ABSTRACT
A biosimilar is a biological medicinal product that is comparable to a reference medicinal product in terms of quality, safety, and efficacy. SB4 was developed as a biosimilar to Enbrel® (etanercept) and was approved as Benepali®, the first biosimilar of etanercept licensed in the European Union (EU). The quality assessment of SB4 was performed in accordance with the ICH comparability guideline and the biosimilar guidelines of the European Medicines Agency and Food and Drug Administration. Extensive structural, physicochemical, and biological testing was performed with state-of-the-art technologies during a side-by-side comparison of the products. Similarity of critical quality attributes (CQAs) was evaluated on the basis of tolerance intervals established from quality data obtained from more than 60 lots of EU-sourced and US-sourced etanercept. Additional quality assessment was focused on a detailed investigation of immunogenicity-related quality attributes, including hydrophobic variants, high-molecular-weight (HMW) species, N-glycolylneuraminic acid (NGNA), and α-1,3-galactose. This comprehensive characterization study demonstrated that SB4 is highly similar to the reference product, Enbrel®, in structural, physicochemical, and biological quality attributes. In addition, the levels of potential immunogenicity-related quality attributes of SB4 such as hydrophobic variants, HMW aggregates, and α-1,3-galactose were less than those of the reference product.
KEYWORDS: Benepali, biosimilar, Brenzys, critical quality attribute, etanercept, Fc fusion protein, SB4
Abbreviations
- ADCC
antibody-dependent cell-mediated cytotoxicity
- CD
circular dichroism
- CDC
complement-dependent cytotoxicity
- CE-SDS
capillary electrophoresis-sodium dodecyl sulfate
- CEX-HPLC
cation exchange–high-performance liquid chromatography
- CHO
Chinese hamster ovary
- CQA
critical quality attribute
- DLS
dynamic light scattering
- DSC
differential scanning calorimetry
- EMA
European Medicines Agency
- EU
European Union
- FDA
Food and Drug Administration
- FRET
fluorescence resonance energy transfer
- FTIR
Fourier transform infrared spectroscopy
- HDX
hydrogen/deuterium exchange
- HIC
hydrophobic interaction chromatography
- HILIC
hydrophilic interaction liquid chromatography
- HMW
high molecular weight
- HPLC
high-performance liquid chromatography
- icIEF
imaged capillary isoelectric focusing
- LC-ESI-MS
liquid chromatography-electrospray ionization-mass spectrometry
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- LC/MS
liquid chromatography-mass spectrometry
- LMW
low molecular weight
- LTα
lymphotoxin α
- MALLS
multi-angle laser light scattering
- MFI
micro-flow imaging
- MOA
mode of action
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- mTNF
membrane-bound tumor necrosis factor
- NANA
N-acetylneuraminic acid
- NGNA
N-glycolylneuraminic acid
- SEC
size exclusion chromatography
- SPR
surface plasmon resonance
- TNFR2
tumor necrosis factor receptor 2
- TNF
tumor necrosis factor
- TSA
total sialic acid
- UPLC
ultra-performance liquid chromatography
- US
United States
- UV
ultraviolet
Introduction
A biosimilar is a biological medicine that contains the same active ingredient of an original, commercialized, biological product. Regulatory authorities such as the European Medicines Agency (EMA), the US Food and Drug Administration (FDA), and Health Canada have established guidelines on the approval requirements for similar biological products (or biosimilars), which must demonstrate similarity in terms of quality, safety, and efficacy with their reference biologics.1,2 In the US, a biosimilar is defined as a product that is highly similar to the reference product “notwithstanding minor differences in clinically inactive components and without clinically meaningful differences in terms of safety, purity, and potency.”3
Antibody-based therapeutics and biosimilar versions are heterogeneous in structure and physicochemical characteristics such as post-translational modifications, which can lead to variability in critical and noncritical quality attributes. Therefore, similarity studies should not only assess similarity, but also identify differences in quality attributes between a biosimilar and its reference product. CQAs for assessment of similarity have been defined on the basis of the mode of action (MOA) of etanercept and results from structure-activity relationship (SAR) studies. Extensive characterization has been performed with more than 60 batches of EU-sourced Enbrel® and US-sourced Enbrel® to demonstrate biosimilarity of SB4 with Enbrel® from either region. The similarity range was separately set by statistical analysis based on the tolerance interval with the given set of available data points from each lot of EU-sourced Enbrel® and US-sourced Enbrel®. Although the ranges for EU- and US-sourced Enbrel® were very similar, they were not identical. The purpose of this extensive characterization, however, was not to compare Enbrel® from 2 regions, but to demonstrate the biosimilarity of SB4 to the reference product from different sources.
For such comparison studies, the similarity ranges should be based on data from the reference product with respect to the CQAs that might affect efficacy, potency and safety; therefore, several CQAs were evaluated by orthogonal methods, including quantitation of total sialic acid (TSA) and peptide mapping and analysis of aggregation by size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC). More than 60 test methods were used for the comparison of quality attributes of SB4 with those of the reference product, and 15 of these test methods were used to establish similarity ranges of 19 quality attributes. The similarity ranges for various quality attributes can be determined by appropriate statistical approaches. For this study, the similarity ranges were established by a 2-tiered tolerance approach (mean ± kSD) with the available data points.4 The statistical analysis generates a tolerance interval (with a k factor) that is guaranteed, within a specified confidence level, to contain a specified proportion of the population. A tolerance interval is interpreted as a probability interval.
Etanercept has been widely used in clinical practice for more than 15 years, and thus it has well-characterized pharmacological, efficacy, and safety profiles.5 It is produced by the Chinese hamster ovary (CHO) cell expression system as a homodimer of the chimeric protein consisting of the extracellular ligand-binding domain of human tumor necrosis factor receptor 2 (TNFR2) and the Fc domain of human IgG1.6 The TNFR2 domain contains 4 cysteine-rich domains, 2 N-glycosylation sites, and 13 potential O-glycosylation sites. The Fc domain contains one N-glycosylation site, the IgG hinge, and the CH2 and CH3 domains. Etanercept binds to circulating TNF with high affinity and acts as a natural antagonist to TNF by preventing the TNF molecule from binding to a cell-bound receptor. The Fc region of IgG as a fusion element of etanercept prolongs its serum half-life.7 Analytical methods were developed based on these MOAs. Critical attributes are highly related with TNF binding and the corresponding neutralization effect, but the effector functions associated with the Fc domain in general (e.g., complement-dependent cytotoxicity [CDC] and antibody-dependent cell-mediated cytotoxicity [ADCC]) are not considered to be critical attributes of etanercept.
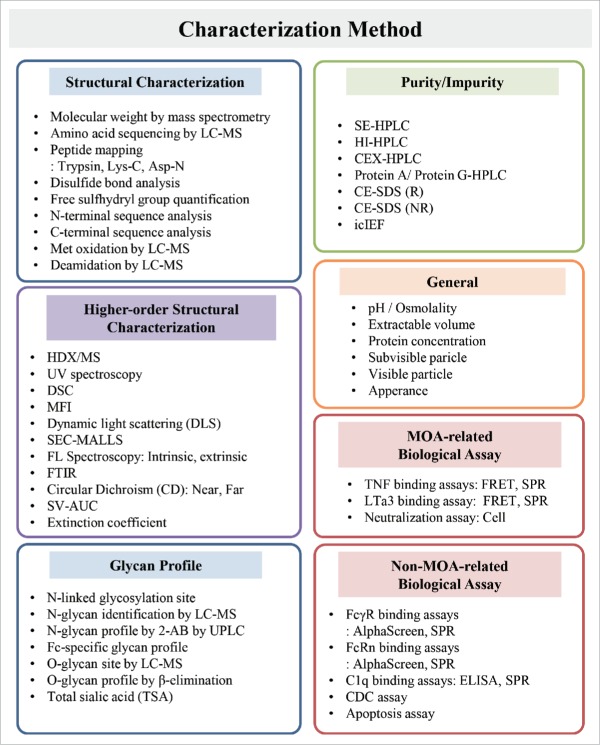
SB4 was developed as a biosimilar of etanercept, in accordance with ICH guidelines and the current FDA and EMA guidelines on the development of biosimilar products. These documents provide guidance on test procedures and acceptance criteria for biotechnological/biological products,8 as well as guidance on quality considerations when similarity is assessed.9 We describe herein a subset of the 42 state-of-the-art methods of structural and physicochemical analysis and the 19 methods of biological analysis that were performed to evaluate the degree of similarity between SB4 and the reference product (Fig. 1 and Table 1).
Figure 1.
Characterization methods classified according to quality attributes.
Table 1.
Summarized attributes and analytical conclusions of the biosimilar SB4 in comparison with the reference product (EU-sourced Enbrel®) following extensive similarity exercises.
| Attribute | Test Method | Key Finding (Conclusion) | |
|---|---|---|---|
| Protein molecular weight | Intact protein measurement by LC-ESI-MS | Similar to the reference product | |
| Amino acid sequence | Peptide mapping by LC-ESI-MS/MS using a combination of digestion enzymes/carboxypeptidase/sialidase/PNGase F | Identical to the reference product | |
| N-terminal sequence | |||
| C-terminal sequence | |||
| Peptide mapping | |||
| Disulfide bonds | |||
| Methionine oxidation | |||
| Free sulfhydryl | Fluorescence detection kit, LC-ESI-MS/MS | ||
| Asparagine deamidation | Peptide mapping, Protein isoaspartyl methyltransferase assay | Minor, non-significant differences and similar to the reference product | |
| Charge heterogeneity | Cation exchange chromatography | Slightly higher acidic variants in SB4, but not significant | |
| Imaged capillary isoelectric focusing | |||
| Glycan profile | N-linked glycosylated site | Peptide mapping after PNGase F treatment | Identical to the reference product |
| N-linked glycan identity | Peptide mapping by procainamide labeling | ||
| N-linked glycosylation quantity | Hydrophilic interaction chromatography by 2-AB labeling | Slight differences in afucosylated glycan content and neutral galactosylated glycan content, but no impact on ADCC and CDC, respectively | |
| O-glycan occupancy | O-glycan occupancy by intact protein measurement, | Slightly lower O-glycan occupancy in SB4, but not significant | |
| O-glycosylated site | Peptide mapping by liquid chromatography-electrospray ionization-tandem mass spectrometry | Identical site to the reference product | |
| Sialic acid content | Ion exclusion chromatography | Similar to the reference product | |
| Higher-order structure | Secondary structure | Fourier transform infrared spectroscopy | |
| Circular dichroism | |||
| Solvent accessibility | Hydrogen/deuterium exchange-mass spectrometry | ||
| Thermostability | Differential scanning calorimetry | ||
| Subvisible particles, μm | Micro-flow imaging | Lower particle concentrations in SB4 | |
| Subvisible aggregates, nm | Dynamic light scattering | Similar to the reference product | |
| Purity | Capillary electrophoresis-sodium dodecyl sulfate | ||
| Size exclusion chromatography | Lower aggregate content in SB4 | ||
| High-molecular-weight species | Size exclusion chromatography –multi-angle laser light scattering | ||
| Sedimentation velocity-analytical ultracentrifugation | |||
| Hydrophobic species | Hydrophobic interaction chromatography | Lower level of Peak 3 (product-related impurities) in SB4 | |
| Biological activities | TNFR-related binding | TNF binding (FRET, SPR) assay | Similar to the reference product |
| LTα binding (FRET,SPR) assay | |||
| TNF neutralization assay | |||
| Fc-related binding (FRET; SPR) | FcRn, FcγRIIa, FcγRIIb, FcγRIa, FcγRIIIa, and FcγRIIIb binding assay | Similar; slightly higher in FcγRIa binding activity but not significant | |
ADCC, antibody-dependent cell-mediated cytotoxicity; CDC, complement-dependent cytotoxicity; FRET, fluorescence resonance energy transfer; LTα, lymphotoxin α; SPR, surface plasmon resonance; TNF, tumor necrosis factor; TNFR, tumor necrosis factor receptor.
Results
Highly sensitive, orthogonal methods were used to compare the physicochemical, biophysical, and biological quality attributes of SB4 with those of the reference product, etanercept. A total of 61 test items were used for these comparisons, and several CQAs were evaluated by different orthogonal methods (Fig. 1, Fig. 2, and Table 1), including those that can detect low but immunogenically relevant amounts of molecular variants, such as HMW aggregates, NGNA, or α-1,3-galactose. Depending on the specification, the similarity range was set as one-sided (upper or lower limit as acceptance criterion) or 2-sided (upper and lower limits as acceptance criteria). Because the acceptance criteria for impurities are defined by a limit and not within a range, one-sided similarity ranges were used for evaluation of the higher-the-better or lower-the-better attributes (e.g., purity of the main peak that resulted from capillary electrophoresis-sodium dodecyl sulfate [CE-SDS], %HMW content seen after size exclusion chromatography [SEC], proportion of overall content of SB4 and the reference product represented by Peak 1, Peak 2, and Peak 3 that were the product of hydrophobic interaction chromatography [HIC]). Two-sided ranges were used for evaluation of attributes that were regarded as better when within a range (e.g., TSA quantitation, imaging capillary isoelectric focusing [icIEF], and tumor necrosis factor [TNF] binding activity). Also, a separate range for each geographical region was calculated with more than 30 batches of EU-marketed Enbrel® and more than 30 batches of US-marketed Enbrel®; the purpose of these calculations was to exclude cross-region variability. Concerning the temporal variability in product quality, the literature contains a report of a shift between the lots of Enbrel® whose expiry dates occurred in or before March 2012 and those whose expiry dates occurred thereafter.10 However, only the latter lots were used for SB4 development, and they all exhibited comparable product quality (data not shown). This approach is in line with the regulatory requirements of both the FDA and EMA, and with their opinions on the best practices in biosimilar development.9,11,12
Figure 2.
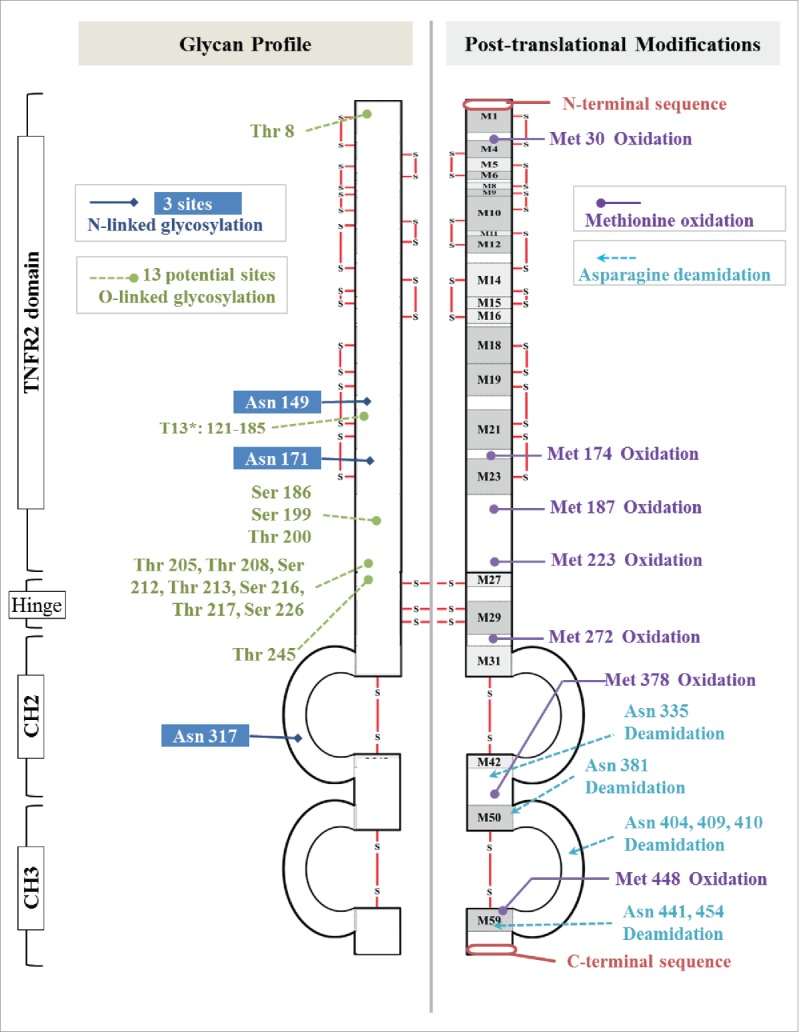
Post-translational modifications in etanercept.
Primary structure and disulfide linkage
According to the EMA and FDA guidelines, the amino acid sequence of a biosimilar must be the same as that of the reference product.9,11,12 Although the amino acid sequence of a protein can be determined indirectly from the DNA that encodes the protein, peptide mapping provides in-depth information about post-translational modifications of the primary sequence. To determine the primary amino acid sequence of SB4, we used the liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) peptide mapping approach, which provided 100% sequence coverage. In addition, we performed disulfide linkage mapping, site-specific N-/O-glycopeptide profile analysis, and N-/C-terminal sequencing, and we evaluated the level of deamidation, oxidation, and N-/C-terminal variants. For these types of analyses, SB4 was digested with different enzymes and under a variety of conditions (e.g., reduced and non-reduced, deglycosylated and non-deglycosylated, and desialylated and non-desialylated conditions). The results of these analyses demonstrated that the examined characteristics of SB4 were similar to those of the reference product.
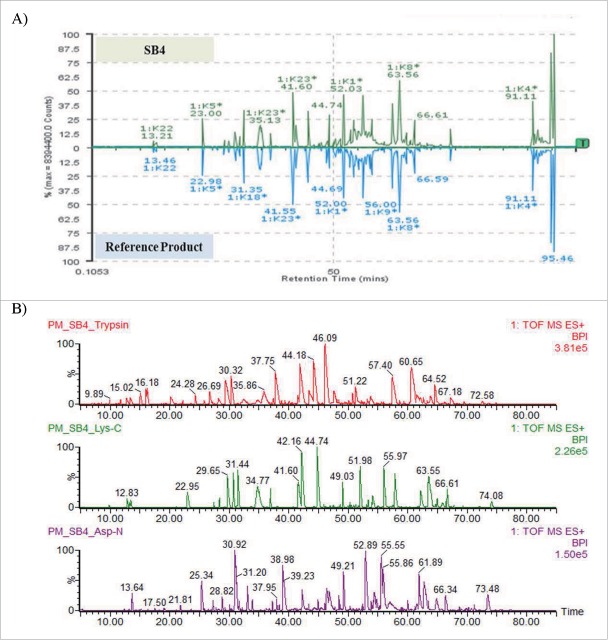
The mirror images of the peptide chromatograms of Lys-C–digested SB4 and reference product showed high similarity in peak intensities and retention times (Fig. 3A). Trace peaks were also comparable, and no new peaks were apparent in the SB4 chromatogram when it was compared with that of the reference product. The amino acid sequences of SB4 and etanercept were identical as determined by the sequencing of peptides generated by 3 enzymes (i.e., Lys-C, trypsin, and Asp-N, Fig. 3B) and by accurate mass determination with complete or partial tandem mass spectrometry (MS/MS) data. This analysis allowed for 100% sequence coverage (data not shown). Similar to the N-terminal amino acid sequence of the reference product (data not shown), the N-terminus of SB4 was heterogeneous: tryptic peptides with an intact N-terminal region (1-LPAQVAFTPYAPEPGSTCR-), an N-terminal region that lacked leucine (1-PAQVAFTPYAPEPGSTCR-), and an N-terminal region that lacked leucine and proline (1-AQVAFTPYAPEPGSTCR-) were detected.
Figure 3.
Identification of the primary sequences of SB4 and the reference product. (A) Mirror images of chromatograms of Lys-C–generated peptides of SB4 and the reference product, etanercept. (B) Comparison of peptide maps resulting from digestion with trypsin, Lys-C, and Asp-N.
Although the N-terminal variants were present, they did not affect biological activity. Results of the TNF binding and TNF neutralization assays showed no correlation between the amount of N-terminal variants present and biological activity (data not shown). Furthermore, the biological activities of the C-terminal regions with or without the terminal lysine residue of SB4 and the reference product were demonstrated to be similar in several biological function assays and the SAR study (data not shown). Also, multiple published articles have reported that the presence of C-terminal variants of monoclonal antibodies is not correlated with the biological activities, pharmacokinetics, and pharmacodynamics of these antibodies, and that this heterogeneity in C-terminal regions is not clinically relevant.13,14
Disulfide mapping of SB4 and the reference product was conducted under non-reducing conditions using trypsin and Asp-N enzymes for digestion. A single chain of etanercept had 13 intrachain disulfide linkages formed by 26 cysteine residues and 3 interchain disulfide linkages between Cys240, Cys246, and Cys249 that promote dimerization. Therefore, all 58 cysteine residues (29 residues per single chain) within the etanercept dimer participate in disulfide linkages.
Of the 9 disulfide-linked peptides that were identified, 4 (M5-M8, M27-M27, M31-M42, and M50-M590) were dipeptides with one disulfide bond, and one (M29-M29) was a dipeptide with 2 disulfide bonds (Fig. 2, Fig. 4 and Table 2). The remaining 4 disulfide-linked peptides were composed of more than 3 peptides. Eleven disulfide bonds were in the TNFR region, 2 were in the Fc region, and 3 were in the hinge region. Identical sets of disulfide-bonded peptides were also detected in the reference product.
Figure 4.
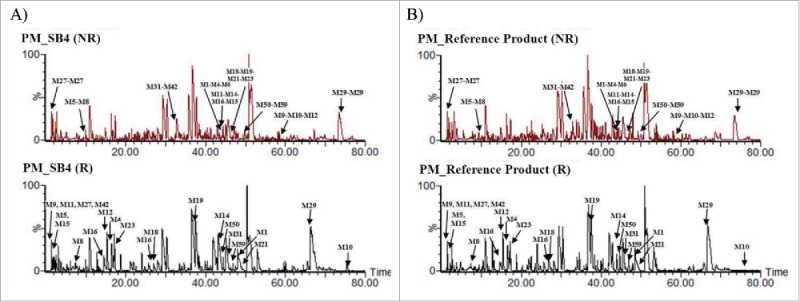
Comparison of disulfide-linked peptides of SB4 and the reference product. (A) Non-reduced (upper panels) and reduced (lower panels) peptide maps of SB4. (B) Non-reduced (upper panels) and reduced (lower panels) peptide maps of the reference product.
Table 2.
Disulfide-linked peptide map of the etanercept biosimilar, SB4.
| No. | Region | Type of Disulfide Bond | Disulfide-Linked Peptides | No. of Disulfide Bonds | Expected m/z (Charge State) |
|---|---|---|---|---|---|
| 1 | TNFR region | Intrachain | M1(1–19)-M4(25–34)-M6(43–47) | 2 | 742.94 (5) |
| 2 | M5(35–42)-M8(50–53) | 1 | 631.27 (2) | ||
| 3 | M9(54–57)-M10(58–77)-M12(81–90) | 2 | 789.33 (5) | ||
| 4 | M11(78–80)-M14(95–108)-M16(114–119)-M15(109–113) | 3 | 788.86 (4) | ||
| 5 | M18(121–135)-M19(136–148)-M21(155–170)-M23(175–185) | 3 | 818.25 (7) | ||
| 6 | Hinge region | Interchain | M27(239–240)-M27(239–240) | 1 | 415.10 (1) |
| 7 | M29(243–268)-M29(243–268) | 2 | 780.26 (7) | ||
| 8 | Fc region | Intrachain | M31(276–284)-M42(341–342) | 1 | 597.30 (2) |
| 9 | M50(381–390)-M59(437–459) | 1 | 641.81 (6) |
TNFR, tumor necrosis factor receptor.
Peptide number was automatically generated by software. These peptides were digested with trypsin and Asp-N after N-deglycosylation.
Other sequence variants generated by post-translational modifications such as oxidation and deamidation were revealed by LC-ESI-MS/MS. The relative oxidation level of residues Met187 and Met272 in SB4, which are sensitive to oxidation, was similar to that in the reference product (Table 3). This finding suggests that the relative susceptibility of the methionine residues to oxidation was consistent between SB4 and the reference product. Also, the relative deamidation levels of 4 SB4 peptides that contained 7 asparagine residues (Asn335, Asn381, Asn404, Ans409, Ans410, Asn441 and Asn454) were similar to those of the reference product (data not shown).
Table 3.
Relative oxidation level of 2 sensitive methionine residues of the biosimilar SB4 compared with those of the reference product, EU-sourced Enbrel®.
| %Oxidation |
|||
|---|---|---|---|
| Product | Sample | Met187 | Met272 |
| SB4 | 1 | 2.9 | 13.9 |
| 2 | 3.1 | 11.9 | |
| 3 | 3.0 | 11.2 | |
| Reference | 1 | 2.0 | 14.8 |
| 2 | 2.2 | 10.5 | |
| 3 | 2.0 | 10.5 | |
Molecular size
We performed LC-ESI-MS to measure the molecular mass of a reduced, N-deglycosylated and desialylated single chain of SB4. There are 13 potential O-glycosylation sites in a single chain of etanercept (Fig. 2), but the major O-glycosylated forms contained 8 to 10 O-glycan units (10 GalNAcβ(1,3)Gal). The average mass of a single chain of SB4 with 10 O-glycan units was determined to be 54,763 Da, which was within 0.01% of its theoretical mass. This average mass was identical to that of the reference product (Fig. 5A).
Figure 5.
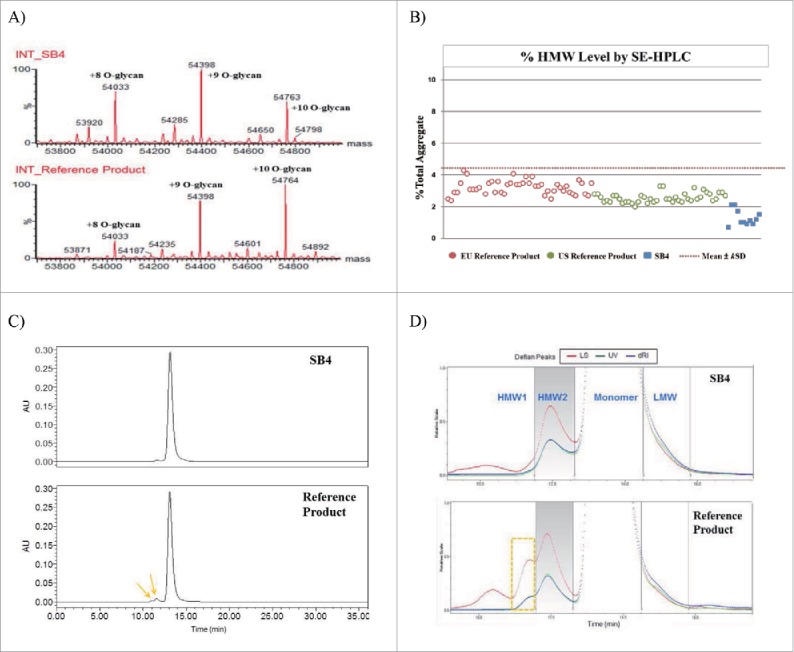
Comparison of protein mass and purity of SB4 and the reference product. (A) Chromatograms of deconvoluted intact protein mass of SB4 and the reference product. Both had been reduced, desialylated, and N-deglycosylated; 8 to 10 O-glycan moieties remained. (B) Comparative dot plot and similarity range of HMW aggregates detected by SEC. (C) Size exclusion chromatograms of SB4 and the reference product (arrows indicate HMW aggregates). (D) Enlarged images of chromatograms obtained by SEC-MALLS of SB4 and the reference product in different detectors (LS, light scattering system; UV, ultraviolet–visible spectroscopy; dRI, differential refractive index).
Based on this result for a reduced, N-deglycosylated, and desialylated single chain of SB4, we estimated the molecular mass of an SB4 single chain. A single polypeptide chain of etanercept containing 3 N-glycan–occupied sites and up to 16 mol/mol chain of sialic acid, has an approximate molecular weight of 65 kDa. Hence, the molecular weight of an SB4 homodimer was estimated to be 130 kDa.
Size heterogeneity in biologic products, especially size heterogeneity resulting from aggregation, is known to enhance immunogenicity and affect safety and efficacy.15,16 Therefore, we evaluated the size heterogeneity, including the presence of aggregates, of SB4 and the reference product. In SEC analysis of SB4, a single HMW peak and LMW shoulder peak were detected along with the monomer peak (approximately 120 kDa as measured by SEC-MALLS). The heterogeneity of HMW aggregate differed between SB4 and the reference product. SB4 consisted of lower quantities of HMW species (Fig. 5B), and these species appeared as a single HMW peak on the chromatogram (Fig. 5C and Fig. 5D). However, SEC analysis of the reference product revealed 2 HMW species (HMW1 and HMW2) and a LMW shoulder partially resolved from the main peak (Fig. 5D). Because of the molecular weight of the monomer, the HMW1 peak of the reference product was considered to be a tetramer whose mass ranged from 493 to 498 kDa, and the HMW2 peak was considered to be a dimer whose mass ranged from 264 to 269 kDa. The molecular weight of the HMW2 peak of SB4 was calculated to be 212 to 233 kDa, which was slightly smaller than the theoretical molecular weight of the dimer, which was 240 kDa (2× the size of monomer as indicated by SEC-MALLS) (Table 4). This difference was considered to be due to the overlap of the HMW2 peak and the monomer peak. Also, the size differences between HMW2 of SB4 (212–233 kDa) and that of the reference product (264–269 kDa) may be caused by the poor resolution of HMW1 from HMW2 in the reference product. As the reference product contained an additional HMW1 peak not seen with SB4, poor resolution of HMW1 and HMW2 species of the reference product affected the exact molecular weight measurement of the HMW2 species. This altered measurement is believed to have resulted in the molecular weight difference between HMW2 of SB4 and the reference product. However, SB4 and the reference product showed similar proportions of HMW: 2.3%–3.1% and 2.9%–3.4%, respectively. Only the reference product had a HMW1 peak, and this peak contributed to the greater HMW content in the reference product. Additionally, since the SEC-MALLS signal (LS) in the HMW1 region does not coincide with any UV or RI signal, it is unlikely to represent protein species.
Table 4.
Results of SEC-MALLS analysis of the biosimilar SB4 and the reference product, EU-sourced Enbrel®.
| MW (kDa) |
%Area |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Product | Sample | HMW1 | HMW2 | Monomer | LMW | HMW1 | HMW2 | Monomer | LMW |
| SB4 | 1 | N/A | 219 | 120 | 96 | N/D | 2.7 | 93.3 | 3.8 |
| 2 | N/A | 212 | 120 | 102 | N/D | 2.3 | 93.7 | 3.9 | |
| 3 | N/A | 233 | 120 | 95 | N/D | 3.1 | 92.5 | 4.4 | |
| Reference | 1 | 498 | 264 | 120 | 91 | 0.7 | 2.7 | 94.1 | 2.5 |
| 2 | 493 | 265 | 120 | 90 | 0.5 | 2.4 | 94.7 | 2.9 | |
| 3 | 494 | 269 | 119 | 91 | 0.7 | 2.7 | 93.7 | 2.9 | |
HMW, high molecular weight; LMW, low molecular weight; MW, molecular weight; N/A, not applicable; N/D, not detected; SEC-MALLS, size exclusion chromatography–multi-angle laser light scattering
The LMW form of SB4 and the reference product had an estimated mass of 95 to 102 kDa and 90 to 91 kDa, respectively. The LMW content as represented by the LMW peak in the SB4 chromatogram was 3.8% to 4.4%. The LMW content of the reference product represented by the corresponding peak ranged from 2.5% to 2.9%. The differences in the molecular weight and content proportions were due to the poor separation of the LMW species from the main monomer. Also, the signal from LMW was distinct enough to allow chromatographic peak integration with the reference product, but not with SB4. Therefore, the %LMW of SB4 was calculated by applying the boundary of retention time that had been identified with the reference product.
Purity and product-related impurities
Degradation products, charge heterogeneity, and other product-related impurities are important quality attributes for safety and potency. Purity and product-related impurities of SB4 were compared with those of the reference product by several methods, including SEC, CE-SDS under reduced and non-reduced conditions, imaged capillary isoelectric focusing (icIEF), and HIC.
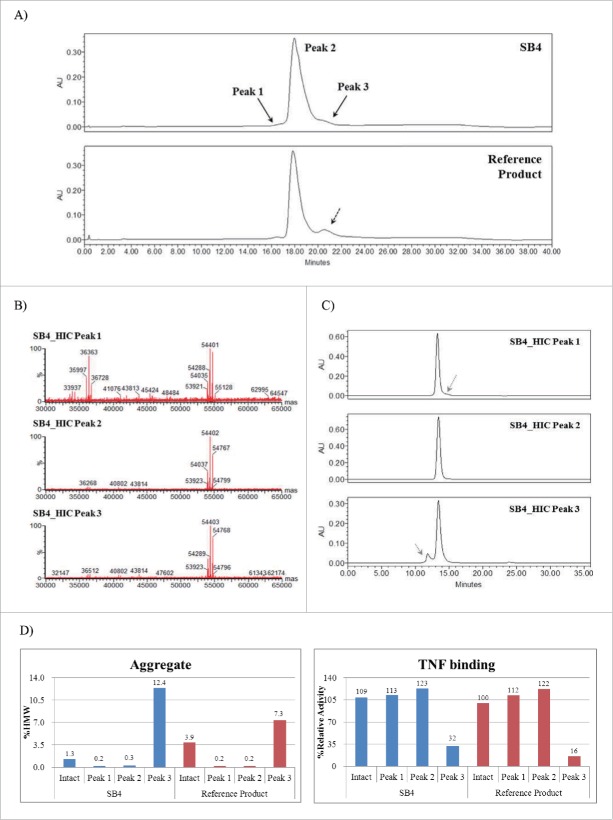
SB4 and the reference product were subjected to HIC, and the resulting chromatogram showed 3 peaks: Peak 1 (a truncated molecule), Peak 2 (an active molecule), and Peak 3 (aggregate and disulfide-scrambled species) (Fig. 6). Each peak on the HIC chromatogram was fractionated, and each fraction was characterized by LC-ESI-MS/MS, SEC, and biological activity assays. Peak 1 was found to contain a truncated single chain (size of 36,363 Da) (Fig. 6B). A truncated and misfolded form of etanercept was reported earlier in US Patent 7294481 (Method for producing recombinant proteins).17 Peak 3 was also found to contain an aggregate (Fig. 6C and Fig. 6D) and disulfide scrambled species (data not shown). The content of Peak 3 derived from SB4 represented 5.4% to 6.9% of its overall content, and that derived from the reference product ranged from 13.5% to 13.6% of its overall content. This result was consistent with that of the SEC analysis, which showed a higher proportion of aggregate in the reference product. The relative TNF binding activity was 32% for the Peak 3 fraction of SB4 and 16% for the Peak 3 fraction of the reference product (Fig. 6D).
Figure 6.
Structure-activity relationship (SAR) results for hydrophobic variants of SB4 and the reference product. (A) Comparative HIC chromatograms of SB4 (upper panel) and the reference product (lower panel). (B) Intact protein mass of each HIC peak. Fragmented protein (36,363 Da) was detected in Peak 1. (C) Size exclusion chromatograms for each HIC fraction of SB4. (D) Relative amounts of HMW aggregate in intact protein and HIC fractions corresponding to Peaks 1–3 and relative TNF binding activities of intact protein and each fraction of interest (Peaks 1–3).
SEC resulted in poor resolution between the monomer and LMW; therefore, the accurate quantitation of LMW impurities could not be achieved by SEC analysis. CE-SDS analysis was used to evaluate purity and LMW impurities in SB4 and the reference product. The electrophoretic profiles of SB4 and the reference product showed similar shapes and patterns of peaks, including those for LMW peaks. This similarity indicated that the purity of the main SB4 peak was similar to that of the reference product under reduced and non-reduced conditions (Fig. 7).
Figure 7.
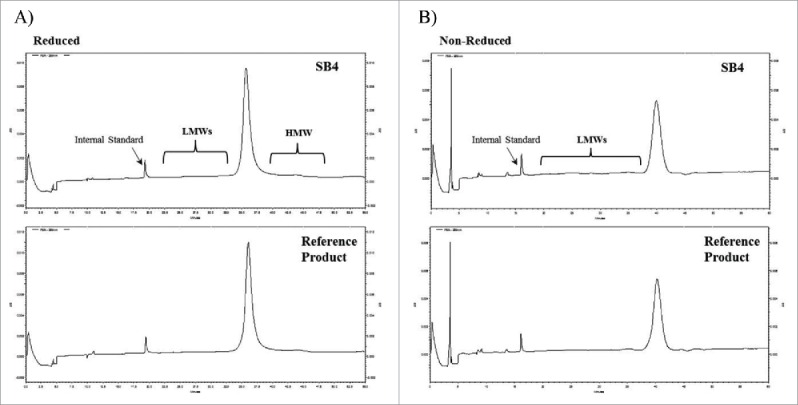
Comparison of CE-SDS electropherograms of SB4 and the reference product under reduced and non-reduced conditions. (A) SB4 (upper panel) and the reference product (lower panel) under reduced conditions. (B) The same 2 products under non-reduced conditions.
Glycosylation
Glycosylation plays a significant role in function, efficacy, clearance, and immunogenicity of a protein. We compared the glycosylation profile of SB4 with that of the reference product. Etanercept has 3 N-glycosylation sites and 13 potential O-glycosylation sites. Two N-linked glycans, most of which are sialylated, are located on the TNFR moiety and one N-linked glycan in the Fc region.18 The O-linked glycan species also contains sialic acid. The glycan species in SB4 and the reference product were enzymatically or chemically released, chromatographically separated, quantified, and identified.
A total of 23 peaks corresponding to N-glycan structures were detected by hydrophilic interaction liquid chromatography (HILIC) with a fluorescence detector (Fig. 8), and 21 species of N-glycan were identified in SB4 by LC-ESI-MS/MS. Each HILIC peak revealed an identical MS/MS spectrum between SB4 and the reference product. This result suggested that the structure of each N-glycan species on SB4 was identical to that of the corresponding species on the reference product and that there was no N-glycan structure specific only to SB4. As reported by DiPaola et al,18 the structure and number of N-linked glycans on the TNFR region and on the Fc region differ. To compare the site-specific profiles of N-linked glycans, the glycopeptide profile obtained by peptide mapping was used. Three similar kinds of profiles were derived for 3 glycosylation sites in both SB4 and the reference product: Asn147 and Asn179 on the TNFR region and Asn317 in the Fc region (Fig. 9). Based on the corresponding mass, we determined that the major forms on SB4 and the reference product are G2 and G2S1 on Asn147, G2F and G2FS1 on Asn179, and G0F and G1F on Asn317. The relative quantities of each species differed slightly between SB4 and the reference product.
Figure 8.
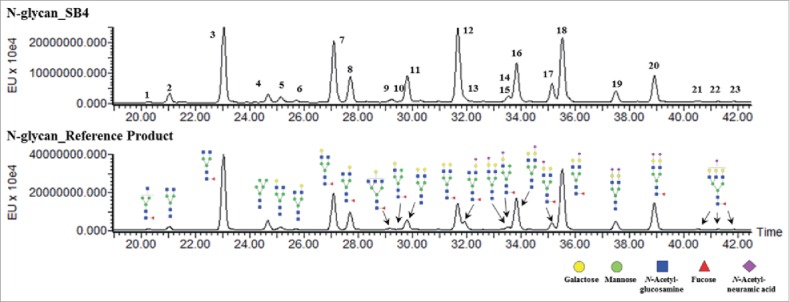
Characterization of N-glycan species of SB4 (upper panel) and the reference product (lower panel). There is no unique peak detectable in SB4 and the reference product.
Figure 9.
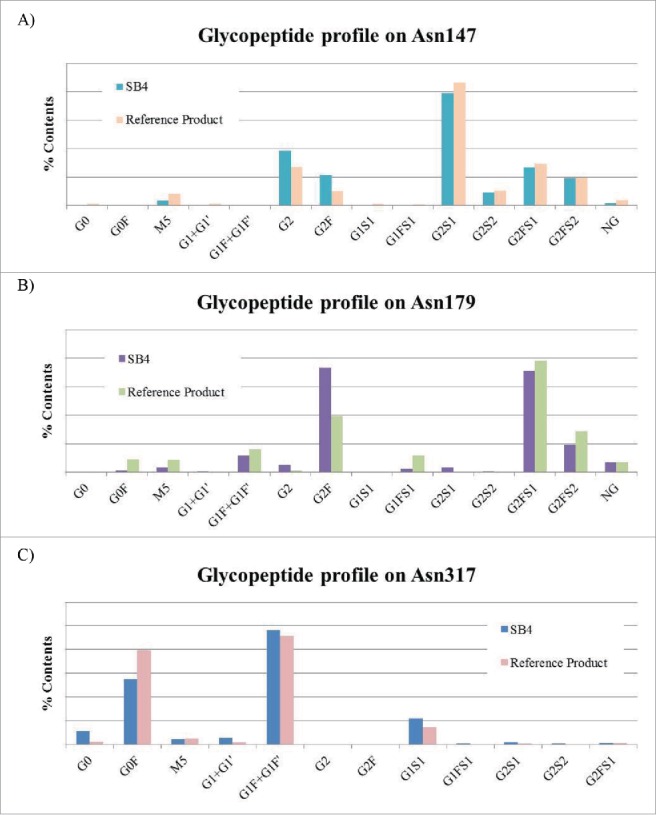
Comparison of the glycopeptide profiles. (A) Glycopeptide profile of Asn147. (B) Glycopeptide profile of Asn179. (C) Glycopeptide profile of Asn317. Results of the site-specific quantitation for the glycopeptides are shown.
In the O-linked glycosylation analysis, most O-glycan peaks with sialic acid that were shown in the chromatogram of the reference product were also detected in SB4. Not only were 3 major O-glycan forms (Peaks 1–3) observed, but also N-glycan forms followed the O-glycan forms on the chromatogram (Fig. 10A). Based on this result, we concluded that the molar concentration of O-glycan was higher than that of N-glycan on both SB4 and the reference product. Peak 1 represented the product derived from the pretreatment for β-elimination, and Peaks 2 and 3 represented monosialylated and disialylated O-glycan, respectively.
Figure 10.
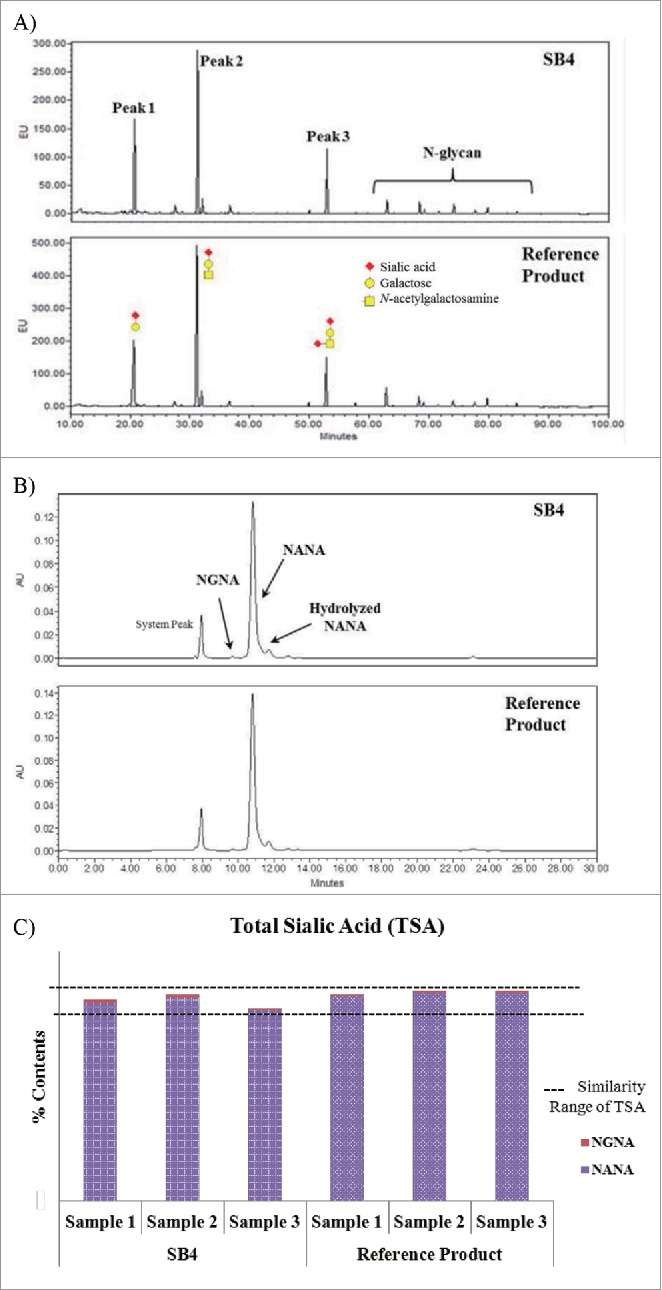
Comparison of O-glycan profiles, TSA chromatograms, and TSA content of SB4 and the reference product. (A) HILIC chromatograms of 2-AB–labeled O-glycan species of SB4 (upper panel) and the reference product (lower panel). (B) Ion exclusion chromatography of sialic acid moieties of SB4 and the reference product. (C) Graphical comparison of TSA content of SB4 and the reference product.
The sialic acid content of a protein is regarded as an important element in determining its half-life in serum.19 The molar concentration of sialic acid was measured by ion exclusion chromatography. The chromatograms showed peaks for SB4 that were identical to those for the reference product (Fig. 10B), and the relative amounts of sialic acid for each peak and the sum of NANA and NGNA (i.e., TSA) were similar (Fig. 10C).
Higher-order structure
Secondary and tertiary structures of relatively small proteins can be reliably evaluated with circular dichroism (CD) and Fourier transform infrared spectroscopy (FTIR); however, large biotherapeutic proteins such as antibodies or Fc fusion proteins such as etanercept are too complex for their secondary structures to be analyzed by these methods. In addition, these large, complex proteins must be folded into a proper 3-dimensional structure to become functional. State-of-the-art methodology has been increasingly requested by regulatory agencies for demonstration of structural similarity between a biosimilar and its reference product.9 Such state-of-the-art methods include hydrogen/deuterium exchange (HDX) and differential scanning calorimetry (DSC), each of which can be used to assess higher-order dynamic structure. Combined with MS, HDX can characterize the solvent accessibility around the surface of a molecule, which reflects overall structural conformation and detects distinguishing structural differences. HDX/MS provides reliable comparative information regardless of protein size.20 DSC can characterize a protein's thermal stability, overall conformation, and folding integrity of each domain.
We compared the structure of SB4 and the reference product by using HDX, DSC, dynamic light scattering (DLS), fluorescence spectroscopy, FTIR, and far-UV CD spectroscopy. The results of all structural analyses of the 2 products were highly similar, but the HDX and DSC methods were regarded as the most sensitive and thus the most informative methods; therefore, the results gathered by these methods are presented here.
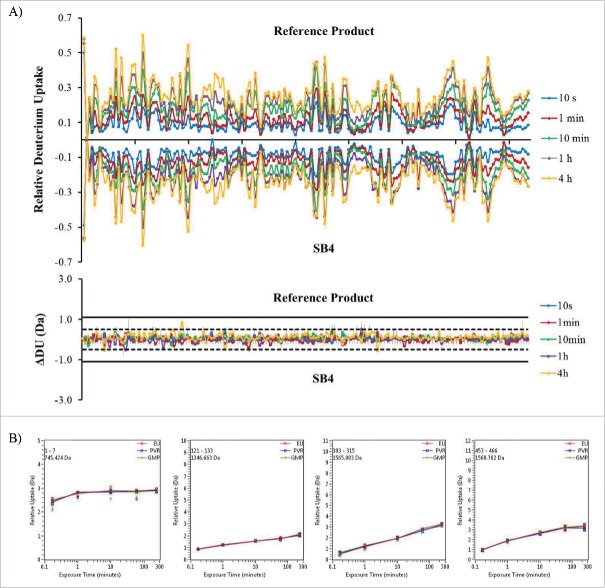
So-called “butterfly plots” of the dynamics of deuterium uptake revealed almost perfect symmetry between SB4 and the reference product (Fig. 11A); this result indicated that SB4 and the reference product conformations were rendered similar in solvent accessibility. The individual uptake rates of peptides from SB4 and the reference product are demonstrated by kinetic curves (Fig. 11B). Analysis of 167 peptides, which covered 92.5% of the SB4 sequence (including the sequences of multiple glycosylated peptides), yielded kinetic curves overlapping those of the reference product.
Figure 11.
Deuterium uptake “butterfly” plots by SB4 and the reference product. (A) Comparison of deuterium uptake over time (10 seconds to 4 hours) by SB4 and the reference product. (B) Representative deuterium uptake rates of peptides including the N-terminus (amino acids 1–7), the middle of the TNFR region (amino acids 121–133), the middle of the Fc region (amino acids 303–315), and C-terminus (amino acids 453–466).
The thermal stabilities of SB4 and etanercept at elevated temperatures were evaluated by DSC. The thermograms of SB4 and the reference product showed 3 transition temperatures that were designated Tm1, Tm2, and Tm3 (Fig. 12). These transition temperatures, which are associated with the unfolding of the TNFR (Tm1), CH2 (Tm2), and CH3 (Tm3) domains,21 were similar for the 2 products: the dominating peaks of the superimposed melting curves for SB4 and the reference product were detected near 56.7°C to 57.0°C for Tm1, 69.3°C to 69.6°C for Tm2, and 82.5°C to 82.8°C for Tm3.
Figure 12.
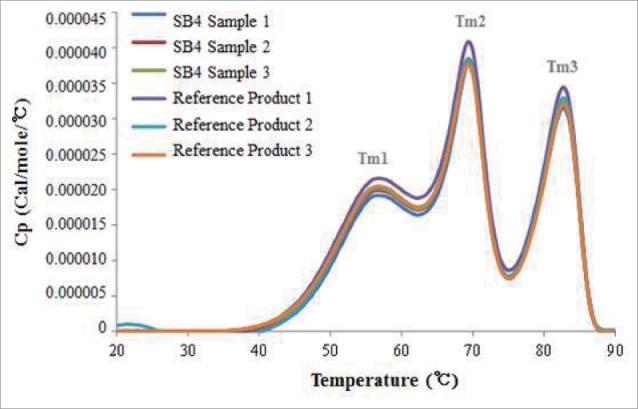
Thermograms from differential scanning calorimetry. Comparative thermograms of SB4 and the reference product show 3 similar, distinctive melting points (Tm).
Biological activities
The in vivo MOA of etanercept is the inhibition of TNF activity. Etanercept reduces inflammation by competitively inhibiting the binding of TNF (a proinflammatory cytokine) and lymphotoxin α (LTα) to TNF receptors on the cell surface, and thus rendering TNF molecules biologically inactive. Based on this MOA, 15 biological assays were employed to evaluate the biological quality of SB4. The assays were arranged into 3 categories: (1) TNFR2-related binding assays, including human TNF binding, LTα binding, and orthologous TNF binding assays; (2) Fc-related binding assays, including those that involved FcγRIa, FcγRIIa, FcγRIIb, FcγRIIIa (V158 allotype), FcγRIIIa (F158 allotype), FcγRIIIb, FcRn, and C1q; and (3) cell-based assays, including those that involved TNF neutralization, CDC, apoptosis, and ADCC. Results of the assays that evaluated TNF binding activity, LTα binding activity, TNF neutralization, and apoptosis are shown in Figs. 13 and 14.
Figure 13.
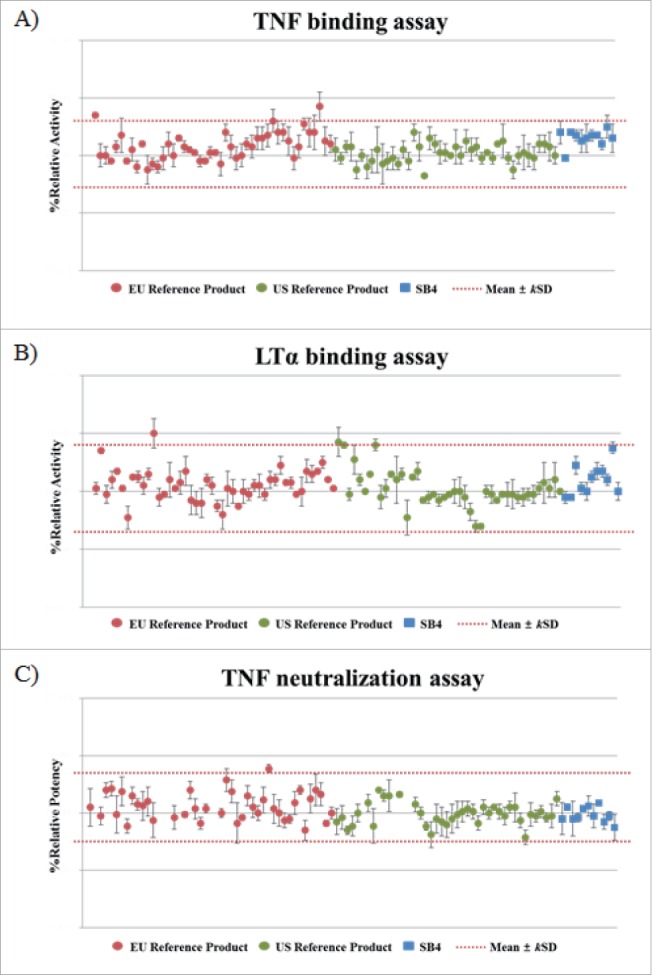
Comparison of the biological activities of SB4 and the reference product obtained from the EU and US reference product. (A) Relative TNF binding activity as shown by FRET assays. (B) Relative LTα binding activity as shown by FRET assays. (C) Relative potency as indicated by a TNF neutralization assay. The dotted line indicates the similarity range (mean±kSD) based on results of etanercept obtained from the EU.
Figure 14.
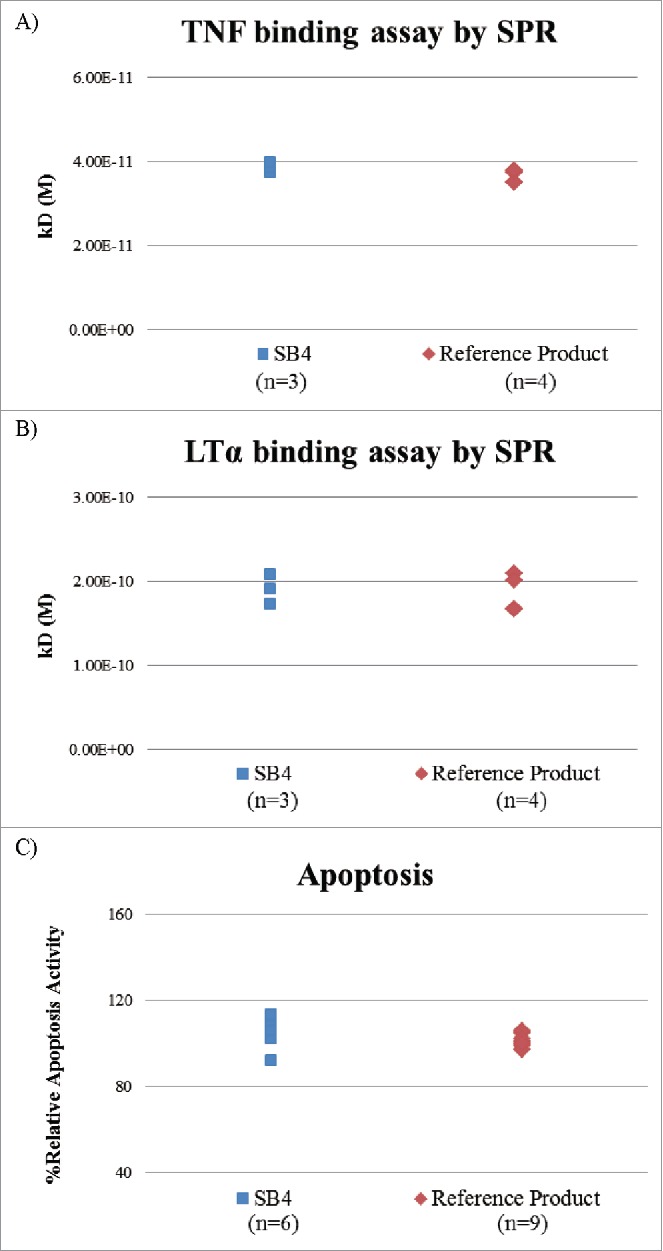
Comparison of TNF- and LTα-binding affinity of SB4 and the reference product by surface plasmon resonance and apoptosis. (A) Absolute binding affinity to TNF. (B) Absolute binding affinity to LTα. (C) Apoptosis activity using mTNF-expressing cell line.
In the fluorescence resonance energy transfer (FRET) assay, the range of relative TNF- and LTα-binding activity of SB4 was similar to that of the reference product from the 46 lots from the EU and from the 43 lots from the US (Fig. 13A and 13B), i.e., the range was within the predetermined similarity range, was steadily updated, and was presented as the mean ± kSD. Evaluation of the potency of SB4 by the TNF neutralization assay using the luciferase reporter gene found the relative potencies of SB4 and etanercept from the 40 lots from the EU and the 40 lots from the US were also similar (Fig. 13C).
To demonstrate the absolute binding affinity and to evaluate the possible FRET labeling-induced interference in relative binding activity, the affinities of TNF and LTα to etanercept were evaluated by surface plasmon resonance (SPR), which does not require labeling. SB4 and the reference produce showed similar binding affinity to TNF and LTα (Fig. 14A and 14B).
Biological activity was also evaluated in an apoptosis assay using a cell line that expressed membrane-bound TNF (mTNF). This assay showed similar activity between SB4 from 6 batches and reference product from 9 lots (Fig. 14C).
Discussion
The focus of ICH Guideline Q5E is the assessment of comparability between different batches of a biological product22 but it is often cited in connection with the assessment of similarity between a proposed biosimilar and its reference product in the early development of biosimilars. Demonstrating biosimilarity, however, generally requires more sophisticated methods and more comprehensive characterization than that used to assess comparability.9,11,12
More than 30 batches of each EU and US lot of reference product were used to establish biosimilarity ranges for use in monitoring any quality shift of the reference product over time. In parallel with the establishment of biosimilarity, 151 lots of the reference product etanercept from different market sources (e.g., EU, US, Canada, Australia, Japan, Switzerland, and Korea) with different expiry dates were steadily purchased and tested. For our study, 42 state-of-the-art methods were used to analyze structural and physicochemical characteristics, and 19 methods were used to analyze biological characteristics (summarized in Table 1). Most requirements to demonstrate biosimilarity of SB4 were satisfied by the use of similarity ranges and side-by-side comparisons. However, a few differences in quality (e.g., the O-glycan occupancy as indicated by intact protein measurement, differences in HMW aggregate levels, and differences in HIC Peak 3 impurity levels) were observed between SB4 and the reference product. These differences in quality were sufficiently justified by the results of a SAR study, which showed that these differences do not negatively influence the key indicators of biological activities of the biosimilar. For example, different peak heights for deconvoluted protein mass of SB4 and the reference product in Fig. 5A resulted from the difference of O-glycan occupancy on the O-glycan site of the TNFR region. The calculated O-glycan occupancy was based on the intact protein mass profile and peak area. The average O-glycan occupancy of the reference product was about 9.3 O-glycan units per chain and that of SB4 was about 8.4 O-glycan units per chain. Importantly, this difference was not a result of the unique structure or the site of O-glycan or a difference in primary sequence. Using LC-ESI-MS/MS, we analyzed the exact O-linked glycosylation site in an O-glycopeptide of SB4 and the reference product and found that each O-glycosylated site of SB4 was identical to that of the reference product.23 LC-ESI-MS/MS analysis is a powerful analytical tool to identify site-specific modification. However, in the case of relative quantitation of different forms of a molecule, this method is not adequate because the detected signal is not correlated with the relative amount of each analyte. Therefore, the relative O-glycan content at specific sites, including the hinge region, was not considered. Although O-glycan may potentially affect binding capacity, multiple critical biological and physicochemical characteristics of SB4 were similar to those of the reference product.24 Compared with the O-glycan occupancy study, the TSA assay was regarded as a more relevant test method in characterizing the biological function because it is well known that the sialic acid content of etanercept can affect product quality and the pharmacokinetic profile. In particular, the NANA content of etanercept affects the protein's half-life, and the NGNA content of the reference product has been implicated in the enhancement of immunogenicity.25 NANA is the major sialic acid present, and its relative quantity in SB4 is comparable to that in the reference product. NGNA is the minor sialic acid, and its relative quantity in SB4 is low (0.2% or below) but comparable to that in the reference product.
The product-related impurities of SB4 and the reference product were detected and quantified by orthogonal state-of-the art methods. The HIC method showed that the level of Peak 3 impurity, which consisted of both aggregates and disulfide-scrambled species, was lower for SB4 than for the reference product. This finding was in agreement with SEC-UV and SEC-MALLS analysis, which demonstrated lower levels of aggregation within SB4 samples than within the reference material. In addition, the difference in the level of Peak 3 appears to have resulted not only from HMW forms but also from disulfide-scrambled species. It is possible that the disulfide-scrambled forms of etanercept may not have resolved in SEC and in other higher-order analytical methods because of greater heterogeneity among the forms due to the cysteine-rich domain of TNFR. As various disulfide-bond scrambled variants of etanercept were eluted in Peak 3 during HIC, the greater amount of Peak 3 in the reference product can be explained not only by the greater HMW content, but also by the greater level of disulfide-scrambled species. We also demonstrated that Peak 3 has much lower biological activity than the active substance (Fig. 6D). According to multiple investigations, an unwanted immune response to a biotherapeutic product may neutralize its biological activity and result in adverse events by inhibiting the product's efficacy and by cross-reacting with an endogenous protein counterpart.14,26-28 These phenomena could be elicited by HMW aggregates and scrambled forms.29-31 Importantly, even if SB4 had lower levels of these potentially immunogenic and lower biologically active forms, the overall ability of SB4 to inhibit TNF was similar to that of the reference product. Based on these results, we hypothesize that the lower amount of Peak 3 impurities in SB4 could be beneficial to safety and immunogenicity.
The other qualities related to immunogenicity are the unique carbohydrate α-1,3-galactose glycan and the sialic acid form NGNA. The α-1,3-galactose glycan and NGNA are not endogenous, but both are linked to glycoproteins expressed in the CHO cell expression system. Within the limits of sensitivity in our N-glycan identification analysis, the α-1,3-galactose glycan of the N-glycan species was not identified in SB4 or the reference product; when the capability of the glycan analytical methods are considered, the relative amount of α-1,3-galactose glycan on either protein was less than approximately 0.1% (i.e., the limit of detection and sensitivity of the procainamide-labeled N-glycan identification method). In the case of NGNA, ion exclusion chromatography was used for the TSA analysis; the relative quantity of NGNA was no more than 0.2% in either SB4 or the reference product (Fig. 10B and 10C). Therefore, we would expect no NGNA-related immunogenicity with SB4.
Post-translational modifications such as oxidation and deamidation are known to affect the quality of a therapeutic protein. Oxidation of methionine residues and deamidation of asparagine residues were analyzed, and a similar level of modification between SB4 and the reference product was observed. Seven methionine residues exist on a single chain of etanercept. Of the 7 methionine residues, only 2 residues (Met187 and Met272) were oxidized in this study, and there was no difference between SB4 (2.9%–3.1% of Met187 and 11.2%–13.9% of Met272) and the reference product (2.0%–2.2% of Met187 and 10.5%–14.8% of Met272). The remaining 5 residues were also analyzed, but each oxidation level of the 5 methionine residues was less than 2% for both SB4 and the reference product. These low levels were not considered significant, and the 2 products showed similar oxidation profiles (Table 3).
Deamidation of all asparagine residues was also compared. Sixteen asparagine residues exist within a single chain of etanercept. Deamidation levels for 7 of the residues were similar between SB4 and the reference product, whereas the deamidation of the remaining residues was determined to be insignificant for both molecules (data not shown).
Based on the mechanism of action of etanercept, 19 biological assay assays (Table 1) were developed and tested. The biological in vitro capability of SB4 was regarded as similar to that of the reference product as demonstrated by the 3 categories of biological test methods: TNFR-related binding activity, Fc-related binding activity, and TNFR-related cell-based activity.
It is known that the fusion protein etanercept exhibits considerably lower activity in ADCC and CDC compared with full-length anti-TNF monoclonal antibodies such as infliximab and adalimumab.32 The ADCC and CDC levels for SB4 and the reference product were also lower than those for infliximab. Based on these results, we considered small differences in N-linked glycosylation between SB4 and the reference product to be non-critical. Therefore, the relative amount of each N-glycan species was not considered significant and is not addressed here in detail.
The precursor of the soluble form of TNF is mTNF, and the affinity of etanercept to soluble TNF is greater than that to mTNF.33 Because of the low affinity for mTNF, the apoptosis assay (using cells that express mTNF) is more sensitive than mere binding assays and better represents the biological activity. Similarity in biological activity between SB4 and the reference product was seen in the apoptosis assays (Fig. 14C), and this similarity in activity was considered to indicate similarity in affinity of SB4 and the reference product to mTNF.
In conclusion, biosimilarity at the quality level was demonstrated on the basis of a very comprehensive similarity exercise. In-depth characterization of differences detected in the charged variants and glycan profiles was performed, and the differences were found to have no effect on biological activity in the in vitro assays. The similarity ranges of 19 quality attributes from 15 test methods were established with the reference product, and most of the quality attributes of SB4 were within the similarity ranges. Quality attributes that were out of range indicated that SB4 contained lower levels of undesirable impurities, such as HMW and misfolded species, than did the reference product. The high degree of similarity and lower levels of undesirable impurities in SB4 provide confidence that SB4 will have a clinical profile (including efficacy) equivalent to that of the originator product, with no additional risk of impurity-related adverse reactions or immunogenicity.34
The results described herein are a part of the totality of evidence concerning the overall similarity of quality, pharmaco-toxicological, pharmacokinetic and pharmacodynamic aspects and clinical efficacy and safety that convinced the EMA to conclude that the benefit-risk profile was positive for SB4.34 SB4 is marketed under the brand names of Brenzys and Benepali.35
Materials and methods
SB4 is formulated with the same active ingredient (50 mg/mL of etanercept) and with the same inactive ingredients (sucrose, sodium chloride, and sodium phosphate) except l-arginine as Enbrel®. All the methods were qualified, or the method performance was assessed as appropriate for the intended purpose.
Peptide mapping
To achieve denaturation and reduction, each sample (200 µg) was mixed with 2 µL of 1 M dithiothreitol and 10 µL of 1 M Tris buffer, pH 8.0, and diluted with 8 M guanidine hydrochloride (final volume, 200 µL). Samples were incubated for 30 minutes at room temperature. Four microliters of 1 M iodoacetamide were then added to each sample, and each sample was incubated for 15 minutes at room temperature and in the dark. After incubation, each sample was loaded onto a 10K molecular weight cut-off spin column and subjected to centrifugation 3 times; after each centrifugation the buffer was replaced with 300 µL of 50 mM Tris HCl, pH 7.8. The sample was digested with Lys-C (Roche, 11047825001) or trypsin (Roche, 11047841001) at 37°C for 16 hours and subsequently with PNGase F (NEB, P0704L) and sialidase A (Prozyme, GK80040) at 37°C for 4 hours. For the disulfide analysis, the reducing step was not performed. After digestion of deglycosylated and desialylated samples with Asp-N (Roche, 11054589001), the deglycosylated digestion products underwent reverse-phase ultra-performance liquid chromatography (UPLC)-mass spectrometry using a BEH300 C18 column (Waters, 186003687/ 1.7 µm, 2.1 mm × 150 mm) at 60°C. Peptides were eluted by a linear gradient of 0%–35% of mobile phase B (mobile phase A, 0.1% formic acid in water; mobile phase B, 0.1% formic acid in acetonitrile) at a flow rate of 0.3 mL/min for 100 minutes and analyzed by the Synapt-G2 system. Data were collected and processed by MassLynx (Waters) v4.1.
Size exclusion chromatography
Sample (100 µg) was directly injected onto a TSK-GEL G3000 SWXl analytical column (Tosoh, 008541, 5 µm / 7.8 mm × 300 mm) at 25°C, which was connected to a Waters HPLC system; monitoring was done by UV (UV) detection (λ = 280 nm). A mobile phase consisting of 100 mM sodium phosphate with 200 mM sodium chloride, pH 6.8, was used. The flow rate was 0.5 mL/min, and monomers and impurities were detected at a UV wavelength of 280 nm. Data were acquired and processed by Empower™3 (Waters) software.
Hydrophobic interaction chromatography
For the analysis of product-related impurities, each sample was diluted with distilled water to a concentration of 1 mg/mL, and 20 µg of sample was injected onto a TSK-GEL Butyl-NPR analytical column (4.6 mm × 35 mm, Tosoh) connected to a Waters HPLC system. Product-related impurities were separated in a linear gradient using mobile phase A that consisted of 1.8 M ammonium sulfate with 0.1 M sodium phosphate, pH 7.0, and mobile phase B that consisted of 0.1 M sodium phosphate, pH 7.0. The flow rate was 1.0 mL/min, and chromatography was monitored by UV detection (λ = 214 nm). Data were acquired and processed by Empower™3 (Waters) software.
Capillary electrophoresis-sodium dodecyl sulfate
Reducing and non-reducing CE-SDS analyses were conducted with a high-performance capillary electrophoresis system (PA 800 plus Pharmaceutical Analysis System; Beckman). For the reducing condition, sample (400 µg) was mixed with 2 µL of a 10 kDa internal standard, 87 µL of SDS-MW sample buffer (Beckman coulter, A10663), and 5 µL of 2-mercaptoethanol and then boiled at 70°C for 10 minutes. For the non-reducing condition, 2-mercaptoethanol was replaced with iodoacetamide. The sample was electrokinetically introduced onto a capillary (Beckman Coulter, bare fused-silica capillary, 50 µm/30.2 cm) by applying voltage at −5 kV for 20 seconds and was separated in the capillary cartridge. Electrophoresis was performed at a constant voltage with an applied field strength of −497 V and monitored by UV detection (λ = 220 nm) through the capillary window and aperture (Beckman Coulter, 144712, 100 × 200 µm). Data were acquired and processed by 32 Karat software with integration capabilities.
N-glycan identification by procainamide labeling and LC-MS
Sample was denatured in 50 mM sodium phosphate, pH 7.5, and 1% NP-40 and treated with PNGase F to release N-glycan. After precipitation with cold ethanol and complete drying, the released N-glycan was regenerated and fluorescently labeled with procainamide (4-amino-N-(2-diethylaminoethyl) benzamide). The labeling mixture consisted of 11 mg procainamide in 100 µL of glacial acetic acid and dimethyl sulfoxide (3:7, v/v) with 6 mg of sodium cyanoborohydride. After incubation at 65°C for 3 hours, the labeled N-glycans were gradually separated by a UPLC system connected to a UPLC BEH glycan column (2.1 mm × 150 mm, 1.7 µm). Labeled N-glycans were detected and identified by a fluorescence detector (Waters) coupled to a mass spectrometer (Synapt-G2, Waters). The mass data were processed with GlycoWorkbench software and MassLynx v4.1.
O-glycan profile characterization using β-elimination
To release O-glycan moieties by β-elimination reaction, the sample (200 µg) was dialyzed in 0.1% trifluoroacetic acid solution and subsequently dried completely. The sample was treated for 18 hours with Orela™ reagent to release O-glycan moieties by β-elimination reaction and acidified with acetic acid. Released free O-glycan was purified by chromatography using a CEX cartridge and dried. After labeling with 2-AB reagent for 3 hours at 65°C, O-glycan was separated by chromatography using an Acquity™ UPLC BEH glycan column (Waters, 186004742 / 2.1×150 mm, 1.7 µm) with mobile phase A that consisted of 50 mM ammonium formate and mobile phase B that consisted of 100% acetonitrile. The flow rate was 0.5 mL/min for 100 minutes. Labeled O-glycan was detected by a fluorescence detector (λex = 330 nm and λem = 420 nm) using a Waters Acquity™ UPLC system. Data were acquired and processed by Empower™3 software.
Quantitation of total sialic acid
The quantity of TSAs, including N-acetylneuraminic acid (NANA) and N-glycolylneuraminic acid (NGNA), in each sample was evaluated by ion exclusion chromatography. Each standard of NANA and NGNA was analyzed separately alongside the carbohydrate sample. Ninety-five micrograms of sample diluted with distilled water were hydrolyzed by adding 0.1 N sulfuric acid at 80°C for 1 hour. NANA and NGNA were isocratically separated on a Rezex RHM-monosaccharide column (00H-0130-K0/ 300 × 7.8 mm) and were monitored with a UV detector (SHIMADZU; λ = 206 nm) using Empower™3 software with integration capabilities. The amounts of NANA and NGNA were calculated on the basis of the calibration curves generated from data for NANA and NGNA standards and were presented as the molar TSA amount per mole of a polypeptide chain.
Hydrogen/deuterium exchange
HDX was adapted to compare higher order structure between samples. Samples were dialyzed in 25 mM sodium phosphate and 100 mM NaCl, pH 6.3, and brought to a concentration of 2.5 mg/mL. HDX was initiated by a 1:10 dilution of sample in D2O buffer at intervals of 10 seconds, 1 minute, 10 minutes, 1 hour, and 4 hours before quenching and injecting into the mass spectrometer. Peptides were digested on an immobilized pepsin column, and the trapped peptide fragments were eluted by a gradient of 5% to 95% acetonitrile in 15 minutes. Mass spectra were collected in MSE mode, and data were analyzed by ProteinLynx Global Server™ (PLGS, Waters) to identify peptides and by DynamX software (Waters) to calculate deuterium uptake and to generate butterfly and difference plots.
Differential scanning calorimetry
A MicroCal VP-Capillary GE Healthcare) was used to analyze the melting temperature I of samples. The sample and the corresponding buffer were heated from 10°C to 95°C (heating rate of 60°C/h). The μ-DSC cell was pressurized to prevent boiling during heating. Samples were diluted to a concentration of approximately 1.0 mg/mL in the placebo buffer prior to the run.
To determine the baseline value, reference and sample cells were filled with water and scanned twice from 10°C to 95°C (heating rate of 60°C/h) to build the thermal history of the µ-DSC cells. Subsequently, the sample was analyzed, with the reference cell filled with formulation buffer and the sample cell filled with the formulation. The baseline value was subtracted from each measurement. Thermal data were normalized for protein concentration. The Tm of the protein was determined from the heating scan. Data were analyzed by Origin 7.0 DSC software.
TNF binding assay
TNF binding by samples was measured by time-resolved FRET assay. A Europium chelate-labeling, fluorophore Cy5-labeling system was used to measure TNF binding activity by competitive inhibition. Fixed concentrations and volumes of Europium chelate-labeled material and Cy5-labeled material were added to the assay plate, and these plates were incubated at ambient temperature with moderate agitation for 1 hour. The signal was measured at a wavelength of 665 nm on a microplate reader using Envision® (PerkinElmer, 2104).
Lymphotoxin α binding assay
The LTα binding assay was identical to the TNF binding assay, but LTα instead of TNF was used.
TNF neutralization assay
The reporter gene assay employed a stable reporter cell line that was engineered to contain regulatory elements upstream of a luciferase reporter gene. Mediated by TNF binding to its receptor, activation of the regulatory gene resulted in expression of the luciferase reporter gene. The expectation was that by inhibiting TNF from binding its receptor, SB4 and the reference product would downregulate indirectly luciferase reporter expression, which was reported as the neutralization effect of the sample. TNF was mixed with the assay standard, the control, or the etanercept sample and then incubated at room temperature for 20 to 40 minutes in a 96-well tissue culture plate. After incubation, cells were transferred to each well of the 96-well tissue culture plate. Cells and each mixture were incubated, and then luciferase activity was measured by the Steady-Glo Luciferase Assay System (Promega) after 24 hours of incubation.
TNF and LTα binding assay by surface plasmon resonance
The SPR technology was used to characterize the affinity and kinetics of the ligand interaction with SB4 and the reference product. TNF and LTα binding assays utilized SPR technology to determine the relative binding affinity of SB4 and the reference product to each molecule. TNF and LTα was immobilized on a CM5 sensor chip using N-hydroxysuccinimide/N-ethyl-N′-(-3-dimethylamino-propyl) carbodiimide (NHS/EDC) at a constant flow rate. Various concentrations of SB4 and the reference product were prepared by 2-fold serial dilution with HBS-EP buffer (10 mM HEPES, pH 7.4; 150 mM NaCl; 3 mM EDTA; 0.005% Tween-20). Samples were injected at a constant flow rate into the flow cell for 180 sec association and 180 sec dissociation. For regeneration, 5 M MgCl2 and 10 mM NaOH were used for TNF and LTα, respectively. Kinetic analysis was performed using a 1:1 interaction model on BIAevaluation™ software.
Apoptosis assay
The relative apoptosis activity (i.e., caspase activity) of SB4 and the reference product was determined in cells expressing mTNF. These cells were incubated with diluted sample at 37°C in a 5% CO2 incubator for 24 h. With the addition of Caspase-Glo®, the relevant luminescence signal, which was induced by caspase acting on a luminogenic peptide substrate, was collected by the microplate reader Envision® (PerkinElmer, 2104), and proportional intensities were reported.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by Samsung Bioepis Co., Ltd. We are grateful to Orlando Jaquez for sharing pearls of wisdom with us as we were writing the manuscript, and we thank Mitch Miller, PharmD, and Julia C. Jones, PharmD, PhD, MWC™, of Med Communications, Inc., for their editing of the manuscript.
References
- 1.Weise M, Bielsky MC, De Smet K, Ehmann F, Ekman N, Narayanan G, Heim H-K, Heinonen E, Ho K, Thorpe R, et al.. Biosimilars-why terminology matters. Nat Biotechnol 2011; 29:690-3; PMID:21822237; http://dx.doi.org/ 10.1038/nbt.1936 [DOI] [PubMed] [Google Scholar]
- 2.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol 2010; 10:345-52; PMID:20414207; http://dx.doi.org/ 10.1038/nri2747 [DOI] [PubMed] [Google Scholar]
- 3.US Food and Drug Administration Biologics price competition and innovation [Internet]. Silver Spring (MD): US Food and Drug Administration; [cited January20, 2016]. Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/ucm216146.pdf. [Google Scholar]
- 4.Howe WG. Two-sided tolerance limits for normal populations-some improvements. J Am Stat Assoc 1969; 64(326):610-20 [Google Scholar]
- 5.Goffe B, Cather JC. Etanercept: An overview. J Am Acad Dermatol 2003; 49:S105-11; PMID:12894133; http://dx.doi.org/ 10.1016/mjd.2003.554 [DOI] [PubMed] [Google Scholar]
- 6.Enbrel® (etanercept) US Full Prescribing Information Immunex Corporation, marketed by Amgen and Wyeth Pharmaceuticals. Thousand Oaks, CA. [Google Scholar]
- 7.Mohler KM, Torrance DS, Smith CA, Goodwin RG, Stremler KE, Fung VP, Madani H, Widmer MB. Soluble tumor necrosis factor (TNF) receptors are effective therapeutic agents in lethal endotoxemia and function simultaneously as both TNF carriers and TNF antagonists. J Immunol 1993; 151(3):1548-61; PMID:8393046 [PubMed] [Google Scholar]
- 8.International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use ICH Tripartite Guideline. Specifications: test procedures and acceptance criteria for biotechnological/biological products Q6B [Internet]. [Recommended for adoption on March 10, 1999. Accessed January20, 2016]. Available from: http://www.gmp-compliance.org/guidemgr/files/3-1-17.pdf [Google Scholar]
- 9.US Food and Drug Administration Guidance for industry: quality considerations in demonstrating biosimilarity of a therapeutic protein product to a reference product [Internet]. Silver Spring (MD): US Food and Drug Administration; [Available April 2015; cited January20, 2016]. Available from: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm291134.pdf [Google Scholar]
- 10.Schiestl M, Stangler T, Torella C, Cepeljnik T, Toll H, Grau R. Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat Biotechnol 2011; 29(4):310-2; PMID:21478841; http://dx.doi.org/ 10.1038/nbt.1839 [DOI] [PubMed] [Google Scholar]
- 11.European Medicines Agency, Committee for Medicinal Products for Human Use Guideline on similar biological medicinal products containing biotechnology-derived proteins as active substance: quality issues (revision 1) [Internet]. London, UK: European Medicines Agency; [Updated May22, 2014; cited February05, 2016]. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2014/06/WC500167838.pdf [Google Scholar]
- 12.European Medicines Agency, Committee for Medicinal Products for Human Use Guideline on similar biological medicinal products [Internet]. London, UK: European Medicines Agency; [Adopted on October 23, 2014; cited February05, 2016]. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2014/10/WC500176768.pdf [Google Scholar]
- 13.Antes B, Amon S, Rizzi A, Wiederkum S, Kainer M, Szolar O, Fido M, Kircheis R, Nechansky A. Analysis of lysine clipping of a humanized Lewis-Y specific IgG antibody and its relation to Fc-mediated effector function. J Chromatogr B: Analyt Technol Biomed Life Sci 2007; 852(1–2):250-6; PMID:17296336; http://dx.doi.org/ 10.1016/j.jchromb.2007.01.024 [DOI] [PubMed] [Google Scholar]
- 14.Dick LW Jr, Qiu D, Mahon D, Adamo M, Cheng K-C. C-terminal lysine variants in fully human monoclonal antibodies: investigation of test methods and possible causes. Biotechnol Bioeng 2008; 100(6):1132-43; PMID:18553400; http://dx.doi.org/ 10.1002/bit.21855 [DOI] [PubMed] [Google Scholar]
- 15.Worobec A, Rosenberg AS. A risk-based approach to immunogenicity concerns of therapeutic protein products, Part 3: effects of manufacturing changes in immunogenicity and the utility of animal immunogenicity studies. BioPharm Int [Internet]. January 1, 2005. [cited February05, 2016] Available from: http://www.biopharminternational.com/risk-based-approach-immunogenicity-concerns-therapeutic-protein-products-part-3-effects-manufacturin [Google Scholar]
- 16.den Engelsman J, Garidel P, Smulders R, Koll H, Smith B, Bassarab S, Seidl A, Hainzl O, Jiskoot W. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm Res 2011; 28:920-33; PMID:20972611; http://dx.doi.org/ 10.1007/s11095-010-0297-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haverick M, Mengisen S, Shameem M, Ambrogelly A. Separation of mAbs molecular variants by analytical hydrophobic interaction chromatography HPLC: overview and applications. mAbs 2014; 6:852-58; PMID:24751784; http://dx.doi.org/ 10.4161/mabs.28693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiPaola M, Li J, Stephens EJ. Development of biosimilars: analysis of etanercept glycosylation as a case study. J Bioanal Biomed 2013; 5:180-86; http://dx.doi.org/; http://dx.doi.org/ 10.4172/1948-593X.1000096 [DOI] [Google Scholar]
- 19.Byrne B, Donohoe GG, O'Kennedy R. Sialic acids: carbohydrate moieties that influence the biological and physical properties of biopharmaceutical proteins and living cells. Drug Discov Today 2007; 12:319-26; PMID:17395092; http://dx.doi.org/ 10.1016/j.drudis.2007.02.010 [DOI] [PubMed] [Google Scholar]
- 20.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci 2011; 100:2071-86; PMID:21491437; http://dx.doi.org/ 10.1002/jps.22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim NA, Lim DG, Lim JY, Kim KH, Jeong SH. Comprehensive evaluation of etanercept stability in various concentrations with biophysical assessment. Int J Pharm 2014; 460:108-18; PMID:24269208; http://dx.doi.org/ 10.1016/j.ijpharm.2013.11.019 [DOI] [PubMed] [Google Scholar]
- 22.International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use ICH Tripartite Guideline. Comparability of biotechnological/biological products subject to changes in their manufacturing process Q5E [Internet]. [Recommended for adoption on November 18, 2004; cited February05, 2016] Available from: http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q5E/Step4/Q5E_Guideline.pdf [Google Scholar]
- 23.Houel S, Hilliard M, Yu YC, McLoughlin N, Martin SM, Rudd PM, Williams JP, Chen W. N- and O-Glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem 2014; 86(1):576-84; PMID:24308717; http://dx.doi.org/ 10.1021/ac402726h [DOI] [PubMed] [Google Scholar]
- 24.Biller M, Mardberg K, Hassan H, Clausen H, Bolmstedt A, Bergstrom T, Olofsson S. Early steps in O-linked glycosylation and clustered O-linked glycans of herpes simplex virus type 1 glycoprotein C: effects of glycoprotein properties. Glycobiology 2000; 10:1259-69; PMID:11159917; http://dx.doi.org/ 10.1093/glycob/10.12.1259 [DOI] [PubMed] [Google Scholar]
- 25.Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J Pharm Sci 2015; 104:1866-84; PMID:25872915; http://dx.doi.org/ 10.1002/jps.24444 [DOI] [PubMed] [Google Scholar]
- 26.Hermeling S, Crommelin DJ, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res 2004; 21:897-903; PMID:15212151; http://dx.doi.org/ 10.1023/B:PHAM.0000029275.41323.a6 [DOI] [PubMed] [Google Scholar]
- 27.Koren E, Smith HW, Shores E, Shankar G, Finco-Kent D, Rup B, Barrett Y-C, Devanarayan V, Gorovits B, Gupta S, et al.. Recommendations on risk-based strategies for detection and characterization of antibodies against biotechnology products. J Immunol Methods 2008; 333:1-9; PMID:18275969; http://dx.doi.org/ 10.1016/j.jim.2008.01.001 [DOI] [PubMed] [Google Scholar]
- 28.Murphy K. Janeway's Immunobiology. 8th ed. New York, NY: Garland Science Publishing, 2011. Chapter 10, The humoral immune response, p. 367-408. [Google Scholar]
- 29.Dintzis RZ, Okajima M, Middleton MH, Greene G, Dintzis HM. The immunogenicity of soluble haptenated polymers is determined by molecular mass and hapten valence. J Immunol 1989; 143:1239-44; PMID:2473123 [PubMed] [Google Scholar]
- 30.Bachmann MF, Rohrer UH, Kündig TM, Bürki K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science 1993; 262(5138):1448-51; PMID:8248784; http://dx.doi.org/ 10.1126/science.8248784 [DOI] [PubMed] [Google Scholar]
- 31.Joubert MK, Hokom M, Eakin C, Zhou L, Deshpande M, Baker MP, Goletz TJ, Kerwin BA, Chirmule N, Narhi LO, et al.. Highly aggregated antibody therapeutics can enhance the in vitro innate and late-stage T-cell immune responses. J Biol Chem 2002; 287:25266-79; http://dx.doi.org/ 10.1074/jbc.M111.330902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitoma H, Horiuchi T, Tsukamoto H, Tamimoto Y, Kimoto Y, Uchino A, To K, Harashima S, Hatta N, Harada M. Mechanisms for cytotoxic effects of anti-tumor necrosis factor agents on transmembrane tumor necrosis factor alpha-expressing cells: comparison among infliximab, etanercept, and adalimumab. Arthritis Rheum 2008; 58:1248-57; PMID:18438840; http://dx.doi.org/ 10.1002/art.23447 [DOI] [PubMed] [Google Scholar]
- 33.Houriuchi T, Mitoma H, Harashima S, Tsukamoto H, Shimoda T. Transmembrane TNF-α: structure, function and interaction with anti-TNF agents. Rheumatology 2010; 49:1215-28; PMID:20194223; http://dx.doi.org/ 10.1093/rheumatology/keq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.European Medicines Agency Human medicines European public assessment report (EPAR): Benepali [Internet]. London, UK: European Medicines Agency; [Available on April 14, 2016; cited April29, 2016]. Available from: http://www.ema.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/004007/human_med_001944.jsp&mid=WC0b01ac058001d124 [Google Scholar]
- 35.European Medicines Agency Committee for Medicinal Products for Human Use. Summary of opinion: Benepali [Internet]. London, UK: European Medicines Agency; [Available on November 19, 2015; cited February05, 2016]. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Summary_of_opinion_-_Initial_authorisation/human/004007/WC500196736.pdf. [Google Scholar]