ABSTRACT
The activation of transcription factors is critical to ensure an effective defense against pathogens. In this study we identify a critical and complementary role of the transcription factors TFEB and TFE3 in innate immune response. By using a combination of chromatin immunoprecipitation, CRISPR-Cas9-mediated genome-editing technology, and in vivo models, we determined that TFEB and TFE3 collaborate with each other in activated macrophages and microglia to promote efficient autophagy induction, increased lysosomal biogenesis, and transcriptional upregulation of numerous proinflammatory cytokines. Furthermore, secretion of key mediators of the inflammatory response (CSF2, IL1B, IL2, and IL27), macrophage differentiation (CSF1), and macrophage infiltration and migration to sites of inflammation (CCL2) was significantly reduced in TFEB and TFE3 deficient cells. These new insights provide us with a deeper understanding of the transcriptional regulation of the innate immune response.
KEYWORDS: autophagy, immune response, macrophages, Tfe3, Tfeb
Introduction
Lysosomes are terminal degradative organelles present in all cell types. Extracellular and membrane bound materials are delivered to lysosomes via endocytosis and intracellular materials reach lysosomes through autophagy.1 Lysosomes' primary roles are in maintaining cellular homeostasis by degrading and recycling macromolecules and organelles; however, more specialized or cell type-specific roles include bone remodeling, unconventional secretion, plasma membrane repair, and growth factor and hormone regulation.2-3 As one of the primary cellular delivery systems to lysosomes, the autophagy pathway has similar physiological roles in regards to recycling extraneous organelles and macromolecules. In addition, autophagy has critical roles in cellular quality control mechanisms, being essential for the efficient removal of defective organelles and aggregated or misfolded proteins, with broad implications in diverse pathological states such as cancer and neurodegeneration.4
The basic helix-loop-helix transcription factor TFEB (transcription factor EB) belongs to the MiT/TFE family together with MITF (microphthalmia-associated transcription factor), TFE3 (transcription factor 3) and TFEC (transcription factor EC).5 It has been shown that TFEB controls lysosomal biogenesis and autophagy by regulating the expression of several lysosomal and autophagic genes.6-7 This transcription factor is normally located in the cytoplasm and translocates to the nucleus in response to nutrient deprivation and metabolic stress.7 TFE3 regulates a similar set of genes and, like TFEB, undergoes cytoplasm-to-nucleus shuttling in response to starvation.8 The mechanism underlying TFEB and TFE3 nuclear translocation is mediated by the phosphorylation status of particular serine residues.7,9-11 In spite of these remarkable similarities between TFEB and TFE3, it is still unclear whether these transcription factors have cooperative, complementary, or partially redundant roles.
Under conditions of abundant intracellular amino acids, TFEB and TFE3 are recruited directly to the lysosomal surface through their interaction with a heterodimeric GTPase complex consisting of GTP-bound RRAGA (Ras-related GTP binding A) or RRAGB (RRAGA/B) and GDP-bound RRAGC or RRAGD (RRAGC/D).12 The mechanistic target of rapamycin (serine/threonine kinase) complex 1 (MTORC1) is a master regulator of cell growth and metabolism, which integrates signals ranging from cellular ATP levels, growth factor signaling, cellular redox status, and nutrient abundance.13 Similarly to TFEB and TFE3, MTORC1 is recruited to lysosomes in nutrient-abundant conditions through the interaction of one of its subunits, RPTOR/raptor, to GTP-bound RRAGA/B and GDP-bound RRAGC/D.14-15 During these conditions, MTORC1 phosphorylates TFEB and TFE3, resulting in their cytosolic sequestration. Conversely, under starvation conditions, TFEB and TFE3 are no longer repressed by MTORC1-mediated phosphorylation, resulting in their nuclear translocation and allowing them to execute lysosomal and autophagy transcriptional programs.9-11 Another recently identified player in the regulation of TFEB activity is the phosphatase PPP3/calcineurin, which dephosphorylates TFEB critical serine residues, thus promoting its nuclear translocation.16
While the response of TFEB and TFE3 to nutrient status, as well as their role in regulating the autophagy-lysosome system, has been well characterized, little is known regarding the role of other types of cellular stress in the induction of TFEB and TFE3 transcriptional activity. Cells of the innate immune system, such as macrophages, are activated in response to pathogens, which are detected by pattern recognition receptors such those in the toll-like receptor (TLR) and nucleotide-binding domain (NLR) families.17-19 Macrophage activation results in drastic changes in the expression of a number of gene sets, including those responsible for inflammatory, chemoattractant, and antimicrobial effectors, among others. This process involves the rapid activation of various transcription factors including NFKB1, interferon-regulatory factor family (IRF) proteins, and CREB1.20 Numerous studies have demonstrated essential roles for the autophagy-lysosome system in macrophages and other cells of the innate immune system in response to pathogen exposure.21 These include antigen processing and presentation, degradation of phagocytosed pathogens, NK and T-cell cytotoxic granule secretion, and TLR signaling.22 In addition, autophagy is implicated in direct engulfment of intracellular pathogens and in modulating inflammatory signaling through the inflammasome complex.23-24
Given the specific and inducible roles of the autophagy-lysosome system in macrophage activation, we hypothesized that lysosome-autophagy regulators TFEB and TFE3 may be involved in this process. Here, we report that in addition to their roles in the starvation response, TFEB and TFE3 are also important for host immune response to infection. Endogenous TFE3 translocated from the cytosol to the nucleus of macrophages and microglia in response to TLR activators and live bacteria through a process that did not require MTORC1 inactivation. Mechanistically, we found that TFE3 occupied promoters of autophagic, lysosomal, and host-immune response genes in LPS (lipopolysaccharide)-stimulated cells; and, mutants lacking TFE3 and TFEB were unable to induce autophagy and lysosomal biogenesis efficiently. Remarkably, transcriptional induction of genes involved in the inflammatory response was greatly hampered in cells deficient in TFEB and TFE3 and these mutant cells were unable to induce secretion of key mediators of the inflammatory response, macrophage differentiation, and macrophage migration to sites of inflammation. Together, our data suggest a new and crucial role of TFEB and TFE3 in regulating host immune response to infection.
Results
TFE3 translocates from the cytosol to the nucleus in response to macrophage activation
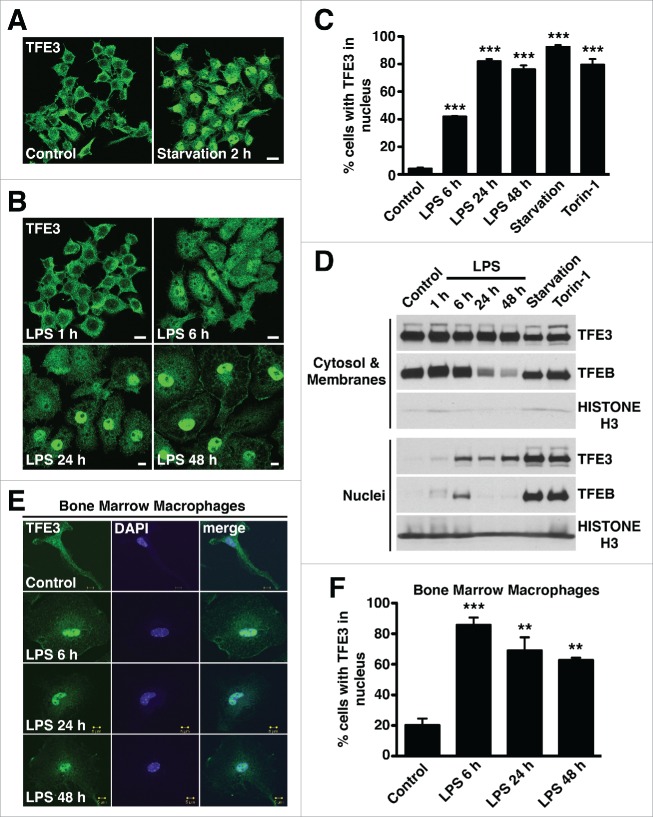
We examined TFE3 nuclear translocation in macrophages in response to a variety of stimuli. In order to determine if macrophages exhibit starvation-induced TFE3 translocation, as previously described by Martina et al.8 in epithelial cells, we starved RAW 264.7 mouse macrophages in EBSS (Earle balanced salt solution) for 2 h. As expected, nutrient-deprived RAW 264.7 cells exhibited a rapid global redistribution of TFE3 from the cytosol to the nucleus (Fig. 1A and C). Similarly, treatment with the MTOR (mechanistic target of rapamycin [serine/threonine kinase]) inhibitor Torin-1 in nutrient replete cells induced a similar level of TFE3 nuclear translocation (Fig. 1C), indicating that MTORC1 activity during nutrient and growth factor abundance actively keeps TFE3 in the cytoplasm of macrophages.
Figure 1.
Induction of TFE3 nuclear translocation in macrophages. (A) RAW 264.7 macrophages starved for 2 h in EBSS display rapid redistribution of TFE3 from the cytosol to nucleus. Scale bar: 10 µm. (B) RAW 264.7 macrophages treated with LPS exhibit a slow and sustained redistribution of TFE3 from the cytosol to nucleus. Nuclear localization peaks by 24 h and is maintained up to 48 h after treatment. Scale bar: 10 µm. (C) Quantification of TFE3 nuclear translocation from panels (Aand B). Treatment with Torin-1 for 3 h served as a positive control for nuclear translocation. *** denotes P value < 0.001 by one-way ANOVA analysis (n = 4, >300 cells per trial). (D) Nuclear-cytoplasmic fractionation of TFE3 and TFEB in Raw 264.7 macrophages. Western blots indicating total TFE3 and TFEB levels in whole cell lysates as well cytosolic/membrane fractions and nuclear fractions. TFE3 is undetectable in the nuclei of untreated cells. Starvation in EBSS for 2 h or treatment with Torin-1 for 3 h induces an increase in TFE3 detectable in the nuclear fraction whereas LPS treatment from 6 to 48 h induces a relatively lower level of nuclear TFE3. Histone H3 serves as specific marker of nuclear fraction. (E) Mouse primary bone marrow macrophages exhibit redistribution of TFE3 from the cytosol to nucleus in response to LPS stimulation. TFE3 translocates to the nucleus by 6 h and is sustained for up to 48 h. Scale bar: 5 µm. (F) Quantification of TFE3 nuclear translocation from panel (E). *** denotes P value < 0.001, and **< 0.01 by one-way ANOVA analysis (n = 3, >390 cells per trial).
Because of the unique roles of the autophagy-lysosome system in macrophages in response to pathogen exposure, we hypothesized that TFE3 may also translocate to the nucleus during the process of macrophage activation. RAW 264.7 cells were treated with LPS, which activates macrophages via toll-like receptor 4 (TLR4). After 6 h of LPS treatment, cells adopted a spread out morphology with membrane projections indicative of their activated status. This change in morphology was accompanied by a measureable increase in the immunofluorescent TFE3 signal seen in the nucleus, with approximately equal distribution spread between the nucleus and cytosol. However, a full nuclear localization to the levels seen in starvation and Torin-1 treatment was not detected until 24 h LPS treatment, which was sustained up to 48 h after treatment (Fig. 1B and C). Thus, LPS treatment requires much longer time to induce TFE3 nuclear translocation compared to nutrient deprivation, indicating that the kinetics of these 2 different mechanisms of TFE3 activation are significantly different. As expected, TFE3 activation in response to LPS was TLR4-dependent since TFE3 nuclear translocation was significantly impaired in TLR4-deleted cells (Fig. S1A to C).
We performed nuclear-cytosolic fractionation to biochemically confirm the presence of nuclear TFE3 after LPS treatment of RAW 264.7 cells. Virtually no TFE3 was detected in the nuclear fraction in unstimulated cells while abundant TFE3 was detected in the nucleus of starved and Torin-1-treated cells, which served as a positive control. Measureable amounts of TFE3 were detected in the nuclear fraction in LPS-treated cells at 6, 24, and 48 h (Fig. 1D). We also detected accumulation of endogenous TFEB in the nucleus at 6 h following LPS stimulation (Fig. 1D). In contrast to TFE3, the amount of nuclear TFEB decreased at 24 and 48 h of LPS treatment, concomitant with a sharp reduction in total TFEB protein levels (see below). These results indicate that both transcription factors respond to macrophage activation.
To verify that the TFE3 nuclear translocation observed in response to LPS treatment in RAW 264.7 cells can also be seen in primary macrophages, we performed the same experiment in mouse bone marrow-derived macrophages (BMDM). As expected, TFE3 nuclear translocation was observed in these cells in response to LPS, however the kinetics were slightly different, with a more rapid induction and a lower level of sustained nuclear TFE3 after 48 h (Fig. 1E and F). Similarly, mouse primary microglia also exhibited a pronounced TFE3 nuclear localization after 6 and 24 h of LPS treatment (Fig. S1D). To rule out that TFE3 translocation is an LPS-specific phenomenon, rather than a general feature of macrophage activation, we tested other stimuli known to activate macrophages. The small molecule R848 is a TLR7 agonist.25 Both LPS and R848 induced a similar level of nuclear TFE3 localization in the mouse alveolar macrophage line, MH-S after 24 h (Fig. S1E). The gram-positive bacterium, Mycobacteria smegmatis, possesses a variety of immunogenic glycopeptidolipids that stimulate TLR2.26 RAW 264.7 cells exposed to M. smegmatis at a multiplicity of infection of 100 exhibited an activated morphology that correlated with TFE3 nuclear translocation that increased up to 24 h after exposure (Fig. S2A and B). All together, our results confirm that TFE3 and TFEB accumulate in the nucleus following macrophage activation.
Nuclear translocation of TFE3 in response to LPS is MTORC1-independent
MTORC1 plays a critical role in the regulation of TFE3 activity in response to nutrient levels. In fully fed cells, MTORC1 phosphorylates TFE3 at serine 321 (S321), thus promoting binding of TFE3 to YWHA/14-3-3 family proteins and retention of the transcription factor in the cytosol. Inactivation of MTORC1 by starvation leads to dissociation of the TFE3-YWHA/14-3-3 complex and transport of TFE3 to the nucleus.8 To understand the mechanism of TFE3 activation in response to LPS, we generated a phospho-specific antibody that recognizes TFE3 only when phosphorylated at S321. Next, we performed subcellular fractionation in RAW 264.7 cells following treatment with either LPS or the MTOR inhibitor Torin-1. Interestingly, we found that nuclear TFE3 was not recognized by our phospho-TFE3 antibody (Fig. 2A). This indicates that, similar to starvation, nuclear translocation of TFE3 in response to LPS requires dephosphorylation of S321. However, because of the relatively slower kinetics and somewhat lower amount of TFE3 that translocates to the nucleus in LPS-treated conditions versus starvation or MTORC1 inhibition, we hypothesized that TFE3 translocation during macrophage activation may be governed by a mechanism other than that caused by MTORC1 inhibition.
Figure 2.
Mechanistic analysis of TFE3 nuclear translocation induced by macrophage activation. (A) Immunoblots of TFE3-Ser321 phosphorylation state in nuclear and cytosolic fractions of RAW 264.7 cells incubated with DMSO (Ctrl.), LPS (24 h), or Torin-1 (2 h). (B) Representative western blots of RAW 264.7 cells treated with LPS for 1, 6, 24 and 48 h. Starvation with EBSS and treatment with Torin-1 were used as positive controls for MTORC1 inhibition. (C) Quantification of 2 independent experiments from (B). (D) MTOR and LAMP1 colocalization in RAW 264.7 macrophages treated with DMSO (Control) or LPS for 1, 6, 24 and 48 h. Starvation with EBSS serves as a positive control of the inactivation and dissociation of MTOR from lysosomal membranes. Insets are 2.6-fold magnification of the selected areas. Scale bars: 10 µm. (E) PPP3/calcineurin inhibition reduces TFE3 nuclear localization in RAW 264.7 macrophages treated with LPS. Pretreatment of cells with the PPP3/calcineurin inhibitor, FK506, reduces the number of cells with nuclear TFE3 after 24 h LPS treatment. * denotes P value < 0.05 by one-way ANOVA analysis (n = 3, > 470 cells per trial).
To test this, we performed western blots on lysates from RAW 264.7 cells treated with LPS or subjected to starvation or MTORC1 inhibition and monitored the phosphorylation status of MTOR, AKT (thymoma viral proto-oncogene), and the MTORC1 substrates RPS6KB and EIF4EBP1. As expected, starvation and Torin-1 treatment led to a complete absence of detectable phospho-RPS6KB and phospho-EIF4EBP1 (Fig. 2B and C). Untreated cells and cells treated with LPS from 6 to 48 h, however, exhibited abundant phospho-MTOR, phospho-AKT, phospho-RPS6KB, and phospho-EIF4EBP signals, thus indicating that MTORC1 remains active as TFE3 translocates to the nucleus (Fig. 2B and C). Sustained MTORC1 activity following LPS treatment was also observed in BMDM and BV2 microglial cells (Fig. S3A and B). Furthermore, starvation of RAW 264.7 cells for 2 h resulted in a clear redistribution of MTORC1 from the lysosomal surface to the cytosol (Fig. 2D). In contrast, MTORC1 remained associated with lysosomes at all times in LPS-treated cells, further suggesting that MTORC1 remains active in activated macrophages (Fig. 2D). Together, our data indicate that LPS-mediated TFE3 translocation is mechanistically distinct from that caused by starvation and MTORC1 inhibition. An alternative explanation would be that LPS inhibits the activity of MTORC1 selectively on specific substrates such as TFE3.
We have recently shown that PPP3/calcineurin, a calcium-activated phosphatase, is locally activated by lysosomal Ca2+ release which dephosphorylates TFEB, allowing active translocation to the nucleus.16 This raises the possibility that a PPP3/calcineurin, or another phosphatase could actively dephosphorylate TFE3, thus allowing nuclear import even under conditions of MTORC1 activity. RAW 264.7 cells pretreated with the PPP3/calcineurin inhibitor, FK506 before LPS stimulation showed a statistically significant decrease in the percentage of cells with nuclear TFE3 after 24 h (Fig. 2E). This suggests that PPP3/calcineurin might participate in TFE3 activation in response to LPS. Nonetheless, the effect of FK506 on TFE3 localization was relatively weak, with approximately 50% of cells still exhibiting nuclear TFE3 after treatment. This may be due to a partial inhibition of PPP3/calcineurin by FK506 or, alternatively, other factors might be involved in regulating TFE3 phosphorylation status in activated macrophages.
TFE3 levels are sustained as TFEB levels decrease during macrophage activation
Since both TFE3 and TFEB respond to nutrient and energy sensing cues and regulate many of the same sets of genes, we wondered if they behaved similarly during RAW 264.7 activation. As shown in our subcellular fractionation experiment, we observed a pronounced decline in TFEB protein levels with increasing time exposed to LPS with a measureable decrease seen at 6 h and a nearly complete absence of detectable TFEB after 48 h, while TFE3 levels remained constant during the same time period (Fig. 1D). Decrease in TFEB protein levels following LPS activation was also observed in other cell types, including microglia-type BV2 cells and primary BMDM (Fig. S4A and B). However, in contrast to RAW 264.7 cells, TFEB levels returned to normal after 24-h LPS treatment in BV2 and BMDM cells (Fig. S4A and B). To assess whether variations in TFEB levels were due to changes in protein stability or to transcriptional regulation, we measured Tfeb mRNA levels by quantitative RT-PCR. Interestingly, we observed a significant decrease in Tfeb mRNA levels at 2 and 6 h of LPS treatment in BV2 and BMDM cells, with a subsequent recovery at 24 h (Fig. S4C and D). Decreased Tfeb levels were also observed in vivo following mouse intraperitoneal LPS injection, and included different organs such as brain, liver, and spleen (Fig. S4E). Interestingly, Tfeb reduction was accompanied in some instances by a concomitant increase in Tfe3 mRNA levels, which might be indicative of a compensatory mechanism between both transcription factors (Fig. S4C, D, and F). All together, our data suggest the presence of coordinated regulation of TFEB and TFE3 levels in activated macrophages and reveal cooperation of these transcription factors in the control of the innate immune response.
TFEB and TFE3 promote lysosome and autophagosome biogenesis in activated macrophages
To confirm that TFE3 and TFEB roles in lysosome biogenesis are relevant in the context of macrophage activation, we measured the rate of newly synthesized LAMP1 (lysosomal-associated membrane protein 1) as a proxy for lysosome biogenesis. Because TFE3 and TFEB are known to regulate transcription of many of the same lysosomal genes, we sought to generate Tfe3 and Tfeb single and double knockout cell lines to determine the relative contribution of each transcription factor to lysosome biogenesis induced by immune activation. To do this, we used a lentiviral CRISPR-Cas9 system to generate genome-edited RAW 264.7 cell lines with endogenous Tfe3 and Tfeb knocked out both alone and in combination (Fig. 3A).
Figure 3.
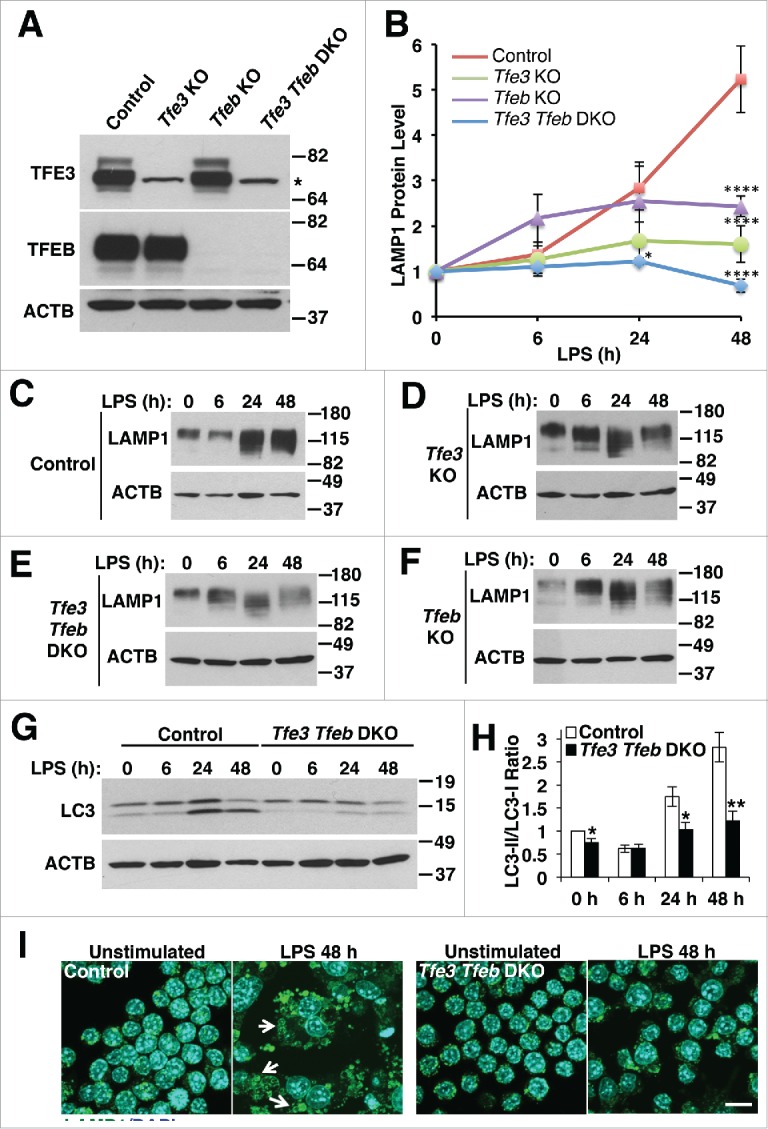
TFE3 and TFEB promote lysosome biogenesis and autophagosome formation in RAW 264.7 macrophages in response to LPS treatment. (A) RAW 264.7 macrophage Tfe3 KO, Tfeb KO and Tfe3 Tfeb double knockout (DKO) clones generated using CRISPR-Cas9 lentiviral transduction. Western blots of individual clones indicate the specificity of the KO cells. * indicates nonspecific cross-reacting band. (B) Relative LAMP1 protein levels quantified from western blots in panels C to F. LAMP1 levels normalized to actin loading control for each experiment. * denotes statistical significance, **** indicates highly significant result compared to control as determined by 2-way ANOVA with the Tukey multiple comparisons test. ((C)to F) Representative western blots of LAMP1 protein levels induced by (C) control, (D) Tfe3 KO, (E) Tfe3 Tfeb DKO, and (F) Tfeb KO clones of RAW 264.7 macrophages treated with LPS for 6 to 48 h. (G) Representative western blot of LC3-I and LC3-II levels in control and Tfe3 Tfeb DKO RAW 264.7 macrophages treated with LPS for 6 to 24 h. (H) Autophagosome formation in control and Tfe3 Tfeb DKO RAW 264.7 macrophages. Quantified by measuring ratio between LC3-II and LC3-I. * denotes P value < 0.05, and ** < 0.01 by the 2-tailed Student t test (n = 3). (I) Control, but not Tfe3 Tfeb DKO RAW 264.7 macrophages undergo distinct morphological changes in response to LPS treatment. Control cells treated with LPS for 48 h exhibit adherent and spread out cell borders and the presence of syncitia along with vacuoles and expanded LAMP1 positive structures (arrows). Scale bar: 10 µm.
As a heavily glycosylated membrane protein, LAMP1 exhibited a characteristic smear as newly synthesized protein accumulated in different parts of the endomembrane system in response to LPS stimulation (Fig. 3B to F). After 48-h LPS treatment, Tfe3 KO, Tfeb KO, and Tfe3 Tfeb double KO (DKO) all showed statistically significant decreases in LAMP1 levels relative to baseline (Fig. 3B to F). Interestingly, the control cells exhibited an approximately 5-fold upregulation of LAMP1 levels at 48 h, whereas the single KO cell lines had an approximately 2-fold upregulation of LAMP1 levels at the same time. Furthermore, the DKO line actually showed a reduction in LAMP1 levels relative to baseline, suggesting no net induction of lysosomal biogenesis caused by LPS in these lines. Only the DKO cell line showed a statistically significant decrease in LAMP1 levels at 24 h compared to controls (Fig. 3B). These data strongly imply that TFE3 and TFEB are at least partially redundant in terms of their ability to induce lysosome biogenesis in response to LPS, but both transcription factors must be present to achieve a maximal response.
Genes controlling autophagosome maturation are transcriptionally co-regulated by the same processes governing lysosome biosynthesis.7 We therefore hypothesized that autophagosome formation in activated macrophages would follow a similar pattern as lysosome biogenesis with DKO cells exhibiting a near complete block of LPS-induced autophagy. Autophagosome formation was assessed by measuring the ratio of lipidated to nonlipidated MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3) referred to as LC3-II (lipidated):LC3-I (nonlipidated). As with lysosome biogenesis, the DKO cells exhibited a strong reduction in the LC3-II:LC3-I ratio at 24 and 48 h after LPS addition. Interestingly, the DKO cells also exhibited a small, but statistically significant decrease in LC3-II:LC3-I in untreated conditions, suggesting a possible reduction in basal autophagy (Fig. 3G and H). Depletion of either Tfe3 or Tfeb was sufficient to reduce autophagy induction in response to LPS. However, this reduction was less pronounced than that observed in the DKO cells (Fig. S5A).
An increase in the LC3-II:LC3-I ratio may be indicative of an increased level of autophagosome biogenesis or a defect in the turnover of LC3-II in autolysosomes. In order to rule out the latter possibility, we performed an autophagic flux assay, in which cells treated with LPS were additionally treated with the lysosomal acidification inhibitor, bafilomycin A1 (BAF). Despite the increased relative LC3-II levels in control cells induced with LPS, there was a further increase in the LC3-II:LC3-I ratio in the BAF-treated samples. Furthermore, there were no statistically significant differences in the autophagic flux between control and DKO cells at any time points (Fig. S5B). These data confirm that autophagy is indeed induced in LPS-stimulated RAW 264.7 cells and that this induction is defective in DKO cells, thus highlighting the importance of TFEB and TFE3 in autophagy upregulation during macrophage activation.
Activated RAW 264.7 cells exhibit characteristic changes after prolonged incubation with LPS, including a spread out and more adherent morphology, dramatically enlarged lysosomes, and the presence of multinucleated cells indicative of cell-cell fusion. After 48-h treatment with LPS, a subpopulation of control RAW 264.7 cells was observed that typified all of these characteristics (Fig. 3I). Lysosomes typically range from 0.1 to 1.0 μm in diameter, however in these LPS-stimulated cells, numerous lysosomes >2 μm were observed. In the case of DKO RAW 264.7 cells, after 48-h LPS treatment, cells exhibited minimal spreading and virtually no cells had multiple nuclei or enlarged lysosomes comparable to those seen in control cells (Fig. 3I). Tfe3 and Tfeb single knockouts both had cell populations qualitatively similar to those described for LPS-activated control cells, suggesting that these 2 genes can partially compensate for the loss of the other, further substantiating the redundant functions of TFE3 and TFEB in certain aspects of macrophage activation (data not shown).
Identification of TFE3 direct-targets using ChIP-seq
To confirm that TFE3 directly binds to the promoter of lysosomal and autophagic genes in activated macrophages, we performed genome-wide analysis of the TFE3 transcriptional network in response to immune stress. For this, we stimulated RAW 264.7 cells with LPS for 24 h and performed a genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) study. Using a MACS 2 algorithm, 1,258 genes, which have peaks surrounding the transcription start site of their promoter, were identified in wild type cells treated with LPS. Importantly, TFE3 ChIP-seq peaks did not accumulate in either the Tfe3 KO cell line or the IgG control. To gain insight into the identities of promoters occupied by TFE3 after LPS stimulation, we used the web tool DAVID to perform gene ontology (GO) analyses. The analysis of “cellular compartment” terms revealed very significant enrichment (using modified Fisher Exact P Value) of genes involved in lysosome (10−19), V-ATPase complex (10−8), endosome (10−6), vacuolar membrane (10−4), and mitochondria (10−4) (Fig. 4A, Table S1). Components associated with melanosomes, membrane bound vesicles, nucleus, Golgi apparatus, and endoplasmic reticulum were also over-represented. Interestingly, we found a high degree of overlap between autophagic and lysosomal genes regulated by TFE3 under starvation or LPS conditions. In fact, over 95% of the autophagic and lysosomal genes modulated by TFE3 in LPS-treated cells also responded to TFE3 following starvation. These results suggest that TFE3 targets mostly the same autophagic and lysosomal genes independently of the stressor (Fig. S6A and Table S2).
Figure 4.
ChIP-seq analysis of TFE3 in LPS-stimulated RAW 264.7 cells. (A to C) GO analysis of the TFE3 binding sites. Numbers in parenthesis indicate the number of TFE3 targets. (A) Categories associated with “cellular compartment.” (B) Categories associated with “biological process.” (C) Categories associated with immune function. (D) Distribution of TFE3 binding sites in lysosomal, autophagy and endocytosis genes. Putative TFE3 binding sites tend to be concentrated within −1000 bp relative to the TSS. (E) Distribution of TFE3 binding sites in immune genes. Putative TFE3 binding sites were concentrated within −1500 bp or greater than −5000 bp relative to the TSS.
Analysis of “biological function” terms showed that proton transport was the most significant response to LPS stimulation (10−6). However, genes involved in lysosome organization, ion transport, cellular response to stress, cell cycle, autophagy, apoptosis, and cell redox homeostasis were also over-represented (Fig. 4B, Table S3). Importantly, 84 terms associated to the immune response were also highly significant (10−5) (Fig. 4B and C, Table S3). A more in-depth analysis of the immune response cluster showed that TFE3 occupied the promoters of genes involved in interleukin and cytokine production, positive regulation of the immune response, and development and activation of the immune response (Fig. 4C).
Previous work showed that genes regulated by TFEB have CLEAR (coordinated lysosomal expression and regulation) motifs (GTCACGTGAC) in their promoter regions to which the transcription factor binds; and, these CLEAR sequences were primarily localized within −1000 bp of the transcription start site (TSS).6,27 It has also been shown that TFE3 binds an E-box sequence (CANNTG) that overlaps with the CLEAR sequence.8,28 Using UCSC Genome Bioinformatics software, analysis of the genomic regions occupied by TFE3 demonstrated that similarly to TFEB, the E-box sequences of lysosomal, autophagy and endocytosis genes were primarily localized within −1000 bp of the genes' respective TSSs (Fig. 4D, Table S4). However, the E-box distance in immune genes was more dispersed. While some genes had E-boxes within −1500 bp of the TSS, a number of them also had these sequences greater than −5000 bp from the TSS (Fig. 4E, Table S4). To confirm our ChIP-seq results, we performed ChIP-qPCR for some of the putative TFE3 targets. As seen in Fig. S6B to E, we detected increased TFE3 binding to CLEAR motifs located in the promoter of several lysosomal and proinflammatory genes upon LPS treatment.
TFEB and TFE3 are required for cytokine production and secretion
Our ChIP-seq analysis unexpectedly identified numerous genes implicated in interleukin, as well as cytokine and chemokine production as putative TFE3 targets. To further assess whether TFEB and TFE3 mediate generation and secretion of cytokines and chemokines we stimulated wild-type or Tfe3 Tfeb DKO cell lines with LPS for 24 h and then assessed cytokine and chemokine secretion using a cytokine blot array. Evaluation of the cytokine secretion profile of LPS-stimulated control and Tfe3 Tfeb DKO macrophages is illustrated in Figure 5A. Notably, we found that the capability of Tfe3 Tfeb DKO cells to produce and secrete several cytokines in response to LPS stimulation was severely impaired compared to wild-type cells (Fig. 5A and B). Some of the components that were significantly compromised in the mutants include key mediators of the inflammatory response including CSF2 (colony-stimulating factor 2 [granulocyte-macrophage]), IL1B (interleukin 1 β), IL2 and IL27, macrophage differentiation including CSF1 (colony-stimulating factor 1 [macrophage]), and macrophage infiltration and migration to sites of inflammation activity of CCL2 (chemokine [C-C motif] ligand 2). While all of the noted cytokines were reduced by 60% to 80%, CCL2 was about 90% reduced in the mutant compared to the wild type (Fig. 5B). It is important to note that not all of the cytokines and chemokines on the array were affected by the loss of TFE3 and TFEB, suggesting the presence of specific immunoregulatory roles of these transcription factors (Fig. 5A).
Figure 5.
TFE3 and TFEB are required for cytokine production and secretion. (A) Cytokine array blots for control and Tfe3 Tfeb DKO RAW 264.7 cells that were stimulated with LPS for 24 h. Cytokine and chemokine levels of the supernatant were detected using the Mouse Cytokine Array Assay kit (R&D Systems) following manufacturer's protocol. (B) Quantification of cytokine and chemokine levels (n = 3). (Cto F) Relative Quantitative Real-Time PCR analysis of (C) Il1b, (D) Il6, (E) Tnf, and (F) Ccl5 mRNA transcript levels in control, Tfe3 KO, Tfeb KO, Tfe3 Tfeb DKO RAW cell lines after 6, 24, and 48 h of LPS stimulation (n = 3). The P values were calculated by the 2-tailed Student t test analysis of each mutant cell line vs. the control cells. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We further assessed cytokine/chemokine production capabilities by measuring mRNA production of Il1b (Fig. 5C), Il6 (Fig. 5D), Tnf (tumor necrosis factor) (Fig. 5E), and Ccl5 (chemokine [C-C motif] ligand 5) (Fig. 5F) in control, Tfe3 KO, Tfeb KO, and Tfe3 Tfeb DKO cell lines in response to LPS stimulation for 6, 24, or 48 h. mRNA transcripts were significantly reduced (∼80% reduction) for all of the tested mediators within 6 h of LPS stimulation and were nearly undetectable after 48 h.
We also confirmed the role of TFEB and TFE3 in inflammation in a model of murine microglial cells (BV2 cells) by knocking down expression of Tfeb, Tfe3, or both, using siRNA. qPCR analysis confirmed efficient depletion of Tfeb and Tfe3 (>60 %) in BV2 cells (Fig. S7A and B). As expected, treatment with LPS for 6 h further reduced Tfeb expression in knockdown cells, while Tfe3 expression remained unchanged (Fig. S7A and B). In agreement with our observations in RAW 264.7 cells, we found that Il1b, Tnf and Il6 mRNA levels were significantly reduced in knockdown cells after LPS treatment compared to control cells (Fig. S7C).
TFEB and TFE3 are required for inflammatory response in BMDM
To further assess the role of TFE3 and TFEB in primary macrophages, we examined LPS responses in BMDM isolated from mice depleted of Tfeb, Tfe3, or both, as well as their control counterparts (Tfebfl/fl). For this we generated Lyz2-Cre;Tfebfl/fl (Tfeb CKO) mice, in which myeloid cells (macrophages, dendritic cells and neutrophils) lack Tfeb expression. In addition, we generated conditional double KO mice, Lyz2-Cre;Tfebfl/fl;Tfe3Fcr/y (Tfe3 KO/Tfeb CKO), by crossing Tfe3Fcr/y(Tfe3 KO; mice with targeted disruption of the Tfe3 gene) with Tfeb CKO mice. Efficient depletion of Tfeb and Tfe3 in BMDM was determined by western blot (Fig. 6A) and qPCR analysis (Fig. 6B). As expected, Tfe3 protein and mRNA were completely absent, while Tfeb levels were significantly reduced. We then assessed lysosomal biogenesis and autophagy induction in BMDM following LPS treatment. As seen in Figure 6C and D, LAMP1 levels were reduced inTfe3 KO and Tfeb CKO cells both at 6 and 24 h post LPS treatment when compared to controls. Decreased LAMP1 levels were more pronounced in Tfe3 KO/Tfeb CKO BMDM, thus suggesting a synergistic effect of the 2 transcription factors. Immunofluorescence analysis for LAMP1 at 6 and 24 h of LPS treatment confirmed a reduction in lysosome size and numbers that, once again, was more evident in double knockout cells (Fig. S8). In addition, we observed a significant reduction in the levels of lipidated LC3-II in single and double knockout BMDM upon LPS treatment (Fig. 6C and E). Therefore, our results confirm a role of TFEB and TFE3 in lysosome biogenesis and autophagy induction in primary macrophages during the inflammatory response.
Figure 6.

TFE3 and TFEB are involved in the immune response in BMDM. (A) Protein levels of TFE3 and TFEB in control and KO BMDM cells. (B) mRNA transcript levels of Tfe3 and Tfeb in BMDM cells. (C) Representative western blot of LAMP1 and LC3 in control and KO BMDM cells treated with LPS for 6 to 24 h and (Dand E) relative quantifications. (Fand G) Cytokines mRNA levels in KO BMDM cells treated with LPS 6 to 24 h compared to control LPS-treated cells. (H) Secreted IL6 and (I) TNF levels in BMDM after 6 to 24 h of LPS treatment. The P values were calculated by the 2-tailed Student t test analysis of each genotype vs. the control cells. **, P ≤ 0.01; ***, P ≤ 0.001.
Next, we monitored expression of several cytokines and chemokines in KO cells by qPCR and ELISA. At 6 and 24 h after LPS treatment, we observed an approximately 30–50% reduction in transcript levels of analyzed cytokines and chemokines, including Il1b, Tnf, Il6 and Ccl5 (Fig. 6F and G). Moreover, absence of either TFE3 or TFEB or the absence of both transcription factors resulted in a modest but significant reduction of IL6 and TNF secretion at 6 and 24 h after LPS stimulation (Fig. 6H and I). These data confirm that TFE3 and TFEB are required for regulated cytokine production in primary macrophages.
To more extensively explore the effect of Tfe3 and Tfeb depletion in BMDM, we analyzed Tfebfl/fl, Tfe3 KO, Tfeb CKO, and Tfe3 KO/Tfeb CKO BMDM after LPS treatment by expression array profiling. Microarray analysis indicated that 568 genes were significantly downregulated and 313 upregulated in Tfe3 KO cells compared to control, 95 genes were downregulated and 48 upregulated in Tfeb CKO, while 927 were downregulated and 704 upregulated in Tfe3 KO/Tfeb CKO. The number of genes differentially expressed in Tfe3 KO/Tfeb CKO macrophages suggests an additive effect of Tfe3 and Tfeb in the global gene expression. The analysis of cellular compartment terms in Tfe3 KO/Tfeb CKO cells showed reduced expression of genes involved in vacuole and vacuolar membrane, lysosome and lysosomal membrane and endosome (Fig. 7A), while genes involved in endoplasmic reticulum, ribosome mitochondria and Golgi apparatus were upregulated (data not shown). Specifically, 108 lysosomal genes were significantly downregulated in Tfe3 KO/Tfeb CKO BMDM compared to control. As expected, this reduction was less pronounced in single KO cells, confirming some level of cooperation between the 2 transcription factors (Fig. S9A and Table S5).
Figure 7.

Expression profiling of stimulated Tfe3 KO/Tfeb CKO BMDM compared to Tfebfl/fl control. (Ato C) GO analysis of microarray data from Tfe3 KO/Tfeb CKO BMDM treated with LPS for 24 h. (A) “Cellular compartment” categories. (B) “Biological processes” categories. (C) “Immune Genes” category.
Analysis of biological processes revealed that most of the downregulated genes in Tfe3 KO/Tfeb CKO BMDM were involved in protein localization, intracellular signaling cascade, cellular response to stress, autophagy, cell death, lysosomal function, as well as in immune response (Fig. 7B). Importantly, over 70 genes associated with the immune response were significantly downregulated in Tfe3 KO/Tfeb CKO cells compared to control (Fig. 7C and Table S6). In addition, 20 immune genes were upregulated in Tfe3 KO/Tfeb CKO BMDM. These included genes implicated in the negative regulation of the immune response as well as genes related to immune system development (Fig. 7C). Once again, reduction in expression of immune genes was more pronounced in DKO than in single KO cells (Fig. S9B). Taken together, our data confirm a role of TFEB and TFE3 in the inflammatory response.
TFEB and TFE3 are required for innate immunity in vivo
To further confirm the physiological relevance of TFEB and TFE3 in inflammation, we analyzed LPS response in vivo. We injected Tfebfl/fl, Tfeb CKO, Tfe3 KO, and Tfe3 KO/Tfeb CKO mice with LPS intraperitoneally and collected liver, spleen, and lung to measure cytokines expression by qPCR. As expected, control mice stimulated with LPS for 4 h showed increased expression of Il1b, Tnf, Ccl3, Ccl4 and Ccl5 in analyzed tissues (liver, spleen, lung) compared to control untreated mice (Fig. 8A to C). Mice depleted of either Tfeb or Tfe3 showed reduced expression of most of the analyzed cytokines and chemokines (Fig. 8A to C). Altogether, our work reveals a novel and important role of TFE3 and TFEB in the transcriptional regulation of the innate immune response.
Figure 8.

Cytokine and chemokine expression in vivo. (Ato C) Relative quantitative analysis of cytokines and chemokines in (A) liver, (B) spleen, and (C) lung from Tfebfl/fl, Tfe3 KO, Tfeb CKO, or Tfe3 KO/Tfeb CKO mice after 4 h LPS treatment. n = 6 to 15. NT indicates nontreated control cells. The P values were calculated by the 2-tailed Student t test analysis of each genotype vs. the control cells. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Discussion
In this study we identify a new and critical role of TFEB and TFE3 in innate immune response. We found that TFE3 was rapidly recruited to the nucleus of activated macrophages where, in co-operation with TFEB, it promoted lysosomal biogenesis and autophagic activity. Furthermore, TFEB and TFE3 were required for the transcription and expression of a number of cytokines, chemokines, and other immune-related genes involved in the innate immune response. Using genomically edited knockout cell lines and in vivo models, we found that TFEB and TFE3 have partially redundant roles in the induction of lysosome biogenesis and in the induction of morphological changes associated to macrophage activation, and they are both required for a rapid and efficient induction of a subset of immune related genes. TFE3 levels appeared stable throughout the activation process whereas TFEB levels fell after the initial activating stimulus, suggesting that the relative levels of TFE3 and TFEB present at different points in the activation process may be used as a control mechanism for the temporal expression of different sets of genes.
In recent years it has become clear that TFEB and TFE3 play an important role in cellular adaptation to nutrient deprivation. The current paradigm establishes that activation of TFEB and TFE3 by starvation promotes transcriptional upregulation of numerous autophagic, lysosomal, and metabolic genes, thus favoring cell survival and adaptation to energy stress. Our current work reveals that these transcription factors also regulate cellular response to other types of stress, in particular, pathogen infection. Interestingly, the mechanism of activation of TFEB and TFE3 may vary depending on type of stress. Thus, starvation-mediated activation involves MTORC1, whereas pathogen-induced activation appears to be MTORC1-independent, or at least does not requires complete MTOR inactivation. This may reflect the different dynamics of the adaptation process. For example, fluctuations in nutrient levels cause MTORC1 inactivation, thus resulting in a rapid TFEB and TFE3 activation. In contrast, macrophages must remain active for long periods of time in order to support a strong immune response. Since prolonged inactivation of MTORC1 is probably not feasible, cells have developed alternative ways to achieve a sustained TFEB and TFE3 activation (e.g. by activation of particular phosphatases such as PPP3/calcineurin).
It is intriguing that cells rely on the same transcription factors to regulate 2 critical but apparently unrelated cellular functions, as is the case of nutrient sensing, energy homeostasis and immune response. One interesting possibility is that cells originally developed the TFEB and TFE3 pathway as a way to detect pathogen infection. It has been described that different bacterial pathogens trigger acute amino acid starvation following infection.29 TFEB and TFE3 could sense the change in amino acid levels induced by bacteria infection and translocate to the nucleus to perform a dual function: first, induction of autophagy as a way to engulf pathogens for delivery to lysosomes and subsequent degradation; and second, cytokine production to promote cell defense and alert neighboring cells of the presence of pathogens. Therefore, the primary role of TFEB and TFE3 may be cell defense and this role has specialized in some tissues to nutrient sensing and adaptation to starvation. Consistent with this idea, a recent study reports that HLH-30, the ortholog of TFEB and TFE3 in C. elegans regulates expression of antibacterial genes and plays an important role in innate immune defense in response to bacterial infection.30 This indicates that the role of TFEB and TFE3 in innate immune response is conserved from simple metazoans to higher organisms. With the acquisition of specialized proteins specialized in pathogen recognition (such as TLR and NLR family members), cells may have developed more complex signaling pathways to couple pathogen infection to TFEB/TFE3 activation.
Previous studies hint a role of TFE3 and TFEB in immune response. TFE3 has been implicated in myeloblast maturation toward the macrophage lineage, mast cell-mediated allergic response, T-cell dependent antibody production, and B-cell activation.31-34 In addition, the reorganization of the endo-lysosomal system induced by TFEB is thought to play a role in antigen presentation in dendritic cells and interferon-independent expression of antiviral genes.35-36 Our work helps to bridge the gaps in knowledge regarding the immune-specific cooperative roles of TFE3 and TFEB in the early and late stages of macrophage activation. We present a model in which the contributions of 2 related, but differentially expressed transcription factors, regulate multiple biological processes critical for a successful host immune response. This response relies on the synergistic effects of both transcription factors to promote a sustained upregulation of lysosomal and autophagy-related genes as well as a rapid and temporally induction of proinflammatory, chemoattractant, and immune specific growth factor mediators. In summary, our study identifies an alternative mode of TFE3 and TFEB activation and adds to the cadre of professional immune regulated transcription factors already known to regulate the innate immune response.
Materials and methods
Cell culture and in vitro differentiation
RAW 264.7 (ATCC, TIB-71), HEK293T (ATCC, CRL-3216), and primary microglia cells were grown in DMEM with GlutaMAX and sodium pyruvate (Gibco, 10569–044) supplemented with 10% fetal bovine serum (FBS; Gibco, 10438–026). MH-S cells were cultured in RPMI-1640 (ATCC, 30–2001) with 10% FBS. BV2 cells were obtained from IRCCS AOU (ICLC, ATL03001). Cells were grown in RPMI-1640 with GlutaMAX (Gibco, 61870–127) with 10% FBS (VWR, 16777–006). Cells were grown at 37°C in a humidified 5% CO2 chamber.
To obtain bone marrow-derived macrophages, BM cells were isolated from femurs and tibias of 8-wk-old female mice. Suspended BM cells were cultured in BM differentiation medium: RPMI-1640 with 2 mM L-glutamine (ATCC, 30–2001) supplemented with 10% FBS, 100 U/ml penicillin (Sigma Aldrich, P4333), 100 µg/ml streptomycin (Gibco, 15140–122), and 40 ng/ml recombinant murine M-CSF (Peprotech, 315–02) or 20% L929 conditioned medium. L-929 cells (ATCC, CCL-1) were grown in DMEM high glucose (VWR, 16777–200) supplemented with 10% FBS (VWR, 16777–006), 1% penicillin, 2 mM L-glutamine (Sigma Aldrich, G7513) for 1 wk and the medium was filtered using a 0.22 µm filter and store at −20°C. Cells were seeded in nontissue culture-treated Petri dishes (BD Biosciences, 351029) and incubated at 37°C in a 5% CO2 atmosphere. Four d after seeding, fresh BM differentiation medium were added to each plate and cells were incubated for an additional 3 d. To isolate BMDM, supernatants were discarded and attached cells were washed with sterile Hank's balanced salt solution (HBSS) (Gibco, 14025–134). Macrophages were detached by adding 2 ml of cell stripper (Corning, 25–056-CI) and incubated at 37°C for 5 min.
Microglia cells were extracted from P1 mouse brains. Every 4 dissected cortices were placed in 15 ml conical tubes with 10 ml of ice cold HBSS for 30 min. After aspirating HBSS, brains were triturated and digested with 4 ml of trypsin-EDTA at 37°C for 15 min. Enzymatic digestion was stopped by adding 4 ml complete microglial media consisting of DMEM with 10% FBS, 0.08% gentamicin (Gibco, 15710–064). Cells were centrifuged at 1500 rpm for 5 min, resuspended in 10 ml microglial complete media, and plated on a 10cm tissue culture dish coated with Poly-D-Lysine. Media was changed at d 3 and cells were harvested at d 10.
Generation of knockout lines
CRISPR-Cas9 guide RNA targeting sequences for mouse Tfeb and Tfe3 were identified bioinformatically using the CRISPR Design Tool available at https://www.genome-engineering.org.37 The targeting sequence search was limited to the first constitutively expressed exon common to all isoforms of the genes. Targeting sequences used were CACGTACTGTCCACCTCGGC for Tfeb and GAGGCGTGAGCGGCGGGAAC for Tfe3. Targeting sequences were cloned in to the lentiCRISPR plasmid (http://www.addgene.org/49535/) described in Shalem et al.38 Lentivirus was produced for the Tfeb and Tfe3 targeting sequences as well as an empty lentiCRISPR vector for control lines. Lentiviral transfer plasmids were cotransfected with VSV-G envelope (https://www.addgene.org/12259/) and packaging plasmids (https://www.addgene.org/12260/) in to HEK293T cells using Lipofectamine LTX (Invitrogen, 15338–500). Media was changed after 24 h and centrifuged and collected 72 h post-transfection.
RAW 264.7 cells at ∼30% confluency were transduced with hexadimethrine bromide (Sigma Aldrich, 107689) and viruses containing control, Tfeb, Tfe3, or both targeting sequences. Media was removed after 24 h and cells selected with 5 μg/ml puromycin (Sigma Aldrich, P8833). Individual clones were isolated by limiting dilution cloning, and knockouts of TFEB and TFE3 were confirmed via western blotting.
Macrophage activation
Macrophages were activated by treating RAW 264.7 or primary macrophages with 1 μg/ml LPS from Escherichia coli 0111:B4 (Sigma Alrcich, L4391) or with 0.2 μg/ml R848 (Enzo Life Sciences, ALX-420-038). BV2 and BMDM cells were activated with 100 ng/ml LPS from Salmonella enterica (Sigma Alrich, L6143).
Mycobacteria smegmatis strain mc(2)155 (ATCC, 700084) was grown to log phase by shaking at 37°C in Middlebrook 7H9 broth (Sigma Aldrich, M0178) supplemented with glycerol. Single cell suspensions were obtained by bath sonication and passage through a 5-μm filter. Bacteria were added to cells with a multiplicity of infection of 100 in prewarmed growth media. For starvation, cells were washed in phosphate-buffered saline (PBS; Gibco, 70011–044) and placed in Earle's balanced salt solution (EBSS; Sigma Aldrich, E2888) or HBSS (Gibco, 14025–134) for 2 h. Torin-1 (Tocris, 4247) was added to cells at 1.25 μM for 2 or 3 h. DMSO or water was used as vehicle controls for all experiments. FK-506 (Cell Signaling Technology, 9974) was used at a concentration 1 μM.
Antibodies
The following antibodies were used in this study: mouse anti-actin (BD Transduction Laboratories, 612656 and Novus Biological, NB600-501), mouse anti-GAPDH (Santa Cruz Biotechnology, sc-365062), rabbit anti-TFE3 (Sigma-Aldrich, HPA023881), rabbit anti-LC3B (Sigma-Aldrich, L7543 and Novus Biological, NB100-2220), rabbit anti-TFEB (Bethyl Laboratories, A303-673A), rat anti-LAMP1 from the Developmental Studies Hybridoma Bank deposited by August, J.T. (DSHB, 1D4B), rat anti-LAMP1 (Santa Cruz Biotechnology, sc-19992), rabbit anti-LAMP1 (Cell Signaling Technology, 9091); rabbit anti-AKT (Cell Signaling Technology, 4691), rabbit anti-phospho-AKT (Cell Signaling Technology, 4058), rabbit anti-RPS6KB (Cell Signaling Technology, 2708), rabbit anti-phospho-RPS6KB (Cell Signaling Technology, 9205), rabbit anti-EIF4EBP1 (Cell Signaling Technology, 9644), rabbit anti-phospho-EIF4EBP (Cell Signaling Technology, 2855), rabbit anti-Histone H3 (Cell Signaling Technology, 9003), rabbit anti-phospho-RPS6 (Cell Signaling Technology, 5364), rabbit anti-TLR4 (Cell Signaling Technology, 14358), mouse anti-RPS6 (Cell Signaling Technology, 2317), HRP conjugated donkey anti-rabbit (Cell Signaling Technology, 7074), donkey anti-mouse (Cell Signaling Technology, 7076), and goat anti-rat secondary antibodies (Southern Biotech, 3010-05), and Alexa Fluor 488 conjugated donkey anti-mouse (Life Technologies, A-21202), goat-anti rat (Life Technologies, A-11006), or and Alexa Fluor 594 conjugated donkey anti-rabbit (Life Technologies, A-21207) or donkey anti-mouse secondary antibodies (Life Technologies, A-21203).
Production of anti-phospho-TFE3 (Ser321) antibody
For antibody production, the synthesis and purification of a phospho-specific TFE3 peptide (AITVSN-pS-CPAELPN; amino acids 315 to 328), and a nonphosphorylated peptide counterpart (AITVSNSCPAELPN) as well as the production of polyclonal antisera was performed by YenZym Antibodies (South San Francisco, CA). Two New Zealand white rabbits were immunized with the phosphopeptide following a 90-d immunization protocol. The antisera were further purified by affinity chromatography against the same phosphopeptide used for immunization. The purified antibody was then affinity-absorbed against the nonphosphorylated peptide counterpart, to separate the phosphopeptide-specific antibody from the crossreactive population. The specificity of anti-phospho-TFE3 antibody was examined by immunoblotting.
Autophagic flux assays
Autophagy was induced in RAW 264.7 cells in duplicate wells with the addition of LPS for varying amounts of time. Two h prior to harvesting, one well in each condition was treated with 100 nM bafilomycin A1 (Sigma Aldrich, B1793). Whole cell lysates were run on western blots and the following calculated to quantify autophagic flux [(LC3-II:LC3-I) BAF-treated/(LC3-II:LC3-I) untreated].
Western blots
Cells were washed with PBS and lysed in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100 (Sigma Aldrich, T9284), with protease inhibitor (Roche, 11836170001) and phosphatase inhibitor cocktails (Roche, 04906837001). Whole cell lysates were homogenized by passing through 25G needles and centrifuged at 16,000 rcf at 4°C. Soluble fractions were mixed with NuPage 4X loading buffer (Life Technologies, NP0007) and 10X reducing agent (Life Technologies, NP0009) and heated at 98°C for 5 min. Equal volumes of lysate were run on Novex 4–20% Tris-Glycine gels (Life Technologies, EC6025) and transferred to 0.2-μm nitrocellulose membranes (GE Healthcare, 10600004). Blots were blocked for 1 h at room temperature in TBS (Quality Biological, 351-086-101) with 0.05% Tween 20 (Sigma Aldrich, P7949) (TBS-T) and 5% nonfat milk. Primary antibodies were incubated overnight at 4°C in TBS-T with either 5% nonfat milk or BSA (Sigma Aldrich, A3294). HRP-conjugated secondary antibodies were incubated 1 h at room temperature. Blots were washed with TBS-T, 3 times, 10 min each after both primary and secondary antibody incubations.
Blots were developed with Western Lighting Plus-ECL (Perkin-Elmer, NEL104001EA) and exposed on Biomax Light Film (Caresteream Health, 876–1520). Blots were quantitated with densitometric analysis using ImageJ (NIH) and normalized to ACTB/β-actin loading controls.
Immunofluorescence
Cells were grown on coverslips and fixed in 4% formaldehyde (Electron Microscopy Sciences, 15710) diluted in PBS for 15 min at room temperature. Slides were washed 3 times with PBS and permeabilized for 20 min in blocking buffer containing 0.1% saponin (Sigma Aldrich, S4521), 0.02% sodium azide, and 10% FBS in PBS. Cells for quantifying nuclear translocation were permeabilized for 10 min in a buffer with 0.1% Triton X-100 and 10% FBS in PBS. Primary and secondary antibodies were incubated in blocking buffer for 1 h at room temperature. Slides were washed in blocking buffer and PBS after primary and secondary antibodies. Coverslips were then mounted with Prolong Gold antifade reagent with DAPI (Life Technologies, P-36931). Images were acquired with an LSM 510 Meta confocal microscope (Zeiss, Oberkochen, Germany) with 63x numerical aperture 1.4 oil immersion objective with a Zeiss AxioCam camera. For TFE3 nuclear translocation in the TLR4 knockdown experiments, ≥350 cells were analyzed per condition using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA). Nuclear TFE3 was calculated by outlining DAPI-positive structures and calculating the average TFE3 staining intensity in each region.
Chromatin immunoprecipitation
For ChIP-seq, RAW 264.7 cells were stimulated for 24 h at 37°C with 1ug/ml LPS and subsequently crosslinked in 1% formaldehyde (Thermo Scientific, 28906) and processed according to the Myer's lab ChIP-seq protocol. Cells were lysed with Buffer 1 (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40 [Sigma Aldrich, NP40S], protease inhibitors) and pelleted. Pellets were resuspended in Buffer 1 and homogenized on ice and pelleted. Pellet was resuspended in Buffer 2 (1X PBS, 1% NP-40, 0.5% sodium deoxycholate [Sigma Aldrich, D6750], 0.1% SDS [Bio-Rad Laboratories, 161–0301], protease inhibitors). The chromatin fraction was sheared by sonication (15 × 30 seconds) in 1.5 ml siliconized microcentrifuge tubes. A 10-ug aliquot of DNA was reverse crosslinked to assess chromatin size (100 to 600 bp). 500 µg of the resulting sheared chromatin samples were cleared overnight at 4°C using DynaIbeads (Invitrogen, 10003D). Also, Tfe3 (5 µg/sample) and nonspecific rabbit IgG antibody (for background control) were incubated with magnetic beads overnight and washed with Buffer 3 (100 mM Tris, pH 7.5, 50 mM LiCl, 1% NP-40, 1% sodium deoxycholate). Precleared chromatin samples were added to washed beads and incubated at 4°C overnight. Beads were washed with Buffer 3 and then eluted and reverse crosslinked with Buffer 4 (1% SDS, 0.1 M NaHCO3) at 65°C overnight.
Analysis of ChIP sequencing
ChIP-Seq data were obtained using an Illumina HiSeq 2000/2500 sequencer (Ilumina, San Diego, CA, USA) and sequencing reads were de-multiplexed by Illumina pipeline, and mapped to the mouse genome (version mm9) using Bowtie algorithm with up to 2 mismatch allowed and reads mapping to more than 20 locations along the genome were discarded.39 ChIP-Seq data generated from IgG were used against the sample data in calling enriched regions and to control for the false discovery rate (FDR). Peaks were called using MACS version 2, with q-value set to 0.05, and peaks were assigned to genes using proximity distance of +/−5 kbp around the TSS as described by Liu et al. and Zhang et al.40-41 Gene ontology terms were categorized based on cellular localization and biological process; and, their P values obtained using DAVID Bioinformatics Resources 6.7. E-box sequences and distance from transcription start sites were analyzed using UCSC Genome Bioinformatics software. ChIP-Seq data and sample annotations were deposited in GEO under accession number GSE75822.
ChIP-qPCR (chromatin immunoprecipitation quantitative PCR)
Chromatin immunoprecipitatin (ChIP) analysis was performed according to the Simple ChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003). The chromatin was immunopreciptated with normal immunoglobulin G (IgG; negative control; Cell Signaling, 2729) and anti-TFE3 antibodies; 2% of the supernatant fraction from the chromatin lacking primary antibody was used as the “input sample” to generate the standard curve. Quantitative real-time PCR was performed using QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems, Life Technologies) in triplicate. Primers were designed to amplify the E-box element in the promoter regions of Ctsa, Ccl3, Tnfsf14, and Il6ra and 2000 base pairs upstream of those promoters. Primer list can be found in Table S7. The thermal profile for this reaction was: 95°C for 3 min and 46 cycles of 95°C for 15 seconds followed by 60°C for 1 min.
Mouse cytokine protein array analysis
Cultured RAW 264.7 cells from control or Tfe3 Tfeb DKO lines were left untreated or incubated with 1ug/ml LPS for 24 h. Cytokines and chemokines from cell culture supernatants were measured using a Mouse Cytokine Array Panel A kit (R&D Systems, ARY006) following the manufacturer's instructions. Quantitative analyses of cytokine or chemokine spots on each blot were performed using ImageJ.
RNA isolation and relative quantitative real-time PCR
WT, Tfe3 KO, Tfeb KO, and Tfe3 Tfeb DKO RAW 264.7 cells were left untreated or incubated with 1ug/ml LPS for 6, 24, or 48 h. RNA was isolated using PureLink RNA Mini Kit (Life Technologies, 12183018A) following the manufacturer's protocol. RNA was reverse transcribed using oligo(dT)20 and SuperScript III First-Strand Synthesis System (Life Technologies, 18080–051). BV2 cells and BMDM were left untreated or incubated with 100 ng/ml LPS for 1, 2, 6 or 24 h. RNA was isolated using RNeasy mini kit (Qiagen, 74106) following manufacturer's protocol. Total RNA from tissues was extracted in TRIzol reagent (Life Technologies, 12183–555) using RNeasy mini kit. RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Life Technologies, 4374967). Relative Quantitative Real-Time PCR was performed using 2 µl cDNA, 1 µl gene-specific primer mix (QuantiTect primer Assays, Qiagen), 5 µl SYBR Green ER qPCR SuperMix (Life Technologies, 11762100), and 2 µl water for a total reaction volume of 10 µl. Samples were run and analyzed using QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems, Life Technologies) in triplicate using the thermal profile of: 50°C for 2 min, 95°C for 10 min, 35 cycles of 95°C for 15 sec and 60°C for 60 sec. Amplification of the reference endogenous genes Gapdh or B2m was used to normalize the sequence of interest. The value is expressed as percent change compared to WT.
ELISA
BMDM cells were stimulated with LPS for 6 to 24 h and culture medium was analyzed by ELISA for IL6 (R&D Systems, M6000B) and TNF following the manufacturer's instructions (R&D Systems, MTA00B).
Gene knockdown
BV2 cells were plated in 6-well plates and transfected with siRNA specific for mouse Tfeb (GE Dharmacon, M-050607-00-0005) and Tfe3 (GE Dharmacon, M-054750-01-0005) at a final concentration of 50 nM for 48 h using Lipofectamine LTX (Life Technologies, 15338–100) following the manufacturer's instructions. RAW 264.7 cells were plated overnight in 6-well plates at a concentration of 5.0 × 105 cells per well and transfected with nontargeting control or Tlr4 ON-TARGETplus siRNA (GE Dharmacon, L-047487-00-0005) at a final concentration of 50 nM. Cells were transfected with Dharmafect 1 according to the manufacturer's instructions (GE Dharmacon, T-2001-01). Cells were split 24 h post transfection and analyzed for knockdown efficiency 72 h post-transfection.
Generation of mouse models
Conditional Tfeb-flox transgenic mouse line generation is described previously by Settembre et al.11 Lyz2-Cre mice are previously described by Clausen et al.42 Tfe3 KO mice are previously described by Steingrimsson et al.43 A summary of the genotypes and abbreviations used in this study are in Table S8. Mice used for experiments were all males and maintained in a C57BL/6 strain background. All experiments were approved by the Committee on Animal Care at Baylor College of Medicine and conform to the legal mandates and federal guidelines for the care and maintenance of laboratory animals.
Microarray
Microarray data were normalized by Robust Multiarray Average method as described by Irizarry et al., using R/BioConductor and Affy packages as described by Gentleman et al and Gautier et al.44-46 In order to detect the statistical significance of differential expression of this gene among the 4 different genotypes, we performed a one-way ANOVA and Tukey multiple comparison test as Post-ANOVA. For each P value, the Benjamini-Hochberg procedure was used to calculate the False Discovery Rate (FDR) to avoid the problem of multiple testing as described by Benjamini et al.47 The selected gene lists were obtained using the following thresholds: FDR < 0.05. The relative abundance of “Biological Process” (BP) Gene Ontology terms in each of the selected lists were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Clustering tool as described by Huang et al.48
Supplementary Material
Abbreviations
- BAF
bafilomycin A1
- BMDM
bone marrow-derived macrophages
- Cas9
CRISPR associated protein 9
- CCL2
chemokine (C-C motif) ligand 2
- ChIP-seq
chromatin immunoprecipitation sequencing
- CLEAR
coordinated lysosomal expression and regulation
- CKO
conditional knockout
- CRISPR
clustered regularly-interspaced short palindromic repeats
- DKO
double knockout
- EBSS
Earle's balanced salt solution
- ELISA
enzyme-linked immunosorbent assay
- FDR
false discovery rate
- GO
gene ontology
- GTP
guanosine triphosphate
- KO
knockout
- LAMP1
lysosomal-associated membrane protein 1
- LPS
lipopolysaccharide
- Lyz2
lysozyme 2
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 β
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MTORC1
MTOR complex 1
- PPP3/calcineurin
protein phosphatase 3
- qPCR
quantitative polymerase chain reaction
- RRAG
Ras-related GTP binding
- RT-PCR
reverse transcription polymerase chain reaction
- TFEB
transcription factor EB
- TFE3
transcription factor E3
- TLR
toll-like receptor
- TSS
transcription start site
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Gustavo Gutierrez (NIAMS) for his assistance in the analysis of the ChIP-seq data and Annamaria Carissimo (TIGEM Bioinformatic Core) for the analysis of microarray data. We are also grateful to Nick Platt (Univ. of Oxford, UK) for helpful suggestions.
Funding
O.B., H.D., J.M., L.S., and R.P. were supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI). L.J., H.Z., and N.R. were supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin diseases (NIAMS) of the NIH and a CRADA between NIH and Genzyme Corporation. N.P., T.H. and A.B. were supported by US National Institutes of Health (R01-NS078072) and by the Beyond Batten Disease Foundation.
References
- [1].Luzio JP, Hackmann Y, Dieckmann NMG, Griffiths GM. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb Perspect Biol 2014; 6:a016840; PMID:25183830; http://dx.doi.org/ 10.1101/cshperspect.a016840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sun-Wada GH, Wada Y, Futai M. Lysosome and lysosome-related organelles responsible for specialized functions in higher organisms, with special emphasis on vacuolar-type proton ATPase. Cell Struct Funct 2003; 28:455-63; PMID:14745137; http://dx.doi.org/ 10.1247/csf.28.455 [DOI] [PubMed] [Google Scholar]
- [3].Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 2009; 10:623-35; PMID:19672277; http://dx.doi.org/ 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- [4].Jiang PD, Mizushima N. Autophagy and human diseases. Cell Res 2014; 24:69-79; PMID:24323045; http://dx.doi.org/ 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994; 8:2770-80; PMID:7958932; http://dx.doi.org/ 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- [6].Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al.. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473-7; PMID:19556463 [DOI] [PubMed] [Google Scholar]
- [7].Settembre C, Di Malta C, Polito VA, Garcia-Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. Tfeb links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor Tfe3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9; PMID:24448649; http://dx.doi.org/ 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of Tfeb. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/ 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/ 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Tuong H, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via Mtor and Tfeb. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/ 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of Tfeb and MITF to lysosomes. J Cell Biol 2013; 200:475-91; PMID:23401004; http://dx.doi.org/ 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Laplante M, Sabatini DM. Mtor Signaling in Growth Control and Disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to Mtorc1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/ 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag Complex Targets Mtorc1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell 2010; 141:290-303; PMID:20381137; http://dx.doi.org/ 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al.. Lysosomal calcium signalling regulates autophagy through calcineurin and Tfeb. Nat Cell Biol 2015; 17:288-99; PMID:25720963; http://dx.doi.org/ 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med 2011; 3:513-27; PMID:21826793; http://dx.doi.org/ 10.1002/emmm.201100160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol 2011; 30:16-34; PMID:21235323; http://dx.doi.org/ 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- [19].O'Neill LAJ, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 2013; 13:453-60; PMID:23681101; http://dx.doi.org/ 10.1038/nri3446 [DOI] [PubMed] [Google Scholar]
- [20].Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol 2009; 9:692-703; PMID:19859064; http://dx.doi.org/ 10.1038/nri2634 [DOI] [PubMed] [Google Scholar]
- [21].Puleston DJ, Simon AK. Autophagy in the immune system. Immunology 2014; 141:1-8; PMID:23991647; http://dx.doi.org/ 10.1111/imm.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Colbert JD, Matthews SP, Miller G, Watts C. Diverse regulatory roles for lysosomal proteases in the immune response. Eur J Immunol 2009; 39:2955-65; PMID:19637232; http://dx.doi.org/ 10.1002/eji.200939650 [DOI] [PubMed] [Google Scholar]
- [23].Jo E-K, Yuk J-M, Shin D-M, Sasakawa C. Roles of autophagy in elimination of intracellular bacterial pathogens. Front Immunol 2013; 4:97; PMID:23653625; http://dx.doi.org/ 10.3389/fimmu.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shi C-S, Shenderov K, Huang N-N, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1 β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255-U74; PMID:22286270; http://dx.doi.org/ 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Grela F, Aumeunier A, Bardel E, Van LP, Bourgeois E, Vanoirbeek J, Leite-de-Moraes M, Schneider E, Dy M, Herbelin A, et al.. The TLR7 Agonist R848 Alleviates Allergic Inflammation by Targeting Invariant NKT Cells To Produce IFN-gamma. J Immunol 2011; 186:284-90; http://dx.doi.org/ 10.4049/jimmunol.1001348 [DOI] [PubMed] [Google Scholar]
- [26].Fujiwara N, Naka T, Ogawa M, Yamamoto R, Ogura H, Taniguchi H. Characteristics of Mycobacterium smegmatis J15cs strain lipids. Tuberculosis 2012; 92:187-92; PMID:22056691; http://dx.doi.org/ 10.1016/j.tube.2011.10.001 [DOI] [PubMed] [Google Scholar]
- [27].Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852-66; PMID:21752829; http://dx.doi.org/ 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- [28].Aksan I, Goding CR. Targeting the microphthalmia basic helix-loop-helix leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol 1998; 18:6930-8; PMID:9819381; http://dx.doi.org/ 10.1128/MCB.18.12.6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LAM, Yang C, Emili A, Philpott DJ, Girardin SE. Amino Acid Starvation Induced by Invasive Bacterial Pathogens Triggers an Innate Host Defense Program. Cell Host Microbe 2012; 11:563-75; PMID:22704617; http://dx.doi.org/ 10.1016/j.chom.2012.04.012 [DOI] [PubMed] [Google Scholar]
- [30].Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves A-MF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate Host Defense Requires Tfeb-Mediated Transcription of Cytoprotective and Antimicrobial Genes. Immunity 2014; 40:896-909; PMID:24882217; http://dx.doi.org/ 10.1016/j.immuni.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zanocco-Marani T, Vignudelli T, Gemelli C, Pirondi S, Testa A, Montanari M, Parenti S, Tenedini E, Grande A, Ferrari S. Tfe3 expression is closely associated to macrophage terminal differentiation of human hematopoietic myeloid precursors. Exp Cell Res 2006; 312:4079-89; PMID:17046750; http://dx.doi.org/ 10.1016/j.yexcr.2006.09.015 [DOI] [PubMed] [Google Scholar]
- [32].Yagil Z, Erlich TH, Ofir-Birin Y, Tshori S, Kay G, Yekhtin Z, Fisher DE, Cheng C, Wong WSF, Hartmann K, et al.. Transcription factor E3, a major regulator of mast cell-mediated allergic response. J Allergy Clin Immunol 2012; 129:1357-U249; PMID:22360977; http://dx.doi.org/ 10.1016/j.jaci.2011.11.051 [DOI] [PubMed] [Google Scholar]
- [33].Huan C, Kelly ML, Steele R, Shapira I, Gottesman SRS, Roman CAJ. Transcription factors Tfe3 and Tfeb are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat Immunol 2006; 7:1082-91; PMID:16936731; http://dx.doi.org/ 10.1038/ni1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Merrell K, Wells S, Henderson A, Gorman J, Alt F, Stall A, Calame K. The absence of the transcription activator Tfe3 impairs activation of B cells in vivo. Mol Cell Biol 1997; 17:3335-44; PMID:9154832; http://dx.doi.org/ 10.1128/MCB.17.6.3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Samie M, Cresswell P. The transcription factor Tfeb acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat Immunol 2015; 16:729-36; PMID:26030023; http://dx.doi.org/ 10.1038/ni.3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hasan M, Koch J, Rakheja D, Pattnaik AK, Brugarolas J, Dozmorov I, Levine B, Wakeland EK, Lee-Kirsch MA, Yan N. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat Immunol 2013; 14:61-71; PMID:23160154; http://dx.doi.org/ 10.1038/ni.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013; 31:827-32; PMID:23873081; http://dx.doi.org/ 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al.. Genome-Scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343:84-7; PMID:24336571; http://dx.doi.org/ 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; PMID:19261174; http://dx.doi.org/ 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 2014; 1150:81-95; http://dx.doi.org/ 10.1007/978-1-4939-0512-6_4 [DOI] [PubMed] [Google Scholar]
- [41].Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137; PMID:18798982; http://dx.doi.org/ 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 1999; 8:265-77; PMID:10621974; http://dx.doi.org/ 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- [43].Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A 2002; 99:4477-82; PMID:11930005; http://dx.doi.org/ 10.1073/pnas.072071099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31:e15; PMID:12582260; http://dx.doi.org/ 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, et al.. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80; PMID:15461798; http://dx.doi.org/ 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gautier L, Cope L, Bolstad BM, Irizarry RA. affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20:307-15; PMID:14960456; http://dx.doi.org/ 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- [47].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B-Method 1995; 57:289-300 [Google Scholar]
- [48].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.