Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Novel missense germ line DDX41 mutations define an earlier age of onset of hematologic malignancies than loss-of-function alleles.
Carriers of DDX41 germ line mutations usually have normal blood counts until a myeloid or lymphoid malignancy develops.
Abstract
Recently our group and others have identified DDX41 mutations both as germ line and acquired somatic mutations in families with multiple cases of late onset myelodysplastic syndrome (MDS) and/or acute myeloid leukemia (AML), suggesting that DDX41 acts as a tumor suppressor. To determine whether novel DDX41 mutations could be identified in families with additional types of hematologic malignancies, our group screened two cohorts of families with a diverse range of hematologic malignancy subtypes. Among 289 families, we identified nine (3%) with DDX41 mutations. As previously observed, MDS and AML were the most common malignancies, often of the erythroblastic subtype, and 1 family displayed early-onset follicular lymphoma. Five novel mutations were identified, including missense mutations within important functional domains and start-loss and splicing mutations predicted to result in truncated proteins. We also show that most asymptomatic mutation carriers have normal blood counts until malignancy develops. This study expands both the mutation and phenotypic spectra observed in families with germ line DDX41 mutations. With an increasing number of both inherited and acquired mutations in this gene being identified, further study of how DDX41 disruption leads to hematologic malignancies is critical.
Introduction
Inherited hematologic malignancies (HMs) typically present at earlier ages than de novo disease.1-4 Recently our group and others identified a class of familial myeloid malignancies due to mutations in DDX41, which encodes a DEAD-box RNA helicase.5 These families exhibit a familial risk of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), but they develop these malignancies at ages typical of de novo disease. More than 65% of familial cases previously identified harbor a heterozygous germ line frameshift mutation, DDX41 c.415_418dupGATG (p.D140Gfs*2), and 50% of MDS or AML diseases that develop in germ line carriers acquire a mutation on the other DDX41 allele, suggesting that DDX41 acts as a tumor suppressor.5 We searched for additional families with novel DDX41 germ line mutations to advance our understanding of this syndrome.
Methods
Patients
All patients in our study signed a written informed consent form to participate in research approved by the institutional review board and conducted in accordance with the Declaration of Helsinki under either the Australian Familial Haematological Cancer Study or The University of Chicago Familial Hematologic Malignancies registry. Study eligibility criteria and cohort phenotype breakdown are provided in the Supplemental Data (available on the Blood Web site).
Sequencing
Sequencing of samples was predominantly performed by whole exome sequencing for Australian samples as previously described6 or by MarrowSeq assay7 for samples collected at The University of Chicago. Some samples were screened by targeted polymerase chain reaction and Sanger sequencing of DDX41. Sample types and coverage for all individuals are shown in supplemental Table 5, and primers used for amplification and sequencing are shown in supplemental Table 4 (Australia) and supplemental Table 3 (Chicago). Reference sequences used throughout for DDX41 are NM_016222.2 and NP_057306.2.
Results and discussion
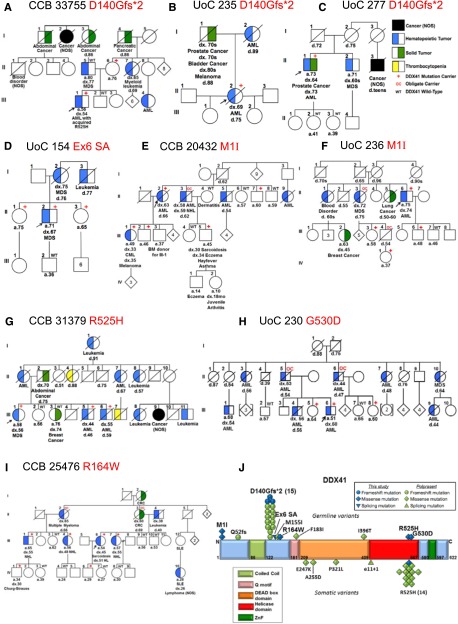
Families with suspected inherited HMs (289 families; see supplemental Data for phenotypic breakdown) were examined by whole exome sequencing; MarrowSeq, a panel-based next-generation sequencing assay; or by targeted Sanger sequencing of frequently mutated exons in DDX41 (see supplemental Table 6 for full details). Heterozygous germ line DDX41 mutations were identified in 9 new families (3%) who had no other detectable predisposition allele. Among these, 3 families carried the recurrent p.D140Gfs*2 mutation5; 1 family carried a germ line p.R525H (c.1574G>A) mutation, previously described only as a somatic mutation at the time of progression to MDS or AML; and 5 carried novel mutations (Figure 1, supplemental Table 1, and supplemental Data). The mean age of onset of MDS or AML in germ line DDX41 mutation carriers in this study was 57 years, younger than the 67 years previously reported (P = .006, two tailed Student t test).5 Combined, the mean age of onset of HMs in all reported germ line DDX41 mutation carriers is 62 years.
Figure 1.
Germ line DDX41 mutations in 9 families with inherited hematologic malignancies. (A-I) Pedigrees with germ line DDX41 mutations. (J) Germ line and somatic DDX41 mutations in hematologic malignancies. BM, bone marrow; CML, chronic myeloid leukemia; CRC, colorectal cancer; HL, Hodgkin lymphoma; NOS, not otherwise specified; SLE, systemic lupus erythematosis; ZnF, zinc finger
All 3 new families carrying the germ line DDX41 p.D140Gfs*2 truncating mutation (Figure 1A-C) exhibited late-onset MDS or AML (mean 67 years). Sequencing of leukemia DNA from 1 individual (33755-III-1) revealed an acquired p.R525H mutation consistent with previous findings (Figure 1A).5
A germ line substitution (c.435-2_435-1delAGinsCA) was identified in the splice acceptor site 5′ to the start of exon 6 in Family 154 (Figure 1D). Polymerase chain reaction of complementary DNA from skin fibroblasts from individual 154-II-2 revealed 2 aberrant splice products generated between exons 4 and 7 (supplemental Figure 1) that introduced frameshift mutations just downstream from the p.D140Gfs*2 mutation and are predicted to produce similarly truncated proteins (p.W146Hfs*29 and p.S145Rfs*17).
Clinical histories in several families suggested that germ line DDX41 mutations may predispose to a wider range of HMs. In 2 families, a novel missense start-loss substitution (c.3G>A, p.M1I) was identified (Figure 1E-F). Single nucleotide polymorphism analysis showed that all individuals in both families who carried the c.3G>A mutation also carried a tightly linked 5′ untranslated region variant, but a second single nucleotide polymorphism in exon 3 was unique to family 20432 (supplemental Table 2), suggesting that these families share an ancestral allele but are only distantly related. Individuals with p.M1I usually developed late onset MDS/AML, but one individual (20432-III-1) developed chronic myeloid leukemia and another (20432-II-3) developed both AML and non-Hodgkin lymphoma (NHL). Family 25476 (Figure 1I) in whom we identified a novel DDX41 missense variant, c.490C>T p.R164W segregating with lymphoid malignancies, adds further support to this observation. In total, 5 of 5 affected individuals with NHL (3 with follicular subtype, early-onset; Table 1), Hodgkin lymphoma, or multiple myeloma carried the p.R164W variant, whereas 3 of 3 unaffected individuals of similar ages did not. Arginine 164 is highly conserved across species (supplemental Figure 2) and is adjacent to the Q motif (Figure 1J), a region involved in adenosine triphosphate binding and hydrolysis,8 likely key for regulating DDX41 helicase activity.
Table 1.
Complete blood count and bone marrow characteristics of individuals with germ line DDX41 mutations
| Panel in Figure 1 | Pedigree | Individual | DDX41 mutation† | Age at time of CBC (y) | Status of HM at time of CBC | WBC (K/μL) | Hgb (g/dL) | MCV (fL) | ANC (K/μL) | Plt (K/μL) | HM diagnosis | BM cellularity | BM dysplasia | Karyotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Malignancy | ||||||||||||||
| A | 33755 | III.1 | p.D140Gfs*2 | 54 | At dx | 1.6 | 13.3 | 108.0 | 0.5 | 128 | AML (M6) | 20% | Erythroid dysplasia | 46,XY[20] |
| B | 235 | II.2 | p.D140Gfs*2 | 69 | At dx | 2.2 | — | — | — | 89 | AML | 15% | No | 46,XX[20] |
| C | 277 | II.1 | p.D140Gfs*2 | 73 | At dx | 1.5 | 8.5 | 112.0 | 0.3 | 62 | AML | 20%-40%; CD5+, CD19+ lymphoid aggregates | Erythroid dysplasia | 46,XY,del(5)(q31.1q34)[6]/ 46,idem,inv(1)(q21q31)[10]/46,XY[4] |
| D | 154 | II.2 | c.435-2_435-1delAGinsCA | 67 | At dx | 3.3 | 12.5 | 92.3 | 1.9 | 102 | MDS (RAEB-2) | 15%-25% | Erythroid dysplasia, rare dysplastic megakaryocyte or myeloid precursor | 46,XY[20] |
| E | 20432 | II.3 | p.M1I‡ | 58 | At dx1 | 0.7 | 8.7 | 103.0 | 0.1 | 34 | AML (M6) | Normal | Erythroid and megakaryocytic dysplasia | 46,XY |
| E | 20432 | II.3 | p.M1I‡ | 59 | At dx2 | 1.2 | 6.5 | 104.0 | 0.08 | 27 | AML now with NHL involvement | Hypocellular; lymphoid aggregates consistent with NHL | Erythroid and megakaryocytic dysplasia | — |
| E | 20432 | III.1 | p.M1I | 33 | 1 mo after dx | 34.7 | 11.0 | 92.0 | 25.3 | 333 | CML | Hypercellular | No | 46,XX, t(9;22)(q34;q11)[16]/ 46,XX[2] |
| F | 236 | II.3 | p.M1I‡ | 72 | At dx | 2.6 | 10.6 | 107.2 | 0.7 | 41 | MDS (RAEB-2) | 5%-10% | Megakaryocytic dysplasia | 46,XX[20] |
| F | 236 | II.6 | p.M1I | 74 | At dx | 4.1 | 13.3 | — | 1.4 | 66 | AML | 15%-20% | No | 47,XY,+8[2]/46,XY[18] |
| G | 31379 | III.1 | p.R525H | 56 | At dx | 3.2 | 15.0 | 102.0 | 1.7 | 171 | MDS (RAEB-2) | Hypercellular | Trilineage dysplasia | 46,XX |
| G | 31379 | III.1 | p.R525H | 58 | 24 mo after dx | 3.2 | 14.0 | 99.0 | 1.6 | 179 | MDS (RAEB-2) | — | Trilineage dysplasia | 46,XX [20] |
| G | 31379 | III.6 | p.R525H | 55 | At dx | 0.4 | 12.0 | 113.1 | 0.2 | 47 | AML | 20% | No | — |
| H | 230 | II.5 | p.G530D‡ | 53 | At dx | — | — | — | — | — | AML | — | — | — |
| H | 230 | II.6 | p.G530D‡ | 44 | At dx | — | — | — | — | — | AML | — | — | — |
| H | 230 | III.6 | p.G530D | 50 | At dx | 2.4 | 14.0 | 99.7 | 0.6 | 151 | AML | 30% | No | 46,XY[20] |
| I | 25476 | III.1 | p.R164W | 55 | At dx | 9.3 | 15.2 | 88.0 | — | 320 | FL (grade 2) | Hypercellular; FL involvement | Mild erythroid dysplasia | 46,XY[15] |
| I | 25476 | III.4 | p.R164W | 48 | At dx | 7.6 | 13.1 | 88.7 | — | 347 | FL (grade 1) | Hypercellular: FL involvement | No | — |
| I | 25476 | III.7 | p.R164W | 55 | At dx | 7.1 | 12.5 | 88.0 | 3.3 | 223 | FL (grade 1) | 30%-50% | No | 46,XX[20] |
| S5§ | 127 | II.11 | p.D140Gfs*2 | 73 | At dx | 3.6 | 12.5 | 97.9 | — | 135 | AML (M6) | 15%-20% | Erythroid dysplasia | 46,XY[20] |
| S5§ | 127 | III.7 | p.D140Gfs*2 | 56 | At dx | 1.1 | 10.0 | 95.8 | 0.2 | 27 | AML | 20% | No | 46,XY,del(20)(q11.21-q13.33)[4]/46,XY[14] |
| Premalignancy | ||||||||||||||
| A | 33755 | III.1 | p.D140Gfs*2 | 54 | 4 mo before AML dx | 2.3 | 15.0 | 102.0 | 1.0 | 145 | ||||
| A | 33755 | III.1 | p.D140Gfs*2 | 52 | 29 mo before AML dx | 5.2 | 15.7 | 92.0 | 3.5 | 225 | ||||
| D | 154 | II.1 | c.435-2_435-1delAGinsCA | 75 | NA | 8.7 | 12.3 | 90.0 | 5.6 | 184 | ||||
| E | 20432 | III.1 | p.M1I | 30 | 29 mo before CML dx | 7.9 | 12.7 | 87.0 | 6.2 | 274 | ||||
| F | 236 | II.6 | p.M1I | 72 | 23 mo before AML dx | — | — | — | — | 123 | ||||
| F | 236 | II.6 | p.M1I | 73 | 15 mo before AML dx | 3.5 | 13.3 | — | 1.5 | 72 | ||||
| H | 230 | III.5 | p.G530D | 63 | NA | 5.7 | 14.2 | 91.3 | 3.6 | 336 | ||||
| H | 230 | III.6 | p.G530D | 50 | 2 mo before AML dx | 2.8 | 14.6 | 97.8 | 1.0 | 185 | ||||
| H | 230 | III.8 | p.G530D | 59 | NA | 3.1 | 12.8 | 100.5 | 1.4 | 180 | ||||
| S5§ | 127 | II.6 | p.D140Gfs*2 | 87 | NA | 8.4 | 14.2 | 94.0 | 4.4 | 296 | ||||
| S5§ | 127 | II.8 | p.D140Gfs*2 | 85 | NA | 5.8 | 12.9 | 93.7 | 3.7 | 126 | None | 20%-30% | No | 46,XY[19] |
| S5§ | 127 | II.10 | p.D140Gfs*2 | 79 | NA | 10.0 | 13.0 | 96.0 | — | 213 | ||||
| S5§ | 127 | II.12 | p.D140Gfs*2 | 75 | NA | 7.5 | 13.9 | 103.0 | 3.7 | 354 | ||||
| S5§ | 127 | III.1 | p.D140Gfs*2 | 60 | NA | 5.1 | 14.6 | 89.6 | — | 189 | ||||
| S5§ | 127 | III.3 | p.D140Gfs*2 | 65 | NA | 9.4 | 15.0 | 90.0 | — | 231 | ||||
| S5§ | 127 | III.8 | p.D140Gfs*2 | 57 | NA | 6.0 | 16.7 | 89.2 | 3.8 | 187 | ||||
| S5§ | 127 | IV.2 | p.D140Gfs*2 | 35 | NA | 8.4 | 14.4 | 91.3 | — | 348 | ||||
| S5§ | 127 | IV.3 | p.D140Gfs*2 | 20 | NA | 4.5 | 13.3 | 94.2 | 2.6 | 165 | ||||
| Controls | ||||||||||||||
| C | 277 | III.2 | WT | 41 | NA | 8.3 | 14.0 | 89.3 | 5.7 | 213 | ||||
| C | 277 | III.3 | WT | 38 | NA | 11.3 | 15.1 | 98.0 | — | 202 | ||||
| H | 230 | III.7 | WT | 65 | NA | 6.2 | 14.2 | 94.4 | — | 262 | ||||
| S5§ | 127 | III.5 | WT | 62 | NA | 4.9 | 13.3 | 88.3 | 2.8 | 213 | ||||
| S5§ | 127 | III.10 | WT | 46 | NA | 6.8 | 13.5 | 90.0 | 4.4 | 313 | ||||
Dash indicates that information is not available.
AML, acute myeloid leukemia; ANC, absolute neutrophil count; BM, bone marrow; CBC, complete blood count; CML, chronic myeloid leukemia; dx, diagnosis; FL, follicular lymphoma; Hgb, hemoglobin; HM, hematologic malignancy; M6, French-American-British subtype; MCV, mean corpuscular volume; MDS, myelodysplastic syndrome; NA, not applicable; NHL, non-Hodgkin lymphoma; Plt, platelet count; RAEB-2, refractory anemia with excess blasts 2; WBC, white blood cell count, WT, wild type.
Numbering for all complementary DNAs and proteins refers to NM_016222.2 and NP_057306.2.
Obligate carrier.
Pedigree 127 is included in supplemental Figure 5.
Finally, 2 families with germ line missense mutations in the helicase domain were identified: 1 carried a p.R525H mutation (Figure 1G and supplemental Figure 3), and 1 carried p.G530D (Figure 1H). Because the p.R525H mutation has been shown to affect the interaction of DDX41 with splicing factors,5 we predict that the nearby p.G530D mutation would have a similarly disruptive capacity. Both families feature high penetrance MDS/AML, with younger ages of HM onset compared with other DDX41 variants (50 vs 63 years; P = .0004, two-tailed Student t test).
Expression of the DDX41 wild type (WT) and mutant proteins in human embryonic kidney cells showed that for WT, R525H, and R164W, the full-length (70 kDa) protein was detected, and localization was predominantly nuclear (supplemental Figure 6), indicating that these mutations do not overtly affect protein translation or localization. In contrast, using a construct engineered to mimic the consequences of the exon 6 splice acceptor mutant, a truncated protein was observed as expected (supplemental Figure 6B). For M1I, disruption of the initiating methionine resulted in a predominant smaller protein (∼55 kDa), indicating the use of an alternate internal translation initiation site (supplemental Figure 6A,C). Interestingly, in addition to full-length protein, the WT and point mutant DDX41 constructs also produced the smaller DDX41 isoform, suggesting that this isoform may occur naturally at some level. Immunofluorescence showed that the smaller isoform was altered in localization, with reduced nuclear and increased cytoplasmic detection (M1I panel, supplemental Figure 6D), consistent with the loss of a predicted nuclear localization signal at the amino terminus (supplemental Figure 7).
Table 1 summarizes the clinical presentation of HMs in DDX41 mutation carriers and blood counts for any family members, when available. The majority of carriers unaffected by or prior to HMs had normal peripheral blood counts well into adulthood (9 of 15 [60%]; see premalignancy characteristics in Table 1), suggesting that haploinsufficiency of DDX41 is sufficient for normal baseline hematopoiesis. Cytopenias or macrocytosis (mean corpuscular volume >100 fL) developed in the remaining 6 at a mean age of 66 years (range, 50-85 years) and led to an HM diagnosis shortly thereafter in 3. Bone marrow morphology in 1 carrier with mild thrombocytopenia and anemia at age 85 years showed normal morphology, cellularity, and karyotype (supplemental Figure 4 A-C). Germ line mutation carriers who develop MDS or AML most often present with leukopenia (10 of 12) with or without macrocytosis or other cytopenias, hypocellular bone marrow with prominent erythroid dysplasia, and a normal karyotype, often leading to erythroleukemia (see Table 1 and supplemental Table 7 for treatment and outcome data). Representative AML morphology is shown in supplemental Figure 4D-G. Thus, complete blood count monitoring alone with bone marrow biopsy reserved for those with progressive cytopenias may be a reasonable approach for clinical follow-up of adult asymptomatic carriers in these families, in addition to previously recommended approaches for assessing cases with a family history.9
Granulomatous and immune disorders presenting prior to HM in germ line mutation carriers were observed in 3 families (Figure 1E,I, supplemental Figure 5, and supplemental Data). Of relevance, DDX41 has been shown to be a cytoplasmic DNA sensor in dendritic cells and therefore has a documented role in the innate immune response.10-12 Dysregulation of such responses may be an initiator of disorders observed in our pedigrees and may be linked to lymphoid malignancy.13,14 In addition, inflammatory skin disorders have previously been associated with RUNX1 mutation–mediated familial HMs.15 Therefore, the co-existence of immune and hematologic disorders in DDX41 germ line mutation carriers highlights the importance of examining a potential interaction between the role of DDX41 in immune function and leukemogenesis.
Our findings show that germ line DDX41 mutations predispose to a variety of HMs (MDS/AML, chronic myeloid leukemia, NHL, and Hodgkin lymphoma). The frequent identification of truncating loss-of-function (LOF) mutations and splicing mutations supports the hypothesis that HM predisposition arises from haploinsufficiency of DDX41. The M1I variant described here, although predicted to be an LOF allele, identified a new, potentially relevant translation isoform of DDX41. There are 2 likely sites of alternative translation initiation consistent with the small isoform (M132 and M155) that include a previously described site of mutation (p.M155I).5 This is suggestive of mutations differentially affecting DDX41 isoforms and may explain the selection for a D140 mutation hotspot as, in this scenario, it could result in isoform switching from the long form to the short form of DDX41, something which is commonly seen in germ line AML-predisposing CEBPA mutations.16
The mean age of HM onset (62 years) of all cases to date emphasizes the importance of obtaining a family history and considering predisposition alleles in all adults with HMs. In addition, the younger age of diagnosis in missense mutant DDX41 families vs LOF mutants, is reminiscent of observations in GATA2 and RUNX1 families in which altered function of these mutant proteins more strongly predisposes to malignant progression.2,17 Therefore, the continued identification of DDX41 mutations will be crucial for a comprehensive understanding of the phenotypic and clinical consequence of mutations in this gene.
Acknowledgments
The authors thank the patients and their family members for their willingness to participate in this research and Dr Andrew Roberts for assistance with clinical information.
This work was supported by The Cancer Research Foundation (L.A.G., J.E.C.), K12 CA139160 (J.E.C.), K08 HL129088 (J.E.C.), Grants No. APP1024215 and APP1023059 from the National Health and Medical Research Council of Australia (H.S.S., C.N.H.), and No. APP565161 from the Cancer Council of South Australia (H.S.S., C.N.H.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.L. performed research, analyzed the data, and wrote the manuscript; A.L.B. analyzed and interpreted the data and wrote the manuscript; L.M.W. performed the research and analyzed the data; C.P., G.R., and M.K.L. contributed to research and edited the manuscript; A.W.S. and J.F. analyzed the data; M.B., C.-E.C., and Y.L. performed research; A.Y., G.K.S., and N.P. analyzed clinical data, assisted with study design, and edited the manuscript; M.F., M.A., L.J., and K.P. collected data and samples; R.J.D. and I.D.L. assisted with study design and edited the manuscript; B.C.M., D.A.P., M.-C.K., T.W., S.K., and A.S assisted with clinical data and samples and reviewed the manuscript; and L.A.G., C.N.H., J.E.C., and H.S.S. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hamish S. Scott, Department of Genetics and Molecular Pathology, Centre for Cancer Biology, SA Pathology, PO Box 14, Rundle Mall, Adelaide, SA 5000, Australia; e-mail: hamish.scott@sa.gov.au; Jane E. Churpek, The University of Chicago, Department of Medicine, Section of Hematology/Oncology, 5841 S Maryland Ave, MC 2115, Chicago, IL 60637-1470; e-mail: jchurpek@bsd.uchicago.edu; Christopher N. Hahn, Department of Genetics and Molecular Pathology, Centre for Cancer Biology, SA Pathology, PO Box 14, Rundle Mall, Adelaide, SA 5000, Australia; e-mail: chris.hahn@sa.gov.au; and Lucy A. Godley, The University of Chicago, Department of Medicine, Section of Hematology/Oncology, 5841 S Maryland Ave, MC 2115, Chicago, IL 60637-1470; e-mail: lgodley@medicine.bsd.uchicago.edu.
References
- 1.Owen C, Barnett M, Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia--a review. Br J Haematol. 2008;140(2):123–132. doi: 10.1111/j.1365-2141.2007.06909.x. [DOI] [PubMed] [Google Scholar]
- 2.Hahn CN, Chong CE, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collin M, Dickinson R, Bigley V. Haematopoietic and immune defects associated with GATA2 mutation. Br J Haematol. 2015;169(2):173–187. doi: 10.1111/bjh.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang MY, Churpek JE, Keel SB, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47(2):180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell. 2015;27(5):658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn CN, Ross DM, Feng J, et al. A tale of two siblings: two cases of AML arising from a single pre-leukemic DNMT3A mutant clone. Leukemia. 2015;29(10):2101–2104. doi: 10.1038/leu.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang MY, Keel SB, Walsh T, et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica. 2015;100(1):42–48. doi: 10.3324/haematol.2014.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordin O, Tanner NK, Doère M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23(13):2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churpek JE, Lorenz R, Nedumgottil S, et al. Proposal for the clinical detection and management of patients and their family members with familial myelodysplastic syndrome/acute leukemia predisposition syndromes. Leuk Lymphoma. 2013;54(1):28–35. doi: 10.3109/10428194.2012.701738. [DOI] [PubMed] [Google Scholar]
- 10.Parvatiyar K, Zhang Z, Teles RM, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13(12):1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KG, Kim SS, Kui L, et al. Bruton’s tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Reports. 2015;10(7):1055–1065. doi: 10.1016/j.celrep.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco A, Rizzo MI, De Virgilio A, et al. Churg-Strauss syndrome. Autoimmun Rev. 2015;14(4):341–348. doi: 10.1016/j.autrev.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Spagnolo P. Sarcoidosis: a Critical Review of History and Milestones. Clin Rev Allergy Immunol. 2015;49(1):1–5. doi: 10.1007/s12016-015-8480-0. [DOI] [PubMed] [Google Scholar]
- 15.Sorrell A, Espenschied C, Wang W, et al. Hereditary leukemia due to rare RUNX1c splice variant (L472X) presents with eczematous phenotype. Int J Clin Med. 2012;3(7) doi: 10.4236/ijcm.2012.37110. doi:10.4236/ijcm.2012.37110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman AD. C/EBPα in normal and malignant myelopoiesis. Int J Hematol. 2015;101(4):330–341. doi: 10.1007/s12185-015-1764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaud J, Wu F, Osato M, et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99(4):1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]