Abstract
The folate cycle is one of the key metabolic pathways used by bacteria to synthesize vital building blocks required for proliferation. Therapeutic agents targeting enzymes in this cycle, such as trimethoprim and sulfamethoxazole, are among some of the most important and continually used antibacterials to treat both Gram-positive and Gram-negative pathogens. As with all antibacterial agents, the emergence of resistance threatens the continued clinical use of these life-saving drugs. In this article, we describe and analyze resistance mechanisms that have been clinically observed and review newer generations of preclinical compounds designed to overcome the molecular basis of the resistance.
Drugs that block the bacterial folate synthesis pathway are important for targeting certain pathogens. Although resistance to these antibiotics is growing, new drugs that target the pathway (especially DHFR) are under development.
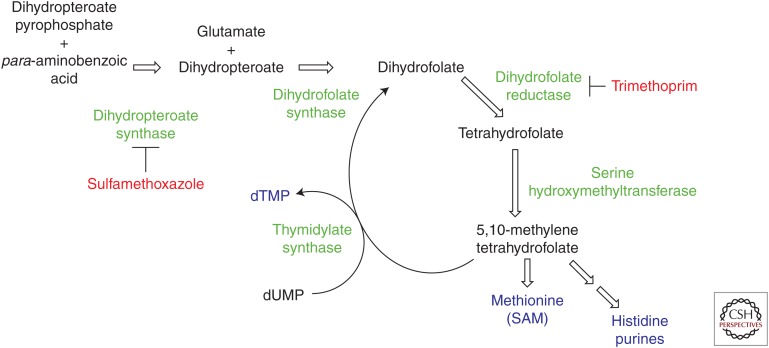
Inhibitors of the folate biosynthetic pathway have been successful drugs since the 1940s when sulfa powder was first applied topically to soldier’s wounds to prevent infection on the battlefield. The folate pathway is essential in the synthesis of one-carbon donors needed for the production of deoxythymidine monophosphate (dTMP), purine nucleotides, and the amino acids methionine and histidine. In bacteria, dihydrofolate is first synthesized from early precursor molecules. Most notably, the bacterial enzyme, dihydropteroate synthase (DHPS) catalyzes the formation of 7,8-dihydropteroate from para-aminobenzoic acid and 6-hydroxymethyl-7,8,-dihydropterin pyrophosphate (Fig. 1). One additional biosynthetic transformation in the bacteria results in the formation of dihydrofolate, which is reduced by the essential enzyme dihydrofolate reductase (DHFR), to tetrahydrofolate. DHFR uses the cofactor, nicotinamide adenine dinucleotide phosphate (NADPH), as the stoichiometric reducing agent bound in a site immediately adjacent to the folate-binding site. These early steps in the formation of dihydrofolate are selective for bacteria as mammalian cells obtain folic acid from dietary sources and then sequentially reduce it to tetrahydrofolate using DHFR. Therefore, antibacterial agents targeting steps before DHFR have inherent selectivity, whereas subsequent targets require the design of species-selective agents. The folate cofactors are then part of a critical cycle from which the essential metabolites dTMP, purine nucleotides, histidine, and methionine required for bacterial growth and division are produced.
Figure 1.
The folate cycle in bacteria. Enzymes are shown in green, key products in blue, and inhibitors in red. Dihydropteroate synthase (DHPS), a critical enzyme in the formation of dihydrofolate, is inhibited by sulfamethoxazole, and dihydrofolate reductase (DHFR) is inhibited by trimethoprim. dTMP, Deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; SAM, S-adenosylmethionine
Blockade of the folate pathway using inhibitors called “antifolates” results in an effective “thymine-less death” for the bacterial cell. The earliest programs to discover antibacterials commenced in the early 1930s and centered on antibacterial activity observed with synthetic dyes. Bayer chemists discovered Prontosil, one of the first systemic antibacterial agents to become widely used (van Miert 1994). Later, it was found that Prontosil is a prodrug and became metabolized to the active agent, sulfanilamide, which was identified as a mimic of the para-aminobenzoic acid substrate and a potent inhibitor of DHPS. Many additional inhibitors of DHPS, such as sulfamethoxazole (Fig. 2), followed and were synthesized to improve pharmacokinetic parameters, reduce side effects, and overcome early resistance (Skold 2010). During the 1940s, George Hitchings and Gertrude Elion determined that substituted 2,4-diaminopyrimidines also interfere with folate metabolism. They synthesized trimethoprim, the first inhibitor of DHFR (Fig. 2). Trimethoprim is very selective for some bacterial DHFR enzymes such as Staphylococcus aureus and Escherichia coli over human DHFR, a highly potent antibacterial and orally bioavailable. One noteworthy feature of trimethoprim (TMP) affinity is its significant cooperativity in binding with the NADPH cofactor. Soon after, Bactrim, a combination of trimethoprim and sulfamethoxazole, became a widely used therapy to treat infections caused by both Gram-positive (skin and soft-tissue infections and pneumonia) and Gram-negative (urinary tract infections) bacteria. To date, trimethoprim and sulfamethoxazole have been one of the few truly synergistic antibacterial combinations and the only clinically used antifolates to treat bacterial infections (Darmstadt 1997; Stevens et al. 2005; Gorwitz et al. 2006; Nathwani et al. 2008; Frei et al. 2010).
Figure 2.
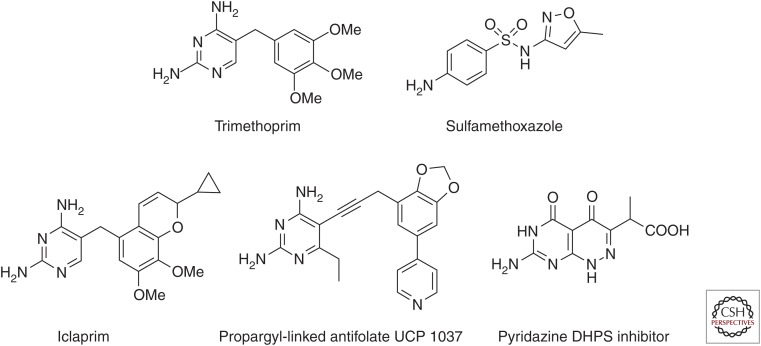
Antifolates. Clinically approved antibacterial antifolates: trimethoprim and sulfamethoxazole. Preclinical and experimental antifolates: dihydrofolate reductase (DHFR) inhibitors iclaprim and propargyl-linked antifolates, and dihydropteroate synthase pyridazine (DHPS) inhibitors.
In this review, we will describe the common mechanisms of resistance to both trimethoprim and sulfamethoxazole, drawing comparisons between resistant proteins when possible to understand the limitations of the enzymes in developing resistance and to pose possibilities to use that knowledge to overcome resistance. New research to develop DHFR and DHPS inhibitors has focused on overcoming current mechanisms of resistance and providing a next generation of antifolates.
RESISTANCE TO TRIMETHOPRIM
During the 1980s, resistance to Bactrim began to occur in both the hospital and community settings (Then et al. 1992; Dale et al. 1997). Two prevalent modes of resistance arose: chromosomal mutations in the dfrB and folP genes of S. aureus, which encode for DHFR and DHPS, respectively, and the horizontal transfer of plasmid-encoded, trimethoprim-resistant dfr genes or sulfamethoxazole-resistant sul genes in both Gram-positive and Gram-negative bacteria (Amyes 1989; Amyes et al. 1992; Skold 2010). A discussion of the chromosomal mutations and plasmid-encoded resistant proteins follows; comparisons of these resistant proteins (Fig. 3) may shed light on common mechanisms to decrease drug affinity.
Figure 3.
Sequence alignment for trimethoprim-sensitive (SaWT and KpWT) and trimethoprim-resistant dihydrofolate reductases (DHFRs). S1 DHFR and S3 DHFR are found in Gram-positive bacteria. DfrA1, DfrA12, and DfrA17 are found in Gram-negative bacteria. Residues colored in red are located in the inhibitor-binding pocket. As Klebsiella pneumoniae and Escherichia coli sequences are almost identical, only one sequence is shown here.
In 1997, Dale et al. (1997) reported that chromosomal mutations in the dfrB gene encoding DHFR in S. aureus were responsible for minimum inhibitory concentration (MIC) changes of ∼256-fold. Of the chromosomal mutations, the active-site mutation, F98Y, posed the greatest problem, conferring >400-fold resistance to trimethoprim (IC50 values for TMP against the wild-type (wt) and F98Y enzymes are 0.01 and 4.1 µM, respectively). The F98Y mutation most frequently occurs with an additional compensatory mutation, either H30N or H149R, that bring the fitness of the enzyme to near wild-type levels and increase the degree of resistance to trimethoprim by 800- to 2400-fold over wt. At the time the study was published, 88% of resistant strains possessed MIC changes of ∼256-fold, suggesting the widespread presence of chromosomal mutations in the dfrB gene.
In parallel, a transposon-encoded trimethoprim-resistant enzyme encoded by the gene dfrA and called S1 DHFR was isolated and shown to confer resistance to strains in Australia and the United States (Amyes 1989). S1 DHFR, which may originate from Staphylococcus epidermis, possesses three key residue substitutions, F98Y, V31I, and G43A, that have been shown to be responsible for the majority of TMP resistance (Fig. 3) (Dale et al. 1995). It is noteworthy that this plasmid-encoded DHFR recapitulates the key F98Y mutation observed in the chromosome. A detailed kinetic and structural comparative study of the wt and S1 enzymes revealed that the S1 enzyme independently reduces the interactions of both TMP and NADPH such that the binding synergy between the two is significantly reduced by 750-fold. A crystal structure of S1 with TMP revealed six molecules in the asymmetric unit, only three of which were the ternary structure. The other three S1 proteins bound only TMP, indicating the altered interactions of the S1 enzyme with NADPH (Heaslet et al. 2009).
In 2005, a second exogenous DHFR encoded by dfrG and called S3 DHFR, was discovered in a collection of 43 clinical isolates of TMP-resistant strains of methicillin-resistant Staphylococcus aureus (MRSA) from Thailand and 244 isolates from Japan (243 of these were TMP-sensitive; one was resistant) (Sekiguchi et al. 2005). TMP-resistant isolates possessing dfrG showed greatly elevated MIC values of at least 512 µg/mL. The dfrG gene, while originally horizontally transferred via plasmid, incorporated into the genome. Isolation and characterization of the S3 enzyme showed that, although KM values for DHF and NADPH were similar to the wt enzyme, TMP affinity was weakened by ∼20,000-fold (IC50 values of the wt and S3 enzyme were 0.013 and 254 µM, respectively). MIC values for TMP using E. coli transformants with the new plasmid were significantly increased, validating that the presence of dfrG confers TMP resistance. All isolates from Thailand were resistant to TMP and possessed dfrG; only one isolate from Japan was resistant to TMP and contained dfrG (Sekiguchi et al. 2005). In a second publication, expansion of dfrG to Africa and transfer to European travelers from Africa was described (Nurjadi et al. 2014).
As Bactrim is first-line treatment for common infections caused by Gram-negative bacteria, surveillance of resistance in this population is also actively monitored. In Gram-negative bacteria, especially the Enterobacteriaceae E. coli and Klebsiella pneumoniae, a different group of trimethoprim-resistant extrachromosomal DHFR proteins are horizontally transferred between bacteria. In Gram-negative bacteria, plasmid-encoded TMP resistance was first reported in 1992 (Amyes et al. 1992) with the description of two types of plasmid-encoded DHFR enzymes. Each type was highly resistant to trimethoprim, conferring MIC values >1 mm, but differed in their protein expression levels, with type 1 enzymes producing ∼10-fold greater levels of protein than the chromosomal enzyme. Two studies, one conducted in Sweden (Brolund et al. 2010) and the other in Western Europe and Canada (Blahna et al. 2006), surveyed the prevalence of type 1 or type 2 integron-encoded trimethoprim-resistant dfr genes. The Swedish study concurred that the type 1 gene dfrA1 is most prevalent and occurred in 34% of resistant isolates. Of the 350 isolates studied in the European/Canadian study, 66% possessed an integron-associated dfr allele. The gene dfrA1 was again the most prevalent (40%), followed by a second gene, dfrA17 (31%).
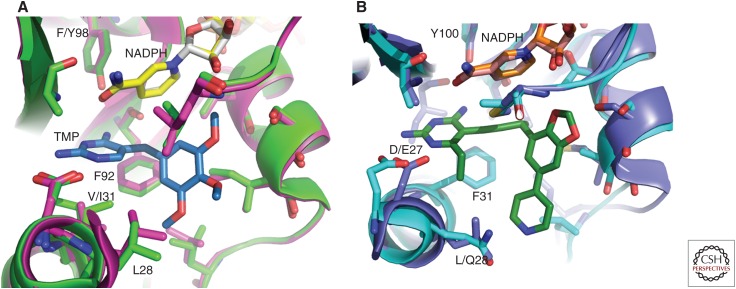
Interestingly, although the sequences of the trimethoprim-resistant DHFR proteins diverge, the proteins share several motifs that distinguish the TMP-resistant proteins from the TMP-sensitive (Fig. 3). For example, position 28 is a leucine in S. aureus (wt), E. coli, and K. pneumoniae (wt) but changes to a tryptophan in S3 DHFR and is consistently a glutamine in all of the TMP-resistant Gram-negative Dfr proteins. This switch from a leucine to a bulky tryptophan or polar glutamine is likely to affect positioning of the diaminopyrimidine ring as well as the hydrophobic trimethoxyphenyl ring (Fig. 4A,B). These effects were in fact observed in the structure of DfrA1 bound to an experimental antifolate (Lombardo et al. 2015).
Figure 4.
Structural comparisons of wild-type and trimethoprim-resistant DHFR. Structural superposition of (A) Staphylococcus aureus (wild-type [wt]) (pink) bound to nicotinamide adenine dinucleotide phosphate (NADPH) (yellow) and trimethoprim (TMP) (blue) with S1 dihydrofolate reductase (DHFR) (green) bound to NADPH and TMP (not shown) (Heaslet et al. 2009), and (B) Klebsiella pneumoniae (wt) (dark blue) bound to NADPH (orange) and a propargyl-linked antifolate (PLA) (not shown) with DfrA1 (cyan) bound to NADPH and a PLA (green) (Lombardo et al. 2015).
Position 31 is valine in S. aureus (wt) but a phenylalanine in K. pneumoniae (wt) or any of the Gram-negative DfrA proteins. The valine in S. aureus and S3 is balanced by Phe 92, which is on the opposite side of the active site, forming hydrophobic interactions that support the diaminopyrimidine ring and linker (Fig. 4A). Interestingly, this residue is an isoleucine in S1 DHFR, which is known to contribute to TMP resistance (Heaslet et al. 2009). In the Gram-negative enzymes, Phe at position 31 is balanced by less bulky residues on the opposite side of the active site (Fig. 4B): Ile in the wt, Ser in DfrA1, Ala in DfrA12, and Ser in DfrA17. These smaller residues (serine and alanine) in the DfrA1, DfrA12, and DfrA17 are likely to decrease affinity with the antifolates relative to the isoleucine in the wild-type.
Interestingly, position 98 is a phenylalanine in S. aureus (wt) and corresponds to a tyrosine in S1 and S3 DHFR that are responsible for TMP-resistance in Gram-positive bacteria (Fig. 4A,B). From the early clinical observations, the chromosomal F98Y mutation is also known to cause significant TMP resistance in S. aureus DHFR. More detailed studies have shown that this residue change greatly affects the position and affinity of the cofactor, NADPH (Heaslet et al. 2009) as well as the kinetics and entropy of binding. Recent structural and biochemical studies investigating the wild-type and F98Y enzymes show that the enzyme displays plasticity in the cofactor binding site, which is enhanced with the F98Y mutation. This plasticity affects a distribution of ternary states that includes an alternate bound configuration of the cofactor (Keshipeddy et al. 2015). In contrast, both the wild-type E. coli and K. pneumoniae DHFR enzymes that are sensitive to TMP as well as the plasmid-encoded TMP-resistant DfrA1, DfrA12, and DfrA17 all natively possess tyrosine at position 98, suggesting that compensatory and subtle differences exist between the Gram-positive and the Gram-negative DHFR enzymes.
RESISTANCE TO SULFAMETHOXAZOLE
Resistance to sulfamethoxazole also arose early after introduction and began to be monitored. As with trimethoprim, resistance to sulfamethoxazole arises through combinations of chromosomal mutations and plasmid-encoded copies of resistant genes. Several point mutations in the folP gene encoding DHPS in S. aureus, have been reported in clinical isolates. Seven of the nine resistant strains analyzed showed at least 13 residues that differ from wild-type; the remaining two isolates have two residue differences. Crystal structures of S. aureus (Hampele et al. 1997), Yersinia pestis (Yun et al. 2012), or Mycobacterium tuberculosis DHPS (Baca et al. 2000) show that most of the mutations implicated in resistance are spread across the surface of the protein or in flexible loops. Resistance mutations F28L/I and P64S (E. coli numbering) are found in loop 1 and loop 2, respectively, close to the active site and affect the para-aminobenzoic acid (PABA) binding site. Yun et al. (2012) further notes that mutations at positions F33 and P69 are likely to affect inhibitor but not substrate binding, which may make these sites more susceptible to resistance-conferring mutations.
In addition to chromosomal gene mutations, sulfamethoxazole resistance can be transferred by integrons carrying the sul1, sul2, or sul3 genes (Skold 1976; Blahna et al. 2006) encoding insensitive forms of DHPS. In fact, a collection of 106 clinical isolates of Stenotrophomonas maltophilia from India, for which TMP-SMX is first-line therapy, showed that 24 out of 106 isolates were resistant to TMP-SMX. The sul1 or sul2 gene was present in 12 or 14 isolates, respectively (Kaur et al. 2015).
Resistance to antibacterial agents can take many forms including efflux, drug-modifying enzymes, changes in cell membrane permeability, and alterations of the drug target. It appears that the majority of resistance to currently used antifolates is heavily dependent on alterations of the drug target and the introduction of mobile elements that carry a resistant copy of the target as opposed to mechanisms that alter drug concentration. This knowledge of the mechanisms of resistance coupled with detailed structural and biochemical analysis underlies the design of next-generation antifolates.
COMPOUNDS IN DEVELOPMENT
Fortunately, there continues to be significant development of new antifolates intended to overcome existing resistance mechanisms. Development has focused on new DHFR inhibitors including iclaprim and the propargyl-linked antifolates as well as new DHPS inhibitors intended to bind and inhibit the pterin pocket of the enzyme, thus avoiding resistance mutations already present.
Iclaprim (Fig. 2), originally developed by Roche and licensed by Arpida for clinical advancement (Hawser et al. 2006), was designed to overcome the resistance conferred by point mutations in the chromosomal gene and to be effective against other TMP-resistant organisms. The compound showed potent activity against MRSA, TMP-resistant MRSA, Streptococcus pyogenes (MIC values between 0.03 µg/mL and 0.06 µg/mL), as well as Gram-negative bacteria such as Enterococcus and Haemophilus influenza (MIC values 0.12 and 0.5 µg/mL, respectively) (Schneider et al. 2003). Furthermore, the propensity of iclaprim to induce the formation of resistance mutations is reported to be very low. Iclaprim advanced through phase I, II, and III clinical trials focused on complicated skin and soft tissue infections. Unfortunately, some cardiotoxicity issues presented during the trials (Sincak and Schmidt 2009) and more importantly, the drug did not meet the regulatory metrics of non-inferiority relative to linezolid in place at the time. Recently, a biotechnology company, Motif Bio (see motifbio.com/iclaprim), received FDA approval to reopen the phase III clinical trials for acute bacterial skin and skin structure infections as well as hospital-acquired bacterial pneumonia.
A series of compounds known as the propargyl-linked antifolates (PLAs; example shown in Fig. 2) is currently under development to inhibit both Gram-positive and Gram-negative bacteria; they are specifically designed to target both trimethoprim-sensitive and trimethoprim-resistant enzymes. Using a structure-based approach founded on the determination of several tens of structures of DHFR from many species, including S. aureus (Frey et al. 2009), mutant forms of S. aureus (Frey et al. 2009, 2010; Keshipeddy et al. 2015), E. coli, K. pneumoniae (Lamb et al. 2014), and plasmid-encoded trimethoprim-resistant DHFR such as DfrA1 (Lombardo et al. 2015), new DHFR inhibitors were designed to overcome mutations that cause TMP resistance as well as whole enzymes that are horizontally transferred for resistance. The development of these compounds has led to very potent inhibitors of wild-type and mutant forms of S. aureus DHFR as well as the growth of the S. aureus (wild-type and mutant) bacteria. Some of the most potent compounds have MIC values of 0.0391 µg/mL against the wild-type and 0.625 µg/mL against S. aureus with the F98Y mutation (Keshipeddy et al. 2015). In addition, many of the compounds show activity against Gram-negative bacteria and show potency against the trimethoprim-resistant DfrA1 (Lombardo et al. 2015). Early investigations into the propensity to incur resistance mutations show that the compounds generally have very low resistance frequencies between 1.2 × 10−9 and 8.9 × 10−10 (Frey et al. 2012).
There has also been considerable effort to develop new inhibitors of DHPS. Switching attention from the para-aminobenzoic acid site where the sulfa compounds bind, medicinal chemistry work has focused on the pocket that binds the partner substrate, dihydropterin pyrophosphate (DHPP) (Zhao et al. 2012). Using a crystal structure of Bacillus anthracis DHPS bound to a lead pterin (Hevener et al. 2012), Lee and coworkers synthesized several pyridazine derivatives intended to optimize different functional groups in the lead (Fig. 2). Using a combination of enzyme assays, ITC, SPR, and crystallography, they show that a demethylated pyridazine core is a pterin mimic for DHPS (Zhao et al. 2012). In parallel work, they show that a compound targeted to the DHPS dimer interface acts as an allosteric inhibitor of the enzyme, opening a new area of design and intervention (Hammoudeh et al. 2014).
CONCLUSION
Despite the fact that trimethoprim and sulfamethoxazole have been in continued use for over 60 years, these classes of antibacterials have not undergone the sustained medicinal chemistry efforts that have led to multiple generations of other important classes such as β-lactams or fluoroquinolones. Although resistance to these agents has appeared, they remain some of the most important first-line therapeutics for common Gram-positive and Gram-negative pathogens. However, increased use coupled with globalization demands the development of new antifolates that will continue to allow this important class to be clinically effective.
ACKNOWLEDGMENTS
The authors acknowledge support from the National Institutes of Health (NIH) (AI104841 and AI111957).
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Amyes S. 1989. The success of plasmid-encoded resistance genes in clinical bacteria. J Med Microbiol 28: 73–83. [DOI] [PubMed] [Google Scholar]
- Amyes S, Towner K, Young H. 1992. Classification of plasmid-encoded dihydrofolate reductases conferring trimethoprim resistance. J Med Microbiol 36: 1–3. [DOI] [PubMed] [Google Scholar]
- Baca A, Sirawaraporn R, Turley S, Sirawaraporn W, Ho W. 2000. Crystal structure of Mycobacterium tuberculosis 6-hydroxymethyl-7,8-dihydropteroate synthase in complex with pterin monophosphate: New insight into the enzymatic mechanism and sulfa-drug action. J Mol Biol 302: 1193–1212. [DOI] [PubMed] [Google Scholar]
- Blahna M, Zalewski C, Reuer J, Kahlmeter G, Foxman B, Marrs C. 2006. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother 57: 666–672. [DOI] [PubMed] [Google Scholar]
- Brolund A, Sundqvist M, Kahlmeter G, Grape M. 2010. Molecular characterisation of trimethoprim resistance in Escherichia coli and Klebsiella pneumoniae during a two-year intervention on trimethoprim use. PLoS ONE 5: e9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale G, Broger C, Hartman P, Langen H, Page M, Then R, Stuber D. 1995. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermis ATCC 14990: The origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol 177: 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale G, Broger C, D’Arcy A, Hartman P, DeHoogt R, Jolidon S, Kompis I, Labhardt A, Langen H, Locher H, et al. 1997. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J Mol Biol 266: 23–30. [DOI] [PubMed] [Google Scholar]
- Darmstadt G. 1997. Oral antibiotic therapy for uncomplicated bacterial skin infections in children. Pediatr Infect Dis J 16: 227–240. [DOI] [PubMed] [Google Scholar]
- Frei C, Miller M, Lewis J, Lawson K, Hunter J, Oramasionwu C, Talbert R. 2010. Trimethoprim-sulfamethoxazole or clindamycin for community-associated MRSA (CA-MRSA) skin infections. J Am Board Fam Med 23: 714–719. [DOI] [PubMed] [Google Scholar]
- Frey K, Liu J, Lombardo M, Bolstad D, Wright D, Anderson A. 2009. Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternative conformation of NADPH that may be linked to trimethoprim resistance. J Mol Biol 387: 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey K, Lombardo M, Wright D, Anderson A. 2010. Towards the understanding of resistance mechanisms in clinically isolated trimethoprim-resistant, methicillin-resistant Staphylococcus aureus dihydrofolate reductase. J Struc Biol 170: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey K, Viswanathan K, Wright D, Anderson A. 2012. Prospectively screening novel antibacterial inhibitors of dihydrofolate reductase for mutational resistance. Antimicrob Agents Chemother 56: 3556–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorwitz RJ, Jernigan DB, Powers JH, Jernigan JA; Participants in the CDC-Convened Experts’ Meeting on Management of MRSA in the Community. 2006. Strategies for clinical management of MRSA in the community: Summary of an experts’ meeting convened by the Centers for Disease Control and Prevention, www.cdc.gov/mrsa/community/clinicians. [Google Scholar]
- Hammoudeh D, Date M, Yun MK, Zhang W, Boyd V, Follis A, Griffith E, Lee R, Bashford D, White S. 2014. Identification and characterization of an allosteric inhibitory site on dihydropteroate synthase. ACS Chem Biol 9: 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampele I, D’Arcy A, Dale G, Kostrewa D, Nielsen J, Oefner C, Page M, Schonfeld HJ, Stuber D, Then R. 1997. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol 268: 21–30. [DOI] [PubMed] [Google Scholar]
- Hawser S, Luciuro S, Islam K. 2006. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol 71: 941–948. [DOI] [PubMed] [Google Scholar]
- Heaslet H, Harris M, Fahnoe K, Sarver R, Putz H, Chang J, Subramanyam C, Barreiro G, Miller JR. 2009. Structural comparison of chromosomal and exogenous dihydrofolate reductase from Staphylococcus aureus in complex with the potent inhibitor trimethoprim. Proteins 76: 706–717. [DOI] [PubMed] [Google Scholar]
- Hevener K, Yun MK, Qi J, Kerr I, Baboglu K, Hurdle J, Blakrishna K, Balakrishna K, White S, Lee R. 2012. Structure-based design of novel pyrimidopyridazine derivative as dihydrofolate synthase inhibitors with increased affinity. J Med Chem 53: 166–177.19899766 [Google Scholar]
- Kaur P, Gautam V, Tewari R. 2015. Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb Drug Resist 21: 380–385. [DOI] [PubMed] [Google Scholar]
- Keshipeddy S, Reeve S, Anderson A, Wright D. 2015. Nonracemic antifolates stereoselectively recruit alternate cofactors and overcome resistance in S. aureus. J Am Chem Soc 137: 8983–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb K, Lombardo M, Alverson J, Priestley N, Wright D, Anderson A. 2014. Crystal structures of Klebsiella pneumoniae dihydrofolate reductase bound to propargyl-linked antifolates reveal features for potency and selectivity. Antimicrob Agents Chem 58: 7484–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M, G-Dayanandan N, Wright D, Anderson A. 2016. Crystal structures of trimethoprim-resistant DfrA1 rationalize potent inhibition by propargyl-linked antifolates. ACS Inf Dis 2: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani D, Morgan M, Masterton R, Dryden M, Cookson B, French G, Lewis D; British Society for Antimicrobial Chemotherapy Working Party on Community-Onset MRSA Infections and Infections. 2008. Guidelines for UK practice for the diagnosis and management of methicillin-resistant Staphylococcus aureus (MRSA) infections presenting in the community. J Antimicrob Chemother 61: 976–994. [DOI] [PubMed] [Google Scholar]
- Nurjadi D, Olalekan A, Layer F, Shittu A, Alabi A, Ghebremedhin B, Schaumber F, Hofmann-Eifler J, Genderen P, Caumes E, et al. 2014. Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J Antimicrob Chemother 69: 2361–2368. [DOI] [PubMed] [Google Scholar]
- Schneider P, Hawser S, Islam K. 2003. Iclaprim, a novel diaminopyrimidine with potent activity on trimethoprim sensitive and resistant bacteria. Bioorg Med Chem Lett 13: 4217–4221. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Tharavichitkul P, Miyoshi-Akiyama T, Chupia V, Fujino T, Araake M, Irie A, Morita K, Kuratsuji T, Kirikae T. 2005. Cloning and characterization of a novel trimethoprim-resistant dihydrofolate reductase from a nosocomial isolate of Staphylococcus aureus CM.S2 (IMCJ1454). Antimicrob Agents Chemother 49: 3948–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincak C, Schmidt J. 2009. Iclaprim, a novel diaminopyrimidine for the treatment of resistant Gram-positive infections. Ann Pharmacother 43: 1107–1114. [DOI] [PubMed] [Google Scholar]
- Skold O. 1976. R-factor mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother 9: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold O. 2010. Sulfonamides and trimethoprim. Exp Rev Anti Infect Ther 8: 1–6. [DOI] [PubMed] [Google Scholar]
- Stevens D, Bisno A, Chambers H, Everett E, Dellinger P, Goldstein E, Gorbach S, Hirschmann J, Kaplan E, Montoya J, et al. 2005. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin Infect Dis 41: 1373–1406. [DOI] [PubMed] [Google Scholar]
- Then R, Kohl I, Burdeska A. 1992. Frequency and transferability of trimethoprim and sulfonamide resistance in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermis. J Chemother 4: 67–71. [DOI] [PubMed] [Google Scholar]
- van Miert A. 1994. The sulfonamide-diaminopyrimidine story. J Vet Pharmacol 4: 309–316. [DOI] [PubMed] [Google Scholar]
- Yun MK, Wu Y, Li Z, Zhao Y, Waddell MB, Ferreia A, Lee R, Bashford D, White S. 2012. Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 335: 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hammoudeh D, Yun M, Qi J, White S, Lee R. 2012. Structure-based design of novel pyrimido[4,5-c] pyridazine derivatives as dihydropteroate synthase inhibitors with increased affinity. ChemMedChem 7: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]