Abstract
Propionic acidemia (PA) is a life-threatening disease caused by the deficiency of a mitochondrial biotin-dependent enzyme known as propionyl coenzyme-A carboxylase (PCC). This enzyme is responsible for degrading the metabolic intermediate, propionyl coenzyme-A (PP-CoA), derived from multiple metabolic pathways. Currently, except for drastic surgical and dietary intervention that can only provide partial symptomatic relief, no other form of therapeutic option is available for this genetic disorder. Here, we examine a novel approach in protein delivery by specifically targeting and localizing our protein candidate of interest into the mitochondrial matrix of the cells. In order to test this concept of delivery, we have utilized cell penetrating peptides (CPPs) and mitochondria targeting sequences (MTS) to form specific fusion PCC protein, capable of translocating and localizing across cell membranes. In vitro delivery of our candidate fusion proteins, evaluated by confocal images and enzymatic activity assay, indicated effectiveness of this strategy. Therefore, it holds immense potential in creating a new paradigm in site-specific protein delivery and enzyme replacement therapeutic for PA.
Abbreviations: PA, propionic acidemia; PCC, propionyl coenzyme-A carboxylase; PP-CoA, propionyl coenzyme-A; CPPs, cell penetrating peptides; MTS, mitochondria targeting sequences; ERT, enzyme replacement therapy; MPP, mitochondrial processing peptidase; LAD, lipoamine dehydrogenase; PCCA, PCCα subunit; PCCB, PCCβ subunit; His-tag, six histidines tag; UPLC-MS/MS, ultra performance liquid chromatography tandem mass spectrometry; CoA, coenzyme-A
Keywords: Propionic acidemia, Enzyme replacement therapy, Protein transduction domains, Mitochondrial targeting sequences, Propionyl coenzyme-A carboxylase
1. Introduction
Metabolic disorders are life-threatening diseases caused by insufficient activities of enzymes required for the catabolism of metabolites that arise from the normal turnover of cellular constituents. One such disease is propionic acidemia (PA), which is an inherited autosomal recessive inborn error of metabolism during the neonatal period [11], [12], [43]. It affects approximately 1 in 30,000 live births worldwide [48] and PA patients may exhibit clinical symptoms such as protein intolerance, vomiting, lethargy, profound metabolic acidosis and mental retardation which can be sufficiently severe to cause death at young age [22]. These symptoms are caused by the deficiency of a mitochondrial biotin-dependent enzyme known as propionyl coenzyme-A carboxylase (PCC). PCC is responsible for the catabolism of propionyl coenzyme-A (PP-CoA), a metabolic intermediate arising from the normal turnover of several essential amino acids, as well as odd chain fatty acids in the matrix of mitochondria. The native PCC holoenzyme is composed of six α (PCCA) and six β (PCCB) subunits with the molecular weights of 72 kDa and 56 kDa respectively [25], [46]. Deficiency in either or both PCC subunits and the consequent accumulation of PP-CoA leads to the pathogenesis of PA [36]. There is currently no cure for the deficiency of PCC and patients are managed by symptomatic treatment such as low-protein-high-energy diet or supplementation with specific mixtures free of propiogenic substrates, vitamins, and trace elements [8]. However, these treatments only partially improve symptoms and the overall outcome of severe forms of PA remains disappointing. Therefore, there is a need for developing an alternative durable and safe treatment for PA patients.
To date, more than 130 proteins and peptides are used as therapeutics in United States and European countries [31]. Enzyme replacement therapy (ERT) is a therapeutic approach which aims to restore the activity of a particular enzyme or protein in patients in cases of deficiency or abnormal production [29]. ERT has been studied in various metabolic enzyme deficiencies such as Gaucher's, Hurler's, Fabry's, Pompe's disease, and in Maroteaux-Lamy syndrome to reverse the pathogenesis of the chief clinical manifestations of these diseases [1], [45], [16], [6], [20], [28], [52]. With the large number of studies on ERT, one can anticipate that ERT will have an expanding role in the treatment of rare metabolic diseases. In this paper, we examine the possibility of developing an enzyme replacement strategy for the relief of PA.
Physicochemical factors such as delivery and stability of protein are classical challenges to the development of ERT for PA patients [31], [34]. One of the key challenges to effective ERT is the plasma membrane of living cells, which is a formidable barrier to permeability by exogenous molecules. In the case of developing protein therapeutics for the treatment of PA, the delivery of an effective therapeutic protein requires the delivery of the protein across the cell membrane, followed by a subsequent targeting of the protein into the mitochondria of the cells poses an additional level of challenge. To overcome the challenge of delivering and targeting proteins into the mitochondria of living cells and stabilize them in ERT, two groups of peptides called cell penetrating peptides (CPPs) and mitochondrial targeting sequences (MTS) were examined. Recent studies used CPP as an approach to overcome the cell's plasma membrane barrier to deliver recombinant enzymes into the cells and their organelles such as mitochondria [9], [24], [26], [47]. The most well-studied CPP is a peptide derived from human immunodeficiency virus transactivator of transcription (HIV-1 TAT), known as the TAT peptide. TAT is an 11-amino acid domain and has been reported to translocate through cell membranes in a receptor-independent fashion either alone or linked to bulky peptides and proteins cargoes [14], [17], [33]. On the other hand, MTS are typically amino-terminal (N-terminal) amphipathic α-helices targeting signal peptides which are around 15–50 amino acid residues in length. They direct mitochondrial proteins to the mitochondrial matrix upon translation and are removed by a mitochondrial processing peptidase (MPP) upon import [4], [18], [19].
Both TAT and MTS have been investigated for their applicability in delivering biologically active fusion proteins to cells and organelles in various murine tissue [3], [15], [50], [55] (for recent reviews see [5], [21], [54]). More recently, Rapoport et al. used protein transduction strategy to develop an approach for restoring the activity of human lipoamine dehydrogenase (LAD) enzymatic complex in a mitochondrial disorder known as LAD deficiency through the utilization of a TAT-MTS-LAD fusion protein [41]. Their result suggested that expressed and purified fusion protein could be successfully delivered into the mitochondria of deficient cell lines and tissue of deficient mice to restore the activity of an essential mitochondrial enzymatic complex [41], [42]. Moreover, numerous groups have successfully exploited the ability of both TAT and MTS fusion domains for transduction of the recombinant mitochondrial proteins across mitochondrial membranes in in vitro and in vivo studies [9], [10], [23], [47]. These pieces of evidences suggested that TAT fusion proteins could cross both cellular and mitochondrial membranes, and that incorporation of a MTS into a TAT fusion protein would allow processing and localization of the exogenous proteins in mitochondria. It is thus suggested that TAT and MTS fusion proteins may represent a viable option as potential mitochondrial protein therapies for direct delivery of ERT into patients. However, targeting of any new protein into mitochondria using the strategy described above requires detailed examination and optimization.
To our knowledge, there has not been any report of functionally active proteins that are specifically targeted or delivered to the mitochondrial matrix as a therapeutic approach for PA. In this paper, we propose the combined utilization of TAT and MTS for transduction of mitochondrial enzyme subunits, PCCA and PCCB, into mitochondria of cells as a first step to the development of a site-specific ERT for PA.
2. Materials and methods
Oligonucleotide primers were ordered from 1st Base (Singapore, Singapore). GelRed® nucleic acid gel stain, MitoView 633 fluorescent mitochondrial dye, and Mix-n-Stain™ CF488A were purchased from Biotium (Hayward, CA). Restriction endonucleases (HindIII-HF, NotI-HF, EcoRI-HF and BamHI-HF) and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA), while KOD DNA polymerase was purchased from Novagen-EMD4 Biosciences (Darmstadt, Germany). Mitochondrial isolation from mammalian cells and enzymatic activity assay were performed as described previously [7].
2.1. Construction of plasmids for expression of fusion protein variants
cDNA of PCC α (pccA) and β (pccB) subunit were ordered to Geneart (San Francisco, CA) to be constructed based on the mature protein sequences reported by Kelson et al. [30]. In order to construct the recombinant pccA and pccB genes fused to MTS-TAT, TAT or MTS domains on their C-terminal site, full length cloned pccA and pccB genes were used as templates in PCR assay. Fusion domains were constructed to be flanked by glycine spacer residues. The details of oligonucleotides used as forward and reverse primers are listed in Supplementary material Tables 1 and 2. All reverse primers were designed to have complementary segments with melting temperature (Tm) at around 68 °C to allow stepwise amplification. Primers were designed using Jellyfish software version 3.3.1 (Field Scientific, Lewisburg, PA). PCR reaction mixture (50 μl) contained 10 nM of each of primers and 0.5 μl of KOD DNA polymerase. The PCR products for pccA and pccB fusion constructs were digested by EcoRI-HF/HindIII-HF and BamHI-HF/NotI-HF endonucleases respectively before ligation into pET28a fusion expression vector (Merck Millipore Division, Merck Pte. Ltd., Darmstadt, Germany), which adds six histidines tag (His-tag) on N-terminal of proteins. pccA and pccB plasmid constructs were transformed into E. coli DH5α competent cells. N-terminal fusion subunits have been previously prepared from pccA and B dodecamer variants, which were constructed by Geneart (NC, USA). All constructs were sequenced by 1st BASE (Singapore, Singapore) for the presence of the intact TAT, MTS and MTS-TAT on their corresponding C or N terminals.
2.2. Recombinant expression and purification of human PCC subunits
E. coli BL21 (DE3) competent cells were transformed with plasmids encoding the N-terminal or C-terminal fusion PCC subunits. The transformed cells were grown at 37 °C in 1 L LB medium containing 50 μg/ml kanamycin with addition of 5 μM biotin for PCCA variants expression. Protein expression was induced by adding isopropyl-β-d thio galactopyranoside (IPTG) to culture with OD600 nm of 0.6–0.8 to a final concentration of 1 mM. Induced cells were allowed to grow overnight at 18 °C before collection. The cells were harvested by centrifugation (7000 rpm for 20 min at 4 °C) and the pellets were sonicated in pulses using denaturing lysis buffer (50 mM NaH2PO4, 30 mM NaCl, 10 mM imidazole, 8 M urea, pH adjusted to 8) for 20 min. The suspensions were clarified by centrifugation (13,000 rpm for 30 min at 4 °C), and the supernatant containing the denatured fusion proteins were loaded on a 5 ml His-Trap nickel columns from GE Healthcare (Uppsala, Sweden). The columns were washed by stepwise addition of increasing imidazole from 10 mM to 500 mM and finally eluted with elution buffer (50 mM NaH2PO4, 30 mM NaCl, 500 mM imidazole, 8 M urea, pH adjusted to 8). The purification procedures were carried out using the ÄKTA Prime Plus system (GE Healthcare, Uppsala, Sweden). Urea and other salts were removed by Hi-prep 26/10 desalting column (GE Healthcare, Uppsala, Sweden). Aliquots of the proteins at 80% purity desalted against phosphate buffered saline (PBS) or TAE buffer (40 mM Tris–HCl, pH 8.0, 0.1 mM EDTA) were snapped frozen in liquid nitrogen and were kept in − 80 °C until use.
2.3. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis
Protein concentrations were calculated by Bradford dye assays. Around 10–30 μg protein per lane were resolved on 12% polyacrylamide SDS-PAGE. Gels were subsequently stained with InstantBlue™ (Expedeon Ltd., Cambridge, UK) to visualize the protein bands. Western blot analysis was performed using anti-PCCB mouse polyclonal (I-DNA Biotechnology Pte-Ltd., Singapore, Singapore), anti-PCCA chicken polyclonal (Sigma-Aldrich, St. Louis, MO) and anti-E1α (Invitrogen, Carlsbad, CA) as primary antibodies and anti-mouse HRP conjugated IgG (goat), and anti-chicken HRP conjugated IgG (bovine) (Santa Cruz Biotechnology, Santa Cruz, CA) as secondary antibodies at 1:1000 dilutions. Densitometric analysis of western blot's bands of target PCC subunits was conducted using National Institutes of Health (NIH) ImageJ 1.47 software.
2.4. Stability assay
The stability assay of original PCC and recombinant fusion subunits was done by using chirascan™ circular dichroism (CD) spectrometer (Applied Photophysics Ltd., Surrey, UK). The measurement was made using temperature ramp rate of 1 °C/min from 10 to 90 °C scanning at intervals of 10 °C. The scan range was from 190 to 260 nm and the bandwidth was 0.5 nm. Proteins were concentrated to final concentrations of a 0.1 mg/ml in PBS or TAE buffers. The CD chromatogram results were expressed as the mean residue ellipticity (MRE [ѳ]) (deg·cm2·dmol− 1) against wavelength (nm). The analyses of secondary structure percentages were carried out by monitoring the CD changes at 222 nm with the changing temperature using CDNN software (Applied Photophysics Ltd., Surrey, UK).
2.5. Cell culture
HeLa cultured cells at passage number less than 15 were kept in a humidified atmosphere with 5% CO2 at 37 °C and maintained in Dulbecco's modified Eagle's medium with 2 mM l-glutamine (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat inactivated fetal bovine serum (FBS) (HyClone, Logan, UT) and 100 μg/ml penicillin/streptomycin (PAN Biotech GmbH, Aidenbach, Germany). PCCA defective (GM22010), PCCB defective (GM22112) and normal (AG15011) lymphocyte cells ordered from Coriell Cell Repositories (Camden, NJ) were kept in passage number less than 10 in RPMI 1640 medium with 2 mM l-glutamine from GIBCO (Grand Island, NY) supplemented with 15% FBS (non-heat inactivated) and 100 μg/ml penicillin/streptomycin.
2.6. Delivery of fusion proteins into HeLa cells and confocal microscopy
Purified proteins at concentrations of 0.5–1 mg/ml were labeled with Mix-n-Stain CF488A green fluorescent dye in accordance with the manufacturer's instructions (Biotium, Hayward, CA). Non-protein and unbound fluorescent dye components were removed by using Zeba spin 40K, 0.5 ml desalting columns (Thermo Fisher Scientific, Waltham, MA) and aliquots of the labeled proteins were kept on ice before use. Cells grown on four-well chamber slides (NUNC Brand Products, Roskilde, Denmark) overnight to 50–70% confluency were treated with fluorescent-labeled fusion proteins variants (0.1 mg/ml final concentration) for 30 min. The cells were subsequently washed with PBS and incubated with red mitochondrial specific fluorescent dye (MitoView 633) for 30 min. The cells were then washed with PBS, fixed in 4% formaldehyde (Sigma Aldrich, St. Louis, MO) for 20 min at room temperature, rewashed and kept in Vectashield® mounting medium (Vector Laboratories, Burlingame, CA). The cells were analyzed for localization of MTS-TAT fusion PCCA and PCCB into mitochondria using objective 60 × of FluoView FV10i confocal microscope systems (Olympus, Tokyo, Japan).
2.7. Delivery of fusion subunits into defective lymphocyte cells for assaying PCC enzymatic activity
GM22010 (PCCA defective lymphocyte) and GM22112 (PCCB defective lymphocyte) cell lines grown in T75 flask (NUNC Brand Products, Roskilde, Denmark) were treated with original, TAT and MTS-TAT fusion subunits (0.1 mg/ml final concentration) for different incubation times. After incubation, the cells were washed with PBS and their mitochondria were isolated through differential centrifugation protocol described previously [7], [37]. The activity of PCC inside the mitochondria was measured with ultra performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) method previously developed by our group [7]. Briefly, 10 μg of mitochondrial lysate was added to the reaction buffer containing 50 mM Tris (pH 8.0), 2 mM ATP, 125 mM KCl, 10 mM MgCl2, 0.5 mg/ml BSA, 10 mM sodium bicarbonate, 3 mmol/l PP-CoA, and 1 mmol/l coenzyme-A (CoA) to initiate the reaction. The reaction was terminated by adding 50 μl of 10% trichloroacetic acid (TCA) to the mixture, diluted with dH2O by dilution factor of 10 and centrifuged at 15,000 g for 15 min. The supernatant containing the CoA-esters was then subjected to UPLC-MS/MS analysis for measuring the concentration of PP-CoA [7] and the reductions of PP-CoA amount were calculated as the activity of enzyme. Calibration standard samples were freshly prepared before each run.
3. Results
3.1. Amplification of C-terminal and N-terminal fusion PCC subunits variants
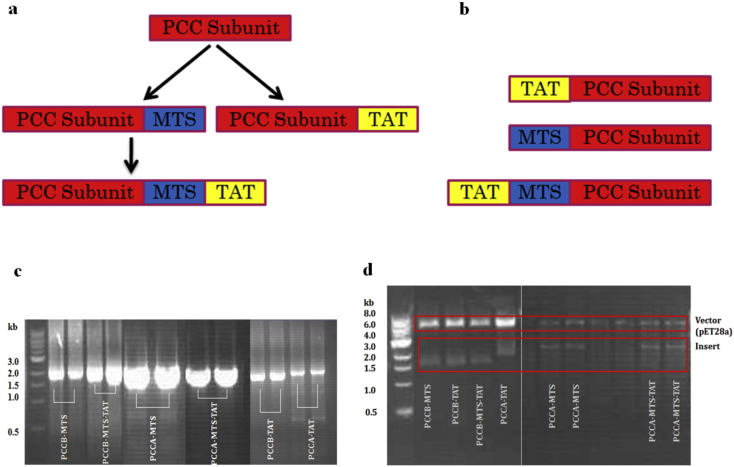
Original cDNA of PCC alpha (pccA) and beta (pccB) subunits were used to amplify the different PCC constructs with the cell penetrating peptide, TAT, and MTS on their ends. Strategy for designing the fusion constructs was presented in Fig. 1a, b. As shown in this figure, fusion gene structures of PCC subunits (pccA and pccB) contained TAT, MTS or MTS-TAT on their 3′ or 5′ end of DNA chain that corresponded to C-terminal or N-terminal of protein chain respectively. Based on the fact that total length of MTS-TAT segment was more than 120 bases (93 bases for MTS and 33 bases for TAT), we have designed several reverse primers with shared segments to amplify the 3′ end of pccA and pccB genes as listed in Supplementary material Table 1. C-terminal types of fusion subunits were designed to compare their delivery and localization effect with N-terminal fusion subunits. The flowchart of C-terminal designs in Fig. 1a shows that fusion MTS-TAT subunits were designed to be amplified from fusion MTS subunits.
Fig. 1.
Structural design and construction of 3′ end PCC fusion subunits variants. (a) 3′ end fusion subunits variants were designed to be amplified from original PCC subunits clone. MTS-TAT fusion subunits could be amplified from MTS fusion subunits. (b) A schematic representation of TAT, MTS, and TAT-MTS fusion 5′ end subunits constructs. Control variants are subunits lacking the MTS or TAT domains. (c) PCR amplification results of pccA and pccB variants by using KOD DNA polymerase. (d) Digestion of cloned 3′ end fusion pccA and pccB subunits in pET28a vector by restriction enzymes. pccA-TAT, pccA-MTS and pccA-MTS-TAT clones were digested by EcoRI-HF and HindIII-HF restriction enzymes. pccB-TAT, pccB-MTS and pccB-MTS-TAT clones were digested by NotI-HF and BamHI-HF restriction enzymes. All digestions were carried out in 37 °C overnight and digestion results visualized by GelRed staining on 10% agarose gel.
Original pccB and pccA constructs and fusion 3′ end variants that had been amplified and inserted into pET28a vector were digested for confirmation of successful insertion (Fig. 1c, d). All fusion variants were amplified successfully (Fig. 1c) from original subunit clones by stepwise amplification PCR using the primers listed in Supplementary material Tables 1 and 2. Fig. 1d demonstrated confirmation of successful insertion of the desired genes into pET28a vector.
3.2. Expression and purification of PCC subunits constructs
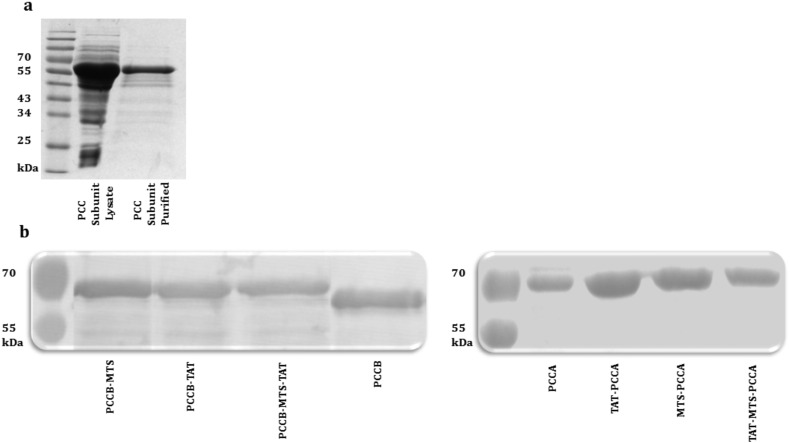
All fusion and original PCC subunit clones were expressed in BL21 competent cells (Supplementary material Fig. 1). The vast majority of the expressed PCC subunits were present in soluble fraction after lysis. Prior works by Nagahara et al. proposed that urea denaturing protein purification is an applicable protocol to denature TAT fusion proteins, purify and increase the efficiency of fusion subunits for cell membrane transduction, since partially folded TAT fusion protein traverse cell membrane more efficiently compared to a completely folded protein [57]. Hence, we adopted the strategy of using urea lysis buffer and denaturing purification protocol for all recombinantly expressed PCCA and PCCB fusion proteins to increase the efficiency of fusion subunits for cell membrane transduction. The 6X His-tag label in pET28a vector assisted in the purification of these proteins using His-Trap ion exchange nickel resin column. The purity and amount of proteins were assessed by SDS-PAGE (Fig. 2a). As shown in Fig. 2a, fusion PCC subunits were efficiently purified under this condition. High concentration of urea and imidazole in eluted purified proteins solution should be removed before treating the mammalian cells with fusion proteins. Although we used both strategies of rapid desalting and dialysis to avoid aggregation while removing urea and imidazole from fusion proteins solution, some of the variants of C-terminal fusion PCCA (PCCA-MTS-TAT and PCCA-MTS) and N-terminal fusion PCCB (TAT-MTS-PCCB, TAT-PCCB, MTS-PCCB) aggregated upon purification and placement of proteins in aqueous solution. Therefore, we did not perform further characterization and stability assay on C-terminal PCCA and N-terminal PCCB variants. Desalting was done in columns equilibrated with PBS or TAE plus 15% glycerol to prevent susceptibility of protein precipitations. Immunoblot assays confirmed the identities of these purified proteins (N-terminal PCCA and C-terminal PCCB), which were selected for subsequent analysis (Fig. 2b). The size of original PCCA and PCCB proteins were around 72 and 56 kDa respectively; however the size of fusion subunits should be around 2–4 kDa higher as shown in this figure. The largest folded protein reported to be delivered using protein transduction is a monoclonal anti-biotin antibody with the molecular mass around 150 kDa [40]. Hence, PCC fusion subunits attached to MTS-TAT domain should be able to transduce across cellular membrane using TAT transduction strategy.
Fig. 2.
Representative of fusion PCC subunits purification and western blot. (a) Reducing SDS-PAGE of a representative PCC subunit lysate is compared with purified one after using nickel affinity column purification method. Gels were stained with InstantBlue™ (Expedeon Ltd., Cambridge, UK). (b) Western blot representative of N-terminal PCCA and C-terminal PCCB original and fusion variants using anti-PCCA chicken polyclonal and anti-PCCB mouse polyclonal as primary antibodies respectively.
3.3. In vitro delivery of fusion subunits into mammalian cells and their mitochondria
3.3.1. Measurement of the secondary structure profile of fusion proteins
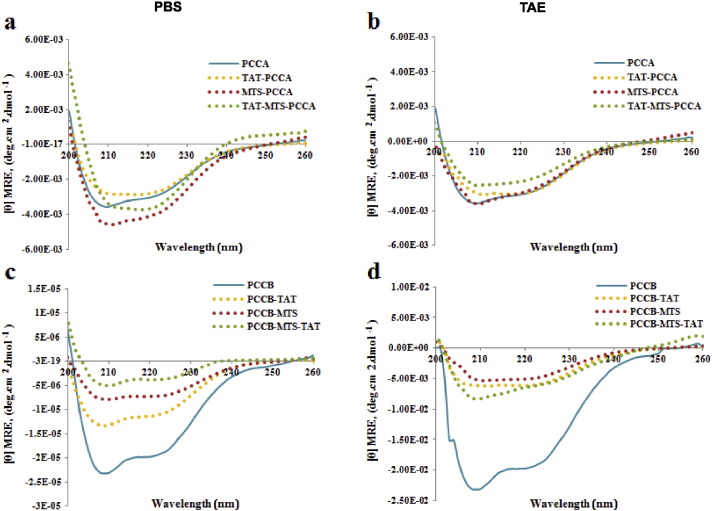
N-terminal PCCA and C-terminal PCCB fusion subunits were further characterized to determine their secondary structure similarity to original PCC subunits and the optimal storage and delivery condition for efficient transduction and mitochondrial localization. We employed measurement of the secondary structure profile of fusion subunits before in vitro delivery by using different conditions to compare the properties of the PCCA and PCCB original subunit with fusion variants. The secondary structure of the variant subunits was assessed by following changes in CD signal. From the CD denaturation curves (Fig. 3a–d) and the data analyzed by the CDNN software (Supplementary material Table 3), α helix and β sheets contents of fusion proteins were estimated and compared with original subunits using the values of the CD signals at 222 nm. The PCCA fusion subunit variants showed very similar denaturation curves and secondary structure contents in TAE compared to in PBS as observed in CD chromatogram (Fig. 3a, b). The percentages of secondary contents (α helix, β sheets and random coil) of fusion subunits were closer to original subunit, especially for TAT-MTS-PCCA (total sum 78.00%) compared with PCCA (total sum 75.90). On the other hand, PCCB fusion variants demonstrated more stable and similar structure with original subunit when in PBS buffer (Fig. 3c, d). However, despite the similarity, the denaturation curves of these variants did not overlay with that of original subunit (Fig. 3c), indicating concentration changes due to formation of some aggregates during the unfolding process. Overall, CD data confirmed that the PCCA and PCCB subunits with a fusion MTS-TAT peptide to their region of N and C terminal respectively, had similar secondary structure to the corresponding original subunits. TAE and PBS buffer were chosen for storage of PCCA and PCCB fusion variants respectively.
Fig. 3.
CD spectra of original PCC subunits and fusion variants. The protein concentrations were 0.1 mg/ml and the path length of the cuvette was 10 mm. (a) PCCA variants in PBS, (b) PCCA variants in TAE, (c) PCCB variants in PBS and (d) PCCB variants in TAE. Buffers were adapted to pH 7. Determination were performed at the beginning of the run at 10 °C, in triplicate for each protein.
3.3.2. Co-localization of fusion proteins in the mitochondria of HeLa cells
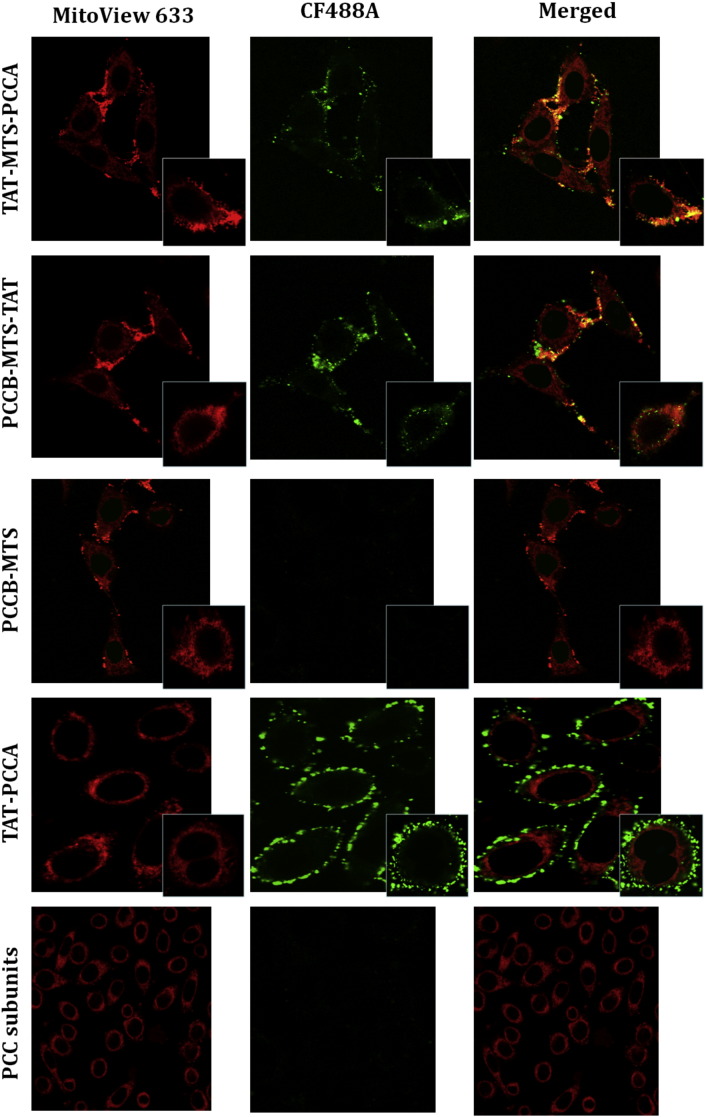
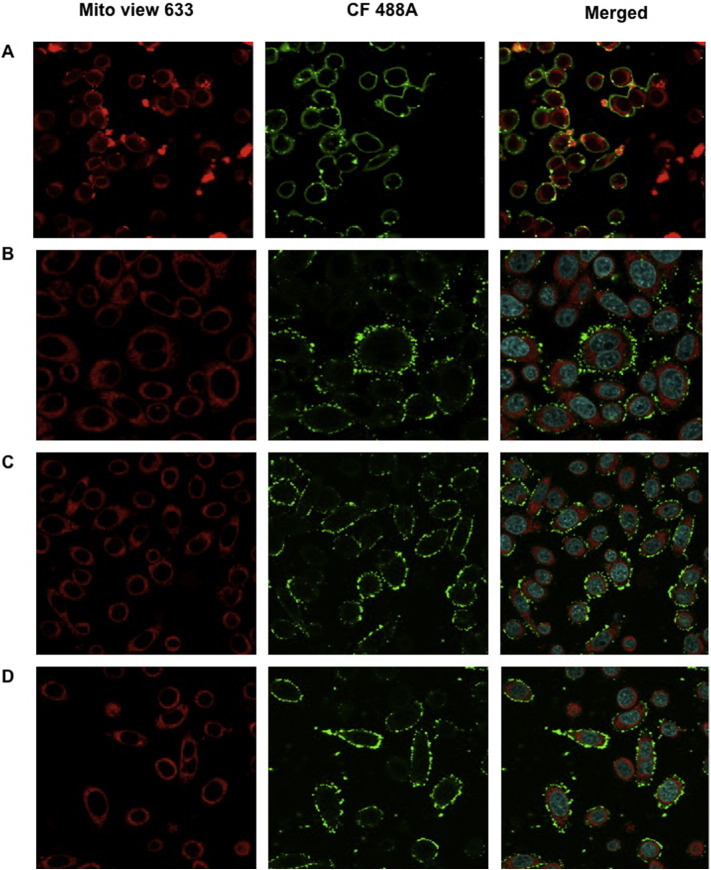
One of our main aims was to evaluate the transduction efficiency of fusion PCC variants into the mitochondria of mammalian cells. Candidate proteins established in this study were optimized to be delivered into cultured HeLa cells to determine their transduction efficiencies as well as localization into mitochondria. Eight candidates had been optimized for delivery assay (PCCA, TAT-PCCA, TAT-MTS-PCCA, MTS-PCCA, PCCB, PCCB-TAT, PCCB-MTS-TAT, PCCB-MTS). For imaging purpose, fusion proteins were labeled with CF488A green dye and added to HeLa cells. Analysis of cells after incubation period by confocal microscopy showed that MTS-TAT fusion proteins transduced into cells after 30 min incubation. Successful co-localization of green-labeled TAT-MTS-PCCA and PCCB-MTS-TAT fusion subunits with red mitochondrial dye inside the mitochondria was shown by orange-brown color in the merged image (Fig. 4). To simplify this figure, confocal images of only two candidate proteins (PCCB-MTS and TAT-PCCA) as control candidates carrying only one fusion peptide were presented. This analysis revealed the localization of PCCB-MTS-TAT and TAT-MTS-PCCA in the mitochondrial matrix, which was absent in the control proteins (TAT-PCCA and PCCB-MTS). The control constructs carrying TAT sequence without MTS informed the effectiveness of the TAT peptide in transducing PCC fusion protein across the plasma and/or organelle membranes but was not able to localize the protein inside the mitochondrial matrix. On the other hand, MTS fusion constructs only incorporated a MTS localization segment for targeting but lack the TAT segment for transduction across cell membrane. However, MTS-TAT fusion constructs (TAT-MTS-PCCA and PCCB-MTS-TAT) with both the transduction domain as well as the localization domain resulted in the import of the proteins into target organelle. Hence, it was demonstrated in this study that both transduction domain (TAT) and localization domain (MTS) are necessary for green labeled MTS-TAT fusion PCC subunits to be translocated into the mitochondria.
Fig. 4.
Fusion PCC subunits co-localization. HeLa cells were treated with CF488A green fluorescent labeled PCCA and PCCB fusion variants (0.1 mg/ml final concentration) for 30 min, washed and incubated with MitoView 633 red fluorescent dye for 30 min before fixation with 4% paraformaldehyde. The cells were analyzed for transduction and co-localization by confocal microscopy. Original magnification is 60 ×. Localization of TAT-MTS-PCCA and PCCB-MTS-TAT subunits (tests) and PCCB-MTS, TAT-PCCA and original PCC subunits (controls) are shown in this figure. Merged photos indicate merging photos of red and green fluorescent filters.
The viability of cells after incubation with TAT fusion proteins (0.1 mg/ml concentration) have been also tested from 30 min up to 24 h by trypan blue staining cell count. We have not seen any adverse effect on viability of HeLa or lymphocyte cells up till the reported period. This corroborates with the study that reported short exposure of TAT fusion proteins into HeLa cells did not exhibit any toxicity effect for 7 h [27].
3.4. Delivery of fusion subunits into the mitochondria of lymphocyte cells
3.4.1. Western blotting analysis
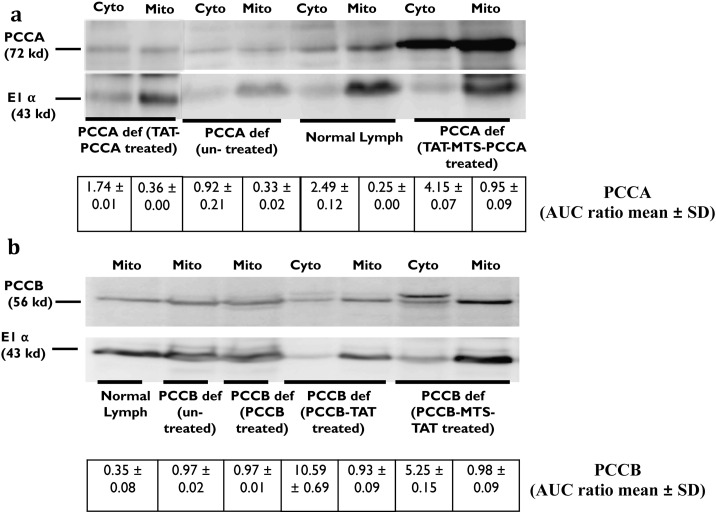
TAT-MTS-PCCA and PCCB-MTS-TAT showed successful co-localization effect in the confocal studies (Section 3.3.2). In order to be developed as a potential ERT lead, the fusion proteins need to be delivered into the mitochondria of PCCA and PCCB defective lymphocyte cells respectively, where they would be assembled with the unaffected native subunits to restore the enzymatic activity of PCC inside the mitochondria. Before conducting the activity assay, the first step was to examine the localization of MTS-TAT fusion proteins in mitochondrial lysate and their ability to be naturally processed in the mitochondria. After incubation, cellular fractions were prepared by separating the mitochondria and the cytosol as described previously [37]. Samples were analyzed by western blotting assay for the presence of the fusion proteins in sub-cellular fractions, using anti-PCCA and anti-PCCB antibodies (Fig. 5a, b). The results indicated the presence of PCCA (72 kDa) and PCCB (56 kDa) in the mitochondrial fractions of the treated cells after 24 h of incubation. The concentration of indigenous PCC in normal cells is low and usually not clearly detectable. However, the intensity of PCCA and PCCB in the mitochondria of treated cells was higher and clearly visible. The purity of the sub-cellular fractions and equal loading of proteins were confirmed by using antibody against the mitochondrial marker, E1α (43 kDa). As shown in Fig. 5a, PCCA protein was detected in mitochondrial fraction at higher intensity in treated cells as compared to untreated and normal lymphocytes. Western blot images were also analyzed by densitometry analysis using ImageJ software (National Institutes of Health (NIH) ImageJ 1.47 software). It was shown that the band corresponding to TAT-MTS-PCCA translocated into the mitochondria of defective cells, had around 3–4 times higher intensity comparing to normal and defective lymphocytes respectively. However, its marker band was almost in the same intensity as normal cells and only twice of defective cells suggesting that the higher PCCA intensity is significant (Fig. 5a, b). There is also high concentration of PCCA protein in cytosolic fraction of treated cells (Fig. 5a), which could have been a portion of the un-localized TAT-MTS-PCCA protein remaining in the cytosol. Fig. 5b showed PCCB band at the higher intensity in defective cells after treating with PCCB-MTS-TAT comparing to untreated cells. Defective cells were also treated with PCCB-TAT and PCCB proteins as controls. As expected, a fraction of PCCB-MTS-TAT was presented in cytosol in its full unprocessed size (60 kDa) slightly higher than its mature size (56 kDa) (Fig. 5b) which represent a fraction of non-processed and un-localized fusion subunit comparing to its localized mitochondrial fraction appear in 56 kDa. Comparing the intensity ratio of each band to its marker in Fig. 5a, b, we concluded that defective cells and normal cells had the same concentration of PCCB in their mitochondrial extraction. In contrast, defective cells treated with PCCB-MTS-TAT showed higher concentration of PCCB in their mitochondrial fraction comparing to untreated defective cells indicating that the exogenous protein has indeed been delivered and localized within the target mitochondria. Moreover, PCCB-TAT control protein lacking the targeting sequences can enter the cells or mitochondria but would not be processed or remain there and diffused out as seen in Fig. 5.
Fig. 5.
Western blot of the fusion and original PCC subunits after delivery to defective lymphocyte cells. Extracted mitochondria and cytosol fractions were analyzed on 12% SDS-PAGE and probed with PCCA and PCCB antibodies. (a) PCCA defective cells treated with TAT-PCCA and TAT-MTS-PCCA are compared with normal lymphocytes and un-treated cells. (b) PCCB defective cells treated with PCCB, PCCB-TAT and PCCB-MTS-TAT are compared with normal lymphocytes and un-treated cells. The purity of the sub-cellular fractions was confirmed using the mitochondrial marker E1α (43 kDa). ImageJ densitometry analysis (National Institutes of Health (NIH) ImageJ 1.47 software) results are presented below each image. Intensity ratio of PCCA and PCCB bands relative to the E1α bands in A and B respectively presented as AUC ratio of corresponding intensity peak (AUC ratio mean ± SD). Anti-chicken HRP conjugated IgG (bovine) and anti-mouse HRP conjugated IgG (goat) (Santa Cruz Biotechnology, CA, USA) were used as secondary antibodies for probing PCCA and PCCB antibodies respectively. PCCA def: PCCA defective lymphocytes, PCCB def: PCCB defective lymphocytes, normal lymph: normal lymphocytes.
3.4.2. Assaying PCC enzymatic activity
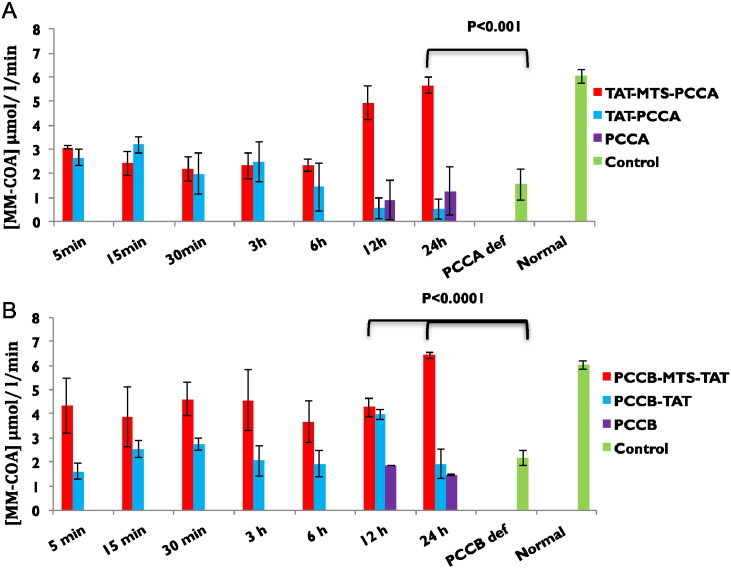
In the earlier Section 3.4.1, we described that western blotting confirmed the existence of delivered and processed fusion PCC subunits in the mitochondria of lymphocyte cells. The next step towards our goal was to assay whether each fusion PCC subunit can restore the enzymatic activity of PCC upon delivery into mitochondria and assembly with indigenous PCC subunit. Therefore, we examined the enzymatic activity assay of PCC by using PCCA and PCCB defective lymphocyte cells incubated for various time periods with the purified TAT-MTS-PCCA and PCCB-MTS-TAT respectively. Mitochondria of treated cells were isolated and prepared for analysis by UPLC-MS/MS activity assay as described previously [7] to test whether MTS-TAT fusion PCC subunits restored enzymatic activity within the mitochondria of cultured cells. PCC defective lymphocyte cells treated with the control subunits (TAT-PCCA, PCCA, PCCB-TAT and PCCB) lacking targeting domain or lacking both transduction and targeting domains, were also analyzed for PCC enzymatic activity. The elevation in PCC enzymatic activity reduced the concentration of PP-CoA in the reaction mixture which corresponded with to increase in methyl malonyl coenzyme-A (MM-CoA) concentration. Both PCCA and PCCB defective cell lines showed much lower PCC activity baseline comparing to normal lymphocytes (Fig. 6a, b) which were 1.53 and 2.16 [MM-CoA] μmol/l/min respectively. The enzymatic activity of PCC within the TAT-MTS-PCCA and PCCB-MTS-TAT treated defective cells increased with incubation time to 5.66 and 6.43 [MM-CoA] μmol/l/min respectively (Fig. 6a, b), which were comparable with the PCC activity in normal lymphocytes (6.03 [MM-CoA] μmol/l/min). PCC enzymatic activity increased from its baseline in defective cells within 30 min of incubation with MTS-TAT fusion subunits and reached its level similar to normal lymphocyte after 24 h of incubation. However, when defective cells were incubated with the control proteins, none of them showed the significant changes in PCC activity even after 24 h incubation, with enzymatic activity, comparable to baseline value for deficient cells. This results concurs with the confocal microscopy results in which incubation of HeLa cells with PCC (MTS-TAT) subunits and the control proteins showed that the MTS-TAT fusion PCCA and PCCB rapidly entered the cells and were detectable after 30 min of incubation (Fig. 4).
Fig. 6.
Schematic representation of the fusion proteins activity. The in vitro enzymatic activity assay of delivered fusion and original subunits was performed as described in Materials and methods. PCCA or PCCB defective lymphocytes were treated with (a) TAT-MTS-PCCA, TAT-PCCA, PCCA, and (b) PCCB-MTS-TAT, PCCB-TAT, PCCB at final concentration of 0.1 mg/ml for different time periods (5 min–24 h). PCC activity was analyzed in isolated mitochondria lysate immediately after the incubation time using the UPLC-MS/MS analysis [7]. A set of control was performed by using un-treated PCCA defective lymphocytes (PCCA def), PCCB defective lymphocytes (PCCB def) and normal lymphocyte cells (normal). Activity assays were conducted at least three times and the values presented are the mean values ± SD. The enzymatic activity values are presented as [MM-CoA] μmol/l/min.
4. Discussion
Currently, modern science offers no cure for patients suffering from mitochondrial disorders such as PA [13], [53]. Although ERT for mitochondrial disorders is significantly more challenging due to the need for organelle-specific delivery, its application to replace a particular activity of an enzyme is becoming a potentially attractive therapeutic approach in the treatment of enzyme deficiency disorders [2], [45], [16], [6], [48]. Therefore, we aimed towards the development of an ERT approach for mitochondrial diseases such as PA in this project.
One of our goals was to facilitate direct and site-specific translocation of PCC across biological membranes and restore the activity of PCC by introducing TAT (as the transduction domain) and MTS (as the targeting domain) fused to the N-terminal or C-terminal of the protein subunits. Since MTS-TAT domain can potentially affect structure of the protein, one critical parameter in delivery of MTS-TAT fusion proteins is the careful design of the fusion protein. Most mitochondrial proteins are synthesized with an N-terminus MTS [44], [51]. It was also observed that MTS may in fact mediate mitochondrial import while attaching to the C-terminal sequences of the passenger polypeptide [3], [38]. If the native protein is known to be tightly associated at the N or C terminal, the fusion MTS-TAT domain may not be exposed properly and it is resulted in poor protein transduction. Therefore, it is necessary to construct and consider delivery of C-terminal MTS-TAT fusion subunits and their comparison with N-terminal fusion ones. Considering this, we designed C-terminal types of fusion subunits to compare their delivery and localization with N-terminal fusion subunits.
To examine the efficiency of delivering a protein into the cells and their mitochondria, four sets of proteins were prepared as follows for comparison of transduction efficiency into mammalian cells: (1) N-terminal PCCA fusion subunits, (2) C-terminal PCCA fusion subunits, (3) N-terminal PCCB fusion subunits and (4) C-terminal PCCB fusion subunits. Only sets 1 and 4 showed minimal protein aggregate during purification procedure to allow subsequent delivery into cells. Occurrence of precipitation of the unfolded proteins in aqueous solution was a major pitfall experienced with TAT fusion proteins [3]. In our project, this problem existed in proteins carrying tag on C-terminal for PCCA and on N-terminal for PCCB. This may be due to interference of the length and charge of the N or C terminal amino acid extension with a tertiary structure of the subunits required for its solubility [49], [56]. Therefore, PCC variants that showed good solubility after purification procedure (sets 1 and 4) were analyzed for conformational changes. The results of CD stability assay showed that PCC subunits with MTS and TAT domain may result in some degrees of conformational changes but little effect on overall secondary structure (Fig. 3b, c).
Next, to verify transduction of our interested proteins into cells and mitochondria, we performed confocal microscopy experiments to examine localization efficiency of fusion subunits in the mitochondria of HeLa cells cultured on chamber slides. Confocal analysis revealed the co-localization effect of both green-labeled MTS-TAT fusion PCC subunits in the mitochondrial matrix, whereas control proteins did not result in any co-localization effect (Fig. 4). It has been demonstrated in some studies that TAT fusion proteins can pass through cellular and organelle membranes including the mitochondrial membranes. Moreover, when MTS was present, the fusion proteins were retained within the mitochondrial matrix of cells and mice tissues [9], [41], [42]. Our results indicated that PCCA and PCCB subunits having MTS-TAT fusion domains on their N-terminal and C-terminal respectively are able to enter cells and their mitochondria rapidly, where they are localized (Fig. 4). Therefore, further activity assays were performed.
The most crucial requirement for the successful treatment of a metabolic disease by ERT is the enzyme's ability to substitute for the deficient endogenous enzyme, including successful assembly into its natural enzymatic complex. In most enzyme studies, cultured lymphocytes or fibroblasts have been used for study of organic acidemia such as PA [32], [35]. There are PCC deficient or defective lymphocytes, which have mutations in one PCC subunits only. In the case of PCC deficiency in one subunit, the replacing subunit needs to be assembled with the other normal endogenous subunit inside the mitochondrial matrix to restore the activity of PCC. Our results successfully indicated that MTS-TAT fusion PCCA and PCCB subunits were able to restore the PCC activity in treated defective lymphocyte cells from patients belonging to two different genotypes (PCCA and PCCB defective). The PCC activity in defective cells was only around 30% of normal value. However, after 24 h of treatment, the enzymatic activity reached to its maximum level that is comparable to un-treated cells (Fig. 6a, b). Given the structure of the PCC, we hypothesized if a singly mutated nonfunctioning subunit causes reduction in activity of the whole PCC enzyme, the exogenous administration of this subunit may be sufficient to restore the activity in that particular defective cell types. It was previously reported that co-transfection of both pccA and pccB wild-type genes was necessary for obtaining maximal enzyme activity in PCCA and PCCB deficient cells [39]. In our study, we demonstrated that incubation for 12–24 h with fusion protein alone was enough to restore the activity to normal control levels. Therefore, these results confirmed our hypothesis that MTS-TAT fusion PCCA or PCCB replacement of defective subunit was sufficient to increase the enzymatic activity of the PCC in PCCA or PCCB defective cells respectively.
In this study, both confocal images and activity results supported our hypothesis that the control subunit with TAT fusion domain may only enter the cells and its organelle but not localize within the mitochondria (Fig. 4, Fig. 5a, b). Consequently, treatment with this TAT fusion subunit did not increase PCC activity in the mitochondria of defective cells as efficient as treatment with MTS-TAT did (Fig. 6a, b). However, MTS allowed the eventual cleavage of the PCC subunits from the TAT sequence, thereby enabling its proper folding and assembling with other native subunits into PCC enzymatic complex. The results suggested that the processing site within the MTS in MTS-TAT fusion subunits is valuable for successful targeting and localization of fusion proteins into the mitochondria. This finding also indicates that both delivery domain (TAT) and targeting domain (MTS) are essential for effective entry into the cell's mitochondria and restoring PCC activity. Length and charge of the N or C terminal amino acid extension can affect a tertiary structure of the precursor, its solubility in the cytosol, its recognition by mitochondrial membranes, and its cleavage by a mitochondrial protease. Once the precursor proteins have been translocated, the MTS is no longer necessary and may actually interfere with further sorting or protein folding and assembly. Our results indicated that the MTS-TAT fusion domain cleaved by MPP upon delivery into mitochondria had no negative influence on PCC subunits processing for assembly and specific enzymatic activity which reached around three times higher than the baseline after 24 h. Western blotting of sub-cellular fractions revealed efficient protein transduction and mitochondrial localization of the fusion proteins (Fig. 5a, b). However, results of un-treated lymphocytes mitochondrial lysate showed PCCB expression level remained unaffected in PCCB defective cells (Fig. 5b). This means that PCCB defective cells have normal level of PCCB but the mutation in PCCB caused this subunit to lose its function. Higher intensity of PCCB in cytosol of PCCB-MTS-TAT treated cells comparing to controls may demonstrated the release of some proteins from mitochondrial fraction to cytosol during the extraction procedure. This hypothesis was supported by the appearance of some mitochondrial E1α with the cytosolic fraction of these cells. Overall, in order to test the concept of delivery, we have utilized three methods including assaying co-localization effect by the confocal images, western blotting and restoring the activity by enzymatic assay. All of these methods demonstrated the ability of MTS-TAT fusion PCCA and PCCB subunits to be delivered into the mitochondria of cells and to be processed there before assembling with the endogenous normal subunits to restore the PCC enzymatic activity.
In conclusion, we believe that the study of PCC activity showed improvement in the activity of treated cell lines. Our results suggest that even a single application of MTS-TAT fusion PCC subunit may considerably improve status of PCC subunit deficiency. Moreover, MTS-TAT fusion PCC subunits could have application for improving clinical presentation of patients and alter their status of PCC deficiency. A key advantage of using MTS-TAT fusion proteins for treating mitochondrial disorders is that it is possible to deliver these proteins into all cell types within the body with no specific targeting.
This project aimed to create a new paradigm in research towards the treatment of PA. Our results provide the preliminary indication that the ERT approach is promising for treatment of PA as demonstrated in this study by replacement of defective subunits of PCC and restoring the activity of whole enzyme. Although in vitro testing gave us reasonable indications to the effectiveness of the candidate proteins, application of ERT to address enzyme deficiencies in metabolic disorders such as PA requires extensive in vivo investigations before it could be potentially applied.
The following are the supplementary data relate to this article.
Acknowledgments
Mahnaz Darvish Damavandi and Tse Siang Kang have been involved in conception and design of the study, analysis and interpretation of data, drafting the article and revising it critically. Han Kiat Ho has been involved in interpretation of data, drafting the article and revising it critically. The authors acknowledge the Faculty of Science, National University of Singapore, Singapore for granting the research start up grant R-148–000–139–750, and the Department of Pharmacy, Faculty of Science, National University of Singapore, Singapore for granting the NUSAGE research grant. This work was also supported by Associazione per la Tutela del Bambino con Malattie Metaboliche, Italy. Mahnaz Darvish Damavandi was supported by the Singapore International Graduate Awards (SINGA) research scholarship. We thank Professor Jan P. Kraus (Department of Pediatrics, University of Colorado, School of Medicine, Aurora, CO, USA) for providing the highly purified PCC enzyme. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ymgmr.2016.06.009.
Appendix A. Supplementary data
Supplementary material
Supplementary Fig. 1.
Reducing SDS-PAGE of fusion PCC subunits showing expression level before and after induction in BL21 competent cells. Induced cells were allowed to grow overnight at 18 °C before collection. Lysates of the cells were run for SDS-PAGE. UI: un-induced. I: induced.
Supplementary Fig. 2.
Fusion PCC-(TAT) control subunits co-localization. HeLa cells were treated with CF488A green fluorescent labeled PCCA and PCCB fusion variants (0.1 mg/ml final concentration) for 30 min, washed and incubated with MitoView 633 red fluorescent dye for 30 min before fixation with 4% paraformaldehyde. The cells were analyzed for transduction and co-localization by confocal microscopy. Original magnification is 60 ×. (A) PCCB-TAT, (B–D) TAT-PCCA.
References
- 1.Amalfitano A., Bengur A.R., Morse R.P., Majure J.M., Case L.E., Veerling D.L., Mackey J., Kishnani P., Smith W., McVie-Wylie A., Sullivan J.A., Hoganson G.E., Phillips J.A., 3rd, Schaefer G.B., Charrow J., Ware R.E., Bossen E.H., Chen Y.T. Recombinant human acid alpha-glucosidase enzyme therapy for infantile glycogen storage disease type II: results of a phase I/II clinical trial. Genet. Med. 2001;3(2):132–138. doi: 10.109700125817-200103000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Barton N.W., Brady R.O., Dambrosia J.M., Di Bisceglie A.M., Doppelt S.H., Hill S.C., Mankin H.J., Murray G.J., Parker R.I., Argoff C.E. Replacement therapy for inherited enzyme deficiency — macrophage-targeted glucocerebrosidase for Gaucher's disease. N. Engl. J. Med. 1991;324(21):1464–1470. doi: 10.1056/NEJM199105233242104. [DOI] [PubMed] [Google Scholar]
- 3.Becker-Hapak M., McAllister S.S., Dowdy S.F. TAT-mediated protein transduction into mammalian cells. Methods. 2001;24(3):247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska A., van der Laan M., Mehnert C.S., Guiard B., Mick D.U., Hutu D.P., Truscott K.N., Wiedemann N., Meisinger C., Pfanner N., Rehling P. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 2010;30(1):307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chugh A., Eudes F., Shim Y.S. Cell-penetrating peptides: nanocarrier for macromolecule delivery in living cells. IUBMB Life. 2010;62(3):183–193. doi: 10.1002/iub.297. [DOI] [PubMed] [Google Scholar]
- 6.Connock M., Burls A., Frew E., Fry-Smith A., Juarez-Garcia A., McCabe C., Wailoo A., Abrams K., Cooper N., Sutton A., O'Hagan A., Moore D. The clinical effectiveness and cost-effectiveness of enzyme replacement therapy for Gaucher's disease: a systematic review. Health Technol. Assess. 2006;10(24):iii–iiv. doi: 10.3310/hta10240. (ix-136) [DOI] [PubMed] [Google Scholar]
- 7.Damavandi M.D., Chan E.C., Kraus J.P., Ho P.C., Kang T.S. Development of an UPLC-MS/MS method for assaying the enzymatic activity of propionyl coenzyme-A carboxylase. Bioanalysis. 2014;6(3):335–348. doi: 10.4155/bio.13.297. [DOI] [PubMed] [Google Scholar]
- 8.de Baulny H.O., Benoist J.F., Rigal O., Touati G., Rabier D., Saudubray J.M. Methylmalonic and propionic acidaemias: management and outcome. J. Inherit. Metab. Dis. 2005;28(3):415–423. doi: 10.1007/s10545-005-7056-1. [DOI] [PubMed] [Google Scholar]
- 9.Del Gaizo V., MacKenzie J.A., Payne R.M. Targeting proteins to mitochondria using TAT. Mol. Genet. Metab. 2003;80(1–2):170–180. doi: 10.1016/j.ymgme.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Del Gaizo V., Payne R.M. A novel TAT-mitochondrial signal sequence fusion protein is processed, stays in mitochondria, and crosses the placenta. Mol. Ther. 2003;7(6):720–730. doi: 10.1016/s1525-0016(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 11.Desviat L.R., Clavero S., Perez-Cerda C., Navarrete R., Ugarte M., Perez B. New splicing mutations in propionic acidemia. J. Hum. Genet. 2006;51(11):992–997. doi: 10.1007/s10038-006-0068-3. [DOI] [PubMed] [Google Scholar]
- 12.Desviat L.R., Perez B., Perez-Cerda C., Rodriguez-Pombo P., Clavero S., Ugarte M. Propionic acidemia: mutation update and functional and structural effects of the variant alleles. Mol. Genet. Metab. 2004;83(1–2):28–37. doi: 10.1016/j.ymgme.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 13.DiMauro S., Schon E.A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348(26):2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 14.Elliott G., O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88(2):223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 15.Endo T., Mitsui S., Nakai M., Roise D. Binding of mitochondrial presequences to yeast cytosolic heat shock protein 70 depends on the amphiphilicity of the presequence. J. Biol. Chem. 1996;271(8):4161–4167. doi: 10.1074/jbc.271.8.4161. [DOI] [PubMed] [Google Scholar]
- 16.Eng C.M., Guffon N., Wilcox W.R., Germain D.P., Lee P., Waldek S., Caplan L., Linthorst G.E., Desnick R.J., G. International Collaborative Fabry Disease Study Safety and efficacy of recombinant human alpha-galactosidase A — replacement therapy in Fabry's disease. N. Engl. J. Med. 2001;345(1):9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 17.Fawell S., Seery J., Daikh Y., Moore C., Chen L.L., Pepinsky B., Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl. Acad. Sci. U. S. A. 1994;91(2):664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gakh O., Cavadini P., Isaya G. Mitochondrial processing peptidases. Biochim. Biophys. Acta. 2002;1592(1):63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 19.Glick B.S., Beasley E.M., Schatz G. Protein sorting in mitochondria. Trends Biochem. Sci. 1992;17(11):453–459. doi: 10.1016/0968-0004(92)90487-t. [DOI] [PubMed] [Google Scholar]
- 20.Harmatz P., Whitley C.B., Waber L., Pais R., Steiner R., Plecko B., Kaplan P., Simon J., Butensky E., Hopwood J.J. Enzyme replacement therapy in mucopolysaccharidosis VI (Maroteaux-Lamy syndrome) J. Pediatr. 2004;144(5):574–580. doi: 10.1016/j.jpeds.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Heitz F., Morris M.C., Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157(2):195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsia Y.E., Scully K.J., Rosenberg L.E. Inherited propionyl-CoA carboxylase deficiency in "ketotic hyperglycinemia". J. Clin. Invest. 1971;50(1):127–130. doi: 10.1172/JCI106466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer S., Thomas R.R., Portell F.R., Dunham L.D., Quigley C.K., Bennett J.P., Jr. Recombinant mitochondrial transcription factor A with N-terminal mitochondrial transduction domain increases respiration and mitochondrial gene expression. Mitochondrion. 2009;9(3):196–203. doi: 10.1016/j.mito.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarver P., Langel U. The use of cell-penetrating peptides as a tool for gene regulation. Drug Discov. Today. 2004;9(9):395–402. doi: 10.1016/S1359-6446(04)03042-9. [DOI] [PubMed] [Google Scholar]
- 25.Fenton W.A., Gravel R.A., Rosenblatt D.S. Vol. 2. 2001. Disorders of propionate and methylmalonate metabolism; pp. 2165–2193. (The Metabolic and Molecular Bases of Inherited Disease). [Google Scholar]
- 26.Joliot A., Prochiantz A. Transduction peptides: from technology to physiology. Nat. Cell Biol. 2004;6(3):189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 27.Jones S.W., Christison R., Bundell K., Voyce C.J., Brockbank S.M., Newham P., Lindsay M.A. Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 2005;145(8):1093–1102. doi: 10.1038/sj.bjp.0706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakkis E.D., Muenzer J., Tiller G.E., Waber L., Belmont J., Passage M., Izykowski B., Phillips J., Doroshow R., Walot I., Hoft R., Neufeld E.F. Enzyme-replacement therapy in mucopolysaccharidosis I. N. Engl. J. Med. 2001;344(3):182–188. doi: 10.1056/NEJM200101183440304. [DOI] [PubMed] [Google Scholar]
- 29.Kang T.S., Stevens R.C. Structural aspects of therapeutic enzymes to treat metabolic disorders. Hum. Mutat. 2009;30(12):1591–1610. doi: 10.1002/humu.21111. [DOI] [PubMed] [Google Scholar]
- 30.Kelson T.L., Ohura T., Kraus J.P. Chaperonin-mediated assembly of wild-type and mutant subunits of human propionyl-CoA carboxylase expressed in Escherichia coli. Hum. Mol. Genet. 1996;5(3):331–337. doi: 10.1093/hmg/5.3.331. [DOI] [PubMed] [Google Scholar]
- 31.Leader B., Baca Q.J., Golan D.E. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 32.Ledley F.D. Perspectives on methylmalonic acidemia resulting from molecular cloning of methylmalonyl CoA mutase. BioEssays. 1990;12(7):335–340. doi: 10.1002/bies.950120706. [DOI] [PubMed] [Google Scholar]
- 33.Mae M., Langel U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr. Opin. Pharmacol. 2006;6(5):509–514. doi: 10.1016/j.coph.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Mahmood I., Green M.D. Pharmacokinetic and pharmacodynamic considerations in the development of therapeutic proteins. Clin. Pharmacokinet. 2005;44(4):331–347. doi: 10.2165/00003088-200544040-00001. [DOI] [PubMed] [Google Scholar]
- 35.Ohura T., Kraus J.P., Rosenberg L.E. Unequal synthesis and differential degradation of propionyl CoA carboxylase subunits in cells from normal and propionic acidemia patients. Am. J. Hum. Genet. 1989;45(1):33–40. [PMC free article] [PubMed] [Google Scholar]
- 36.Neufeld E.F. The Metabolic and Molecular Bases of Inherited Disease. 2001. The mucopolysaccharidoses; pp. 3421–3452. [Google Scholar]
- 37.Palacino J.J., Sagi D., Goldberg M.S., Krauss S., Motz C., Wacker M., Klose J., Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004;279(18):18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 38.Pastukh V., Shokolenko I.N., Wilson G.L., Alexeyev M.F. Mutations in the passenger polypeptide can affect its partitioning between mitochondria and cytoplasm: mutations can impair the mitochondrial import of DsRed. Mol. Biol. Rep. 2008;35(2):215–223. doi: 10.1007/s11033-007-9073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Cerda C., Clavero S., Perez B., Rodriguez-Pombo P., Desviat L.R., Ugarte M. Functional analysis of PCCB mutations causing propionic acidemia based on expression studies in deficient human skin fibroblasts. Biochim. Biophys. Acta. 2003;1638(1):43–49. doi: 10.1016/s0925-4439(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 40.Pooga M., Kut C., Kihlmark M., Hallbrink M., Fernaeus S., Raid R., Land T., Hallberg E., Bartfai T., Langel U. Cellular translocation of proteins by transportan. FASEB J. 2001;15(8):1451–1453. doi: 10.1096/fj.00-0780fje. [DOI] [PubMed] [Google Scholar]
- 41.Rapoport M., Saada A., Elpeleg O., Lorberboum-Galski H. TAT-mediated delivery of LAD restores pyruvate dehydrogenase complex activity in the mitochondria of patients with LAD deficiency. Mol. Ther. 2008;16(4):691–697. doi: 10.1038/mt.2008.4. [DOI] [PubMed] [Google Scholar]
- 42.Rapoport M., Salman L., Sabag O., Patel M.S., Lorberboum-Galski H. Successful TAT-mediated enzyme replacement therapy in a mouse model of mitochondrial E3 deficiency. J. Mol. Med. (Berl) 2011;89(2):161–170. doi: 10.1007/s00109-010-0693-3. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Pombo P., Perez-Cerda C., Perez B., Desviat L.R., Sanchez-Pulido L., Ugarte M. Towards a model to explain the intragenic complementation in the heteromultimeric protein propionyl-CoA carboxylase. Biochim. Biophys. Acta. 2005;1740(3):489–498. doi: 10.1016/j.bbadis.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Sato T., Esaki M., Fernandez J.M., Endo T. Comparison of the protein-unfolding pathways between mitochondrial protein import and atomic-force microscopy measurements. Proc. Natl. Acad. Sci. U. S. A. 2005;102(50):17999–18004. doi: 10.1073/pnas.0504495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiffmann R., Murray G.J., Treco D., Daniel P., Sellos-Moura M., Myers M., Quirk J.M., Zirzow G.C., Borowski M., Loveday K., Anderson T., Gillespie F., Oliver K.L., Jeffries N.O., Doo E., Liang T.J., Kreps C., Gunter K., Frei K., Crutchfield K., Selden R.F., Brady R.O. Infusion of alpha-galactosidase A reduces tissue globotriaosylceramide storage in patients with Fabry disease. Proc. Natl. Acad. Sci. U. S. A. 2000;97(1):365–370. doi: 10.1073/pnas.97.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scriver C.R., Beaudet A.L., Sly W.S., Valle D. McGraw Hill; New York: 1995. The Metabolic and Molecular Bases of Inherited Disease; pp. 1362–1363. [Google Scholar]
- 47.Shokolenko I.N., Alexeyev M.F., LeDoux S.P., Wilson G.L. TAT-mediated protein transduction and targeted delivery of fusion proteins into mitochondria of breast cancer cells. DNA Repair (Amst) 2005;4(4):511–518. doi: 10.1016/j.dnarep.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Ugarte M., Perez-Cerda C., Rodriguez-Pombo P., Desviat L.R., Perez B., Richard E., Muro S., Campeau E., Ohura T., Gravel R.A. Overview of mutations in the PCCA and PCCB genes causing propionic acidemia. Hum. Mutat. 1999;14(4):275–282. doi: 10.1002/(SICI)1098-1004(199910)14:4<275::AID-HUMU1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 49.Viebrock A., Perz A., Sebald W. The imported preprotein of the proteolipid subunit of the mitochondrial ATP synthase from Neurospora crassa. Molecular cloning and sequencing of the mRNA. EMBO J. 1982;1(5):565–571. doi: 10.1002/j.1460-2075.1982.tb01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogtle F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139(2):428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 51.Waltner M., Hammen P.K., Weiner H. Influence of the mature portion of a precursor protein on the mitochondrial signal sequence. J. Biol. Chem. 1996;271(35):21226–21230. [PubMed] [Google Scholar]
- 52.Wraith J.E., Clarke L.A., Beck M., Kolodny E.H., Pastores G.M., Muenzer J., Rapoport D.M., Berger K.I., Swiedler S.J., Kakkis E.D., Braakman T., Chadbourne E., Walton-Bowen K., Cox G.F. Enzyme replacement therapy for mucopolysaccharidosis I: a randomized, double-blinded, placebo-controlled, multinational study of recombinant human alpha-l-iduronidase (laronidase) J. Pediatr. 2004;144(5):581–588. doi: 10.1016/j.jpeds.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 53.Zeviani M. Mitochondrial disorders. Suppl. Clin. Neurophysiol. 2004;57:304–312. [PubMed] [Google Scholar]
- 54.Zhang X., Zhang X., Wang F. Intracellular transduction and potential of Tat PTD and its analogs: from basic drug delivery mechanism to application. Expert Opin. Drug Deliv. 2012;9(4):457–472. doi: 10.1517/17425247.2012.663351. [DOI] [PubMed] [Google Scholar]
- 55.Ziegler A., Nervi P., Durrenberger M., Seelig J. The cationic cell-penetrating peptide CPP(TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence. Biochemistry. 2005;44(1):138–148. doi: 10.1021/bi0491604. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann R., Hennig B., Neupert W. Different transport pathways of individual precursor proteins in mitochondria. Eur. J. Biochem. 1981;116(3):455–460. doi: 10.1111/j.1432-1033.1981.tb05357.x. [DOI] [PubMed] [Google Scholar]
- 57.Nagahara H., Vocero-Akbani A.M., Snyder E.L., Ho A., Latham D.G., Lissy N.A., Becker-Hapak M., Ezhevsky S.A., Dowdy S.F. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 1998;4(12):1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material