ABSTRACT
Although implicated in neurodegeneration, autophagy has been characterized mostly in yeast and mammalian non-neuronal cells. In a recent study, we sought to determine if SPHK1 (sphingosine kinase 1), implicated previously in macroautophagy/autophagy in cancer cells, regulates autophagy in neurons. SPHK1 synthesizes sphingosine-1-phosphate (S1P), a bioactive lipid involved in cell survival. In our study, we discovered that, when neuronal autophagy is pharmacologically stimulated, SPHK1 relocalizes to the endocytic and autophagic organelles. Interestingly, in non-neuronal cells stimulated with growth factors, SPHK1 translocates to the plasma membrane, where it phosphorylates sphingosine to produce S1P. Whether SPHK1 also binds to the endocytic and autophagic organelles in non-neuronal cells upon induction of autophagy has not been demonstrated. Here, we determined if the effect in neurons is operant in the SH-SY5Y neuroblastoma cell line. In both non-differentiated and differentiated SH-SY5Y cells, a short incubation of cells in amino acid-free medium stimulated the formation of SPHK1-positive puncta, as in neurons. We also found that, unlike neurons in which these puncta represent endosomes, autophagosomes, and amphisomes, in SH-SY5Y cells SPHK1 is bound only to the endosomes. In addition, a dominant negative form of SPHK1 was very toxic to SH-SY5Y cells, but cultured primary cortical neurons tolerated it significantly better. These results suggest that autophagy in neurons is regulated by mechanisms that differ, at least in part, from those in SH-SY5Y cells.
KEYWORDS: autophagy, neurodegeneration, neurons, SH-SY5Y cells, sphingosine kinase 1
Autophagy involves an intricate machinery of organelle biogenesis, trafficking and fusion. Biogenesis of autophagosomes begins with an autophagosome precursor,1 the phagophore, which originates from multiple sources, including the endoplasmic reticulum (ER).2 The autophagosome then fuses with a lysosome for degradation of the sequestered contents. The autophagosome can also fuse with early or late endosomes, forming a hybrid organelle, an amphisome. The amphisome also fuses with the lysosome for turning over its contents.4 Mechanisms that regulate these fusion events are complex and not fully understood.
Although autophagy is implicated in neurodegenerative diseases, this process has been mainly studied in yeast, non-neuronal cells, and cell lines. Whether the mechanisms that govern the induction and regulation of autophagy in neurons are the same as in other cell types is not understood. Some proteins in post-mitotic neurons may be missing in dividing immortalized cells and vice versa. Differences between highly polarized neurons and non-polar cells may be also very important. For example, autophagosomes are continuously generated in the distal axon from the ER in neurons under basal conditions.5-8 When autophagy is stimulated, autophagosomes form frequently in the soma and proximal processes.9 Remarkably, autophagy-deficient fibroblasts, a commonly used model in the autophagy field, are viable.10 Yet, mice deficient in autophagy exhibit massive neuronal loss in the brain, suggesting that autophagy is important in neural survival.11 In addition, neurons appear to be less responsive to classical non-neuronal autophagy enhancers, such as amino acid starvation.12 However, withdrawal of insulin from culture media strongly induces autophagy in neurons.13,14 These observations underscore the significance of using primary neurons to study autophagy in neurodegeneration and the importance of the differences in autophagy between neurons and non-neuronal cells in taking efforts to target autophagy in neurons therapeutically.
In a recent paper, we investigated if SPHK1/SK1 (sphingosine kinase 1), which regulates autophagy in a breast cancer cell line,15 does so in primary neurons. SPHK1 generates sphingosine-1-phosphate (S1P), which regulates a wide variety of cellular processes, including cell survival and autophagy.15-17 We discovered that, in primary neurons, pharmacological stimulation of neuronal autophagy caused the formation of SPHK1-positive puncta (Fig. 1), which are positive for endosomal or autophagosomal markers or both.13 The 3 pools of organelles suggest that endosomes and autophagosomes fuse in neurons in which autophagy is induced. A dominant-negative form of SPHK1 inhibits autophagosome synthesis in neurons, suggesting that SPHK1 plays a role in the biogenesis of autophagosomes.13
Figure 1.
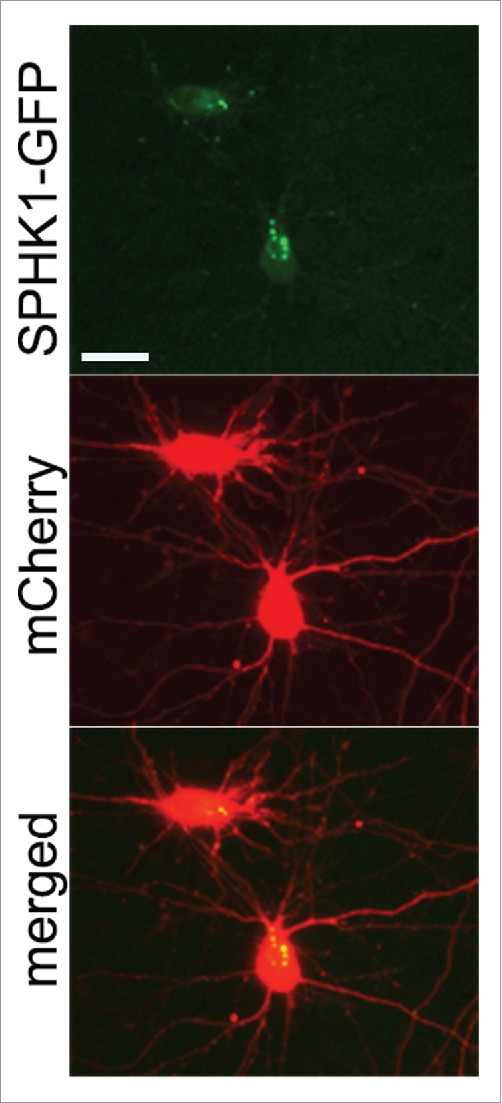
In primary rat cortical neurons, transfected SPHK1-GFP often exhibits a punctate appearance (top panel), whereas the distribution of a co-transfected marker of morphology mCherry (middle panel) is diffuse. Bar: 25 μm.
Upon stimulation of non-neuronal cells with growth factors, SPHK1 translocates to the plasma membrane, where it phosphorylates sphingosine to produce S1P.18 SPHK1 also associates with endosomes,19 phagosomes in macrophages,20,21 and endocytic tubules,22 induced by cholesterol extraction. Where SPHK1 localizes to in cell lines undergoing autophagy is not known. First, to investigate that the effect we observe in neurons is operant in non-neuronal cells, we expressed SPHK1-GFP in non-differentiated and differentiated SH-SY5Y cells, which have been commonly used in the field of neurodegeneration. Differentiation of SH-SY5Y cells with retinoic acid decreases proliferation rate and drives the phenotype toward cholinergic neuron-like cells.23 In most cells, SPHK1-GFP had a diffuse cytoplasmic distribution (Fig. 2A), consistent with observations in various cell lines.24 In rare cells, however, we observed SPHK1-GFP puncta in the soma or cell processes (Fig. 2A, B). Cells co-expressing mCherry revealed comparatively diffuse distribution of the red signal, thereby excluding the possibility that the GFP-positive puncta were the result of membrane blebbing in dying cells (Fig. 2B).
Figure 2.
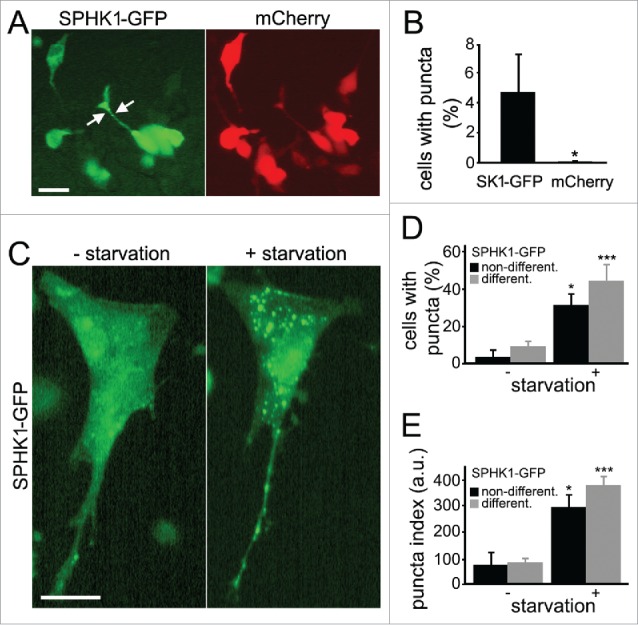
Stimulation of autophagy caused the formation of SPHK1-positive puncta in SH-SY5Y cells. (A) Ectopically expressed SPHK1-GFP exhibited a punctate appearance (left panel) in rare SH-SY5Y cells, whereas the distribution of a co-transfected marker of morphology, mCherry (right panel) was diffuse. Bar: 50 µm. (B) Percentage of SH-SY5Y cells with SPHK1-GFP- and mCherry-positive puncta under control conditions. *, P < 0.01 (t-test). (C) Stimulation of autophagy with amino acid withdrawal promoted the formation of SPHK1-GFP-positive puncta. Live cells transfected with SPHK1-GFP were observed with epifluorescence before (left panel) and after (right panel) amino acid withdrawal (2 h; HBSS solution). Bar: 50 µm. (D) Percentage of differentiated and nondifferentiated SH-SY5Y cells with SPHK1-GFP-positive puncta before and after amino acid withdrawal (2 h; HBSS solution). *, P < 0.01; ***, P < 0.0001 (t-test). (E) The puncta index was estimated by measuring the standard deviation of SPHK1-GFP fluorescence intensity in a region corresponding to the cell soma and major processes before and after amino acid withdrawal (2 h; HBSS solution). *, P < 0.01; ***, P < 0.0001 (t-test). a.u., arbitrary units.
To determine if autophagy stimulation would affect SPHK1 distribution, we expressed SPHK1-GFP in non-differentiated and differentiated SH-SY5Y cells and subjected the cells to starvation by amino acid withdrawal (Fig. 2C, D, E), which in many cell types leads to autophagy enhancement and the synthesis of new autophagosomes.25 In live SH-SY5Y cells, a short incubation of cells in amino acid–free medium stimulated the formation of SPHK1-GFP-positive puncta (Fig. 2C, D, E).
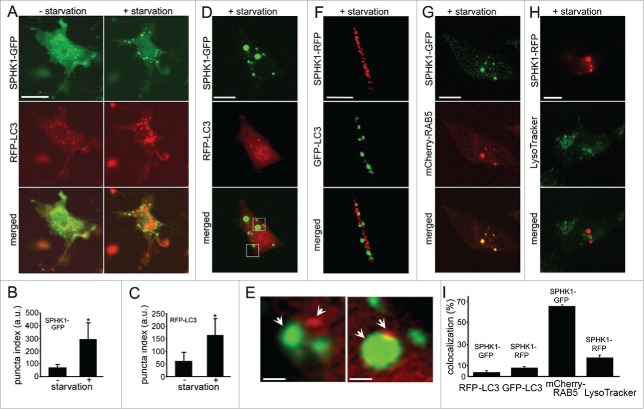
Since neurons and SH-SY5Y cells formed SPHK1-GFP puncta when autophagy was stimulated, we hypothesized that the puncta might represent endosomes, autophagosomes and amphisomes, similar to the situation in neurons.13 To test this, we co-expressed MAP1LC3/LC3 fused to TagRFP (RFP-LC3), along with SPHK1-GFP in SH-SY5Y cells. Live cells transfected with RFP-LC3 and SPHK1-GFP were subjected to amino acid withdrawal (Fig. 3A). As expected, both RFP-LC3 and SPHK1-GFP formed puncta (Fig. 3A, B, C). We looked at cells by confocal microscopy; starved cells were fixed, and the colocalization of RFP-LC3 and SPHK1-GFP was assessed (Fig. 3D). We found that, unlike neurons in which many SPHK1-GFP and RFP-LC3 structures colocalize,13 in SH-SY5Y cells SPHK1-GFP- and RFP-LC3-positive organelles were distinct structures (Fig. 3E, left panel). However, in some rare cases, we also observed a docking event between SPHK1-GFP- and RFP-LC3-positive organelles (Fig. 3E, right panel). To make sure that we did not detect organelles positive for both SPHK1-GFP and RFP-LC3 due to the quenching of the GFP signal when SPHK1-GFP- and RFP-LC3-positive organelles fuse, we used a SPHK1-TagRFP (TagRFP is pH-insensitive) construct (SPHK1-RFP). SH-SY5Y cells were transfected with SPHK1-RFP and GFP-LC3, starved, and the colocalization of SPHK1-RFP and GFP-LC3 was assessed. As expected, SPHK1-RFP did not localize to autophagic GFP-LC3-positive organelles (Fig. 3F). We, therefore, conclude that SPHK1 in neurons and in SH-SY5Y cells undergoing autophagy differentially colocalize with autophagosomal organelles.
Figure 3.
SPHK1 colocalizes with endosomes in SH-SY5Y cells. (A) Live SH-SY5Y cells transfected with SPHK1-GFP and RFP-LC3 were observed before (left panel) and after (right panel) 2 h-incubation in HBSS. Note that even before starvation, some RFP-LC3 autophagosomes were observed, suggesting SPHK1-GFP expression stimulates autophagy. Bar: 50 µm. (B) The puncta index was measured by calculating the standard deviation of SPHK1-GFP fluorescence in a region corresponding to the cell soma before and after starvation. *, P < 0.01 (t-test). (C) The puncta index was measured by calculating the standard deviation of RFP-LC3 fluorescence in a region corresponding to the cell soma before and after starvation. *, P < 0.01 (t-test). (D) Starved SH-SY5Y cells transfected with SPHK1-GFP and RFP-LC3 were fixed and observed with confocal microscopy. Bar: 10 µm. (E) The image D is zoomed in to show that no colocalization was observed between SPHK1-GFP- and RFP-LC3-positive organelles (left panel). A rare example of SPHK1-GFP and RFP-LC3 puncta docking (right panel). Bar: 0.2 µm. (F) Differentiated SH-SY5Y cells were transfected with SPHK1-RFP and GFP-LC3, starved (2 h; HBSS solution) and fixed. A neurite observed with confocal microscopy represents no colocalization between SPHK1-RFP and GFP-LC3. Bar: 5 µm. (G) Starved SH-SY5Y cells transfected with SPHK1-GFP and mCherry-RAB5 were fixed and observed by confocal microscopy. Bar: 10 µm. (H) Live and starved SH-SY5Y cells transfected with SPHK1-RFP were stained with 400 nM LysoTracker Green and imaged. Bar: 10 µm. (I) Colocalization of SPHK1-GFP and RFP-LC3 puncta, SPHK1-RFP and GFP-LC3, SPHK1-GFP and mCherry-RAB5, or SPHK1-RFP and LysoTracker Green was measured with the Coloc_2 plugin (ImageJ/Fiji).
Next, we tested whether the SPHK1-positive organelles in SH-SY5Y cells are LC3-negative endosomes or possibly lysosomes. First, we transfected SH-SY5Y cells with SPHK1-GFP and mCherry-RAB5, a marker of early endosomes, incubated them in amino acid-free medium to stimulate the formation of puncta, and then fixed and analyzed with confocal imaging (Fig. 3G, I). Colocalization analysis revealed that starvation stimulated the association of SPHK1-GFP with endosomes (Fig. 3I). Interestingly, SH-SY5Y cells developed enlarged SPHK1- and RAB5-positive endosomes, which were also observed in SH-SY5Y cells expressing SPHK1-GFP and RFP-LC3 (Fig. 3A, D). Indeed, RAB5 and lipid-modifying enzymes, such as phosphatidylinositol 3-kinase, promote homotypic endosomal fusion events, resulting in enlargement of endosomal vesicles.26-28 To confirm that SPHK1 does not associate with lysosomes, cells were transfected with SPHK1-RFP, starved, and stained with a green LysoTracker dye. As expected, SPHK1-RFP-positive puncta and LysoTracker Green-positive puncta were 2 distinct types of organelles (Fig. 3H, I).
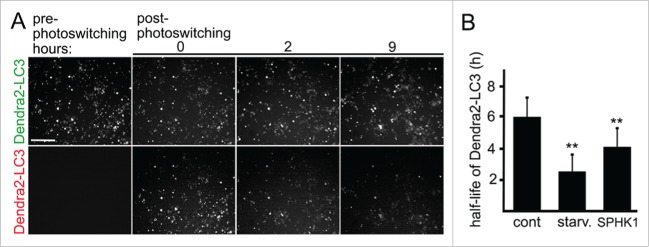
We recently demonstrated that overexpressed SPHK1 increases autophagic flux in primary cortical neurons.13 We also observed more RFP-LC3 puncta in SPHK1-GFP-expressing SH-SY5Y cells before starvation, suggesting that SPHK1 upregulated autophagy (Fig. 3A). To confirm that SPHK1 enhances, not inhibits, autophagic flux in SH-SY5Y cells, we used a photoswitchable automated platform that we recently developed to monitor the degradation of LC3.13,29,30 This system is based on the fluorescent properties of the Dendra2 protein derived from an octocoral.31 Upon brief irradiation with short-wave visible light, this fluorescent protein undergoes an irreversible conformational change (“photoswitch”). Photoswitching changes the spectral properties of Dendra2 from a protein that absorbs blue light and emits green fluorescence to one that absorbs green light and emits red fluorescence. Photoswitched Dendra-2 maintains these spectral properties until the cell degrades the protein, destroying its fluorescence. Three cohorts of SH-SY5Y cells were nucleofected with Dendra2-LC3. The first cohort was co-nucleofected with an empty plasmid. The second cohort was co-nucleofected with an empty plasmid and starved before the experiment. The third cohort was co-transfected with non-tagged SPHK1. SH-SY5Y cells were then photoswitched, and the decay of red fluorescence measured (Fig. 4A, B). The half-life of Dendra2-LC3 in SH-SY5Y cells was significantly shorter than in neurons.29,30 Withdrawal of amino acids, which we used as a positive control, increased the flux, as expected (Fig. 4B).15 We discovered that cells that overexpressed SPHK1 had enhanced degradation of photoswitched fluorescence (Fig. 4B). We also attempted to measure the flux in cells transfected with a dominant negative form of SPHK1 (dnSPHK1); however, dnSPHK1 was toxic, making analyses toilsome to do.
Figure 4.
SPHK1 enhances autophagic flux in SH-SY5Y cells. (A) Photoswitchable marker Dendra2-LC3 for measuring autophagic flux in live cells. Brief irradiation with short-wavelength visible light caused Dendra2 to undergo an irreversible conformational change and emit red fluorescence that can be tracked until altered molecules are cleared. Optical pulse-chase of SH-SY5Y cells nucleofected with Dendra2-LC3 allowed measuring the half-life of the autophagy marker in live cells. Note the decay of red fluorescence. (10x; scale bar: 200 μm). (B) The half-life of Dendra2-LC3 calculated from individual cell Dendra2-LC3 under different conditions. The half-life was reduced by starvation or by SPHK1 expression. **, P < 0.01 (ANOVA).
We recently found that SPHK1 has a direct role at the very early stages of the autophagic pathway.13 In primary neurons, SPHK1 is necessary for the biogenesis of autophagosomes. SPHK1 promotes the formation of pre-autophagosomal BECN1/Beclin 1-positive structures, and dnSPHK1 inhibits synthesis of autophagosomes. We, therefore, sought to confirm these findings in non-differentiated and differentiated SH-SY5Y cells. However, unlike neurons, dnSPHK1-GFP was very toxic to both non-differentiated and differentiated SH-SY5Y cells. We attempted several times to starve dnSPHK1-GFP- and RFP-LC3-transfected SH-SY5Y cells, but even brief starvation (>5 min) led to the detachment of SH-SY5Y cells from the substrate, making analysis impossible. These observations (e.g., dnSPHK1-GFP is toxic to a cancer cell line, and starvation greatly enhances dnSPHK1-GFP's toxicity) underscore the importance of SPHK1 for the survival of cancer cells during starvation.15
We were struck by the fact that dnSPHK1-GFP was so toxic to SH-SY5Y cells. We did not observe such toxicity of the dominant negative form in neurons. We decided to quantitatively analyze the toxicity of dnSPHK1 in neurons. We transfected 3 cohorts of primary cortical neurons: i) with mApple + GFP, ii) mApple + dnSPHK1-GFP, or iii) with mApple + dnSPHK1-GFP. The first 2 cohorts were treated with a vehicle, and the third was treated with an autophagy inducer, 10-NCP.9,13,29 Neurons were then imaged every 24 h with an automated microscope.13,30,32-34 Cumulative hazard plots revealed that neurons expressing dnSPHK1-GFP that were treated with 10-NCP displayed decreased survival relative to control neurons (Fig. 5). Although the toxicity indeed increased, a significant portion of neurons still survived, indicating that SPHK1-dependent autophagy was less important for neuronal survival than for cancerous SH-SY5Y cells (Fig. 5).
Figure 5.
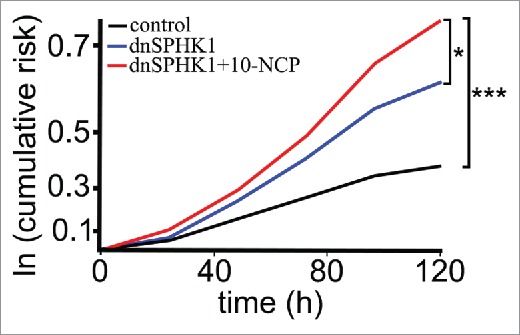
Survival analysis of neurons expressing a dominant negative form of SPHK1 in cultured cortical neurons. Primary cortical neurons transfected with mApple (a morphology and viability marker) and GFP or with mApple and dnSPHK1-GFP were tracked with an automated microscope. Some dnSPHK1-GFP-expressing neurons were stimulated with 10-NCP to induce autophagy. Cumulative risk for death was calculated with JMP software. 10-NCP increased the risk for death (i.e., reduced survival) of neurons expressing dnSPHK1. *, P = 0.01; ***, P < 0.001 (log-rank test).
Our findings demonstrate that SPHK1-mediated autophagy in cultured cortical neurons differs from that in cancerous SH-SY5Y neuroblastoma cells. In SH-SY5Y cells, upon induction of autophagy, SPHK1 preferably interacts with the endocytic pathway; in neurons, SPHK1 is associated with endosomes, autophagosomes, and amphisomes. In cancer cells, SPHK1 is critical for autophagy-dependent survival during starvation; in neurons, SPHK1 appears to be less significant for cell survival than in cancer cells, but still, the SPHK1 pathway is important for the degradation of misfolded proteins.13 Remarkably, sphk1 knockout mice are viable, although they exhibit several immune system abnormalities, and memory problems.35,36 Our findings are in an agreement with the data that the deletion of Sphk1 significantly attenuates the formation of cancers in mouse models.37,38 Whether SPHK1-deficient mice are prone to develop neurodegeneration still remains to be answered.
Materials and methods
Plasmids and chemicals
pGW1-mCherry, pGW1-SPHK1-GFP, pGW1-dnSPHK1-GFP, pGW1-TagRFP-LC3 (RFP-LC3), and pGW1-SPHK1-TagRFP (SPHK1-RFP) were previously described.13 pmCherry-RAB539 was from Addgene (27679; deposited by Dr. Christien Merrifield, Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom). Phosphate-buffered saline (PBS; D-1408), retinoic acid (R-2625), glycine (G-8898), and dimethyl sulfoxide (D-8779) were from Sigma. 10-(4′-(N-diethylamino)butyl)-2-chlorophenoxazine hydrochloride (10-NCP) was from EMD Millipore (124020). Triton X-100 was from Santa Cruz Biotechnology (sc-29112).
Cell line cultures and transfection
SH-SY5Y neuroblastoma cells were maintained in a 1:1 mixture of Dulbecco's modified Eagle's medium (HyClone, SH30022.01) and Ham's F12 medium (HyClone, SH30026.01) supplemented with 10% fetal bovine serum (Sigma, B-9433) and penicillin-streptomycin (Gibco, 15240–062). Before an experiment, SH-SY5Y cells were replated into 24-well tissue-culture plates (105/well). After 24 h, cells were transfected with Lipofectamine 2000 (Thermo Fisher Scientific, 12566014), according to the manufacturer's protocol, or were nucleofected with the Neon Transfection System (Thermo Fisher Scientific, Waltham, MA), according to the manufacturer's protocol.
Treatments
SH-SY5Y cells were starved in Hank's balanced salt solution (HBSS) for 2 h. In some experiments, SH-SY5Y cells were differentiated with 10 μM retinoic acid for 5 d in low-serum medium.
Fluorescence microscopy
Live-cell imaging was performed with a Nikon TE2000E-PFS microscope (a long-working-distance Nikon CFI S Plan Fluor 20× [NA 0.45] objective, a 300 W Xenon Lambda LS illuminator [Sutter Instruments, Novato, CA]) and the EVOS microscopy system (Thermo Fisher Scientific). Fixed cells were analyzed with a Zeiss LSM510 confocal microscope.
Image analysis
Puncta formation and puncta indexes were analyzed as described.9,13 Briefly, the redistribution of SPHK1-GFP into punctate structures was reflected by the puncta index, which is the standard deviation of the intensities measured among pixels within the cellular region of interest before and after treatment. Diffuse localization corresponds to a low puncta index, and punctate localization corresponds to a high puncta index.
The decays of photoswitched and non-photoswitched Dendra2 fluorescence were calculated by measuring “red” fluorescence intensity over a region of interest (fluorescence of non-photoswitched “green” molecules served as a guide for drawing the region of interest). The background-subtracted red intensities were plotted against time and transformed into log values.13,29,30
Colocalization with lysosomes
Since there is a possibility that SPHK1-GFP does not apparently colocalize with lysosomes due to quenching of the GFP signal, SPHK1-RFP13 was used to test if SPHK1-TagRFP colocalizes with LysoTracker Green DND-26 (Thermo Fisher Scientific, L-7526). Cells were transfected with pGW1-SPHK1-TagRFP and starved in HBSS for 2 h. Neurons were then treated with 400 nM Lysotracker Green DND-26 for 10 min and imaged.
Survival analysis
Cortical neurons were transfected with mApple (a morphology and viability marker; a gift from Dr. Kurt Thorn, University of California, San Francisco) + GFP or with mApple + dnSPHK1-GFP. Some neurons were treated with vehicle (dimethyl sulfoxide) or with 0.5 μM 10-NCP9,13 and imaged every 24 h for 1 wk. An image of the fiducial field with neurons on the plate was collected at the first time-point and used as a reference image for tracking the same neurons over time. Each time the same plate was imaged thereafter, the fiducial image was aligned with the reference image. Neurons that died during the imaging interval were assigned a survival time. These event times were used to obtain the exponential cumulative survival graphs and analyzed for statistical significance by log-rank test. Statistical analysis was performed with JMP (SAS Institute, Cary, NC).13,30,34
Ethics statement
Rats were maintained in accordance with guidelines and regulations of the University of Texas, Houston (protocol number #AWC-13-122). All experimental protocols were approved by the University of Texas, Houston.
Abbreviations
- 10-NCP
10-(4′-(N-diethylamino)butyl)-2-chlorophenoxazine hydrochloride
- dnSPHK1
dominant negative sphingosine kinase 1
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HBSS
Hank's balanced salt solution
- LC3
microtubule associated protein 1 light chain 3
- PBS
phosphate-buffered saline
- RFP
red fluorescent protein
- SPHK1
sphingosine kinase 1
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the University of Texas McGovern Medical School, Houston (A.T.), and the Hereditary Disease Foundation (A.T.). Support also was provided by 3R01 NS039074 (S.F.), NS083390 (S.F.) and the Taube/Koret Center for Neurodegenerative Disease Research.
References
- [1].Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol 2013; 25:455-60; PMID:23578367; http://dx.doi.org/ 10.1016/j.ceb.2013.03.004 [DOI] [PubMed] [Google Scholar]
- [2].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/ 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141:656-67; PMID:20478256; http://dx.doi.org/ 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I'm confused. Autophagy 2014; 10:549-51; PMID:24657946; http://dx.doi.org/ 10.4161/auto.28448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell 2014; 30:71-85; PMID:25026034; http://dx.doi.org/ 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy 2012; 8:858-60; PMID:22617438; http://dx.doi.org/ 10.4161/auto.20055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 2014; 84:292-309; PMID:25374356; http://dx.doi.org/ 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 2012; 196:407-17; PMID:22331844; http://dx.doi.org/ 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A 2010; 107:16982-7; PMID:20833817; http://dx.doi.org/ 10.1073/pnas.1004498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett 2006; 580:2623-9; PMID:16647067; http://dx.doi.org/ 10.1016/j.febslet.2006.04.008 [DOI] [PubMed] [Google Scholar]
- [11].Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al.. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; PMID:16625205; http://dx.doi.org/ 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- [12].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101-11; PMID:14699058; http://dx.doi.org/ 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moruno Manchon JF, Uzor NE, Dabaghian Y, Furr-Stimming EE, Finkbeiner S, Tsvetkov AS. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci Rep 2015; 5:15213; PMID:26477494; http://dx.doi.org/ 10.1038/srep15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem 2009; 284:2363-73; PMID:19017649; http://dx.doi.org/ 10.1074/jbc.M806088200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 2006; 281:8518-27; PMID:16415355; http://dx.doi.org/ 10.1074/jbc.M506182200 [DOI] [PubMed] [Google Scholar]
- [16].Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol 2012; 22:50-60; PMID:22001186; http://dx.doi.org/ 10.1016/j.tcb.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Sphingolipids in macroautophagy. Methods Mol Biol 2008; 445:159-73; PMID:18425450; http://dx.doi.org/ 10.1007/978-1-59745-157-4_11 [DOI] [PubMed] [Google Scholar]
- [18].Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA). J Biol Chem 2002; 277:35257-62; PMID:12124383; http://dx.doi.org/ 10.1074/jbc.M203033200 [DOI] [PubMed] [Google Scholar]
- [19].Hayashi S, Okada T, Igarashi N, Fujita T, Jahangeer S, Nakamura S. Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J Biol Chem 2002; 277:33319-24; PMID:12077123; http://dx.doi.org/ 10.1074/jbc.M201442200 [DOI] [PubMed] [Google Scholar]
- [20].Kusner DJ, Thompson CR, Melrose NA, Pitson SM, Obeid LM, Iyer SS. The localization and activity of sphingosine kinase 1 are coordinately regulated with actin cytoskeletal dynamics in macrophages. J Biol Chem 2007; 282:23147-62; PMID:17519232; http://dx.doi.org/ 10.1074/jbc.M700193200 [DOI] [PubMed] [Google Scholar]
- [21].Thompson CR, Iyer SS, Melrose N, VanOosten R, Johnson K, Pitson SM, Obeid LM, Kusner DJ. Sphingosine kinase 1 (SPHK1) is recruited to nascent phagosomes in human macrophages: inhibition of SPHK1 translocation by Mycobacterium tuberculosis. J Immunol 2005; 174:3551-61; PMID:15749892; http://dx.doi.org/ 10.4049/jimmunol.174.6.3551 [DOI] [PubMed] [Google Scholar]
- [22].Shen H, Giordano F, Wu Y, Chan J, Zhu C, Milosevic I, Wu X, Yao K, Chen B, Baumgart T, et al.. Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat Cell Biol 2014; 16:652-62; PMID:24929359; http://dx.doi.org/ 10.1038/ncb2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol 2013; 1078:9-21; PMID:23975817; http://dx.doi.org/ 10.1007/978-1-62703-640-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, Spiegel S. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol 1999; 147:545-58; PMID:10545499; http://dx.doi.org/ 10.1083/jcb.147.3.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci 1999; 112 (Pt 21):3667-75; PMID:10523503 [DOI] [PubMed] [Google Scholar]
- [27].Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 1998; 394:494-8; PMID:9697774; http://dx.doi.org/ 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- [28].Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell 1991; 64:915-25; PMID:1900457; http://dx.doi.org/ 10.1016/0092-8674(91)90316-Q [DOI] [PubMed] [Google Scholar]
- [29].Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, et al.. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 2014; 10(8):677-85; PMID:24974230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, Finkbeiner S. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol 2013; 9:586-92; PMID:23873212; http://dx.doi.org/ 10.1038/nchembio.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 2006; 24:461-5; PMID:16550175; http://dx.doi.org/ 10.1038/nbt1191 [DOI] [PubMed] [Google Scholar]
- [32].Mitra S, Tsvetkov AS, Finkbeiner S. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in huntington disease. J Biol Chem 2009; 284:4398-403; PMID:19074152; http://dx.doi.org/ 10.1074/jbc.M806269200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mitra S, Tsvetkov AS, Finkbeiner S. Protein turnover and inclusion body formation. Autophagy 2009; 5:1037-8; PMID:19838079; http://dx.doi.org/ 10.4161/auto.5.7.9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tsvetkov AS, Ando DM, Finkbeiner S. Longitudinal imaging and analysis of neurons expressing polyglutamine-expanded proteins. Methods Mol Biol 2013; 1017:1-20; PMID:23719904; http://dx.doi.org/ 10.1007/978-1-62703-438-8_1 [DOI] [PubMed] [Google Scholar]
- [35].Kanno T, Nishizaki T, Proia RL, Kajimoto T, Jahangeer S, Okada T, Nakamura S. Regulation of synaptic strength by sphingosine 1-phosphate in the hippocampus. Neuroscience 2010; 171:973-80; PMID:20950672; http://dx.doi.org/ 10.1016/j.neuroscience.2010.10.021 [DOI] [PubMed] [Google Scholar]
- [36].Niessen F, Schaffner F, Furlan-Freguia C, Pawlinski R, Bhattacharjee G, Chun J, Derian CK, Andrade-Gordon P, Rosen H, Ruf W. Dendritic cell PAR1-S1P3 signalling couples coagulation and inflammation. Nature 2008; 452:654-8; PMID:18305483; http://dx.doi.org/ 10.1038/nature06663 [DOI] [PubMed] [Google Scholar]
- [37].Heffernan-Stroud LA, Helke KL, Jenkins RW, De Costa AM, Hannun YA, Obeid LM. Defining a role for sphingosine kinase 1 in p53-dependent tumors. Oncogene 2012; 31:1166-75; PMID:21765468; http://dx.doi.org/ 10.1038/onc.2011.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kohno M, Momoi M, Oo ML, Paik JH, Lee YM, Venkataraman K, Ai Y, Ristimaki AP, Fyrst H, Sano H, et al.. Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol Cell Biol 2006; 26:7211-23; PMID:16980623; http://dx.doi.org/ 10.1128/MCB.02341-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 2011; 9:e1000604; PMID:21445324; http://dx.doi.org/ 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]