Abstract
Poly(rC)-binding proteins (PCBPs) are multifunctional adapters that mediate interactions between nucleic acids, iron cofactors, and other proteins, affecting the fates and activities of the components of these interactions. Here, we show that PCBP2 forms a complex with the Hippo pathway components Salvador (Sav1), Mst1, Mst2, and Lats1 in human cells and mouse tissues. Hippo is a kinase cascade that functions to phosphorylate and inactivate the transcriptional coactivators YAP and TAZ, which control cell growth and proliferation. PCBP2 specifically interacts with the scaffold protein Sav1 and prevents proteolytic cleavage of the Mst1 kinase, resulting in increased signaling through Hippo and suppressed activity of YAP and TAZ. Human breast epithelial cells lacking PCBP2 exhibit impaired proteasomal degradation of TAZ. They accumulate TAZ in both the nucleus and the cytosol, increase expression of YAP and TAZ connective tissue growth factor (CTGF) and Cyr61 target genes, and exhibit anchorage-independent growth. Thus, PCBP2 can function as a component of the Hippo complex, enhancing signaling, suppressing activity of YAP and TAZ, and altering the growth characteristics of cells.
INTRODUCTION
The poly(rC)-binding proteins (PCBPs) are a family of four multifunctional adapter proteins that can bind RNA, proteins, and metal ions and mediate the interaction of those components with other cellular proteins (1–3). PCBPs contain three tandem repeats of the ancient and conserved RNA-binding domains found in heterogeneous ribonucleoprotein K, called KH domains. The KH domains bind single-stranded RNA and DNA. PCBP1 and PCBP2 (also called hnRNP E1 and E2 or αCP-1 and -2) are the most abundant members of the family and are ubiquitously expressed in all tissues. PCBP1 and PCBP2 bind to a variety of C-rich sequences in cellular and viral RNAs, affecting splicing (4), polyadenylation (5), stability, translation (1, 2), and localization (6).
PCBPs, especially PCBP2, can affect other processes through protein-protein interactions. PCBP2 interacts with MAVS, an adaptor protein that activates innate immune responses to viral RNA (7). PCBP2 contributes to microRNA (miRNA) processing in cells by interacting with both Dicer and precursor miRNA to enhance processing into mature miRNA (8). PCBP1 and PCBP2 function in cellular iron trafficking by binding iron and delivering it directly to iron-dependent enzymes via metal-mediated protein-protein interactions (9). PCBP1 and PCBP2 deliver iron to ferritin, an iron storage protein, (10, 11) and enzymes that contain mononuclear or dinuclear iron centers, including prolyl hydroxylase, which regulates hypoxia-inducible factors (12), and deoxyhypusine hydroxylase (13), which functions in the modification of lysine to hypusine in eukaryotic initiation factor 5A. PCBP2 also binds to DMT1, an integral membrane protein that transports iron to the cytosol, potentially linking iron uptake with cytosolic distribution (14).
Because PCBP1 and PCBP2 function as adaptors in many cellular processes, we sought to identify the broad set of proteins that bind to PCBP1 and PCBP2 by using affinity purification and mass spectrometry. We identified components of the Hippo signaling pathway in PCBP-containing complexes. The Hippo pathway is a signaling cascade first characterized in Drosophila melanogaster, where inactivating mutations in multiple components of the pathway resulted in similar phenotypes of tissue overgrowth (15). The pathway is both highly conserved and divergent in mammals. The core components consist of the Mst1 and Mst2 kinases, which bind to Salvador (Sav1), a scaffold protein that also serves to activate the kinases. Mst1/Mst2 can phosphorylate and activate the downstream kinases Lats1 and Lats2 and their associated scaffold protein, Mob1. In turn, Lats1 and Lats2 can phosphorylate and inactivate the transcriptional coactivators YAP and TAZ, which regulate the expression of many genes involved in cell growth and proliferation. Thus, activation of Hippo signaling serves to limit the activity of YAP and TAZ, which limits their capacity to promote cell proliferation, growth, and differentiation. While signal transduction through the Hippo pathway is relatively linear in flies, in mammals it exhibits considerably more complexity and variability. We determined that PCBP2 specifically associates with components of this kinase cascade and regulates the activities of its downstream transcriptional coactivators (16).
MATERIALS AND METHODS
Cell culture.
MCF10A cells were cultured in Dulbecco's modified Eagle's medium containing nutrient mixture F-12 (DMEM–F-12) (Life Technologies) supplemented with 5% horse serum (Sigma), 10 μg/ml insulin, 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), and 20 ng/ml epidermal growth factor (EGF) (Sigma). Flp-In T-Rex 293 cells expressing doxycycline-inducible, Flag-tagged versions of PCBPs were maintained in DMEM containing 10% fetal bovine serum (FBS) (HyClone Serum), 100 μg/ml hygromycin (Life Technologies), and 15 μg/ml blasticidin (Sigma). MCF7 and MDA-MB-231 cells (a gift from Chuxia Deng, NIH) and HEK293 cells (ATCC) were cultured in DMEM–F-12 supplemented with 10% FBS (Serum Source International). PCBP2 (12), Mst1/2, and Lats1/2 were depleted using Stealth Select RNAi (Life Technologies). TAZ and YAP were depleted using small interfering RNA (siRNA) from Origene. A scrambled-sequence LoGC siRNA pool (Life Technologies) was used as a negative control. Two sequential transfections spaced 24 h apart were performed with 100 pmol of siRNA by using Lipofectamine RNAiMax transfection reagent, and cells were harvested 72 h posttransfection. Anchorage-independent growth was assessed by seeding 5,000 MCF10A control-transfected cells or PCB2-depleted cells onto ultra-low-attachment plates in mammosphere formation medium (complete MCF10A culture medium supplemented with B27 [20 μl/ml], fibroblast growth factor [FGF] [20 ng/ml], EGF [20 ng/ml], and antibiotic/antimycotic) (17). The images were taken after 6 days of incubation using an Olympus IX81 microscope. Colonies >50 μm in diameter were scored as mammospheres. Where indicated, doxycycline was used at 1 μg/ml, desferrioxamine (Dfo) at 100 μM, ferric ammonium citrate at 100 μg/ml, bortezomib at 25 nM, and cycloheximide at 40 μM. Mouse studies were conducted according to protocols approved by the NIDDK Animal Care and Use Committee.
Immunoprecipitation (IP) and mass spectrometry.
HEK293 cell lines expressing Flag-PCBP1, Flag-PCBP2, or the empty vector were grown in medium supplemented with 1 μg/ml doxycycline and 100 μM ferric ammonium citrate or 100 μM desferrioxamine B. Ten 15-cm plates were harvested, and Flag immunoprecipitates were prepared as described previously (18). Where indicated, 50 μM RNase A was added to lysis and wash buffers. High-salt washes were performed by increasing sodium chloride in the wash buffer to 500 μM. Immune complexes were eluted with Flag peptide, and the eluates were precipitated in 10% cold trichloroacetic acid. The precipitated eluates underwent tryptic digestion and mass spectrometry analysis as described previously (18, 19).
RNA extraction and qRT-PCR.
RNA was isolated from cells using the RiboPure RNA purification kit (Life Technologies). Reverse transcription was performed using a High-Capacity cDNA reverse transcription kit (Life Technologies) and 500 ng of total RNA. Quantitative real-time PCR (qRT-PCR) was performed using 1% of the cDNA template with SYBR green dye on an ABI 7500 system according to the manufacturer's protocols. Primers (Table 1) were designed using ABI software and validated for melting temperatures and melting profiles; the efficiency was >90%. The mRNA values were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) using the ΔΔCT method. RNA immunoprecipitations from MCF10A cells were performed using antibodies (PCBP2-RN025P and PCBP1-RN024P) and an assay kit (RN1001) from Medical and Biological Laboratories according to the manufacturers' instructions. RNase inhibitor (RNase Out; Life Technologies) was added to the lysis buffer at 200 U/ml.
TABLE 1.
Primer sequences for quantitative real-time PCR
| Gene product | Primer | Sequence |
|---|---|---|
| P2 | Forward | TCGGCCAGAAAGGAGAATCAGTTAA |
| Reverse | GACAATTCCCTTCTGAGATGTTGATAC | |
| Mst1 | Forward | ACGTTTCAGAGCCCAGAGTT |
| Reverse | TCGAGGACCATCAAAGTCG | |
| TAZ | Forward | ACCTGTACGCCAACACAGTG |
| Reverse | ACACGGAGTACTTGCGCTCA | |
| CTGF | Forward | TGCATCCGTACTCCCAAAATC |
| Reverse | TGCTGGTGCAGCCAGAAAG | |
| Cyr61 | Forward | GAGTGGGTCTGTGACGAGGAT |
| Reverse | GGTTGTATAGGATGCGAGGCT | |
| GAPDH | Forward | GGTGAAGGTCGGAGTCAACGG |
| Reverse | GAGGTCAATGAAGGGGTCATTG |
Immunoprecipitation and Western blotting.
Total cell lysates were prepared from cells using Cytobuster protein extraction reagent (Millipore) with a cocktail of protease and phosphatase inhibitors on ice (Active Motif). Uniform concentrations of protein were run in a 4 to 12% Tris-Bis gel, followed by Western blotting. For immunoprecipitations, cells or tissues were lysed in IP lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 0.1% NP-40, 5% glycerol) with protease inhibitors (Active Motif). Anti-Flag beads (Biolegend; 2 h at 4°C) were used for Flag-tagged PCBPs. Empty-vector-transfected cells were used as a negative control. For endogenous proteins in MCF10A cells and tissues, PCBP2 or Lats1 antibody was incubated with lysate overnight at 4°C. Protein G or protein A coupled to magnetic beads was added for 4 h at 4°C. Normal mouse IgG or normal rabbit IgG, respectively, was used as a negative control. The samples were boiled for 10 min before analysis by Western blotting. Nuclear and cytosolic extracts were prepared according to the protocol (Active Motif). The following primary antibodies were diluted in antibody diluents (Life Technologies): PCBP2 (1:1,000; Abnova), TAZ (1:1,000; Abcam), Sav (1:500; Cell Signaling), Mst1 (1:1,000; Cell Signaling), Mst2 (1:5,000; OriGene), Lats1 (1:500; Cell Signaling), phospho-Lats1 (1:500; Cell Signaling), YAP (1:500; Santa Cruz), phospho-YAP (1:1,000; Cell Signaling), Flag (1:5,000; Biolegend), GAPDH raised in mice (1:5,000; Millipore), and GAPDH raised in rabbits (1:5,000; Cell Signaling). Secondary antibodies were conjugated to infrared dyes (1:10,000; LiCor). The band densities of proteins in SDS-polyacrylamide gels were measured using the Odyssey system (LiCor).
Immunofluorescence.
Cells seeded on glass coverslips (Abcam) were transfected with either negative-control or PCBP2 siRNA. After 72 h, the cells were fixed with 4% cold paraformaldehyde (Fisher Scientific) for 15 min, permeabilized for 15 min with 0.5% saponin, and immunostained with antibodies against TAZ (1:100; Abcam) in Tris-buffered saline (TBS) containing 5% donkey serum plus 0.1% saponin (blocking buffer) for 1.5 h at room temperature. TBS with 0.1% saponin was used as a washing buffer after the permeabilization. The secondary antibodies (Alexa Fluor 488–donkey anti-mouse antibody; Invitrogen) were incubated for 1 h. Fluorescence images of the immunostained cells were obtained using laser scanning confocal microscopy (LSM700).
Luciferase activity assays.
PCBP2 was depleted as described above in MCF10A cells, and then connective tissue growth factor (CTGF) promoter-luciferase or CTGF mutant promoter-luciferase plasmid (gifts from Kun-Liang Guan, UCSD) were cotransfected with pRL-TK renilla luciferase (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Firefly luciferase activity was assayed using the Dual-Luciferase reporter system (Promega). Samples were read in POLARstar Omega (BMG Labtech). Activity was reported as the ratio to the renilla luciferase control.
PCBP2 mutations.
Full-length PCBP2 (isoform a) was constructed from the PCBP2 plasmid (isoform b; Origene) (10). The KH1, KH2, and KH3 domains and linker region (2) were deleted by PCR using Phusion high-fidelity DNA polymerase (NEB). Primer sequences for mutations are provided in Table 2. The primers directed amplification of the whole plasmid, omitting the KH1, KH2, KH3, or linker region. T4 DNA ligase (NEB) was used for ligation of the purified PCR product. PCBP2 mutations were confirmed by sequencing and Western blotting.
TABLE 2.
Primer sequences for PCBP2 deletion mutations
| Mutation | Primer | Sequence |
|---|---|---|
| Full-length PCBP2 | Forward | TCCCAGTCCCCCCCGAAGGGCGTGAC |
| Reverse | GAGAGTCTCCAACATGACCACGCAGATC | |
| KH1 | Forward | GACATAAGCAGCTCTATGACC |
| Reverse | GACATTTAATCCACCTTCAATCAC | |
| KH2 | Forward | CTCTCCCAGTCCCCCCCGAA |
| Reverse | TGGGTCTACTGGCAGCTGTG | |
| KH3 | Forward | TCCTCGGAGACGGGTGGCAT |
| Reverse | CTGAGCAGATGCATCCAAACC | |
| Variable region | Forward | ACTACTTCTCATGAACTCACC |
| Reverse | AGTCTCCAACATGACCACGCA |
Statistical analysis.
Data were analyzed with Prism (GraphPad). The results are reported as means and standard errors of the mean (SEM). Differences between two groups were determined by unpaired Student t tests. Differences among groups were determined by one-way analysis of variance (ANOVA).
RESULTS
PCBP1 and PCBP2 form complexes with components of Hippo.
We purified protein complexes containing PCBPs from cell lines stably expressing Flag epitope-tagged PCBP1 or PCBP2 and identified the interacting proteins by mass spectrometry (18). To identify iron-dependent interactions, cells were grown in iron-chelated or iron-supplemented medium prior to affinity purification. Cell lines without Flag-tagged proteins were included as specificity controls. Ferritin, a protein known to interact with PCBP1 and PCBP2 (10, 11), was detected in iron-treated samples (Tables 3,4, and 5). The three core components of the Hippo signaling pathway, Sav1, Mst1, and Mst2 (16, 20, 21), were the highest-scoring proteins specifically interacting with PCBP2 in our analysis. Peptides representing each of the proteins were detected in all experimental data sets with 45 to 60% peptide coverage. Detection of Sav1, Mst1, and Mst2 was unaffected by degradation of RNA, produced by the addition of RNase A to some purifications, suggesting that PCBP2 did not depend on RNA for its association with Hippo.
TABLE 3.
Hippo components detected by mass spectrometry in cells expressing Flag-PCBP2a
| Protein | No. of spectra/no. of unique peptides in: |
|||
|---|---|---|---|---|
| Dfo-treated cells |
Fe-treated cells |
|||
| Treatment 1 | Treatment 2 | Treatment 1 | Treatment 2 | |
| PCBP2 | 1,810/45 | 602/26 | 1,305/37 | 1,436/25 |
| PCBP1 | 581/30 | 239/18 | 307/24 | 780/26 |
| Sav1 | 77/24 | 13/8 | 66/25 | 13/3 |
| Mst1 | 67/32 | 56/16 | 62/29 | 67/16 |
| MST2 | 84/39 | 75/20 | 95/41 | 67/14 |
| Ferritin H | 0 | 0 | 3/2 | 0 |
| Ferritin L | 0 | 0 | 2/2 | 0 |
Treatment 1, immunoprecipitation with RNase A followed by low-salt wash; treatment 2, immunoprecipitation without RNase A followed by high-salt wash.
TABLE 4.
Hippo components detected by mass spectrometry in cells expressing Flag-PCBP1a
| Protein | No. of spectra/no. of unique peptides in: |
|||||
|---|---|---|---|---|---|---|
| Dfo-treated cells |
Fe-treated cells |
|||||
| Treatment 1 | Treatment 2 | Treatment 3 | Treatment 1 | Treatment 2 | Treatment 3 | |
| PCBP1 | 2,561/52 | 2,784/52 | 2,377/33 | 1,993/42 | 2,800/54 | 3,061/43 |
| PCBP2 | 331/16 | 520/15 | 682/15 | 248/11 | 436/17 | 904/15 |
| Sav1 | 2/2 | 0 | 0 | 3/3 | 0 | 0 |
| Mst1 | 0 | 0 | 0 | 4/3 | 0 | 0 |
| Mst2 | 0 | 0 | 0 | 4/3 | 0 | 0 |
| Ferritin H | 0 | 0 | 0 | 3/3 | 9/6 | 5/2 |
| Ferritin L | 0 | 0 | 0 | 3/2 | 3/3 | 0 |
Treatment 1, immunopreciptation without RNase A followed by low-salt wash; treatment 2, immunoprecipitation with RNase A followed by low-salt wash; treatment 3, immunoprecipitation without RNase A followed by high-salt wash.
TABLE 5.
Hippo components detected by mass spectrometry in cells without Flag-tagged proteinsa
| Protein | No. of spectra/no. of unique peptides with: |
||||||
|---|---|---|---|---|---|---|---|
| Empty vector in: |
Flag | ||||||
| Dfo-treated cells |
Fe-treated cells |
||||||
| Treatment 1 | Treatment 2 | Treatment 3 | Treatment 1 | Treatment 2 | Treatment 3 | ||
| PCBP2 | 0 | 18/7 | 10/7 | 21/7 | 16/9 | 0 | 0 |
| PCBP1 | 38/13 | 27/20 | 0 | 36/13 | 25/16 | 12/6 | 0 |
| Sav1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mst1 | 7/6 | 0 | 0 | 0 | 0 | 0 | 8/4 |
| Mst2 | 6/5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ferritin H | 0 | 0 | 0 | 0 | 2/2 | 0 | 0 |
| Ferritin L | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Treatment 1, immunopreciptation without RNase A followed by low-salt wash; treatment 2, immunoprecipitation with RNase A followed by low-salt wash; treatment 3, immunoprecipitation without RNase A followed by high-salt wash.
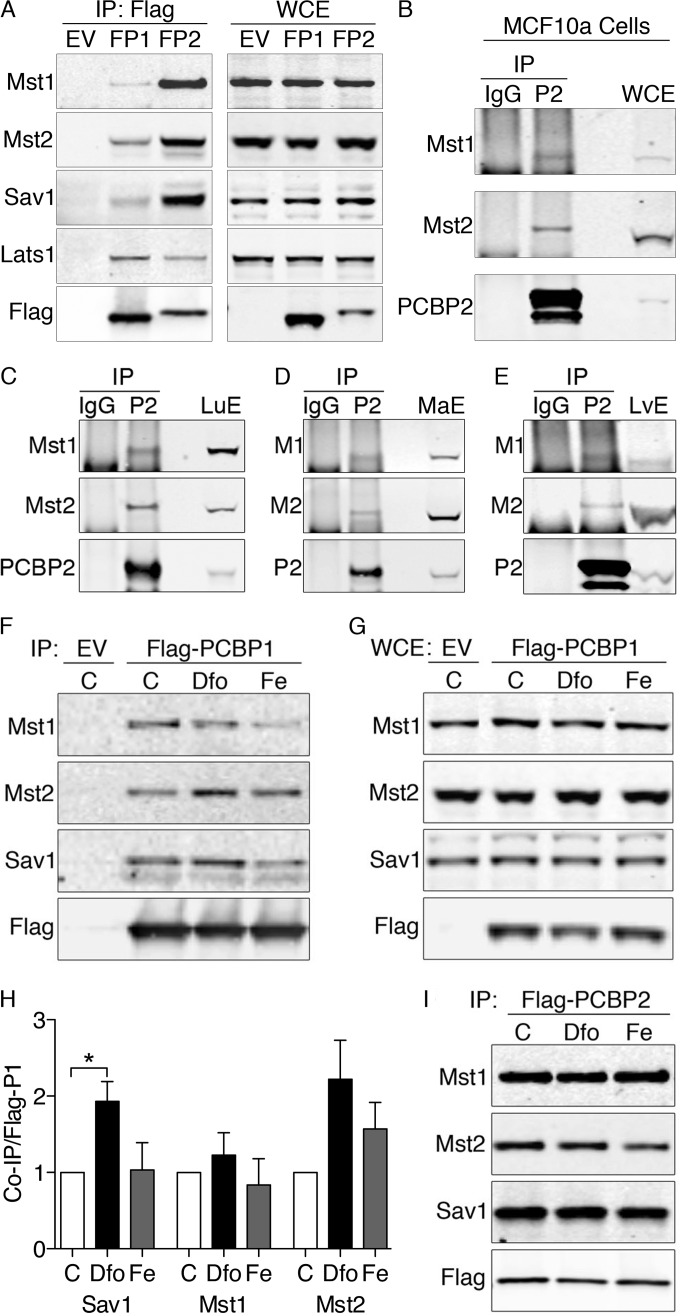
To confirm the interactions detected by mass spectrometry, we examined PCBP1 and PCBP2 immunoprecipitates by Western blotting. Immune complexes from Flag-PCBP1 cells contained small amounts of Sav1, Mst1, and Mst2, while complexes from Flag-PCBP2 cells contained large amounts of Sav1, Mst1, and Mst2, suggesting a more stable interaction (Fig. 1A). Lats1 was also present in immune complexes from both PCBP1 and PCBP2 cell lines. These interacting proteins were not detected in immune complexes from a cell line without a Flag-tagged protein (Fig. 1A, EV), confirming the specificity of the interactions. In some immortalized cell lines, the Hippo signaling pathway does not control growth or cell division, although Hippo components are expressed. MCF10A, an immortalized human cell line derived from normal breast epithelium, contains a functional Hippo pathway that regulates cell growth and division (22). Immunoprecipitation of endogenous PCBP2 in these cells revealed coprecipitation of both Mst1 and Mst2, which were not present in control IgG precipitates (Fig. 1B). Thus, PCBP2 associates with the Hippo signaling complex in cells that rely on this pathway to regulate growth.
FIG 1.
Physical interactions between Hippo signaling complex, PCBP1, and PCBP2 in human cells and murine tissues. (A) Physical interactions between PCBP1, PCBP2, and Hippo components in HEK293 cell lines. HEK293 stable cell lines containing empty vector (EV), inducible Flag-PCBP1 (FP1), or Flag-PCBP2 (FP2) were induced for 18 h. The lysates were subjected to IP with anti-Flag resin, and immune complexes (IP: Flag) and whole-cell extracts (WCE) were examined by Western blotting using antibodies against Mst1, Mst2, Sav1, total Lats1 (tLats1), and Flag. (B) Physical interactions between PCBP2 and the Hippo complex in MCF10A cells. MCF10A cell lysates were subjected to IP with anti-PCBP2 antibody or normal mouse IgG. Immune complexes and WCE were examined by Western blotting for Mst1, Mst2, and PCBP2. Sav1 is not detectable due to the interference of IgG heavy chain. Experiments were replicated three times. (C to E) Binding of Mst1 and Mst2 to PCBP2 in lung and liver. Mouse lung extracts (C), mammary gland extracts (D), and liver extracts (E) were subjected to IP with anti-PCBP2 and normal mouse IgG. Immune complexes and tissue extracts (LuE, MaE, and LvE) were analyzed by Western blotting for Mst1 and Mst2. (F to I) Effects of iron supplementation or chelation on binding of Hippo components to PCBPs. Cells expressing Flag-PCBP1 (F and G) or Flag PCBP2 (I) were treated overnight with iron (Fe) (100 μg/ml) or the iron chelator Dfo (100 μM). The lysates were subjected to IP and Western blotting as for panel A. (H) Quantitation of Hippo components coprecipitated with Flag-PCBP1. Experiments were replicated three times; *, P < 0.05. The error bars indicate SEM.
PCBP2 binds to Mst1 and Mst2 in mouse tissues.
Mst1 and Mst2 are required for normal lung development in the mouse (23), and misregulation of Hippo signaling through YAP contributes to adenocarcinoma of the lung in humans (24). Therefore, we examined whether PCBP2 formed a complex with Hippo components in the mouse lung. PCBP2 immune complexes isolated from mouse lung extracts demonstrated strong interactions with Mst1 and Mst2 (Fig. 1C). No Mst1 or Mst2 was detected in nonimmune IgG precipitates, indicating the interactions were specific for PCBP2. In accordance with complex formation in MCF10A cells, Mst1 and Mst2 were detected in complex with PCBP2 in mouse mammary tissues (Fig. 1D). Mst1 and Mst2 also regulate liver size and hepatocyte proliferation in the mouse (25–27). PCBP2 immune complexes purified from mouse liver extracts demonstrated the presence of both Mst1 and Mst2 (Fig. 1E). Thus, PCBP2 forms complexes with Hippo components in mouse tissues that rely on Hippo to control growth and development.
We next examined the effects of iron on these interactions. While the weak interactions between PCBP1 and Mst1, Mst2, and Sav1 were affected by iron (Fig. 1F to H), the stronger interactions with PCBP2 remained unaffected (Fig. 1I).
Sav1 mediates the interaction between PCBP2 and Hippo components.
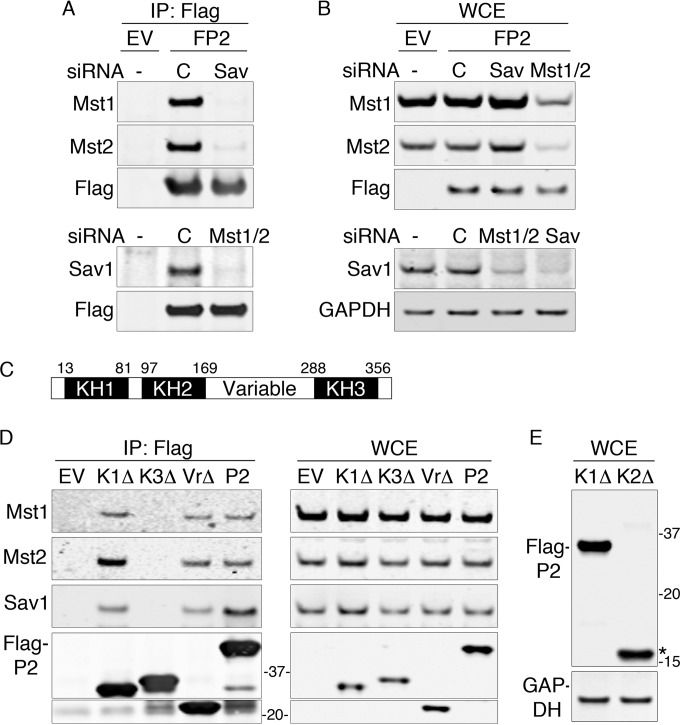
We determined which of the core components of the Hippo complex mediated the interaction with PCBP2 by depleting Sav1 or Mst1 and Mst2 in Flag-PCBP2 cells with specific siRNAs and analyzing Flag-PCBP2 immune complexes for the remaining components of the Hippo complex. Depletion of Sav1 resulted in complete loss of Mst1 and Mst2 from Flag-PCBP2 complexes, while a nontargeting siRNA had no effect (Fig. 2A, top). Western blot analysis of whole-cell extracts confirmed depletion of Sav1 and demonstrated no change in the levels of Mst1 and Mst2 with Sav1 depletion (Fig. 2B), indicating that Sav1 was necessary to mediate the interaction with PCBP2. In a reciprocal experiment, we depleted Mst1 and Mst2 and also found that interactions between PCBP2 and Sav1 were lost (Fig. 2A, bottom). However, Mst1 and Mst2 depletion also resulted in a dramatic loss of Sav1 in whole-cell extracts (Fig. 2B, bottom), suggesting that Mst1 and/or Mst2 was required for the accumulation of Sav1 in HEK cells.
FIG 2.
Sav1-mediated interaction between Hippo and the KH3 domain of PCBP2. (A and B) Loss of Hippo-PCBP2 interactions in cells lacking Sav1. Flag-PCBP2 cell lines were treated with nontargeting siRNA or siRNA against Sav1 or Mst1 and Mst2. Immune complexes (A) or whole-cell extracts (B) from empty vector or Flag-PCBP2 lysates were analyzed by Western blotting. The experiments were replicated three times. (C) Domain organization of PCBP2. hnRNP K-homology domains (KH1 to -3) and the variable domain are indicated. The numbers are amino acid residues marking the borders of domains. (D) Requirement for the KH3 domain for Hippo interactions. HEK 293T cells were transfected with plasmids encoding Flag-tagged PCBP2 (P2), mutant PCBP2 lacking the indicated domain (K1Δ, K3Δ, and VrΔ), or the empty vector (EV). The lysates were subjected to anti-Flag IP. Immune complexes (left) and whole-cell extracts (right) were analyzed by Western blotting. (E) Degradation of mutant PCBP2 lacking the KH2 domain. Cells were transfected as for panel D with plasmids encoding the PCBP2 KH1Δ or KH2Δ mutant and examined by Western blotting. The asterisk indicates the proteolytic fragment of Flag-PCBP2 KH2Δ.
Structural characterization of PCBP2-Hippo interactions.
We mapped the domain(s) of PCBP2 involved in interactions with Hippo components. Structurally, PCBP2 contains two amino-terminal KH domains and a single carboxy-terminal KH domain separated by an unstructured variable domain (Fig. 2C). Mutant versions of Flag-PCBP2 lacking each of the KH domains or the variable domain were expressed in HEK293 cells and examined for their capacity to coprecipitate Mst1, Mst2, and Sav1 (Fig. 2D). Versions of PCBP2 lacking KH1 or the variable domain coprecipitated Mst1, Mst2, and Sav1 similarly to the full-length PCBP2. In contrast, PCBP2 lacking the KH3 domain exhibited no interaction with any of the Hippo components, despite being expressed at levels similar to those of the full-length PCBP2. PCBP2 lacking the KH2 domain did not accumulate in cells except as a proteolytic fragment (Fig. 2E) and was not further tested. These data indicated that the carboxyl-terminal KH3 domain was required for interaction with Hippo, likely via the scaffold protein Sav1.
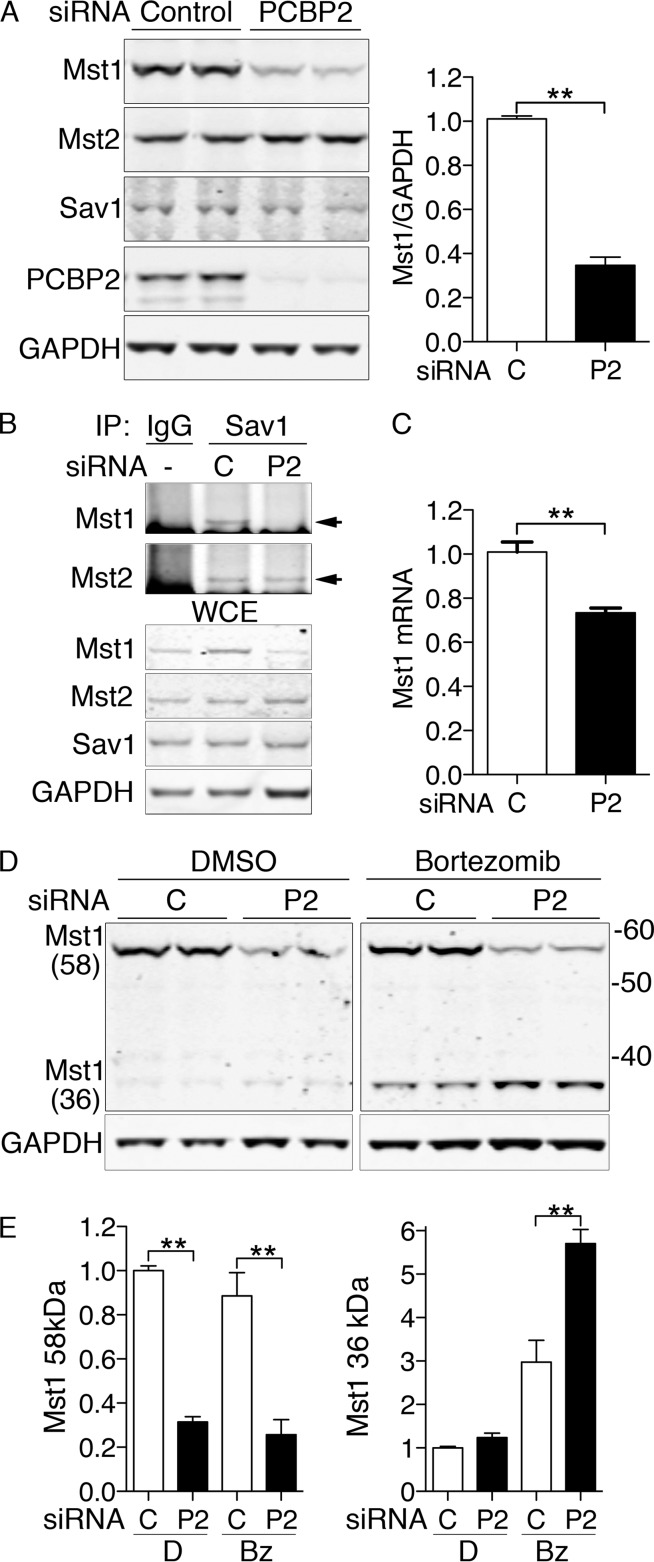
Mst1 is lost from the Hippo complex in MCF10A cells lacking PCBP2.
We questioned whether PCBP2 had an effect on the composition or stability of the Sav1/Mst1/Mst2 complex in MCF10A cells. Therefore, we depleted PCBP2 and analyzed components of the complex. When PCBP2 was depleted with siRNA (Fig. 3A), levels of Sav1 and Mst2 were unaffected, but the level of Mst1 was markedly reduced (Fig. 3A and B). Although Mst1 and Mst2 can interact with scaffold/regulator proteins other than Sav1 (20), we determined that the loss of Mst1 in PCBP2-depleted cells affected the population of Mst1 associated with the Hippo complex. Both Mst1 and Mst2 were detected in Sav1 complexes from control cells (Fig. 3B). In contrast, Mst1 was not detectable in Sav1 complexes from PCBP2-depleted cells, although Mst2 was still present. We found only a small decrease in Mst1 mRNA in PCBP2-depleted cells (Fig. 3C), which was unlikely to fully account for the loss of Mst1 protein.
FIG 3.
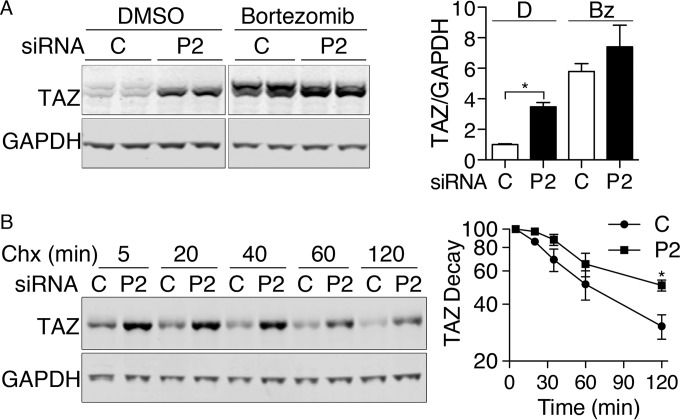
Degradation of Mst1 with PCBP2 depletion in MCF10A cells. (A) Loss of Mst1 protein with PCBP2 depletion. MCF10A cells were transfected with nontargeting control or PCBP2-specific siRNAs and then analyzed by Western blotting. (Left) Paired biological replicates. (Right) Mst1 quantification. Ratios are normalized to control. (B) Loss of Mst1 from the Hippo complex with PCBP2 depletion. PCBP2 was depleted as for panel A. The lysates were subjected to IP with anti-Sav1 or rabbit IgG. (Top) Immune complexes were examined for Mst1 and Mst2. The arrows indicate specific Mst1/2 signal. (Bottom) WCE were analyzed by Western blotting for Mst1, Mst2, Sav1, and GAPDH. (C) Minor change in Mst1 mRNA transcripts with PCBP2 depletion. RNAs from control and PCBP2-depleted MCF10A cells were extracted, and Mst1 transcripts were measured by real-time PCR. Mst1 transcripts are presented relative to control-transfected cells. (D and E) Increased cleavage and degradation of MST1 with PCBP2 depletion. Cells were treated with either nontargeting or PCBP2-specific siRNAs for 48 h, followed by either diluent (dimethyl sulfoxide [DMSO]) or proteasome inhibitor (bortezomib; 25 nM) for 24 h. Mst1 was detected by Western blotting, and paired biological replicates are shown. (E) Mass is indicated in kDa. The long (58- Da) and short (36-kDa) forms of Mst1 were quantitated and analyzed separately. All experiments were replicated three times. The error bars indicate SEM. **, P < 0.0001.
To determine whether PCBP2 contributed to the stability of Mst1, we treated cells with either nontargeting or PCBP2-specific siRNAs, followed by either diluent or bortezomib to inhibit proteasome-mediated degradation (Fig. 3D and E). Inhibition of the proteasome revealed the accumulation of a 36-kDa Mst1 species, particularly in cells lacking PCBP2. Levels of the 36-kDa Mst1 were 2-fold higher in PCBP2-depleted cells than in control cells. Mst1 is detected in cells and tissues as either a full-length protein of 59 kDa or a truncated 36-kDa amino-terminal fragment with full catalytic activity (27). The shorter form lacks both the autoinhibitory domain and the SARAH domain, which mediates interaction of Mst1 with Sav1 (20). These data indicated that, in cells lacking PCBP2, a larger fraction of Mst1 was cleaved into the short form and rapidly degraded.
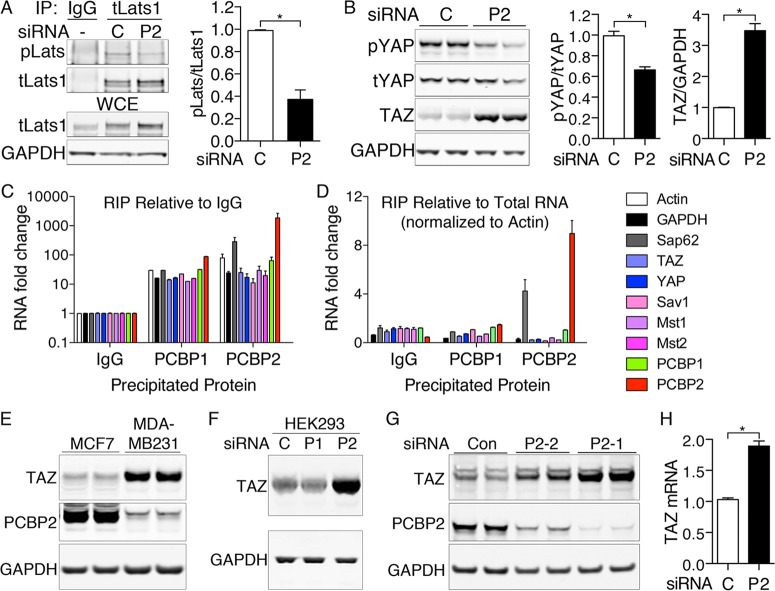
PCBP2 depletion reduces phosphorylation of Lats and YAP and produces accumulation of TAZ.
Lats1 and Lats2 are downstream kinases in the Hippo pathway that can be phosphorylated by the Sav1-Mst1/Mst2 complex (28). We questioned whether PCBP2 affected the activity of the complex by measuring levels of phosphorylated Lats1 (phospho-Lats1) in anti-total Lats immune complexes (Fig. 4A). Levels of phospho-Lats1 were significantly reduced in PCBP2-depleted versus control cells, while levels of total Lats remained unchanged or slightly elevated.
FIG 4.
Loss of Lats and YAP phosphorylation and increased TAZ levels with PCBP2 depletion. (A) Loss of Lats1 phosphorylation with PCBP2 depletion. PCBP2 was depleted in MCF10A cells, and the lysates were subjected to IP with anti-total Lats1 or normal rabbit IgG. Immune complexes and WCE were examined for phospho-Lats (pLats) and total Lats1 (tLats1) (left) and quantified (right). (B) Increased TAZ protein and decreased YAP phosphorylation with PCBP2 depletion. PCBP2 was depleted, and levels of phospho-YAP, total YAP, and TAZ were measured by Western blotting in duplicate (left) and quantified (right). (C and D) Lack of specific interaction between PCBP2 and Hippo-related transcripts. PCBP1 or PCBP2 was immunoprecipitated from MCF10A cells, and coprecipitating RNA was analyzed by RT-PCR. Rabbit IgG precipitations served as a negative control. The relative abundance of precipitated transcripts relative to IgG (C) or relative to total mRNA, normalized to actin (D), are shown. Normalization to GAPDH rather than actin yielded similar results. (E) Expression of TAZ and PCBP2 in breast cancer cell lines. MCF7 and MDA-MB-231 cells were grown under identical conditions, and the lysates were analyzed by Western blotting. (F). Increased TAZ protein with PCBP2 depletion in HEK293 cells. HEK293 cells were transfected with the indicated siRNAs and analyzed by Western blotting. (G) Increased TAZ in cells depleted of PCBP2 by alternative siRNA. MCF10A cells were transfected with control nontargeting siRNA or one of two siRNAs against PCBP2 (P2-1 or P2-2). TAZ or PCBP2 levels were detected by Western blotting. (H) TAZ mRNA levels with PCBP2 depletion. TAZ transcripts were measured by real-time PCR and are presented relative to the control. The experiments were replicated three times. The error bars indicate SEM. *, P < 0.0001.
Both YAP and TAZ are substrates for the kinases of the Hippo pathway, and phosphorylation results in their retention in the cytosol (22) and/or degradation via the proteasome (29). We examined the role of PCBP2 in YAP and TAZ phosphorylation and stability. Cells depleted of PCBP2 exhibited a small reduction in the amount of phospho-YAP, with little change in the total amount of YAP but a larger, 3.5-fold increase in total levels of TAZ (Fig. 4B).
We examined whether the RNA-binding activity of PCBP2 could account for the effects of depletion on Hippo signaling. We isolated PCBP1- and PCBP2-RNA complexes from HEK293 cells and measured the binding of control and Hippo-related mRNAs. Although both PCBP1 and PCBP2 interacted with all the tested transcripts at a level above that with negative-control IgG precipitates (Fig. 4C), only transcripts for PCBP2 and Sap62 (previously identified PCBP2-interacting RNAs) exhibited selective enrichment in the PCBP2-specific pool (Fig. 4C and D) (30). Thus, RNA immunoprecipitation offered no evidence that effects on Hippo signaling were mediated through PCBP2-RNA interactions.
Breast cancers and cell lines derived from breast cancers frequently display increased TAZ levels and TAZ activity (31–33). We examined PCBP2 and TAZ levels in breast cancer cell lines that form less invasive (MCF7) and more invasive (MDA-MB-231) tumors in vitro. TAZ levels were lower and PCBP2 levels were higher in the less invasive, more differentiated MCF7 line, whereas the opposite was observed in the more invasive, less differentiated MDA-MB-231 line, consistent with a role for PCBP2 in Hippo signaling and TAZ activity (Fig. 4E).
We confirmed that PCBP2 depletion also produced TAZ accumulation in HEK293 cells (Fig. 4F). An alternative siRNA against PCBP2 also produced partial depletion of PCBP2 and an increase in TAZ, indicating the TAZ increase was unlikely to represent an off-target effect (Fig. 4G). TAZ mRNA increased with PCBP2 depletion (Fig. 4H), but this change did not fully explain the increase in TAZ protein. Because PCBP2 depletion affected TAZ to a much greater extent than YAP, we focused on TAZ.
Accumulation of TAZ due to increased stability.
The elevation of TAZ in PCBP2-depleted cells could be due to the increased stability of hypophosphorylated TAZ in the setting of reduced Hippo signaling. If this were occurring, blocking the degradation of TAZ would tend to equalize the levels of TAZ in control and PCBP2-depleted cells. We inhibited proteasome-mediated degradation in MCF10A cells and found TAZ increased to similar levels in both control and PCBP2-depleted cells (Fig. 5A), indicating that the increase in TAZ associated with PCBP2 depletion was primarily due to increased stability of the TAZ protein. Phosphorylated TAZ exhibits slightly slower electrophoretic migration than unphosphorylated TAZ (33). In control cells, TAZ was detected as two species with slightly different migration rates (Fig. 5A and B), with the slower-migrating form likely representing phospho-TAZ. The TAZ that accumulated in PCBP2-depleted cells, with or without bortezomib treatment, was predominately the faster-migrating form, suggesting that the TAZ accumulating in PCBP2-depleted cells was hypophosphorylated. We measured the rate of TAZ degradation in PCBP2-depleted cells and found the half-life was approximately 120 min versus 60 min in control cells (Fig. 5B), indicating that the half-life of TAZ was prolonged by PCBP2 depletion.
FIG 5.
TAZ stabilization with PCBP2 depletion. (A) Equalization of TAZ levels in control and PCBP2-depleted cells by proteasome inhibition. Cells were treated with siRNAs for 48 h, followed by DMSO or bortezomib for 24 h. TAZ levels were examined by immunoblotting (left) and quantified (right). (B) Prolonged half-life of TAZ with PCBP2 depletion. Cells were treated with siRNAs, followed by cycloheximide (Chx) for the indicated times. The decay of TAZ was measured by immunoblotting (left) and quantified (right). All experiments were replicated three or four times. The error bars indicate SEM. *, P < 0.002.
PCBP2 depletion activates TAZ.
Phosphorylation of TAZ through the Hippo pathway results in the retention of TAZ in the cytosol and/or its degradation by the proteasome, while loss of phosphorylation results in stabilization and nuclear accumulation (22, 29). We examined the effects of PCBP2 on subcellular localization of TAZ by indirect immunofluorescence (Fig. 6A). In control cells, TAZ was detected at low levels in both the cytosol and nuclei. In contrast, PCBP2-depleted cells exhibited TAZ signal that was slightly higher in the cytosol and markedly higher in nuclei than in control cells. To quantitatively examine the change in nuclear TAZ with PCBP2 depletion, we separated siRNA-transfected cell lysates into nuclear-enriched and cytosolic fractions (Fig. 6B). Consistent with the immunofluorescence data, PCBP2-depleted cells exhibited 3- to 4-fold-increased levels of TAZ in both nuclear and cytosolic fractions compared to control cells. This increase in nuclear TAZ would be predicted to affect transcription.
FIG 6.
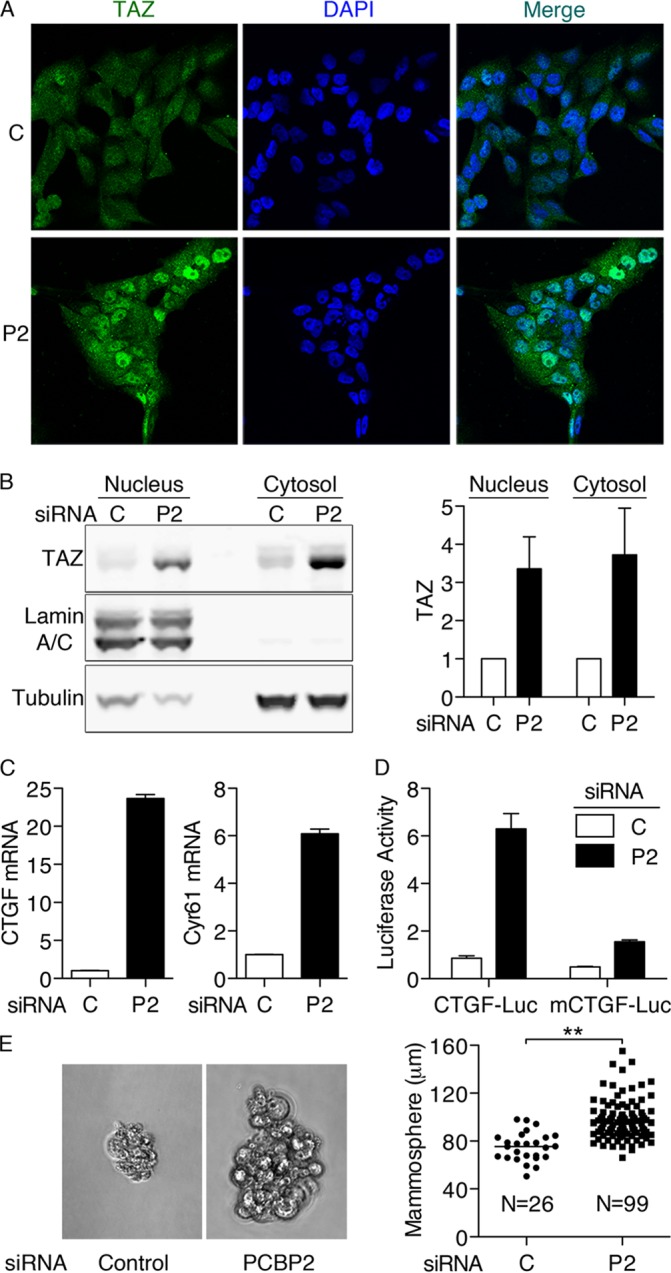
Nuclear localization of TAZ and transcriptional activation of TAZ target genes with PCBP2 depletion. (A) Nuclear localization of TAZ with PCBP2 depletion. MCF10A cells were treated with control (C) or PCBP2 (P2) siRNA and then analyzed by indirect immunofluorescence against TAZ (green) with DAPI staining of nuclei (blue). The cells were imaged with identical exposure times. (B) TAZ expression in cytosolic and nuclear fractions. Nuclear and cytosolic extracts prepared from PCBP2-depleted cells were subjected to Western blotting against TAZ, Lamin A/C (nucleus), and tubulin (cytosol) (left) and quantified (right). (C) CTGF and Cyr61 mRNA levels increase with PCBP2 depletion. Transcripts were measured by RT-PCR. (D) CTGF promoter-luciferase reporter activity with PCBP2 depletion. MCF10A cells were treated with siRNAs, followed by cotransfections with TK-Renilla and CTGF-luciferase (CTGF-Luc) or its mutant (mCTGF-Luc). Luciferase activity was normalized with TK-Renilla and is presented relative to the control (P = 0.0002). (E) Anchorage-independent growth of MCF10A cells with PCBP2 depletion. MCF10A cells were depleted of PCBP2, and then, 5,000 cells were seeded onto ultra-low-attachment plates with mammosphere formation medium. After 6 days, the cells were imaged (left), and the sizes and the numbers of all cell aggregates larger than 50 μm were recorded (right). Cumulative data from 3 experiments are shown. Horizontal bars indicate the means. All experiments were replicated three times; the error bars indicate SEM. **, P < 0.0001.
TAZ and YAP coactivate transcription by binding to transcription factors of the TEA domain (TEAD) family, which directly interact with promoter regions of target genes (34, 35). CTGF and cysteine-rich angiogenic inducer 61 (Cyr61) are two highly regulated targets of TAZ/TEAD in cells. We measured 24-fold and 6-fold inductions of CTGF and Cyr61 mRNAs, respectively, in MCF10A cells depleted of PCBP2, indicating that TAZ and/or YAP was highly activated in PCBP2-depleted cells (Fig. 6C). Again, an alternative siRNA against PCBP2 also produced elevated CTGF mRNA (data not shown). To further examine the activation of TAZ and YAP, we transfected cells with plasmids containing either the CTGF promoter fused to a luciferase reporter or a similar plasmid in which the three TEAD-binding sites in the promoter were mutated. PCBP2-depleted cells exhibited strong, 6.3-fold activation of the wild-type reporter compared to PCBP2-expressing cells, while cells containing the mutated promoter construct exhibited only 1.3-fold activation (Fig. 6D). Thus, the transcriptional activation of CTGF and Cyr61 was due to the specific activation of TAZ/YAP in complex with TEAD.
Overexpression of TAZ in MCF10A cells results in enhanced proliferation and anchorage-independent growth (32). We examined the capacity for PCBP2-depleted cells to form colonies (mammospheres) under ultra-low-attachment conditions. Cells expressing PCBP2 exhibited very weak anchorage-independent growth (Fig. 6E), forming small colonies with an average size of 75 μm at a rate of 8.7 per 5,000 plated cells. In contrast, cells depleted of PCBP2 formed colonies with an average size of 100 μm at a rate of 33 per 5,000 cells. Thus, depletion of PCBP2 led to increased TAZ/YAP activity, which was associated with enhanced anchorage-independent growth.
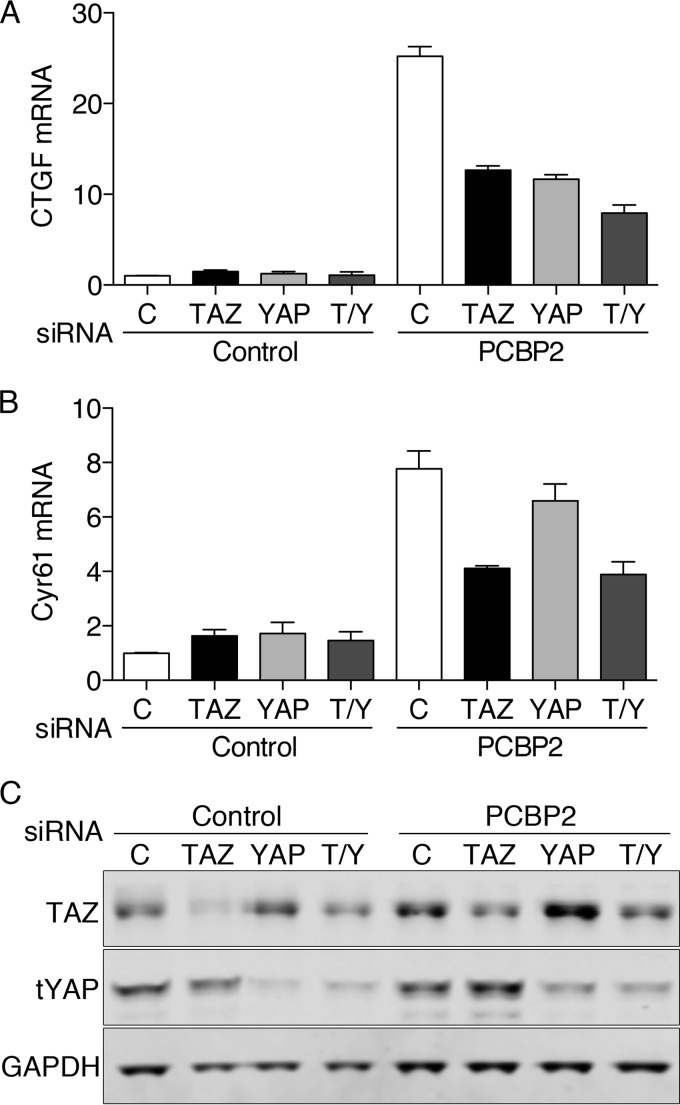
PCBP2 affects Hippo signaling through both YAP and TAZ.
PCBP2-depleted cells exhibited changes in transcription and growth (Fig. 6) in the setting of changes in both YAP phosphorylation and TAZ degradation (Fig. 4 and 5). We sought to determine whether the PCBP2-dependent changes were mediated by YAP, TAZ, or both. YAP and TAZ were depleted using siRNA in cells expressing endogenous PCBP2 or depleted of PCBP2. We then measured transcript levels of CTGF and Cyr61 (Fig. 7A to C). Depletion of either YAP, TAZ, or both had very small effects on CTGF and Cyr61 in cells expressing endogenous PCBP2, indicating that basal activities of YAP and TAZ were low in these cells. In PCBP2-depleted cells, CTGF and Cyr61 transcripts were again elevated. Codepletion of PCBP2 and either TAZ or YAP resulted in a 50% reduction in CTGF transcripts compared to the TAZ- and YAP-expressing cells, and depletion of both TAZ and YAP resulted in a 69% reduction (Fig. 7A). Cyr61 transcripts exhibited a slightly different pattern upon codepletion of PCBP2 and either TAZ or YAP (Fig. 7B). YAP depletion had only a small effect, decreasing Cyr61 by 15%. Depletion of both TAZ and YAP had an effect similar to depletion of TAZ alone. Western blotting confirmed partial depletion of YAP and TAZ in PCBP2-codepleted cells (Fig. 7C). These results suggested that the PCBP2-dependent changes in CTGF mRNA levels were largely mediated through changes in both TAZ and YAP activity and that TAZ primarily controlled the changes in Cyr61.
FIG 7.
Requirement for YAP and TAZ for PCBP2-mediated changes in transcription. MCF10A cells were treated with control or PCBP2 siRNA, followed by control, TAZ, or YAP siRNA. (A and B) Transcript levels of CTGF (A) and Cyr61 (B) were measured and are expressed relative to the control. (C) TAZ and YAP siRNA specificity and knockdown efficiency were measured by Western blotting. The error bars indicate SEM.
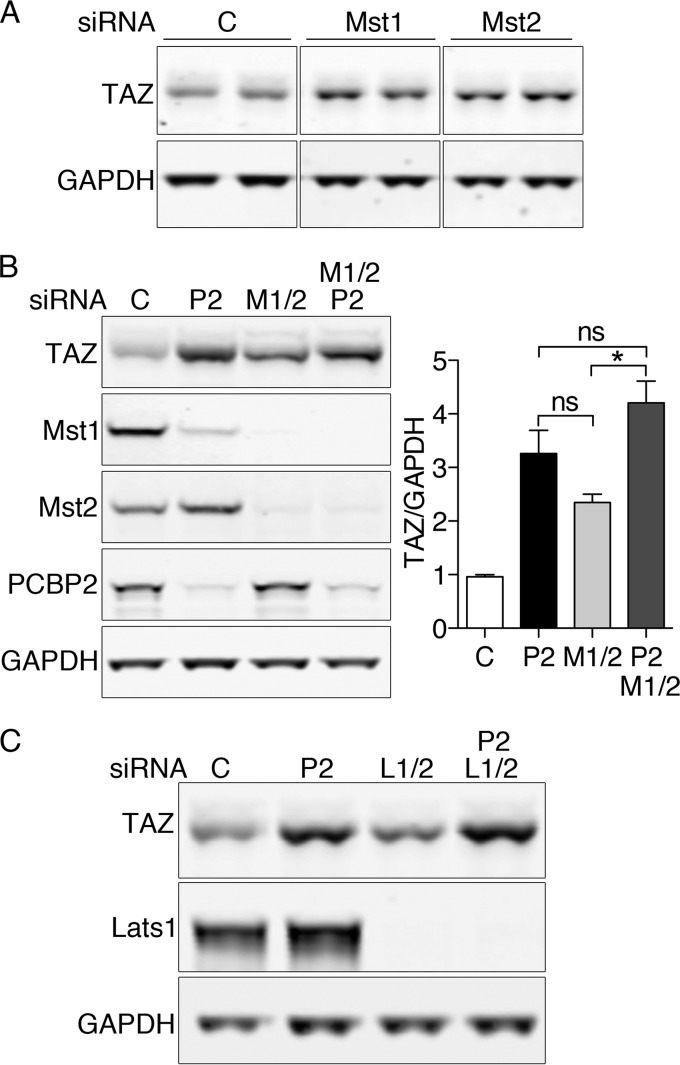
PCBP2-dependent changes in TAZ are mediated through Mst1 and Mst2.
Our studies indicated that depletion of PCBP2 was associated with a loss of Mst1 and an increase in TAZ protein and activity. We sought to determine whether the increase in TAZ was caused by the loss of Mst1 and Mst2 activity. Depletion of Mst1 or Mst2 individually was associated with small increases in TAZ (Fig. 8A), confirming that both kinases contribute to TAZ regulation. Because depletion of Mst1 alone did not produce the same increase in TAZ as PCBP2 depletion, Mst1 alone could not account for all of the observed effects of PCBP2 depletion. If the effects of PCBP2 on TAZ were mediated solely through Mst1 and Mst2, then depletion of PCBP2 and Mst1/2 together would be similar to the deletion of PCBP2 alone. We compared TAZ levels in cells depleted of PCBP2, Mst1, or Mst2, or all three together, and found that, although TAZ levels were slightly higher in cells codepleted of PCBP2, Mst1, and Mst2 than in cells depleted of only PCBP2, the difference was not significant (Fig. 8B). This result suggests that the effects of PCBP2 depletion on TAZ are largely mediated through Mst1 and -2 (not through Mst1 alone), although PCBP2 may have some effects on TAZ that are independent of Mst1/2.
FIG 8.
Requirement for both Mst1 and Mst2 for PCBP2-mediated changes in TAZ. (A) Individual contributions of both Mst1 and Mst2 to TAZ regulation. MCF10A cells were treated with control or Mst1- or Mst2-specific siRNA and then analyzed by Western blotting for TAZ and GAPDH. (B) Similar effects on TAZ expression with PCBP2 and Mst1/2 depletion. Cells were treated with the indicated siRNAs and analyzed by Western blotting (left) with quantification (right). All experiments were replicated three times. The error bars indicate SEM. *, P = 0.005; ns, not significant. (C) Lack of effect of Lats1/2 depletion on TAZ levels. Cells were treated with the indicated siRNAs and analyzed by Western blotting.
Mst1 and Mst2 signaling in Drosophila and some mammalian cells is mediated through Lats1 and Lats2, while Mst1/2 signals can bypass Lats1 and Lats2 in certain mouse tissues (25–27). We performed an epistasis analysis of the role of Lats1 and Lats2 in the PCBP2-dependent regulation of TAZ (Fig. 8C). Depletion of Lats1 and Lats2 had no effect on the levels of TAZ, and the codepletion of PCBP2 with Lats1 and Lats2 resulted in an increase in TAZ that was identical to the increase when PCBP2 alone was depleted. These data indicated that Lats1 and Lats2 did not contribute to the basal regulation of TAZ in MCF10A cells.
DISCUSSION
In this study, we have demonstrated that PCBP2 binds to core components of the Hippo signaling complex and affects basal signaling through Hippo. PCBP2 associates with Mst1, Mst2, and Sav1 in human cells and in mouse mammary tissue, liver, and lung, which rely on this pathway to control growth. PCBP2 stabilizes Mst1 within the Hippo complex by inhibiting Mst1 cleavage into a short form that is rapidly degraded. Loss of PCBP2 impairs the turnover of TAZ, leading to accumulation of nuclear TAZ and increased TEAD-mediated transcription of TAZ target genes. Enhanced TAZ activity in the absence of PCBP2 leads to stimulation of anchorage-independent growth. In this study, we employed a cell line, MCF10A, in which endogenously expressed PCBP2 and Hippo components control growth, and used siRNA to produce genetic deficiencies of Pcbp2 and Hippo components. Thus, potential artifacts produced by protein overexpression do not complicate the interpretation of our data. All of these data indicate that PCBP2 plays a biologically meaningful role in the activity of the Hippo signaling complex.
PCBP2 affects Hippo signaling through multiple mechanisms.
Cells lacking PCBP2 exhibited accumulation of TAZ that was largely due to its impaired turnover in the proteasome. Epistasis analysis indicated that this PCBP2-dependent effect on TAZ was primarily mediated through both Mst1 and Mst2, as depletion of both kinases simultaneously produced accumulations of TAZ similar to those produced by depletion of PCBP2 alone and depletion of Mst1/2 and PCBP2. Mst1 and Mst2 did not exhibit identical sensitivities to PCBP2, however, as only Mst1 exhibited cleavage and degradation. The amino-terminal kinase domains of both Mst1 and Mst2 are followed by autoinhibitory domains and protein-protein interaction (SARAH) domains. Caspase cleavage sites exist within these autoinhibitory domains; cleavage at these sites results in loss of the autoinhibitory and SARAH domains, which prevents association with Sav1 or other potential scaffold proteins (15, 20). PCBP2 may function directly to stabilize the Mst1-Sav1 heterodimer and sterically protect Mst1 from cleavage. PCBP2 could also function indirectly by inhibiting the expression or activation of proteases that might cleave Mst1. PCBP2 depletion by siRNA is associated with activation of apoptotic markers in HeLa and glioma cells (36, 37); thus, both mechanisms may be at work.
In contrast, Mst2 levels and binding to Sav1 were unaffected by depletion of PCBP2, suggesting that PCBP2 enhances the activity of the complex, apart from its effects on Mst1 stability. PCBP2 may link or modulate the activities of specific upstream regulators to the Hippo complex. Upstream signals known to activate Hippo are diverse, and it is likely that more signals have yet to be identified. These signals include agonists for G-protein-coupled receptors, mechanical stressors, and cell-cell contacts (16). Alternatively, PCBP2 may enhance the coupling of kinase activity to downstream targets.
Epistasis links Mst1/2 but not Lats1/2 to PCBP2-mediated TAZ activity.
Although the effects of PCBP2 on TAZ were largely mediated through Mst1/2 activity, these effects appeared to be unlinked to Lats, as depletion of Lats1/2 had no effect on the level of TAZ. The Hippo signaling pathway is not linear in mammalian cells and tissues, as it is in Drosophila. Not only can Mst1/2 associate with more than one type of scaffold/regulator (Sav1 or Rassf1 to -5), they can phosphorylate more than one type of downstream kinase (Lats1/2 and Ndr1/2) or directly phosphorylate transcription factors (FOXO1 and FOXO3) (15, 20). Genetic models of Mst1 and -2 deficiency in mice indicate that Hippo signaling can bypass Lats1/2 in the liver (25–27). Basal phosphorylation of Lats1/2 in mouse liver is not affected by deletion of Mst1/Mst2, although these mice exhibit diminished YAP phosphorylation, increased hepatocyte proliferation, and tumor formation in the liver. Our data indicate that Lats1/2 are not required for the basal regulation of TAZ in MCF10A cells, but Lats1/2 may be recruited under conditions where Hippo signaling is stimulated. Studies in breast cancer cell lines indicate that signaling through the G-protein-coupled estrogen receptor activates YAP and TAZ via inhibition of Lats1/2 kinase activity, an effect that does not appear to involve Mst1/2 (33). Our data indicated that the basal level of Hippo signaling in dividing MCF10A cells was sufficient to keep YAP- and TAZ-dependent transcriptional activity low, as target gene expression was minimally affected by YAP or TAZ depletion. PCBP2 may function to preserve this basal level of Hippo signaling in MCF10A cells. Modulating levels of PCBP2 in cells or tissues may also help to set this basal level of signaling.
PCBP2 affects TAZ to a greater extent than YAP.
Our data indicated that depletion of PCBP2 in MCF10A cells had a greater effect on TAZ stability than on YAP phosphorylation. TAZ may have a greater role than YAP in the development of some cancers, especially breast cancer. TAZ tends to be expressed at high levels in highly invasive breast cancer cell lines (31) and tumors (32). Here, we demonstrated an inverse relationship between PCBP2 expression levels and TAZ levels in two breast cancer cell lines, MCF7 and MDA-MB-231. While correlation does not prove causation, these results are consistent with a role for PCBP2 in regulating the activity of Hippo signaling in breast cancer cells. MCF10A cells engineered to overexpress TAZ exhibit enhanced migration and anchorage-independent growth (22, 31). In contrast, knockdown of TAZ in breast cancer cell lines inhibits proliferation (31, 32).
PCBP2 and its paralog, PCBP1, have been identified in many protein-protein interactions, altering the fate of metals, RNA, and proteins in the cell. Despite their structural similarity, they exhibit specific interactions and functions; both are independently essential to murine development (38). Murine embryos carrying constitutive deletion alleles of PCBP2 die in midgestation with cardiovascular and hematopoietic defects. Transcriptome analysis of liver tissue from these embryos revealed upregulation of numerous genes involved in development, including the TAZ target Cyr61. Further studies in conditional-deletion models may clarify the role of PCBP2 in regulating murine cell growth and division.
ACKNOWLEDGMENTS
We thank K. L. Guan for plasmids and C. Deng, C. Li, X. Xu, and J. T. Lahusen for cell lines and technical assistance.
These studies were supported by the Intramural Research Program, NIDDK, NIH. A. A. Vashisht and J. A. Wohlschlegel are supported by NIH GM089778.
We have no financial or nonfinancial competing interests.
F.L., K.Z.B., and A.A.V. carried out experiments. F.L., K.Z.B. J.A.W., and C.C.P. designed experiments and analyzed data. F.L. and C.C.P. prepared figures and wrote the manuscript.
REFERENCES
- 1.Chaudhury A, Chander P, Howe PH. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makeyev AV, Liebhaber SA. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278. doi: 10.1017/S1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostareck-Lederer A, Ostareck DH. 2012. Precision mechanics with multifunctional tools: how hnRNP K and hnRNPs E1/E2 contribute to post-transcriptional control of gene expression in hematopoiesis. Curr Protein Pept Sci 13:391–400. doi: 10.2174/138920312801619484. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Huang XH, Dong L, Hu D, Ge C, Zhan YQ, Xu WX, Yu M, Li W, Wang X, Tang L, Li CY, Yang XM. 2010. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer 9:72. doi: 10.1186/1476-4598-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji X, Wan J, Vishnu M, Xing Y, Liebhaber SA. 2013. alphaCP poly(C) binding proteins act as global regulators of alternative polyadenylation. Mol Cell Biol 33:2560–2573. doi: 10.1128/MCB.01380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimura K, Kano F, Murata M. 2008. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Lin L, Li Z, Ye X, Xiong K, Aryal B, Xu Z, Paroo Z, Liu Q, He C, Jin P. 2012. Iron homeostasis regulates the activity of the microRNA pathway through poly(C)-binding protein 2. Cell Metab 15:895–904. doi: 10.1016/j.cmet.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philpott CC, Ryu MS. 2014. Special delivery: distributing iron in the cytosol of mammalian cells. Front Pharmacol 5:173. doi: 10.3389/fphar.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidgens S, Bullough KZ, Shi H, Li F, Shakoury-Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A, Stemmler TL, Philpott CC. 2013. Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J Biol Chem 288:17791–17802. doi: 10.1074/jbc.M113.460253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Bencze KZ, Stemmler TL, Philpott CC. 2008. A cytosolic iron chaperone that delivers iron to ferritin. Science 320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandal A, Ruiz JC, Subramanian P, Ghimire-Rijal S, Sinnamon RA, Stemmler TL, Bruick RK, Philpott CC. 2011. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab 14:647–657. doi: 10.1016/j.cmet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey AG, Nandal A, Park JH, Smith PM, Yabe T, Ryu MS, Ghosh MC, Lee J, Rouault TA, Park MH, Philpott CC. 2014. Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc Natl Acad Sci U S A 111:8031–8036. doi: 10.1073/pnas.1402732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanatori I, Yasui Y, Tabuchi M, Kishi F. 2014. Chaperone protein involved in transmembrane transport of iron. Biochem J 462:25–37. doi: 10.1042/BJ20140225. [DOI] [PubMed] [Google Scholar]
- 15.Pan D. 2010. The hippo signaling pathway in development and cancer. Dev Cell 19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu FX, Guan KL. 2013. The Hippo pathway: regulators and regulations. Genes Dev 27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesuna F, Lisok A, Kimble B, Raman V. 2009. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia 11:1318–1328. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. 2009. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlschlegel JA. 2009. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol 497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 20.Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. 2012. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev Biol 23:770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat SJ, Chernoff J. 2015. Regulation of mammalian Ste20 (Mst) kinases. Trends Biochem Sci 40:149–156. doi: 10.1016/j.tibs.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. 2008. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol Cell Biol 28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C, Yao E, Chuang PT. 2015. A conserved MST1/2-YAP axis mediates Hippo signaling during lung growth. Dev Biol 403:101–113. doi: 10.1016/j.ydbio.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HY, Yu SL, Ho BC, Su KY, Hsu YC, Chang CS, Li YC, Yang SY, Hsu PY, Ho H, Chang YH, Chen CY, Yang HI, Hsu CP, Yang TY, Chen KC, Hsu KH, Tseng JS, Hsia JY, Chuang CY, Yuan S, Lee MH, Liu CH, Wu GI, Hsiung CA, Chen YM, Wang CL, Huang MS, Yu CJ, Chen KY, Tsai YH, Su WC, Chen HW, Chen JJ, Chen CJ, Chang GC, Yang PC, Li KC. 2015. R331W Missense mutation of oncogene YAP1 is a germline risk allele for lung adenocarcinoma with medical actionability. J Clin Oncol 33:2303–2310. doi: 10.1200/JCO.2014.59.3590. [DOI] [PubMed] [Google Scholar]
- 25.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. 2010. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang CY, Gao B, Jiang J, Yang Y. 2010. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A 107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 29.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, Li T, Chan SW, Lim CJ, Hong W, Zhao S, Xiong Y, Lei QY, Guan KL. 2010. The Hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem 285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waggoner SA, Liebhaber SA. 2003. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol Cell Biol 23:7055–7067. doi: 10.1128/MCB.23.19.7055-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. 2008. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 32.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Wang S, Wang Z, Feng X, Liu P, Lv XB, Li F, Yu FX, Sun Y, Yuan H, Zhu H, Xiong Y, Lei QY, Guan KL. 2015. Estrogen regulates Hippo signaling via GPER in breast cancer. J Clin Invest 125:2123–2135. doi: 10.1172/JCI79573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh D, Srivastava GP, Xu D, Schulz LC, Roberts RM. 2008. A link between SIN1 (MAPKAP1) and poly(rC) binding protein 2 (PCBP2) in counteracting environmental stress. Proc Natl Acad Sci U S A 105:11673–11678. doi: 10.1073/pnas.0803182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han W, Xin Z, Zhao Z, Bao W, Lin X, Yin B, Zhao J, Yuan J, Qiang B, Peng X. 2013. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest 123:2103–2118. doi: 10.1172/JCI61820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghanem LR, Kromer A, Silverman IM, Chatterji P, Traxler E, Penzo-Mendez A, Weiss MJ, Stanger BZ, Liebhaber SA. 2015. The poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol Cell Biol 36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]