Abstract
Amyloid-β (Aβ) peptides aggregate to form polymorphic amyloid fibrils and a variety of intermediate assemblies, including oligomers and protofibrils, both in vitro and in human brain tissue. Since the beginning of the 21st century, considerable progress has been made to characterize the molecular structures of Aβ aggregates. Full molecular structural models based primarily on data from measurements using solid-state nuclear magnetic resonance (ssNMR) have been developed for several in vitro Aβ fibrils and one metastable protofibril. Partial structural characterization of other aggregation intermediates has been achieved. One full structural model for fibrils derived from brain tissue has also been reported. Future work is likely to focus on additional structures from brain tissue and on further clarification of nonfibrillar Aβ aggregates.
Amyloid-β peptides can assemble into a variety of aggregates (e.g., fibrils), but structural studies are challenging because of their biochemical characteristics. Recently, progress has been made using solid-state NMR.
Aggregation of amyloid-β (Aβ) peptides in brain tissue is widely believed to be the root cause of neurodegeneration in Alzheimer’s disease (AD) (Hardy 2006). Aβ peptides are produced by a series of enzymatic cleavages of the amyloid precursor protein (APP), a transmembrane protein that is expressed primarily in neural tissue (Arai et al. 1991; Haass and Selkoe 1993). Most Aβ peptides in human brain tissue are either 40 or 42 residues in length (Aβ40 and Aβ42), and are typically produced in an approximate 5:1 abundance ratio but with more pronounced aggregation of Aβ42 (Gravina et al. 1995). Aβ aggregation is traditionally considered to be an extracellular process, although there is also evidence that Aβ aggregation can occur intracellularly (Pensalfini et al. 2014).
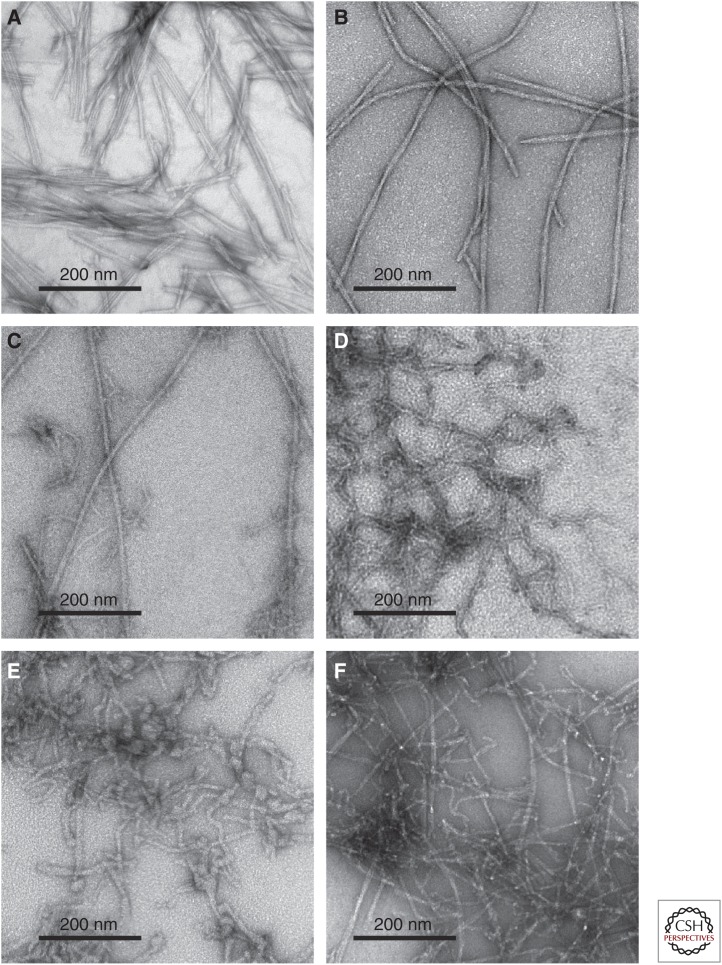
Multiple aggregated forms of Aβ have been identified (Tycko 2014). Aβ peptides have nonzero solubilities in aqueous solutions, with the precise concentration of unaggregated molecules at thermodynamic equilibrium being dependent on the peptide length (e.g., Aβ40 vs. Aβ42), temperature, pH, and other conditions. Typical values are well below 100 nm at 37°C and pH 7 (Brännström et al. 2013; Qiang et al. 2013). At Aβ concentrations above this solubility limit, the predominant aggregated structural state at thermodynamic equilibrium is the amyloid fibril (i.e., a state in which many Aβ monomers self-assemble to form straight, unbranched filaments that are typically 5–10 nm in width and 0.1–10 μm in length, and that contain “cross-β” supramolecular structural motifs [see below]). Transmission electron microscope (TEM) images of amyloid fibrils are shown in Figure 1A–C. Aβ fibrils are the main proteinaceous components of the Aβ plaques that develop in AD brain tissue and also account for most of the total mass of Aβ in brain tissue (Kuo et al. 1996; Lesne et al. 2013).
Figure 1.
Negatively stained transmission electron microscope (TEM) images of amyloid-β (Aβ) aggregates. (A) Aβ40 fibrils prepared in vitro with “striated ribbon” morphologies. (B) Aβ40 fibrils prepared in vitro with “twisted” morphologies. (C) Aβ40 fibrils derived from Alzheimer’s disease (AD) brain tissue and prepared by seeding synthetic Aβ40 with amyloid-enriched brain extract. (D) Metastable D23N–Aβ40 protofibrils prepared in vitro. (E) Aβ40 aggregates observed before the appearance of mature fibrils in vitro. A variety of morphologies are seen, including worm-like protofibrils and globular oligomers. (F) Polymorphic Aβ42 aggregates, including both fibrils and protofibrils.
In addition to amyloid fibrils, Aβ peptides can also self-assemble into a variety of oligomeric (Lambert et al. 1998; Walsh et al. 2002; Kayed et al. 2003; Lesne et al. 2006; Glabe 2008; Tomic et al. 2009; Yu et al. 2009; Sandberg et al. 2010; Ladiwala et al. 2012; Lesne et al. 2013; Liu et al. 2015) and “protofibrillar” (Goldsbury et al. 2000, 2005; Williams et al. 2005; Wu et al. 2010; Lasagna-Reeves et al. 2011) states (see below), as shown in Figure 1D–F. In experiments performed in vitro to characterize the Aβ aggregation process, these oligomers and protofibrils are generally transient or metastable species, which ultimately convert to fibrils as thermodynamic equilibrium is approached. At equilibrium, nonfibrillar aggregated species have small (possibly nearly zero) populations. However, the approach to true thermodynamic equilibrium can be very slow depending on experimental conditions, so that nonfibrillar species can remain abundant for days, weeks, or even longer periods.
Considerable uncertainty currently exists regarding the identities of aggregated Aβ species that are the most significant neurotoxic agents in AD. This uncertainty arises in part from uncertainty regarding primary neurotoxicity mechanisms and also from uncertainty regarding the precise identities of Aβ aggregates that actually develop in AD brain tissue (see below). It seems possible that multiple distinct forms of aggregated Aβ contribute to neurodegeneration in AD through multiple distinct mechanisms (Deshpande et al. 2006).
The remainder of this review focuses on the molecular structures of Aβ aggregates. Detailed molecular structural information is important for several reasons: (1) as a prerequisite for a fundamental understanding of the molecular and intermolecular interactions that drive aggregation and fibril formation; (2) as a prerequisite for a fundamental understanding of relationships among the various types of Aβ aggregates and the aggregation “pathway”; (3) as a basis for the development of chemical compounds that bind selectively to Aβ aggregates for purposes of diagnostic imaging; (4) as a basis for the development of compounds that interact with Aβ aggregates, to inhibit or retard aggregation, or possibly to guide the aggregation process toward relatively nontoxic species; and (5) as a basis for understanding interactions of Aβ aggregates with metal ions, membrane surfaces, cell-surface receptors, or other entities that may play roles in neurotoxicity mechanisms.
Detailed structural characterization of Aβ aggregates has not been straightforward because they are inherently noncrystalline and insoluble and are, therefore, not amenable to direct studies by traditional methods of X-ray crystallography and multidimensional liquid-state nuclear magnetic resonance (NMR). In addition, macroscopic samples of Aβ aggregates tend to be structurally heterogeneous, and nonfibrillar species are transient or have low abundances. Nonetheless, considerable progress has been made through the application of novel solid-state NMR (ssNMR) methods (Griffiths et al. 1995; Lansbury et al. 1995; Benzinger et al. 1998; Antzutkin et al. 2000, 2002, 2003; Balbach et al. 2000, 2002; Petkova et al. 2002, 2004, 2005, 2006; Jaroniec et al. 2004; Paravastu et al. 2008; Van Melckebeke et al. 2010; Bayro et al. 2011; Qiang et al. 2012; Lu et al. 2013; Schütz et al. 2014), electron microscopy (Jiménez et al. 1999; Goldsbury et al. 2000, 2005; Meinhardt et al. 2009), hydrogen/deuterium exchange (Lührs et al. 2005; Kheterpal and Wetzel 2006; Olofsson et al. 2007), electron paramagnetic resonance (EPR) (Török et al. 2002), X-ray fiber diffraction (McDonald et al. 2012), X-ray crystallography of amyloid-like peptide microcrystals (Nelson et al. 2005; Sawaya et al. 2007; Laganowsky et al. 2012), and other biophysical and biochemical techniques (Kheterpal et al. 2001; Kodali et al. 2010; Klinger et al. 2014). For Aβ fibrils, complete structural models that are based primarily on ssNMR data now exist (Petkova et al. 2006; Paravastu et al. 2008; Bertini et al. 2011; Schütz et al. 2014; Sgourakis et al. 2015; Xiao et al. 2015), including one complete model for Aβ40 fibrils derived from human brain tissue (Lu et al. 2013). A detailed model of a metastable Aβ protofibril has also been developed from ssNMR data (Qiang et al. 2012). Complete models for oligomeric Aβ assemblies have not yet been developed, but a substantial body of experimental data has been obtained for samples with a variety of morphologies, prepared under a variety of in vitro conditions (Chimon and Ishii 2005; Chimon et al. 2007; Ahmed et al. 2010; Scheidt et al. 2011, 2012; Lopez del Amo et al. 2012; Tay et al. 2013; Lendel et al. 2014; Sarkar et al. 2014; Huang et al. 2015; Parthasarathy et al. 2015; Potapov et al. 2015).
EARLY STRUCTURAL STUDIES
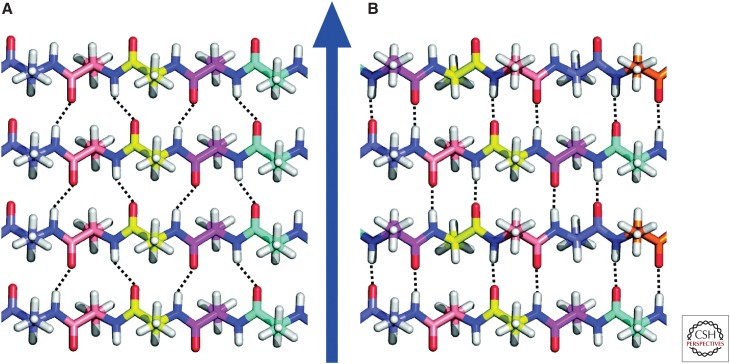
X-ray fiber diffraction data first showed that amyloid fibrils contain cross-β structural motifs (Eanes and Glenner 1968). A cross-β motif is one in which certain segments of the peptide or protein sequence adopt extended, β-strand conformations and assemble into ribbon-like β-sheets (Astbury et al. 1959), with the β-strands running approximately perpendicular to the fibril growth axis and with hydrogen bonds between β-strands running approximately parallel to the growth axis, as shown in Figure 2. The spacing between β-strands within a β-sheet is necessarily d = 0.47–0.48 nm, as dictated by the fixed length of the interstrand hydrogen bonds. This well-defined spacing leads to a characteristic, sharp maximum in X-ray scattering intensity at the angle θ = sin−1(λ/2d), where λ is the X-ray wavelength, directed along the fibril growth axis (i.e., in the meridional direction) in fiber diffraction measurements on aligned fibrils. In addition, the diffraction data typically show a broader maximum in scattering intensity perpendicular to the growth axis (i.e., in the equatorial direction) at a scattering angle that corresponds to a 0.8–1.0 nm spacing. This equatorial feature is commonly attributed to the existence of multiple β-sheet layers within the fibrils.
Figure 2.
Amyloid fibrils contain β-sheets that are organized into cross-β motifs. (A) Idealized representation of an in-register parallel cross-β structure, which is formed by identical β-strand segments from different peptide molecules with intermolecular hydrogen bonds (dotted lines) between residue k in one β-strand and residues k−1 and k+1 in an adjacent β-strand. (B) Idealized representation of an antiparallel cross-β structure. The blue arrow represents the fibril growth direction.
As depicted in Figures 2 and 3, a variety of cross-β motifs are conceivable, comprised of either parallel β-sheets or antiparallel β-sheets, with each peptide or protein molecule contributing one or more β-strands to each β-sheet and with one or more β-sheet layers. From fiber-diffraction data alone, it has not been possible to distinguish parallel β-sheets from antiparallel β-sheets, to determine the identities of β-strand-forming segments, or to determine the number of β-sheet layers within a fibril. Nonetheless, the fact that amyloid fibrils contain cross-β structures is a fundamental property that greatly simplifies the development of structural models from ssNMR data (see below).
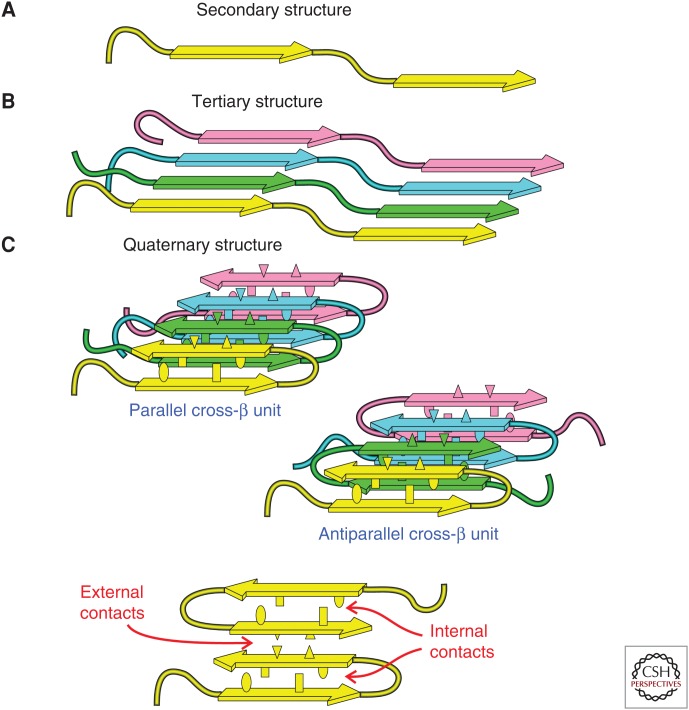
Figure 3.
Levels of molecular structure within an amyloid fibril. (A) Secondary structure refers to the identities of β-strand and non-β-strand segments (arrows and tubes). Non-β-strand segments can be either ordered or disordered. (B) Tertiary structure refers to the organization of β-strand segments into parallel or antiparallel β-sheets. (C) Quaternary structure refers to the stacking or interactions among β-sheets, which is mediated by amino acid side chains. A set of molecules that are hydrogen-bonded to one another in β-sheets constitutes one cross-β unit. A fibril structure may contain multiple cross-β units. Internal quaternary contacts involve interactions among side chains within one cross-β unit (ellipses and rectangles). External quaternary contacts involve interactions among side chains from different cross-β units (triangles).
Methods of ssNMR were first applied to amyloid fibrils by Griffin, Lansbury, and colleagues, who studied fibrils formed by peptides representing residues 34–42 of Aβ (Aβ34–42) and residues 20–29 of the islet amyloid polypeptide (Griffiths et al. 1995; Lansbury et al. 1995). Experiments on Aβ34–42 fibrils led to a model in which the cross-β motif in these fibrils was comprised of antiparallel β-sheets. These experiments involved preparation of Aβ34–42 peptides with 13C labels at specific backbone carbonyl and α-carbon sites, and measurements of nuclear magnetic dipole–dipole couplings between the carbonyl and α-carbon labels using the “rotational resonance” (R2) technique (Raleigh et al. 1988). The strength of magnetic dipole–dipole couplings is inversely proportional to the inverse cube of distances between 13C labels, allowing R2 data to serve as a strong restraint on molecular structure. The conclusion that Aβ34–42 fibrils contain antiparallel β-sheets followed from the detection of dipole–dipole couplings that were consistent with intermolecular distances <0.6 nm between carbonyl and α-carbon labels for certain nonsequential residue pairs (Lansbury et al. 1995).
R2 is one example of a class of ssNMR techniques (called “dipolar recoupling” techniques) that are used for quantitative distance measurements in ssNMR studies of biological and nonbiological systems (Raleigh et al. 1988; Gullion and Schaefer 1989; Tycko and Dabbagh 1990; Gregory et al. 1995; Jaroniec et al. 2001; Tycko 2007). Dipolar recoupling techniques generally consist of trains of radio frequency (RF) pulses that are synchronized with rapid rotation of the sample about an axis inclined at the “magic angle,” θm = 54.7356°, to the external magnetic field of the NMR spectrometer. Magic angle spinning (MAS) is an essential component of most biomolecular ssNMR measurements because MAS produces a dramatic enhancement of spectral resolution and signal-detection sensitivity. Dipolar recoupling techniques restore dipole–dipole couplings that are otherwise averaged out by MAS, allowing internuclear distances to be measured while retaining the resolution and sensitivity advantages of MAS. (In the specific case of R2, no RF pulses are applied, but the MAS rate is adjusted to match the difference in NMR frequencies of the two 13C-labeled sites.)
Around 1998, several molecular structural models for fibrils formed by full-length Aβ peptides were proposed (Chaney et al. 1998; Lazo and Downing 1998; George and Howlett 1999; Li et al. 1999; Tjernberg et al. 1999). These models were all qualitatively different from one another, reflecting the fact that no atomic-level structural restraints for full-length Aβ fibrils were available from experimental measurements. All of these models involved antiparallel β-sheets, supported by observations from infrared spectroscopy (Hilbich et al. 1991) and the ssNMR data for Aβ34–42 fibrils. None of these models has subsequently been found to be correct.
In 1998, Benzinger et al. (1998) reported evidence from ssNMR for parallel β-sheets in fibrils formed by Aβ10–35 (i.e., residues 10–35 of full-length Aβ), a peptide that had been chosen as a model for full-length Aβ in earlier work (Lee et al. 1995). A series of Aβ10–35 samples with 13C labels at single carbonyl sites was prepared. Dipolar recoupling data then indicated intermolecular 13C–13C distances of ∼0.5 nm for all fibril samples. These data ruled out the possibility of any antiparallel β-sheet structure and were most consistent with an in-register parallel β-sheet structure (i.e., one in which identical β-strand segments from different molecules form β-sheets, with amino acid residue k of each molecule being hydrogen-bonded to residues k−1 and k+1 of neighboring molecules, as depicted in Fig. 2A).
Although the results of Benzinger et al. (1998) were initially controversial, subsequent ssNMR data (Antzutkin et al. 2000, 2002; Balbach et al. 2002) provided support for in-register parallel β-sheets in Aβ40, Aβ42, and Aβ10–35 fibrils. In-register parallel β-sheets are now known to be the most common type of cross-β structure in “mature” (i.e., thermodynamically stable) amyloid fibrils that are formed by polypeptide chains that are more than about 20 residues in length (Margittai and Langen 2008; Tycko and Wickner 2013). Antiparallel β-sheets have been found in fibrils formed by shorter peptides (Balbach et al. 2000; Petkova et al. 2004; Bu et al. 2007) and in metastable Aβ aggregates (see below).
DEVELOPMENT OF FULL STRUCTURAL MODELS FOR AMYLOID FIBRILS FROM ssNMR DATA: GENERAL CONSIDERATIONS
As shown schematically in Figure 3, development of a full molecular structural model for an amyloid fibril requires experimental restraints at several structural levels. First, one must obtain restraints on the conformation of individual molecules (i.e., “secondary structure,” in the usual terminology of biochemistry and structural biology). In particular, the segments of the amino acid sequence that are conformationally ordered must be distinguished from disordered segments, and β-strand segments must be distinguished from non-β-strand segments. In ssNMR spectra, ordered segments exhibit sharp, strong 13C and 15N NMR lines, whereas disordered segments exhibit broader or weaker lines (depending on the hydration level of the sample). Residues in β-strand segments exhibit characteristic patterns of 13C NMR chemical shifts, with backbone carbonyl and α-carbon lines being shifted to lower chemical shift values and β-carbon lines being shifted to higher chemical shift values relative to “random coil” chemical shifts. (Only glycine α-carbon lines do not show this behavior.) Identification of ordered/disordered segments and β-strand/non-β-strand segments is performed most efficiently by acquisition of two- or three-dimensional (2D or 3D) ssNMR spectra from fibril samples in which various combinations of residues (or all residues) are uniformly 15N, 13C-labeled, allowing the conformations at many sites to be evaluated simultaneously from measurements on a single sample. This strategy was first demonstrated on amyloid fibrils by Balbach et al. (2000). Additional conformational restraints can be obtained from other types of ssNMR measurements (Jaroniec et al. 2002; Antzutkin et al. 2003; Hu et al. 2012), which are especially useful for defining the conformations in non-β-strand segments and at glycine residues.
Next, one must determine whether the cross-β motif is comprised of parallel or antiparallel β-sheets, as well as the precise intermolecular alignment (i.e., the registry of interstrand hydrogen bonds) within the β-sheets. In the specific context of amyloid fibrils, this aspect of supramolecular organization can be termed “tertiary structure.” As discussed above, dipolar recoupling measurements on samples in which single sites are 13C-labeled can provide the necessary restraints on intermolecular distances (Benzinger et al. 1998; Balbach et al. 2002; Tycko 2007). Measurements of 15N–13C dipole–dipole couplings on selectively labeled samples are also valuable, especially for characterizing antiparallel β-sheets (Petkova et al. 2004; Qiang et al. 2012). When multiple residues are uniformly labeled, the intermolecular alignment within antiparallel β-sheets can also be determined from 2D 13C–13C ssNMR spectra obtained with “proton-mediated” transfers of spin polarization between 13C nuclei (Petkova et al. 2004; Qiang et al. 2012). Other ssNMR strategies for characterizing β-sheet structures have been demonstrated by other research groups (Van Melckebeke et al. 2010; Bayro et al. 2011; Schütz et al. 2014). EPR measurements on spin-labeled peptides and proteins have also produced important information about both the secondary and tertiary structure in fibrils formed by a variety of peptides and proteins (Török et al. 2002; Cobb et al. 2007; Margittai and Langen 2008).
When a polypeptide contains more than one β-strand segment, β-sheets formed by different segments can stack against one another. One must then identify the faces of each β-sheet that form intersheet contacts and the specific amino acid side chains that participate in intersheet interactions. In the specific context of amyloid fibrils, this aspect of supramolecular organization can be termed “quaternary structure.” Restraints on intersheet contacts can be obtained from 2D or 3D ssNMR spectra of fibril samples that contain uniformly labeled residues when these spectra are acquired under experimental conditions that lead to long-range inter-residue cross-peak signals (Petkova et al. 2006; Paravastu et al. 2008; Van Melckebeke et al. 2010; Qiang et al. 2012; Lu et al. 2013; Schütz et al. 2014). Such cross-peaks result from transfers of nuclear spin polarization between 13C, 15N, or 1H sites on side chains of residues that are brought together by intersheet interactions. The polarization transfers are driven by inter-residue dipole–dipole couplings and can be observed when, for example, 13C–13C distances between side chains are up to ∼0.8 nm.
The cross-β motif formed by one set of molecules that are hydrogen-bonded to one another in β-sheets can be called one “cross-β unit” (CBU). A single fibril can contain two or more CBUs, which are typically arranged in a symmetric manner about the fibril growth axis. The presence of more than one CBU within Aβ40 and Aβ42 fibrils is indicated by quantitative measurements of the fibril mass per length (MPL) using scanning TEM (STEM) (Goldsbury et al. 2000, 2005; Antzutkin et al. 2002), energy-filtered TEM (Paravastu et al. 2009), or tilted-beam, dark-field TEM (Chen et al. 2009). Given that full-length Aβ peptides have a molecular mass of 4.3 kDa and that the spacing between molecules in an in-register parallel β-sheet is 0.47 nm, the MPL of one CBU must be 4.3 kDa/0.47 nm = 9.0 kDa/nm. Experimental measurements on Aβ fibrils prepared under various conditions indicate minimum MPL values of ∼18 or 27 kDa/nm (Goldsbury et al. 2005; Petkova et al. 2005; Chen et al. 2009), consistent with two or three CBUs in the minimal fibril structure, which can be called the “protofilament.” Thicker fibrils with higher MPL values can form by self-association of protofilaments, but fibrils containing only one CBU apparently do not exist in isolation.
The existence of multiple CBUs within a protofilament complicates the interpretation of intersheet contacts discussed above because these contacts can be either within one CBU (“internal quaternary contacts”) or between different CBUs (“external quaternary contacts”). Assignment of individual long-range inter-residue cross-peaks (or similar data) to internal or external quaternary contacts can only be done reliably after multiple long-range cross-peaks have been observed, so that only one set of assignments is physically possible and internally consistent.
Once experimental restraints on secondary, tertiary, and quaternary structures (as defined above) have been obtained, one can use computational methods to find a final set of structures that are fully consistent with all restraints. The range of variations among these structures can be used as an indication of the precision of the final structural model. Although a larger number of restraints generally leads to greater precision, it should be recognized that accurate structural models for amyloid fibrils do not require a large number of restraints per amino acid residue, as may be the case for globular protein structures. Once the secondary and tertiary structures are determined, fewer than 10 unambiguous, long-range, inter-residue cross-peaks may be sufficient to define the quaternary structure uniquely and accurately (although perhaps not with high precision). This is because a cross-β structure requires that all β-strands in the structure be nearly perpendicular to the fibril growth axis and that all interstrand hydrogen bonds be nearly parallel to the growth axis. In addition, neighboring molecules within one CBU are related by approximate translational symmetry along the growth axis, and different CBUs are related by approximate rotational or screw symmetry about the growth axis. Symmetry restraints are often imposed to facilitate structural calculations (Lu et al. 2013; Schütz et al. 2014; Sgourakis et al. 2015).
On the other hand, certain aspects of amyloid fibril structures can be nearly impossible to determine from experimental restraints. In particular, when two β-sheets stack against one another within one CBU, it is well known that amino acid side chains in the interface tend to optimize their packing by interdigitation, leading to displacement or “staggering” of β-strands in one β-sheet relative to the other (Buchete et al. 2005; Petkova et al. 2006; Bedrood et al. 2012). The presence of staggering can be inferred from the effects of isotopic dilution on ssNMR measurements of side-chain–side-chain distances or contacts (Petkova et al. 2006; Paravastu et al. 2008). However, the direction of this displacement, which can be toward either end of the fibril, and the amplitude of displacement, which can be more than 0.47 nm, is not readily determined.
MOLECULAR STRUCTURES OF TWO DISTINCT, SELF-PROPAGATING Aβ40 FIBRIL POLYMORPHS FORMED IN VITRO
Petkova et al. (2002) used the strategy described above to develop a preliminary model for Aβ40 fibrils, with several characteristic features: (1) the Aβ40 conformation includes a disordered N-terminal segment (residues 1–9), two β-strand segments (residues 10–22 and 30–40), and an intervening bend or loop (residues 23–29); (2) the two β-strand segments form separate in-register parallel β-sheets, so that the CBU is a double-layered parallel β-sheet structure, with internal quaternary contacts involving favorable hydrophobic interactions among side chains of the two β-strands; (3) oppositely charged side chains of D23 and K28 form salt bridges within the core of the CBU; and (4) the protofilament consists of two CBUs, with external quaternary contacts involving hydrophobic residues of the C-terminal β-strand as well as side-chain interdigitation, as in “steric zippers” identified by Eisenberg and colleagues (Nelson et al. 2005; Sawaya et al. 2007).
This preliminary model was a significant advance over earlier modeling attempts in that it was consistent with a substantial set of experimental observations, including the existing data from ssNMR (Balbach et al. 2002; Petkova et al. 2002), electron microscopy (Antzutkin et al. 2002), EPR (Török et al. 2002), hydrogen/deuterium exchange (Kheterpal et al. 2000), and limited proteolysis (Kheterpal et al. 2001). Important features of this model have been retained in subsequent structural models, including the overall conformation of Aβ40 monomers, the identities of exposed and buried segments, the in-register parallel β-sheet organization, the presence of D23–K28 salt bridges, the existence of two CBUs within the protofilament, and the hydrophobic fibril core that results from these structural features.
However, it became clear that a single Aβ40 fibril structure could not explain all observations, especially that 13C NMR chemical shifts for individual sites varied from one fibril sample to another (Petkova et al. 2002), that MPL values of 18 kDa/nm and 27 kDa/nm were observed in different samples (Goldsbury et al. 2005), and that ssNMR data restraining the conformation in residues 23–29 could not be fit well by a single conformation (Antzutkin et al. 2003). Then, Petkova et al. (2005) showed that the molecular structure of Aβ40 fibrils is reproducibly dependent on the precise details of growth conditions. Depending on the presence or absence of gentle agitation of the Aβ40 solution during fibril growth, with all other conditions being equal, fibrils with two distinct morphologies in TEM images, distinct sets of NMR chemical shifts, and distinct MPL values could be prepared. Moreover, once a batch of “parent” fibrils with a particular structure was prepared de novo, subsequent samples of “daughter” and “granddaughter” fibrils, with the same structures as their parents, could be prepared by seeded fibril growth (Petkova et al. 2005). Thus, the molecular structure in Aβ fibrils is not determined uniquely by amino acid sequence.
Once the phenomenon of self-propagating molecular-level polymorphism in Aβ fibrils was established, protocols for preparing relatively homogeneous samples were developed (Petkova et al. 2006; Paravastu et al. 2008). Structural models for the two Aβ40 fibril polymorphs shown in Figure 1A and B were then developed from ssNMR data. As shown in Figure 4, the two polymorphs possess quite similar secondary structures, identical tertiary structures, similar internal quaternary contacts, and the same Aβ40 segments exposed on the protofilament surface. The most striking difference is in their symmetry. Fibrils grown with gentle agitation contain protofilaments comprised of two CBUs, with approximate twofold rotational symmetry about the fibril growth axis (Fig. 4A). These protofilaments tend to self-associate to form “striated ribbons” in negatively stained TEM images (Fig. 1A). Fibrils grown under quiescent conditions contain protofilaments with three CBUs and approximate threefold rotation symmetry (Fig. 4B). The fibrils are less prone to self-association, so that negatively stained TEM images typically show individual protofilaments, with a gradual twist along their length (Fig. 1B). The D23–K28 salt bridges discussed above are present in the twofold symmetric fibrils, but not in the threefold symmetric fibrils, as revealed by differences in 15N–13C dipole–dipole couplings between D23 and K28 side chains (Paravastu et al. 2008). This difference is one manifestation of significant conformational differences in residues 23–29.
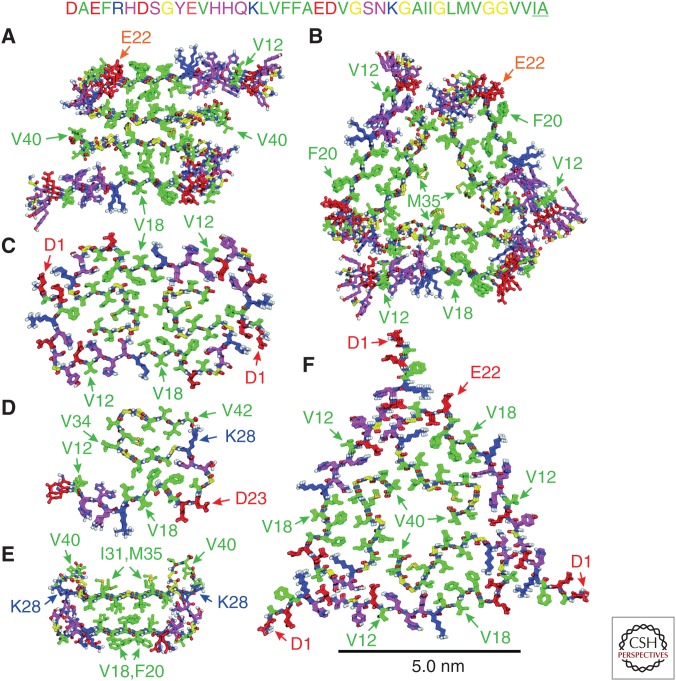
Figure 4.
Molecular structures of Aβ aggregates developed from ssNMR data. (A) Twofold symmetric Aβ40 fibril with the morphology in Figure 1A from Protein Data Bank (PDB) file 2LMN. (B) Threefold symmetric Aβ40 fibril with the morphology in Figure 1B from PDB file 2LMP. (C) E22Δ–Aβ40 fibril from PDB file 2MVX. (D) Aβ42 fibril from PDB file 2MXU. (E) D23N–Aβ40 protofibril from PDB file 2LNQ. (F) Brain-derived Aβ40 fibril with the morphology in Figure 1C from PDB file 2M4J. All models are viewed in cross section, with the fibril or protofibril growth axis perpendicular to the page. Three or four molecular repeats along the growth axis are shown in each case. Carbon atoms of hydrophobic, negatively charged, positively charged, and glycine residues are green, red, blue, and yellow, respectively. Carbon atoms of other residues are magenta. In C, D, and F (but not A, B, and E), conformational and translational symmetry restraints were imposed during structure calculations. The amino acid sequences of wild-type Aβ40 and Aβ42 (including the two underlined C-terminal residues) are shown at the top.
Atomic coordinates for twofold and threefold symmetric Aβ40 fibrils formed in vitro can be found in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB codes 2LMN and 2LMO for twofold symmetric fibrils; PDB codes 2LMP and 2LMQ for threefold symmetric fibrils). For each polymorph, the two sets of coordinates involve different directions of staggering. As discussed above, isotopic dilution experiments indicate that internal quaternary contacts in both polymorphs are staggered, but the direction cannot be determined from existing data.
OTHER STRUCTURAL MODELS FOR IN VITRO Aβ FIBRILS DEVELOPED FROM ssNMR DATA
An additional twofold symmetric structural model for Aβ40 fibrils was subsequently developed from ssNMR data by Bertini et al. (2011). The details of internal and external quaternary contacts are different in this model compared with the model in Figure 4A, although the identities of amino acid side chains that are involved in these contacts are the same. Residues 1–9 are not fully disordered in fibrils studied by Bertini et al. (2011). Significant differences in growth conditions and the presence of a nonnative methionine residue at the N terminus in Aβ40 samples prepared by Bertini et al. (2011) most likely account for the structural differences.
A threefold symmetric structural model for fibrils formed by the Asp23Asn “Iowa” mutant of Aβ40 (D23N–Aβ40) (Grabowski et al. 2001) has been reported by Sgourakis et al. (2015). This model is very similar to the model for wild-type Aβ40 fibrils in Figure 4B. Differences include the presence of external quaternary contacts between V40 and K28 in the D23N–Aβ40 fibrils and conformational differences in residues 23–29.
Figure 4C shows a twofold symmetric structural model for fibrils formed by the Glu22-deletion “Osaka” mutant of Aβ40 (E22Δ–Aβ40) (Tomiyama et al. 2008), which was developed by Schütz et al. (2014). The entire E22Δ–Aβ40 sequence was found to be structurally ordered, as indicated by the detection of sharp, strong NMR signals from all residues. Significant differences in secondary and quaternary structures were found, compared with the twofold symmetric, wild-type Aβ40 structure in Figure 4A, and D23–K28 salt bridges were found to be absent. The in-register parallel β-sheet structure formed by residues 30–40 is divided into three separate sections by turn-like conformations at glycine residues 33 and 37.
Figure 4D shows a partial structural model for fibrils formed in vitro by Aβ42, developed by Xiao et al. (2015). In this case, residues 12–15, 17–19, 26–28, 30–33, and 38–40 form separate parallel β-sheets, with salt bridges between K28 and A42. This is a partial model because only one CBU is included, although MPL data for Aβ42 fibrils indicate two or more CBUs (Antzutkin et al. 2002). The structural differences between Aβ42 and Aβ40 fibrils identified by Xiao et al. (2015) help explain the fact that Aβ42 fibril fragments do not act as efficient seeds for the growth of Aβ40 fibrils, at least for polymorphs that have been examined in vitro (Lu et al. 2013; Cukalevski et al. 2015). The absence of efficient cross-seeding suggests that Aβ40 fibrils and Aβ42 fibrils may develop and propagate independently of one another in brain tissue.
MOLECULAR STRUCTURES OF TRANSIENT AND METASTABLE Aβ AGGREGATES
In studies of Aβ aggregation in vitro, TEM and atomic-force microscope images commonly show a variety of aggregated states that are not true mature fibrils. These nonfibrillar states often appear before mature fibrils and may persist for hours or days but eventually disappear as self-assembly proceeds to its thermodynamic end point. Hence, they are considered to be intermediate stages in Aβ aggregation. Intermediates include Aβ oligomers with roughly spherical appearances and variable sizes and Aβ protofibrils, which are worm-like in appearance, with greater curvature and shorter lengths than mature fibrils. Nonfibrillar species are of great interest in the research community because of experimental evidence that they may be important neurotoxic species in AD (Lambert et al. 1998; Walsh et al. 2002; Kayed et al. 2003; Williams et al. 2005; Lesne et al. 2006, 2013; Glabe 2008; Tomic et al. 2009; Lasagna-Reeves et al. 2011; Liu et al. 2015).
A variety of oligomeric and protofibrillar states have been partially characterized by ssNMR (Ahmed et al. 2010; Scheidt et al. 2011, 2012; Lopez del Amo et al. 2012; Tay et al. 2013; Lendel et al. 2014; Sarkar et al. 2014; Huang et al. 2015; Parthasarathy et al. 2015; Potapov et al. 2015). The main finding, originally reported by Ishii and coworkers (Chimon and Ishii 2005; Chimon et al. 2007), is that the Aβ conformation in nonfibrillar intermediates is remarkably similar to the conformation in fibrils. Most data on supramolecular structure indicate that in-register parallel β-sheets are not present in nonfibrillar intermediates and that the predominant intermolecular alignment may be antiparallel (Tay et al. 2013; Huang et al. 2015; Potapov et al. 2015). Ishii and coworkers have reported dipolar recoupling data that support parallel intermolecular alignment in large, spherical Aβ40 intermediates (Chimon et al. 2007; Parthasarathy et al. 2015).
To date, only one detailed molecular structural model for an intermediate has been reported, namely, a model for protofibrils formed by D23N–Aβ40 (Qiang et al. 2012). As shown in Figure 4E, D23N–Aβ40 protofibrils contain a molecular conformation that is similar to the conformation in Aβ40 fibrils, with residues 16–22 and 30–34 forming β-strands that interact through hydrophobic side-chain–side-chain contacts. However, the supramolecular structure is qualitatively different, with an alternating, antiparallel alignment of D23–Aβ40 molecules that creates a double-layered antiparallel CBU. MPL data indicate that the protofibrils contain only one CBU. The model for D23N–Aβ40 protofibrils in Figure 4E (PDB code 2LNQ) is supported by NMR chemical shifts, long-range cross-peaks, intermolecular 13C–13C and 15N–13C dipole–dipole couplings, and multiple proton-mediated 2D 13C–13C spectra (Qiang et al. 2012).
D23N–Aβ40 protofibrils eventually convert to fibrils with threefold symmetry and in-register parallel β-sheets (Tycko et al. 2009; Qiang et al. 2011) but can persist for many weeks. Metastability of protofibrils can be attributed to the favorable hydrophobic interactions in the protofibril core and to their qualitatively different supramolecular structure. Spontaneous internal conversion of protofibrils to fibrils would require a dramatic rearrangement of all intermolecular hydrogen bonds and would be additionally hindered by numerous steric clashes. Conversion to fibrils can only occur through gradual shrinkage and dissolution of the metastable protofibrils and gradual elongation of the more thermodynamically stable fibrils, as discussed elsewhere (Qiang et al. 2013; Tycko 2014). Because direct, internal conversion of protofibrils to fibrils cannot occur, protofibrils are “off-pathway” intermediates in the Aβ self-assembly process. It appears likely that most intermediates that have been examined by in vitro experiments are also off-pathway, with the possible exception of the oligomers studied by Ishii and colleagues (Chimon et al. 2007; Parthasarathy et al. 2015).
AGGREGATED Aβ IN BRAIN TISSUE
In light of the molecular-level polymorphism of Aβ fibrils that has been established by in vitro studies, as well as the observation of multiple self-assembly intermediates, it is important to ask which Aβ structures actually develop in brain tissue. Direct structural characterization of aggregated Aβ in brain tissue by ssNMR is not possible because ssNMR measurements require milligram-scale quantities of isotopically labeled material. However, fibril structures in brain tissue can be amplified and labeled by seeded fibril growth, using amyloid-containing brain extract as the source of seeds and labeled Aβ (either synthetic or recombinant) as the source of monomers that add to the brain-derived seeds (Paravastu et al. 2009). Using this approach, Lu et al. (2013) investigated Aβ40 fibrils derived from brain tissue of two AD patients. Surprisingly, ssNMR spectra of brain-derived fibrils indicated highly homogenous structures, with the same predominant structures in different regions of the cerebral cortex from each patient. However, spectra of Aβ40 fibrils from patient 1 were dramatically different from spectra of Aβ40 fibrils from patient 2, indicating significant differences in secondary structure. Moreover, the two patients had different clinical histories, raising the possibility that differences in fibril structure may correlate with differences in disease development.
A complete structural model for fibrils derived from patient 1, shown in Figure 4F, was developed from a large set of ssNMR data (PDB code 2M4J) (Lu et al. 2013). These fibrils have threefold symmetry and share many aspects of secondary, tertiary, and quaternary structure with Aβ40 fibrils prepared in vitro. Significant differences from the in vitro threefold symmetric Aβ40 fibrils studied by Paravastu et al. (2008) (Fig. 4B) include the presence of D23–K28 salt bridges, structurally ordered N-terminal segments, and breaks in the C-terminal β-strand at glycine residues 33 and 37. Some of these features are also present in ssNMR-based structural models for fibrils formed in vitro by E22Δ–Aβ40 and Aβ42 (Figs. 4C,D). Thus, Aβ fibrils that develop in brain tissue and can be amplified with the protocols of Lu et al. (2013) are qualitatively similar to fibrils prepared in vitro but differ in specific details. Although a structural model for fibrils derived from patient 2 was not developed by Lu et al. (2013), ssNMR spectra and MPL data indicate threefold symmetry but with differences in internal quaternary contacts and in the conformation in residues 28–32.
Structural variations discussed above may provide a basis for the development of compounds for diagnostic imaging (Klunk et al. 2004; Wong et al. 2010) that distinguish among Aβ fibril polymorphs in living patients. If significant correlations between variations in fibril structure and variations in disease development (e.g., progression from mild cognitive impairment to AD) are established by future studies, such imaging compounds may be valuable in clinical practice. Similarly, if certain fibril polymorphs are shown to have higher pathogenicity, specific structural differences among polymorphs may provide a basis for the development of compounds that direct Aβ aggregation toward relatively innocuous structures.
CONCLUDING REMARKS
As described above, we now have a good understanding of molecular structures and structural variations within Aβ fibrils, which was largely absent 15 years ago. Although not all possible structures (including all disease-associated mutants, Aβ peptide lengths, and fibril polymorphs) have been characterized, it now seems most important to focus on those that develop in brain tissue. A critical question for which the current answer appears to be “possibly yes” (Heilbronner et al. 2013; Lu et al. 2013; Stöhr et al. 2014; Watts et al. 2014; Cohen et al. 2015) is whether structural variations are clinically relevant. Further experiments are needed using ssNMR measurements on brain-seeded fibrils as described above or perhaps using techniques that provide less detailed structural information but require less material (Bergh et al. 2015). In addition, further clarification of molecular structures within nonfibrillar Aβ aggregates is an important goal for future experiments. Because nonfibrillar aggregates are apparently not amenable to amplification and isotopic labeling by seeded growth, structural studies of nonfibrillar aggregates from brain tissue may depend on methods other than ssNMR.
ACKNOWLEDGMENTS
Research on Aβ structures in Dr. Tycko’s laboratory is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, a component of the National Institutes of Health.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. 2010. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat Struct Mol Biol 17: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. 2000. Multiple quantum solid state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci 97: 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. 2002. Supramolecular structural constraints on Alzheimer’s β-amyloid fibrils from electron microscopy and solid state nuclear magnetic resonance. Biochemistry 41: 15436–15450. [DOI] [PubMed] [Google Scholar]
- Antzutkin ON, Balbach JJ, Tycko R. 2003. Site-specific identification of non-β-strand conformations in Alzheimer’s β-amyloid fibrils by solid state NMR. Biophys J 84: 3326–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H, Lee VM, Messinger ML, Greenberg BD, Lowery DE, Trojanowski JQ. 1991. Expression patterns of β-amyloid precursor protein (β-APP) in neural and nonneural human tissues from Alzheimer’s-disease and control subjects. Ann Neurol 30: 686–693. [DOI] [PubMed] [Google Scholar]
- Astbury WT, Beighton E, Parker KD. 1959. The cross-β configuration in supercontracted proteins. Biochim Biophys Acta 35: 17–25. [DOI] [PubMed] [Google Scholar]
- Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. 2000. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer’s β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry 39: 13748–13759. [DOI] [PubMed] [Google Scholar]
- Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, Tycko R. 2002. Supramolecular structure in full-length Alzheimer’s β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid state nuclear magnetic resonance. Biophys J 83: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayro MJ, Debelouchina GT, Eddy MT, Birkett NR, MacPhee CE, Rosay M, Maas WE, Dobson CM, Griffin RG. 2011. Intermolecular structure determination of amyloid fibrils with magic-angle spinning and dynamic nuclear polarization NMR. J Am Chem Soc 133: 13967–13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrood S, Li YY, Isas JM, Hegde BG, Baxa U, Haworth IS, Langen R. 2012. Fibril structure of human islet amyloid polypeptide. J Biol Chem 287: 5235–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. 1998. Propagating structure of Alzheimer’s β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci 95: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh J, Zetterström P, Andersen PM, Brännström T, Graffmo KS, Jonsson PA, Lang LS, Danielsson J, Oliveberg M, Marklund SL. 2015. Structural and kinetic analysis of protein-aggregate strains in vivo using binary epitope mapping. Proc Natl Acad Sci 112: 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Gonnelli L, Luchinat C, Mao JF, Nesi A. 2011. A new structural model of Aβ40 fibrils. J Am Chem Soc 133: 16013–16022. [DOI] [PubMed] [Google Scholar]
- Brännström K, Öhman A, Lindhagen-Persson M, Olofsson A. 2013. Ca2+ enhances Aβ polymerization rate and fibrillar stability in a dynamic manner. Biochem J 450: 189–197. [DOI] [PubMed] [Google Scholar]
- Bu ZM, Shi Y, Callaway DJE, Tycko R. 2007. Molecular alignment within β-sheets in Aβ14–23 fibrils: Solid state NMR experiments and theoretical predictions. Biophys J 92: 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchete NV, Tycko R, Hummer G. 2005. Molecular dynamics simulations of Alzheimer’s β-amyloid protofilaments. J Mol Biol 353: 804–821. [DOI] [PubMed] [Google Scholar]
- Chaney MO, Webster SD, Kuo YM, Roher AE. 1998. Molecular modeling of the amyloid β 1–42 peptide from Alzheimer’s disease. Protein Eng 11: 761–767. [DOI] [PubMed] [Google Scholar]
- Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. 2009. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci 106: 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimon S, Ishii Y. 2005. Capturing intermediate structures of Alzheimer’s β-amyloid, Aβ(1–40), by solid state NMR spectroscopy. J Am Chem Soc 127: 13472–13473. [DOI] [PubMed] [Google Scholar]
- Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. 2007. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol 14: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. 2007. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci 104: 18946–18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Kim C, Haldiman T, Elhag M, Tatsuoka C, Zhang SL, Haines J, Lerner A, Safar J. 2015. Distinct strains of amyloid β prions implicated in rapidly progressive Alzheimer disease. Prion 9: S76–S77. [Google Scholar]
- Cukalevski R, Yang XT, Meisl G, Weininger U, Bernfur K, Frohm B, Knowles TPJ, Linse S. 2015. The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem Sci 6: 4215–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Mina E, Glabe C, Busciglio J. 2006. Different conformations of amyloid β induce neurotoxicity by distinct mechanisms in human cortical neurons. J Neurosci 26: 6011–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes ED, Glenner GG. 1968. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem 16: 673–677. [DOI] [PubMed] [Google Scholar]
- George AR, Howlett DR. 1999. Computationally derived structural models of the β-amyloid found in Alzheimer’s disease plaques and the interaction with possible aggregation inhibitors. Biopolymers 50: 733–741. [DOI] [PubMed] [Google Scholar]
- Glabe CG. 2008. Structural classification of toxic amyloid oligomers. J Biol Chem 283: 29639–29643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbury CS, Wirtz S, Müller SA, Sunderji S, Wicki P, Aebi U, Frey P. 2000. Studies on the in vitro assembly of Aβ 1–40: Implications for the search for Aβ fibril formation inhibitors. J Struct Biol 130: 217–231. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. 2005. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol 352: 282–298. [DOI] [PubMed] [Google Scholar]
- Grabowski TJ, Cho HS, Vonsattel JPG, Rebeck GW, Greenberg SM. 2001. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann Neurol 49: 697–705. [DOI] [PubMed] [Google Scholar]
- Gravina SA, Ho LB, Eckman CB, Long KE, Otvos L, Younkin LH, Suzuki N, Younkin SG. 1995. Amyloid-β protein (Aβ) in Alzheimer’s-disease brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Aβ-40 or Aβ-42(43). J Biol Chem 270: 7013–7016. [DOI] [PubMed] [Google Scholar]
- Gregory DM, Mitchell DJ, Stringer JA, Kiihne S, Shiels JC, Callahan J, Mehta MA, Drobny GP. 1995. Windowless dipolar recoupling: The detection of weak dipolar couplings between spin 1/2 nuclei with large chemical shift anisotropies. Chem Phys Lett 246: 654–663. [Google Scholar]
- Griffiths JM, Ashburn TT, Auger M, Costa PR, Griffin RG, Lansbury PT. 1995. Rotational resonance solid state NMR elucidates a structural model of pancreatic amyloid. J Am Chem Soc 117: 3539–3546. [Google Scholar]
- Gullion T, Schaefer J. 1989. Rotational-echo double-resonance NMR. J Magn Reson 81: 196–200. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. 1993. Cellular processing of β-amyloid precursor protein and the genesis of amyloid β-peptide. Cell 75: 1039–1042. [DOI] [PubMed] [Google Scholar]
- Hardy J. 2006. Has the amyloid cascade hypothesis for Alzheimer’s disease been proved? Curr Alzheimer Res 3: 71–73. [DOI] [PubMed] [Google Scholar]
- Heilbronner G, Eisele YS, Langer F, Kaeser SA, Novotny R, Nagarathinam A, Aslund A, Hammarström P, Nilsson KPR, Jucker M. 2013. Seeded strain-like transmission of β-amyloid morphotypes in APP transgenic mice. EMBO Rep 14: 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. 1991. Aggregation and secondary structure of synthetic amyloid βA4 peptides of Alzheimer’s disease. J Mol Biol 218: 149–163. [DOI] [PubMed] [Google Scholar]
- Hu KN, Qiang W, Bermejo GA, Schwieters CD, Tycko R. 2012. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N–15N and carbonyl 13C–13C dipolar recoupling data. J Magn Reson 218: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Zimmerman MI, Martin PK, Nix AJ, Rosenberry TL, Paravastu AK. 2015. Antiparallel β-sheet structure within the C-terminal region of 42-residue Alzheimer’s amyloid-β peptides when they form 150-kDa oligomers. J Mol Biol 427: 2319–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. 2001. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate 13C–15N distance measurements in uniformly 13C, 15N-labeled peptides. J Am Chem Soc 123: 3507–3519. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, Filip C, Griffin RG. 2002. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon–nitrogen distances in uniformly 13C, 15N-labeled solids. J Am Chem Soc 124: 10728–10742. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. 2004. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci 101: 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez JL, Guijarro JL, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. 1999. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. Embo J 18: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486–489. [DOI] [PubMed] [Google Scholar]
- Kheterpal I, Wetzel R. 2006. Hydrogen/deuterium exchange mass spectrometry—A window into amyloid structure. Acc Chem Res 39: 584–593. [DOI] [PubMed] [Google Scholar]
- Kheterpal I, Zhou S, Cook KD, Wetzel R. 2000. Aβ amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc Natl Acad Sci 97: 13597–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. 2001. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry 40: 11757–11767. [DOI] [PubMed] [Google Scholar]
- Klinger AL, Kiselar J, Ilchenko S, Komatsu H, Chance MR, Axelsen PH. 2014. A synchrotron-based hydroxyl radical footprinting analysis of amyloid fibrils and prefibrillar intermediates with residue-specific resolution. Biochemistry 53: 7724–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang YM, Blomqvist G, Holt DP, Bergstrom M, Savitcheva I, Huang GF, Estrada S, et al. 2004. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh compound-B. Ann Neurol 55: 306–319. [DOI] [PubMed] [Google Scholar]
- Kodali R, Williams AD, Chemuru S, Wetzel R. 2010. Aβ(1–40) forms five distinct amyloid structures whose β-sheet contents and fibril stabilities are correlated. J Mol Biol 401: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YM, Emmerling MR, Vigo-Pelfrey C, Kasunic TC, Kirkpatrick JB, Murdoch GH, Ball MJ, Roher AE. 1996. Water-soluble Aβ(N-40, N-42) oligomers in normal and Alzheimer disease brains. J Biol Chem 271: 4077–4081. [DOI] [PubMed] [Google Scholar]
- Ladiwala ARA, Litt J, Kane RS, Aucoin DS, Smith SO, Ranjan S, Davis J, Van Nostrand WE, Tessier PM. 2012. Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity. J Biol Chem 287: 24765–24773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao ML, Pensalfini A, Soriaga AB, Landau M, Teng PK, et al. 2012. Atomic view of a toxic amyloid small oligomer. Science 335: 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, et al. 1998. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci 95: 6448–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RGS, et al. 1995. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol 2: 990–998. [DOI] [PubMed] [Google Scholar]
- Lasagna-Reeves CA, Glabe CG, Kayed R. 2011. Amyloid-β annular protofibrils evade fibrillar fate in Alzheimer disease brain. J Biol Chem 286: 22122–22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo ND, Downing DT. 1998. Amyloid fibrils may be assembled from β-helical protofibrils. Biochemistry 37: 1731–1735. [DOI] [PubMed] [Google Scholar]
- Lee JP, Stimson ER, Ghilardi JR, Mantyh PW, Lu YA, Felix AM, Llanos W, Behbin A, Cummings M, Van Criekinge M, et al. 1995. 1H NMR of Aβ amyloid peptide congeners in water solution. Conformational changes correlate with plaque competence. Biochemistry 34: 5191–5200. [DOI] [PubMed] [Google Scholar]
- Lendel C, Bjerring M, Dubnovitsky A, Kelly RT, Filippov A, Antzutkin ON, Nielsen NC, Härd T. 2014. A hexameric peptide barrel as building block of amyloid-β protofibrils. Angew Chem Int Ed Engl 53: 12756–12760. [DOI] [PubMed] [Google Scholar]
- Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. 2006. A specific amyloid-β protein assembly in the brain impairs memory. Nature 440: 352–357. [DOI] [PubMed] [Google Scholar]
- Lesne SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA, Ashe KH. 2013. Brain amyloid-β oligomers in ageing and Alzheimer’s disease. Brain 136: 1383–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LP, Darden TA, Bartolotti L, Kominos D, Pedersen LG. 1999. An atomic model for the pleated β-sheet structure of Aβ amyloid protofilaments. Biophys J 76: 2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Reed MN, Kotilinek LA, Grant MKO, Forster CL, Qiang W, Shapiro SL, Reichl JH, Chiang ACA, Jankowsky JL, et al. 2015. Quaternary structure defines a large class of amyloid-β oligomers neutralized by sequestration. Cell Rep 11: 1760–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez del Amo JM, Fink U, Dasari M, Grelle G, Wanker EE, Bieschke J, Reif B. 2012. Structural properties of EGCG-induced, nontoxic Alzheimer’s disease Aβ oligomers. J Mol Biol 421: 517–524. [DOI] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. 2013. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 2005. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc Natl Acad Sci 102: 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R. 2008. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: Molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys 41: 265–297. [DOI] [PubMed] [Google Scholar]
- McDonald M, Box H, Bian W, Kendall A, Tycko R, Stubbs G. 2012. Fiber diffraction data indicate a hollow core for the Alzheimer’s Aβ 3-fold symmetric fibril. J Mol Biol 423: 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fändrich M. 2009. Aβ(1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol 386: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Öhman A. 2007. Amide solvent protection analysis demonstrates that amyloid-β(1–40) and amyloid-β(1–42) form different fibrillar structures under identical conditions. Biochem J 404: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau WM, Tycko R. 2008. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci 105: 18349–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. 2009. Seeded growth of β-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci 106: 7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy S, Inoue M, Xiao YL, Matsumura Y, Nabeshima Y, Hoshi M, Ishii Y. 2015. Structural insight into an Alzheimer’s brain-derived spherical assembly of amyloid β by solid-state NMR. J Am Chem Soc 137: 6480–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensalfini A, Albay R, Rasool S, Wu JW, Hatami A, Arai H, Margol L, Milton S, Poon WW, Corrada MM, et al. 2014. Intracellular amyloid and the neuronal origin of Alzheimer neuritic plaques. Neurobiol Dis 71: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. 2002. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci 99: 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. 2004. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J Mol Biol 335: 247–260. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. 2005. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science 307: 262–265. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Yau WM, Tycko R. 2006. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry 45: 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov A, Yau WM, Ghirlando R, Thurber KR, Tycko R. 2015. Successive stages of amyloid-β self-assembly characterized by solid state nuclear magnetic resonance with dynamic nuclear polarization. J Am Chem Soc 137: 8294–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau WM, Tycko R. 2011. Structural evolution of Iowa mutant β-amyloid fibrils from polymorphic to homogeneous states under repeated seeded growth. J Am Chem Soc 133: 4018–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau WM, Luo YQ, Mattson MP, Tycko R. 2012. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci 109: 4443–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Kelley K, Tycko R. 2013. Polymorph-specific kinetics and thermodynamics of β-amyloid fibril growth. J Am Chem Soc 135: 6860–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh DP, Levitt MH, Griffin RG. 1988. Rotational resonance in solid state NMR. Chem Phys Lett 146: 71–76. [Google Scholar]
- Sandberg A, Luheshi LM, Sollvander S, de Barros TP, Macao B, Knowles TPJ, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, et al. 2010. Stabilization of neurotoxic Alzheimer amyloid-β oligomers by protein engineering. Proc Natl Acad Sci 107: 15595–15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B, Mithu VS, Chandra B, Mandal A, Chandrakesan M, Bhowmik D, Madhu PK, Maiti S. 2014. Significant structural differences between transient amyloid-β oligomers and less-toxic fibrils in regions known to harbor familial Alzheimer’s mutations. Angew Chem Int Ed Engl 53: 6888–6892. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447: 453–457. [DOI] [PubMed] [Google Scholar]
- Scheidt HA, Morgado I, Rothemund S, Huster D, Fändrich M. 2011. Solid state NMR spectroscopic investigation of a β protofibrils: Implication of Aβ-sheet remodeling upon maturation into terminal amyloid fibrils. Angew Chem Int Ed Engl 50: 2837–2840. [DOI] [PubMed] [Google Scholar]
- Scheidt HA, Morgado I, Huster D. 2012. Solid state NMR reveals a close structural relationship between amyloid-β protofibrils and oligomers. J Biol Chem 287: 22822–22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Güntert P, Böckmann A, Glockshuber R, Meier BH. 2014. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl 53: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourakis NG, Yau WM, Qiang W. 2015. Modeling an in-register, parallel “Iowa” Aβ fibril structure using solid state NMR data from labeled samples with Rosetta. Structure 23: 216–227. [DOI] [PubMed] [Google Scholar]
- Stöhr J, Condello C, Watts JC, Bloch L, Oehler A, Nick M, DeArmond SJ, Giles K, DeGrado WF, Prusiner SB. 2014. Distinct synthetic Aβ prion strains producing different amyloid deposits in bigenic mice. Proc Natl Acad Sci 111: 10329–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay WM, Huang DT, Rosenberry TL, Paravastu AK. 2013. The Alzheimer’s amyloid-β(1–42) peptide forms off-pathway oligomers and fibrils that are distinguished structurally by intermolecular organization. J Mol Biol 425: 2494–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjernberg LO, Callaway DJ, Tjernberg A, Hahne S, Lilliehöök C, Terenius L, Thyberg J, Nordstedt C. 1999. A molecular model of Alzheimer amyloid β-peptide fibril formation. J Biol Chem 274: 12619–12625. [DOI] [PubMed] [Google Scholar]
- Tomic JL, Pensalfini A, Head E, Glabe CG. 2009. Soluble fibrillar oligomer levels are elevated in Alzheimer’s disease brain and correlate with cognitive dysfunction. Neurobiol Dis 35: 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama T, Nagata T, Shimada H, Teraoka R, Fukushima A, Kanemitsu H, Takuma H, Kuwano R, Imagawa M, Ataka S, et al. 2008. A new amyloid-β variant favoring oligomerization in Alzheimer’s-type dementia. Ann Neurol 63: 377–387. [DOI] [PubMed] [Google Scholar]
- Török M, Milton S, Kayed R, Wu P, McIntire T, Glabe CG, Langen R. 2002. Structural and dynamic features of Alzheimer’s Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem 277: 40810–40815. [DOI] [PubMed] [Google Scholar]
- Tycko R. 2007. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys 126: 064506. [DOI] [PubMed] [Google Scholar]
- Tycko R. 2014. Physical and structural basis for polymorphism in amyloid fibrils. Prot Sci 23: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Dabbagh G. 1990. Measurement of nuclear magnetic dipole–dipole couplings in magic angle spinning NMR. Chem Phys Lett 173: 461–465. [Google Scholar]
- Tycko R, Wickner RB. 2013. Molecular structures of amyloid and prion fibrils: Consensus versus controversy. Acc Chem Res 46: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R, Sciarretta KL, Orgel J, Meredith SC. 2009. Evidence for novel β-sheet structures in Iowa mutant β-amyloid fibrils. Biochemistry 48: 6072–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Böckmann A, Meier BH. 2010. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid state NMR spectroscopy. J Am Chem Soc 132: 13765–13775. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. 2002. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416: 535–539. [DOI] [PubMed] [Google Scholar]
- Watts JC, Condello C, Stöhr J, Oehler A, Lee J, DeArmond SJ, Lannfelt L, Ingelsson M, Giles K, Prusiner SB. 2014. Serial propagation of distinct strains of Aβ prions from Alzheimer’s disease patients. Proc Natl Acad Sci 111: 10323–10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AD, Sega M, Chen ML, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. 2005. Structural properties of Aβ protofibrils stabilized by a small molecule. Proc Natl Acad Sci 102: 7115–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DF, Rosenberg PB, Zhou Y, Kumar A, Raymont V, Ravert HT, Dannals RF, Nandi A, Brasic JR, Ye WG, et al. 2010. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir F 18). J Nucl Med 51: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C. 2010. Fibrillar oligomers nucleate the oligomerization of monomeric amyloid β but do not seed fibril formation. J Biol Chem 285: 6071–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YL, Ma BY, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. 2015. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol 22: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LP, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, Hillen H, Barghorn S, Ebert U, Richardson PL, et al. 2009. Structural characterization of a soluble amyloid β-peptide oligomer. Biochemistry 48: 1870–1877. [DOI] [PubMed] [Google Scholar]