Abstract
HDM2 and HDMX are key negative regulatory factors of the p53 tumor suppressor under normal conditions by promoting its degradation or preventing its trans activity, respectively. It has more recently been shown that both proteins can also act as positive regulators of p53 after DNA damage. This involves phosphorylation by ATM on serine residues HDM2(S395) and HDMX(S403), promoting their respective interaction with the p53 mRNA. However, the underlying molecular mechanisms of how these phosphorylation events switch HDM2 and HDMX from negative to positive regulators of p53 is not known. Our results show that these phosphorylation events reside within intrinsically disordered domains and change the conformation of the proteins. The modifications promote the exposition of N-terminal interfaces that support the formation of a new HDMX-HDM2 heterodimer independent of the C-terminal RING-RING interaction. The E3 ubiquitin ligase activity of this complex toward p53 is prevented by the p53 mRNA ligand but, interestingly, does not affect the capacity to ubiquitinate HDMX and HDM2. These results show how ATM-mediated modifications of HDMX and HDM2 switch HDM2 E3 ubiquitin ligase activity away from p53 but toward HDMX and itself and illustrate how the substrate specificity of HDM2 E3 ligase activity is regulated.
INTRODUCTION
The activity of the RING-E3 ubiquitin ligase HDM2 is tightly regulated by posttranslational modifications and protein-protein interactions that control its subcellular localization and interacting partners (1, 2). The best-characterized activity of HMD2 is the N-terminal interaction between its hydrophobic pocket and the conserved BOX-I domain of p53, which under normal cellular conditions promotes p53 polyubiquitination. The nonredundant HDM2 paralogue HDMX (also called HDM4) is, like HDM2, upregulated in human cancers (3–5). Despite the similarity with the C-terminal RING domain of HDM2 (approximately 75%), HDMX does not harbor E3 ubiquitin ligase activity toward p53, and its negative activity is instead linked to suppression of p53 trans activity. Mice lacking either mdm2 (the hdm2 mouse orthologous) or mdmx (the hdmx mouse orthologous) die early during development in a p53-dependent manner (6–8). It has been shown more recently that HDMX assists HDM2-mediated degradation of p53 by stabilizing HDM2 via a C-terminal RING-RING interaction and also by promoting HMD2-mediated polyubiquitination of p53 in the cytoplasm (9, 10).
The ataxia telangiectasia mutated (ATM) kinase is a key regulator of p53 activity during the double-stranded DNA damage response that helps to induce p53 expression and activity. The direct and indirect phosphorylation of residues within the p53 BOX-I domain of p53 prevents the interaction with HDM2 and is linked to an increase in p53 activity (11–13). The phosphorylation on HDM2 at serine 395 (394 in mouse MDM2) is crucial for p53 stabilization, and animals carrying the S394A mutation have an impaired response to irradiation (14). The corresponding mutation in the human protein prevents the RING domain of HMD2 from binding the p53 mRNA and avoids the capacity of HMD2 to induce p53 synthesis following DNA damage (14). HMDX assists HMD2-mediated degradation of p53 under normal conditions and switches to support HMD2-mediated induction of p53 synthesis during genotoxic stress (9, 15). This switch in activity is ATM dependent via phosphorylation on serine 403, which allows HMDX to bind the nascent p53 mRNA and form an RNA platform to which HMD2 binds (16). Hence, HDM2 and HDMX are functionally dependent on each other during both the downregulation of p53 under normal conditions and the upregulation of p53 following ATM activation (3–5, 9, 17, 18).
The phosphorylation events on HDM2(S395) and HDMX(S403) fall in intrinsically disordered regions (IDR) that serve as regulatory links between the acidic and the RING domains. Multidomain proteins often possess IDRs that connect folded domains (19). It has recently been shown that more than 30% of eukaryotic proteins possess predicted IDRs and that these are engaged in molecular recognition and signaling and are involved in posttranslational modifications (20, 21). For example, phosphorylation sites share biochemical characteristics with IDRs such as amino acid composition, sequence complexity, amino acid charge, or hydrophobicity (22). Along these lines, IDRs have a much higher frequency of known phosphorylation sites than ordered regions, suggesting a strong preference for locating phosphorylation sites in regions that are intrinsically disordered. Phosphorylation on IDRs can affect the global protein conformation by modulating the disposition of the folded domains, thereby regulating the function of the protein.
Here, we investigated how allosteric interactions control the ability of HDM2 and HDMX to switch between positive and negative regulation of p53 depending on cellular conditions. We show how ATM-mediated phosphorylation events on HDM2 and HDMX create new interfaces between the two proteins and how the interaction with the p53 mRNA switches HDM2's E3 ubiquitin ligase activity away from p53 and toward HDM2/HDMX. These data not only help to identify novel drug targets to control HDM2 activity toward p53 but also illustrate the molecular mechanisms of the substrate specificity of E3 ubiquitin ligases under different cellular conditions.
MATERIALS AND METHODS
Plasmids and antibodies.
Point mutations of HDMX and HDM2 were generated by site-directed mutagenesis. The cDNA for HDM2(1-432), HDM2(1-354), HDM2(1-301), HDMX(1-255), HDMX-N-terminal, HDM2-N-terminal, HDMX-C-terminal, HDM2-C-terminal, HDMX-Ac domain, and HDM2-Ac domain were PCR amplified and cloned in pET28a, pET41, pDUET, and/or pCDNA. Proteins were detected with 4B2, CM1, 5.1-HDMX, and 2A10. YB25-anti-HDM2 is an in-house rabbit anti-C-terminal region of HDM2, and chicken-HDMX is an in-house chicken anti-HDMX; mouse anti-His (Invitrogen) also was used. Immunoprecipitations (IPs) were performed using protein G-Sepharose beads (Sigma) and developed with ECL mix (GE Healthcare).
Purification of recombinant protein.
Histidine-tagged recombinant proteins HDM2, HDMX, HDM2(1-432), HDM2(1-354), HDM2(1-301), HDMX(1-255), HDMX-N-terminal, HDM2-N-terminal, HDMX-C-terminal, and HDM2-C-terminal were produced in Escherichia coli BL21(DE3) cells cultured at 37°C and purified using HiTrap nickel columns (GE Healthcare). Lysates in buffer A (50 mM Tris, 200 mM NaCl, 40 mM imidazole, 10 μM ZnSO4, 10% glycerol) were complemented with 10 mg of phenylmethanesulfonyl fluoride (PMSF) (Sigma). The pellet was resuspended in 10 ml of urea buffer (8 M urea, 50 mM Tris, pH 8.5, 150 mM NaCl, 10 mM imidazole) and incubated for 2 h at room temperature (RT). After centrifugation, the supernatant was loaded into the nickel column and washed with the following buffers: 25 ml of urea buffer, 25 ml of urea buffer plus 0.2% Triton X-100, and 25 ml of buffer A plus 10 mM β-mercaptoethanol. Recombinant proteins were eluted from nickel columns with buffer B (50 mM Tris, pH 8.5, 200 mM NaCl, 10 μM ZnSO4, 300 mM imidazole). Proteins were dialyzed against phosphate buffer with 10 μM ZnSO and concentrated in Amicon centrifugal filter units (Merck Millipore). p53 was induced at 15°C overnight and purified on a HiTrap nickel column from the soluble fraction. Proteins were quantified and analyzed by SDS-PAGE. For spectroscopy experiments, HDM2 and HDM2(S395D) were additionally purified by size-exclusion chromatography (Superdex 75).
CD spectroscopy.
The circular dichroism (CD) spectra of HDM2 and HDM2(S395D) were recorded using a Jasco J-810 spectropolarimeter in a wavelength range of 200 to 280 nm for far-UV CD and 250 to 310 nm for near-UV CD. The protein concentration was 0.5 mg/ml in phosphate buffer, pH 8, and quartz cuvettes of 0.1-cm path length were used.
Intrinsic fluorescence.
The emission fluorescence spectra of the proteins at 0.5 mg/ml in phosphate buffer, pH 8, were recorded in a Perkin-Elmer LS-55 spectrofluorometer in quartz cuvettes of 1-cm path length at room temperature. The excitation wavelengths were 280 and 295 nm. Blanks without protein were recorded and subtracted from the experimental spectra.
Limited proteolysis.
Thirty micrograms of HDM2 and HDM2(S395D) was incubated with 0 to 1,000 ng of proteinase K (Life Technologies) at 30°C for 5 min. The reaction was arrested by the addition of PMSF (3 mM final concentration). Samples were analyzed by SDS-PAGE.
Enzyme-linked immunosorbent assay (ELISA).
Ninety-six-well plates (Thermo Scientific) were coated with 10 ng/μl of protein in 0.1 M NaHCO3 (200 μl/well) overnight at 4°C. After incubation, plates were washed 6 times with 200 μl 0.1% PBS-Tween 20, blocked for 1 h at RT with 0.1% PBS-Tween 20 containing 3% bovine serum albumin (BSA), and then washed 6 times with 200 μl 0.1% PBS-Tween 20. The second bound protein was diluted (from 0 to 10 or 20 μg/μl) and added to the plate. For ternary complexes, p53 protein (10 ng/μl) or p53 mRNA (10 ng/μl) were mixed with the second bound protein and incubated for 2 h at 4°C prior to loading in the plates (RNaseOUT was added if p53 mRNA was present). Plates were washed 6 times with 200 μl 0.1% PBS-Tween 20 and incubated with the primary specific antibody (1:1,000) for 1 h at RT. After the plates were washed 6 times with 200 μl 0.1% PBS-Tween 20, secondary antibody was incubated for 1 h at RT. Plates were washed and incubated with 100 μl/well of ECL mix (GE Healthcare), and luminescence was measured in a Cary eclipse fluorescence spectrophotometer.
In vitro ubiquitination assays.
Ubiquitination reaction mixtures contained the following: 25 mM HEPES (pH 8.0); 10 mM MgCl2; 4 mM ATP; 0.5 mM dithiothreitol (DTT); 0.05% (vol/vol) Triton X-100; 0.25 mM benzamidine; 10 mM creatine phosphate; 3.5 U/ml creatine kinase, ubiquitin, or His-tagged ubiquitin (2 μg); E1 (100 nM), E2 (1 μM), and the proteins of interest; and p53 at 5 μg, HDMX/S403D at 10 μg, and p53 mRNA at 100 ng when applied. Reactions were started with the addition of HDM2 or S395D, reaction mixtures were incubated for 15 min at 30°C, and products were analyzed with 4% to 12% SDS-PAGE followed by immunoblotting.
In vitro transcription.
To produce RNA transcripts, the plasmid DNA was linearized by restriction digestion downstream of the insert, followed by gel purification using a DNA gel extraction kit prior to transcription reactions. One microgram of linearized template DNA was used in the reaction mixture containing 5× transcription buffer, nucleoside triphosphate mix (10 mM each), 50 U RiboLock, and 30 U of T7 RNA polymerase in diethyl pyrocarbonate-treated water for a final volume of 50 μl. The reaction mixture was incubated for 2 h at 37°C. Two microliters of DNase 1 was added and incubated for 15 min more to remove the template. RNA transcripts were purified by standard phenol-chloroform extraction and ethanol precipitation.
PLA.
The proximal ligation assay (PLA) method was performed according to the manufacturer's instructions. Briefly, H1299 cells were grown on coverslips and transfected with the indicated constructs, fixed in 4% paraformaldehyde for 10 min before permeabilization, and blocked in 1× PBS containing 0.1% saponin and 3% BSA for 30 min. Primary (anti-HDM2 and anti-HDMX) and secondary (mouse and rabbit) antibodies were incubated for 1 h each at 37°C. After the cells were washed, PLA probes were added, followed by hybridization, ligation for 30 min at 37°C, and amplification for 100 min at 37°C. DNA (blue) and HDMX-HDM2 (red) were visualized after incubation with the detection solution. Slides were analyzed by fluorescence microscopy.
Far Western analysis.
E. coli total cell lysate expressing HDMX was lysed in buffer A, and 40 μl from the total fraction was loaded into SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was washed with 1× PBS–0.1% Tween 20 for 7 days at 4°C in order to renature the protein. After blocking with 1× PBS–3% milk, the membrane was incubated with different constructs [HDM2, HDM2(1-432), HDM2(1-354), and HDM2(1-301)] at a concentration of 20 mg/ml in 1× PBS–3% milk overnight at 4°C. The membrane was washed with 1× 0.1% PBS–3× Tween 20 for 15 min, incubated with 2A10 anti-HDM2 antibody, and developed using a standard Western blotting (WB) technique.
RESULTS
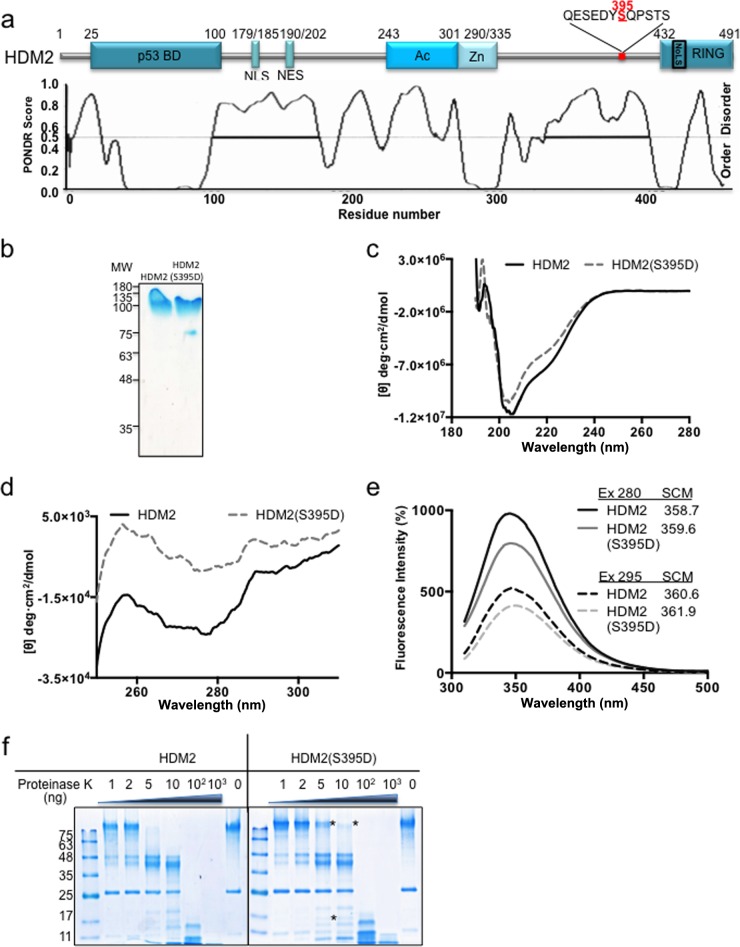
Phosphorylation of HDM2 on serine 395 induces conformational changes in HDM2.
Phosphorylation of HDM2(S395) takes place in a predicted disordered region in the C terminus of HDM2 that connects the Zn finger with the RING domain (23) (Fig. 1a). This phosphorylation event is mediated by the ATM kinase and switches HDM2 from a negative to a positive regulator of p53, which is required for activation of p53 following DNA damage (14, 15, 24). Previous work has shown that the binding of the full-length HDM2 to the p53 mRNA requires residue 395 phosphorylation, while the RING domain (positions 396 to 491) alone binds to HDM2 (15). Hence, p53 mRNA binding is likely the result of a conformational change in HDM2. We set out to investigate the underlying allosteric changes in HDM2 that allow it to bind the p53 mRNA and switch its activity toward p53. The phosphomimetic mutant HDM2(S395D) has been shown to mimic the effect of ATM-mediated phosphorylation of HDM2 both in vitro and in animal models (14). We structurally characterized wild-type (WT) HDM2 and HDM2(S395D) using in vitro-purified recombinant proteins (Fig. 1b). The far-UV circular dichroism spectra of the two proteins are slightly different, indicating that phosphorylation induces structural modifications that are reflected in the secondary structure of the protein (Fig. 1c). In addition, the near-UV circular dichroism analysis shows stronger differences, suggesting substantial changes in the ternary structure (Fig. 1d). In concordance with this, the intrinsic fluorescence spectra of HDM2 and HDM2(S395D) show a significant quenching in the fluorescence intensity with a discrete difference of 1 nm in the spectral center of mass (Fig. 1e). The conformational change in the structure of HDM2(S395D) was also observed using limited proteolysis by proteinase K (25, 26). The wild type and HDM2(S395D) mutant were treated with proteinase K concentrations ranging from 1 to 1,000 ng at 30°C for 5 min (Fig. 1f). HDM2 protein demonstrated a higher susceptibility to proteolysis than HDM2(S395D), indicating global conformational changes that expose cleavage sites differentially. Taken together, these data suggest that the serine 395 phosphorylation event in the IDR near the RING domain alters the global conformation of the protein.
FIG 1.
HDM2 conformational rearrangement promoted by phosphomimetic mutant HDM2(S395D). (a) Cartoon illustrating major domains of HDM2. p53 BD, the p53 binding domain; NLS, nuclear localization signal; NES, nuclear export signal; Ac, acidic domain; Zn, zinc finger; RING, really interesting new gene domain. The predicted intrinsically disordered regions of HDM2 are shown in the lower panel. Serine 395 is indicated in red and is located at the end of a disordered region. (b) Recombinant purified HDM2 and HDM2(S395D). MW, molecular weights in thousands. (c) Far-UV CD spectra of the HDM2 and phosphomimetic mutant HDM2(S395D). (d) Near-UV CD spectra of the HDM2 and phosphomimetic mutant HDM2(S395D). (e) The intrinsic fluorescence emission spectra of HDM2 and phosphomimetic mutant HDM2(S395D). The spectral center of mass (SCM) at excitation (Ex) wavelengths of 280 and 295 nm were calculated. All spectra shown were obtained after subtracting the blank (no enzyme) from the experimental values. (f) Limited proteolysis of HDM2 and the phosphomimetic mutant S395D by proteinase K. Asterisks represent the main differences between the wild type and the mutant, showing that HDM2(S395D) is less susceptible to proteinase K. One representative experiment of 3 is shown.
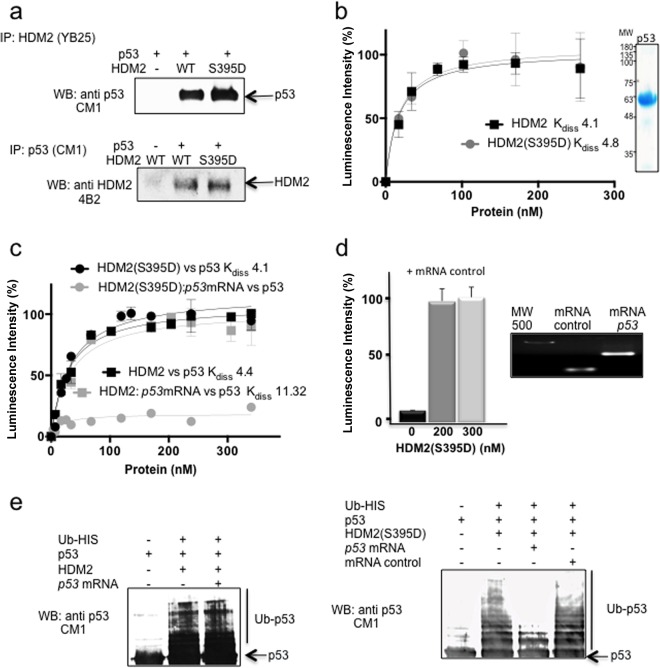
p53 mRNA-HDM2(S395D) interaction induced after phosphorylation inhibits p53 ubiquitination.
ATM-dependent phosphorylation of HDM2 at serine 395 and the consequent binding to the p53 mRNA not only promotes p53 synthesis but also prevents its degradation (15, 24, 27, 28). To test how this event prevents HDM2-mediated ubiquitination of p53, we performed protein-protein interaction pulldown assays in which the supernatant of transformed E. coli expressing recombinant HDM2 or HDM2(S395D) was mixed with the supernatant of recombinant p53 and incubated with antibodies against either p53 (CM-1) or HMD2 (YB25). The result shows that both HDM2 and HDM2(S395D) interact equally with p53 (Fig. 2a). This was further confirmed using ELISA in which plates were coated with 10 ng/μl recombinant p53 and increasing concentrations of HDM2 or HDM2(S395D) were added. Both proteins were shown to bind with similar high affinities to p53, with Kd (dissociation constant) values of 4.1 and 4.8 nM, respectively (Fig. 2b). Hence, the effect of serine 395 phosphorylation in preventing p53 ubiquitination cannot be explained by a loss of affinity to p53.
FIG 2.
p53 mRNA-HDM2 interaction is required to inhibit p53-HDM2 interaction and p53 ubiquitination. (a) Coimmunoprecipitation of HDM2 (upper) or p53 (lower). Total lysate from E. coli transformed with p53 was mixed with the total lysates from E. coli transformed with HDM2 or HDM2(S395D). (b) ELISA using a fixed amount of recombinant purified p53 (10 ng/μl) and increasing amounts of HDM2 or HDM2(S395D) (0 to 20 ng/μl). HDM2 and HDM2(S395D) show similar affinity toward p53, with Kd values of 4.1 and 4.8, respectively. The data shown represent the averages and standard deviations (SD) from five independent experiments. Purified p53 is shown. (c) Same as panel b but in the presence or absence of 10 ng p53 mRNA. In the presence of p53 mRNA, HDM2(S395D) lost the interaction with the p53 protein. The data represent the averages and SD from five independent experiments. In the lower panel the interaction with an mRNA control is shown. (d) p53 and HDM2(S395D) interaction in the presence or absence of the mRNA control. The integrity of the mRNAs used is also shown. (e) In vitro ubiquitination of recombinant p53 with HDM2 or HDM2(S395D) in the presence or absence of in vitro-synthesized 100 ng p53 mRNA or 100 ng of mRNA control. The left panel shows the wild-type HDM2 reactions, while the right panel shows HDM2(S395D). The presence of the p53 mRNA completely prevents the ubiquitination of p53.
On a molecular level, the interaction of HDM2 with p53 mRNA takes place in the C-terminal RING of HDM2, whereas the HDM2-p53 protein interaction takes place in the N terminus of HDM2 (29–31). When we tested whether HDM2(S395D) is able to bind p53 mRNA and protein simultaneously using ELISA with plates coated with 10 ng/μl p53 protein, we observed that the presence of 10 ng of in vitro-synthesized p53 mRNA indeed blocks the interaction between p53 and HDM2(S395D) (Fig. 2c). As the wild-type HDM2 protein does not bind the p53 mRNA, its interaction with p53 was not affected by the presence of the p53 mRNA. In Fig. 2d we show that the HDM2(S395D) p53 interaction is not affected by the presence of a control mRNA (Fig. 2d).
To further investigate the role of HDM2(S395D) phosphorylation in the stabilization of p53 after DNA damage, we performed an in vitro p53 ubiquitination assay using HDM2 and HDM2(S395D). Both proteins were able to ubiquitinate p53 in vitro (Fig. 2e). However, when we added 100 ng of p53 mRNA to the reaction mixture, the p53 ubiquitination by HDM2(S395D), but not that by wild-type HDM2, was completely blocked (Fig. 2e, third lanes of both images), confirming that the binding of HDM2(S395D) with the p53 mRNA but not with a control mRNA is responsible for inhibiting the interaction and preventing p53 ubiquitination (Fig. 2e, right, fourth lane). Hence, in addition to the phosphorylation of p53 N-terminal residues, these results offer an additional mechanism to help explain why HDM2, following phosphorylation by ATM, does not target p53 for degradation.
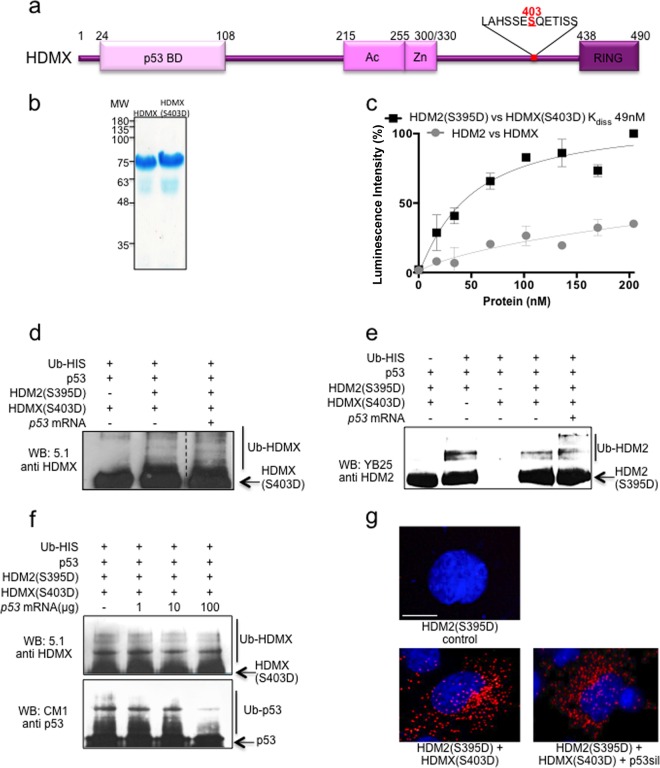
p53 mRNA-HDM2(S395D) interaction switches HMD2 E3 ligase substrate specificity from p53 toward HDMX and itself.
HDMX is a paralogous protein of HDM2 that shares the same structured domains with HDM2 and has high sequence similarity (Fig. 3a). Despite having a RING domain with 75% similarity to HDM2, HDMX exhibits no E3 ubiquitin ligase activity. It is, however, able to bind the p53 protein, block its transactivation activity, and work in collaboration with HDM2 to degrade p53 by forming a heterodimer through their RING domains to downregulate p53 (10, 32). Additionally, HDMX is a substrate of HDM2 E3 ligase activity (33, 34). The HDMX(S403D) mutant has, like HDM2, been shown to act as an ATM phosphomimetic (16). First, we tested if the HDMX-HDM2 interaction changes after ATM-mediated phosphorylation on HMD2 and HDMX. ELISA of the HDM2-HDMX interaction showed that HDM2(S395D) and HDMX(S403D) had 3-fold higher affinity than wild-type proteins (Kd of 49 nM) (Fig. 3b and c). We next tested if the presence of p53 mRNA also inhibits HDM2 E3 ligase activity toward HDMX by performing an in vitro ubiquitination assay using the HDM2(S395D) and HDMX(S403D) mutants in the presence of p53 protein and in vitro-synthesized p53 mRNA. It might have been expected that the binding of the p53 mRNA to the RING domain of HDM2 would block its E3 ligase activity, but to our surprise, HDMX was still ubiquitinated by HDM2 (Fig. 3d). Furthermore, we also found that HDM2(S395D) is autoubiquitinated in the presence of p53 mRNA (Fig. 3e). These results support a model whereby HDM2(S395D) binds to and ubiquitinates p53, but once the p53 mRNA is present, HDM2(S395D) no longer binds p53 and the E3 ligase activity instead shifts toward itself and HDMX. We used the phosphomimetic HDM2 and HDMX mutants and p53 protein with increasing amounts of p53 mRNA in order to see the effect of the mRNA on the substrate specificity of HDM2 E3 ubiquitin ligase activity. Figure 3f shows that even with 1 ng of p53 mRNA there is an effect on p53 ubiquitination, whereas even with 100 ng of p53 mRNA HDMX is still ubiquitinated.
FIG 3.
HDM2(S395D)-p53 mRNA interaction does not inhibit ubiquitination of HDMX(S403D) and HDM2(S395D). (a) Cartoon illustrating major domains of HDMX. p53 BD, the p53 binding domain; Ac, acidic domain; Zn, zinc finger; RING, really interesting new gene domain. Serine 403 is indicated in red. (b) Recombinant purified HDMX and HDMX(S403D). (c) ELISA using a fixed amount of recombinant purified HDMX or phosphomimetic mutant HDMX(S403D) (10 ng/μl) and increasing amounts of HDM2 or HDM2(S395D) (0 to 20 ng/μl), respectively. The WT HDM2-HDMX proteins show very low affinity compared to that of the two proteins that mimic ATM-dependent phosphorylation. The figure represents averages and SD from five independent experiments. (d) In vitro HDM2(S395D) ubiquitination of recombinant HDMX(S403D) in the presence of p53 protein and with or without 100 ng p53 mRNA. The ubiquitination of HDMX(S403D) is not inhibited by the presence of p53 mRNA. (e) In vitro HDM2(S395D) autoubiquitination assay in the presence of p53 protein and HDMX(S403D) with or without p53 mRNA. In the presence of p53 mRNA, polyubiquitination increases, probably due to the inhibition of the interaction with p53 protein, which in the absence of mRNA is still able to interact with HDM2(S395D) and become ubiquitinated. (f) p53 mRNA dose-dependent in vitro ubiquitination of p53 and HDMX(S403D). (g) PLA using anti-HDMX and anti-HDM2. H1299 cells were transfected with the indicated constructs. Cell nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI) (blue).
Using the proximity ligase assay (PLA), which gives a specific signal if two different antibody epitopes are localized within 40 nm of each other, we tested the interaction between HDMX and HDM2 in the presence of the p53 mRNA in H1299 cells. Cells were transfected with HDM2(S395D) and HDMX(S403D) together with a silent p53 construct (p53sil) that lacks the initiation AUG codons at positions +1 and +120 and is not able to produce the p53 protein. This showed that the p53 mRNA is not able to block the HDM2(S395D)-HDMX(S403D) interaction in cellulo (Fig. 3g). These findings support the in vitro data and suggest that the p53 mRNA does not inhibit the E3 ubiquitin ligase activity of HDM2 but instead controls its substrate specificity.
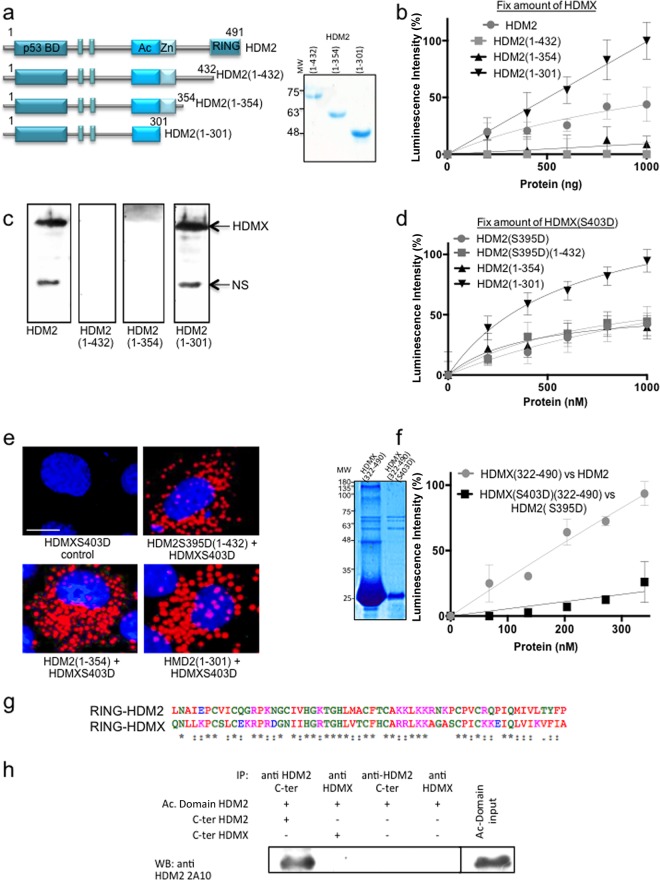
The HDMX-HDM2 interaction is independent of RING domains after DNA damage.
These data indicate that ATM-mediated phosphorylation of HDMX and HDM2 increases their affinity for each other and exposes their respective RNA binding pockets that together change the substrate specificity of the HDM2 E3 ubiquitin ligase activity. Based on these results, we wanted to know if the heterodimer interphase formed after the ATM-dependent phosphorylation between HDMX and HDM2 could be different from the RING-RING interaction previously described. We carried out a deletion series of the HDM2 C-terminal domain to pinpoint the region of interaction. The first construct, named HDM2(1-432) (amino acids 1 to 432), lacks the RING C-terminal domain; the second construct, HDM2(1-354) (amino acids 1 to 354), lacks the RING domain plus the IDR where S395 phosphorylation takes place. Finally, the smallest construct additionally lacked the Zn finger domain: HDM2(1-301) (amino acids 1 to 301). The three constructs were well expressed and purified (Fig. 4a).
FIG 4.
HDMX-HDM2 interaction is independent of RING domains after DNA damage. (a) Diagrams of HDM2 stop constructs in the C terminus. Construct HDM2(1-432) (amino acids 1 to 432) lacks the RING C-terminal domain; construct HDM2(1-354) (amino acids 1 to 354) lacks the RING domain plus the IDR where S395 phosphorylation takes place. Construct HDM2(1-301) also is lacking the Zn finger domain (amino acids 1 to 301). Recombinant purified proteins are also shown. (b) ELISA using HDMX (10 ng/μl) and increasing amounts of HDM2, HDM2(1-432), HDM2(1-354), and HDM2(1-301) (from 0 to 10 ng/μl). Neither HDM2(1-432) nor HDM2(1-354) binds to HDMX. However, the smallest one, HDM2(1-301), binds even more strongly than full-length HDM2. (c) Far Western analysis of the interaction using the three constructs and full-length HDMX. NS, nonspecific. (d) Same as panel b but using HDMX(S403D), HDM2(S395D), and HDM2(S395D)(1-432) instead of the wild-type version of each protein. In this case, all of the constructs bind to HDMX(S403D) with affinity similar to that of the full-length HDM2(S395D), and HDM2(1-301) binds more strongly. (e) PLA using anti-HDMX and anti-HDM2. H1299 cells were transfected with the indicated constructs. Cell nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (f) ELISA using HDMX or HDMX(S403D) (10 ng/μl) and increasing concentrations of the C-terminal HDM2 or HDM2(S395D) (amino acids 322 to 491), respectively. The C terminus in the wild-type version binds the full-length HDMX more strongly than the phosphomimetic versions. The recombinant purified C-terminal domains are shown. (g) Alignment of the HDMX(432-491) and HDM2(431-490) RING domains shows more than 75% similitude and more than 45% identity (ESPript; http://espript.ibcp.fr). (h) Coimmunoprecipitation of C-terminal (C-ter) domain of HDMX(322-490) and Ac domain(241-335). The positive control is the Ac domain of HDM2 with the C terminus of HDM2(322-491).
We first tested the affinity of HDMX to increasing concentrations of the full-length HDM2 or the HDM2(1-432), HDM2(1-354), or HDM2(1-301) construct (Fig. 4b). We observed that deletion of the RING domain HDM2(1-432) (amino acids 1 to 432) abolished the interaction with HDMX. Similarly, a further deletion of HDM2(1-354), which includes the phosphorylation site for ATM (residue 395), showed a similar profile, in line with diverse studies that demonstrate that the lack of the HDM2 RING domain results in an inhibition of the interaction with HDMX (9, 32, 35–39). Surprisingly, a further deletion including the Zn domain [HDM2(1-301)] showed strong affinity to HDMX. Using far Western blotting we also observed that HDM2 and HDM2(1-301) proteins, but not HDM2(1-432) and HDM2(1-354) proteins, bind the full-length HDMX (Fig. 4c). We then tested the affinity of the phosphomimetic mutant HDMX(S403D) to HDM2(S395D) and HDM2(S395D)(1-432). Under these conditions, all HDM2 constructs interacted with HDMX(S403D), but again HDM2(1-301) showed the highest affinity (Fig. 4d).
Using the PLA and H1299 cells, we observed that these different interactions take place mainly in the cytoplasm (Fig. 4e). Additionally, we tested the interaction between the HDMX(322-490) C-terminal domain (amino acids 322 to 490) and the full-length HDM2 and the phosphomimetic mutant S403D(322-490) C-terminal domain with the S395D full-length protein. Surprisingly, the interaction between the phosphomimetic mutants was weak compared with that of the two wild-type proteins (Fig. 4f). As shown in Fig. 3c, where the full-length versions of the proteins were used, the interaction between the phosphomimetic proteins was three times higher than that of the wild-type versions of the proteins, supporting the hypothesis that the RING domain of HDMX is not involved in the new architecture of the heterodimer after ATM phosphorylation.
Recently, Cheng et al. shown that the HDM2 RING domain is activated by intramolecular interaction with its acidic domain (40); thus, we hypothesized that the acidic domain in the construct HDM2(1-301) is able to recognize the RING domain of HDMX, since both RING domains have sequences with greater than 75% similarity (41) (Fig. 4g). However, coimmunoprecipitation using the acidic domain of HDM2(241-335) (amino acids 241 to 335) and the C-terminal domain of HDMX(322-490) (amino acid 322 to 490) showed no interaction (Fig. 4h).
The results show that HDM2 and HDMX interact via a novel N-terminal interface controlled by the Zn domain, and deletion of the C terminus down to residue 301 [HDM2(1-301)] results in a protein with strong affinity for HDMX. The results also suggest that the new interface is formed after DNA damage because of its dependence on the level of phosphorylation of HDM2 and HDMX.
HDM2(S395D) and HDMXS(403D) interact through the p53 binding domain in the N termini of both proteins.
To study if the RING domain of HDMX is implicated or not in the N-terminal interaction site and to further determine the location of the N-terminal HDM2-HDMX interface, we generated an HDMX construct lacking the RING domain, IRD, and Zn finger domain named HDMX(1-255) (amino acids 1 to 255) (Fig. 5a). When transfected in H1299 cells together with the different HDM2 constructs, we observed that HDMX(1-255) interacts with all of the HDM2 constructs tested using the PLA. However, the HDM2(1-301)–HMDX(1-255) interaction appears to be stronger, with more PLA interactions (Fig. 5b). Using the recombinant HDMX(1-255) and HDM2(1-301) proteins, we performed an ELISA to estimate the Kd of this interaction at 46.9 nM (Fig. 5c), which is nearly the same as that observed for the two full-length phosphomimetic mutants shown in Fig. 3c (Kd of 49 nM). This suggests that under ATM phosphorylation conditions, the heterodimer is formed in a RING-RING-independent interaction fashion.
FIG 5.
HDM2(S395D) and HDMXS(403D) interact through the N-terminal domain of both proteins. (a) Cartoon illustrating the constructs HDM2(1-301) and HDMX(1-255). Recombinant purified HDMX(1-255) is shown. (b) PLA using anti-HDMX and anti-HDM2. H1299 cells were transfected with the indicated constructs. Cell nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI) (blue). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (c) Constructs of HDMX and HMD2 lacking the Zn finger and the RING domain were tested by ELISA. The Kd (46.9 nM) of the interaction is similar to the one calculated for the interaction between the phosphomimetic mutants. (d) The N-terminal HDMX domain was tested for its ability to recognize different HDM2 domains. In a pDUET plasmid we cloned the N terminus of HDM2(1-100) with a His tag and the acidic domain of HDM2(241-335) without any tag. The total lysate of pDUET construct was pulled down with GST-HDMX(1-110) to observe if the N terminus of HDMX interacts with one of the two domains of HDM2 cloned in pDUET. The membrane was probed with anti-HDM2 4B2 (epitope 29-50), showing the N terminus of HDM2, and anti-His, showing both N-termini, because GST-HDMX(1-110) also has a His tag, and 2A10 (epitope 255-262), which did not react because the acidic domain of HDM2 does not interact with GST-HDMX(1-110).
Finally, we wanted to find the domains involved in the new heterodimer. We used an N-terminal HDMX domain [HDMX(1-110)] carrying a glutathione S-transferase (GST) tag to see if we could pull down the interacting domain of HDM2. Figure 5d shows that the HDMX N-terminal domain is able to interact with the N-terminal domain of HDM2 [HDM2(1-100)] but not with the HDM2 acidic domain HDM2(243-335) (Fig. 5d). Taken together, these results suggest that ATM phosphorylation in the IDR of HDM2 or HDMX opens up an N-terminal interacting site that allows the formation of a second HDMX-HDM2 heterodimer.
Overall, our results demonstrate the ability of HDMX and HDM2 to form heterodimers with different architectures to gain versatility of functions in response to changing cellular conditions.
DISCUSSION
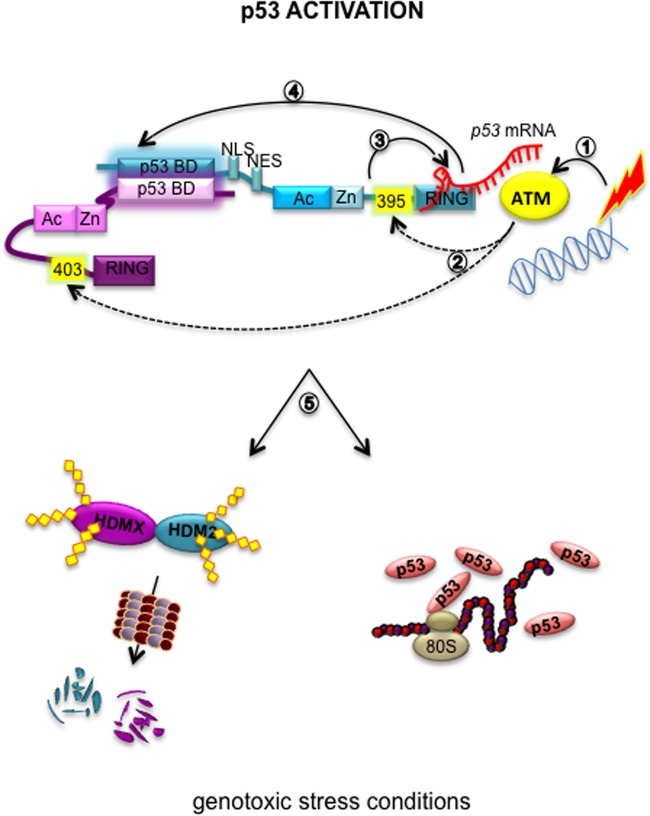
HDM2 is considered a hub protein due to its capacity to interact with a large number of different partners, of which p53 is the most well described. As a negative regulator, HDM2 can inhibit p53 via protein-protein interaction, repressing the transcriptional activity of p53, the same mechanism used by HDMX, or through its E3 ubiquitin ligase activity and p53 polyubiquitination. Despite not having its own E3 ligase activity, HDMX assists HMD2-mediated ubiquitination of p53 and can suppress p53 trans activity (10). One of the mechanisms by which HDM2-mediated suppression of p53 is prevented following DNA damage involves ATM-mediated phosphorylation of p53 on serine 15, releasing it from the HDM2 interaction (11–13). An additional mechanism to ensure optimal levels of p53 protein after genotoxic stress is also mediated by ATM and relates to phosphorylation of HDM2 at S395 and HDMX at S403 and not only stimulates p53 synthesis but also prevents its degradation (15, 16). Under the same conditions, HDMX is rapidly degraded by HDM2 (33, 34). The results presented here might help to explain these different observations and further support the notion that heterodimerization between HDM2 and HDMX under normal and genotoxic conditions is important to control HDM2's E3 ubiquitin ligase activity. Under normal conditions the heterodimer is formed through the RING domains of both proteins. Following DNA damage, phosphorylation events on HMD2 serine 395 and HMDX serine 403 instead results in a new interface that, together with the binding of the p53 mRNA, prevents the interaction with the p53 protein and instead turns HMD2 E3 ligase activity toward HMDX and itself (Fig. 6).
FIG 6.
Proposed model of the allosteric effects on the HDM2-HDMX heterodimer by p53 mRNA. p53 mRNA binding on HDM2 promotes an allosteric change that allows p53 activation. (1) Under genotoxic stress conditions ATM is activated. (2) ATM phosphorylates HDM2(S395) and HDMX(S403). (3) The phosphorylation of S395 allows p53 mRNA binding to the RING domain. (4) The p53 mRNA interaction opens up a new binding site between HDMX and HDM2 in the N-terminal region in a RING-RING-independent manner. (5) These events allow the proper ubiquitination of HDMX and HDM2 itself to ensure the p53 stabilization and activation after DNA damage. Dotted lines represent ATM-dependent phosphorylations.
The results presented support a model in which after DNA damage, the phosphorylation event by ATM at serine 395, located in the IDR of HDM2 between the Zn and the RING domains, induces a conformational change that exposes a p53 mRNA interacting interface. This in turn induces a second allosteric modification which promote an HDM2(N-terminal)-HDMX(N-terminal) interaction that controls HDM2 E3 ligase substrate specificity.
It will be interesting to determine if the E2 ubiquitin-conjugating enzyme recruited by the two heterodimers is different and specific to polyubiquitination of p53 or if it is able to target HDMX and HDM2 for proteasomal degradation. There are at least 38 different E2 genes in humans (42, 43), and it is well known that a single E2 protein can interact with several different E3s. It is also possible that one E3 could interact with diverse E2 proteins (44), providing more versatility to the system of protein degradation. The E2-E3 pair could play a role in substrate specificity under different cellular conditions. Another question that arises from this work is the role of intramolecular interactions that has recently been described for HDMX (45, 46) and for HDM2 (40). These intramolecular interactions could add another level of regulation to the way the domains are exposed to the solvent to allow highly accurate control of the interactome of HDM2.
Altogether, the data in this work suggest that the architecture of the heterodimer formed by HDMX and HDM2 differs under normal or genotoxic stress conditions. This rearrangement involves different interacting domains resulting in different specificities. The plasticity of these multidomain proteins facilitates interaction with many different partners in order to perform multiple functions but also enables different intramolecular interactions among the interacting proteins, providing an additional level of regulation. In this context, posttranslational modifications will define some of these interactions, reducing the natural flexibility of the protein and regulating the formation of multimeric complexes to give the appropriate cellular response depending on environmental conditions.
ACKNOWLEDGMENTS
We thank Yolanda Rebolloso-Gómez and Celina González-Gallegos for technical support.
We have no conflicts of interest to declare.
Funding Statement
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Meek DW, Hupp TR. 2010. The regulation of MDM2 by multisite phosphorylation–opportunities for molecular-based intervention to target tumours? Semin Cancer Biol 20:19–28. doi: 10.1016/j.semcancer.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Fahraeus R, Olivares-Illana V. 2014. MDM2's social network. Oncogene 33:4365–4376. doi: 10.1038/onc.2013.410. [DOI] [PubMed] [Google Scholar]
- 3.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, Li KC, Hong TM, Yang PC. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 4.Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, Schlegel U, Reifenberger G. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59:6091–6096. [PubMed] [Google Scholar]
- 5.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 6.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 7.Montes de Oca Luna R, Amelse LL, Chavez-Reyes A, Evans SC, Brugarolas J, Jacks T, Lozano G. 1997. Deletion of p21 cannot substitute for p53 loss in rescue of mdm2 null lethality. Nat Genet 16:336–337. doi: 10.1038/ng0897-336. [DOI] [PubMed] [Google Scholar]
- 8.Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 9.Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Wang J, Jiang X. 2011. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 286:23725–23734. doi: 10.1074/jbc.M110.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677. [DOI] [PubMed] [Google Scholar]
- 12.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 11:3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shieh SY, Ikeda M, Taya Y, Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334. [DOI] [PubMed] [Google Scholar]
- 14.Gannon HS, Woda BA, Jones SN. 2012. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell 21:668–679. doi: 10.1016/j.ccr.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, Fahraeus R. 2012. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell 21:25–35. doi: 10.1016/j.ccr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Malbert-Colas L, Ponnuswamy A, Olivares-Illana V, Tournillon AS, Naski N, Fahraeus R. 2014. HDMX folds the nascent p53 mRNA following activation by the ATM kinase. Mol Cell 54:500–511. doi: 10.1016/j.molcel.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 17.Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, Wahl GM. 2006. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell 9:273–285. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Wade M, Wang YV, Wahl GM. 2010. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Z, Mizianty MJ, Kurgan L. 2014. Genome-scale prediction of proteins with long intrinsically disordered regions. Proteins 82:145–158. doi: 10.1002/prot.24348. [DOI] [PubMed] [Google Scholar]
- 20.Peng Z, Xue B, Kurgan L, Uversky VN. 2013. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ 20:1257–1267. doi: 10.1038/cdd.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terakawa T, Higo J, Takada S. 2014. Multi-scale ensemble modeling of modular proteins with intrinsically disordered linker regions: application to p53. Biophys J 107:721–729. doi: 10.1016/j.bpj.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform 10:30–40. [PubMed] [Google Scholar]
- 24.Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fahraeus R. 2008. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol 10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 25.Reyes-Vivas H, Martinez-Martinez E, Mendoza-Hernandez G, Lopez-Velazquez G, Perez-Montfort R, Tuena de Gomez-Puyou M, Gomez-Puyou A. 2002. Susceptibility to proteolysis of triosephosphate isomerase from two pathogenic parasites: characterization of an enzyme with an intact and a nicked monomer. Proteins 48:580–590. doi: 10.1002/prot.10179. [DOI] [PubMed] [Google Scholar]
- 26.De La Mora-De La Mora I, Torres-Larios A, Mendoza-Hernandez G, Enriquez-Flores S, Castillo-Villanueva A, Mendez ST, Garcia-Torres I, Torres-Arroyo A, Gomez-Manzo S, Marcial-Quino J, Oria-Hernandez J, Lopez-Velazquez G, Reyes-Vivas H. 2013. The E104D mutation increases the susceptibility of human triosephosphate isomerase to proteolysis. Asymmetric cleavage of the two monomers of the homodimeric enzyme. Biochim Biophys Acta 1834:2702–2711. doi: 10.1016/j.bbapap.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. 2008. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naski N, Gajjar M, Bourougaa K, Malbert-Colas L, Fahraeus R, Candeias MM. 2009. The p53 mRNA-Mdm2 interaction. Cell Cycle 8:31–34. [DOI] [PubMed] [Google Scholar]
- 29.Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon IJ, Gabizon R, Mattia M, Zupnick A, Brown LM, Friedler A, Prives C. 2010. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol 17:982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Marechal V, Levine AJ. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol 13:4107–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. 1996. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med 2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 32.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC, Prives C, Shiloh Y, Jochemsen AG, Taya Y. 2005. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. 2003. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 35.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 447:5–9. [DOI] [PubMed] [Google Scholar]
- 36.Waning DL, Lehman JA, Batuello CN, Mayo LD. 2011. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem 286:216–222. doi: 10.1074/jbc.M110.183012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X. 2011. p53 regulation: teamwork between RING domains of Mdm2 and MdmX. Cell Cycle 10:4225–4229. doi: 10.4161/cc.10.24.18662. [DOI] [PubMed] [Google Scholar]
- 38.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. 2006. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol 363:433–450. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Kawai H, Wiederschain D, Yuan ZM. 2003. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 23:4939–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Q, Song T, Chen L, Chen J. 2014. Autoactivation of the MDM2 E3 ligase by intramolecular interaction. Mol Cell Biol 34:2800–2810. doi: 10.1128/MCB.00246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye YH, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Ye Y. 2008. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci 65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambivero CT, Cilenti L, Main S, Zervos AS. 2014. Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP. Cell Signal 26:2921–2929. doi: 10.1016/j.cellsig.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Bista M, Petrovich M, Fersht AR. 2013. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci U S A 110:17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Borcherds W, Wu S, Becker A, Schonbrunn E, Daughdrill GW, Chen J. 2015. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc Natl Acad Sci U S A 112:4624–4629. doi: 10.1073/pnas.1420833112. [DOI] [PMC free article] [PubMed] [Google Scholar]