Abstract
The cyclin-dependent kinase (CDK) inhibitor p21 is an unstructured protein regulated by multiple turnover pathways. p21 abundance is tightly regulated, and its defect causes tumor development. However, the mechanisms that underlie the control of p21 level are not fully understood. Here, we report a novel mechanism by which a component of the SCF ubiquitin ligase, Fbl12, augments p21 via the formation of atypical ubiquitin chains. We found that Fbl12 binds and ubiquitinates p21. Unexpectedly, Fbl12 increases the expression level of p21 by enhancing the mixed-type ubiquitination, including not only K48- but also K63-linked ubiquitin chains, followed by promotion of binding between p21 and CDK2. We also found that proteasome activator PA28γ attenuates p21 ubiquitination by interacting with Fbl12. In addition, UV irradiation induces a dissociation of p21 from Fbl12 and decreases K63-linked ubiquitination, leading to p21 degradation. These data suggest that Fbl12 is a key factor that maintains adequate intracellular concentration of p21 under normal conditions. Our finding may provide a novel possibility that p21's fate is governed by diverse ubiquitin chains.
INTRODUCTION
The inhibitor of cyclin-dependent protein kinase (CDK) p21Waf1/Cip1 (here referred to as p21) plays important roles in the regulation of cellular functions such as the cell cycle, DNA repair, and apoptosis. The relationship between p21 functions and cellular progression has been extensively studied (1). Normally, p21 inhibits CDKs through direct binding, leading to the suppression of cellular progression. p21 also suppresses proliferating cell nuclear antigen (PCNA), which is a crucial factor for DNA replication and repair. Thus, p21 acts as a master modulator that governs the cell cycle, and defects in p21 increase the risk of developing cancer.
p21 abundance is tightly controlled at all stages from transcription and translation to posttranslational regulation via such processes as mRNA clearance, translational rate, and protein degradation. Defects in these mechanisms that result in aberrant p21 levels can cause cancer development (1, 2). This suggests that intracellular p21 abundance is intrinsically determined through the regulation of transcriptional and posttranslational modification. The transcriptional regulation of p21 is mediated by a variety of factors, such as p53, E2F1, Klf6, Myc, and AP4 (3–7), indicating that transcriptionally regulated p21 expression is governed by several pathways in a context-dependent manner. In addition, posttranslational modification is also associated with cellular p21 protein levels. Recent studies have determined that p21 phosphorylation at several sites is involved in its stability. For instance, Akt phosphorylates p21 at the Thr145 residue, resulting in p21 accumulation in the cytoplasm (8–10), and JNK1 and p38α stabilize p21 through phosphorylation of Ser130 (11, 12). Therefore, it is clear that p21 is controlled not only by transcriptional regulation but also by posttranslational modification.
p21 is an unstructured protein that is easily degraded by the proteasome under basal conditions (13). In order to protect itself from this default degradation, newly synthesized p21 associates with target proteins to prevent this default degradation (14–16). It has also been shown that carcinogenic factors, including exposure to UV, change expression of p21. Interestingly, substantial studies have shown that increased p21 expression significantly associates with metastasis, recurrence, and survival in humans (17, 18). Therefore, the maintenance of p21 abundance at an adequate level is critically important, and abnormal expression results in an increase of risk of various disorders (1). So far, it has been reported that several E3 ubiquitin ligases, including SCFskp2, Cul2LRR1, Cul4CDT2, APC/CCDC20, and MKRN1, enhance p21 ubiquitination, leading to proteasome degradation (14, 19–23). Interestingly, a p21 mutant with all lysines mutated to arginines is still ubiquitinated. This suggests that p21 ubiquitination occurs not only on intramolecular lysines but also on the N-terminal methionine (24). Moreover, p21 has been reported to interact with the proteasome α7 subunit of 20S proteasome and proteasome activator PA28γ (25–27) to promote proteasomal degradation of p21, independently of ubiquitination. Despite the topic of p21 degradation and ubiquitination, the physiological significance of p21 ubiquitination associated with its degradation remains unclear.
In this study, we report that Fbl12 and PA28γ regulate the p21 expression level. We found that Fbl12 associates with both p21 and PA28γ, resulting in a complex formation. In addition, mixed-type ubiquitination of p21 induced by Fbl12 is positively associated with the amount of p21 via attenuation of the degradation rate. In addition, this effect was suppressed by PA28γ, resulting in a decrease in p21 expression level. Furthermore, UV irradiation promotes p21 degradation irrespective of Fbl12 expression. Thus, our findings provide the novel mechanisms by which both Fbl12 and PA28γ mediate p21 turnover via the control of mixed-type ubiquitin chain.
MATERIALS AND METHODS
Materials.
The following antibodies were used for immunoblot analyses: Flag (M2; Sigma), Myc (9E10; Santa Cruz), hemagglutinin (HA; Y-11; Santa Cruz), green fluorescent protein (GFP; catalog number 598; MBL), glutathione S-transferase (GST; B-14; Santa Cruz), tubulin (DM1A; Sigma), His (GE Healthcare catalog number 27-4710-01), p21 (F-5; Santa Cruz), PA28γ (47/Psme3; BD Transduction Lab.), β5 (ab3330; abcam), Fbl12 (ab96831; abcam), Lys48-specific ubiquitin (Apu2; Merck Millipore), and Lys63-specific ubiquitin (Apu3; Merck Millipore). The following antibodies were used for immunocytochemistry: GFP (catalog number 598; MBL), p21 (F-5; Santa Cruz), and PA28γ (47/Psme3; BD Transduction Lab.). Lipofectamine 2000 was purchased from Invitrogen. The siFbl12 (EHU054981) and anti-Flag M2–agarose beads were purchased from Sigma. Hoechst 33342 was purchased from Life Technologies. Talon metal affinity resin was purchased from Clontech. Glutathione-Sepharose was purchased from GE Healthcare. MG132 was purchased from the Peptide Institute. Polyethyleneimine was purchased from Polyscience. Cycloheximide (CHX) was purchased from Wako.
Cell culture, transfection, and stimulation.
HEK 293, HEK 293T, HeLa, and HCT116 cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Wako) containing 5 to 10% fetal bovine serum as well as penicillin (100 units) and streptomycin (100 mg) (P/S). To generate Fbxl12-deficient cells, HEK 293 cells were transfected with pSpCas9(BB)-2A-Puro-Fbl12 plasmid and incubated in the presence of 1.0 μg/ml of puromycin. Cells were transfected using Lipofectamine 2000, Lipofectamine RNAi MAX reagent (Life Technologies), and polyethyleneimine (Polyscience) according to the manufacturers' instructions. HEK 293 and HeLa cells were irradiated by 12-μW/cm2 UV and then subjected to each analysis.
Plasmid construction.
pCAGEN-His-Ub, pCAGEN-His-Ub K48R, pCAGEN-His Ub K63R, pCAGEN-His-Ub 48K, and pCAGEN-His-Ub 63K constructs were provided by Y. Gotoh (University of Tokyo, Japan). pcDNA3-Flag-Fbl12, pcDNA3-Flag-Fbl12ΔF, and pcDNA3-Flag-Skp1 constructs were described previously (28). p21 and Skp2 cDNAs were amplified by PCR and subcloned into pcDNA-Flag. The Fbl12 and Skp1 cDNAs were subcloned into pRSFDuet-1. The Fbl12 cDNA was subcloned into the EcoRI and XhoI sites of pCS4-Myc and pCS4-EGFP (where EGFP is enhanced GFP). The PA28γ and CDK2 cDNAs were amplified from the human HCT116 cDNA library and subcloned into the BglII sites of pCS4-Myc. The p21 cDNA was amplified from Flag-p21 and subcloned into the BglII sites of pCS4 and pCS4-EGFP. The p21ΔNLS cDNA was amplified from Flag-p21 and subcloned into the BglII sites of pCS4-EGFP. The p21 NLS oligonucleotides were annealed and inserted into the EcoRI and XbaI sites of pCS4-EGFP. The p21 and PA28γ cDNA were subcloned into the BamHI sites of pGEX-6p-1. pSpCas9(BB)-2A-Puro was purchased from Addgene. The oligonucleotides of Fbl12 were designed and inserted into pSpCas9(BB)-2A-Puro by ligation into the BbsI sites. The primers used were as follows: PA28γ forward, 5′-AGGATCCGCCACCATGGCCTCGTTGCTGAAGGTG-3′, and reverse, 5′-GGGATCCTCAGTACAGAGTCTCTGCATTGCTGCTCCG-3′; p21 forward, 5′-TAGGATCCGCCACCATGTCCAATCCTGGTGATG-3′, and reverse, 5′-GCGGATCCTCAGGGTTTTCTCTTGCAGAAGACC-3′; p21ΔNLS forward, 5′-TAGGATCCGCCACCATGTCCAATCCTGGTGATG-3′, and reverse, 5′-TAGGATCCTCATCGGCCCTGAGATGTTCCGG-3′; and CDK2 forward, 5′-GGGATCCGCCACCATGGAGAACTTCCAAAAGG-3′, and reverse, 5′-AGGATCCTCAGAGTCGAAGATGGGGTACTGGCTTGG-3′. The following oligonucleotides of p21 NLS were used: sense, 5′-TAATTCAAACGGAGGCAGACCAGCCTGACAGATTTCTATCACTCCAAGCGCAGATTGGTCTTCTGCAAGAGAAAACCCTGAT-3′, and antisense, 5′-CTAGATCAGGGTTTTCTCTTGCAGAAGACCAATCTGCGCTTGGAGTGATAGAAATCTGTCAGGCTGGTCTGCCTCCGTTTG-3′. The following oligonucleotides of Fbl12 for CRISPR/Cas9 were used: sense, 5′-CACCGACCTGACGCTCTACACGATG-3′, and antisense, 5′-AAACCATCGTGTAGAGCGTCAGGTC-3′.
RT-qPCR.
Total RNAs were prepared by Isogen II (Nippon Gene). The cDNA were synthesized by SuperScript III reverse transcriptase (Life Technologies). Quantitative reverse transcription-PCR (RT-qPCR) was performed with the Thunderbird SYBR qPCR mix (Toyobo) and appropriate primers and then analyzed by using the Thermal Cycler Dice Real Time system (TaKaRa). The primers used were as follows: Fbl12 forward, 5′-CGGTGGCTGTGGCGACATGTC-3′, and reverse, 5′-CAGGTAGCCACCCATCCGCA-3′; p21 forward, 5′-TACCCTTGTGCCTCGCTCAG-3′, and reverse, 5′-GGAGAAGATCAGCCGGCGTT-3′; and actin forward, 5′-TGGACATCCGCAAAGACCTG-3′, and reverse, 5′-GGAGGAGCAATGATCTTGATCTTC-3′.
Immunoblot analysis.
Cells were lysed in extraction buffer (0.5% NP-40, 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM, EDTA, 1 mM dithiothreitol [DTT]) and centrifuged at 14,000 rpm for 5 min. The cleared lysates were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane, probed with primary antibodies, and detected with horseradish peroxidase (HRP)-conjugated secondary antibodies and chemiluminescence reagent (ECL Plus Western blotting detection reagents; GE Healthcare). Immunoblotting data were quantified using ImageJ software.
Coimmunoprecipitation and MS analysis.
The cell lysates (see “Immunoblot analysis” above) were mixed with either anti-Flag–agarose beads (Sigma) or protein G-agarose beads (Thermo Scientific) containing either anti-Myc (9E10; Santa Cruz) or anti-GFP (catalog number 598; MBL) antibodies for 3 h at 4°C. The immunoprecipitants were washed and subjected to immunoblot analysis with the indicated antibodies. All experiments have been performed more than twice, and data are reproducible. The mass spectrometry analysis has been described previously (29). Briefly, Flag-tagged Fbl12 was expressed in HEK 293 cells, immunoprecipitated using anti-Flag antibody, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. These analyses were performed four times. All proteins identified by MS analyses in every experiment are listed in Table 1.
TABLE 1.
Proteins identified by MS analysis of Fbl12 immunoprecipitates in this study
| Protein name | Description |
|---|---|
| PSME3 (PA28γ) | Proteasome activator subunit 3; isoform 1 |
| SKP1 | S-phase kinase-associated protein 1; isoform b |
| CUL1 | Cullin 1 |
| TCEB1 | Elongin C |
| UBR5 | Ubiquitin protein ligase E3 component n-recognin 5 |
| AKAP8L | A kinase anchor protein 8-like |
| PPM1G | Protein phosphatase 1g |
| PPP5C | Protein phosphatase 5, catalytic subunit |
| PRKAR1A | Cyclic AMP-dependent protein kinase, regulatory subunit alpha 1 |
| GPRASP2 | G protein-coupled receptor associated sorting protein 2 |
| IPO5 | Importin 5 |
| NAP1L1|NAP1L4 | Nucleosome assembly protein 1-like 1 or 4 |
| NAP1L4 | Nucleosome assembly protein 1-like 4 |
| DDB1 | Damage-specific DNA binding protein 1 |
| TIMM13 | Translocase of inner mitochondrial membrane 13 |
| NIP30 | Hypothetical protein LOC80011 |
| RNF219 | Ring finger protein 219 |
| SAPS3 | SAPS domain family, member 3 |
| LOC152667|NIP30 | NIP30-like family |
| TTC9C | Tetratricopeptide repeat domain 9C |
| FKBP5 | FK506 binding protein 5 |
| MAGED1 | Melanoma antigen family D, 1 |
| CACYBP | Calcyclin binding protein; isoform 2 |
| CALM1|CALM2|CALM3 | Calmodulin 1 or 2 or 3 |
| CALM1|CALM2|CALM3|CALML3 | Calmodulin 1 or 2 or 3 or calmodulin-like 3 |
| CALU | Calumenin |
| RCN2 | Reticulocalbin 2, EF-hand calcium binding domain |
GST pulldown assay.
The recombinant GST, GST-p21, GST-PA28γ, and His-Fbl12/Skp1 were purified from Escherichia coli BL21-Gold (DE3). The recombinant proteins were mixed with glutathione-Sepharose beads (GE Healthcare) in buffer (0.5% NP-40, 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM DTT) and incubated for 3 h at 4°C. The precipitants were washed and subjected to immunoblot analysis with the indicated antibodies.
His tag pulldown assay.
Cells were transfected with the indicated constructs and lysed in extraction buffer (6 M guanidinium-HCl, 50 mM sodium phosphate buffer [pH 8.0], 300 mM NaCl, and 5 mM imidazole). Cell lysates were sonicated briefly and were then incubated with Talon metal affinity resin (Clontech) for 4 h at 4°C. The precipitants were washed with buffer (50 mM sodium phosphate buffer [pH 8.0], 300 mM NaCl, and 5 mM imidazole) and then subjected to immunoblot analysis. All experiments have been performed more than twice, and data are reproducible.
Immunocytochemistry.
HeLa cells plated on 15-mm coverslips and grown in 12-well plates were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature. The coverslips were washed in PBS, blocked with 5% bovine serum albumin (BSA) in PBS with 0.4% Triton X-100, and then incubated with the indicated primary antibodies for 1 h at room temperature or overnight at 4°C. Following a PBS wash, samples were incubated with secondary antibodies (Alexa Fluor 594–anti-mouse IgG [1:500], Alexa Fluor 488–anti-rabbit IgG [1:500]) for 30 min at room temperature in blocking solution. Cells were imaged using a fluorescence microscope (Biorevo BZ-9000; Keyence). The images were quantified by using ImageJ software and Prism (GraphPad Software).
Cell proliferation assay.
HEK 293 cells were transfected with either plasmid. The transfected cells were plated on 96-well plates and incubated for 4 days. Cellular proliferation was analyzed using microplate reader model 680 (Bio-Rad) and the Cell Counting Kit-8 (Dojindo) according to the manufacturers' instructions.
RESULTS
Fbl12 promotes p21 ubiquitination.
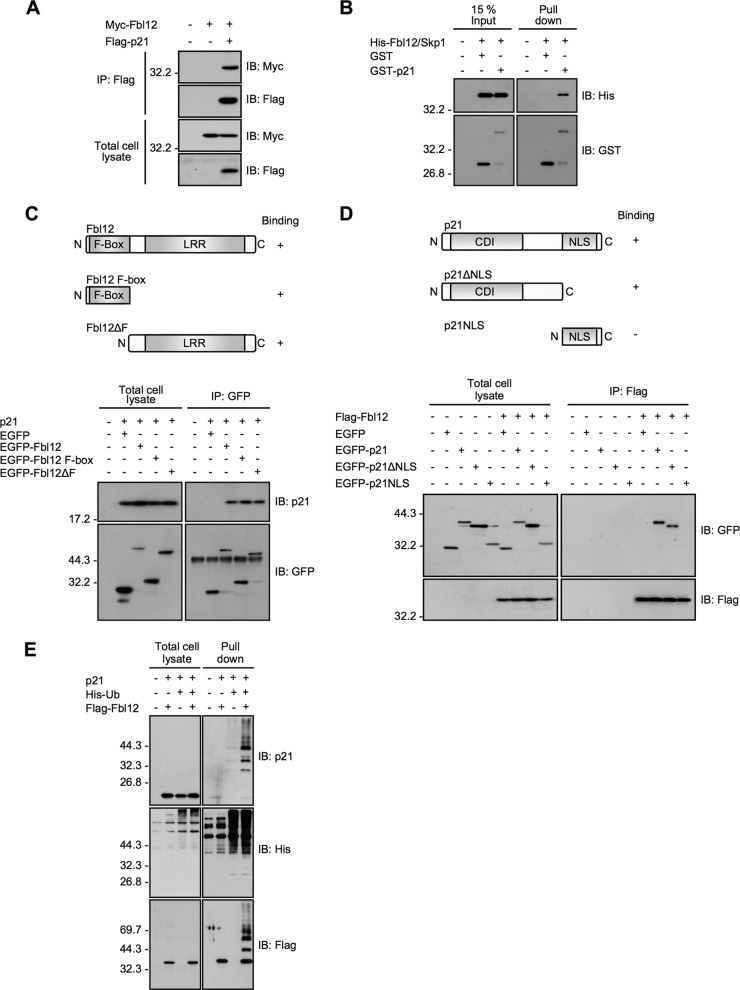
A previous study has shown that Fbl12 binds and ubiquitinates p57Kip2 during transforming growth factor β1 (TGF-β1)-mediated inhibition of osteoblastic cell differentiation (28). It has also been reported that SCFFbl12 enhances ubiquitination of target proteins implicated in the cell cycle, DNA repair, and embryonic differentiation (28, 30–32). In addition, recent large-scale screening identified a somatic mutation of Fbl12 in renal carcinoma (33). Thus, it is likely that SCFFbl12 regulates cellular proliferation and differentiation; however, it has been unclear whether SCFFbl12 mediates ubiquitination of another Cip/Kip protein, p21. To examine whether Fbl12 interacts with p21, we first introduced Flag-p21 and Myc-Fbl12 plasmids into HEK 293T cells. Immunoprecipitation of Flag-p21 resulted in coimmunoprecipitation of Myc-Fbl12 (Fig. 1A). We then tested whether this interaction is direct. To investigate this, we purified the recombinant proteins and performed the GST pulldown assay. As it is difficult to solubilize the recombinant Fbl12, we copurified Fbl12 with Skp1, which is reported to stabilize the conformation of F-box proteins (34). GST-p21 binds to His-Fbl12/Skp1 proteins efficiently in vitro (Fig. 1B). These data cannot rule out the possibility that p21 associates with Skp1; however, we speculate that p21 binds to Fbl12 directly since p21 is capable of interaction with Fbl12ΔF (see below). To further examine which region of Fbl12 is responsible for this interaction, we used the deletion mutants of Fbl12. Unexpectedly, the two deletion mutants that lacked the leucine-rich repeat and F-box region were capable of interacting with p21 (Fig. 1C), suggesting that p21 associates with both F-box and leucine-rich repeat regions. Next, to map the binding site of p21, we subdivided p21 into two fragments. p21ΔNLS interacted with Fbl12; on the other hand, the NLS region of p21 could not associate with Fbl12. These data suggest that the CDK inhibitor domain (CDI) and linker region are responsible for this interaction (Fig. 1D). As Fbl12 forms the SCF ubiquitin ligase complex, we next sought to determine if expression of Fbl12 promotes p21 ubiquitination. To examine this, we transfected cells with histidine-tagged ubiquitin (His-Ub) and precipitated ubiquitinated proteins using Talon metal affinity resins under denatured conditions. The amount of ubiquitinated p21 was markedly increased when Fbl12 was expressed (Fig. 1E), suggesting that SCFFbl12 promotes p21 ubiquitination in cells.
FIG 1.
Fbl12 promotes p21 ubiquitination. (A) Coimmunoprecipitation of Myc-Fbl12 and Flag-p21 in HEK 293T cells. (B) GST pulldown of His-Fbl12/Skp1 and either GST or GST-p21. The precipitated proteins were analyzed by immunoblotting. (C, top) Schematic structure of Fbl12. F-Box, F-box domain; LRR, leucine-rich repeat. (Bottom) Coimmunoprecipitation of p21 and either EGFP-Fbl12, EGFP-Fbl12 F-box, or EGFP-Fbl12ΔF. (D, top) Schematic structure of p21. CDI, CDK inhibitor domain; NLS, nuclear localization signal. (Bottom) Coimmunoprecipitation of Flag-Fbl12 and either EGFP-p21, EGFP-p21ΔNLS, or EGFP-p21NLS. (E) Ubiquitinated proteins were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting.
Fbl12 increases p21 expression levels associated with mixed-type ubiquitination.
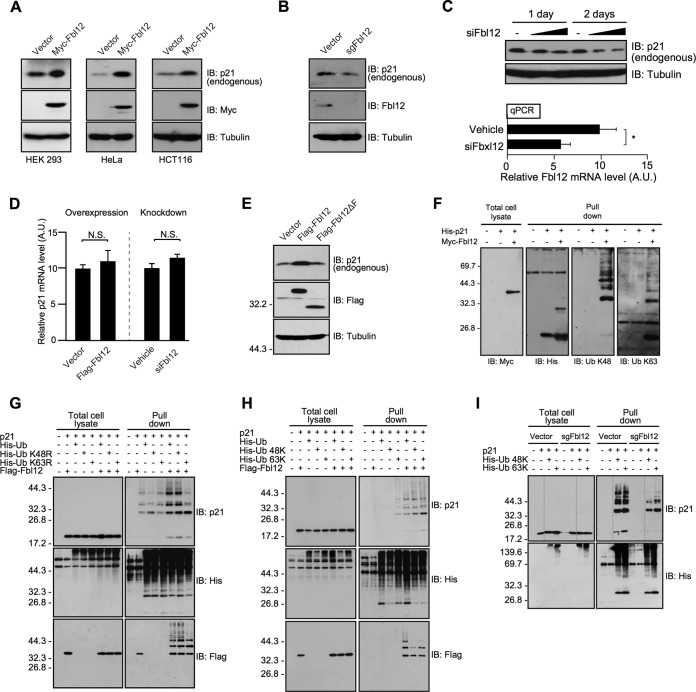
Since it is known that the ubiquitination plays important roles in the selective protein degradation, we examined whether Fbl12 controls p21 expression level via the protein degradation. Unexpectedly, overexpression of Fbl12 in HEK 293, HeLa, and HCT116 cells increased the amount of endogenous p21. These data imply that Fbl12 positively regulates p21 abundance in cells (Fig. 2A). We next investigated whether Fbl12 is necessary for an upregulation of p21 under normal conditions. To investigate this, we developed a clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 construct, which produces single-guide Fbl12 RNA (sgFbl12) (35), and disrupted the Fbxl12 gene by sgFbl12 construct. The endogenous p21 is slightly reduced in Fbxl12-deficient cells, suggesting that Fbl12 affects the amount of p21 in cells (Fig. 2B). To further confirm whether Fbl12 is involved in the intracellular p21 level, we used an endoribonuclease-prepared small interfering RNA (siRNA) pool (siFbl12). The knockdown of endogenous Fbl12 by siFbl12 seemed to decrease the amount of p21 (Fig. 2C). We next sought to determine whether Fbl12 regulates either p21 transcription or mRNA stability in cells. To test this, we quantified the p21 mRNA levels using qRT-PCR. The amount of p21 mRNA was not significantly affected by either overexpression or knockdown of Fbl12, implying that Fbl12 has little effect on the regulation of mRNA levels (Fig. 2D). Next, to investigate whether the increase in the level of p21 was dependent on the SCFFbl12 ubiquitin ligase activity, we used the Flag-Fbl12ΔF mutant, which is unable to form a functional SCF complex. Expression of Fbl12 clearly increased p21 expression level. On the other hand, Fbl12ΔF did not substantially increase p21 compared to full-length Fbl12 (Fig. 2E), although Fbl12ΔF is capable of interacting with p21 (Fig. 1C). Therefore, it is likely that upregulation of p21 by Fbl12 is dependent on its ubiquitination. Our data raise the question of why the p21 expression level was increased despite its ubiquitination. To answer this question, we analyzed the linkage mode of the polyubiquitin chain using linkage-specific antibodies. We transfected His-p21 into HEK 293 cells, precipitated it by affinity resins, and followed this with detection of ubiquitin linkage by immunoblot analysis. Interestingly, expression of Fbl12 enhanced not only K48-linked but also K63-linked ubiquitination, suggesting that SCFFbl12 promotes a mixed type of ubiquitination. To further confirm this, we used mutant ubiquitin. Consistent with results shown in Fig. 2F, expression of His-Ub K48R, of which residue Lys48 is mutated to arginine, promoted p21 ubiquitination as well as wild-type (WT) His-Ub (Fig. 2G). These data suggest that Fbl12 has an effect on the formation of not only the K48-linked ubiquitin chains but also other linkage modes. Interestingly, His-Ub K63R slightly but significantly attenuated this ubiquitination (Fig. 2G). In addition, expression of Ub 48K and 63K, of which all lysine residues are mutated to arginine except at Lys48 and Lys63, respectively, formed a polyubiquitin chain on p21 by expression of Fbl12 (Fig. 2H). Furthermore, both 48K- and 63K-like ubiquitinations of p21 are slightly decreased in Fbxl12-deficient cells (Fig. 2I). These results suggest that expression of Fbl12 promotes the formation of a mixed type of polyubiquitin chain containing both K48 and K63 linkage modes, leading to an increase of p21 expression.
FIG 2.
Fbl12 increases p21 expression levels associated with mixed-type ubiquitination. (A) HEK 293, HeLa, and HCT116 cells were transfected with either Myc-Fbl12 or control vector. Cell lysates were subjected to immunoblot analysis. (B) Fbxl12-deficient cell lysates were analyzed by immunoblotting. (C, top) HEK 293 cells were treated with siFbl12 (200 ng/ml or 400 ng/ml) and incubated for 1 or 2 days. Cell lysates were subjected to immunoblot analysis. (Bottom) The qPCR analysis of Fbl12 mRNA in esiRNA-treated HEK 293 cells. Results were normalized to actin expression (n = 3; data are means ± standard errors of the means [SEM]; *, P < 0.05 by Student's t test). A.U., arbitrary units. (D) qPCR analysis of p21 mRNA in either Flag-Fbl12-expressing cells (left) or siFbl12-treated cells (right). Results were normalized to actin expression (n = 3; means ± SEM; N.S., not significant by Student's t test). (E) HEK 293 cells were transfected with either Flag-Fbl12 or Flag-Fbl12ΔF. Cell lysates were subjected to immunoblot analysis. (F) His-p21 were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting. (G and H) Ubiquitinated proteins were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting. (I) Ubiquitinated proteins were purified from denatured Fbxl12-deficient cell lysates using Talon metal affinity resin and analyzed by immunoblotting.
Fbl12 suppresses default degradation of p21.
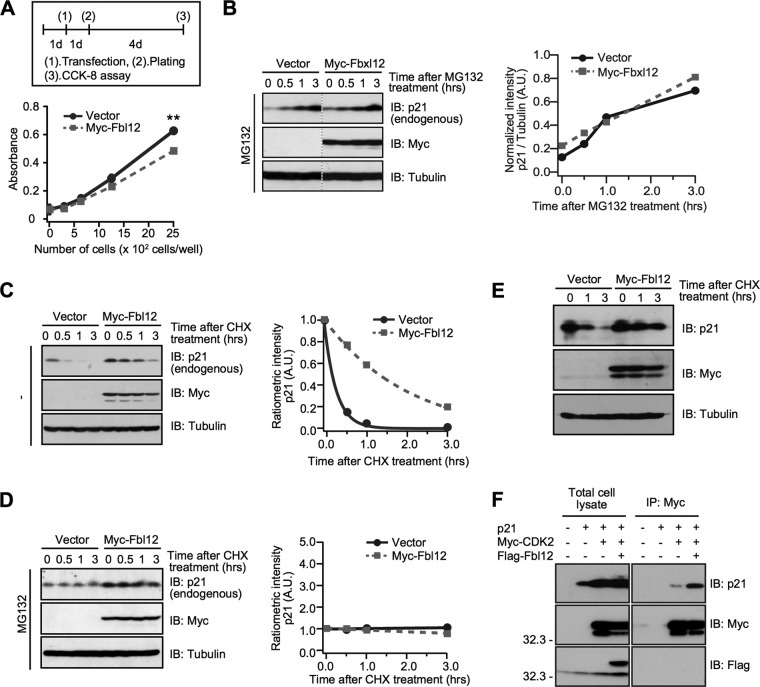
As p21 is known to be a CDK inhibitor, we investigated whether Fbl12 impedes the cellular proliferation. As we expected, Fbl12 expression slightly but significantly delayed the proliferation (Fig. 3A), suggesting that Fbl12-induced p21 upregulation is involved in cell growth. Next, to examine the mechanisms of how Fbl12 augments the expression level, we analyzed the synthesis rate using a proteasome inhibitor. Interestingly, the amount of endogenous p21 was clearly increased after treatment with MG132. Moreover, Fbl12 expression had little effect on the synthesis rate (Fig. 3B), suggesting that the p21 synthesis rate is rapid under basal conditions independently of Fbl12. We then examined the degradation rate of endogenous p21. The half-life of p21 in Fbl12-expressing cells was clearly extended compared to that in control cells (Fig. 3C). In addition, p21 degradation was completely blocked by treatment with proteasome inhibitor, MG132, regardless of Fbl12 expression (Fig. 3D), demonstrating that Fbl12 attenuates the proteasome-dependent p21 degradation. We further confirmed whether Fbl12 affects the half-life of overexpressed p21. Overexpression of Fbl12 attenuates the degradation rate of exogenous p21. Moreover, Fbl12 did not affect the upper limit of overexpressed p21 (Fig. 3E), implying that an upper limit of p21 expression is already dictated innately. Probably, overexpression of p21 reaches the saturation condition in cells. Next, we investigated the mechanisms of how Fbl12 regulates the amount of p21. Previously, it has been reported that the p21 binding proteins block the default degradation after new synthesis of p21 (14–16). Additionally, Fbl12 negatively regulates cellular proliferation. Therefore, we hypothesized that Fbl12 modulates the binding affinity of p21 with a target protein involved in the cell cycle, leading to a delay of degradation. To examine this, we tested whether Fbl12 regulates the interaction between p21 and CDK2, which is one of the targets of p21 (36). Interestingly, expression of Fbl12 clearly enhanced their binding ability (Fig. 3F), demonstrating that the mixed-type ubiquitination synthesized by Fbl12 promotes their interaction. This finding may support the idea that binding proteins, such as CDK2, protect p21 from proteasome-dependent degradation.
FIG 3.
Fbl12 suppresses default degradation of p21. (A) HEK 293 cells were transfected with either control or Myc-Fbl12 plasmids. The cellular proliferation was analyzed using the Cell Counting Kit-8 (n = 3; means ± SEM; **, P < 0.01 by Student's t test). (B, left) HEK 293 cells were transfected with either Myc-Fbl12 or control vector. Cells were incubated with 20 μM MG132 for the indicated times and were then subjected to immunoblot analyses. (Right) Data were quantified using ImageJ software. These data are representative of three independent experiments. (C, left) HEK 293 cells were transfected with either Myc-Fbl12 or control vector. Cells were incubated in the absence or presence of 50 μg/ml cycloheximide (CHX) for the indicated times and were then subjected to immunoblot analyses. (Right) Data were quantified by using ImageJ software. These data are representative of three independent experiments. (D, left) HEK 293 cells were transfected with either Myc-Fbl12 or control vector. Cells were incubated in the absence or presence of 50 μg/ml CHX together with 20 μM MG132 for the indicated times and were then subjected to immunoblot analyses. (Right) Data were quantified using ImageJ software. These data are representative of three independent experiments. (E) HEK 293 cells were transfected with the indicated vectors. Cells were incubated in the absence or presence of 50 μg/ml CHX for the indicated times and were then subjected to immunoblot analyses. (F) Coimmunoprecipitation of Flag-Fbl12, p21, and Myc-CDK2 in HEK 293 cells. Ectopic expression of Fbl12 promoted the association of p21 with CDK2.
PA28γ associates with SCFFbl12.
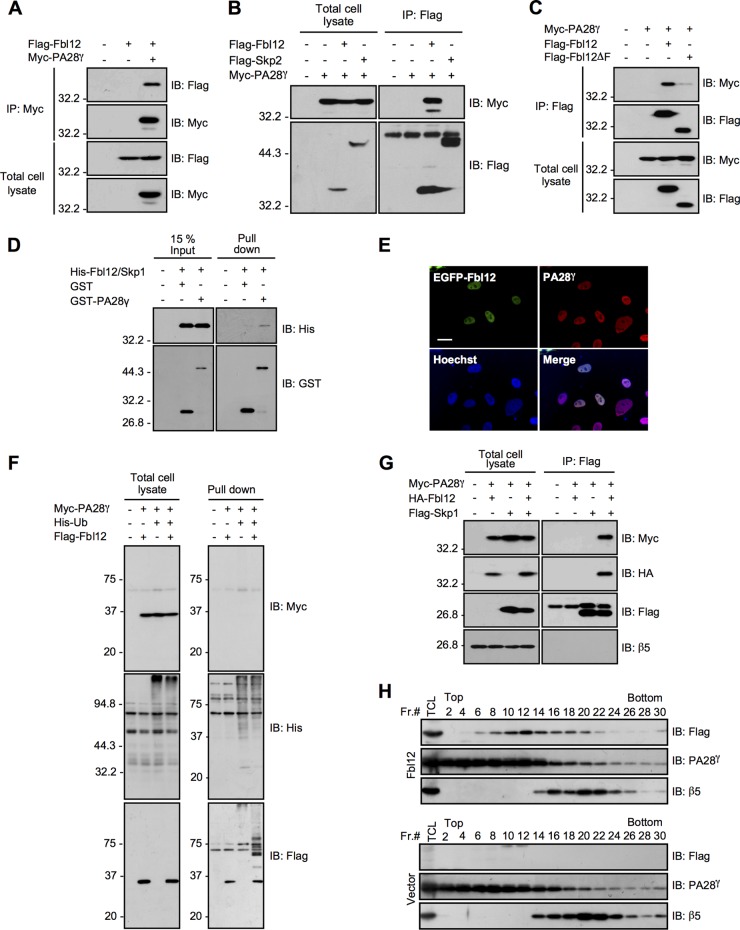
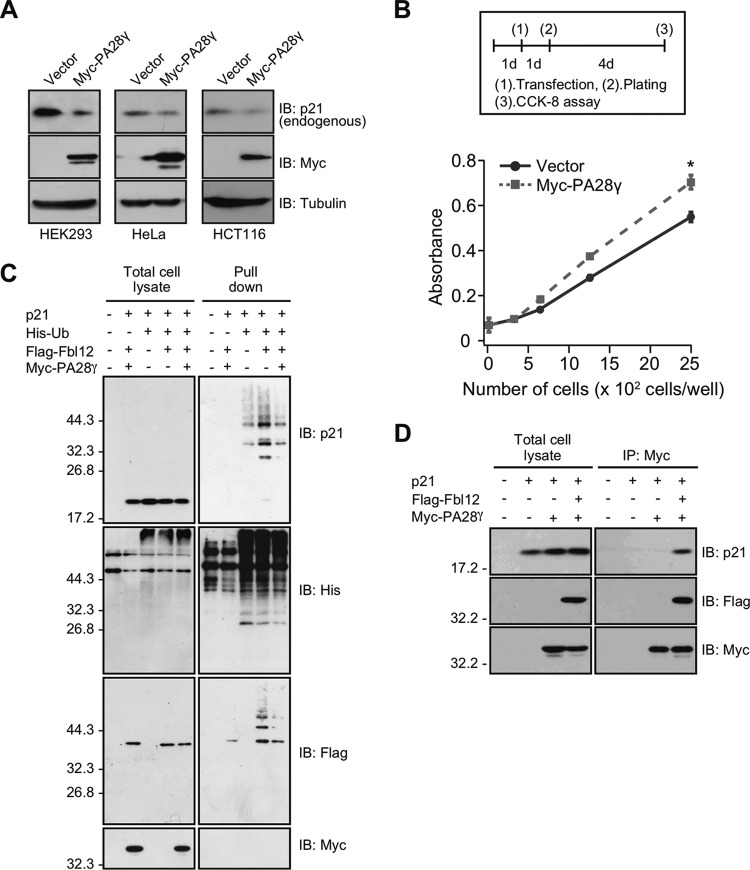
To understand the precise mechanisms by which SCFFbl12 governs the amount of p21 in cells, we performed proteomics analysis by using an LC-MS/MS system to identify the other Fbl12 binding proteins. Consequently, we identified 27 proteins as Fbl12-binding targets (Table 1). One of the most interesting proteins was PA28γ. PA28γ forms a homohexameric ring and acts as a proteasome activator (37). Recent studies have reported that PA28γ directly associates with p21, leading to p21 degradation independently of ubiquitination (25, 26). Thus, it is likely that PA28γ is involved in the control of p21 expression levels in concert with Fbl12. Before pursuing this hypothesis, we verified the interaction between Fbl12 and PA28γ. Myc-PA28γ was significantly associated with Flag-Fbl12 (Fig. 4A). In the reverse experiment, immunoprecipitation of Flag-Fbl12 but not of Flag-Skp2 (a structural homologue of Fbl12) caused coimmunoprecipitation of Myc-PA28γ (Fig. 4B). We next determined the binding region using deletion mutants. We found that the F-box domain of Fbl12 is required for this interaction (Fig. 4C). Furthermore, recombinant GST-PA28γ was associated with His-Fbl12/Skp1 in vitro, suggesting that PA28γ is able to bind Fbl12 directly (Fig. 4D). To ask whether these proteins colocalize in culture cells, we performed immunocytochemistry after ectopic expression of EGFP-Fbl12. Both EGFP-Fbl12 and endogenous PA28γ colocalized in the nucleus, supporting the finding that Fbl12 associates with PA28γ in the same subcellular region (Fig. 4E). We then examined whether SCFFbl12 ubiquitinates PA28γ as well as p21. The ubiquitination level of Myc-PA28γ was comparable with that of the control sample, indicating that SCFFbl12 does not enhance ubiquitination of PA28γ (Fig. 4F). As the F-box domain, which is essential to associate with Skp1 and form the SCF ubiquitin ligase complex, was important to interact with PA28γ (Fig. 4C), we next asked whether Skp1 and the PA28γ-20S proteasome are part of the same protein complex. When HA-Fbl12 was expressed in cells, Myc-PA28γ was coprecipitated with Flag-Skp1. However, β5, which is a core subunit of the 20S proteasome, was not included in this complex (Fig. 4G). These results suggest that SCFFbl12 is associated with PA28γ, which is free from the 20S proteasome. To further confirm this observation, we performed a glycerol density gradient centrifugation. Most PA28γ was found in the lighter fraction, and a small concentration of PA28γ was found in the heavier fraction, including the β5 subunit. Intriguingly, the fractions containing Fbl12 were different from β5 (Fig. 4H), supporting the idea that SCFFbl12 associates with a certain amount of PA28γ, which is free from the 20S proteasome.
FIG 4.
PA28γ associates with SCFFbl12. (A) Coimmunoprecipitation of Flag-Fbl12 and Myc-PA28γ. (B) Coimmunoprecipitation of Flag-tagged F-box proteins (Fbl12 or Skp2) and Myc-PA28γ. (C) Coimmunoprecipitation of Myc-PA28γ and either Flag-Fbl12 or Flag-Fbl12ΔF. (D) GST pulldown of His-Fbl12/Skp1 and either GST or GST-PA28γ. The precipitated proteins were analyzed by immunoblotting. (E) Fluorescent images of HeLa cells expressing EGFP-Fbl12. Cells were stained with anti-GFP and anti-PA28γ antibodies. Bar, 20 μm. (F) Ubiquitinated proteins were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting. (G) Coimmunoprecipitation of Flag-Skp1, HA-Fbl12, and Myc-PA28γ in HEK 293T cells. (H) Cell lysates separated by a glycerol density gradient centrifugation were subjected to immunoblot analysis. Flag-Fbl12 and PA28γ form a complex smaller than 20S.
PA28γ attenuates SCFFbl12-dependent p21 ubiquitination.
Previous studies have reported that PA28γ-20S proteasome promotes p21 degradation independently of ubiquitination. Also, it has been reported that the defects in PA28γ delay the degradation rate under normal conditions (25, 26). To verify these results, we assessed if PA28γ mediates both p21 degradation and amplification, at least in our system. We found that overexpression of PA28γ decreased p21 expression levels in several cell lines (Fig. 5A) and attenuated cellular proliferation (Fig. 5B), demonstrating that PA28γ is involved in regulating cellular proliferation via p21 expression level. Since p21 ubiquitination regulated by Fbl12 is implicated in expression level and proliferation, we investigated if PA28γ has an effect on SCFFbl12-induced p21 ubiquitination. As we described before, overexpression of Fbl12 increased p21 ubiquitination. However, this effect was suppressed by PA28γ (Fig. 5C), suggesting that PA28γ attenuates SCFFbl12-induced p21 ubiquitination. These observations prompted us to investigate whether PA28γ controls p21 expression via incorporation into the SCFFbl12 complex. To assess this, we performed immunoprecipitation assays. In contrast to the previous research, PA28γ did not show a clear binding to p21. However, this binding was dramatically increased when Fbl12 was coexpressed in cells (Fig. 5D), implying that PA28γ decreases p21 expression associated with the regulation of SCFFbl12.
FIG 5.
PA28γ attenuates SCFFbl12-induced p21 ubiquitination. (A) HEK 293, HeLa, and HCT116 cells were transfected with either Myc-PA28γ or control vector. Cell lysates were subjected to immunoblot analysis. (B) HEK 293 cells were transfected with either control or Myc-Fbl12 plasmids. Cellular proliferation was analyzed using the Cell Counting Kit-8 (n = 3; means ± SEM; *, P < 0.05 by Student's t test). (C) Ubiquitinated proteins were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting. Expression of PA28γ reduced SCFFbl12-induced p21 ubiquitination. (D) Coimmunoprecipitation of Flag-Fbl12, p21, and Myc-PA28γ in HEK 293T cells. Ectopic expression of Fbl12 promoted the association of p21 with PA28γ.
UV stimulation induces p21 degradation through disassembly of protein complex.
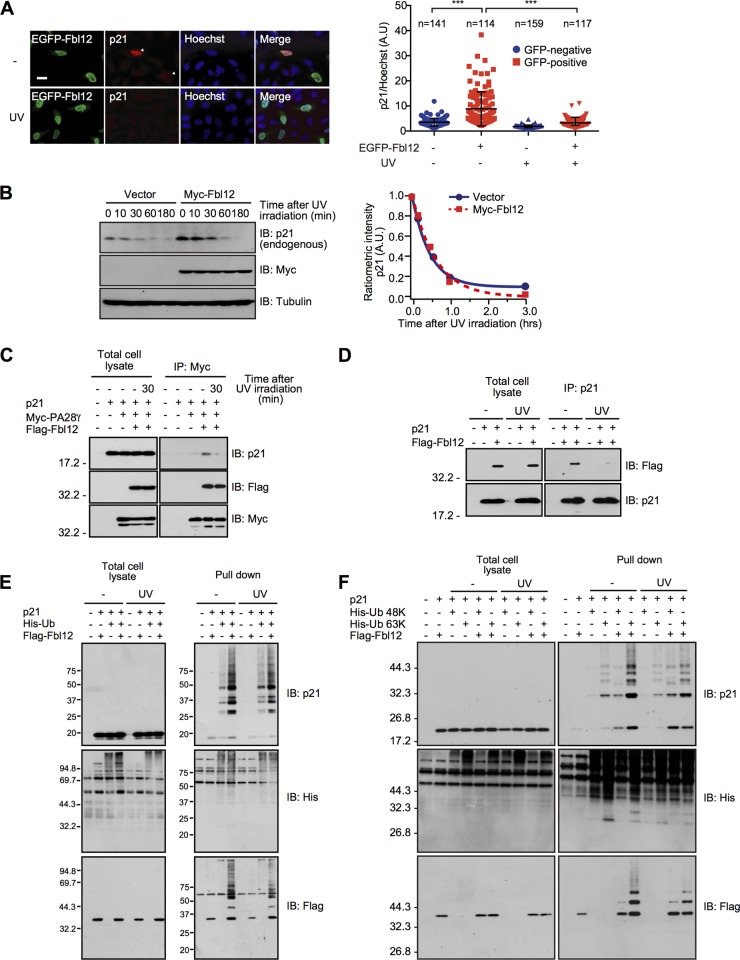
Previous study has shown that UV irradiation induces p21 expression through p53 activation (3). On the other hand, several studies have shown that UV irradiation triggers rapid degradation of p21 (38–41). Thus, we examined whether Fbl12 suppresses UV-induced p21 turnover. Stimulation with UV for 1 h markedly decreased the amount of endogenous p21 even in the presence of Fbl12 expression in HeLa cells (Fig. 6A). In addition, Fbl12 had little effect on the degradation rate of p21 in response to UV irradiation (Fig. 6B). Meanwhile, we did not observe a decrease in the amount of overexpressed p21 by the UV irradiation in our system (Fig. 6C to F). Probably, the p21 synthesis rate through an expression vector is higher than through an endogenous p21 promoter. Taken together, our data suggest that expression of Fbl12 does not attenuate UV-induced p21 degradation. The question arises as to why UV irradiation promotes p21 degradation despite the existence of Fbl12. To answer this question, we investigated whether UV stimulation alters the protein complex status. Consistent with our data shown in Fig. 5D, Fbl12 promotes an association of p21 with PA28γ. On the other hand, this effect was attenuated by UV irradiation (Fig. 6C). As PA28γ remained bound to the SCFFbl12 complex after UV stimulation, we hypothesized that the binding ability between p21 and Fbl12 was decreased in response to UV stimulation. To confirm this idea, we conducted immunoprecipitation assays with or without UV irradiation. We found that p21 was released from Fbl12 after UV stimulation (Fig. 6D), suggesting that UV stimulation promotes disassembly of the PA28γ-SCFFbl12-p21 complex, resulting in promotion of p21 degradation. Finally, we examined whether UV stimulation regulates the ubiquitination status of p21 in our system. Stimulation with UV for 1 h had little effect on the change of p21 ubiquitination in the absence of Fbl12 expression (Fig. 6E). Interestingly, this stimulation slightly reduced the K63-linked ubiquitination even in the Fbl12-expressing cells (Fig. 6F), demonstrating that UV stimulation may attenuate Fbl12-induced K63-linked ubiquitination. Taken together, these data suggest that UV irradiation induces disassembly of protein complex and attenuates K63-linked ubiquitination, leading to rapid degradation of p21.
FIG 6.
UV stimulation induces p21 degradation through disassembly of protein complex. (A, left) Fluorescent images of HeLa cells expressing EGFP-Fbl12. Cells were stimulated by 12-μW/cm2 UV and were then subjected to immunocytochemistry using anti-GFP and anti-p21 antibodies. Arrowheads indicate Fbl12-expressing cells. Bar, 20 μm. (Right) Ratiometric measurement of p21 to Hoechst fluorescence observed in cells expressing EGFP-Fbl12 before and after stimulation with UV (n > 100; means ± SD; ***, P < 0.001 by one-way analysis of variance [ANOVA]). (B, left) HEK 293 cells were transfected with either Myc-Fbl12 or control vector. Cells were stimulated by 12-μW/cm2 UV and were then subjected to immunoblot analysis. (Right) Expression level was quantified using ImageJ software. (C) Coimmunoprecipitation of Flag-Fbl12, p21, and Myc-PA28γ in HEK 293 cells. UV irradiation promoted the dissociation of p21 from the Fbl12-PA28γ complex. (D) Coimmunoprecipitation of Flag-Fbl12 and p21 in HEK 293 cells. UV irradiation promoted the dissociation of p21 from Fbl12. (E and F) HEK 293 cells were stimulated with UV in the presence or absence of Fbl12 expression. Ubiquitinated proteins were purified from denatured cell lysates using Talon metal affinity resin and analyzed by immunoblotting.
DISCUSSION
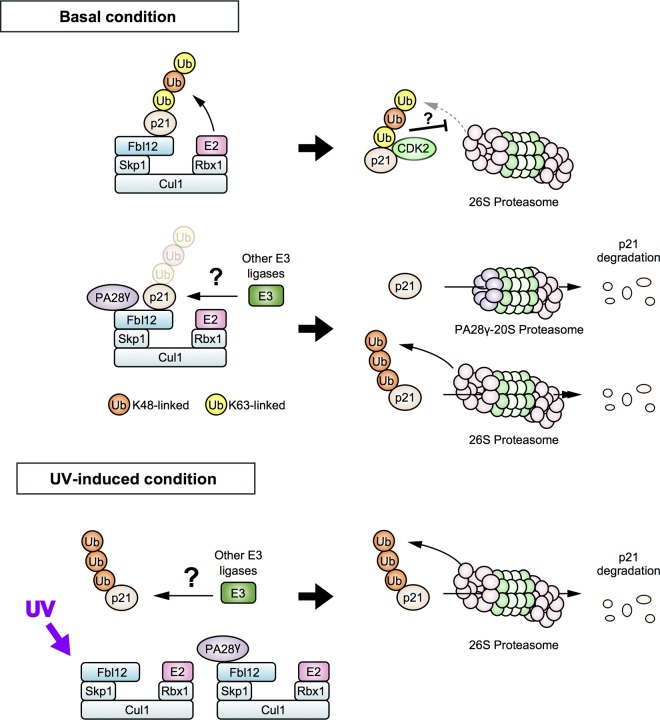
We have shown here that Fbl12 increases p21 expression levels through their interactions. SCFFbl12 enhances p21 mixed-type ubiquitination, and this effect was suppressed by PA28γ. Consequently, expression of Fbl12 extended the half-life of p21. Furthermore, UV stimulation promotes disassembly of the PA28γ-SCFFbl12-p21 complex and reduces mixed-type ubiquitination of p21, resulting in its degradation. These data demonstrate that Fbl12 controls the intracellular concentration of p21 and that Fbl12-induced mixed-type ubiquitination is a key event that prevents default degradation of p21 via binding proteins (Fig. 7).
FIG 7.
Model for the regulation of p21 turnover. (Top) SCFFbl12 promotes mixed-type ubiquitination of p21 and attenuates the default degradation under basal conditions, and this effect is cancelled by PA28γ. (Bottom) UV stimulation promotes disassembly of PA28γ-SCFFbl12-p21 complex and induces rapid degradation of p21. Another E3 ligase may ubiquitinate free p21 in cells.
Recent studies have shown that p21 associates with binding proteins immediately after the synthesis, resulting in its stabilization (14–16). Probably, this binding hampers K48-linked ubiquitination or recognition by proteasome. In our study, we found that SCFFbl12 enhances mixed-type ubiquitination, including K63 linkage ubiquitin chain, leading to an increase of p21 expression. Although the mechanisms inferred from our findings have not been clarified, several studies have posited a relationship between the SCF complex and the K63-linked ubiquitin chain. For example, one of the F-box proteins, Fbxl21, ubiquitinates CRY to control the circadian rhythms (42, 43). It was also reported that SCFFbxl21 mediates the formation of K63- and K11-linked polyubiquitin chains, implying that the linkage modes of ubiquitin chains might be a determinant of CRY stability (42). Additionally, SCFβ-TrCP competes with SCFFbxw7 and leads to K63-linked ubiquitin chain conjugation on c-Myc that eventually leads to the blockage of c-Myc from proteasomal degradation (44). Given that the conjugation of ubiquitin chains of more than four molecules is thought to be necessary for the recognition of 26S proteasome (45), insertion of K63-linked ubiquitination into a K48-linked ubiquitin chain may hamper the preferred conformation of the polyubiquitin chain, resulting in the decrease of affinity for 26S proteasome. Since we observed p21 degradation being attenuated by Fbl12 overexpression along with the rapid p21 synthesis (Fig. 3B), the p21 degradation rate might be surpassed by its synthesis rate. Consequently, p21 expression level appears to increase in an Fbl12-dependent manner. Alternatively, it is possible that the K63-linked ubiquitination competes with K48-linked ubiquitination on the same lysine site(s) of target proteins in a manner similar to that of c-Myc.
It has been shown that the E2s, which are ubiquitin-conjugating enzymes, are important for the determination of ubiquitin linkage mode (46). Usually, SCF complex seems to conjugate K48-linked ubiquitination with target protein via the recruitment of specific E2 enzymes, such as UBE2R1 and UBE2D2. On the other hand, recent studies have reported that the heterodimer of UBE2N with UBE2V1 plays important roles in the formation of K63-linked ubiquitin chain (46). The mechanisms of how the SCF complex mediates mixed-type ubiquitination remain unclear. It has been known that Ubc4, which is a yeast homologue of UBE2D2, is involved in the formation of not only K48-linked ubiquitination but also K63-linked ubiquitination under stress conditions (47). In addition, UBE2D2 is responsible for SCFFbl12-induced p57 ubiquitination (28). Thus, UBE2D2 is thought to be a primary candidate that controls SCFFbl12-dependent mixed-type ubiquitination. However, our preliminary experiments have shown that the combination between SCFFbl12 and UBE2D2 had little effect on the enhancement of p21 ubiquitination in our in vitro ubiquitination assay (data not shown). This suggests that another specific E2 enzyme is responsible for this cascade reaction in cells. Alternatively, it is plausible that other unknown factors play important roles in the regulation of linkage specificity and activity of SCFFbl12. Indeed, expression of PA28γ decreases the ubiquitination level of not only p21 but also, possibly, autoubiquitinated Fbl12 (Fig. 5C). Probably, PA28γ alters the structural distance from E2 enzyme to the substrate on the SCFFbl12 complex, resulting in a decrease of ubiquitin ligase activity. The biological events that underlie PA28γ-regulated p21 turnover are yet to be elucidated. Previously, several groups have shown that PA28γ promotes ubiquitin-independent degradation of p21 through binding (25, 26). We report here that an increment of PA28γ negatively regulates the amount of p21. As the PA28γ level has been reported to be upregulated by NF-κB activation (48), the regulation of the amount of PA28γ could mediate a pivotal process that controls p21 expression regulated by stress responses.
Recently, we reported that UV irradiation induces Fbl12ΔF transcription via the alternative promoter, leading to an involvement of Fbl12 regulation (49). Since overexpression of Fbl12ΔF does not affect the cellular proliferation, it is possible that Fbl12-related pathways are linked to UV-induced DNA damage response. Interestingly, we occasionally observed a punctate structure colocalized with Fbl12 in the nucleus after UV irradiation (data not shown). It has also been reported that PA28γ translocates to Cajal bodies in response to UV, leading to an enhancement of their degradation (50). These results have suggested that the SCFFbl12-PA28γ protein complex is a potent mediator that regulates nuclear integrity following DNA damage response. Since a defect in the Fbxl12 gene generated by CRISPR/Cas9 had little effect on the proliferation under basal condition (data not shown), our finding could be involved not only in cellular proliferation but also in DNA damage responses as well as Fbl12ΔF. One potential mechanism is that Fbl12 activity is augmented by stress stimulation such as UV. Intriguingly, Fbl12 is thought to be phosphorylated at Ser-124 (http://www.phosphosite.org/). This observation suggests that Fbl12 activity is modulated by phosphorylation in response to stress stimulation. However, the precise mechanisms that underlie the control of this mechanisms mediated by SCFFbl12-PA28γ proteins remain unclear. This will need to be investigated more thoroughly in the future.
In conclusion, we found that Fbl12 binds and ubiquitinates p21. This ubiquitination is associated with the stability of p21. In addition, we found that Fbl12 regulates default degradation under basal conditions but not under UV-stimulated conditions. Therefore, our findings elucidate novel mechanisms that underlie the regulation of p21 expression level in cells.
ACKNOWLEDGMENTS
We thank Toru Natsume (AIST, Japan) for helping with the proteomics experiments, Takuma Aihara for critical reading of the manuscript, and the members of the Chiba laboratory for helpful discussions and technical support.
REFERENCES
- 1.Abbas T, Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gsponer J, Futschik ME, Teichmann SA, Babu MM. 2008. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 4.Gartel AL, Najmabadi F, Goufman E, Tyner AL. 2000. A role for E2F1 in Ras activation of p21(WAF1/CIP1) transcription. Oncogene 19:961–964. doi: 10.1038/sj.onc.1203411. [DOI] [PubMed] [Google Scholar]
- 5.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng Chan FJAM, Ferrari AC, Martignetti JA, Friedman SL. 2001. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee S, Conrad SE. 2005. c-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. J Biol Chem 280:17617–17625. doi: 10.1074/jbc.M502278200. [DOI] [PubMed] [Google Scholar]
- 7.Jung P, Menssen A, Mayr D, Hermeking H. 2008. AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci U S A 105:15046–15051. doi: 10.1073/pnas.0801773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Dowbenko D, Lasky LA. 2002. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem 277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 9.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol 3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 10.Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. 2001. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol 21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnouin K, Dubuisson ML, Child ES, Fernandez de Mattos S, Glassford J, Medema RH, Mann DJ, Lam EW. 2002. H2O2 induces a transient multi-phase cell cycle arrest in mouse fibroblasts through modulating cyclin D and p21Cip1 expression. J Biol Chem 277:13761–13770. doi: 10.1074/jbc.M111123200. [DOI] [PubMed] [Google Scholar]
- 12.Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. 2002. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem 277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- 13.Asher G, Reuven N, Shaul Y. 2006. 20S proteasomes and protein degradation “by default.” Bioessays 28:844–849. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 14.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem 278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 15.Yamada K, Ono M, Perkins ND, Rocha S, Lamond AI. 2013. Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Mol Cell 49:922–933. doi: 10.1016/j.molcel.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A. 2005. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell 17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 17.Ferrandina G, Stoler A, Fagotti A, Fanfani F, Sacco R, De Pasqua A, Mancuso S, Scambia G. 2000. p21WAF1/CIP1 protein expression in primary ovarian cancer. Int J Oncol 17:1231–1235. [DOI] [PubMed] [Google Scholar]
- 18.Sarbia M, Stahl M, zur Hausen A, Zimmermann K, Wang L, Fink U, Heep H, Dutkowski P, Willers R, Muller W, Seeber S, Gabbert HE. 1998. Expression of p21WAF1 predicts outcome of esophageal cancer patients treated by surgery alone or by combined therapy modalities. Clin Cancer Res 4:2615–2623. [PubMed] [Google Scholar]
- 19.Yu ZK, Gervais JL, Zhang H. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A 95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starostina NG, Simpliciano JM, McGuirk MA, Kipreos ET. 2010. CRL2(LRR-1) targets a CDK inhibitor for cell cycle control in C. elegans and actin-based motility regulation in human cells. Dev Cell 19:753–764. doi: 10.1016/j.devcel.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. 2007. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell 27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. 2009. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115:71–82. doi: 10.1016/S0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell 26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell 26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday MJ. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J 20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M, Nakamoto T, Nishimori S, Tanaka K, Chiba T. 2008. A new ubiquitin ligase involved in p57KIP2 proteolysis regulates osteoblast cell differentiation. EMBO Rep 9:878–884. doi: 10.1038/embor.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. 2002. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem 74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- 30.Postow L, Funabiki H. 2013. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 12:587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallampalli RK, Kaercher L, Snavely C, Pulijala R, Chen BB, Coon T, Zhao J, Agassandian M. 2013. Fbxl12 triggers G1 arrest by mediating degradation of calmodulin kinase I. Cell Signal 25:2047–2059. doi: 10.1016/j.cellsig.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama M, Nita A, Yumimoto K, Nakayama KI. 2015. FBXL12-mediated degradation of ALDH3 is essential for trophoblast differentiation during placental development. Stem Cells 33:3327–3340. doi: 10.1002/stem.2088. [DOI] [PubMed] [Google Scholar]
- 33.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida Y, Murakami A, Tanaka K. 2011. Skp1 stabilizes the conformation of F-box proteins. Biochem Biophys Res Commun 410:24–28. doi: 10.1016/j.bbrc.2011.05.098. [DOI] [PubMed] [Google Scholar]
- 35.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- 37.Mao I, Liu J, Li X, Luo H. 2008. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci 65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fotedar R, Bendjennat M, Fotedar A. 2004. Role of p21WAF1 in the cellular response to UV. Cell Cycle 3:134–137. [PubMed] [Google Scholar]
- 39.Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. 2003. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Zeng SX, Lu H. 2006. UV induces p21 rapid turnover independently of ubiquitin and Skp2. J Biol Chem 281:26876–26883. doi: 10.1074/jbc.M605366200. [DOI] [PubMed] [Google Scholar]
- 41.Soria G, Podhajcer O, Prives C, Gottifredi V. 2006. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 42.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 43.Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, Takahashi JS. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popov N, Schulein C, Jaenicke LA, Eilers M. 2010. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol 12:973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 45.Saeki Y, Isono E, Oguchi T, Shimada M, Sone T, Kawahara H, Yokosawa H, Toh-e A. 2004. Intracellularly inducible, ubiquitin hydrolase-insensitive tandem ubiquitins inhibit the 26S proteasome activity and cell division. Genes Genet Syst 79:77–86. doi: 10.1266/ggs.79.77. [DOI] [PubMed] [Google Scholar]
- 46.Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnason T, Ellison MJ. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol 14:7876–7883. doi: 10.1128/MCB.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Luan Y, Xiang D, Tan X, Chen H, Deng Q, Zhang J, Chen M, Huang H, Wang W, Niu T, Li W, Peng H, Li S, Li L, Tang W, Li X, Wu D, Wang P. 2016. The 11S proteasome subunit PSME3 is a positive feedforward regulator of NF-kappaB and important for host defense against bacterial pathogens. Cell Rep 14:737–749. doi: 10.1016/j.celrep.2015.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuruta F, Kim J, Fukuda T, Kigoshi Y, Chiba T. 2015. The intronic region of Fbxl12 functions as an alternative promoter regulated by UV irradiation. Biochem Biophys Rep 3:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cioce M, Boulon S, Matera AG, Lamond AI. 2006. UV-induced fragmentation of Cajal bodies. J Cell Biol 175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]