Abstract
Traditional views of synthetic biology often treat the cell as an unstructured container in which biological reactions proceed uniformly. In reality, the organization of biological molecules has profound effects on cellular function: not only metabolic, but also physical and mechanical. Here, we discuss a variety of perturbations available to biologists in controlling protein, nucleotide, and membrane localization. These range from simple tags, fusions, and scaffolds to heterologous expression of compartments and other structures that confer unique physical properties to cells. Next, we relate these principles to those guiding the spatial environments outside of cells such as the extracellular matrix. Finally, we discuss new directions in building intercellular organizations to create novel symbioses.
Cells are highly structured, spatially separating incompatible and functionally distinct processes. Various engineering strategies (e.g., heterologous expression of compartments) can be used to modify cellular organization.
Spatial organization of biological molecules is foundational to life. The localization of proteins to distinct regions of the cell—be they as simple as the cytosol, membrane, and nucleoid—restricts and directs enzymatic interactions. The division of cellular contents into compartments further separates otherwise incompatible chemical and enzymatic microenvironments. And finally, the organization of proteins into polymeric structures endows cells with the ability to control their mechanical properties and exert forces in their environments.
Synthetic biology is typically understood to mean endowing cells with novel functions based on genetic manipulations. Of course, modifying transcription necessarily modifies the protein environment in the cell as well. However, simply expressing proteins that are assumed to act independently of their environment severely limits the potential of this discipline to generate new cellular behaviors. Here, we describe a range of engineered modifications to cellular organization. Some of these have reached mainstream use in studying and perturbing cellular functions, whereas others have unrealized potential.
DIRECTING PROTEIN LOCALIZATION
A fundamental source of enzymatic organization within cells is the direction of proteins to specific compartments or locations within the cell (Blobel and Sabatini 1971). These can include the nucleus (Silver et al. 1984), mitochondria (Rapaport 2003), chloroplasts (Tian and Okita 2014), vacuoles (Washida et al. 2009), or, in the case of Gram-negative bacteria, the periplasm (Danese and Silhavy 1998). As the responsible elements are amino-terminal sequences on the proteins, this localization mechanism can be readily exploited to direct proteins to novel locations. For example, this technology has recently been combined with variable splicing to create reporters with differential localization to plant cell chloroplasts, peroxisomes, and the cytosol (Voges et al. 2013).
In addition to directing proteins to specific localizations in the cell, their oligomerization state can be modulated. Glutathione S-transferase tags typically used for affinity purification form dimers (Parker et al. 1990), and this property alters protein function such as by increasing the binding affinity of a DNA-binding protein (Niedziela-Majka et al. 1998). This property has been exploited as a test of the functional role of the oligomeric state (Lee et al. 2003).
Although induced hetero- or homodimerization is a common technique in molecular and cell biology, these tools and approaches have, nevertheless, been expanded in interesting ways. In addition to localizing individual proteins, they have been used to move whole organelles. A synthetic heterodimerization “crutch” linking the endoplasmic reticulum (ER) to mitochondria was used in a screen to identify factors that naturally colocalize these organelles (Kornmann et al. 2009). Dimerization can also be inducible, as well as constitutive. The FK506-binding protein (FKBP) can be used as a tag that dimerizes on withdrawal of its chemical ligand; mutants can reversibly convert between monomers and dimers (Rollins et al. 2000). Furthermore, engineered phytochrome interaction factor (PIF) pairs are induced to dimerize with light; this permits optogenetic control of protein spatial localization, even in regions of single cells (Levskaya et al. 2009). And, finally, dimerizing tags are available in a variety of affinities, including those that form covalent bonds. The SpyCatcher and SpyTag system is a derivative of a bacterial fibronectin-binding protein that irreversibly forms an isopeptide bond on interaction (Zakeri et al. 2012).
In addition to directing the localization of single proteins, tags can be used to make larger aggregates, which can, in turn, be used to probe cellular function. For example, FM repeats (tandem repeats of FKBP domains) have been fused to enzymes in the Golgi apparatus, causing them to polymerize into aggregates on withdrawal of the ligand. The accumulation of these polymerized enzymes in the cis- or trans-Golgi have been used to investigate models of cisternal maturation (Rivera et al. 2000; Lavieu et al. 2013; Rizzo et al. 2013).
SCAFFOLDS
Directing the localization of proteins can be used to not only probe cellular function, but also enhance enzymatic activity, or to achieve a metabolic or signaling effect. Multiple proteins can be scaffolded, or coordinated with specified geometry, to enhance their activity. The exact nature of enzymatic enhancement on scaffolds is likely to be a combination of several factors, including a high local concentration of substrates and their “channeling” within a shared hydration shell. The latter has been proposed after introducing an inert bridge protein between glucose oxidase and horseradish peroxidase on a DNA scaffold in vitro, thereby increasing the enzymatic output compared with similar scaffolds lacking the bridge (Fu et al. 2012).
Scaffolding to enhance enzymatic activity occurs naturally in cells. For example, fatty acid synthetase is a single protein with seven enzymatic centers; their direct translational fusion is likely to confer metabolic benefits (Smith 1994). Multiple polypeptides can also be brought together by protein–protein interactions; in Bacillus subtilis, the nonribosomal peptide synthetases and polyketide synthetase enzymes assemble to form an ∼2.5 MD complex, and these complexes are further localized together in one focus in the cell (Straight et al. 2007).
From a synthetic biology perspective, the simplest engineered enzymatic enhancement is by direct fusion of one protein to another. This approach was used to achieve 15-fold enhancement of resveratrol production in yeast and human cells (Zhang et al. 2006b). Scaffolds, however, offer the additional flexibility of linking components by protein–protein interactions. An early example is a modification of the bacterial cellulosome, which is an assembly that organizes cellulose-degrading enzymes. This work enhanced the natural cellulolytic activity of Clostridium by combining enzymes and binding domains from different species (Fierobe et al. 2001). A synthetic protein scaffold with more distantly related components was created in 2009 by recruiting yeast metabolic enzymes to metazoan proteins that naturally interact to function in signaling. This improved melvalonate production between threefold and more than 70-fold in Escherichia coli, depending on the concentration of enzymes expressed—at lower concentrations of enzymes, scaffolds were more beneficial, presumably because of a greater relative increase in effective concentration (Dueber et al. 2009). These protein scaffolds are limited in the potential spatial organization of the interacting domains, and by the fact that their assembly is not, by nature, polymeric.
In contrast, the modular properties of nucleic acids can be used to create structures that offer both theoretically infinite size and fine control of enzymatic orientation. RNA can be constructed scaffolds in varying dimensionalities—zero-, one-, and two-dimensional scaffolds can be constructed by varying the base pairing between modular segments (Delebecque et al. 2012). The number of interacting proteins is limited only by the number of aptamers (and aptamer-binding proteins fused to the enzymes of interest). Much like protein scaffolds, RNA scaffold can achieve >50-fold enzymatic enhancements in E. coli, although the limited stability of RNA may affect scaffold performance over long timescales (Delebecque et al. 2012).
The potential problems with RNA stability may be alleviated with the use of DNA scaffolds instead, although this platform presents its own inherent limitations. For example, DNA topology is limited to a single dimension, and supercoiling is also a potential problem. Nevertheless, DNA scaffolds have been used to generate up to fivefold enhancements in resveratrol, 1,2-propanediol, and mevalonate production in E. coli (Conrado et al. 2012). Each enzyme was fused to a zinc-finger protein; the diversity of this protein family suggests that many enzymes could be arranged in a specific array.
MEMBRANE BARRIERS
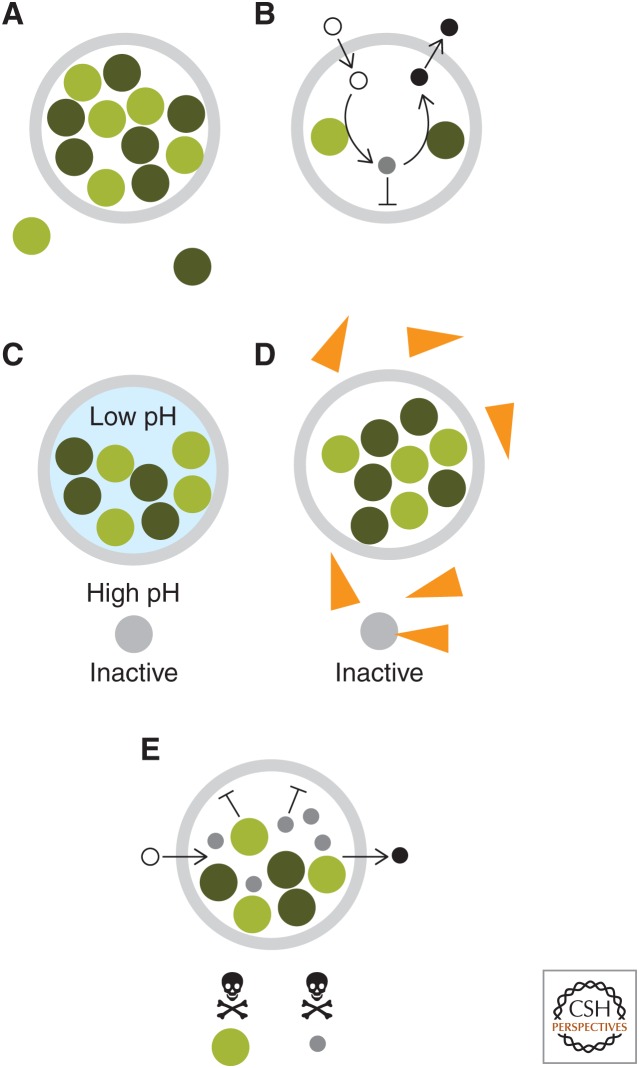
Although scaffolds may organize high concentrations of enzymes in specific orientations, the surrounding environment remains essentially identical to the cellular compartment in which they form. This could be disadvantageous for several reasons (Fig. 1). First, reactions may produce toxic products, byproducts, or intermediates, such as the formaldehyde formed as an intermediate of methanol metabolism (Fassel et al. 1992). Second, enzymes may need to be insulated from certain substrates to prevent nonproductive side reactions—for example, RuBisCO can catabolize a reaction with molecular oxygen rather than carbon dioxide, drastically reducing enzymatic efficiency. Third, enzymes may require a specific chemical environment in terms of pH, redox potential, or ionic strength that is distinct from the rest of the cytosol, such as is created in mitochondria.
Figure 1.
Potential benefits of compartmentalization (with gray circles representing intracellular compartments). (A) Concentration of enzymes. (B) Increased concentration of intermediates through a selectively permeable diffusion barrier. (C) Maintenance of a chemical microenvironment essential for enzymatic function. (D) Protection of enzymes from molecules from deactivators or competitors. (E) Isolation of toxic enzymes of intermediates from the cell.
Because most compartments in eukaryotes are membrane based, it is no surprise that many engineering attempts have focused on membrane encapsulation. The most readily implemented form of this technology is the use of targeting sequences to localize enzymes to existing organelles. For example, up to five enzymes in an isobutanol synthesis pathway have been directed to yeast mitochondria; there, they provide modest (approximately threefold) enhancements overexpression in the cytoplasm (Avalos et al. 2013).
In addition, new membrane topologies can also be created from the ER, often through the overexpression of foreign membrane-associated proteins. In tobacco plants, viral proteins form whorled membrane structures termed Z-membranes that appear to derive from the ER (Gong et al. 1996). By making fusion to these viral proteins, various enzymes can be localized on the cytoplasmic surface of these membranes. In mammalian tissue culture cells, ER derivatives having a cuboid or crystalline morphology (termed organized smooth ER) can be generated by expression of certain proteins (Yamamoto et al. 1996; Snapp et al. 2003). This morphology can be produced with chimeric proteins that inducibly dimerize, enabling the topology to be dynamically controlled (Lingwood et al. 2009).
Vesicles have also been generated from other membrane sources, for example, from the cell membrane of E. coli (Fig. 2). This was accomplished by overexpressing foreign bacterial glucosyltransferases (Eriksson et al. 2009) or by expressing eukaryotic calveolin (Walser et al. 2012). The latter strategy produces morphologically normal calveolae 50 nm in diameter that encapsulate contents derived from the periplasm. Furthermore, by making fusions to the calveolin, the vesicles can be functionalized with proteins that allow them to be targeted to specific cell types after purification.
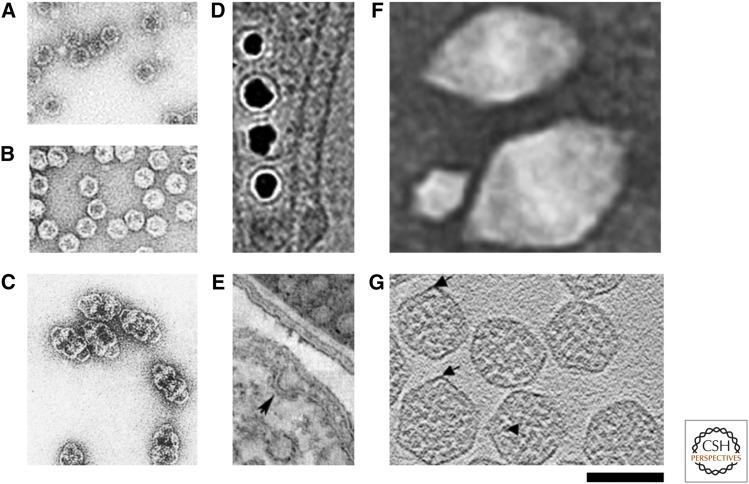
Figure 2.
Transmission electron micrographs (TEMs) of cellular compartments, scaled to their relative size. (A) Lumazine synthase (Zhang et al. 2006a). (B) Encapsulin (Sutter et al. 2008). (C) Vaults (Kedersha and Rome 1986). (D) Magnetosomes near the cell membrane (Komeili et al. 2006). (E) Calveolae: budding from the cell membrane is indicated with an arrow (Walser et al. 2012). (F) Gas vesicles (Pfeifer 2012). (G) α carboxysomes: arrows indicate vertexes, triangle indicates RuBisCO (Schmid et al. 2006). Scale bar, 100 µm.
In addition to directing encapsulation within membrane-bound compartments, we are also able to control release of contents from membrane-bound compartments, such as the cell itself. This takes the shape of inducible bacterial lysis, reviewed in Rice and Bayles (2008). These strategies can involve the expression of lysozyme and pore-forming proteins in response to varied triggers (Pasotti et al. 2011). However, bacteria can also be made to kill specific foreign strains of bacteria with the use of specialized protein machines. For example, pyocins are contractile apparati resembling phage tails that kill target bacteria by depolarizing bacterial membranes (Uratani and Hoshino 1984). Pyocins can be engineered to kill varied bacteria by substitution of their tail fiber proteins from strain-specific phage (Williams et al. 2008). R bodies, proteinaceous toxin delivery devices resembling needles that can extend up to 20-μm long, function to mix the cytoplasms of the expressing bacteria with that of a predatory eukaryote in response to the low-pH environment of a phagosome (reviewed in Pond et al. 1989). As R-bodies can break foreign membranes to release encapsulated protein cargo (Polka and Silver 2016), they could be used to release or mix contents from multiple cells, synthetic vesicles, or compartments.
MICROCOMPARTMENTS
Although further afield from the traditional conception of eukaryotic organelles, proteinaceous compartments may offer a more flexible, robust, and practical platform for engineering compartmentalization. Perhaps the most versatile are microcompartments: icosahedral protein structures, typically between 100 and 200 nm in diameter, found in diverse bacteria. They naturally encapsulate and insulate enzymes from the cellular environment with faceted, selectively permeable shells composed of pentameric and hexameric proteins. Three of the best-studied microcompartment classes include Pdu (propanediol use) microcompartments; Eut (ethanolamine use) microcompartments; and carboxysomes, which contain carbonic anhydrase and RuBisCO. Carboxysomes form two major families: α and β carboxysomes, which differ in the type of RuBisCO incorporated and also by the importance of the matrix in organizing the shell (Schmid et al. 2006; Rae et al. 2013). Microcompartments are now known to be more widespread than originally thought, with 17% of sequenced genomes bearing sequences thought to encode microcompartments of these and other types (Jorda et al. 2013).
Microcompartments are of particular interest because they can be assembled from a small number of protein components and are, therefore, more portable to heterologous expression systems. For example, Pdu compartments from Citrobacter freundii have been made in E. coli by heterologous expression (Parsons et al. 2008); the empty shell can also be made by expressing only five proteins (Parsons et al. 2010). Because the lumen components were not transferred in the latter study, the compartments are hollow, leaving the opportunity for them to be filled with alternate cargo, as discussed below. Pdu compartments from Salmonella entericum have also been made in E. coli (Sargent et al. 2013), and empty Eut shells have been reported to form in E. coli as well (Choudhary et al. 2012). Finally, α carboxysomes from Halothiobacillus neapolitanis have been expressed in E. coli from a single operon (Bonacci et al. 2012). Complete β carboxysomes have, so far, resisted heterologous expression, perhaps because of a more complicated genetic organization spanning several operons. However, expression of β carboxysome shell proteins in E. coli does produce capsid-like structures (albeit smaller than carboxysomes) that associate with heterologous cargoes tagged with targeting sequences (Cai et al. 2016).
Heterologous expression of carboxysomes, even with their native contents, is of particular interest in light of the goal of improving carbon fixation in plants. Carbon fixation is a metabolic bottleneck for many plants, and the introduction of carboxysomes into the cells of agriculturally important organisms could theoretically increase crop yields by 36%–60% (McGrath and Long 2014). Although the cyanobacterial RuBisCO is faster than plant RuBisCO, it is also more sensitive to molecular oxygen, and thus is detrimental to the growth of its host without encapsulation. Thus, encapsulating this enzyme will be necessary to reap any metabolic benefits of cyanobacterial RuBisCO. Progress has been made toward this end in the expression of carboxysome components in tobacco, with shell and RuBisCO both being successfully expressed (Lin et al. 2014a,b). However, a complete carboxysome in plants has yet to be achieved.
For microcompartments to have novel functions, heterologous cargoes must be targeted to them. In general, this is accomplished by exploiting natural mechanisms of targeting cargoes to the microcompartment lumen. Across many families of microcompartments, short regions at the termini of cargo proteins are thought to form small α helices that bind to the interior surfaces of shell proteins (Fan et al. 2012). The best-developed platform for heterologous targeting is the Pdu microcompartment system. The terminus of PduP from Salmonella enterica is necessary and sufficient to drive cargoes to the microcompartment. Green fluorescent protein (GFP) tagged with Pdu1–18 cannot only localize to the compartments with fluorescence microscopy, but it is also protected from immunoprecipitation from cell lysates after disruption; this suggests it is located inside the compartment rather than simply associated with the exterior. In addition to PduP, a second enzyme, PduD, also contains an 18-amino-acid sequence sufficient for targeting heterologous cargoes to the S. enterica Pdu compartment. PduD is also required for recruiting the other two members of its natural complex, PduCDE, suggesting that not only direct fusions, but also protein–protein interactions, could be used to localize enzymes to the S. enterica compartment (Fan and Bobik 2011).
Targeting of engineered cargoes should ideally be functional in a heterologous system. PduC, D, and V can all direct GFP to colocalize with heterologously expressed Pdu compartments in E. coli (Parsons et al. 2010). Also in this system, PduP1–18 and PduD1–18 can be used to target two enzymes involved in ethanol production to the compartment, thereby increasing ethanol yield (Lawrence et al. 2014). It is possible that the enzymes are located on the outside, rather than the inside of the compartment, but nonetheless the Pdu structure succeeds as a scaffold. Finally, a targeting system exists in β carboxysomes. The carboxyl terminus of CcmN is responsible for directing it to the Synechococcus elongatus carboxysome (Kinney et al. 2012). The peptide is also necessary for the formation of the β carboxysome shell, although it does not exist in α carboxysomes. Although the CcmN peptide has not yet been shown to localize heterologous cargo, proteins can be targeted to the lumen of the carboxysome by fusion to full-length, native cargoes, like RuBisCO. These fusions, at least in the case of fluorescent proteins, do not impact carboxysome structure, function (Savage et al. 2010), or the establishment of an internal oxidizing microenvironment (Chen et al. 2013), implying that carboxysomes could accommodate small enzymatic cargoes and retain their function.
To optimize enzymatic activity in any engineered compartment, the chemical microenvironments may need to be engineered. The concentration of small molecules inside microcompartments is thought to be controlled by the size and charge of the pores formed in the middle of shell protein hexamers; by exchanging residues in the pores, the selectivity of the permeability barrier could, theoretically, be changed. As a step toward this goal, chimeric carboxysome shells have been made. In these experiments, a shell protein (CsoS1) from Prochlorococcus marinus colocalized with S. elongatus carboxysomes, and even structurally complemented a deletion of their major shell protein (Cai et al. 2014). Although CsoS1 shares <50% similarity with S. elongatus shell proteins, this study shows that components of α (P. marinus) and β (S. elongatus) carboxysomes can be interchangeable, expanding future engineering possibilities.
In addition to the permeability of the shell, the functionality of an engineered microcompartment would be influenced by its size: not only through the number of molecules that could be encapsulated, but also in the surface to volume ratio, which may influence the compartment’s microenvironment. There is a large natural range in microcompartment diameters—from 40-nm Haliangium ochraceum compartments to 500-nm β carboxysomes in Spirulina platensis (Rae et al. 2013; Lassila et al. 2014). In addition, the size of at least one species of β carboxysome can be modulated genetically. In S. elongatus, the protein CcmM is localized to the lumen of carboxysome, where it organizes RuBisCO into a solid matrix. CcmM is present in two isoforms; deleting the short isoform drives the formation of small carboxysomes (Long et al. 2010), whereas fusing a tag to CcmM drives the formation of larger carboxysomes (Long et al. 2007).
ENCAPSULINS
Although microcompartments have shells made up of multiple proteins, a second type of bacteria compartment, encapsulins, have only one shell protein and are characterized by smaller diameters, ∼30 nm. They typically encapsulate proteins involved in oxidative stress responses, such as ferritin-like proteins or peroxidases.
The first encapsulin was discovered in Pyrococcus furiosus. This compartment is made out of only one protein that functions both as its shell and enzymatic cargo (Namba et al. 2005). Related compartments were later found in other organisms, including Thermatoga maritima; this encapsulin is composed of 60 monomers of a single subunit that form a cage 24 nm in diameter (Sutter et al. 2008). The crystal structure of the shell protein shows structural similarity to viral capsids, but like microcompartments, there is no homology on the sequence level. These investigators also expressed the two-component encapsulin operon from Brevibacterium linens in E. coli and found that the cargo, a single oligomeric complex, is localized within the lumen of the compartments, supporting the hypothesis that a carboxy-terminal sequence targets cargoes to encapsulin lumens.
The purified encapsulin from Rhodococcus jostii can be assembled and disassembled in vitro in a pH-dependent fashion, and the assembly of the encapsulin enhances the enzymatic activity of the peroxidase cargo (Rahmanpour and Bugg 2013). The Rhodococcus erythropolis encapsulin is capable of both being expressed heterologously in E. coli and of packaging heterologous cargo. Proteins tagged with the carboxy-terminal 37 amino acids of its natural cargo are protected from trypsin proteolysis in the presence of the shell (Tamura et al. 2015).
Myxococcus xanthus also produces an encapsulin compartment; the native compartment copurifies with not one, but three cargo proteins. This suggests that these structures could enclose multiple proteins and perhaps support encapsulation of whole enzymatic pathways (McHugh et al. 2014). This encapsulin can be expressed in E. coli, although the heterologously expressed protein forms some 18-nm compartments, as well as those of the native 32-nm size. Furthermore, encapsulins are also being explored as a platform for targeted drug delivery (Moon et al. 2014).
LUMAZINE SYNTHASE
A third type of proteinaceous compartment found in bacteria is composed of lumazine synthase, an enzyme involved in riboflavin biosynthesis. In some bacteria, yeast, and plants, this protein forms pentamers (Baldi et al. 2000; Meining et al. 2000), but in other species these pentamers are further assembled into capsids, namely, in B. subtilis (Ladenstein et al. 1994) and Aquifex aeolicus (Zhang et al. 2001). The capsids from the latter system, termed AaLS, can be expressed and purified from E. coli; they assemble, in vitro, into compartments either 16 or 30 nm, depending on pH and buffer conditions (Zhang et al. 2006a). They have no native cargo.
To facilitate encapsulation of heterologous cargoes, the internal surface of lumazine synthase has been mutated to increase electrostatic interactions by charge complementarity. This strategy has been useful for encapsulating proteins bearing a positively charged stretch of 10 arginine residues on the carboxyl terminus coexpressed with these modified AaLS compartments in E. coli (Seebeck et al. 2006). Furthermore, this approach can raise the tolerated expression level of human immunodeficiency virus (HIV) protease, which is toxic to E. coli (Wörsdörfer et al. 2011b). This selection formed the basis of a screen for mutants of AaLS that could better protect its host, presumably through improving encapsulation owing to increased negative charges on its interior surface. These improved capsids, called AaLS-13, can be isolated and loaded in vitro with monomers of GFP that have been mutated to bear 29 additional positively charged residues (Wörsdörfer et al. 2011a), or even with similarly engineered human ferritin (Beck et al. 2014). Ferritin is itself a small microcompartment, ∼12 nm in diameter, that binds and stores iron. Thus, the encapsulation of human ferritin into A. aeolicus lumazine synthase generates a nested synthetic organization. It is also reminiscent of natural encapsulins that can bear ferritin cargoes, but here the interaction is dependent on engineered properties of the compartments, which come from different domains of life.
Lumazine synthase capsids have been engineered to produce vaccines because they can act as scaffolds for antigen presentation and are themselves immunogenic (Alfano et al. 2015; Du and Wang 2015). They have also been fused to a chelating agent for use as a contrasting agent for magnetic resonance imaging (MRI) (Song et al. 2015).
VAULTS
Not all protein compartments have a bacterial origin. Vaults, lemon-shaped barrels 70-nm long and 40 nm in diameter, are found in the cytoplasm of eukaryotic cells, but their function is unclear. They are naturally composed of three proteins and an untranslated RNA (Kedersha and Rome 1986; Kickhoefer et al. 1993), but can be purified from insect cells by expression of the major structural protein alone (Stephen et al. 2001). A domain called INT, found on one of the nonessential vault proteins (Kickhoefer et al. 1999), is sufficient to target heterologous cargoes to vaults expressed in insect cells (Kickhoefer et al. 2005). These INT-tagged proteins can be visualized in cryo-EM reconstructions as two rings of additional density encircling each half of the barrel. INT tagging also has biochemical effects. Targeting a fluorescent protein with vaults with INT reduces the speed with which fluorescence decreases in response to high ionic strength, suggesting that they are partially protected from solution. Furthermore, INT-tagged luciferase is functional, yet displays reduced kinetics, suggesting that enzymes can function inside vaults (Kickhoefer et al. 2005). From an engineering perspective, this approach has been used to package a Chlamydia trachomatis protein into vaults for vaccine development (Zhu et al. 2014). Manganese peroxidase packaged within vaults is more stable, suggesting potential benefits for remediation of pollutants (Wang et al. 2015).
Because vaults are dynamic structures, cargoes can be loaded in vitro (Poderycki et al. 2006); the two halves of the vault can even completely exchange (Yang et al. 2010). These approaches can be used to package inorganic compounds, like Ni-NTA-nanogold, into the vaults (Goldsmith et al. 2009).
Vaults proteins can also be engineered to gain new functions. For example, an amphipathic helix was fused to the amino terminus of the major vault protein, causing lipids to be encapsulated in the vault (Buehler et al. 2014). The lipid-containing vaults bind with greater avidity to fat-soluble drugs. Purified vaults are readily taken up by cells (Kickhoefer et al. 2005), but fusing a viral endosomolytic peptide enhances the delivery of DNA and other molecules to the cytoplasm (Han et al. 2011).
MAGNETOSOMES
In nature, the function of complex structures is not limited to isolating enzymatic activity. Packing of specialized materials—metals and gas—can serve important roles in natural cellular migration, and have been exploited for diverse practical purposes as well.
Magnetosomes are intracellular structures that enable some bacteria to position themselves using the Earth’s magnetic field. Morphologically, they are invaginations of the cell membrane that enclose magnetite crystals 50 nm in diameter; these are spatially distributed through the cell by an actin homolog (Komeili et al. 2006). Although they are encoded by large islands of genes, magnetosomes from Magnetospirillum gryphiswaldense have been heterologously expressed in Rhodospirillum rubrum, with their order and morphology preserved (Kolinko et al. 2014).
MamC, a protein that localizes to the membrane, has been tagged with cameloid antibody fragments (i.e., “nanobodies”) in M. gryphiswaldense. Because this fusion enables interactions between magenetosomes and the nanobody target protein both in vitro and in vivo, it can be used to generate an intracellular “nanotrap” that localizes natural proteins to the magnetosome in living cells (Pollithy et al. 2011).
Magnetosomes also have many possible applications after purification, for example, their patterning on a surface could be used to create a magnetic memory device (Galloway et al. 2012). Furthermore, a phosphohydrolase has also been fused to MamC in Magnetospirillum magneticum to create magnetosomes that could be purified and used in vitro to degrade a pesticide (Ginet et al. 2011). Magnetosomes from this organism can also be functionalized with multiple enzymes with defined stoichiometry for cellulose hydrolysis, among many other fusion partners (Honda et al. 2015).
Magnetic particles in yeast have also been produced, although these inclusions lack the structure of magnetosomes (Nishida and Silver 2012). The particles will form naturally in Saccharomyces cerevisiae in the presence of ferric citrate, but the yield and size of the particles can be increased by knocking out a yeast iron transporter and heterologously expressing human ferritin.
GAS VESICLES
Gas vesicles constitute another type of genetically encoded, specialized compartment. Used to regulate bacterial buoyancy during migration to appropriate parts of the water column, gas vesicles are protein-enclosed structures that exclude water and so contain only gas. They have a variety of physical morphologies, including spindles, cylinders, and bicones, and they can grow to sizes ranging from 100 nm to 2 µm in length, depending on the species (Pfeifer 2012).
Bacillus megaterium gas vesicles have been heterologously expressed in E. coli (Li and Cannon 1998). Because gas vesicles make cells float, they have been proposed as a tool to facilitate harvests of bacteria in an industrial setting through the formation of a pellicle. Purified gas vesicles can also be used to display antigenic peptides (Stuart et al. 2001), to oxygenate tissue culture media (Sundararajan and Ju 2006), and to serve as contrast agents for ultrasound imaging (Shapiro et al. 2014).
CELL MORPHOLOGY AND THE CYTOSKELETON
In eukaryotic cells, long-range order is achieved through dynamic cytoskeletal proteins and reaction-diffusion systems. If these systems could be controlled, cell shape, migration, and cell size could all be potentially regulated. Currently, the eukaryotic actin and microtubule cytoskeletons can be exploited to direct cargoes to specific positions within cells (reviewed in Goodman et al. 2012). For example, a DNA-binding domain fused to a portion of a microtubule motor, a dynein light chain, can enhance transfection affinity, presumably because the reagent enables the microtubule cytoskeleton to move the DNA toward the nucleus (Toledo et al. 2012). Furthermore, targeting peroxisomes to motors can direct these organelles toward either the nucleus or the periphery of the cell, depending on the polarity of the motors (Kapitein et al. 2010a,b).
Although bacteria and archaea lack ubiquitous, persistent load-bearing cytoskeletons in the traditional sense, they do have protein polymers and reaction-diffusion systems that regulate division, cell-wall deposition, and DNA segregation. In addition, these proteins also control the position of compartments like carboxysomes (Savage et al. 2010) and magnetosomes (Komeili et al. 2006). Therefore, these proteins could play an important role in specifying the intracellular location of synthetic structures in the future.
THE EXTRACELLULAR ENVIRONMENT
Biological organization does not stop at the plasma membrane. In bacteria, the extracellular environment provides a means for protection against toxins and mechanical forces. This matrix can also be modified for scaffolding applications, and to create novel structural and mechanical properties. For example, curli protein forms amyloid fibrils and is a major component of the extracellular matrix made by E. coli and related bacteria. By placing curli protein under an inducible promoter, biofilm production can be induced with an external signal (Chen et al. 2014). Curli fibrils have also been tagged with metal-binding peptides to enable them to bind gold nanoparticles and quantum dots to make “nanowires,” and the proportion of tagged curli subunits can be regulated temporally with different inducers (Chen et al. 2014). Additionally, curli fibrils can be covalently decorated, via SpyCatcher/SpyTag, with any protein of interest (Nguyen et al. 2014).
The extracellular matrix of eukaryotic cells can also be modified to influence behavior. For example, a SpyCatcher/SpyTag cross-linked elastin network that incorporates leukemia-inhibitory factor has been used as a growth medium for mouse embryonic stem cells; this medium maintains stem-cell pleuripotency (Sun et al. 2014). In theory, this matrix could be genetically encoded.
In addition to matrixes, bacteria make other extracellular structures with biological relevance that are amenable to engineering. For example, Geobacter sulfurreducens makes extracellular magnetite particles, the size and composition of which can be altered by changing the composition of available metals (Byrne et al. 2013). A second example is exosomes, extracellular vesicles up to 200 nm in diameter, that naturally package messenger RNA and proteins. Exosomes are used to communicate between cells, and have been engineered to incorporate heterologous cargoes (Marcus and Leonard 2013). Similarly, in Gram-negative bacteria, outer membrane vesicles are produced naturally for many purposes, including carrying toxins and protecting bacteria from antibiotics and phage (reviewed in Kulp and Kuehn 2010; Avila-Calderón et al. 2015). They are also the focus of active engineering efforts, for example, in the production of vaccines (Chen et al. 2010).
SYMBIOSIS
Finally, compartments within cells can be cells themselves. For example, diatoms such as Hemiaulus hauckii internalize cyanobacteria like Richelia intracellularis; these contain dedicated nitrogen-fixing cells called heterocysts. The specialized compartments allow the cyanobacteria, and thus the diatom, to reap the benefits of both nitrogen fixation and oxygenic photosynthesis (Carpenter et al. 1999; Schouten et al. 2013).
Recapitulating endosymbiotic interactions is a goal of particular interest because it could lead to the development of synthetic organelles. For example, cyanobacteria have been engineered to escape from lysosomes; they have been shown to survive inside zebrafish and mammalian macrophages (Agapakis et al. 2011). Notably, similarly treated E. coli were severely cytotoxic in these experiments, suggesting that S. elongatus may be inherently better suited to engineering stable endosymbiotic interactions in the future.
Cells do not need to engulf—or be engulfed by—their symbiont to take advantage of inherently dissociated cellular components and capacities. Organization into a consortium allows for the division of labor among different cells, which effectively act as individual compartments. In the example of Chlorochromatium aggregatum, the consortium is made up of a single β-proteobacterium surrounded by green sulfur bacteria (Overmann 2010). Although the nonmotile green sulfur bacteria can live alone, they benefit from the specialized motility of the flagellated β-proteobacterium that shows chemotaxis toward light, despite the bacterium not being photosynthetic (Frostl and Overmann 1998). The motility acquired through symbiosis gives the green sulfur bacteria an advantage over other nonmotile competition. Presumably, the β-proteobacteria also benefits through the provision of photosynthetically fixed carbon.
Often this division of labor endows consortia with capacities far beyond those of individual species. Visually stunning examples of this are microbial mats in which colored striations represent different metabolic capacities: gold, green, pink, peach, or dim green layers correspond to organisms growing together capitalizing on different photosynthetic machinery with metabolisms that differ in ability to withstand exposure to oxygen, reduce sulfate, and fix nitrogen (Stal et al. 1985).
Symbiotic consortia appeal to engineers for two reasons: (1) division of labor lends itself well to modularity, and (2) it is widely believed that consortia are more stable than monocultures. In this vein, synthetic biologists have engineered consortia made up of different strains of the same organism, as well as symbioses involving multiple species. There have been many applied examples of the utility of symbiosis, especially in isobutanol and lipid production from sunlight or cellulosic feedstocks (Ortiz-Marquez et al. 2012; Do Nascimento et al. 2013; Minty et al. 2013). Although at their simplest these artificial consortia are merely novel combinations of existing wild-type strains, there are many engineering interventions that can improve their stability. For example, engineered auxotrophies force interdependence (Shou et al. 2007; Wintermute and Silver 2010; Müller et al. 2014), and quorum-sensing systems can be used to facilitate communication between consortium members (Brenner et al. 2007; Weber et al. 2007; Chen et al. 2015).
It is worth noting that symbiosis can also involve metazoan partners. Consider the bioluminescence provided by Vibrio fischeri in the bobtail squid (Naughton and Mandel 2012), amino acids provided by bacterial endosymbionts in aphids (Hansen and Moran 2011), organic carbon and oxygen provided by photosynthetic algae in corals (Roth 2014), and our own microbiomes. These and other examples of natural symbiosis will provide fertile ground for future engineering efforts.
CONCLUDING REMARKS
The ultimate goal of these engineering efforts is to control not only the contents of cells, but also the spatial relationship of these contents to one another. Given that the activity of proteins is dependent on both their potential interacting partners and their local chemical environment, cellular organization must be manipulated to explore the full range of potential biological activity. To gain this mastery, synthetic biologists must adopt into their toolkit some microscopy and biochemical methods associated with traditional cell biology.
ACKNOWLEDGMENTS
We are grateful to Arash Komeili and Ertan Ozyamak (UC Berkeley) for helpful discussions regarding magnetosomes. We thank the National Science Foundation (MCB-1409586, DGE1144152), Jane Coffin Childs Fund, the Wyss Institute for Biologically Inspired Engineering, Department of Energy (DE-SC0012658), and Defense Advanced Research Projects Agency Living Foundries (HR0011-14-C-0072).
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Agapakis CM, Niederholtmeyer H, Noche RR, Lieberman TD, Megason SG, Way JC, Silver PA. 2011. Towards a synthetic chloroplast. PLoS ONE 6: e18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano EF, Lentz EM, Bellido D, Dus Santos MJ, Goldbaum FA, Wigdorovitz A, Bravo-Almonacid FF. 2015. Expression of the multimeric and highly immunogenic Brucella spp. lumazine synthase fused to bovine rotavirus VP8d as a scaffold for antigen production in tobacco chloroplasts. Front Plant Sci 6: 1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos JL, Fink GR, Stephanopoulos G. 2013. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31: 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Calderón ED, Araiza-Villanueva MG, Cancino-Diaz JC, López-Villegas EO, Sriranganathan N, Boyle SM, Contreras-Rodríguez A. 2015. Roles of bacterial membrane vesicles. Arch Microbiol 197: 1–10. [DOI] [PubMed] [Google Scholar]
- Baldi PC, Velikovsky CA, Braden BC, Giambartolomei GH, Fossati CA, Goldbaum FA. 2000. Structural, functional and immunological studies on a polymeric bacterial protein. Braz J Med Biol Res 33: 741–747. [DOI] [PubMed] [Google Scholar]
- Beck T, Tetter S, Künzle M, Hilvert D. 2014. Construction of Matryoshka-type structures from supercharged protein nanocages. Angew Chem Int Ed Engl 54: 937–940. [DOI] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD. 1971. Ribosome-membrane interaction in eukaryotic cells. In Biomembranes (ed. Manson LA), pp. 193–195. Springer, Berlin. [Google Scholar]
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. 2012. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci 109: 478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, Karig DK, Weiss R, Arnold FH. 2007. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci 104: 17300–17304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler DC, Marsden MD, Shen S, Toso DB, Wu X, Loo JA, Zhou ZH, Kickhoefer VA, Wender PA, Zack JA, et al. 2014. Bioengineered vaults: Self-assembling protein shell-lipophilic core nanoparticles for drug delivery. ACS Nano 8: 7723–7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JM, Coker VS, Moise S, Wincott PL, Vaughan DJ, Tuna F, Arenholz E, van der Laan G, Pattrick RAD, Lloyd JR, et al. 2013. Controlled cobalt doping in biogenic magnetite nanoparticles. J R Soc Interface R Soc 10: 20130134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. 2014. Engineering bacterial microcompartment shells: Chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4: 444–453. [DOI] [PubMed] [Google Scholar]
- Cai F, Bernstein SL, Wilson SC, Kerfeld CA. 2016. Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol 170: 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EJ, Montoya JP, Burns J, Mulholland MR, Subramaniam A, Capone DG. 1999. Extensive bloom of a N2-fixing diatom/cyanobacterial association in the tropical Atlantic Ocean. Mar Ecol Prog Ser 185: 273–283. [Google Scholar]
- Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, Putnam D. 2010. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci 107: 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AH, Robinson-Mosher A, Savage DF, Silver PA, Polka JK. 2013. The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PloS ONE 8: e76127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Deng Z, Billings AN, Seker UOS, Lu MY, Citorik RJ, Zakeri B, Lu TK. 2014. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater 13: 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR. 2015. Emergent genetic oscillations in a synthetic microbial consortium. Science 349: 986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. 2012. Engineered protein nano-compartments for targeted enzyme localization. PLoS ONE 7: e33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnšek J, Tomšič N, Avbelj M, Gaber R, et al. 2012. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res 40: 1879–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 32: 59–94. [DOI] [PubMed] [Google Scholar]
- Delebecque CJ, Silver PA, Lindner AB. 2012. Designing and using RNA scaffolds to assemble proteins in vivo. Nat Protoc 7: 1797–1807. [DOI] [PubMed] [Google Scholar]
- Do Nascimento M, Dublan Mde L, Ortiz-Marquez JC, Curatti L. 2013. High lipid productivity of an Ankistrodesmus-Rhizobium artificial consortium. Bioresour Technol 146: 400–407. [DOI] [PubMed] [Google Scholar]
- Du ZQ, Wang JY. 2015. A novel lumazine synthase molecule from Brucella significantly promotes the immune-stimulation effects of antigenic protein. Genet Mol Res 14: 13084–13095. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD. 2009. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27: 753–759. [DOI] [PubMed] [Google Scholar]
- Eriksson HM, Wessman P, Ge C, Edwards K, Wieslander A. 2009. Massive formation of intracellular membrane vesicles in Escherichia coli by a monotopic membrane-bound lipid glycosyltransferase. J Biol Chem 284: 33904–33914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Bobik TA. 2011. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193: 5623–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci 109: 14995–15000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassel TA, Buchholz LA, Collins ML, Remsen CC. 1992. Localization of methanol dehydrogenase in two strains of methylotrophic bacteria detected by immunogold labeling. Appl Environ Microbiol 58: 2302–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierobe HP, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich JP, Bayer EA. 2001. Design and production of active cellulosome chimeras selective incorporation of dockerin-containing enzymes into defined functional complexes. J Biol Chem 276: 21257–21261. [DOI] [PubMed] [Google Scholar]
- Frostl J, Overmann J. 1998. Physiology and tactic response of the phototrophic consortium “Chlorochromatium aggregatum.” Arch Microbiol 169: 129–135. [DOI] [PubMed] [Google Scholar]
- Fu J, Liu M, Liu Y, Woodbury NW, Yan H. 2012. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J Am Chem Soc 134: 5516–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JM, Bramble JP, Rawlings AE, Burnell G, Evans SD, Staniland SS. 2012. Biotemplated magnetic nanoparticle arrays. Small 8: 204–208. [DOI] [PubMed] [Google Scholar]
- Ginet N, Pardoux R, Adryanczyk G, Garcia D, Brutesco C, Pignol D. 2011. Single-step production of a recyclable nanobiocatalyst for organophosphate pesticides biodegradation using functionalized bacterial magnetosomes. PLoS ONE 6: e21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith LE, Pupols M, Kickhoefer VA, Rome LH, Monbouquette HG. 2009. Utilization of a protein “shuttle” to load vault nanocapsules with gold probes and proteins. ACS Nano 3: 3175–3183. [DOI] [PubMed] [Google Scholar]
- Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW. 1996. Z-membranes: Artificial organelles for overexpressing recombinant integral membrane proteins. Proc Natl Acad Sci 93: 2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman BS, Derr ND, Reck-Peterson SL. 2012. Engineered, harnessed, and hijacked: Synthetic uses for cytoskeletal systems. Trends Cell Biol 22: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Kickhoefer VA, Nemerow GR, Rome LH. 2011. Targeted vault nanoparticles engineered with an endosomolytic peptide deliver biomolecules to the cytoplasm. ACS Nano 5: 6128–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci 108: 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Tanaka T, Yoshino T. 2015. Stoichiometrically controlled immobilization of multiple enzymes on magnetic nanoparticles by the magnetosome display system for efficient cellulose hydrolysis. Biomacromolecules 16: 3863–3868. [DOI] [PubMed] [Google Scholar]
- Jorda J, Lopez D, Wheatley NM, Yeates TO. 2013. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci Publ Protein Soc 22: 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. 2010a. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol 20: 290–299. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, van der Zwan WA, Wulf PS, Keijzer N, Hoogenraad CC. 2010b. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys J 99: 2143–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH. 1986. Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J Cell Biol 103: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Searles RP, Kedersha NL, Garber ME, Johnson DL, Rome LH. 1993. Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J Biol Chem 268: 7868–7873. [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. 1999. The 193-Kd vault protein, Vparp, is a novel poly(Adp-ribose) polymerase. J Cell Biol 146: 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, et al. 2005. Engineering of vault nanocapsules with enzymatic and fluorescent properties. Proc Natl Acad Sci 102: 4348–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 287: 17729–17736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, Pósfai M, Tompa É, Plitzko JM, Brachmann A, et al. 2014. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol 9: 193–197. [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311: 242–245. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64: 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenstein R, Ritsert K, Huber R, Richter G, Bacher A. 1994. The lumazine synthase/riboflavin synthase complex of Bacillus subtilis. Eur J Biochem 223: 1007–1017. [DOI] [PubMed] [Google Scholar]
- Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA. 2014. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J Mol Biol 426: 2217–2228. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Zheng H, Rothman JE. 2013. Stapled Golgi cisternae remain in place as cargo passes through the stack. eLife 2: e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, et al. 2014. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3: 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LA, Hoewyk DV, Orr-Weaver TL. 2003. The Drosophila cell cycle kinase PAN GU forms an active complex with PLUTONIUM and GNU to regulate embryonic divisions. Genes Dev 17: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461: 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Cannon MC. 1998. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J Bacteriol 180: 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MAJ, Hanson MR. 2014a. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J Cell Mol Biol 79: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR. 2014b. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Schuck S, Ferguson C, Gerl MJ, Simons K. 2009. Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J Biol Chem 284: 12041–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD. 2007. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 282: 29323–29335. [DOI] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD. 2010. Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol 153: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus ME, Leonard JN. 2013. FedExosomes: Engineering therapeutic biological nanoparticles that truly deliver. Pharm Basel Switz 6: 659–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JM, Long SP. 2014. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, Heymann JB, Winkler DC, Lam AS, Wall JS, Steven AC, et al. 2014. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J 33: 1896–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meining W, Mörtl S, Fischer M, Cushman M, Bacher A, Ladenstein R. 2000. The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 Å resolution reveals the binding mode of a phosphonate intermediate analogue. J Mol Biol 299: 181–197. [DOI] [PubMed] [Google Scholar]
- Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. 2013. Design and characterization of synthetic fungal–bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci 110: 14592–14597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Lee J, Min J, Kang S. 2014. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules 15: 3794–3801. [DOI] [PubMed] [Google Scholar]
- Müller MJI, Neugeboren BI, Nelson DR, Murray AW. 2014. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci 111: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K, Hagiwara K, Tanaka H, Nakaishi Y, Chong KT, Yamashita E, Armah GE, Ono Y, Ishino Y, Omura T, et al. 2005. Expression and molecular characterization of spherical particles derived from the genome of the hyperthermophilic euryarchaeote Pyrococcus furiosus. J Biochem (Tokyo) 138: 193–199. [DOI] [PubMed] [Google Scholar]
- Naughton LM, Mandel MJ. 2012. Colonization of Euprymna scolopes squid by Vibrio fischeri. J Vis Exp 2012: e3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PQ, Botyanszki Z, Tay PKR, Joshi NS. 2014. Programmable biofilm-based materials from engineered curli nanofibres. Nat Commun 5: 4945. [DOI] [PubMed] [Google Scholar]
- Niedziela-Majka A, Rymarczyk G, Kochman M, Ozyhar A. 1998. GST-Induced dimerization of DNA-binding domains alters characteristics of their interaction with DNA. Protein Expr Purif 14: 208–220. [DOI] [PubMed] [Google Scholar]
- Nishida K, Silver PA. 2012. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol 10: e1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Marquez JCF, Do Nascimento M, Dublan Mde A, Curatti L. 2012. Association with an ammonium-excreting bacterium allows diazotrophic culture of oil-rich eukaryotic microalgae. Appl Environ Microbiol 78: 2345–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmann J. 2010. The phototrophic consortium “Chlorochromatium aggregatum”—A model for bacterial heterologous multicellularity. Adv Exp Med Biol 675: 15–29. [DOI] [PubMed] [Google Scholar]
- Parker MW, Bello ML, Federici G. 1990. Crystallization of glutathione S-transferase from human placenta. J Mol Biol 213: 221–222. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lünsdorf H, Rambach A, et al. 2008. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 283: 14366–14375. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. 2010. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 38: 305–315. [DOI] [PubMed] [Google Scholar]
- Pasotti L, Zucca S, Lupotto M, Cusella De Angelis MG, Magni P. 2011. Characterization of a synthetic bacterial self-destruction device for programmed cell death and for recombinant proteins release. J Biol Eng 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F. 2012. Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol 10: 705–715. [DOI] [PubMed] [Google Scholar]
- Poderycki MJ, Kickhoefer VA, Kaddis CS, Raval-Fernandes S, Johansson E, Zink JI, Loo JA, Rome LH. 2006. The vault exterior shell is a dynamic structure that allows incorporation of vault-associated proteins into its interior. Biochemistry (Mosc) 45: 12184–12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polka JK, Silver PA. 2016. A tunable protein piston that breaks membranes to release encapsulated cargo. ACS Synth Biol 10.1021/acssynbio.5b00237. [DOI] [PubMed] [Google Scholar]
- Pollithy A, Romer T, Lang C, Müller FD, Helma J, Leonhardt H, Rothbauer U, Schüler D. 2011. Magnetosome expression of functional camelid antibody fragments (nanobodies) in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 77: 6165–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond FR, Gibson I, Lalucat J, Quackenbush RL. 1989. R-body-producing bacteria. Microbiol Rev 53: 25–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77: 357–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanpour R, Bugg TDH. 2013. Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J 280: 2097–2104. [DOI] [PubMed] [Google Scholar]
- Rapaport D. 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep 4: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev 72: 85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Wang X, Wardwell S, Courage NL, Volchuk A, Keenan T, Holt DA, Gilman M, Orci L, Cerasoli F, et al. 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 287: 826–830. [DOI] [PubMed] [Google Scholar]
- Rizzo R, Parashuraman S, Mirabelli P, Puri C, Lucocq J, Luini A. 2013. The dynamics of engineered resident proteins in the mammalian Golgi complex relies on cisternal maturation. J Cell Biol 201: 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins CT, Rivera VM, Woolfson DN, Keenan T, Hatada M, Adams SE, Andrade LJ, Yaeger D, van Schravendijk MR, Holt DA, et al. 2000. A ligand-reversible dimerization system for controlling protein–protein interactions. Proc Natl Acad Sci 97: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS. 2014. The engine of the reef: Photobiology of the coral-algal symbiosis. Front Microbiol 5: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F, Davidson FA, Kelly CL, Binny R, Christodoulides N, Gibson D, Johansson E, Kozyrska K, Lado LL, MacCallum J, et al. 2013. A synthetic system for expression of components of a bacterial microcompartment. Microbiology 159: 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DF, Afonso B, Chen AH, Silver PA. 2010. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327: 1258–1261. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol 364: 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten S, Villareal TA, Hopmans EC, Mets A, Swanson KM, Sinninghe Damsté JS. 2013. Endosymbiotic heterocystous cyanobacteria synthesize different heterocyst glycolipids than free-living heterocystous cyanobacteria. Phytochemistry 85: 115–121. [DOI] [PubMed] [Google Scholar]
- Seebeck FP, Woycechowsky KJ, Zhuang W, Rabe JP, Hilvert D. 2006. A simple tagging system for protein encapsulation. J Am Chem Soc 128: 4516–4517. [DOI] [PubMed] [Google Scholar]
- Shapiro MG, Goodwill PW, Neogy A, Yin M, Foster FS, Schaffer DV, Conolly SM. 2014. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat Nanotechnol 9: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Ram S, Vilar JMG. 2007. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci 104: 1877–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA, Keegan LP, Ptashne M. 1984. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci 81: 5951–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. 1994. The animal fatty acid synthase: One gene, one polypeptide, seven enzymes. FASEB J 8: 1248–1259. [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol 163: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Kang YJ, Jung H, Kim H, Kang S, Cho H. 2015. Lumazine synthase protein nanoparticle-Gd(III)-DOTA conjugate as a T1 contrast agent for high-field MRI. Sci Rep 5: 15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal LJ, van Gemerden H, Krumbein WE. 1985. Structure and development of a benthic marine microbial mat. FEMS Microbiol Lett 31: 111–125. [Google Scholar]
- Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. 2001. Assembly of vault-like particles in insect cells expressing only the major vault protein. J Biol Chem 276: 23217–23220. [DOI] [PubMed] [Google Scholar]
- Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. 2007. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci 104: 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart ES, Morshed F, Sremac M, DasSarma S. 2001. Antigen presentation using novel particulate organelles from halophilic archaea. J Biotechnol 88: 119–128. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang WB, Mahdavi A, Arnold FH, Tirrell DA. 2014. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc Natl Acad Sci 111: 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan A, Ju LK. 2006. Use of cyanobacterial gas vesicles as oxygen carriers in cell culture. Cytotechnology 52: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Boehringer D, Gutmann S, Günther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol 15: 939–947. [DOI] [PubMed] [Google Scholar]
- Tamura A, Fukutani Y, Takami T, Fujii M, Nakaguchi Y, Murakami Y, Noguchi K, Yohda M, Odaka M. 2015. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol Bioeng 112: 13–20. [DOI] [PubMed] [Google Scholar]
- Tian L, Okita TW. 2014. mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr Opin Plant Biol 22C: 77–85. [DOI] [PubMed] [Google Scholar]
- Toledo MAS, Janissen R, Favaro MTP, Cotta MA, Monteiro GA, Prazeres DMF, Souza AP, Azzoni AR. 2012. Development of a recombinant fusion protein based on the dynein light chain LC8 for non-viral gene delivery. J Control Release 159: 222–231. [DOI] [PubMed] [Google Scholar]
- Uratani Y, Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol 157: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges MJ, Silver PA, Way JC, Mattozzi MD. 2013. Targeting a heterologous protein to multiple plant organelles via rationally designed 5′ mRNA tags. J Biol Eng 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser PJ, Ariotti N, Howes M, Ferguson C, Webb R, Schwudke D, Leneva N, Cho KJ, Cooper L, Rae J, et al. 2012. Constitutive formation of caveolae in a bacterium. Cell 150: 752–763. [DOI] [PubMed] [Google Scholar]
- Wang M, Abad D, Kickhoefer VA, Rome LH, Mahendra S. 2015. Vault nanoparticles packaged with enzymes as an efficient pollutant biodegradation technology. ACS Nano 9: 10931–10940. [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Kaneko S, Crofts N, Sakulsingharoj C, Kim D, Choi SB, Hamada S, Ogawa M, Wang C, et al. 2009. Identification of cis-localization elements of the maize 10-kDa δ-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J Cell Mol Biol 60: 146–155. [DOI] [PubMed] [Google Scholar]
- Weber W, Baba MDE, Fussenegger M. 2007. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc Natl Acad Sci 104: 10435–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74: 3868–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA. 2010. Emergent cooperation in microbial metabolism. Mol Syst Biol 6: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörsdörfer B, Pianowski Z, Hilvert D. 2011a. Efficient in vitro encapsulation of protein cargo by an engineered protein container. J Am Chem Soc 134: 909–911. [DOI] [PubMed] [Google Scholar]
- Wörsdörfer B, Woycechowsky KJ, Hilvert D. 2011b. Directed evolution of a protein container. Science 331: 589–592. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Masaki R, Tashiro Y. 1996. Formation of crystalloid endoplasmic reticulum in COS cells upon overexpression of microsomal aldehyde dehydrogenase by cDNA transfection. J Cell Sci 109: 1727–1738. [DOI] [PubMed] [Google Scholar]
- Yang J, Kickhoefer VA, Ng BC, Gopal A, Bentolila LA, John S, Tolbert SH, Rome LH. 2010. Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano 4: 7229–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. 2012. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci 109: E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Meining W, Fischer M, Bacher A, Ladenstein R. 2001. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 Å resolution: Determinants of thermostability revealed from structural comparisons. J Mol Biol 306: 1099–1114. [DOI] [PubMed] [Google Scholar]
- Zhang X, Konarev PV, Petoukhov MV, Svergun DI, Xing L, Cheng RH, Haase I, Fischer M, Bacher A, Ladenstein R, et al. 2006a. Multiple assembly states of lumazine synthase: A model relating catalytic function and molecular assembly. J Mol Biol 362: 753–770. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li SZ, Li J, Pan X, Cahoon RE, Jaworski JG, Wang X, Jez JM, Chen F, Yu O. 2006b. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J Am Chem Soc 128: 13030–13031. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Jiang J, Said-Sadier N, Boxx G, Champion C, Tetlow A, Kickhoefer VA, Rome LH, Ojcius DM, Kelly KA. 2014. Activation of the NLRP3 inflammasome by vault nanoparticles expressing a chlamydial epitope. Vaccine 33: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]