Abstract
Recent developments in studies of tumor heterogeneity have provoked new thoughts on cancer management. There is a desperate need to understand influence of the tumor microenvironment on cancer development and evolution. Applying principles and quantitative methods from ecology can suggest novel solutions to fulfil this need. We discuss spatial heterogeneity as a fundamental biological feature of the microenvironment, which has been largely ignored. Histological samples can provide spatial context of diverse cell types coexisting within the microenvironment. Advanced computer-vision techniques have been developed for spatial mapping of cells in histological samples. This has enabled the applications of experimental and analytical tools from ecology to cancer research, generating system-level knowledge of microenvironmental spatial heterogeneity. We focus on studies of immune infiltrate and tumor resource distribution, and highlight statistical approaches for addressing the emerging challenges based on these new approaches.
Advanced computing techniques have been developed for the spatial analysis of diverse cell types in histological tumor samples. This has enabled ecological theories to be applied to the study of tumor microenvironments.
Cancer is an evolutionary and ecological process (Merlo et al. 2006). Concerted efforts to study cancer evolution have enabled us to map the landscape of cancer genetic diversity, to track cancer evolution over time and space, and to decipher the genetic drivers behind it (Gerlinger et al. 2012; de Bruin et al. 2014; Hobor et al. 2014; Misale et al. 2014; Arena et al. 2015; Siravegna et al. 2015; Yates et al. 2015; Williams et al. 2016). Besides genetic drivers, evolutionary forces can shape diversity through the interplay between genetic variants and environmental factors. There is accumulating evidence to support the influence from the microenvironment on cancer progression and evolution (Weinberg 2008; Junttila and de Sauvage 2013; Marusyk et al. 2014; Williams et al. 2016). Genetic variations among neoplastic subclones place them in competition with each other, allowing them to occupy specialized niches in a manner analogous to diverse species in ecosystems (Greaves 2015; Nawaz and Yuan 2015). To gain fitness advantages, cancer cells can actively engage in constructing ecological niches by modifying their surrounding environments, such as modulating immune checkpoint pathways for immune evasion, co-opting fibroblasts to provide growth factors, and stimulating angiogenesis to obtain nutrients (Merlo et al. 2006; Greaves and Maley 2012). In turn, the environment shapes cancer cell phenotypes by providing selective pressure through a myriad of mechanisms, including nutrient supply via adjacent blood vessels, immune regulation, and tissue remodeling (Weinberg 2008; Junttila and de Sauvage 2013; Greaves 2015). These cancer–microenvironment interactions can have significant implications for cancer development and evolution.
As such, there is a desperate need to understand the roles of microenvironmental factors during cancer progression and evolution (Junttila and de Sauvage 2013; Greaves 2015). Decades of research on a related topic in ecology have revealed insights on mechanisms, analytical approaches, and experimental pitfalls that may aid our studies of ecological processes in tumors. In this article, we summarize key messages from ecological theories and methods that are relevant for understanding microenvironmental heterogeneity in human solid tumors. Specifically, we outline (1) spatial heterogeneity as a fundamental feature of the tumor microenvironment and its clinical implications, (2) advanced computer-vision techniques applied to histology that enable spatial analysis of complex tumors, (3) experimental and analytical tools required to achieve a systematic understanding of microenvironmental spatial heterogeneity, (4) clinical significance of microenvironmental spatial heterogeneity with regards to immune infiltrate and tumor resource distribution, and (5) statistical methods for addressing challenges emerged from these new approaches.
CLINICAL SIGNIFICANCE OF MICROENVIRONMENTAL SPATIAL HETEROGENEITY
Spatial Heterogeneity Is a Fundamental Feature of the Tumor Microenvironment
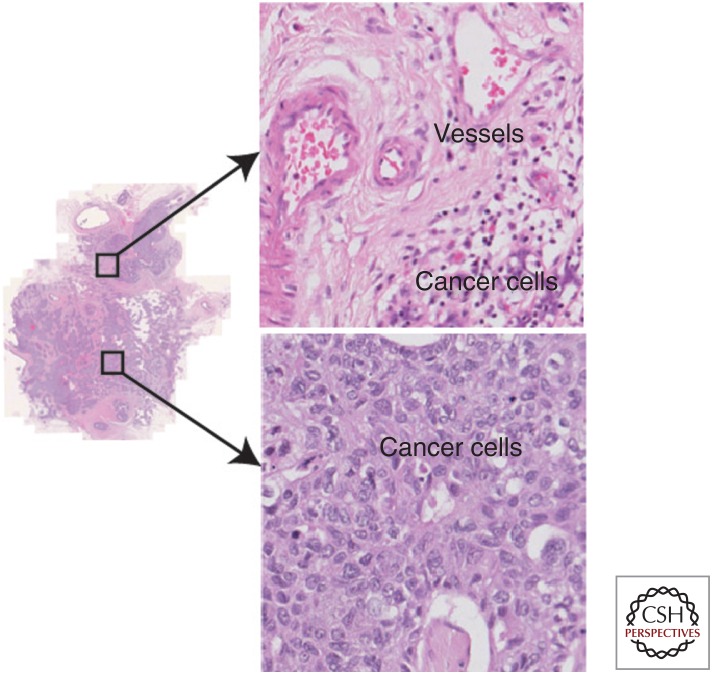
It is important to recognize that the orchestrated influence of microenvironmental components on cancer is often accompanied by strong regional differences (Gillies et al. 2012; Junttila and de Sauvage 2013). Evidence of spatial variations has been well documented in pathological observations (Clemente et al. 1996; Galon et al. 2006; Kruger et al. 2013). This is analogous to the environmental impacts that have been frequently observed in natural ecosystems. For example, riparian and desert regions coexist within a small spatial distance in the Arizona desert. As a result, diverse plant species and phenotypes emerged with strong regional variations. Similarly, high spatial heterogeneity has been observed in tumors, such as coexisting vascular and hypoxic regions (Fig. 1) (Alfarouk et al. 2013). Evidence of cancer genotype variation under different microenvironments has emerged. In glioblastoma, cancer cells with epidermal growth factor receptor (EGFR) amplification have been observed in poorly vascularized areas, whereas platelet-derived growth factor receptor (PDGFRA)-amplified cancer cells were enriched near endothelial cells (Little et al. 2012). This spatial association between genetically different cancer cells and blood vessels may be attributed to environmental adaptation, or the ability of cancer cells to modify their environments. In both cases, a sufficient knowledge of the spatial variability in the microenvironment would be useful for identifying the driving factors of tumor heterogeneity.
Figure 1.
Spatial heterogeneity of the tumor microenvironment illustrated with an ovarian cancer histological hematoxylin and eosin (H&E) tumor section, where regional differences with respect to vessel distribution can be seen.
The importance of spatial structure in biological systems has long been recognized by ecologists (Tilman and Kareiva 1997). For example, a geographical survey revealed environmental traits that allow successful adaptation and establishment of invasive plant species (Matesanz et al. 2015). Analysis of spatial dispersal patterns of zebra mussels in northern America identified the most efficient way for them to spread, thereby providing useful means for intervention (Johnson and Padilla 1996). These examples of geographical expansion of invasive species show how studying spatial structure can shed light on the driving factors of an ecological process and suggest potential interventions with parallels to studies of cancer prognosis as well as treatment.
A New Generation of Diagnostic, Prognostic, and Predictive Biomarkers
There is strong clinical and experimental evidence to support the importance of tumor microenvironment in cancer progression and mediation of drug resistance (Gatenby and Gillies 2008; Gillies et al. 2012). For example, molecular subtyping has repeatedly revealed new prognostic subtypes related to the microenvironment (Finak et al. 2008; Tothill et al. 2008; Gentles et al. 2015); the presence of tumor-infiltrating immune cells, cancer-associated fibroblasts, and vascular invasion has been shown to be highly predictive of prognosis and treatment response across different types of cancers (Hwang et al. 2008; Anderberg et al. 2009; Denkert et al. 2010; Nakasone et al. 2012). Nevertheless, the spatial dimensions of the tumor microenvironment have only begun to attract attention recently (Galon et al. 2006; Heindl et al. 2015; Nawaz and Yuan 2015). Spatial locations of immune cells have been shown to correlate with clinical outcome in different cancers. In colorectal cancer, a prognostic factor that incorporates type, density, and location of immune cells outperformed traditional histopathological methods to stage cancer (Galon et al. 2006). In estrogen receptor (ER)-negative/human epidermal growth factor receptor 2 (Her2)-negative (Loi et al. 2013) and Her2-negative (Issa-Nummer et al. 2013) breast cancer patients, a high degree of immune infiltration in tumor stroma was found to be associated with increased survival and complete response rates, respectively. Recent developments in computer vision has enabled ecological statistics to be directly applied to histological samples, providing quantitative spatial heterogeneity measures of immune infiltrate that are predictive of prognosis in breast cancer (Maley et al. 2015; Nawaz et al. 2015; Yuan 2015) and follicular lymphoma (Nelson et al. 2015). These novel tumor features were shown to be independent of clinical variables and immune cell counts. A new generation of biomarkers beyond traditional clinical parameters and cell counting is on the horizon.
New Opportunities in Cancer Therapy
Applying principles from spatial ecology and complexity of resource networks can suggest novel solutions to the problem of therapeutic resistance in cancer management. It can further lead to other clinical innovations including the development of efficient treatment strategies. The problem of therapeutic resistance can be fundamentally attributed to tumor heterogeneity. The emergency of drug resistance may be partly explained by complex structures of the tumor microenvironment. For instance, spatial heterogeneity of nutrient resources replicated in an ecologically designed microfluidic device to mimic the bone marrow environment can facilitate rapid emergence of chemotherapy resistance in multiple myeloma cells (Wu et al. 2015). A substantial part of adaptive strategies of resistant cells is to regulate ancient genes, suggesting that phenotypic diversity may be more rapidly achieved in this way to adapt to extreme selective pressures.
Therefore, an understanding of microenvironmental heterogeneity can provide the basis of effective therapeutic strategies. In ecology, it is known that the most efficient way to kill a species is by destroying its niche environment, and this idea has been proposed for cancer therapeutics (Walther et al. 2015). Effective uses of therapies to disrupt cancer cell niches in their own environment, such as antiangiogenic drugs and immunotherapy, have the potential to transform cancer management (Formenti and Demaria 2013; Wood et al. 2014; Brahmer et al. 2015). To successfully apply this strategy in personalized medicine, it is critical to recognize that diverse environments can coexist within the same tumor, as discussed before. Therefore, an assessment of the spatial heterogeneity of microenvironment in the first place can be a step toward predicting treatment resistance and avoiding selection of resistant populations.
COMPUTER VISION TO ENABLE RAPID MAPPING OF MICROENVIRONMENTAL SPATIAL STRUCTURE
Histology and imaging are excellent resources for obtaining tumor spatial structure in large quantities. Such spatial data, once quantitatively analyzed, will aid the identification of clinically relevant features, potentially yielding predictions more powerful than measurements of cell abundance that ignore the spatial context. With appropriate methodologies, studies of pathohistological tumor sections can reveal the spatial context of cancer–microenvironment interactions at single-cell resolution, whereas powerful imaging techniques allow us to track the spatiotemporal changes in the microenvironment over the course of treatment. The remaining part of this article will focus on recent developments in analysis of histological samples. Much more spatial- and texture-oriented analyses have been proposed for imaging data and are discussed extensively elsewhere (Gatenby et al. 2013; Hu et al. 2015).
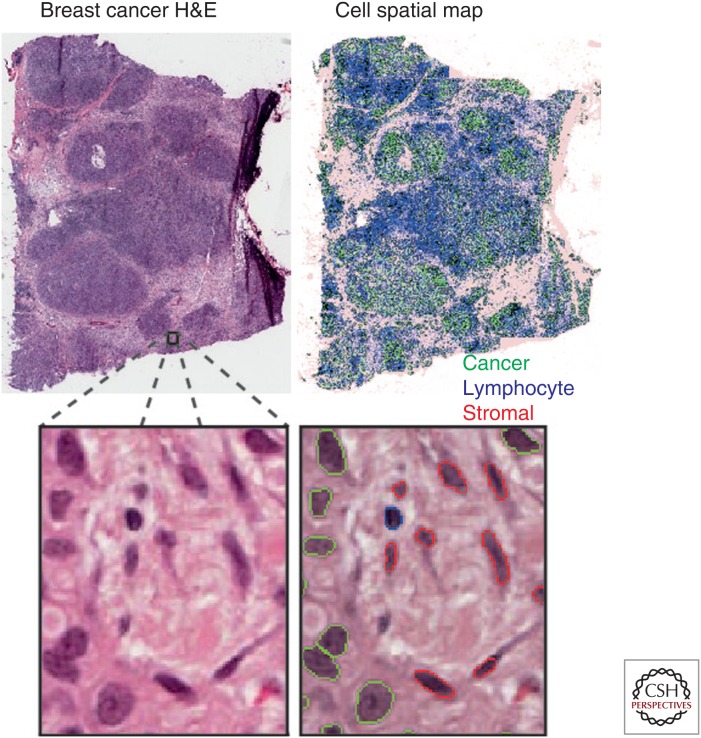
With advancing computing techniques, remarkable progress in image analysis has been made on objective assessment of cellular context in digitized cancer histological sections. The use of machine learning methods enables automated identification of various cell types, tumor components, and regions based on human expert input, namely, supervised learning (Holmes et al. 2009; Basavanhally et al. 2010; Tuominen et al. 2010; Balsat et al. 2011, 2014; Beck et al. 2011; Doyle et al. 2012; Yuan et al. 2012; Lu et al. 2014). The computer compares a new cell with what human experts call a cancer, stromal, or other cell types and determines its type based on morphological similarity (Fig. 2). As a result, rapid mapping of the identities and spatial locations of millions of cells is now possible. Just as large areas of land can be mapped for population density variation, a tumor sample can be processed to map changes in density of its constituent cells (Fig. 2). Such methods thus offer a new opportunity for studying the spatial structure of tumors. Nevertheless, there are many accompanying challenges. It is well known that image-analysis methods can be sensitive to sample quality and variability; therefore, it is imperative that methods are developed to accommodate the significant amount of variation in histological samples (McCann et al. 2015). Comprehensive reviews in this specialized field are available (Gurcan et al. 2009; Kothari et al. 2013). In this review, we will focus on the next step following image analysis—spatial analysis of the tumor microenvironment.
Figure 2.
Spatial mapping of cancer and normal cells in histological images using automated image analysis techniques. Shown are a breast cancer hematoxylin and eosin (H&E)-stained image and a spatial distribution map of cell types identified by automated image analysis, including cancer cells (green), stromal cells (red), and lymphocytes (blue).
QUANTITATIVE ANALYSIS OF SPATIAL HETEROGENEITY IN THE TUMOR MICROENVIRONMENT
The first step in understanding heterogeneity is to identify patterns. In ecology, spatial statistics (Ripley 1984) has been widely applied to capture patterns of species and/or habitats. It is recognized that, in many situations, direct measurements of ecological processes can be impossible (McIntire and Fajardo 2009). Thus, a rapidly emerging concept, “space as a surrogate,” has been proposed for maximizing inference about ecological processes through the analysis of spatial patterns, rather than relying on time-series data (McIntire and Fajardo 2009). Many recent studies have successfully examined spatial patterns to understand a diverse array of ecological processes where experimental manipulation or direct measurements are difficult to obtain or are not feasible (de Knegt et al. 2009; Sanders et al. 2013; Smith et al. 2013). This bears high similarity with the situation in cancer research, where the majority of data have been gathered using biopsy and surgical resection samples. Experimental manipulation directly on human tissues without subjecting them to further modification and selection is almost impossible. Tissue-engineered models, such as tumor spheroids and organoids, although highly successful for expanding our knowledge on drug resistance, can lack critical interactions between cancer and the microenvironment, such as limited release of cytokines (Villasante and Vunjak-Novakovic 2015). A key benefit of using “space as a surrogate” in studies of cancer is the amount of spatial data a single tumor can provide alone. With thousands or millions of cells as spatial points, a statistically significant spatial pattern is more likely to be generated by biological processes than noise or biases. Here, we discuss current progress in establishing the spatial heterogeneity of tumor microenvironments and how systematic studies have contributed to our understanding of tumor ecology.
Spatial Heterogeneity of Immune Infiltrate
Interactions between cancer cells and immune cells are an important component of the ecological conditions in which cancer cells exist and evolve (Greaves and Maley 2012). As discussed above, an array of studies has established the clinical significance of immune cell infiltrate in a number of cancer types (Galon et al. 2006; Issa-Nummer et al. 2013; Loi et al. 2014; Denkert et al. 2015). The spatial interactions among immune and cancer cells generate complex ecological dynamics that can ultimately influence tumor progression and response to treatment (Demaria et al. 2005; Fridman et al. 2012; Denkert et al. 2015; Gentles et al. 2015). Ecology can provide a framework for understanding these complex dynamics beyond cell abundance and predicting clinical outcomes. Several ecological methods have been applied to studying spatial patterns of immune infiltrate, where strong predictors of clinical outcome have been identified for different breast cancer subtypes.
Colocalization of Cancer and Immune Cells
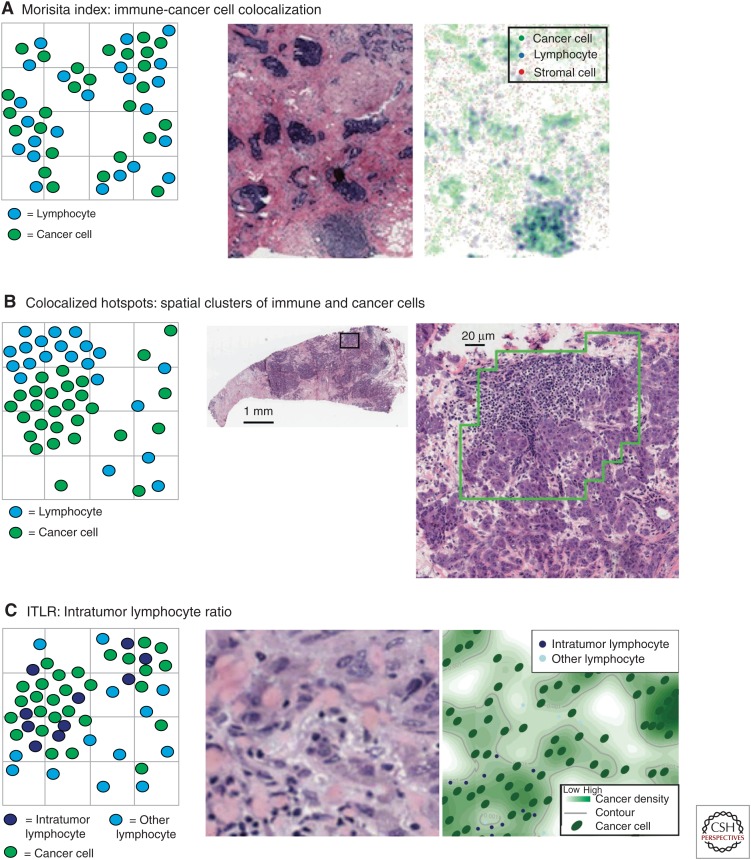
Identical amounts and types of immune cells in two tumors do not necessarily equate to the same effect of immune infiltrate. Immune cell distribution can vary dramatically in different tumors. How immune cells distribute relative to cancer cells may have profound clinical implications. The Morisita–Horn index is a measure of similarity among community structure in ecology (Morisita 1959; McIntosh et al. 2004; Scalon et al. 2011). It can be used to quantify the extent of colocalization between two or more species given their spatial structures. For example, it was used to study predator–prey interactions by establishing a positive association between predator body size and prey diversity (Radloff and Du Toit 2004). In breast tumors, this index has been used for quantifying colocalization of immune and cancer cells (Fig. 3A) (Maley et al. 2015). Mathematically, the Morisita–Horn index uses the proportional distribution of two or more variables as input. To study the bivariate relationship between cancer and immune cells, the index is defined as
where pil and pic are the proportion of all immune cells and cancer cells within a tumor, respectively, at a region i, and 1 ≤ i ≤ R, where R is the total number of regions into which a sample has been divided (Fig. 4). We will discuss how tumor regions were defined in the next section. The value of the Morisita–Horn index ranges from 0 indicating no similarity or colocalization to 1 for the two structures being identical or perfectly colocalized.
Figure 3.
Schematic representation of cell spatial patterns captured by three statistical methods with histology image examples: (A) Morisita index, (B) Getis–Ord hotspot analysis, and (C) intratumor lymphocyte ratio (ITLR).
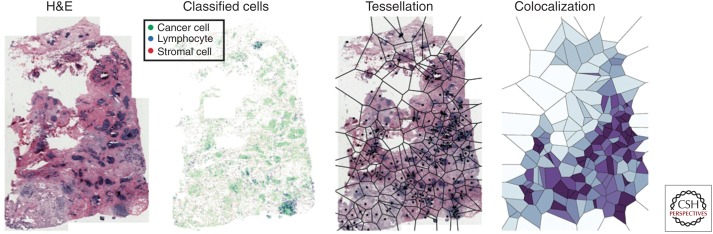
Figure 4.
Quantifying cancer-immune cell colocalization using histological images. From left to right: hematoxylin and eosin (H&E) image of a breast cancer; classified cells in this image (cancer in green, lymphocyte in blue, and stromal cells in red); Voronoi tessellation over this image using random cancer cells as seeds; measuring cell colocalization based on the proportional data in the Voronoi grids (high colocalization in dark purple, and low colocalization in light blue).
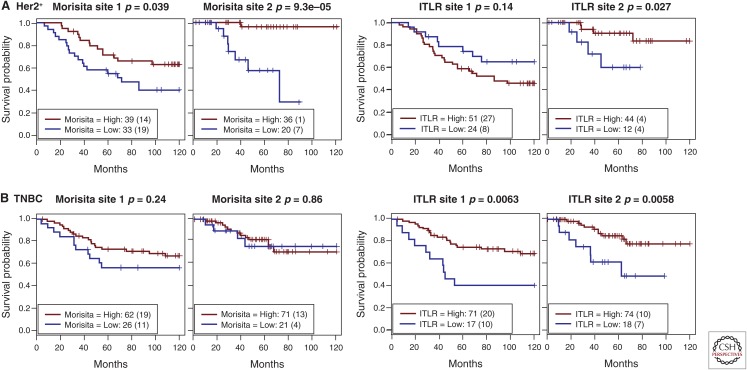
Because the Morisita index measures colocalization, the opportunity to directly relate this quantitative index with clinical outcome may provide a clue as to the extent to which cancer cells have evaded antitumor immune response or recruited immune cells with protumor effect. If a low Morisita score (low levels of colocalization of immune and cancer cells) is associated with a poor clinical outcome, this might suggest that cancer cells have evolved immune evasion strategies in these patients. A high Morisita score (high levels of colocalization) associated with a good prognosis might indicate effective immune predation. On the other hand, a high Morisita score associated with a poor prognosis might indicate mutualistic interactions or co-option of immune cells. When the Morisita–Horn index was applied to 1026 breast cancer samples following image analysis of the histological specimens, it was observed that a high degree of colocalization between cancer and immune cells was associated with significantly increased probability of 10-year, disease-specific survival in Her2-positive breast cancers (Maley et al. 2015). This association likely suggests that the presence of immune cells is indicative of effective predation by the immune system in Her2-positive cancer. But this effect is not evident in other subtypes of breast cancer, possibly because of a less clearly defined antitumor effect (e.g., effective predation) of the immune cells or other unknown reasons.
The Morisita–Horn index has many advantages over other community similarity indices. Community similarity indices have been evaluated in terms of their dependencies on sample size, species diversity, and other confounding factors (Wolda 1981). The Morisita index was found to be among the most robust to sample size and species diversity when compared with other similarity measures. It was recommended because of the small effect of sample size and diversity, and, if logarithmic transformation of data is required, the Morisita–Horn transformation can be used. Thus, the Morisita–Horn index as an ecological measure presents a robust option for quantifying spatial patterns in tumors.
Immune-Cancer Hotspots
Another type of spatial pattern is spatial clustering. Many methods have been proposed to identify such a pattern. For example, statistical tests such as the Ripley’s K function (Ripley 1976) can be used to confirm the presence of spatial clustering. Alternatively, there are methods to identify specific regions where spatial clustering exists, such as the hotspot analysis (Getis and Ord 1992). An advantage of the second type of method is that specific regions of interest can be identified, and this type of method has already been applied to the analysis of tumor microenvironment. Getis–Ord hotspot analysis (Getis and Ord 1992) was used to detect significant levels of immune cell clustering, or “immune hotspots,” in histology sections (Fig. 3B) (Nawaz et al. 2015). Mathematically, z scores are evaluated for each region for a specific cell type in a sample, given by
where S and U are two normalizing factors:
where R is the total number of regions, cj is the cell count for region j, c̄ is the mean value of c for all regions in the image, and wi,j indicates a neighborhood relationship between region i and j:
The z scores indicate whether statistically significant clusters of specific cell types are found for each spatial region. The same analysis was separately applied to cancer and immune cells. In ER-negative breast cancer, abundance of cancer or immune hotspots was not associated with clinical outcome. However, when combined, a so-called immune-cancer hotspot score was defined as the fractional area within a tumor, with an overlap of cancer and immune hotspots. This was found to be significantly associated with favorable prognosis in ER-negative breast cancer (Nawaz et al. 2015).
Intratumor Lymphocyte Ratio (ITLR)
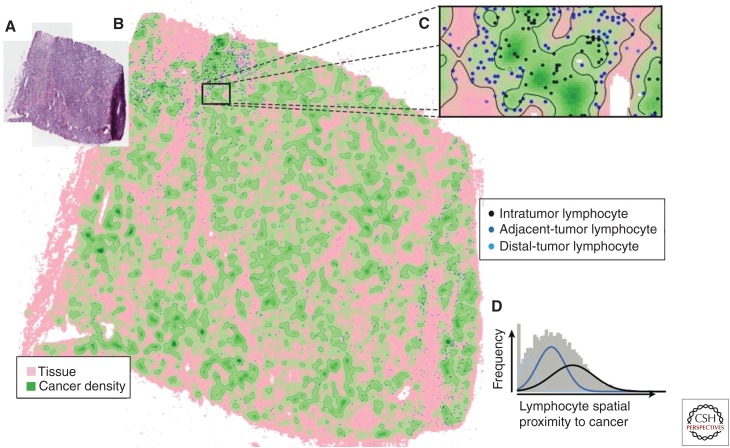
A quantitative ratio to represent the degree of infiltration of immune cells into the tumor has been proposed (Yuan 2015). Unsupervised Gaussian mixture clustering (Fraley and Raftery 2003) was used to detect different types of lymphocytes based on their spatial proximity to cancer cells (Fig. 5). The cluster with the shortest distance to cancer cells was classified as intratumor lymphocytes (Fig. 3C). This was used to define a quantitative measure for a tumor, the ITLR as
where nITL is the number of intratumor lymphocytes and nc is the total number of cancer cells in a histological sample. In ER-negative/Her2-negative breast cancer, high ITLR was found to be associated with good disease-specific survival (Yuan 2015).
Figure 5.
Quantifying intratumor immune infiltration with intratumor lymphocyte ratio (ITLR). (A) Building a cancer density map using a kernel estimator, and (B) cancer density map with lymphocytes as spatial points. The density of cancer cells at the location of a lymphocyte can be used as a direct measurement of spatial proximity of this lymphocyte to cancer. (C) A higher resolution map of a tumor region. (D) Clustering lymphocytes based on their spatial relationships to cancer using Gaussian mixture clustering revealed three subclasses of lymphocytes.
Comparison of Different Immune Measures
All of the above-mentioned immune spatial measures were found to be independent of existing clinical parameters in breast cancer (Maley et al. 2015; Nawaz et al. 2015; Yuan 2015). They were further compared with pathologist’s scoring of immune abundance and a quantitative score of lymphocyte ratio that was also obtained from automated image analysis (Maley et al. 2015; Nawaz et al. 2015; Yuan 2015). Lymphocyte ratio, as a measure of the presence of immune cells in a sample without accounting for its spatial distribution, is defined as the fraction of cells in a sample that are immune cells, that is,
where nl is the number of immune cells, nc is the number of cancer cells, and ns is the number of stromal cells in a sample. All spatial measures were found to be stronger prognostic factors than pathological score and lymphocyte ratio, and, in the respective breast cancer subtypes, they were found to be prognostic (Maley et al. 2015; Nawaz et al. 2015; Yuan 2015). This highlights the importance of examining not just cell abundance but also spatial patterns that can be indicative of active immune response.
Despite high correlations between some of these spatial measures, they appear to hold specific prognostic value in different breast cancer subtypes. For example, the Morisita index and ITLR were highly correlated in 180 ER-negative/Her2-negative breast cancers from the METABRIC study (Curtis et al. 2012) (new data for this review; Pearson’s correlation coefficient r = 0.50, p < 0.001). However, whereas ITLR was associated with survival in the ER-negative/Her2-negative but not the Her2-positive subtype, the Morisita index was found to be prognostic in the Her2-positive subtype but not other subtypes (Fig. 6). Generally speaking, the Morisita index measures the degree of immune-cancer cell colocalization within a tumor, while ITLR measures the amount of immune cells infiltrated into tumor nests. Therefore, a tumor with a low amount of intratumor lymphocytes that are highly colocalized with cancer cells will have a high Morisita index but low ITLR. Hence, unlike the Morisita index, which is a global measure of trend in lymphocyte distribution, ITLR concerns regional abundance of lymphocytes that colocalized with cancer cells. This is also true for the hotspot score, where only spatial coclustering of cancer and immune cells is quantified. More evenly spread cancer and immune cell distributions in a sample, despite greater intermixing with potentially high Morisita and ITLR score, could, in fact, lead to a low hotspot score. Therefore, different spatial measures, although interrelated, can be used to identify unique spatial arrangement patterns and should be considered separately in studies of different cancer types and subtypes.
Figure 6.
Comparison of prognostic value of two immune spatial measures, the Morisita–Horn index and intratumor lymphocyte ratio (ITLR), in two independent patient cohorts (site 1 and 2) in breast cancer subtype (A) Her2-positive (Her2+), and (B) triple-negative breast cancer (TNBC), defined as ER-negative/Her2-negative.
Spatial Heterogeneity of Tumor Resource
During disease progression, perfusion variability of resources such as nutrient and oxygen in the microenvironments can generate significant selective pressure, leading to accelerated cancer evolution and disease progression (Gatenby et al. 2013). As discussed above, tumor resource heterogeneity often occurs as a result of irregular vasculature that creates hypoxic or arid zones (Alfarouk et al. 2013). The clinical and therapeutic consequences of tumor resource heterogeneity have received substantial research investigations. Texture analysis of magnetic resonance images (MRIs) has been used to identify spatial heterogeneity and regional variations that are associated with microenvironmental features, including cell density, tissue stiffness, blood flow, and nutrient dispersion (Gatenby et al. 2013; Chaudhury et al. 2015). Using digital pathology, the spatial distribution of ER-positive and ER-negative cells were investigated in relation to vascular density and tissue necrosis in breast cancer histology specimens (Lloyd et al. 2014). A strong association between ER expression and vascular area was identified, suggesting that environmental variables were likely to be responsible for spatial heterogeneity in estrogen distribution and thus directly relevant for antiestrogen treatment. More recently, combined theoretical modeling and histology analysis of breast cancer showed considerable regional variations in cancer proliferation phenotype accompanied by environmental conditions such as vascularity and immune response (Lloyd et al. 2016). Besides spatial variations, temporal heterogeneity in the microenvironment can also impose greater selective pressure than constant conditions. Hypoxia is commonly recognized as a harsh environmental condition; however, breast cancer cell lines exposed to intermittent hypoxia evolved an even higher degree of resistance to etoposide compared with cells under chronic hypoxia or normoxia (Verduzco et al. 2015). Here, in the interest of quantitative statistical studies, we discuss a spatial analysis method that has been applied to histological analysis and can be used to dissect the resource heterogeneity of tumor microenvironment.
Fractal Dimensions
To measure a complexity pattern such as the vasculature, the use of fractal dimensions (Mandelbrot 1983) has been proposed (Losa 1995; Cross 1997; Lennon et al. 2015). For example, fractal dimensions may be used to identify features of oncogenic vascular systems that may contribute to the origins of cancer (Baum 2015). Recently, fractal dimensions have been applied to analyzing oral cancer histology samples (Bose et al. 2015). Fractals, as mathematical geometry that concern self-similarity, are often measured over a range of dimensions. For example, the box-counting method estimates fractal dimensions by counting the number of boxes with a range of sizes needed to cover the spatial geometry under study (Cross 1994). Let N be the number of b-by-b boxes required to cover a spatial point pattern S, then the fractal dimension of S, dim(S), is defined as
The more complex the geometric pattern, the more boxes are needed at each scale and, hence, the higher the fractal dimension score. Using oral cancer histology samples, fractal dimensions were measured using the box-counting method, and a high score of fractal dimensions in the cell pattern was reported to be associated with improved disease-specific survival, lymphocytic infiltration, and tumor proliferation (Bose et al. 2015). Besides histological analysis, fractals analysis has also demonstrated its uses in biomarker discovery based on dynamic contrast-enhanced MRI (Rose et al. 2009). As measures of global heterogeneity, fractals have shown superior prognostic power compared to region-based measures that do not sufficiently explore the relationships between tumor regions (Rose et al. 2009). We anticipate the application of fractal dimensions in histology by using specific markers, such as hypoxia markers, to contribute to our understanding of resource heterogeneity in the microenvironment.
Challenges in Spatial Analysis of Histology Samples
Spatial Tessellation
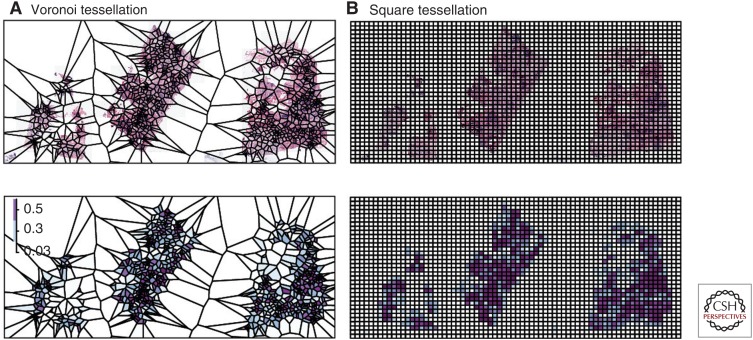
Histological sections can often contain up to millions of cells. It is thus a nontrivial task to discern spatial patterns from data at this scale. This challenge can also be found in ecology, where spatial data are sometimes acquired at a large scale. Tessellation effectively reduces complex problems to individual local structures, thus has been used widely in ecology. A tessellation is a mosaic set of spatially separated polygons. Commonly used tessellation models include Voronoi (Getis 1986) and rigid squares (Fig. 7). Voronoi tessellation is generated by seeds/spatial points to create polygons that contain all their closest neighbors. It has been suggested that because Voronoi tessellation mimics naturally emerged patterns, it is therefore particularly useful for studies of biological processes in nature (Getis 1994). For example, Voronoi tessellation has been used to predict plant harvest based on the Voronoi parameters of spatial patterns of plants. In the pioneering work of Mead (1966), measures of the Voronoi polygon were found to best predict carrot monoculture yield. These measures include area and two shape features of the polygons, and plants that grow close to the centroids of large isodiametric polygons tend to have a better yield.
Figure 7.
Different spatial tessellation methods to provide spatial resolution for histological sample analysis: (A) Voronoi tessellation for a hematoxylin and eosin (H&E) slide and corresponding immune cell density heatmap as polygons, and (B) square tessellation for an H&E slide and corresponding immune cell density heatmap as polygons.
Because of its desirable property, Voronoi tessellation has widespread applications, including those in histological image analysis. For example, it was used to extract architectural features of cells in histological image analysis of breast cancer, prostate cancer, B-cell lymphoma, and Barrett’s esophagus (Doyle et al. 2007; Basavanhally et al. 2010; Muldoon et al. 2010; Guidolin et al. 2015). For spatial analysis in histology, the benefits of two tessellation configurations on colocalization measures have been investigated: a Voronoi lattice versus a rigid square lattice (Maley et al. 2015). To generate Voronoi polygons, cancer cells were randomly sampled and used as “seeds” for tessellation, and the number of cancer cells and immune cells were computed for each polygon (Fig. 7A). Meanwhile, square tessellations were generated to have a similar amount of polygons as the Voronoi tessellation (Fig. 7B). Voronoi tessellation resulted in a more normally distributed set of cells in the polygons compared with the distributions of cells resulting from square tessellation (Maley et al. 2015). Combinations of tessellation and spatial analysis methods were further analyzed (Maley et al. 2015). Both the Morisita–Horn index and Pearson correlation have been widely used in ecology to study the similarity of structures between two communities, for example, to compare Salmonella colonization routes (Lim et al. 2014). When applied to quantification of spatial colocalization of cancer and immune cells, the Morisita–Horn index displayed high statistical significance for both types of tessellation in terms of association with survival in breast cancer, whereas Pearson correlation was associated with survival only when used in conjunction with Voronoi tessellation. This is not surprising because the Pearson correlation is known to be sensitive to data with skewed distribution. Therefore, the choice of spatial analysis methods should be carefully evaluated based on the use of spatial tessellation schemes.
Spatial Scale
Spatial heterogeneity is scale dependent. This phenomenon has been well documented in a number of studies in ecology, emphasizing that a scale needs to be chosen that is appropriate for the ecological process under study (Gardner et al. 1987; Turner et al. 1989). In histology analysis, the influence on spatial analysis by the use of different spatial scales along with spatial methods has been investigated. Cancer-immune cell colocalization was measured using the Morisita–Horn and Pearson correlation methods using square and Voronoi tessellation of eight different spatial scales, where larger scale indicates larger regions (Maley et al. 2015). Changes in their prognostic value according to the spatial scales were evaluated. The Morisita–Horn index was more robust to a spatial scale compared with a Pearson correlation. Hence, there is a need to evaluate robustness of the spatial index over different spatial configurations and to choose an appropriate scale in histological studies.
CONCLUDING REMARKS
In this review, we discussed how a desire to understand the interactions between cancer cells and the microenvironment has fueled a developing interest in studying tumors from a novel perspective: ecology. Within a Darwinian framework, analysis of tumor spatial heterogeneity can reveal distinct features in cancer habitats that indicate a number of different ecological processes. Studies of these ecological processes occurring in tumors can benefit from application of spatial statistics tools routinely used in ecological studies. Histology samples provide an abundance of data as input for these methods because of preserved spatial context. Thus, spatial analysis empowered by large-scale analysis of archival histology samples could facilitate studies of ecological interactions in human tumors with far-reaching implications. It can aid in the identification of patients at higher risk of progression or treatment resistance who may benefit from new treatments. We listed examples where spatial analysis of tumor histological specimen revealed associations between cancer prognosis and immune infiltration or resource distribution. Development of robust analytical tools capable of handling challenges presented in histological samples could play a key role in propelling this niche area into mainstream research and clinical uses. Nevertheless, histology on its own can be limited by the two-dimensional representation of a three-dimensional tumor. Radio-imaging modalities can step in to address this problem (Chaudhury et al. 2015). Fabricated devices as those developed in Wu et al. (2015) could be used to test hypotheses in controlled environments. Further, integrating a variety of assays including histology, imaging, genomics, and in vitro systems will provide multiple layers of information for the spatial and molecular structure of the tumor, revealing new cancer–microenvironment interactions that exist at different spatial scales.
ACKNOWLEDGMENTS
The author acknowledges support from the Institute of Cancer Research, Wellcome Trust (Grant No. 105104/Z/14/Z), and National Health Service (NHS) funding to the National Institute for Health Research (NIHR) Biomedical Research Centre and Italian Association for Cancer Research (AIRC).
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Alfarouk KO, Ibrahim ME, Gatenby RA, Brown JS. 2013. Riparian ecosystems in human cancers. Evol Appl 6: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg C, Li H, Fredriksson L, Andrae J, Betsholtz C, Li X, Eriksson U, Pietras K. 2009. Paracrine signaling by platelet-derived growth factor-CC promotes tumor growth by recruitment of cancer-associated fibroblasts. Cancer Res 69: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena S, Bellosillo B, Siravegna G, Martinez A, Canadas I, Lazzari L, Ferruz N, Russo M, Misale S, Gonzalez I, et al. 2015. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res 21: 2157–2166. [DOI] [PubMed] [Google Scholar]
- Balsat C, Blacher S, Signolle N, Beliard A, Munaut C, Goffin F, Noel A, Foidart JM, Kridelka F. 2011. Whole slide quantification of stromal lymphatic vessel distribution and peritumoral lymphatic vessel density in early invasive cervical cancer: A method description. ISRN Obstet Gynecol 2011: 354861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsat C, Signolle N, Goffin F, Delbecque K, Plancoulaine B, Sauthier P, Samouelian V, Beliard A, Munaut C, Foidart JM, et al. 2014. Improved computer-assisted analysis of the global lymphatic network in human cervical tissues. Mod Pathol 27: 887–898. [DOI] [PubMed] [Google Scholar]
- Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, Bhanot G, Madabhushi A. 2010. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans Biomed Eng 57: 642–653. [DOI] [PubMed] [Google Scholar]
- Baum M. 2015. Why does the weeping willow weep? Reconceptualizing oncogenesis in breast cancer. N Engl J Med 373: 1267–1269. [DOI] [PubMed] [Google Scholar]
- Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. 2011. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 3: 108–113. [DOI] [PubMed] [Google Scholar]
- Bose P, Brockton NT, Guggisberg K, Nakoneshny SC, Kornaga E, Klimowicz AC, Tambasco M, Dort JC. 2015. Fractal analysis of nuclear histology integrates tumor and stromal features into a single prognostic factor of the oral cancer microenvironment. BMC Cancer 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. 2015. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 373: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury B, Zhou M, Goldgof DB, Hall LO, Gatenby RA, Gillies RJ, Patel BK, Weinfurtner RJ, Drukteinis JS. 2015. Heterogeneity in intratumoral regions with rapid gadolinium washout correlates with estrogen receptor status and nodal metastasis. J Magn Res Imaging 42: 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente CG, Mihm MC Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. 1996. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 77: 1303–1310. [DOI] [PubMed] [Google Scholar]
- Cross SS. 1994. The application of fractal geometric analysis to microscopic images. Micron 25: 101–113. [DOI] [PubMed] [Google Scholar]
- Cross SS. 1997. Fractals in pathology. J Pathol 182: 1–8. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. 2014. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Knegt HJ, van Langevelde F, Coughenour MB, Skidmore AK, de Boer WF, Heitkönig IMA, Knox NM, Slotow R, van der Waal C, Prins HHT. 2009. Spatial autocorrelation and the scaling of species–environment relationships. Ecology 91: 2455–2465. [DOI] [PubMed] [Google Scholar]
- Demaria S, Kawashima N, Yang AM, Devitt ML, Babb JS, Allison JP, Formenti SC. 2005. Immune-mediated inhibition of metastases after treatment with local radiation and CTLA-4 blockade in a mouse model of breast cancer. Clin Cancer Res 11: 728–734. [PubMed] [Google Scholar]
- Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, et al. 2010. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 28: 105–113. [DOI] [PubMed] [Google Scholar]
- Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, Pfitzner BM, Salat C, Loi S, Schmitt WD, et al. 2015. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2–positive and triple-negative primary breast cancers. J Clin Oncol 33: 983–991. [DOI] [PubMed] [Google Scholar]
- Doyle S, Hwang M, Shah K, Madabhushi A, Feldman M, Tomaszeweski J. 2007. Automated grading of prostate cancer using architectural and textural image features. In ISBI 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. pp. 1284–1287. [Google Scholar]
- Doyle S, Feldman M, Tomaszewski J, Madabhushi A. 2012. A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng 59: 1205–1218. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. 2008. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14: 518–527. [DOI] [PubMed] [Google Scholar]
- Formenti SC, Demaria S. 2013. Combining radiotherapy and cancer immunotherapy: A paradigm shift. J Natl Cancer Inst 105: 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C, Raftery AE. 2003. Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. J Classif 20: 263–286. [Google Scholar]
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. 2012. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer 12: 298–306. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. 2006. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313: 1960–1964. [DOI] [PubMed] [Google Scholar]
- Gardner RH, Milne BT, Turnei MG, O’Neill RV. 1987. Neutral models for the analysis of broad-scale landscape pattern. Landscape Ecol 1: 19–28. [Google Scholar]
- Gatenby RA, Gillies RJ. 2008. A microenvironmental model of carcinogenesis. Nat Rev Cancer 8: 56–61. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Grove O, Gillies RJ. 2013. Quantitative imaging in cancer evolution and ecology. Radiology 269: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getis A. 1986. Book review: Spatial data-analysis by example, Vol. 1, Point pattern and quantitative data (ed. Upton GJG, Fingleton B). Am Cartogr 13: 363–365. [Google Scholar]
- Getis A. 1994. Book review: Spatial tessellations—Concepts and applications of Voronoi diagrams (ed. Okabe A, Boots B, Sugihara K). Geogr Anal 26: 88–90. [Google Scholar]
- Getis A, Ord JK. 1992. The analysis of spatial association by use of distance statistics. Geogr Anal 24: 189–206. [Google Scholar]
- Gillies RJ, Verduzco D, Gatenby RA. 2012. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer 12: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M. 2015. Evolutionary determinants of cancer. Cancer Discov 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin D, Marinaccio C, Tortorella C, Ruggieri S, Rizzi A, Maiorano E, Specchia G, Ribatti D. 2015. A fractal analysis of the spatial distribution of tumoral mast cells in lymph nodes and bone marrow. Exp Cell Res 339: 96–102. [DOI] [PubMed] [Google Scholar]
- Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. 2009. Histopathological image analysis: A review. IEEE Rev Biomed Eng 2: 147–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl A, Nawaz S, Yuan Y. 2015. Mapping spatial heterogeneity in the tumor microenvironment: A new era for digital pathology. Lab Invest 95: 377–384. [DOI] [PubMed] [Google Scholar]
- Hobor S, Van Emburgh BO, Crowley E, Misale S, Di Nicolantonio F, Bardelli A. 2014. TGFα and amphiregulin paracrine network promotes resistance to EGFR blockade in colorectal cancer cells. Clin Cancer Res 20: 6429–6438. [DOI] [PubMed] [Google Scholar]
- Holmes S, Kapelner A, Lee PP. 2009. An interactive java statistical image segmentation system: GemIdent. J Stat Softw 30: i10. [PMC free article] [PubMed] [Google Scholar]
- Hu LS, Ning S, Eschbacher JM, Gaw N, Dueck AC, Smith KA, Nakaji P, Plasencia J, Ranjbar S, Price SJ, et al. 2015. Multi-parametric MRI and texture analysis to visualize spatial histologic heterogeneity and tumor extent in glioblastoma. PLoS ONE 10: e0141506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, Logsdon CD. 2008. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res 68: 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa-Nummer Y, Darb-Esfahani S, Loibl S, Kunz G, Nekljudova V, Schrader I, Sinn BV, Ulmer HU, Kronenwett R, Just M, et al. 2013. Prospective validation of immunological infiltrate for prediction of response to neoadjuvant chemotherapy in HER2-negative breast cancer—A substudy of the neoadjuvant GeparQuinto trial. PloS ONE 8: e79775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LE, Padilla DK. 1996. Geographic spread of exotic species: Ecological lessons and opportunities from the invasion of the zebra mussel Dreissena polymorpha. Biol Conserv 78: 23–33. [Google Scholar]
- Junttila MR, de Sauvage FJ. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501: 346–354. [DOI] [PubMed] [Google Scholar]
- Kothari S, Phan JH, Stokes TH, Wang MD. 2013. Pathology imaging informatics for quantitative analysis of whole-slide images. JAMIA 20: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger JM, Wemmert C, Sternberger L, Bonnas C, Dietmann G, Gancarski P, Feuerhake F. 2013. Combat or surveillance? Evaluation of the heterogeneous inflammatory breast cancer microenvironment. J Pathol 229: 569–578. [DOI] [PubMed] [Google Scholar]
- Lennon FE, Cianci GC, Cipriani NA, Hensing TA, Zhang HJ, Chen CT, Murgu SD, Vokes EE, Vannier MW, Salgia R. 2015. Lung cancer—A fractal viewpoint. Nat Rev Clin Oncol 12: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CH, Voedisch S, Wahl B, Rouf SF, Geffers R, Rhen M, Pabst O. 2014. Independent bottlenecks characterize colonization of systemic compartments and gut lymphoid tissue by Salmonella. PLoS Pathog 10: e1004270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S, Jurgensmeier JM, Jones C. 2012. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res 72: 1614–1620. [DOI] [PubMed] [Google Scholar]
- Lloyd MC, Alfarouk KO, Verduzco D, Bui MM, Gillies RJ, Ibrahim ME, Brown JS, Gatenby RA. 2014. Vascular measurements correlate with estrogen receptor status. BMC Cancer 14: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA. 2016. Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Res 10.1158/0008-5472.CAN-15-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, et al. 2013. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol 31: 860–867. [DOI] [PubMed] [Google Scholar]
- Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, et al. 2014. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann Oncol 25: 1544–1550. [DOI] [PubMed] [Google Scholar]
- Losa GA. 1995. Fractals in pathology: Are they really useful? Pathologica 87: 310–317. [PubMed] [Google Scholar]
- Lu H, Papathomas TG, van Zessen D, Palli I, de Krijger RR, van der Spek PJ, Dinjens WN, Stubbs AP. 2014. Automated selection of hotspots (ASH): Enhanced automated segmentation and adaptive step finding for Ki67 hotspot detection in adrenal cortical cancer. Diagn Pathol 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Koelble K, Natrajan R, Aktipis A, Yuan Y. 2015. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Res 17: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbrot BB, Wheeler JA. 1983. The fractal geometry of nature. Am J Phys 51: 286–287. [Google Scholar]
- Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. 2014. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514: 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesanz S, Horgan-Kobelski T, Sultan SE. 2015. Evidence for rapid ecological range expansion in a newly invasive plant. AoB Plants 7: plv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MT, Ozolek JA, Castro CA, Parvin B, Kovacevic J. 2015. Automated histology analysis: Opportunities for signal processing. IEEE Signal Process Mag 32: 78–87. [Google Scholar]
- McIntire EJB, Fajardo A. 2009. Beyond description: The active and effective way to infer processes from spatial patterns. Ecology 90: 46–56. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Peckarsky BL, Taylor BW. 2004. Predator-induced resource heterogeneity in a stream food web. Ecology 85: 2279–2290. [Google Scholar]
- Mead R. 1966. A Relationship between individual plant-spacing and yield. Ann Bot 30: 301–309. [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, Maley CC. 2006. Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924–935. [DOI] [PubMed] [Google Scholar]
- Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, Russo M, Buscarino M, Lazzari L, Sartore-Bianchi A, et al. 2014. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 6: 224–226. [DOI] [PubMed] [Google Scholar]
- Morisita M. 1959. Measuring of the dispersion of individuals and analysis of the distributional patterns. Mem Fac Sci Kyushu Univ Ser 2: 215–235. [Google Scholar]
- Muldoon TJ, Thekkek N, Roblyer D, Maru D, Harpaz N, Potack J, Anandasabapathy S, Richards-Kortum R. 2010. Evaluation of quantitative image analysis criteria for the high-resolution microendoscopic detection of neoplasia in Barrett’s esophagus. J Biomed Optics 15: 026027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, et al. 2012. Imaging tumor–stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 21: 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz S, Yuan Y. 2015. Computational pathology: Exploring the spatial dimension of tumor ecology. Cancer Lett 10.1016/j.canlet.2015.11.018. [DOI] [PubMed] [Google Scholar]
- Nawaz S, Heindl A, Koelble K, Yuan Y. 2015. Beyond immune density: Critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod Pathol 28: 1621. [DOI] [PubMed] [Google Scholar]
- Nelson LS, Mansfield JR, Lloyd R, Oguejiofor K, Salih Z, Menasce LP, Linton KM, Rose CJ, Byers RJ. 2015. Automated prognostic pattern detection shows favourable diffuse pattern of FOXP3+ Tregs in follicular lymphoma. Br J Cancer 113: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff FG, Du Toit JT. 2004. Large predators and their prey in a southern African savanna: A predator’s size determines its prey size range. J Animal Ecol 73: 410–423. [Google Scholar]
- Ripley BD. 1976. The second-order analysis of stationary point processes. J Appl Probab 13: 255–266. [Google Scholar]
- Ripley BD. 1984. Spatial statistics—Developments 1980–3. Int Stat Rev 52: 141–150. [Google Scholar]
- Rose CJ, Mills SJ, O’Connor JPB, Buonaccorsi GA, Roberts C, Watson Y, Cheung S, Zhao S, Whitcher B, Jackson A, et al. 2009. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med 62: 488–499. [DOI] [PubMed] [Google Scholar]
- Sanders D, Sutter L, van Veen FJF. 2013. The loss of indirect interactions leads to cascading extinctions of carnivores. Ecol Lett 16: 664–669. [DOI] [PubMed] [Google Scholar]
- Scalon JD, Avelar MBL, Alves GdF, Zacarias MS. 2011. Spatial and temporal dynamics of coffee-leaf-miner and predatory wasps in organic coffee field in formation. Ciência Rural 41: 646–652. [Google Scholar]
- Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, et al. 2015. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21: 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Hurley BJ, Toms CN, Mackey AD, Solangi M, Kuczaj SA. 2013. Hurricane impacts on the foraging patterns of bottlenose dolphins Tursiops truncatus in Mississippi Sound. Marine Ecol Prog Ser 487: 231–244. [Google Scholar]
- Tilman D, Kareiva PM. 1997. Spatial ecology: The role of space in population dynamics and interspecific interactions. Princeton University Press, Princeton, NJ. [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. 2008. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 14: 5198–5208. [DOI] [PubMed] [Google Scholar]
- Tuominen VJ, Ruotoistenmaki S, Viitanen A, Jumppanen M, Isola J. 2010. ImmunoRatio: A publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 12: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, O’Neill R, Gardner R, Milne B. 1989. Effects of changing spatial scale on the analysis of landscape pattern. Landscape Ecol 3: 153–162. [Google Scholar]
- Verduzco D, Lloyd M, Xu L, Ibrahim-Hashim A, Balagurunathan Y, Gatenby RA, Gillies RJ. 2015. Intermittent hypoxia selects for genotypes and phenotypes that increase survival, invasion, and therapy resistance. PLoS ONE 10: e0120958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasante A, Vunjak-Novakovic G. 2015. Tissue-engineered models of human tumors for cancer research. Exp Opin Drug Discov 10: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther V, Hiley CT, Shibata D, Swanton C, Turner PE, Maley CC. 2015. Can oncology recapitulate paleontology? Lessons from species extinctions. Nat Rev Clin Oncol 12: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. 2008. Coevolution in the tumor microenvironment. Nat Genet 40: 494–495. [DOI] [PubMed] [Google Scholar]
- Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. 2016. Identification of neutral tumor evolution across cancer types. Nat Genet 48: 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolda H. 1981. Similarity indexes, sample-size and diversity. Oecologia 50: 296–302. [DOI] [PubMed] [Google Scholar]
- Wood SL, Pernemalm M, Crosbie PA, Whetton AD. 2014. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 40: 558–566. [DOI] [PubMed] [Google Scholar]
- Wu A, Zhang Q, Lambert G, Khin Z, Gatenby RA, Kim HJ, Pourmand N, Bussey K, Davies PCW, Sturm JC, et al. 2015. Ancient hot and cold genes and chemotherapy resistance emergence. Proc Natl Acad Sci 112: 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. 2015. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J R Soc Interface 10.1098/rsif.2014.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, et al. 2012. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 4: 157–143. [DOI] [PubMed] [Google Scholar]