ABSTRACT
Newly emerging highly pathogenic avian influenza (HPAI) H5N2, H5N3, H5N5, H5N6, H5N8 and H5N9 viruses have been spreading in poultry and wild birds. The H5N6 viruses have also caused 10 human infections with 4 fatal cases in China. Here, we assessed the cross-neutralization and cross-protection of human and mouse monoclonal antibodies against 2 viruses: a HPAI H5N8 virus, A/chicken/Netherlands/14015526/2014 (NE14) and a HPAI H5N6 virus, A/Sichuan/26221/2014 (SC14). The former was isolated from an infected chicken in Netherlands in 2014 and the latter was isolated from an infected human patient in Sichuan, China. We show that antibodies FLA5.10, FLD21.140, 100F4 and 65C6, but not AVFluIgG01, AVFluIgG03, S139/1 and the VRC01 control, potently cross-neutralize the H5N8 NE14 and H5N6 SC14 viruses. Furthermore, we show that a single injection of >1 mg/kg of antibody 100F4 at 4 hours before, or 20 mg/kg antibody 100F4 at 72 hours after, a lethal dose of H5N8 NE14 enables mice to withstand the infection. Finally, we show that a single injection of 0.5 or 1 mg/kg antibody 100F4 prophylactically or 10 mg/kg 100F4 therapeutically outperforms a 5-day course of 10 mg/kg/day oseltamivir treatment against lethal H5N8 NE14 or H5N6 SC14 infection in mice. Our results suggest that further preclinical evaluation of human monoclonal antibodies against newly emerging H5 viruses is warranted.
KEYWORDS: Cross-protection, highly pathogenic avian influenza virus, human monoclonal antibody, hemagglutinin, newly emerging H5 viruses, oseltamivir
Abbreivations
- HPAI
highly pathogenic avian influenza
- NE14
A/chicken/Netherlands/14015526/2014
- SC14
A/Sichuan/26221/2014
- SZ06
A/Shenzhen/406H/06
- AH05
A/Anhui/1/2005
- VN04
A/Vietnam/CL26/2004
- VN05
A/Vietnam/CL115/2005
- HA
Hemagglutinin
- NA
Neuraminidase
- mAbs
Monoclonal antibodies
- HAU
Hemagglutinin unit
- PFU
Plaque formation unit
- TCID50
50% tissue culture infective dose
- MLD50
50% mouse lethal dose
- MN
Microneutralization
- PN
HA and NA pseudotype-based neutralization
- RLA
relative luciferase activity
- i.p.
Intraperitoneally
- i.n.
Intranasally
Introduction
Since 1996, the highly pathogenic avian influenza (HPAI) H5N1 virus has spread in a variety of domestic and wild birds, and was sporadically transmitted to humans in Asia, Europe and Africa. As of February 2016, the World Organization for Animal Health had highlighted thousands of HPAI H5N1 outbreaks in poultry and wild birds in various countries.1 As of February 25, 2016, 846 human H5N1 infections had been confirmed, resulting in 449 deaths.2
During the initial circulation and spread before 2008, the hemagglutinin (HA) genes of the HPAI H5N1 viruses evolved into 10 phylogenetically distinctive clades (clades 0 to 9), and clades 2 and 7 have further evolved into many subclades, but with no evidence of gene exchange between influenza viruses. Since 2008, HA genes from HPAI H5N1 viruses were found to be re-assorted with neuraminidase (NA) and various other genes of low pathogenic avian influenza viruses. As a result, newly emerging HPAI H5N2, H5N3, H5N5, H5N6, H5N8 and H5N9 viruses have been spreading in poultry and wild birds in various countries of Asia, Europe and North America.1-5 The H5N6 viruses have also caused 10 human infections with 4 fatal cases in China.2,6 Thus, the incursions of the newly emerging HPAI H5 viruses constitute a substantial threat to animals and humans.
The newly emerging HPAI H5 viruses isolated from domestic and wild birds contain the HA gene from an ancestral HPAI H5N1 A/Goose/Guangdong/1/1996 lineage,5 which raises the possibility that currently available anti-H5N1 vaccine candidates7-13 and monoclonal antibodies (mAbs)14-22 may provide sufficient cross-protection against the newly emerging HPAI H5 viruses. Indeed a recent study showed that human immune sera elicited with H5N1 vaccines exhibit considerable cross-reactivity against a newly emerging H5N8 virus.23 In the study reported here, we assessed the cross-neutralization of a HPAI H5N8 virus, A/chicken/Netherlands/14015526/2014 (NE14) and a HPAI H5N6 virus, A/Sichuan/26221/2014 (SC14), by 7 anti-HA mAbs (100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and S139/1) along with a control antibody VRC01. We and others had previously isolated the antibodies from memory B cells of HPAI H5N1-infected individuals or from vaccinated animals. 16-18,24,25
100F4 and 65C6, which were isolated from an individual infected with HPAI H5N1 A/Shenzhen/406H/06 H5N1 strain (SZ06, subclade 2.3.4), potently neutralized all clades and subclades of HPAI H5N1 viruses except for subclade 7.2.18,26 AVFluIgG01 and AVFluIgG03 were isolated from an individual infected with HPAI H5N1 A/Anhui/1/2005 strain (AH05, subclade 2.3.4). AVFluIgG01 neutralizes most H5N1 strains tested, but with potency much lower than 65C6 and 100F4, whereas AVFluIgG03 only neutralizes 11 of 17 viruses tested with comparable potency to 65C6 and 100F4.17,27 FLA5.10 was isolated from donor CL26 infected with HPAI H5N1 A/Vietnam/CL26/2004 strain (VN04, clade 1), and FLD21.140 was isolated from donor CL115 infected with HPAI H5N1 A/Vietnam/CL115/2005 strain (VN05, clade 1). Both neutralize HPAI H5N1 viruses from clade 1; FLD21.140 also neutralizes strains from clade 2.16,28 S139/1 isolated from H3 HA-immunized BALB/c mice binds HA from strains of subtypes 1, 2, 3, 5, 9 and 13 and neutralizes viruses from subtypes 1, 2, 3 and 13.24 Antibody VRC01 recognizes the CD4-binding site of HIV-1 gp120, and was used here as a negative control.25 In addition, we tested the cross-protection of antibody 100F4 against the H5N8 NE14 virus in mice prophylactically and therapeutically. Finally, oseltamivir, a neuraminidase (NA) inhibitor, is an antiviral medication used to treat flu caused by influenza A and influenza B viruses and to prevent flu after exposure.29 However, no studies on its treatment against the newly emerging H5 viruses have been reported. Therefore, in this study we compared the in vivo efficacy against the H5N8 NE14 and H5N6 SC14 between antibody 100F4 and oseltamivir.
Results
Generation of recombinant HPAI H5N8 and H5N6 viruses for in vitro and in vivo testing
To select representative challenge viruses, we downloaded HA sequences of 37 HPAI H5 strains including H5N1, H5N2, H5N3, H5N5, H5N6 and H5N8 strains from GISAID (http://platform.gisaid.org) or NCBI (http://www.ncbi.nlm.nih.gov). Using the neighbor-joining method (MEGA5.1), we then constructed a phylogenetic tree (Fig. 1A). These newly emerging H5 strains cover all 3 subgroups of group A and group B from various countries in Asia, Europe and North America and from various species.30 The representative strains (circled in orange) were selected for the amino acid sequence comparison. Fig. 1B shows that H5 HA (subclade 2.3.4.4) in H5N8 NE14 strain (highlighted in red bold font) and in H5N6 SC14 strain (highlighted in blue bold font) contain mutations shared by the majority of newly emerging H5 strains.
Figure 1.
The selection of A/chicken/Netherlands/14015526/2014 H5N8 (NE14) and A/Sichuan/26221/2014 H5N6 (SC14) challenge strains. (A) Phylogenetic tree of HA among 37 HPAI H5 strains using the neighbor-joining method. Newly emerging HPAI H5N2, H5N6 and H5N8 strains covered all 3 subgroups of group A and group B from various countries in Asia, Europe and North America and from various species.30 The representative strains (circled in orange) were selected for the amino acid sequence comparison. (B) Amino acid sequence alignment among selected H5 strains compared with the sequence of H5N1 A/Shenzhen/406H/2006 (SZ06) virus. The NE14 H5N8 and SC14 H5N6 strains are shown in red and blue bold font, respectively. (C) The summary of hemagglutination units (HAU), plaque forming unit (PFU), the 50% tissue culture infective dose (TCID50), and the 50% mouse lethal dose (MLD50) of the NE14 and SC14 viruses.
We next generated recombinant H5N8 NE14 and H5N6 SC14 virus by co-transfecting 293 T and the Madin-Darby canine kidney (MDCK) cell mixture with gene segments encoding HA and NA proteins from H5N8 NE14 or H5N6 SC14 strain, and the remaining 6 gene segments encoding NP, PA, PB1, PB2, M and NS from A/WSN/1933 strain. The resulting H5N8 NE14 and H5N6 viruses were propagated in MDCK cells. Fig. 1C summarizes hemagglutination units (HAU), plaque forming unit (PFU), 50% tissue culture infective dose (TCID50), and 50% mouse lethal dose (MLD50) of the H5N8 NE14 and H5N6 SC14 viruses to be used for in vitro and in vivo testing (see below).
Assessment of cross-neutralization of mAbs against the H5N8 NE14 and H5N6 SC14 viruses
We then tested the cross-neutralization of H5N8 NE14 and H5N6 SC14 viruses by mAbs 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140, S139/1 and a control antibody VRC0116-18,24,25 measured by microneutralization (MN) and plaque reduction assays. As measured by the MN assay, FLD21.140 at concentrations of 0.3125 μg/ml or higher or FLA5.10, 100F4 and 65C6 at concentrations of 1.25 μg/ml or higher completely neutralize the H5N8 NE14 virus infection (Table 1). In contrast, AVFluIgG01, AVFluIgG03, S139/1 or VRC01 control antibody even at concentrations 20 μg/ml do not have any neutralization activity. Similar cross-neutralization results with the same panel of antibodies were observed against H5N6 SC14 virus (Table 1B).
Table 1.
Cross-neutralization of the H5N8 NE14 (A) or H5N6 SC14 (B) virus by monoclonal antibodies measured by microneutralization assay.
A
| Antibody concentration (µg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibody | 20 | 10 | 5 | 2.5 | 1.25 | 0.625 | 0.3125 | 0.15625 | 0 |
| 100F4 | − | − | − | − | − | + | ++++ | ++++ | ++++ |
| 65C6 | − | − | − | − | − | ++++ | ++++ | ++++ | ++++ |
| AVFluIgG01 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| AVFluIgG03 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| FLA5.10 | − | − | − | − | − | + | +++ | ++++ | ++++ |
| FLD21.140 | − | − | − | − | − | − | − | ++ | ++++ |
| S139/1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| VRC01 |
++++ |
++++ |
++++ |
++++ |
++++ |
++++ |
++++ |
++++ |
++++ |
| B | |||||||||
| 100F4 | − | − | − | − | − | +/− | ++++ | ++++ | ++++ |
| 65C6 | − | − | − | − | − | ++ | +++ | ++++ | ++++ |
| AVFluIgG01 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| AVFluIgG03 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| FLA5.10 | − | − | − | − | − | − | ++ | ++++ | ++++ |
| FLD21.140 | − | − | − | − | − | − | +++ | +++ | ++++ |
| S139/1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| VRC01 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
“-” To “++++” represent CPE scores from no to full-blown.
Similar results were also obtained from the plaque reduction assay. FLA5.10 and FLD21.140 at concentrations of 0.625 μg/ml or higher or antibodies 100F4 and 65C6 at concentrations of 1.25 μg/ml or higher completely neutralize the H5N8 NE14 virus infection, whereas AVFluIgG01, AVFluIgG03, S139/1 or VRC01 control antibody even at 20 μg/ml do not have any neutralization activity (Table 2A). FLA5.10, FLD21.140, 100F4 and 65C6 at concentrations of 2.5 μg/ml or higher completely neutralize the H5N6 SC14 virus infection, whereas AVFluIgG01, AVFluIgG03, S139/1 or VRC01 control antibody even at 20 μg/ml do not have any neutralization activity (Table 2B). Thus, we conclude that antibodies FLA5.1, FLD21.140, 100F4 and 65C6 potently cross-neutralize both H5N8 NE14 and H5N6 SC14 viruses.
Table 2.
Cross-neutralization of the H5N8 NE14 (A) or H5N6 SC14 (B) virus by monoclonal antibodies measured by plaque reduction assay.
A
| Antibody concentration (µg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibody | 20 | 10 | 5 | 2.5 | 1.25 | 0.625 | 0.3125 | 0.15625 | 0 |
| 100F4 | 0 | 0 | 0 | 0 | 0 | 2.7 ± 0.6* | 19.3 ± 2.5 | 59.3 ± 3.5 | 59.0 ± 7.5 |
| 65C6 | 0 | 0 | 0 | 0 | 0 | 8.0 ± 2.0 | 42.7 ± 5.0 | 59.3 ± 5.5 | 59.0 ± 7.5 |
| AVFluIgG01 | 59.3 ± 4.0 | 54.3 ± 3.2 | 53.3 ± 4.2 | 57.0 ± 4.6 | 62.3 ± 4.0 | 56.3 ± 7.4 | 57.0 ± 3.6 | 59.3 ± 6.7 | 59 ± 7.5 |
| AVFluIgG03 | 54.0 ± 6.1 | 54.3 ± 4.0 | 56.7 ± 4.2 | 58.0 ± 6.2 | 59.0 ± 6.6 | 58.3 ± 3.8 | 57.7 ± 2.5 | 61.0 ± 2.6 | 59.0 ± 7.5 |
| FLA5.10 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0 ± 1.0 | 4.7 ± 2.5 | 59.0 ± 7.5 |
| FLD21.140 | 0 | 0 | 0 | 0 | 0 | 0 | 0.7 ± 0.6 | 1.0 ± 1.0 | 59.0 ± 7.5 |
| S139/1 | 63.7 ± 6.0 | 56.0 ± 5.3 | 55.7 ± 4.0 | 60.7 ± 2.1 | 59.7 ± 5.7 | 60.3 ± 1.2 | 64.3 ± 1.5 | 60.7 ± 5.1 | 59.0 ± 7.5 |
| VRC01 |
55.0 ± 4.0 |
56.7 ± 1.5 |
58.7 ± 1.5 |
62.0 ± 6.2 |
61.3 ± 3.1 |
61.7 ± 6.4 |
61.0 ± 4.6 |
63 ± 5.6 |
59.0 ± 7.5 |
| B | |||||||||
| 100F4 | 0 | 0 | 0 | 0 | 1.0 ± 1.7* | 11.0 ± 4.6 | 55.0 ± 4.6 | 60.0 ± 7.9 | 62.0 ± 3.5 |
| 65C6 | 0 | 0 | 0 | 0 | 16.0 ± 3.6 | 15.0 ± 4.4 | 27.0 ± 5.2 | 53.0 ± 3.6 | 62.0 ± 3.5 |
| AVFluIgG01 | 65.0 ± 3.6 | 64.3 ± 6.1 | 61.7 ± 4.2 | 66.0 ± 6.0 | 61.0 ± 6.2 | 59.0 ± 6.2 | 63.0 ± 8.5 | 60.0 ± 9.0 | 62.0 ± 3.5 |
| AVFluIgG03 | 63.0 ± 4.4 | 61.7 ± 3.1 | 60.0 ± 5.2 | 60.0 ± 5.3 | 60.7 ± 5.5 | 64.0 ± 7.5 | 62.0 ± 6.9 | 62.7 ± 3.5 | 62.0 ± 3.5 |
| FLA5.10 | 0 | 0 | 0 | 0 | 9.0 ± 5.2 | 6.0 ± 5.2 | 57.0 ± 3.0 | 54.7 ± 4.9 | 62.0 ± 3.5 |
| FLD21.140 | 0 | 0 | 0 | 0 | 10.0 ± 9.6 | 19.0 ± 6.9 | 26.0 ± 8.7 | 58.3 ± 2.1 | 62.0 ± 3.5 |
| S139/1 | 64.7 ± 4.0 | 65.3 ± 5.5 | 62.3 ± 2.5 | 67.3 ± 4.2 | 59.7 ± 3.8 | 60.7 ± 3.8 | 64.0 ± 6.6 | 63.0 ± 3.6 | 62.0 ± 3.5 |
| VRC01 | 59.7 ± 3.1 | 63.7 ± 2.1 | 61.0 ± 7.5 | 63.3 ± 3.2 | 62.0 ± 2.6 | 61.3 ± 4.2 | 63.7 ± 2.1 | 64.3 ± 5.5 | 62.0 ± 3.5 |
Mean ± SD of the number of plaques in triplicates.
Since the PN assay is considered a more sensitive and quantitative assay than the plaque reduction and MN assays, we also tested cross-neutralization of antibodies 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and S139/1 against H5N8 NE14 (subclade 2.3.4.4) and H5N1 SZ06 (subclade 2.3.4) pseudotypes using the PN assay. Fig. 2 shows that while S139/1 does not have any neutralization activity against H5N8 NE14 and H5N1 SZ06 pseudotypes, 100F4 and 65C6 neutralize both H5N8 NE14 and H5N1 SZ06 pseudotypes extremely well (IC50 ranging from 0.014 to 0.019 μg/ml). AVFluIgG01 and AVFluIgG03, while neutralizing H5N1 SZ06 pseudotype well (IC50 0.057 and 0.008 μg/ml, respectively), have no or very little neutralization activity against H5N8 NE14 pseudotype. In contrast, while FLA5.10 and FLD21.140 neutralize H5N1 SZ06 pseudotype relatively poorly (IC50 4.6 and 1.9 μg/ml, respectively), they neutralize H5N8 NE14 pseudotype well (IC50 0.03 and 0.014 μg/ml, respectively). Thus, we conclude that AVFluIgG01, AVFluIgG03 and S139/1 have no or very little cross-neutralization activity against H5N8 NE14 and H5N6 SC14 viruses, even though AVFluIgG01 and AVFluIgG03 neutralize H5N1 SZ06 virus well.
Figure 2.
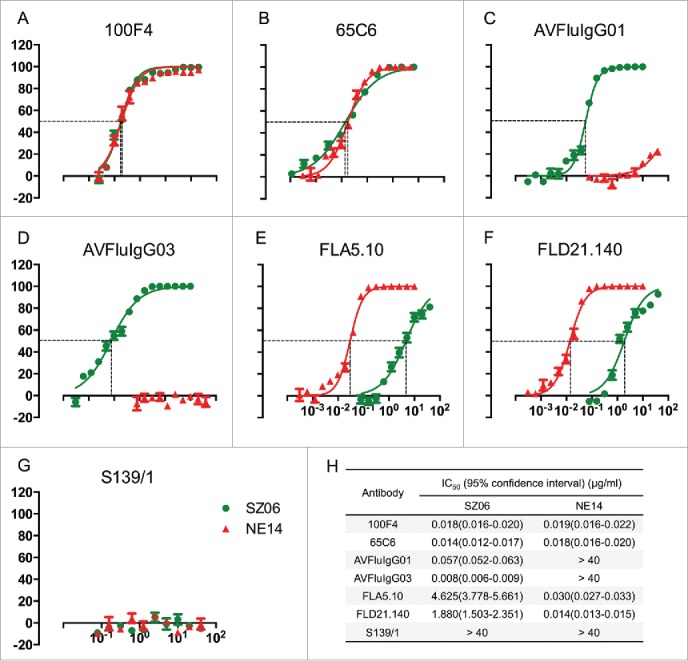
Neutralization activity of 7 monoclonal antibodies against H5N1 SZ06 and H5N8 NE14 pseudotypes using PN assay. (A to G) The titration of neutralization activity of antibodies 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and S139/1 against H5N1 SZ06 (green color line) and H5N8 NE14 (red color line) pseudotypes, respectively. (H) The summary of IC50 values of antibodies 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and S139/1 against H5N1 SZ06 (green line) and H5N8 NE14 (red line) pseudotypes.
Comparison of amino acid conservation of neutralization epitopes among newly emerging H5 strains
The total antibody-contact residues of 100F4, 65C6 and AVFluIgG03 neutralization epitopes have been recently solved by analyzing co-crystals of HA1-Fab complexes of these antibodies.27 The key residues of these neutralization epitopes, as well as the AVFluIgG01 neutralization epitope, were mapped by various mutagenesis assays, such as indirect immunofluorescence assay, yeast surface display and PN assay.17,27 FLA5.10 and FLD21.140 epitopes have only been mapped by the whole genome phage display library and random peptide phage display library, but not by co-crystallization.28 Therefore, it is likely that the amino acid sequences defined by these mapping technologies only consist of the portion of the neutralization epitopes.
To determine the amino acid conservation of these neutralization epitopes among newly emerging HPAI H5 strains, including H5N8 NE14 and H5N6 SC14 strains, we aligned the total and the key residues of these epitopes among representative newly emerging HPAI H5 strains, along with the parental HPAI H5N1 SZ06, AH05, VN04 or VN05 strain (Fig. 3A). The total and key residues, as well as mutations in the total and key residues, among these H5 strains are summarized in Fig. 3B. Of note, although along the course of time mutations accumulate in these epitopes, no mutations were found in 2 key residues of 100F4 epitope in all H5 strains compared. Because of this, we chose antibody 100F4 for the following in vivo studies.
Figure 3.
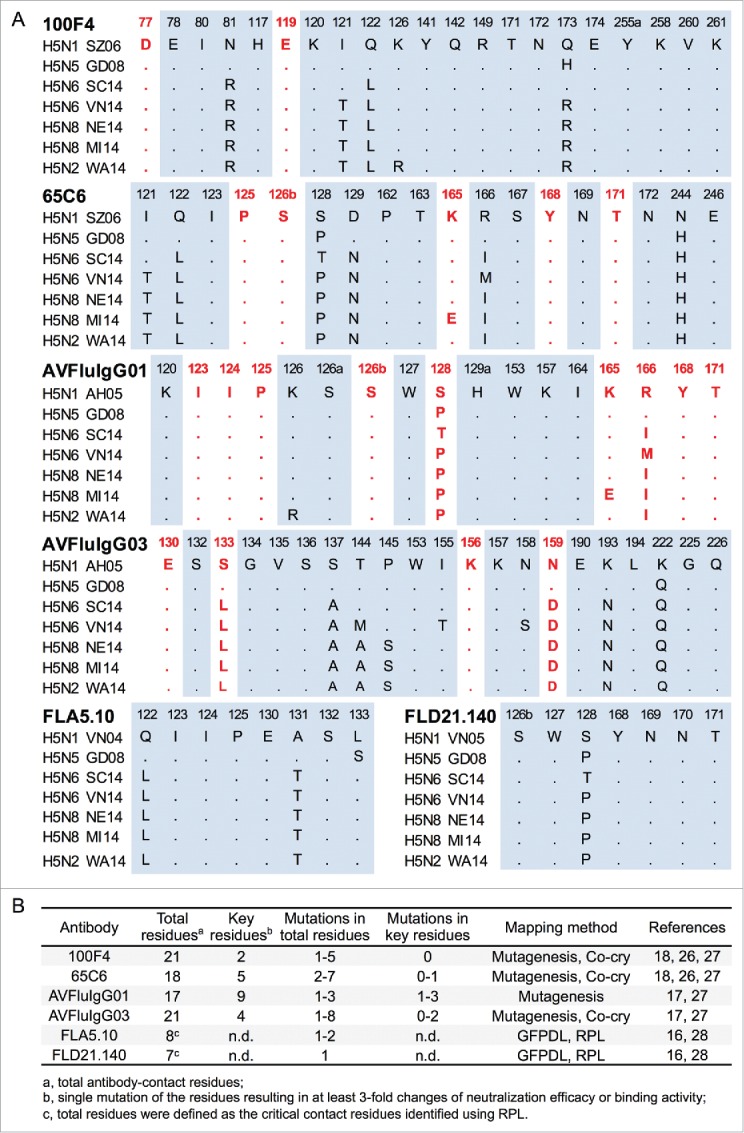
The comparison of the total and the key residues of 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10 and FLD21.140 epitopes. (A) The amino acid sequence alignment of the total and the key residues of 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10 and FLD21.140 epitopes among selected HPAI H5 strains compared to parental HPAI H5N1 virus. The key residues of these epitopes were defined as a single amino acid mutation that results in a 3-fold or more decrease in neutralization or binding activity of a given antibody. Key residues of each epitope are highlighted in red bold font. (B) The summary of the total and the key residues, as well as the mutations in the total and the key residues of 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10 and FLD21.140 epitopes among selected HPAI H5 strains. n.d. stands for not determined. Co-cry: co-crystallization. GFPDL: Gene-Fragmented Phage Display Libraries. RPL: Random Peptide phage display Library.
Potent cross-protection of antibody 100F4 against HPAI H5N8 virus
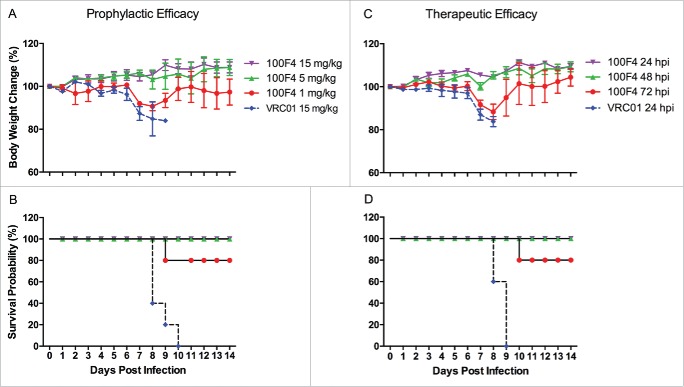
Figs. 4A and B show the prophylactic efficacy of antibody 100F4. Mice inoculated with 15 mg/kg control antibody VRC01 exhibited severe sickness on day 6 post challenge and all mice died. In contrast, all mice inoculated with 1 mg/kg antibody 100F4 were sick at 7 to 8 d post challenge and one mouse died, while the remaining 4 mice survived. Mice inoculated with 5 or 15 mg/kg antibody 100F4 exhibited no signs of sickness and weight loss, and all survived.
Figure 4.
In vivo efficacy of antibody 100F4 against the H5N8 NE14 virus. (A and B) Prophylactic efficacy of antibody 100F4. Time course of body weight changes (A). Survival rate of each group, which was calculated as percent survival within each experimental group (n = 5 mice per group) (B). (C and D) Therapeutic efficacy of antibody 100F4. Time course of body weight changes (C). Survival rate of each group, which was calculated as percent survival within each experimental group (n = 5 mice per group) (D).
Figs. 4C and D show the therapeutic efficacy of antibody 100F4. Mice injected with 20 mg/kg control antibody VRC01 at 24 hours post infection exhibited severe sickness and weight loss, and all died. In contrast, all mice injected with 20 mg/kg antibody 100F4 at 24 and 48 hours post infection survived with no weight loss. After injection with 20 mg/kg antibody 100F4 at 72 hours post infection, all 5 mice were sick at 7 to 8 d post challenge and one mouse died, while the remaining 4 mice survived. Taken together, our in vivo challenge results demonstrated that antibody 100F4 can efficiently cross-protect against HPAI H5N8 infection prophylactically and therapeutically.
Comparison of in vivo efficacy between antibody 100F4 and oseltamivir against H5N8 NE14 and H5N6 SC14 strains
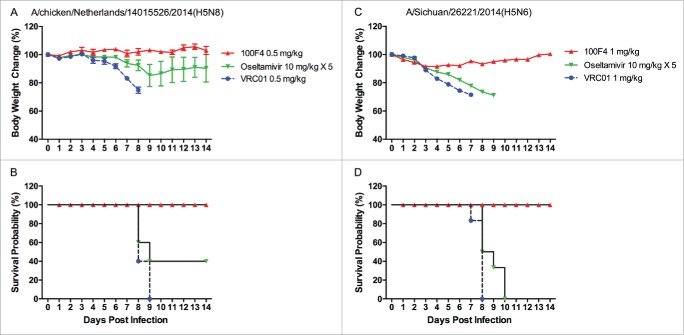
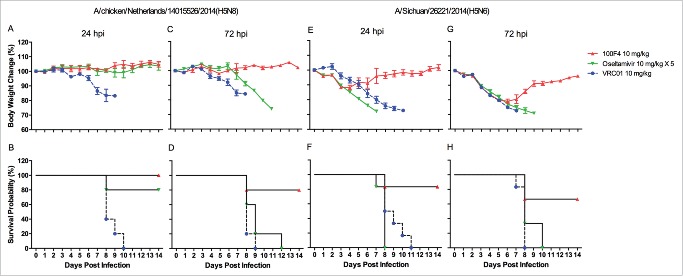
Oseltamivir, marketed under the brand name Tamiflu®, is an antiviral medication used to treat flu caused by influenza A and influenza B viruses, and to prevent the development of flu after exposure.29,31 As a NA inhibitor, oseltamivir mimics the natural substrate of NA, sialic acid, and binds to the NA active site. By so doing, it prevents NA from cleaving the sialic acid residues on host cell receptors, and thereby prevents the release of newly synthesized viruses. Clinically, oseltamivir is effective against all NA subtypes and associated with little toxicity.29,31 Therefore, we first compared prophylactic efficacy between antibody 100F4 and oseltamivir against H5N8 NE14 or H5N6 SC14 strains. Figs. 5A and B show that when challenged with H5N8 NE14 strain, mice injected with 0.5 mg/kg VRC01 control antibody exhibited severe sickness and all mice died, whereas all mice injected with 0.5 mg/kg antibody 100F4 exhibited no signs of sickness and all survived. In contrast, mice administered a 5-day course of 10 mg/kg/day oseltamivir lost significant weight, and only 2 of 5 mice survived. Figs. 5C and D show that, when challenged with H5N6 SC14 strain, mice injected with 1 mg/kg VRC01 control antibody exhibited severe sickness and all mice died, whereas all mice injected with 1 mg/kg antibody 100F4 exhibited slight weight loss and all survived. In contrast, mice administered a 5-day course of 10 mg/kg/day oseltamivir lost significant weight and all died.
Figure 5.
Prophylactic efficacy of antibody 100F4 against the H5N8 NE14 or H5N6 SC14 virus in mice compared to oseltamivir treatment. (A) The time course of body weight changes. (B) The survival rate of different antibody and oseltamivir treatment groups when challenged with H5N8 NE14 strain. Survival rate was calculated as percent survival within each experimental group (n = 5 mice per group). (C) The time course of body weight changes. (D) The survival rate of different antibody and oseltamivir treatment groups when challenged with H5N6 SC14 strain. Survival rate was calculated as percent survival within each experimental group (n = 6 mice per group).
We then compared the therapeutic efficacy between antibody 100F4 and oseltamivir against H5N8 NE14 or H5N6 SC14 strain. Figs. 6A and B show that, when infected with H5N8 NE14 strain, all mice injected with VRC01 at 24 hours post infection died, whereas all mice injected with antibody 100F4 at 24 hours post infection survived with no weight loss. In contrast, 4 of 5 mice administered a 5-day course of oseltamivir starting at 24 hours post infection survived with no significant weight loss. Figs. 6C and D show that 4 of 5 mice survived when injected with antibody 100F4 at 72 hours post infection, but no mice that were injected with VRC01 or administered a 5-day course of oseltamivir starting at 72 hours post infection survived.
Figure 6.
Therapeutic efficacy of antibody 100F4 against H5N8 NE14 or H5N6 SC14 virus compared to oseltamivir treatment in mice. (A) The time course of body weight changes. (B) The survival rate of different antibody and oseltamivir treatment groups delivered at 24 hours after the H5N8 NE14 infection. (C) The time course of body weight changes. (D) The survival rate of different antibody and oseltamivir treatment groups delivered at 72 hours after the H5N8 NE14 virus infection. Survival rate was calculated as percent survival within each experimental group (n = 5 mice per group). (E) The time course of body weight changes. (F) The survival rate of different antibody and oseltamivir treatment groups delivered at 24 hours after the H5N6 SC14 infection. (G) The time course of body weight changes. (H) The survival rate of different antibody and oseltamivir treatment groups delivered at 72 hours after the H5N6 SC14 virus infection. Survival rate was calculated as percent survival within each experimental group (n = 6 mice per group).
When infected with H5N6 SC14 strain, all mice injected with VRC01 at 24 hours post infection died, whereas 5 of 6 mice injected with antibody 100F4 at 24 hours post infection survived with less than 15% weight loss (Fig. 6E, F). In contrast, all mice administered a 5-day course of oseltamivir starting at 24 hours post infection died. Figs. 6G and H show that when injected with antibody 100F4 at 72 hours post infection, 4 of 6 mice survived with less than 23% weight loss, but no mice injected with VRC01 or administered a 5-day course of oseltamivir starting at 72 hours post infection survived. Taken together, these results demonstrated that a single injection of antibody 100F4 outperforms a 5-day course of oseltamivir treatment both prophylactically and therapeutically.
Discussion
The continuous spread among poultry and wild birds, and the potential zoonotic transmission of newly emerging HPAI H5 viruses to humans, pose a major public health threat because few specific treatment options exist. In this study, we assessed the cross-neutralization by mAbs 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and S139/1, tested the cross-protection of antibody 100F4, and compared the in vivo efficacy between antibody 100F4 and oseltamivir against the newly emerging H5N8 NE14 and H5N6 SC14 viruses.
One important finding of this study is that antibodies 100F4, 65C6, FLA5.10 and FLD21.140, but not antibodies AVFluIgG01, AVFluIgG03, and S139/1, potently cross-neutralize newly emerging H5N8 NE14 and H5N6 SC14 viruses (Table 1 and 2, Fig. 2). Because it was previously shown that antibodies 100F4 and 65C6 neutralize all clades and subclades of HPAI H5N1 viruses except for subclade 7.218,26 and FLD21.140 neutralizes strains from both clades 1 and 2,16,28 potent cross-neutralization of the H5N8 NE14 or H5N6 SC14 virus by these antibodies was expected. However, potent cross-neutralization by antibody FLA5.10, as well as the failure of antibodies AVFluIgG01, AVFluIgG03 to neutralize the H5N8 NE14 or H5N6 SC14 virus (subclade 2.3.4.4) is somewhat unexpected. Previous studies showed that the antibody FLA5.10, which was isolated from an individual infected with HPAI VN04 (clade 1), only neutralized HPAI H5N1 strains from clade 1, but not clade 2.16,28 In contrast, antibodies AVFluIgG01 and AVFluIgG03, which were isolated from an individual infected with HPAI H5N1 AH05 virus (subclade 2.3.4), neutralized many H5N1 clades including subclade 2.3.417,27 (Fig. 3 in this study). Here, however, we show that it is antibodies FLA5.10 and FLD 21.140, but not antibodies AVFluIgG01 and AVFluIgG03, that cross-neutralize the H5N8 NE14 and H5N6 SC14 viruses (subclade 2.3.4.4), strongly suggesting that it can be difficult to predict which antibodies will cross-neutralize these newly emerging HPAI H5 viruses.
Another important finding of this study is that antibody 100F4 exhibits potent prophylactic and therapeutic efficacy in vivo. A single intraperitoneal injection of antibody 100F4 at 1 mg/kg or higher cross-protected mice from infection with lethal H5N8 NE14 virus challenge (Fig. 4), and its in vivo prophylactic and therapeutic efficacy outperforms a 5-day course of 10 mg/kg/day treatment of oseltamivir against H5N8 NE14 or H5N6 SC14 virus (Figs. 5 and 6). Thus, antibody 100F4 could have potential as a treatment of human pandemic or zoonotic HPAI H5 cases. Although antibodies can be more costly to produce compared to small molecule drugs such as oseltamivir, given the high degrees of mortality associated with HPAI H5 infection, the higher cost of antibody therapy may be justified if it is more efficacious and saves lives. In addition, the demonstration of prophylactic efficacy of antibody 100F4 against both HPAI H5N8 NE14 and H5N6 SC14 strains provides information that may be useful in 100F4 epitope-based vaccine design.
Although no antibody-based therapies for influenza have been approved to date, the approach represents a plausible intervention for cases of severe influenza. For example, transfusion of human blood products from patients who recovered from the 1918 “Spanish flu” resulted in a 50% reduction in influenza mortality (from 37% to 16%) during the pandemic.32 Passive immunization by vertical acquisition of specific antibodies was found to be associated with influenza immunity in early infancy in humans.16,33-35 Transfusion of convalescent-phase plasma from a patient who recovered from HPAI H5N1 infection resulted in a dramatic reduction of viral load and complete recovery.36 Compared to convalescent-phase plasma samples and plasma samples from other species, human mAbs offer better therapeutic options because large quantities of antibody products that are free of adventitious agents associated with preparations of human plasma samples can be manufactured, and human mAbs should have no or minimal immunogenicity.
In conclusion, our findings that antibodies 100F4, 65C6, FLA5.10 and FLD21.140 potently cross-neutralize the HPAI H5N8 NE14 and H5N6 viruses, and that a single injection of antibody 100F4 outperforms a 5-day course of oseltamivir treatment against the lethal H5N8 NE14 or H5N6 SC14 virus infection in mice, indicate further evaluation of mAbs 100F4, 65C6, FLA5.10 and FLD21.140 against newly emerging HPAI H5 infection is warranted.
Materials and methods
Ethics statement
The experimental protocol (CULATR-3064-13) was approved by the Animal Use Committee and the Safety Committee on BSL-3 Facility and Infectious Agents Li Ka Shing Faculty of Medicine, the University of Hong Kong. All infection experiments were conducted at the biosafety level 3 (BSL-3) facilities complying with the Ethics Committee regulations of University of Hong Kong in accordance with EC directive 86/609/CEE.
Cell lines and oseltamivir
The packaging cell line 293 T was purchased from Invitrogen (Waltham, MA USA) and maintained in complete Dulbecco's Modified Eagle Medium [i.e. high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml); Invitrogen Life Technologies]. The MDCK cell line was maintained in complete DMEM. Oseltamivir (Tamiflu®, Hoffman-La Roche, Catalog number, B3017) was purchased from a local hospital.
Production and purification of human mAbs by 293T cells
The gene segments encoding the variable regions of the heavy and light chains of human mAbs 100F4, 65C6, AVFluIgG01, AVFluIgG03, FLA5.10, FLD21.140 and VRC01 were cloned into the human antibody expression vector containing the constant region of human IgG1 and human κ or λ chain. The gene segments encoding the variable regions of the heavy and light chains of mouse mAb S139/1 were cloned into mouse antibody expression vector containing the constant region of mouse IgG2a and mouse κ chain. Full-length human IgG1 or mouse IgG2a were expressed by transient transfection of 293 T cells, purified by affinity chromatography using Protein A agarose (Pierce, Thermo), and titered by BCA Protein Assay Kit (Thermo Scientific).27
Generation of recombinant H5N8 NE14 and H5N6 viruses
Recombinant H5N8 and H5N6 viruses were generated by co-transfecting 293 T and MDCK cell mixture with gene segments encoding HA and NA proteins from NE14 or SC14 virus (subclade 2.3.4.4) and the remaining 6 gene segments encoding NP, PA, PB1, PB2, M and NS from A/WSN/1933 strain as described by Hoffmann et al.37 The resulting H5N8 NE14 or H5N6 SC14 virus was propagated in MDCK cells. The TCID50 and the MLD50 were determined by serial titration of viruses in MDCK cells and in BALB/c mice as previously described.38
Generation of H5N1 and H5N8 pseudotypes
The method to generate the codon optimized H5 HA from SZ06 (subclade 2.3.4) and NE14 (subclade 2.3.4.4) and N1 NA from A/Thailand/(KAN-1)/04 and N8 NA from NE14 and the method to produce influenza HA and NA pseudotypes were the same as described previously.39 Briefly, 4.5 × 106 293T cells were co-transfected with 14 μg of pHR'CMV-luc, 14 μg of pCMVΔ8.2, 2 μg of pCMV/R-HA, and 0.5 μg of pCMV/R-NA using a calcium phosphate precipitation method. After overnight incubation, the cells were washed once with phosphate-buffered saline (PBS) and cultured in 10 ml of complete DMEM. The HA and NA pseudotype-containing supernatants were harvested after 48 hours and stored in −800C in aliquots until use. The relative luciferase activity (RLA) of these pseudotype stocks was determined as described previously.39
HA and NA pseudotype-based neutralization (PN) assay
PN assay was performed as previously described.39 Briefly, MDCK cells (3×103 cells per well) were seeded onto 96-well culture plate in complete DMEM overnight. Serially diluted human or mouse mAbs were incubated with H5N1 SZ06 and H5N8 NE14 pseudotypes (ranging from 200,000 to 400,000 RLA) at the final volume of 100 μl at 370C for 1 hour. The mixture was added onto MDCK cells. After overnight incubation, cells were washed with PBS and cultured in complete DMEM medium. RLA was measured in 48 hours by a BrightGlo Luciferase assay according to the manufacturer's instruction (Promega).
Microneutralization assay
Neutralization activity of mAbs against the recombinant H5N8 NE14 or H5N6 SC14 virus was analyzed in an MN assay based on the methods of the WHO Global Influenza Program.40 Briefly, the antibodies were serially 2-folded diluted (starting at 20 μg/ml) prior to mixing with 100 TCID50 of virus for 1 hour at 37°C and the mixture were added to monolayers of MDCK cells. The results were recorded after 72 hours and scored as previously described.39
Plaque reduction assay
MDCK cells (5×105 cells per well) in complete DMEM were seeded into 6-well plates. When the cells grew into monolayer, medium was removed and replaced with 700 μl serum-free DMEM. The recombinant H5N8 NE14 or H5N6 SC14 virus (60 PFU virus per well) was incubated with 2-fold diluted mAbs (starting at 20 μg/ml) with a final volume of 100 µl at 37°C for 1 hour. The mixture was then added onto the MDCK monolayer. After the incubation, 0.8% low-melting point agarose in MEM (3 ml per well) was added. After 72 hours' incubation, the agarose overlay was discarded and the cells were stained with 0.5% (w/v) crystal violet for 1 hour. Crystal violet was then removed, cells were washed with H2O, and the number of plaques was counted. The assay was performed in triplicate.
Animal experiments
To test prophylactic efficacy of antibody 100F4, female BALB/c mice at age of 8 weeks were randomly divided into 4 groups (5 mice per group). Mice in groups 1, 2 and 3 were intraperitoneally (i.p.) injected with 1, 5, 15 mg/kg antibody 100F4, respectively. Mice in group 4 were i.p. injected with a control antibody VRC01 (15 mg/kg). Four hours later mice were intranasally (i.n.) challenged with 10 MLD50 of the NE14 H5N8 virus. After the challenge, mice were monitored and recorded daily for signs of illness for 14 d. Mice that lost 30% or more of their initial body weight were euthanized and counted as dead.
To test therapeutic efficacy, 20 female BALB/c mice were i.n. infected with 10 MLD50 of the NE14 H5N8 virus. At 24, 48 and 72 hours post infection, 5 mice per group were i.p. injected with 20 mg/kg antibody 100F4 or control antibody VRC01 at 24 hours post infection. Mice were monitored and recorded daily for signs of illness for 14 d. Mice that lost 30% or more of their initial body weight were euthanized and counted as dead.
To compare prophylactic efficacy between antibody 100F4 and oseltamivir, 8 week old female BALB/c mice (5 or 6 mice per group) were i.p. injected with 0.5 or 1 mg/kg antibody 100F4 or 0.5 or 1 mg/kg VRC01 control 4 hours before being challenged with 5 MLD50 of the H5N8 NE14 or H5N6 SC14 virus or with 10 mg/kg oseltamivir orally administered daily for 5 d starting 4 hours before the same challenge. After the challenge survival, weight loss and clinical signs of sickness were monitored daily for 14 d. Mice that lost 30% or more of their initial body weight were euthanized.
To compare their therapeutic efficacy, 30 female BALB/c mice were i.n. infected with 5 MLD50 of the H5N8 NE14 virus. Another 36 female BALB/c mice were i.n. infected with 5 MLD50 of the H5N6 SC14 virus. At 24 and 72 hours post infection, 5 or 6 mice per group were i.p. injected with 10 mg/kg antibody 100F4 or VRC01 or orally administered daily with 10 mg/kg oseltamivir for 5 d. The survival, weight loss and clinical signs of sickness were monitored for 14 d.
Statistical analysis
The PN assay data were collected from 3 independent experiments. Titration curves were generated using sigmoid dose-response of nonlinear fit from GraphPad. The IC50 and 95% confidence intervals were determined by the best-fit values. The data of the plaque reduction assay were analyzed by mean ± SD using Microsoft Excel. The response of each mouse was counted as an individual data point for statistical analysis. The data obtained from animal studies were examined using one-way analysis of variance from GraphPad.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank other members of the Unit of Anti-Viral Immunity and Genetic Therapy, Institut Pasteur of Shanghai, Chinese Academy of Sciences for helpful discussion during the course of this study.
Funding
This work was supported by 12–5 Mega Project (2013ZX10004003-003) from the Ministry of Science and Technology in China and a grant from Li Kai-Shing Founda- 495 tion in Hong Kong.
References
- 1.OIE Update on highly pathogenic avian influenza in animals (type H5 and H7). Paris, France: World Organization for Animal Health, 2016 [Google Scholar]
- 2.WHO Influenza at the human-animal interface, monthly risk assessment summary. Geneva, Switzerland: World Health Organization, 2016 [Google Scholar]
- 3.de Vries E, Guo H, Dai M, Rottier PJ, van Kuppeveld FJ, de Haan CA. Rapid emergence of highly pathogenic avian influenza subtypes from a subtype H5N1 hemagglutinin variant. Emerg Infect Dis 2015; 21:842-6; PMID:25897518; http://dx.doi.org/ 10.3201/eid2105.141927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall JS, Dusek RJ, Spackman E. Rapidly Expanding Range of Highly Pathogenic Avian Influenza Viruses. Emerg Infect Dis 2015; 21:1251-2; PMID:26079209; http://dx.doi.org/ 10.3201/eid2107.150403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhagen JH, Herfst S, Fouchier RA. Infectious disease. How a virus travels the world. Science 2015; 347:616-7; PMID:25657235; http://dx.doi.org/ 10.1126/science.aaa6724 [DOI] [PubMed] [Google Scholar]
- 6.WHO Archive of avian influenza disease outbreak news. Geneva, Switzerland: World Health Organization, 2016 [Google Scholar]
- 7.Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 2007; 317:825-8; PMID:17690300; http://dx.doi.org/ 10.1126/science.1135165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MW, Cheng TJ, Huang Y, Jan JT, Ma SH, Yu AL, Wong CH, Ho DD. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc Natl Acad Sci U S A 2008; 105:13538-43; PMID:18765801; http://dx.doi.org/ 10.1073/pnas.0806901105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz JA, Buonocore L, Suguitan AL Jr., Silaghi A, Kobasa D, Kobinger G, Feldmann H, Subbarao K, Rose JK. Potent vesicular stomatitis virus-based avian influenza vaccines provide long-term sterilizing immunity against heterologous challenge. J Virol 2010; 84:4611-8; PMID:20181720; http://dx.doi.org/ 10.1128/JVI.02637-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, et al.. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis 2011; 11:916-24; PMID:21975270; http://dx.doi.org/ 10.1016/S1473-3099(11)70240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducatez MF, Bahl J, Griffin Y, Stigger-Rosser E, Franks J, Barman S, Vijaykrishna D, Webb A, Guan Y, Webster RG, et al.. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc Natl Acad Sci U S A 2011; 108:349-54; PMID:21173241; http://dx.doi.org/ 10.1073/pnas.1012457108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou F, Wang G, Buchy P, Cai Z, Chen H, Chen Z, Cheng G, Wan XF, Deubel V, Zhou P. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J Virol 2012; 86:6970-8; PMID:22496212; http://dx.doi.org/ 10.1128/JVI.06930-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Zhou F, Buchy P, Zuo T, Hu H, Liu J, Song Y, Ding H, Tsai C, Chen Z, et al.. DNA prime and virus-like particle boost from a single H5N1 strain elicits broadly neutralizing antibody responses against head region of H5 hemagglutinin. J Infect Dis 2014; 209:676-85; PMID:23911711; http://dx.doi.org/ 10.1093/infdis/jit414 [DOI] [PubMed] [Google Scholar]
- 14.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al.. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 2008; 3:e3942; PMID:19079604; http://dx.doi.org/ 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al.. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16:265-73; PMID:19234466; http://dx.doi.org/ 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, Hien TT, Sallusto F, Ha do Q, Farrar J, et al.. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med 2007; 4:e178; PMID:17535101; http://dx.doi.org/ 10.1371/journal.pmed.0040178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Lu X, Li C, Wang M, Liu Q, Li Z, Hu X, Li J, Liu F, Li Q, et al.. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 2009; 4:e5476; PMID:19421326; http://dx.doi.org/ 10.1371/journal.pone.0005476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, et al.. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol 2012; 86:2978-89; PMID:22238297; http://dx.doi.org/ 10.1128/JVI.06665-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al.. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850-6; PMID:21798894; http://dx.doi.org/ 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- 20.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al.. Highly conserved protective epitopes on influenza B viruses. Science 2012; 337:1343-8; PMID:22878502; http://dx.doi.org/ 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, et al.. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 2013; 87:12619-35; PMID:24049169; http://dx.doi.org/ 10.1128/JVI.01577-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du L, Jin L, Zhao G, Sun S, Li J, Yu H, Li Y, Zheng BJ, Liddington RC, Zhou Y, et al.. Identification and structural characterization of a broadly neutralizing antibody targeting a novel conserved epitope on the influenza virus H5N1 hemagglutinin. J Virol 2013; 87:2215-25; PMID:23221567; http://dx.doi.org/ 10.1128/JVI.02344-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries RD, De Gruyter HL, Bestebroer TM, Pronk M, Fouchier RA, Osterhaus AD, Sutter G, Kreijtz JH, Rimmelzwaan GF. Induction of influenza (H5N8) antibodies by modified vaccinia virus Ankara H5N1 vaccine. Emerg Infect Dis 2015; 21:1086-8; PMID:25988813; http://dx.doi.org/ 10.3201/eid2106.150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog 2009; 5:e1000350; PMID:19300497; http://dx.doi.org/ 10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al.. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329:856-61; PMID:20616233; http://dx.doi.org/ 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian M, Hu H, Zuo T, Wang G, Zhang L, Zhou P. Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol 2013; 87:3571-7; PMID:23269809; http://dx.doi.org/ 10.1128/JVI.01292-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, Shi X, Liu X, Fan S, Ren H, et al.. Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun 2015; 6:8855; PMID:26635249; http://dx.doi.org/ 10.1038/ncomms9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khurana S, Suguitan AL Jr., Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, Manischewitz J, King LR, Subbarao K, Golding H. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med 2009; 6:e1000049; PMID:19381279; http://dx.doi.org/ 10.1371/journal.pmed.1000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med 2005; 353:1363-73; PMID:16192481; http://dx.doi.org/ 10.1056/NEJMra050740 [DOI] [PubMed] [Google Scholar]
- 30.Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. J Virol 2015; 89:6521-4; PMID:25855748; http://dx.doi.org/ 10.1128/JVI.00728-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscona A. Medical management of influenza infection. Annu Rev Med 2008; 59:397-413; PMID:17939760; http://dx.doi.org/ 10.1146/annurev.med.59.061506.213121 [DOI] [PubMed] [Google Scholar]
- 32.Luke TC, Kilbane EM, Jackson JL, Hoffman SL. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann Intern Med 2006; 145:599-609; PMID:16940336; http://dx.doi.org/ 10.7326/0003-4819-145-8-200610170-00139 [DOI] [PubMed] [Google Scholar]
- 33.Puck JM, Glezen WP, Frank AL, Six HR. Protection of infants from infection with influenza A virus by transplacentally acquired antibody. J Infect Dis 1980; 142:844-9; PMID:7462695; http://dx.doi.org/ 10.1093/infdis/142.6.844 [DOI] [PubMed] [Google Scholar]
- 34.Sweet C, Bird RA, Jakeman K, Coates DM, Smith H. Production of passive immunity in neonatal ferrets following maternal vaccination with killed influenza A virus vaccines. Immunology 1987; 60:83-9; PMID:3817868 [PMC free article] [PubMed] [Google Scholar]
- 35.Sweet C, Jakeman KJ, Smith H. Role of milk-derived IgG in passive maternal protection of neonatal ferrets against influenza. J Gen Virol 1987; 68 (Pt 10):2681-6; PMID:3668509; http://dx.doi.org/ 10.1099/0022-1317-68-10-2681 [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med 2007; 357:1450-1; PMID:17914053; http://dx.doi.org/ 10.1056/NEJMc070359 [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 2000; 97:6108-13; PMID:10801978; http://dx.doi.org/ 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding H, Tsai C, Gutierrez RA, Zhou F, Buchy P, Deubel V, Zhou P. Superior neutralizing antibody response and protection in mice vaccinated with heterologous DNA prime and virus like particle boost against HPAI H5N1 virus. PLoS One 2011; 6:e16563; PMID:21305045; http://dx.doi.org/ 10.1371/journal.pone.0016563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, Zhou B, Wang S, Lu S, Buchy P, et al.. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 2009; 27:6777-90; PMID:19732860; http://dx.doi.org/ 10.1016/j.vaccine.2009.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Network WGIS Manual for the laboratory diagnosis and virological surveillance of influenza. Geneva, Switzerland: World Health Organization, 2011 [Google Scholar]