Abstract
It is poorly understood how a single protein, p53, can be responsive to so many stress signals and orchestrates very diverse cell responses to maintain/restore cell/tissue functions. The uncovering that TP53 gene physiologically expresses, in a tissue-dependent manner, several p53 splice variants (isoforms) provides an explanation to its pleiotropic biological activities. Here, we summarize a decade of research on p53 isoforms. The clinical studies and the diverse cellular and animal models of p53 isoforms (zebrafish, Drosophila, and mouse) lead us to realize that a p53-mediated cell response is, in fact, the sum of the intrinsic activities of the coexpressed p53 isoforms and that unbalancing expression of different p53 isoforms leads to cancer, premature aging, (neuro)degenerative diseases, inflammation, embryo malformations, or defects in tissue regeneration. Cracking the p53 isoforms’ code is, thus, a necessary step to improve cancer treatment. It also opens new exciting perspectives in tissue regeneration.
The myriad cellular activities of p53 may be explained, in part, by the splice variants produced in a tissue-specific manner from the TP53 gene. Aberrant expression of these variants leads to cancer, degenerative diseases, and aging.
p53 is a central sensor of cell signals and a master regulator of cell response to damage (Lane and Levine 2010). But how can so many distinct extracellular and intracellular signals modulate p53 activity? How can only one protein bind specifically to so many different DNA sequences (p53 response elements) (el-Deiry et al. 1992; Bourdon et al. 1997; Khoury and Bourdon 2011; Simeonova et al. 2012) and directly regulate expression of thousands of genes? How does p53 select the target genes to trigger on time the appropriate cell responses to so many different cellular damages? Why is it so difficult to link TP53 mutation status to prognosis and cancer treatment?
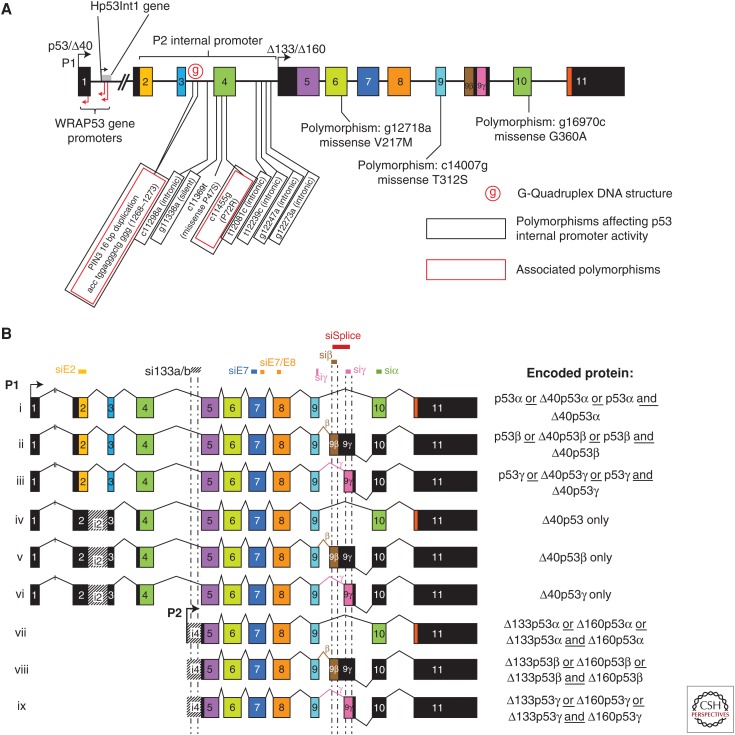
The TP53 gene is highly conserved in multicellular organisms (Lane et al. 2010). It is located on human chromosome 17p13.1 and is composed of 13 exons of which the first is noncoding (Fig. 1A). TP53 contains the Hp53Int1 and WRAP53 genes within exon-1/intron-1 (Reisman et al. 1996; Mahmoudi et al. 2009). It presents multiple genetic polymorphisms defining more than 100 distinct TP53 haplotypes, some of which are correlated with an increased risk of cancer (Dumont et al. 2003; Garritano et al. 2010; Wu et al. 2013). Although it is unequivocally established that TP53 is the most frequently mutated gene in human cancer, it is still difficult in the clinic to link TP53 mutation status to cancer treatment and clinical outcome, suggesting that the p53 pathway is not entirely understood. The discovery that the TP53 gene encodes several different splice variants may explain the discrepancy.
Figure 1.
TP53 locus and p53mRNAs. All introns/exons are represented to scale. Black boxes represent noncoding sequences, whereas coding sequences are colored. (A) The human TP53 gene’s locus structure. The TP53 gene, which is composed of 11 exons and two cryptic exons (9β and 9γ), encodes several p53 isoforms attributable to alternative promoters (↱ P1 and P2) and alternative retention of the cryptic exons. The noncoding exon-1 and intron-1 contain different promoters for the WRAP53 gene (antisense-coded) and intron-1 contains the Hp53Int1 gene. A G-quadruplex DNA structure located within intron-3 modulates splicing of intron-2 and activities of the internal p53 promoter P2. Several polymorphisms (including Pin3 and R72P) change activities of the internal p53 promoter (P2). (B) p53 mRNAs. The TP53 gene encodes nine different mRNAs attributable to the alternative promoters (↱ P1 and P2) and splicing (^). The promoter P1, located upstream of exon-1, encodes for intron-2 spliced (i, ii, and iii) or intron-2 retained (iv, v, and vi) mRNAs. The intron-2 spliced mRNAs can encode the full length (ATG1) and/or the Δ40 (ATG40) proteins, depending on the cell context, whereas the mRNA retaining intron-2 can only encode the Δ40 proteins. The P2 initiation transcription site is located in intron-4 and encodes for three transcripts (vii, viii, and ix), which encode the Δ133 and the Δ160 forms. Small interfering RNAs (siRNAs) targeting the different p53 isoforms are represented on top of the corresponding exons or introns.
p53 splice variants were first identified in the late 1980s in human and mouse (Matlashewski et al. 1984; Wolf et al. 1985). Thereafter, an alternative splicing of TP53 intron-9 has been described (Arai et al. 1986; Flaman et al. 1996). To date, in human, nine p53 mRNAs (Fig. 1B) encoding 12 different p53 protein isoforms have been described (Bourdon et al. 2005; Marcel et al. 2010a), p53α (also named full-length p53, FLp53, canonical p53, TAp53α), p53β (or p53i9), p53γ, Δ40p53α (or ΔNp53, p44 or p47), Δ40p53β, Δ40p53γ, Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, and Δ160p53γ (Fig. 2A).
Figure 2.
Human p53 protein isoforms. All exons and domains are represented to scale. (A) Schematic of the 12 p53 isoforms. The color of the protein domains matches the corresponding exon. p53α is composed of two transactivation domains (TA-1 and TA-2), a proline-rich domain (PRD), the DNA-binding domain (DBD), the hinge domain (HD), the oligomerization domain (OD), and the α regulatory domain. Compared with p53α, Δ40 forms lack the TA-1 because of alternative translation initiation at ATG40. The Δ133 and Δ160 protein isoforms are transcribed from P2 and lack TA-1, TA-2, PRD, and part of the DBD. Regarding the carboxy-terminal isoforms, β and γ forms are attributable to an alternative splicing of intron-9 (brown and pink). As they contain the entire exon-9, they encode the beginning of the OD. The theoretical molecular weight of each protein isoform is indicated. The epitope boundaries of different p53 antibodies are represented by dotted lines. It is important to note that Sapu and CM1 polyclonal antibodies contain multiple epitopes within the first 80 amino acids of p53α and several epitopes in the α-carboxy-terminal regulatory domain, explaining the enhanced detection of p53α compared with the other p53 protein isoforms. (B) Amino-acid sequence of human p53 isoforms. The color of the amino-acid sequence matches the encoding exon. The translation initiation methionines (M) are indicated in red. The α, β, or γ peptide sequences are indicated. The underlined amino-acid sequences relate to the conserved cysteine domains in the DBD. Three nuclear localization sequences (NLS) are represented in black boxes. One is common to all isoforms (exon-9) and two are specific of the α forms. The OD encoded by exon-9 and -10 is shown in a red box.
THE HUMAN p53 ISOFORMS
For decades, it was thought that one gene encodes one protein. The sequencing of the human genome changed this dogma and revealed that ∼98% of the human genes are alternatively spliced and contain multiple initiation sites of transcription (promoter). The TP53 gene is no exception. To date, it is reported that human TP53 differentially expresses in normal tissue at least nine mRNAs in a tissue-dependent manner. They are a result of alternative promoter usage (P1 and P2) and alternative splicing of intron-2 and intron-9 (Fig. 1B). Furthermore, depending on the cell type, the translation of the p53 mRNAs can be initiated at different codons. For the mRNAs transcribed from the proximal promoter (P1), translation can be initiated at codons 1 and/or 40, whereas the mRNAs transcribed from the internal promoter (P2) translation can be initiated at codons 133 and/or 160.
The fully spliced p53 transcript (i) encodes the canonical p53 protein (p53α) but also encodes the Δ40p53α isoform thanks to an internal ribosomal entry site (IRES) (Yin et al. 2002; Candeias et al. 2006; Ray et al. 2006). This transcript also exists with two different alternative splicings of exon-9 retaining thus the exon-9β or -9γ (ii/iii) and encoding, respectively, the p53β and/or Δ40p53β, and the p53γ and/or Δ40p53γ. Both exon-9β and exon-9γ contain stop codons so that exon-10 and -11 are noncoding in β and γ p53 mRNA splice variants. The second type of transcript, also expressed from the promoter P1, conserves intron-2 (iv/v/vi). Retention of this intron leads to several stop codons when translation is initiated from codon 1, thus preventing synthesis of full-length p53 proteins. However, translation can still be initiated at codon 40 so that this group of mRNAs encodes exclusively Δ40p53α, Δ40p53β, or Δ40p53γ. The mRNAs vii/viii/ix are transcribed from the internal promoter P2 located within intron-4. Their translation can start either at the codons 133 or 160 encoding, thus, the Δ133 and Δ160 p53 protein isoforms (α, β, or γ, respectively).
At the protein level, p53α (canonical p53) was the first p53 protein isoform to be identified. It is a 393-amino-acid-long protein with seven functional domains (Fig. 2A,B). The amino terminus is composed of two transactivation (TA) domains, TA-1 and TA-2, which are required to induce a distinct subset of p53-target genes. p53α also contains a proline-rich domain (PRD), a DNA-binding domain (DBD), and a hinge domain (HD). The carboxyl terminus is composed of an oligomerization domain (OD) and a negative regulation domain (α). The negative regulation domain, rich in lysine (K-rich), undergoes many different posttranslational modifications (phosphorylation, methylation, acetylation, sumoylation, ubiquitinylation, neddylation, etc.), which regulate p53α activity and stability (Meek and Anderson 2009). In addition, the hinge and α domains contain several nuclear localization signals (NLSs). The p53 DBD protein sequence is highly conserved through evolution. It contains several conserved cysteines and histidines that coordinate Zn2+ or Mg2+ essential for p53 conformation and DNA-binding activity (Fig. 2B) (Pavletich et al. 1993; Cho et al. 1994; Xue et al. 2009).
The Δ40, Δ133, and Δ160 protein isoforms are, respectively, lacking the first 39, 132, and 159 amino acids. The Δ40 isoforms have lost the TA-1 but still retain the TA-2 and the complete DBD. The Δ133 isoforms lack both TA domains and a small part of the first conserved cysteine box of the DBD, whereas the Δ160 isoforms lack both TA domains and the entire first conserved cysteine box of the DBD, retaining the three other conserved cysteine boxes of the DBD.
All p53 isoforms, whether transcribed from the P1 or P2 promoter, can alternatively splice exon-9β and -9γ to produce the β- and γ-carboxy-terminal protein domains, replacing part of the oligomerization domain and the entire α domain. The β-carboxy-terminal protein is composed of 10 amino acids, although the γ-carboxy-terminal protein domain is composed of 15 amino acids. Of note, the first seven amino acids of the oligomerization domain are present in the α, β, and γ p53 isoforms.
SCIENTIFIC TOOLS TO INVESTIGATE p53 ISOFORM BIOLOGICAL ACTIVITIES
(RT-q)PCR
The most reliable method to identify and quantify p53 splice variants is (RT-q)PCR (quantitative reverse transcription polymerase chain reaction). Several protocols and a set of primers have been designed and published to detect and quantify p53 isoform mRNAs in cell lines and tumor samples (Khoury et al. 2013). However, there is not always a strict correlation between the expression levels of p53 mRNA variants and p53 protein isoforms because p53 protein isoforms are also regulated at the posttranscriptional level.
Small Interfering RNAs (siRNAs)
We and others have developed several siRNAs and shRNAs targeting specifically distinct exons or introns of human TP53, which enable knockdown differentially and specifically subsets of human p53 splice variants (Fig. 1B; Table 1). The siRNAs are essential tools to determine the biological activities of endogenous p53 protein isoforms (Bourdon et al. 2005; Fujita et al. 2009; Aoubala et al. 2011; Terrier et al. 2011, 2012, 2013; Marcel et al. 2012, 2014; Wei et al. 2012; Bernard et al. 2013; Mondal et al. 2013).
Table 1.
p53 isoform-specific small interfering RNAs (siRNAs)
| Position on sequence (nt) GenBank accession number (NG_017013) | Exon location | Exon sequence | Targeted forms | |
|---|---|---|---|---|
| si E2 | 15968–15986 | Exon 2 | GCAGUCAGAUCCUAGCGUC | p53α, p53β, p53γ, Δ40p53α, Δ40p53β, Δ40p53γ |
| si 133a | 17212–17230 | Intron 4 | GGAGGUGCUUACACAUGUU | Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, Δ160p53γ |
| si 133b | 17271–17289 | Intron 4 | CUUGUGCCCUGACUUUCAA | Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, Δ160p53γ |
| si E7 | 18322–18340 | Exon 7 | GCAUGAACCGGAGGCCCAU | All |
| si E7/E8 | 18363–18370/ 18714–18724 |
Exon 7 and 8 | GACTCCAGTGGTAATCTAC | All |
| si β | 19211–19229 | Exon 9β | GGACCAGACCAGCUUUCAA | p53β, Δ40p53β, Δ133p53β, Δ160p53β |
| si γ | 19009–19016/ 19285–19295 |
Exon 9 and 9γ | CCCUUCAGAUGCUACUUGA | p53γ, Δ40p53γ, Δ133p53γ, Δ160p53γ |
| si Splice | 19284–19302 | Exon 9β and 9γ | GAUGCUACUUGACUUACGA | p53β, Δ40p53β, Δ133p53β, Δ160p53β, p53γ, Δ40p53γ, Δ 133p53γ, Δ160p53γ |
| si α | 21846–21864 | Exon 10 | GUGAGCGCUUCGAGAUGUU | p53α, Δ40p53α, Δ133p53α, Δ160p53α |
Antibodies
Several mouse monoclonal antibodies are available to detect endogenous p53 protein isoforms by immunofluorescence, immunohistochemistry (IHC) on paraffin-embedded tissue, and western blotting (Marcel et al. 2013). The epitope of each p53 antibody has been determined by ELISA on an epitope-mapping peptide library (Figs. 2A, 3B; Table 2). The mouse monoclonal antibodies DO-1 and DO-7 recognize the same epitope within the TA-1 domain and can only detect p53α, p53β, and p53γ. The mouse monoclonal antibody 1801 recognizes an epitope within the TA-2 domain and, thus, detects p53α, p53β, p53γ, Δ40p53α, Δ40p53β, and Δ40p53γ. In some cell lines and tissue, 1801 cross-reacts with a protein not related to p53. The mouse monoclonal 421 and BP53.10 recognize a similar epitope within the α domain and, thus, detect p53α, Δ40p53α, Δ133p53α, and Δ160p53α. The other mouse monoclonal antibodies (DO-11, 240, DO-12, and 1620) recognize distinct epitopes within the DBD and should, thus, theoretically detect all p53 protein isoforms. Because p53 protein isoforms can be modified by posttranslational modifications, which alter their migration on SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis), it is recommended to transfect cells with siRNA targeting distinct p53 splice variants to validate that the bands detected are p53 protein isoforms (Marcel et al. 2013). Furthermore, posttranslational modifications within epitopes can prevent binding of p53 antibodies. Detection by DO-1 is impaired by phosphorylation of serine-20 (Craig et al. 1999). Similarly, it is well established that binding of 421 antibody is prevented by posttranslational modification of lysine-372, serine-376/378, threonine-377, and histidine-380 (Pospísilová et al. 2000). The antibody BP53-10 could be used to circumvent this problem as it is less sensitive to posttranslational modifications surrounding its epitope.
Figure 3.
Localization of the human p53 small interfering RNA (siRNA) and p53 antibodies’s epitopes. (A) p53 isoform-specific siRNAs are represented on the human TP53 gene. (B) p53 antibodies’ epitopes on human p53 protein. The underlined amino-acid sequences correspond to the epitopes of the named antibodies (mAb1801, mAb240, mAb DO-11, and mAb DO-12). mAb DO-1 and DO-7 have the same epitope. mAb1620 antibody has a conformational epitope that is represented by green boxes. The mAb 421 epitope is shown in a red box, whereas the mAb BP53.10 epitope is underlined.
Table 2.
p53 isoform-specific antibodies
| Epitope | ||||
|---|---|---|---|---|
| Amino acids | Exon | Sequence | Recognized forms | |
| DO-1 | 20–25 | 2 | SDLWKL | p53α, p53β, p53γ |
| DO-7 | 20–25 | 2 | SDLWKL | p53α, p53β, p53γ |
| 1801 | 46–55 | 4 | SPDDIEQWFT | p53α, p53β, p53γ, Δ40p53α, Δ40p53β, Δ40p53γ |
| 1620 | 145–157 + 201–212 |
5/6 | LWVDST PPPGTRV + LRVEYLDDRN TF | ND |
| DO-11 | 181–190 | 5/6 | RCSDSDGLAP | All |
| 240 | 211–220 | 6 | TFRHSVVVPY | All |
| DO-12 | 256–267 | 7/8 | TLEDSSGNLLGR | All |
| SAPU | 11–65/ 290–301/ 363–393 |
2/3/4/ 8/9/ 10/11 |
EPPLSQ-DPGPDEAPR/RKKGEPHHELPP/ RAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
All |
| CM1 | 1–90/ 379–393 |
2/3/ 4/11 |
MEEPQSDPSV… …TPAAPAPAPS/ RHKKLMFKTEGPDSD |
p53α, p53β, p53γ, Δ40p53α, Δ40p53β, Δ40p53γ, Δ133p53α, Δ160p53α |
| 421 | 372–379 | 11 | KKGQSTSR | p53α, Δ40p53α, Δ133p53α, Δ160p53α |
| BP53.10 | 374–378 | 11 | GQSTS | p53α, Δ40p53α, Δ133p53α, Δ160p53α |
ND, not determined.
The rabbit (CM1) and sheep (SAPU) polyclonal antibodies have been raised against bacterially produced recombinant human p53α. They recognize several distinct epitopes within the amino-terminal and α domains so that the detection of p53α is strongly enhanced compared with the other p53 isoforms, thus preventing any expression-level comparisons between p53α and the other isoforms. CM1 and SAPU antibodies have a lower affinity for the Δ133/Δ160 and β/γ forms that lack the amino terminus or carboxyl-terminus epitopes, respectively. Only SAPU recognizes an epitope within the HD, which is common to all p53 protein isoforms. It can, thus, detect Δ133p53β/γ and Δ160p53β/γ, whereas CM1 cannot.
THE p53 ISOFORM RELEVANCE IN CLINIC
In recent years, numerous studies have shown that p53 isoforms are abnormally expressed in breast cancer, melanoma, renal cell carcinoma (RCC), acute myeloid leukemia (AML), colon carcinoma, head and neck tumors (HNSSCs), hepatic cholangiocarcinoma, ovarian cancer, lung tumor, and glioblastoma (Bourdon et al. 2005, 2011; Anensen et al. 2006; Boldrup et al. 2007; Avery-Kiejda et al. 2008; Marabese et al. 2008; Fujita et al. 2009; Song et al. 2009; Hofstetter et al. 2010; Nutthasirikul et al. 2013; Takahashi et al. 2013).
Several studies investigated whether p53 isoforms were associated with cancer patient prognosis. First, it was reported that the evolution from colorectal adenoma to carcinoma was potentially driven by an imbalance of p53β/Δ133p53α ratio in favor of the latter (Fujita et al. 2009). In AML, distinct p53 isoform biosignatures correlate with clinical outcome (Ånensen et al. 2012), whereas in cholangiocarcinoma, down-regulation of the TAp53 isoforms (p53α/p53β/p53γ) combined with up-regulation of the Δ133 forms is associated with a shortened overall survival (Nutthasirikul et al. 2013).
Importantly, several studies in different human cancers have reported that the prognostic value of TP53 mutation status is improved when combined with p53 isoform expression. Wild-type (WT) TP53 mucinous or serous ovarian cancer patients expressing Δ40p53α have a better clinical outcome than WT TP53 mucinous or serous ovarian cancer patients not expressing Δ40p53α. Reciprocally, mutant TP53 serous ovarian cancer patients expressing Δ133p53α have a better disease-free and overall survival than mutant TP53 serous ovarian cancer patients not expressing Δ133p53α (Hofstetter et al. 2011, 2012; Chambers and Martinez 2012). Furthermore, in WT TP53 ovarian cancers, p53β expression is associated with higher risk of recurrence because it is a marker of serous and poorly differentiated cancer (Hofstetter et al. 2010).
In breast cancer, it was reported that p53β expression is associated with smaller tumors and longer disease-free survival in mutant TP53 tumors, whereas Δ40p53 isoforms expression was found to be associated with the aggressive triple negative subtype (Avery-Kiejda et al. 2014). In another study, it was shown that mutant TP53 breast cancer patients expressing p53γ have as a good prognosis as WT TP53 breast cancer patients, whereas mutant TP53 breast cancer patients devoid of p53γ expression have a particularly poor prognosis (Bourdon et al. 2011).
Altogether, it suggests that the prognostic values of p53β, p53γ, Δ40p53α, or Δ133p53α depend on the TP53 mutations status and the cancer type.
Other studies have investigated whether p53 isoforms play active roles in cancer formation and treatment. Silent TP53 mutation or mutations in noncoding regions, such as IRES sequences, introns, or splicing sites, are associated with cancer formation probably because they lead to unbalanced p53 isoform expression despite expressing WT p53 proteins (Hofstetter et al. 2010; Grover et al. 2011; Khan et al. 2013). Furthermore, the pathogenic bacteria Helicobacter pylori has recently been shown to induce expression of Δ133 and Δ160 isoforms in gastric epithelial cells, increasing their survival and probably promoting cancer formation (Wei et al. 2012). Moreover, p53 isoforms are involved in response to chemotherapy in vivo in AML and melanoma (Anensen et al. 2006; Avery-Kiejda et al. 2008; Ånensen et al. 2012).
In conclusion, p53 isoforms expression is associated to clinical outcome of cancer patients. For a given cancer type, the accuracy of patient prognosis is greatly improved by combining p53 isoforms expression and TP53 mutation status, which may be used to predict response to treatment for some cancer types.
ANIMAL MODELS: IN SEARCH OF THE PHYSIOLOGICAL RELEVANCE OF ISOFORMS
The structure of the TP53 gene is highly conserved through evolution (Bourdon et al. 2005; Chen et al. 2005). Several p53 isoform animal models were, thus, generated to investigate their biological relevance and roles in carcinogenesis.
Zebrafish
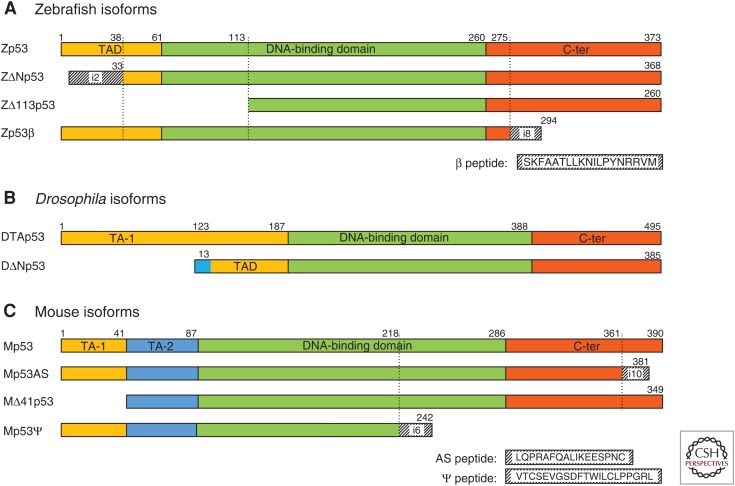
The zebrafish TP53 gene (Zp53) has a dual gene structure as its human counterpart (Storer and Zon 2010). In addition to the Zp53α protein, corresponding to the human p53α (Cheng et al. 1997), the gene also encodes Zp53β, ZΔNp53α, and ZΔ113p53α. In addition, in some zebrafish strains, the Zp53 gene expresses from the internal promoter TA2Zp53, TA3Zp53, TA4Zp53, and TA5Zp53 because of a polymorphism (Fig. 4A).
Figure 4.
p53 isoforms in animal models. Like human p53 protein isoforms, zebrafish, Drosophila, and mouse p53 proteins are also composed of several functional domains: the transactivation domain (TAD, yellow), the DNA-binding domain (DBD, green) and the carboxy-terminal regulatory domain containing the oligomerization domain (C-ter, orange). For each species, all domains are represented to scale compared with the full-length protein of the corresponding species. Hatched protein domains are encoded by cryptic exons within introns (the corresponding intron is indicated). (A) Zebrafish p53 isoforms. In the ZΔNp53 isoform, the first 33 amino acids are encoded by a cryptic exon in intron-2. In Zp53β, the last 19 amino acids are encoded by a cryptic exon in intron-8. Of note, Zp53β is not homologous to human p53β. (B) Drosophila p53 protein isoforms. Like in the human TP53 gene family, DΔNp53 (also named Dp53) protein is encoded by an mRNA transcribed from an internal promoter. DΔNp53 is homologous to Δ40p53/Δ133p53/ΔNp63/ΔNp73 proteins. Only DTAp53 protein contains the conserved box I (FxxLW) corresponding to the transactivation domain (TA-1) of the p53 protein family. (C) Mouse p53 isoforms. In Mp53AS protein (alternatively spliced [AS]), the last 26 amino acids of Mp53α are substituted by 17 others encoded by a cryptic exon in intron-10. Mp53AS protein is homologous to human p53β. MΔ41p53 produced by alternative splicing of intron-2 is homologous to human Δ40p53α. The Mp53Ψ is produced by alternative splicing of intron-6 adding 21 new amino acids as indicated. Up to now, no homologous protein to Mp53Ψ has been found in other organisms or in human cells expressing the WT TP53 gene.
ZΔNp53α is produced through retention of intron-2, similarly to the human Δ40p53 forms. However, ZΔNp53α is not completely identical to the human form as the translation is initiated in intron-2 and substitutes the 38 first amino acids of canonical p53 by 33 different amino acids constituting a different TA domain (Davidson et al. 2010). ZΔNp53α accumulates in response to γ-ray and is able to form a protein complex with Zp53α through the OD. When coexpressed with Zp53α, overexpression of ZΔNp53α induces developmental malformations (hypoplasia, head malformation, somites, and eyes). This phenotype does not appear after overexpression of ZΔNp53α mutated in the oligomerization domain or in Zp53 morpholino-depleted embryo, suggesting that ZΔNp53α acts through oligomerization with Zp53 isoforms during development.
ZΔ113p53α is homologous to human Δ133p53α. It is produced from the internal promoter, which is transactivated by Zp53α (Chen et al. 2005; Marcel et al. 2010b). ZΔ113p53α oligomerizes with Zp53α inhibiting apoptosis by differentially modulating Zp53 target gene expression (Ou et al. 2014). ZΔ113p53α induces expression of cell-cycle arrest genes, such as p21 and cyclin-G1 and of antiapoptotic genes, such as Bcl-XL, while inhibiting expression of proapoptotic genes like Reprimo or Bax (Chen et al. 2009). Furthermore, ZΔ113p53α is strongly induced after DNA double-strand break (DSB) and activates the DNA–DSB repair pathways, notably by up-regulating the transcription of repair genes such as RAD51, RAD52, or lig4 (Gong et al. 2015).
Recently, Shi et al. reported that a natural polymorphism, a 4-bp deletion within intron-4, creates four new upstream translation initiation sites in frame to the ZΔ113p53 open reading frame. This leads to four protein isoforms with a putative TA domain named TA2p53, TA3p53, TA4p53, and TA5p53. They lack the 93 first amino acids of Zp53α that are, respectively, replaced by 65, 45, 18, and 9 amino acids (Shi et al. 2015). These isoforms coexpressed with Zp53α confer resistance to irradiation. In the same publication, Shi et al. also reported the alternative splicing of Zp53 intron-8 leading to Zp53β. However, Zp53β is not homologous to human p53β.
Drosophila
The Drosophila melanogaster TP53 gene (Dmp53) is the single Drosophila ortholog of the mammalian p53 family and has a dual gene structure with an internal promoter. It encodes proteins homologous to p53, p63, and p73 (Lu et al. 2009), and it is thought that, as the unique member of the p53 family, it is likely that Dmp53 carries the ancestral functions of all p53 family genes. Dmp53 transcribes four different mRNA variants but, so far, only two p53 protein isoforms could be detected: DTAp53 and DΔNp53 (Fig. 4B). DTAp53 contains, in the first 40 amino acids, the highly conserved TA domain present in mammalian p53α (Bourdon et al. 2005). DΔNp53 (also called Dp53), expressed from the internal promoter, lacks the 123 first amino acids of DTAp53, which are replaced by 13 different ones encoded by a cryptic exon. DΔNp53 does not contain the conserved TA domain but is still able to transactivate. DΔNp53 was the first Drosophila p53 isoform cloned and was thus called Dp53 as the investigators thought they had cloned full-length Drosophila p53 (Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000). However, DΔNp53 is homologous to the amino-terminally truncated isoforms of the p53 family proteins (Δ40p53, ΔNp63, or ΔNp73). The literature on Drosophila p53 is, thus, confusing as most genetic studies do not discriminate between DTAp53 and DΔNp53 isoforms, reporting phenotype associated with Dmp53 gene as being mediated by Dp53 protein.
Another source of confusion is that mutant p53 flies referred both to p53-null flies and to transgenic flies overexpressing single-point missense mutant of Dmp53 (UAS-Dmp53R155H or UAS-Dmp53H159N). However, p53-null mutation leads to complete loss of p53 protein expression and, thus, p53 activity, whereas missense mutation leads to overexpression of mutant p53 proteins that are biochemically and biologically very active as they directly interact with numerous proteins modifying, thus, gene expression and cell responses. p53-null mutation can lead to different phenotypes than missense p53 mutation as shown in Drosophila and mammalian models (Brodsky et al. 2000; Ollmann et al. 2000; Jassim et al. 2003; Lang et al. 2004; Olive et al. 2004; Wells et al. 2006; de la Cova et al. 2014; Simón et al. 2014)
The Dmp53 gene is involved in apoptosis through the Reaper–Hid–Grim cascade by directly regulating Reaper expression in response to damage (Brodsky et al. 2000, 2004). However, its physiological activity is not limited to apoptotic pathways. Recent studies unravel the primordial roles of Drosophila p53 protein isoforms in tissue regeneration by directly controlling apoptosis-induced proliferation (AiP), compensatory proliferation (CP), cell differentiation, organ size, system growth, and cell competition (Peterson et al. 2002; Jassim et al. 2003; Wells et al. 2006; Stieper et al. 2008; Mendes et al. 2009; Fan et al. 2010; Mesquita et al. 2010; Morata et al. 2011; Wells and Johnston 2012; de la Cova et al. 2014; Simón et al. 2014). This is consistent with Dmp53 being involved in life-span control (Bauer et al. 2005, 2010; Waskar et al. 2009; for review, see Martín et al. 2009; Kashio et al. 2014; Mollereau and Ma 2014).
However, the contribution of each Drosophila p53 protein isoform in those biological phenomena is still poorly understood. Recently, Dichtel-Danjoy et al. (2013) investigated the respective roles of DTAp53 and DΔNp53 overexpression in apoptosis and AiP in the developing wing imaginal disc. AiP is a phenomenon in which cells undergoing apoptosis after a stress event (i.e., “undead cells”) secrete mitogens, such as Wingless (Wg), Decapentaplegic (Dpp), or Hedgehog (Hh) (homologous to human Wnt, TGF-β, or Hedgehog, respectively) to promote proliferation of neighboring cells to restore the normal organ and body size. Thus, at the same time that the stress event eliminates a large number of cells, it also generates a stimulus to proliferation. AiP is directly controlled by Dmp53 (Wells et al. 2006; Dichtel-Danjoy et al. 2013; Simón et al. 2014). Both DTAp53 and DΔNp53 protein isoforms were capable of activating apoptosis but they used different molecular mechanisms. However, only DΔNp53 (not DTAp53) can induce Wg expression in “undead” cells and enhanced proliferation of neighboring cells in Dmp53-null flies, suggesting that DΔNp53 is the main isoform that regulates AiP. DTAp53 and DΔNp53 seem, thus, to have distinct biological functions.
Mouse
The mouse TP53 gene (MsTP53) has a dual gene structure as its human counterpart. To date, four mouse p53 protein isoforms have been published: Mp53α, Mp53AS, MΔ41p53α, and Mp53Ψ (Fig. 4C). Mp53α is the equivalent to the canonical human p53α. Mp53AS is a carboxy-terminal splicing variant because of retention of part of intron-10. It encodes a 2-kDa shorter protein with different carboxy-terminal amino acids, which has a strong homology with the human β p53 proteins. Mp53AS can bind DNA and modulate p53-target-gene expression in a promoter-dependent manner. Mp53AS can induce apoptosis when expressed in absence of Mp53α (Almog et al. 1997). However, when Mp53AS and Mp53α are coexpressed, cells do not induce apoptosis in response to damage (Wolf et al. 1985; Almog et al. 2000). Mp53AS and Mp53α oligomerize together and bind DNA to regulate gene expression (Wu et al. 1994). Therefore, Mp53AS and Mp53α harbor completely different/opposite activities when they are expressed alone or in combination.
Recently, two knockin p53 mice, Mp53Δ24 and Mp53Δ31-expressing carboxy-terminal truncated mutants of p53 have been generated. The Mp53Δ24 mice contain a stop mutation at codon 367 in the beginning of exon-11 so that the mice express Mp53Δ24, MΔ41p53Δ24 but also the WT Mp53AS and Mp53Ψ proteins, because alternative splicing of intron-10 can still occur (Hamard et al. 2013). The Mp53Δ31 mice contain a stop mutation at codon 360 at the end of exon-10 so that the mice express Mp53Δ31, MΔ41p53Δ31, and WT Mp53Ψ. The Mp53Δ31 mice do not express Mp53AS (Simeonova et al. 2013). In both studies, the MsTP53 gene has been extensively sequenced to ensure the absence of any additional mutations in MsTP53 exons and introns. Interestingly, the Mp53Δ24 and Mp53Δ31 mice present different phenotypes despite being of similar mixed genetic background (BL6/129Sv). The Mp53Δ24 and Mp53Δ31 mice have different blood cell counts and different sizes of heart, thymus, spleen, testis, and cerebellum. Mp53Δ31 mice also develop oral leukoplakia (100%) and pulmonary fibrosis (87%), traits not reported for Mp53Δ24 mice. One of the most intriguing differences is in the repopulating capabilities of hematopoietic stem cells (HSCs) from embryos of the Mp53Δ24 and Mp53Δ31 mice. WT matching mixed genetic background mice (BL6/129Sv) were lethally irradiated to destroy the bone marrow cells. The irradiated mice were then transplanted with HSCs from Mp53Δ24, Mp53Δ31, or WT MsTP53 mice (BL6/129Sv). In both laboratories, the transplantation of HSC from WT MsTP53 mice rescued 100% of the irradiated mice by regenerating the bone marrow, although none of the irradiated and nontransplanted mice survived, indicating that both laboratories master this assay. Importantly, the transplantation of HSC from Mp53Δ31 mice completely fails to repopulate the bone marrow and to rescue, thus, the irradiated mice (0% rescue, n = 5), whereas the transplantation of HSC from Mp53Δ24 mice rescue ∼70% (n = 6) of the irradiated mice by repopulating the bone marrow (Hamard et al. 2013; Simeonova et al. 2013).
Knowing the potent transcriptional activities of Mp53AS and its tissue-specific expression, it is likely that the different phenotypes of the Mp53Δ24 and Mp53Δ31 mice are attributable to differential expression of Mp53AS. Further investigations will be required to determine the physiological roles of Mp53AS in tissue regeneration in mice.
Recently, an additional isoform attributed to the alternative splicing of intron-6, Mp53Ψ (also named p53Ψ) has been described. This results from the physiological insertion of the last 55 nucleotides of intron-6, which rapidly leads to a stop codon. Mp53Ψ is thus a very short p53 isoform containing the TA and proline domains but no DBD (Senturk et al. 2014). Mp53Ψ may act as a prometastatic factor in promoting epithelial–mesenchymal transition and regulating mitochondrial activity together with cyclophilin-D. Interestingly, Mp53Ψ seems to be expressed after damage in specific stem cells during tissue regeneration in mice. In liver injured with CCL4, the lesions get smaller concomitantly to higher expression of Mp53Ψ. This is a fascinating result as it indicates that the Msp53 gene is involved in tissue regeneration in mammalians. Furthermore, it opens new perspectives as p53 isoform expression could be manipulated to control organ regeneration in mammalians.
Mp53Ψ mRNA expression is unequivocal in regenerating lung or liver mouse tissues; further investigations will be required to confirm physiological p53Ψ expression in WT human cells. So far, endogenous human p53Ψ protein could only be detected in one cancer cell line (Hop62) that bears a mutation in the acceptor-splicing site of human TP53 intron-6. Such a mutation abolishes the canonical splicing of intron-6 and promotes the insertion of the last 49 nucleotides of human TP53 intron-6 leading to a p53Ψ-like protein as shown by the investigators (Senturk et al. 2014). The development of human/mouse p53Ψ-specific antibodies and siRNA would greatly help to assess p53Ψ expression and study its biological activities.
The MΔ41p53α isoform has also been genetically investigated. MΔ41p53α (also named p44) is the mouse counterpart of human Δ40p53α. MΔ41p53α overexpression in WT Mp53 mice leads to a smaller animal with premature aging and shorter life span. These effects have been attributed to abnormal insulin-like growth factor (IGF) signaling, driving a reduced cellular proliferation with increased senescence (Maier et al. 2004; Gambino et al. 2013), inhibition of embryonic stem cell (ESC) differentiation (Ungewitter and Scrable 2010), neurodegeneration (Pehar et al. 2010, 2014), and impaired β-cell proliferation (Hinault et al. 2011), which suggests that the MΔ41p53 transgenic mice have altered tissue-regeneration capabilities.
To genetically investigate the biological activities of Δ133p53 isoforms, Slatter et al. (2011) have generated knockin mutant TP53 mice that are deleted of exon-3 and -4 (MsΔ122p53), expressing ubiquitously Δ133p53-like protein (Δ122p53α) and probably Δ122p53AS in a tissue-dependent manner because of alternative splicing of intron-10. The homozygote MsΔ122p53 mice show enhanced proinflammatory phenotype, features of autoimmune disease, and early tumor onset (Campbell et al. 2012; Sawhney et al. 2015). It was thus thought that Δ122p53 proteins were oncogenic. However, it was recently genetically shown that Δ122p53 proteins enhance the tumor-suppressor activities of an attenuated p53 mutant deleted of the proline domain (MΔpro) (Slatter et al. 2015).
Altogether, the different animal models are consistent in demonstrating that the p53 isoforms are potent and essential components of the p53 pathway, being involved in cancer, aging, tissue regeneration, glucose metabolism, embryo development, immune system, and bacterial infection (Wei et al. 2012). The balance between the different p53 isoforms is crucial in determining cell-fate outcome.
REGULATION OF HUMAN p53 ISOFORM EXPRESSION/ACTIVITIES
p53 isoform expression is regulated at the transcriptional level by modulating the TP53 promoter activities and the alternative splicing of intron-2 and intron-9.
In addition to the epigenetic events that regulate the tissue-specific activity of the p53 promoters, the internal TP53 promoter activity is also influenced by several polymorphisms, particularly the 16-bp insertion in intron-3 (pin3) and the R72P polymorphism in exon-4 altering thus Δ133p53 isoform expression (Fig. 1A) (Bellini et al. 2010; Marcel et al. 2012). In addition, the internal TP53 promoter is transactivated by p53α, diverse p63/p73 isoforms, and p68 (Moore et al. 2010; Aoubala et al. 2011; Marcel et al. 2012).
SRSF1 and SRSF3, a highly conserved family of splicing factors frequently deregulated in cancer, regulate the alternative splicing of TP53 intron-9 inhibiting retention of exon-9β/9γ (Tang et al. 2013; Marcel et al. 2014). Interestingly, TG003, a small drug inhibitor of the Cdc2-like kinases (Clks) that activate SRSF1 and SRSF3, increases endogenous p53β/γ expression inhibiting cell proliferation in a p53 isoform-dependent manner (Marcel et al. 2014).
The alternative splicing of TP53 intron-2 leads to Δ40p53. Its expression is influenced by a G-quadruplex structure within intron-3, which is strengthened by the pin3 polymorphism that contains further G-quadruplex structures (Marcel et al. 2011).
p53 isoform expression is also regulated at the translational and posttranslational levels. The Δ40p53 isoforms can be translated from IRES controlled by IRES transactivating factors (ITAFs), such as polypyrimidine-tract-binding protein (PTB), dyskerin, DAP5, annexin A2, and PTB-associated splicing factor (PSF) (Grover et al. 2008; Sharathchandra et al. 2012; Weingarten-Gabbay et al. 2014).
Camus et al. (2012) have reported that all p53 isoforms are ubiquitinated and degraded by the proteasome. However, although MDM-2 binds to all p53 protein isoforms, it can only promote the ubiquitination and degradation of p53β. MDM2 binds probably Δ133p53 isoforms (α, β, or γ) through the DBD (Wallace et al. 2006) and/or the α-carboxy-terminal domain (Poyurovsky et al. 2010) because the Δ133p53 isoforms have lost the MDM2-binding domain present in the amino-terminal end of p53α, p53β, and p53γ.
In addition to proteasome degradation, Δ133p53α is also degraded by autophagy during replicative senescence. Pharmacological inhibition of autophagy by bafilomycin A1 restores Δ133p53α expression levels. The autophagic degradation of Δ133p53α induces senescence and is inhibited by direct interaction of the chaperone-associated E3 ubiquitin ligase STUB1/CHIP with Δ133p53α (Horikawa et al. 2014).
Altogether, expression of each p53 isoform is tightly and differentially regulated, enabling dynamic and accurate triggering of the appropriate cellular response to damage. The diverse regulations of p53 isoforms expression could be used as therapeutic targets to trigger controlled cell-fate outcome.
p53 ISOFORM BIOLOGICAL ACTIVITIES
p53α, p53β (Mp53AS), p53γ, Δ40p53α (MΔ41p53α), and Δ133p53α (ZΔ113p53α and MΔ122p53) have been shown to differentially regulate gene expression and to be biochemically and biologically active either alone or in combination. Δ133p53β has recently been shown to regulate cell stemness; however, its molecular mechanism is still unknown (Arsic et al. 2015).
Over the past 10 years, using diverse human cell lines and animal models, all data have consistently indicated that the balance of expression among the p53 isoforms define the p53-mediated cell response to trigger after damage, virus/bacterial infection, or cell signals (Maier et al. 2004; Chen et al. 2009; Fujita et al. 2009; Medrano et al. 2009; Davidson et al. 2010; Pehar et al. 2010, 2014; Ungewitter and Scrable 2010; Aoubala et al. 2011; Hinault et al. 2011; Slatter et al. 2011, 2015; Terrier et al. 2012; Wei et al. 2012; Bernard et al. 2013; Dichtel-Danjoy et al. 2013; Gambino et al. 2013; Mondal et al. 2013; Silden et al. 2013; Horikawa et al. 2014; Marcel et al. 2014; Takahashi et al. 2014; Arsic et al. 2015; Gong et al. 2015). The p53 isoforms thus play primordial roles in cell-cycle progression, programmed cell death, senescence, inflammation, stem-cell renewal and differentiation, aging, neurodegeneration, glucose metabolism, angiogenesis, embryo development, and cancer.
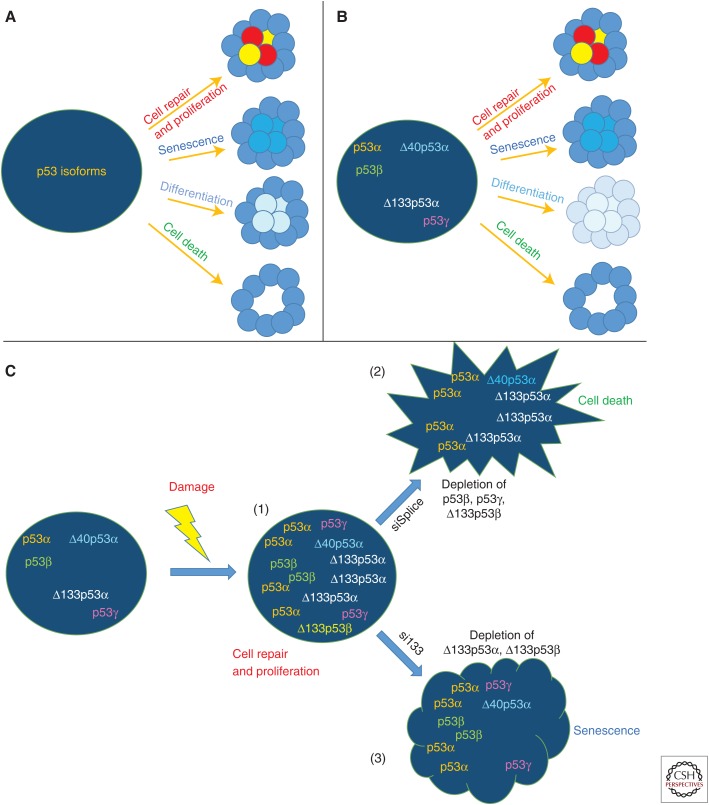
The manipulation of p53 isoforms using splicing-factor inhibitors, autophagy inhibitors, DNA damage, p53 isoform overexpression, and/or siRNA targeting, specifically the p53 isoforms, enables the triggering of different cell-fate outcomes in response to the same damage (Aoubala et al. 2011; Horikawa et al. 2014; Marcel et al. 2014; Gong et al. 2015). Mechanistically, p53 isoforms oligomerize so that, for example, the oligomers composed of p53β and p53α regulate different p53-responsive genes than the ones composed of p53γ and p53α or Δ133p53α and p53α (Fujita et al. 2009; Aouba et al. 2011; Bernard et al. 2013; Marcel et al. 2014; Solomon et al. 2014). p53 isoform expression is cell-type-specific and several p53 isoforms are always concomitantly coexpressed or are mutually exclusive (Fig. 5). Hence, the p53-mediated cell response would be the sum of the activities of each oligomer of the coexpressed p53 isoforms in a given tissue. It implies that none of the p53 isoforms, including canonical p53α, is able to abolish the activity or expression of the other coexpressed p53 isoforms. It is thus impossible to define a p53 isoform as oncogene or tumor suppressor because its activity depends on the cell context. Hence, although p53β in T cells or normal human fibroblasts prevents cell growth and induces senescence (Fujita et al. 2009; Mondal et al. 2013), Marcel et al. (2014) showed that endogenous p53β/γ in MCF-7 cells inhibit cell proliferation by promoting G1-cell-cycle arrest and cell death under standard culture conditions, but promote cell proliferation after treatment with TG003. In the Δ133p53-like mouse model (MsΔ122p53), although Δ133p53α/Δ122p53 inhibits WT p53 tumor suppressor activity, it has recently been reported that Δ122p53 proteins enhance the tumor-suppressor activities of an attenuated p53 mutant deleted of the proline domain (MΔpro) (Slatter et al. 2015). This is consistent with clinical studies. Mutant TP53 serous ovarian cancer patients expressing Δ133p53α have better disease-free and overall survival than mutant TP53 serous ovarian cancer patients not expressing Δ133p53α (Hofstetter et al. 2011, 2012; Chambers and Martinez 2012).
Figure 5.
Biological model. (A) p53 isoforms define cell-fate outcomes in response to intracellular and extracellular cell signals—cell survival/proliferation, senescence, differentiation, or programmed cell death. (B) The cell type (epigenetic) defines p53 isoform expression and the possibilities of cell-fate outcomes. (C) In response to damage, p53 isoforms are differentially regulated and define cell-fate outcome. (1) In this theoretical cell, the coexpressed p53 isoforms orchestrate cell repair and allow cell proliferation in response to damage. However, it is important to note that the cell response to the same damage can be drastically changed by manipulating expression of only a subset of p53 isoforms using p53 isoform-specific small interfering RNA (siRNA). (2) Hence, after depletion of p53β, p53γ, and Δ133p53β using the siRNA siSplice, the cell response to the same damage is to induce cell death, (3) although, after depletion of Δ133p53α and Δ133p53β using the siRNA si133, the cell response to the same damage is to induce senescence. (This model is based on data in Wu et al. 1994, Almog et al. 2000, Yin et al. 2002, Chen et al. 2009, Fujita et al. 2009, Ungewitter and Scrable 2010, Aoubala et al. 2011, Terrier et al. 2012, Marcel et al. 2014, Slatter et al. 2015, and Gong et al. 2015.)
CONCLUSION AND INSIGHTS
Based on the data gathered over the past 10 years by several laboratories using diverse human cell lines and animal models, we can now assert that p53 isoforms are physiologically active and potent proteins. However, their activities are cell-type-dependent. The misregulation of p53 isoform expression leads to cancer, premature aging, (neuro)degenerative diseases, inflammation, and embryo malformations. The data leads us then to realize that a p53-mediated cell response is not driven by a single protein, canonical p53, but is in fact the sum of the activities of the coexpressed p53 isoforms in a given tissue. Furthermore, a p53-mediated cell response is broader than just preventing cancer formation by controlling cell proliferation and cell death. In fact, the effects of p53 isoform expression misregulation are consistent with a physiological role of p53 isoforms in organ maintenance/regeneration as observed in Drosophila, zebrafish, and mouse p53 isoform animal models (Jassim et al. 2003; Chen et al. 2005, 2009; Medrano et al. 2009; Davidson et al. 2010; Dichtel-Danjoy et al. 2013; Hamard et al. 2013; Simeonova et al. 2013; De la Cova et al. 2014; Kashio et al. 2014; Senturk et al. 2014; Simón et al. 2014; Gong et al. 2015). The balance between the different p53 isoform is crucial in predicting cell-fate outcome. Therefore, deciphering the p53 isoforms combinatorics (p53 isoforms’ code) offers fascinating and important new perspectives to treat cancer, degenerative diseases and aging.
Cracking the p53 isoforms’ code may seem a daunting task because of the high number of p53 isoforms and posttranslational modifications. However, the p53 isoforms are rarely coexpressed all together at once; some isoforms are mutually exclusive, whereas others are always coexpressed so that the number of p53 isoform combinations is limited. Cracking the p53 isoforms’ code seems thus an achievable and necessary task to accurately predict response to cancer treatment and to control stem-cell renewal and differentiation in the treatment of degenerative diseases and aging. It offers new exciting perspectives in regenerative medicine.
ACKNOWLEDGMENTS
J-C.B. is a fellow of Breast Cancer Now (Grant No. 2012MaySF127). All of the p53 antibodies described in this review (except SAPU) are commercially available or can be requested from Dr. Borek Vojtesek (vojtesek@mou.cz). The SAPU antibody can be requested from J.-C.B.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Almog N, Li R, Peled A, Schwartz D, Wolkowicz R, Goldfinger N, Pei H, Rotter V. 1997. The murine C′-terminally alternatively spliced form of p53 induces attenuated apoptosis in myeloid cells. Mol Cell Biol 17: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almog N, Goldfinger N, Rotter V. 2000. p53-dependent apoptosis is regulated by a C-terminally alternatively spliced form of murine p53. Oncogene 19: 3395–3403. [DOI] [PubMed] [Google Scholar]
- Anensen N, Oyan AM, Bourdon JC, Kalland KH, Bruserud O, Gjertsen BT. 2006. A distinct p53 protein isoform signature reflects the onset of induction chemotherapy for acute myeloid leukemia. Clin Cancer Res 12: 3985–3992. [DOI] [PubMed] [Google Scholar]
- Ånensen N, Hjelle SM, Van Belle W, Haaland I, Silden E, Bourdon JC, Hovland R, Taskén K, Knappskog S, Lønning PE, et al. 2012. Correlation analysis of p53 protein isoforms with NPM1/FLT3 mutations and therapy response in acute myeloid leukemia. Oncogene 31: 1533–1545. [DOI] [PubMed] [Google Scholar]
- Aoubala M, Murray-Zmijewski F, Khoury MP, Fernandes K, Perrier S, Bernard H, Prats AC, Lane DP, Bourdon JC. 2011. p53 directly transactivates Δ133p53α, regulating cell fate outcome in response to DNA damage. Cell Death Differ 18: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai N, Nomura D, Yokota K, Wolf D, Brill E, Shohat O, Rotter V. 1986. Immunologically distinct p53 molecules generated by alternative splicing. Mol Cell Biol 6: 3232–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsic N, Gadea G, Lagerqvist EL, Busson M, Cahuzac N, Brock C, Hollande F, Gire V, Pannequin J, Roux P. 2015. The p53 isoform Δ133p53β promotes cancer stem cell potential. Stem Cell Rep 4: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery-Kiejda KA, Zhang XD, Adams LJ, Scott RJ, Vojtesek B, Lane DP, Hersey P. 2008. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin Cancer Res 14: 1659–1668. [DOI] [PubMed] [Google Scholar]
- Avery-Kiejda KA, Morten B, Wong-Brown MW, Mathe A, Scott RJ. 2014. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 35: 586–596. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. 2005. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol 15: 2063–2068. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Chang C, Bae G, Morris SN, Helfand SL. 2010. Dominant-negative Dmp53 extends life span through the dTOR pathway in D. melanogaster. Mech Ageing Dev 131: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini I, Pitto L, Marini MG, Porcu L, Moi P, Garritano S, Boldrini L, Rainaldi G, Fontanini G, Chiarugi M, et al. 2010. ΔN133p53 expression levels in relation to haplotypes of the TP53 internal promoter region. Hum Mutat 31: 456–465. [DOI] [PubMed] [Google Scholar]
- Bernard H, Garmy-Susini B, Ainaoui N, Van Den Berghe L, Peurichard A, Javerzat S, Bikfalvi A, Lane DP, Bourdon JC, Prats AC. 2013. The p53 isoform, Δ133p53α, stimulates angiogenesis and tumour progression. Oncogene 32: 2150–2160. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Bourdon JC, Coates PJ, Sjöström B, Nylander K. 2007. Expression of p53 isoforms in squamous cell carcinoma of the head and neck. Eur J Cancer 43: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. 1997. Further characterisation of the p53 responsive element—Identification of new candidate genes for trans-activation by p53. Oncogene 14: 85–94. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19: 2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Khoury MP, Diot A, Baker L, Fernandes K, Aoubala M, Quinlan P, Purdie CA, Jordan LB, Prats AC, et al. 2011. p53 mutant breast cancer patients expressing p53γ have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res 13: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103–113. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell HG, Slatter TL, Jeffs A, Mehta R, Rubio C, Baird M, Braithwaite AW. 2012. Does Δ133p53 isoform trigger inflammation and autoimmunity? Cell Cycle 11: 446–450. [DOI] [PubMed] [Google Scholar]
- Camus S, Ménendez S, Fernandes K, Kua N, Liu G, Xirodimas DP, Lane DP, Bourdon JC. 2012. The p53 isoforms are differentially modified by Mdm2. Cell Cycle 11: 1646–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias MM, Powell DJ, Roubalova E, Apcher S, Bourougaa K, Vojtesek B, Bruzzoni-Giovanelli H, Fåhraeus R. 2006. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 25: 6936–6947. [DOI] [PubMed] [Google Scholar]
- Chambers SK, Martinez JD. 2012. The significance of p53 isoform expression in serous ovarian cancer. Future Oncol 8: 683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. 2005. Loss of function of def selectively up-regulates Δ113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev 19: 2900–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, Peng J. 2009. p53 isoform Δ113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev 23: 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Ford BL, O’Neal PE, Mathews CZ, Bradford CS, Thongtan T, Barnes DW, Hendricks JD, Bailey GS. 1997. Zebrafish (Danio rerio) p53 tumor suppressor gene: cDNA sequence and expression during embryogenesis. Mol Mar Biol Biotechnol 6: 88–97. [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. 1994. Crystal structure of a p53 tumor suppressor–DNA complex: Understanding tumorigenic mutations. Science 265: 346–355. [DOI] [PubMed] [Google Scholar]
- Craig AL, Blaydes JP, Burch LR, Thompson AM, Hupp TR. 1999. Dephosphorylation of p53 at Ser20 after cellular exposure to low levels of non-ionizing radiation. Oncogene 18: 6305–6312. [DOI] [PubMed] [Google Scholar]
- Davidson WR, Kari C, Ren Q, Daroczi B, Dicker AP, Rodeck U. 2010. Differential regulation of p53 function by the N-terminal ΔNp53 and Δ113p53 isoforms in zebrafish embryos. BMC Dev Biol 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cova C, Senoo-Matsuda N, Ziosi M, Wu DC, Bellosta P, Quinzii CM, Johnston LA. 2014. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab 19: 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy ML, Ma D, Dourlen P, Chatelain G, Napoletano F, Robin M, Corbet M, Levet C, Hafsi H, Hainaut P, et al. 2013. Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ 20: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont P, Leu JIJ, Della Pietra AC III, George DL, Murphy M. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49. [DOI] [PubMed] [Google Scholar]
- Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H, Bergmann A. 2010. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ 17: 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D, Iggo R, Frebourg T. 1996. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 12: 813–818. [PubMed] [Google Scholar]
- Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR, et al. 2009. p53 isoforms Δ133p53 and p53β are endogenous regulators of replicative cellular senescence. Nat Cell Biol 11: 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino V, De Michele G, Venezia O, Migliaccio P, Dall’ Olio V, Bernard L, Minardi SP, Della Fazia MA, Bartoli D, Servillo G, et al. 2013. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell 12: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F, Brugiéres L, Vargas FR, Brentani RR, Ashton-Prolla P, et al. 2010. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: Evidence for a founder effect. Hum Mutat 31: 143–150. [DOI] [PubMed] [Google Scholar]
- Gong L, Gong H, Pan X, Chang C, Ou Z, Ye S, Yin L, Yang L, Tao T, Zhang Z, et al. 2015. p53 isoform Δ113p53/Δ133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell Res 25: 351–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R, Ray PS, Das S. 2008. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 7: 2189–2198. [DOI] [PubMed] [Google Scholar]
- Grover R, Sharathchandra A, Ponnuswamy A, Khan D, Das S. 2011. Effect of mutations on the p53 IRES RNA structure: Implications for de-regulation of the synthesis of p53 isoforms. RNA Biol 8: 132–142. [DOI] [PubMed] [Google Scholar]
- Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, Senturk E, Gillespie V, Aaronson SA, Merad M, et al. 2013. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev 27: 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinault C, Kawamori D, Liew CW, Maier B, Hu J, Keller SR, Mirmira RG, Scrable H, Kulkarni RN. 2011. Δ40 isoform of p53 controls β-cell proliferation and glucose homeostasis in mice. Diabetes 60: 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter G, Berger A, Fiegl H, Slade N, Zorić A, Holzer B, Schuster E, Mobus VJ, Reimer D, Daxenbichler G, et al. 2010. Alternative splicing of p53 and p73: The novel p53 splice variant p53δ is an independent prognostic marker in ovarian cancer. Oncogene 29: 1997–2004. [DOI] [PubMed] [Google Scholar]
- Hofstetter G, Berger A, Schuster E, Wolf A, Hager G, Vergote I, Cadron I, Sehouli J, Braicu EI, Mahner S, et al. 2011. Δ133p53 is an independent prognostic marker in p53 mutant advanced serous ovarian cancer. Br J Cancer 105: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter G, Berger A, Berger R, Zorić A, Braicu EI, Reimer D, Fiegl H, Marth C, Zeimet AG, Ulmer H, et al. 2012. The N-terminally truncated p53 isoform Δ40p53 influences prognosis in mucinous ovarian cancer. Int J Gynecol Cancer 22: 372–379. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Fujita K, Jenkins LMM, Hiyoshi Y, Mondal AM, Vojtesek B, Lane DP, Appella E, Harris CC. 2014. Autophagic degradation of the inhibitory p53 isoform Δ133p53α as a regulatory mechanism for p53-mediated senescence. Nat Commun 5: 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassim OW, Fink JL, Cagan RL. 2003. Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. EMBO J 22: 5622–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ. 2000. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci 97: 7301–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashio S, Obata F, Miura M. 2014. Interplay of cell proliferation and cell death in Drosophila tissue regeneration. Dev Growth Differ 56: 368–375. [DOI] [PubMed] [Google Scholar]
- Khan D, Sharathchandra A, Ponnuswamy A, Grover R, Das S. 2013. Effect of a natural mutation in the 5′ untranslated region on the translational control of p53 mRNA. Oncogene 32: 4148–4159. [DOI] [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. 2011. p53 isoforms: An intracellular microprocessor? Genes Cancer 2: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MP, Marcel V, Fernandes K, Diot A, Lane DP, Bourdon JC. 2013. Detecting and quantifying p53 isoforms at mRNA level in cell lines and tissues. Methods Mol Biol 962: 1–14. [DOI] [PubMed] [Google Scholar]
- Lane D, Levine A. 2010. p53 research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2: a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Brown C, Madhumalar A, Ghadessy FJ, Verma C. 2010. Mdm2 and p53 are highly conserved from placozoans to man. Cell Cycle 9: 540–547. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. 2009. p53 ancestry: Gazing through an evolutionary lens. Nat Rev Cancer 9: 758–762. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Méndez-Vidal C, Wiman KG, Farnebo M. 2009. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell 33: 462–471. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. 2004. Modulation of mammalian life span by the short isoform of p53. Genes Dev 18: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabese M, Marchini S, Marrazzo E, Mariani P, Cattaneo D, Fossati R, Compagnoni A, Signorelli M, Moll UM, Codegoni AM, et al. 2008. Expression levels of p53 and p73 isoforms in stage I and stage III ovarian cancer. Eur J Cancer 44: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Perrier S, Aoubala M, Ageorges S, Groves MJ, Diot A, Fernandes K, Tauro S, Bourdon JC. 2010a. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett 584: 4463–4468. [DOI] [PubMed] [Google Scholar]
- Marcel V, Vijayakumar V, Fernández-Cuesta L, Hafsi H, Sagne C, Hautefeuille A, Olivier M, Hainaut P. 2010b. p53 regulates the transcription of its Δ133p53 isoform through specific response elements contained within the TP53 P2 internal promoter. Oncogene 29: 2691–2700. [DOI] [PubMed] [Google Scholar]
- Marcel V, Tran PLT, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou MP, Hall J, Mergny JL, Hainaut P, Van Dyck E. 2011. G-quadruplex structures in TP53 intron 3: Role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 32: 271–278. [DOI] [PubMed] [Google Scholar]
- Marcel V, Petit I, Murray-Zmijewski F, Goullet de Rugy T, Fernandes K, Meuray V, Diot A, Lane DP, Aberdam D, Bourdon JC. 2012. Diverse p63 and p73 isoforms regulate Δ133p53 expression through modulation of the internal TP53 promoter activity. Cell Death Differ 19: 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Khoury MP, Fernandes K, Diot A, Lane DP, Bourdon JC. 2013. Detecting p53 isoforms at protein level. Methods Mol Biol 962: 15–29. [DOI] [PubMed] [Google Scholar]
- Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. 2014. Modulation of p53β and p53γ expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ 21: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín FA, Peréz-Garijo A, Morata G. 2009. Apoptosis in Drosophila: Compensatory proliferation and undead cells. Int J Dev Biol 53: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Matlashewski G, Lamb P, Pim D, Peacock J, Crawford L, Benchimol S. 1984. Isolation and characterization of a human p53 cDNA clone: Expression of the human p53 gene. EMBO J 3: 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. 2009. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging 30: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. 2009. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol 1: a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes CS, Levet C, Chatelain G, Dourlen P, Fouillet A, Dichtel-Danjoy ML, Gambis A, Ryoo HD, Steller H, Mollereau B. 2009. ER stress protects from retinal degeneration. EMBO J 28: 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita D, Dekanty A, Milán M. 2010. A dp53-dependent mechanism involved in coordinating tissue growth in Drosophila. PLoS Biol 8: e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Ma D. 2014. The p53 control of apoptosis and proliferation: Lessons from Drosophila. Apoptosis 19: 1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal AM, Horikawa I, Pine SR, Fujita K, Morgan KM, Vera E, Mazur SJ, Appella E, Vojtesek B, Blasco MA, et al. 2013. p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes. J Clin Invest 123: 5247–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HC, Jordan LB, Bray SE, Baker L, Quinlan PR, Purdie CA, Thompson AM, Bourdon JC, Fuller-Pace FV. 2010. The RNA helicase p68 modulates expression and function of the Δ133 isoform(s) of p53, and is inversely associated with Δ133p53 expression in breast cancer. Oncogene 29: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Shlevkov E, Pérez-Garijo A. 2011. Mitogenic signaling from apoptotic cells in Drosophila. Dev Growth Differ 53: 168–176. [DOI] [PubMed] [Google Scholar]
- Nutthasirikul N, Limpaiboon T, Leelayuwat C, Patrakitkomjorn S, Jearanaikoon P. 2013. Ratio disruption of the Δ133p53 and TAp53 isoform equilibrium correlates with poor clinical outcome in intrahepatic cholangiocarcinoma. Int J Oncol 42: 1181–1188. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 119: 847–860. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91–101. [DOI] [PubMed] [Google Scholar]
- Ou Z, Yin L, Chang C, Peng J, Chen J. 2014. Protein interaction between p53 and Δ113p53 is required for the anti-apoptotic function of Δ113p53. J Genet Genomics 41: 53–62. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Chambers KA, Pabo CO. 1993. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev 7: 2556–2564. [DOI] [PubMed] [Google Scholar]
- Pehar M, O’Riordan KJ, Burns-Cusato M, Andrzejewski ME, del Alcazar CG, Burger C, Scrable H, Puglielli L. 2010. Altered longevity-assurance activity of p53:p44 in the mouse causes memory loss, neurodegeneration and premature death. Aging Cell 9: 174–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehar M, Ko MH, Li M, Scrable H, Puglielli L. 2014. P44, the “longevity-assurance” isoform of P53, regulates tau phosphorylation and is activated in an age-dependent fashion. Aging Cell 13: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. 2002. reaper is required for neuroblast apoptosis during Drosophila development. Development 129: 1467–1476. [DOI] [PubMed] [Google Scholar]
- Pospísilová S, Brázda V, Amrichová J, Kamermeierová R, Palecek E, Vojtesek B. 2000. Precise characterisation of monoclonal antibodies to the C-terminal region of p53 protein using the PEPSCAN ELISA technique and a new non-radioactive gel shift assay. J Immunol Methods 237: 51–64. [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon IJL, Gabizon R, Mattia M, Zupnick A, et al. 2010. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol 17: 982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S. 2006. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 7: 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Bálint É, Loging WT, Rotter V, Almon E. 1996. A novel transcript encoded within the 10-kb first intron of the human p53 tumor suppressor gene (D17S2179E) is induced during differentiation of myeloid leukemia cells. Genomics 38: 364–370. [DOI] [PubMed] [Google Scholar]
- Sawhney S, Hood K, Shaw A, Braithwaite AW, Stubbs R, Hung NA, Royds JA, Slatter TL. 2015. α-Enolase is upregulated on the cell surface and responds to plasminogen activation in mice expressing a Δ133p53α mimic. PLoS ONE 10: e0116270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturk S, Yao Z, Camiolo M, Stiles B, Rathod T, Walsh AM, Nemajerova A, Lazzara MJ, Altorki NK, Krainer A, et al. 2014. p53Ψ is a transcriptionally inactive p53 isoform able to reprogram cells toward a metastatic-like state. Proc Natl Acad Sci 111: E3287–E3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharathchandra A, Lal R, Khan D, Das S. 2012. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol 9: 1429–1439. [DOI] [PubMed] [Google Scholar]
- Shi H, Tao T, Huang D, Ou Z, Chen J, Peng J. 2015. A naturally occurring 4-bp deletion in the intron 4 of p53 creates a spectrum of novel p53 isoforms with anti-apoptosis function. Nucleic Acids Res 43: 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silden E, Hjelle SM, Wergeland L, Sulen A, Andresen V, Bourdon JC, Micklem DR, McCormack E, Gjertsen BT. 2013. Expression of TP53 isoforms p53β or p53γ enhances chemosensitivity in TP53(null) cell lines. PLoS ONE 8: e56276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonova I, Lejour V, Bardot B, Bouarich-Bourimi R, Morin A, Fang M, Charbonnier L, Toledo F. 2012. Fuzzy tandem repeats containing p53 response elements may define species-specific p53 target genes. PLoS Genet 8: e1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonova I, Jaber S, Draskovic I, Bardot B, Fang M, Bouarich-Bourimi R, Lejour V, Charbonnier L, Soudais C, Bourdon JC, et al. 2013. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Rep 3: 2046–2058. [DOI] [PubMed] [Google Scholar]
- Simón R, Aparicio R, Housden BE, Bray S, Busturia A. 2014. Drosophila p53 controls Notch expression and balances apoptosis and proliferation. Apoptosis 19: 1430–1443. [DOI] [PubMed] [Google Scholar]
- Slatter TL, Hung N, Campbell H, Rubio C, Mehta R, Renshaw P, Williams G, Wilson M, Engelmann A, Jeffs A, et al. 2011. Hyperproliferation, cancer, and inflammation in mice expressing a Δ133p53-like isoform. Blood 117: 5166–5177. [DOI] [PubMed] [Google Scholar]
- Slatter TL, Hung N, Bowie S, Campbell H, Rubio C, Speidel D, Wilson M, Baird M, Royds JA, Braithwaite AW. 2015. Δ122p53, a mouse model of Δ133p53α, enhances the tumor-suppressor activities of an attenuated p53 mutant. Cell Death Dis 6: e1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon H, Sharon M, Rotter V. 2014. Modulation of alternative splicing contributes to cancer development: Focusing on p53 isoforms, p53β and p53γ. Cell Death Differ 21: 1347–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Huo S, Lü J, Liu Z, Fang X, Jin X, Yuan M. 2009. Expression of p53 isoforms in renal cell carcinoma. Chin Med J 122: 921–926. [PubMed] [Google Scholar]
- Stieper BC, Kupershtok M, Driscoll MV, Shingleton AW. 2008. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev Biol 321: 18–26. [DOI] [PubMed] [Google Scholar]
- Storer NY, Zon LI. 2010. Zebrafish models of p53 functions. Cold Spring Harb Perspect Biol 2: a001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Giannini C, Sarkaria JN, Schroeder M, Rogers J, Mastroeni D, Scrable H. 2013. p53 isoform profiling in glioblastoma and injured brain. Oncogene 32: 3165–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Markovic SN, Scrable HJ. 2014. Dominant effects of Δ40p53 on p53 function and melanoma cell fate. J Invest Dermatol 134: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM, Stauffer JK, Zheng ZM, Harris CC. 2013. Downregulation of splicing factor SRSF3 induces p53β, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 32: 2792–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier O, Josset L, Textoris J, Marcel V, Cartet G, Ferraris O, N’guyen C, Lina B, Diaz JJ, Bourdon JC, et al. 2011. Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway. Virol J 8: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier O, Marcel V, Cartet G, Lane DP, Lina B, Rosa-Calatrava M, Bourdon JC. 2012. Influenza A viruses control expression of proviral human p53 isoforms p53β and Δ133p53α. J Virol 86: 8452–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier O, Diederichs A, Dubois J, Cartet G, Lina B, Bourdon JC, Rosa-Calatrava M. 2013. Influenza NS1 interacts with p53 and alters its binding to p53-responsive genes, in a promoter-dependent manner. FEBS Lett 587: 2965–2971. [DOI] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. 2010. Δ40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev 24: 2408–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. 2006. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell 23: 251–263. [DOI] [PubMed] [Google Scholar]
- Waskar M, Landis GN, Shen J, Curtis C, Tozer K, Abdueva D, Skvortsov D, Tavaré S, Tower J. 2009. Drosophila melanogaster p53 has developmental stage-specific and sex-specific effects on adult life span indicative of sexual antagonistic pleiotropy. Aging 1: 903–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Noto J, Zaika E, Romero-Gallo J, Correa P, El-Rifai W, Peek RM, Zaika A. 2012. Pathogenic bacterium Helicobacter pylori alters the expression profile of p53 protein isoforms and p53 response to cellular stresses. Proc Natl Acad Sci 109: E2543–E2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, Kimchi A. 2014. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 33: 611–618. [DOI] [PubMed] [Google Scholar]
- Wells BS, Johnston LA. 2012. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol 361: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Yoshida E, Johnston LA. 2006. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 16: 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Harris N, Goldfinger N, Rotter V. 1985. Isolation of a full-length mouse cDNA clone coding for an immunologically distinct p53 molecule. Mol Cell Biol 5: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Lee L, Miner Z, Kulesz-Martin M. 1994. Wild-type alternatively spliced p53: Binding to DNA and interaction with the major p53 protein in vitro and in cells. EMBO J 13: 4823–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Zhang Z, Chu H, Xu M, Xue Y, Zhu H, Zhang Z. 2013. Intron 3 sixteen base pairs duplication polymorphism of p53 contributes to breast cancer susceptibility: Evidence from meta-analysis. PLoS ONE 8: e61662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wang S, Feng X. 2009. Influence of magnesium ion on the binding of p53 DNA-binding domain to DNA-response elements. J Biochem (Tokyo) 146: 77–85. [DOI] [PubMed] [Google Scholar]
- Yin Y, Stephen CW, Luciani MG, Fåhraeus R. 2002. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol 4: 462–467. [DOI] [PubMed] [Google Scholar]