Biotinylated cRNA was hybridized on Mouse 2.0 ST GeneChips that were stained, washed and scanned following standard procedures. Cell files were normalized using robust multiarray analysis (RMA) implemented Affymetrix Expression Console. The normalized expression data were then imported and analyzed for differential gene expression using BRB-ArrayTools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team.
qRT-PCR
RT–PCRs were performed as described recently43. The extracted RNA (1μg per sample) was transcribed into cDNA using the High Capacity RNA-to-cDNA Kit (Life Technologies) following the provided protocol. A total of 1 μl cDNA was transferred into a 96-well Multiply® PCR plate (Sarstedt, Germany) and 11.5 μl ABsolute™ QPCR® SYBR Green master mix (Thermo Fisher).
Statistical analysis
Data were tested for normality applying the Kolmogorov–Smirnov test. If normality was given, an unpaired t-test was applied. If the data did not meet the criteria of normality, the Mann–Whitney U-test was applied.
Supplementary Material
Suppl. Fig. 1: Single cell RNA sequencing of pvMΦ, microglia and monocytes.
a) Work flow for obtaining and analysing single-cell RNA-seq from mouse cortical cells and monocytes from dissection to single-cell RNA-seq and biclustering.
b) Representative image of single cells captured in the chip. All cells were checked individually by light microscopy and chambers with no or damaged cells (right squares) were omitted from subsequent analysis.
c) Bar graphs for selected markers for individual pvMΦ (black), cortical microglia (grey) and monocytes (dark grey) evaluated by single cell RNA-seq. Bars represent means ± s.e.m. of single cells.
Suppl. Fig. 2: Irradiation induces engraftment of bone marrow-derived CNS macrophages and microglia.
a) Direct fluorescence microscopic visualization revealed numerous GFP+ donor-derived Iba-1+ pvMΦ, mMΦ, cpMΦ and few microglia 20 weeks after transfer of bone marrow from actin-GFP mice into lethally irradiated wild-type mice. Representative images are shown. Arrows indicate double positive cells. Asterisks point to single Iba-1 (red) positive cells. Nuclei were stained by DAPI (blue). Scale bar = 25 μm.
b) Quantification of donor-derived GFP+Iba-1+ cells. Each symbol represents one mouse with quantification of at least three tissue sections. Data represent means ± s.e.m. of at least five animals per group.
Suppl. Fig. 3: Macrophage development in CX3CR1GFP/WT mice.
Sagittal brain sections from E9.5 to adulthood (P60) depicting meningeal fibroblasts (ER-TR7, red), endothelial cells (CD31, red) and myeloid cells (GFP, green) together with nuclear staining (DAPI, blue). As early as E9.5 dpc elongated GFP+ macrophages first appear in the meninges (arrow) adjacent to the neuroectodermal border. Cx3cr1GFP+ macrophages start seeding the neuroectoderm at 9.5 dpc to become parenchymal microglia (asterisk). GFP+ pvMΦ and cpMΦ are clearly detectable starting from E12.5 onwards (arrows). Scale bar: 25 μm.
Suppl. Fig. 4: CNS macrophages do not require Batf3.
Localisation and presence of pvMΦ, mMΦ and cpMΦ in adult wild-type (WT) and Batf3−/− mice evaluated using Iba-1 immunohistochemistry. Representative figure are presented (upper images) and quantification thereof (lower images). Each symbol represents one mouse with quantification of a minimum of three tissue sections. Data represent means ± s.e.m. of at least four cells in three animals per group. N.s. = not significant. Significant differences were determined by an unpaired t-test.
Suppl. Fig. 5: Graphical abstract of experimental findings.
Supplementary Movie 1
3D-rendering (Imaris, Bitplane) of the confocal image stack represented in Fig. 3h that illustrates the distinct location and morphology of pvMΦ, mMΦ and microglial cells in the intact spinal cord of a Cx3cr1CreER:R26-tomato mice (tomato, red) at 8 weeks after TAM and injected with dextran-AF647 (blood vessel, blue).
Supplementary Movie 2
Confocal projection of the dorsal spinal cord as shown in Fig. 3h followed by in vivo 2-photon timelapse of the myeloid cells in the same area labeled by tdTomato (white) in Cx3cr1CreER:R26-tomato mice at 8 weeks after TAM.
Acknowledgments
We thank Maria Oberle, Margarethe Ditter and Tina el Gaz for excellent technical assistance and Tuan Leng Tay for critical reading of the manuscript. MP was supported by the DFG (SFB 992, SFB 1160, FOR1336, PR 577/8-1), the Fritz-Thyssen Foundation, the European Union’s Seventh Framework Program FP7 under Grant agreement 607962 (nEUROinflammation) and the Gemeinnützige Hertie Foundation (GHST). JP, IB and SJ were supported by the DFG (FOR1336). MP and MK are further supported by the BMBF-funded Competence Network on Multiple Sclerosis (KKNMS). RZ was supported by the DFG (SFB 1160, ZE872/3-1).
Footnotes
AUTHOR CONTRIBUTIONS
TG, MJCJ, PW, FP, NH, KF, OS, KK, LA, MK and GL conducted experiments and analyzed the data. RZ, SE, FG, JP, FR, IB, MK, SL and SJ analyzed the data, contributed to the in vivo studies and provided mice or reagents. TG and MP supervised the project and wrote the manuscript.
Reference List
- 1.Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- 2.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 3.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 4.Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Bra in Res. 1999;117:145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 5.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 7.Kierdorf K, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 8.Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickey WF, Kimura H. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science. 1988;239:290–292. doi: 10.1126/science.3276004. [DOI] [PubMed] [Google Scholar]
- 10.Hickey WF, Vass K, Lassmann H. Bone marrow-derived elements in the central nervous system: an immunohistochemical and ultrastructural survey of rat chimeras. J Neuropathol Exp Neurol. 1992;51:246–256. doi: 10.1097/00005072-199205000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, et al. Perivascular, but not parenchymal, cerebral engraftment of donor cells after non-myeloablative bone marrow transplantation. Exp Mol Pathol. 2013;95:7–17. doi: 10.1016/j.yexmp.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr CM, Manning J, Lewis CA, Rossi FM, Krieger C. Submyeloablative conditioning with busulfan permits bone marrow-derived cell accumulation in a murine model of Alzheimer’s disease. Neurosci Lett. 2015;588:196–201. doi: 10.1016/j.neulet.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Mildner A, et al. Microglia in the adult brain arise from Ly-6C(hi)CCR2(+) monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 14.Mildner A, et al. Distinct and Non-Redundant Roles of Microglia and Myeloid Subsets in Mouse Models of Alzheimer’s Disease. J Neurosci. 2011;31:11159–11171. doi: 10.1523/JNEUROSCI.6209-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kierdorf K, Katzmarski N, Haas CA, Prinz M. Bone marrow cell recruitment to the brain in the absence of irradiation or parabiosis bias. PLoS One. 2013;8:e58544. doi: 10.1371/journal.pone.0058544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 17.Jamieson AR, et al. Exploring nonlinear feature space dimension reduction and data representation in breast Cadx with Laplacian eigenmaps and t-SNE. Med Phys. 2010;37:339–351. doi: 10.1118/1.3267037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford AL, Goodsall AL, Hickey WF, Sedgwick JD. Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4+ T cells compared. J Immunol. 1995;154:4309–4321. [PubMed] [Google Scholar]
- 19.Bechmann I, et al. Turnover of rat brain perivascular cells. Exp Neurol. 2001;168:242–249. doi: 10.1006/exnr.2000.7618. [DOI] [PubMed] [Google Scholar]
- 20.Priller J, et al. Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat Med. 2001;7:1356–1361. doi: 10.1038/nm1201-1356. [DOI] [PubMed] [Google Scholar]
- 21.Gomez PE, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molawi K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211:2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldmann T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 25.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 26.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 28.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 29.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013 doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 32.Chorro L, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J Exp Med. 2009;206:3089–3100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunis G, et al. IFN-gamma-dependent activation of the brain’s choroid plexus for CNS immune surveillance and repair. Brain. 2013;136:3427–3440. doi: 10.1093/brain/awt259. [DOI] [PubMed] [Google Scholar]
- 35.Engelhardt B, Wolburg-Buchholz K, Wolburg H. Involvement of the choroid plexus in central nervous system inflammation. Microsc Res Tech. 2001;52:112–129. doi: 10.1002/1097-0029(20010101)52:1<112::AID-JEMT13>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 36.Axtell RC, Steinman L. Gaining entry to an uninflamed brain. Nat Immunol. 2009;10:453–455. doi: 10.1038/ni0509-453. [DOI] [PubMed] [Google Scholar]
- 37.Adolfsson J, et al. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 38.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kivisakk P, et al. Localizing central nervous system immune surveillance: meningeal antigen-presenting cells activate T cells during experimental autoimmune encephalomyelitis. Ann Neurol. 2009;65:457–469. doi: 10.1002/ana.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anandasabapathy N, et al. Flt3L controls the development of radiosensitive dendritic cells in the meninges and choroid plexus of the steady-state mouse brain. J Exp Med. 2011;208:1695–1705. doi: 10.1084/jem.20102657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misgeld T, Nikic I, Kerschensteiner M. In vivo imaging of single axons in the mouse spinal cord. Nat Protoc. 2007;2:263–268. doi: 10.1038/nprot.2007.24. [DOI] [PubMed] [Google Scholar]
- 42.Romanelli E, et al. Cellular, subcellular and functional in vivo labeling of the spinal cord using vital dyes. Nat Protoc. 2013;8:481–490. doi: 10.1038/nprot.2013.022. [DOI] [PubMed] [Google Scholar]
- 43.Goldmann T, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–1629. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1: Single cell RNA sequencing of pvMΦ, microglia and monocytes.
a) Work flow for obtaining and analysing single-cell RNA-seq from mouse cortical cells and monocytes from dissection to single-cell RNA-seq and biclustering.
b) Representative image of single cells captured in the chip. All cells were checked individually by light microscopy and chambers with no or damaged cells (right squares) were omitted from subsequent analysis.
c) Bar graphs for selected markers for individual pvMΦ (black), cortical microglia (grey) and monocytes (dark grey) evaluated by single cell RNA-seq. Bars represent means ± s.e.m. of single cells.
Suppl. Fig. 2: Irradiation induces engraftment of bone marrow-derived CNS macrophages and microglia.
a) Direct fluorescence microscopic visualization revealed numerous GFP+ donor-derived Iba-1+ pvMΦ, mMΦ, cpMΦ and few microglia 20 weeks after transfer of bone marrow from actin-GFP mice into lethally irradiated wild-type mice. Representative images are shown. Arrows indicate double positive cells. Asterisks point to single Iba-1 (red) positive cells. Nuclei were stained by DAPI (blue). Scale bar = 25 μm.
b) Quantification of donor-derived GFP+Iba-1+ cells. Each symbol represents one mouse with quantification of at least three tissue sections. Data represent means ± s.e.m. of at least five animals per group.
Suppl. Fig. 3: Macrophage development in CX3CR1GFP/WT mice.
Sagittal brain sections from E9.5 to adulthood (P60) depicting meningeal fibroblasts (ER-TR7, red), endothelial cells (CD31, red) and myeloid cells (GFP, green) together with nuclear staining (DAPI, blue). As early as E9.5 dpc elongated GFP+ macrophages first appear in the meninges (arrow) adjacent to the neuroectodermal border. Cx3cr1GFP+ macrophages start seeding the neuroectoderm at 9.5 dpc to become parenchymal microglia (asterisk). GFP+ pvMΦ and cpMΦ are clearly detectable starting from E12.5 onwards (arrows). Scale bar: 25 μm.
Suppl. Fig. 4: CNS macrophages do not require Batf3.
Localisation and presence of pvMΦ, mMΦ and cpMΦ in adult wild-type (WT) and Batf3−/− mice evaluated using Iba-1 immunohistochemistry. Representative figure are presented (upper images) and quantification thereof (lower images). Each symbol represents one mouse with quantification of a minimum of three tissue sections. Data represent means ± s.e.m. of at least four cells in three animals per group. N.s. = not significant. Significant differences were determined by an unpaired t-test.
Suppl. Fig. 5: Graphical abstract of experimental findings.
Supplementary Movie 1
3D-rendering (Imaris, Bitplane) of the confocal image stack represented in Fig. 3h that illustrates the distinct location and morphology of pvMΦ, mMΦ and microglial cells in the intact spinal cord of a Cx3cr1CreER:R26-tomato mice (tomato, red) at 8 weeks after TAM and injected with dextran-AF647 (blood vessel, blue).
Supplementary Movie 2
Confocal projection of the dorsal spinal cord as shown in Fig. 3h followed by in vivo 2-photon timelapse of the myeloid cells in the same area labeled by tdTomato (white) in Cx3cr1CreER:R26-tomato mice at 8 weeks after TAM.


