Abstract
The cryptochrome/photolyase protein family possesses a conserved triad of tryptophans that may act as a molecular wire to transport electrons from the protein surface to the FAD cofactor for activation and/or signaling-state formation. Members from the animal (and animal-like) cryptochrome subclade use this process in a light-induced fashion in a number of exciting responses, such as the (re-)setting of circadian rhythms or magnetoreception; however, electron-transfer pathways have not been explored in detail yet. Therefore, we present an in-depth time-resolved optical and electron-paramagnetic resonance spectroscopic study of two cryptochromes from Chlamydomonas reinhardtii and Drosophila melanogaster. The results do not only reveal the existence of a fourth, more distant aromatic amino acid that serves as a terminal electron donor in both proteins, but also show that a tyrosine is able to fulfill this very role in Chlamydomonas reinhardtii cryptochrome. Additionally, exchange of the respective fourth aromatic amino acid to redox-inactive phenylalanines still leads to light-induced radical pair formation; however, the lifetimes of these species are drastically reduced from the ms- to the μs-range. The results presented in this study open up a new chapter, to our knowledge, in the diversity of electron-transfer pathways in cryptochromes. Moreover, they could explain unique functions of animal cryptochromes, in particular their potential roles in magnetoreception because magnetic-field effects of light-induced radical pairs strongly depend on distance and orientation parameters.
Introduction
Cryptochromes (Crys) are blue-light photoreceptors found in all domains of life (1, 2). They participate in a wide range of regulatory functions such as inhibition of hypocotyl growth, blue-light-dependent flowering, synchronization of the internal clock by ambient light (3, 4), and magnetoreception (5, 6, 7, 8). Recently, Crys have also been used in the emerging field of optogenetics; several constructs demonstrate the potential of Crys for light-controlled modulation of protein functions (9, 10). In addition, Crys are closely related to the photolyase DNA repair enzymes (Pl) (1, 2, 11, 12, 13). Both have similar tertiary structures despite a C-terminal extension that is supposed to control signal transduction in Crys. Crys and Pls harbor a noncovalently bound light- and redox-active FAD as an essential cofactor. Based on structural and functional similarities the Cry/Pl family can be further subdivided into several subclades: class-I to class-III cyclobutane pyrimidine dimer Pls, plant Crys, DASH (Drosophila, Arabidopsis, Synechocystis, Homo) Crys, Cry-pros, and animal (animal-like) Crys/(6,4) Pls (2, 14, 15).
The photoreceptor mechanism of Crys can be separated into at least two parts: the primary photochemistry triggered by blue-light excitation of FAD, and the subsequent secondary light-independent processes followed by downstream signal transduction. In the dark, the FAD cofactor resides in the fully oxidized ground state (FADox) (16, 17, 18, 19). Upon light excitation, the singlet-state excited flavin abstracts an electron from a close-by tryptophan that is part of the so-called conserved “Trp-triad” (20, 21, 22), thus forming an anionic semiquinone radical (FAD•−) and concomitantly a short-lived tryptophan radical. Subsequently, stepwise sequential electron transfer (ET) takes place along the Trp-triad until the terminal surface-exposed tryptophanyl cation radical is formed, which corresponds to the [FAD•−···TrpH•+] ≡ RP1 radical pair. RP1 is further stabilized by deprotonation, thus forming the secondary [FAD•−···Trp•] radical pair, RP2 (20, 23). RP2 either deactivates by backward electron transfer to regenerate the ground state moieties or the Trp• radical is reduced by an exogenous electron donor; the latter process results in a net reduction of the FAD cofactor. In combination with FAD reduction, protonation of the FAD•− occurs in Pls and plant/DASH Crys. Unlike in Pls, no further (or only partial) reduction of the FAD radical to the fully reduced redox state (FADH−) was observed both in plant and in animal Crys (16, 17, 18, 24).
In general, the theory of a conserved Trp-triad was widely accepted during the last decade, although its biological relevance is still under discussion (25, 26, 27), and various exceptions have been discovered in the past few years. For example, an alternative nonconserved tryptophan that serves as a terminal electron donor in Synechocystis sp. Cry DASH was identified (28). Furthermore, in 2012 a new class of prokaryotic cryptochromes was discovered, which lacks the conserved Trp-triad (14). In a recent study of a (6-4) photolyase the existence of a fourth tryptophan involved in ET was proposed based on amino acid sequence similarities (29).
In light of the previously mentioned notions that Cry electron-transfer pathways upon light excitation are essential but may be variable, the focus of this contribution is to provide an in-depth spectroscopic characterization of the fast events in two proteins, Chlamydomonas reinhardtii (CraCry) and Drosophila melanogaster Cry (DmCry), from the animal (and animal-like) Cry subclade. DmCry is by far the best-investigated protein among the animal Crys: its direct involvement in circadian timing (30) and magnetoreception has been proven (8, 31), although the molecular mechanisms of both processes remain largely unexplored, and immediate magnetic field effects have not yet been published. On the other hand, the exact function of CraCry is still under discussion; studies point in the direction of its function as a red light transcriptional regulator, although the exact mechanism of action remains to be elucidated (32).
To obtain spectroscopic information at a molecular level, steady-state photoreduction experiments of CraCry, and two complementary time-resolved techniques with nanosecond time resolutions, transient electron-paramagnetic resonance (trEPR) and transient (optical) absorption spectroscopy (TA), were applied to probe the involvement of the Trp-triads comprising the residues W399, W376, and W322 in CraCry, and W420, W397, and W342 in DmCry (also denoted as TrpA, TrpB, and TrpC) in the primary photochemistry of animal (and animal-like) Crys. Additionally, various point mutants were investigated to corroborate our conclusions gathered from the wild-type (WT) proteins.
Materials and Methods
Preparation of CraCry mutants
Mutant plasmids were generated via the Round-the-horn site-directed mutagenesis method using phosphorylated back-to-back primers. The plasmids (pET28a vector) were sequenced by GATC (Konstanz, Germany). Expression and purification of all CraCry variants were carried out following published procedures (32). To get rid of remaining nucleases bound to the protein, additional purification through a heparin affinity column was carried out: after application to the column, the protein was washed with four column volumes of buffer containing 50 mM sodium phosphate pH 7.8, and 20% (v/v) glycerol. Elution was performed using a salt gradient reaching a concentration of 2 M sodium chloride. As a last purification step, size-exclusion chromatography was performed in a buffer containing 50 mM NaH2PO4 pH 7.8, and 100 mM NaCl. After a final buffer exchange, the protein was obtained in a buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl, and 35% (v/v) glycerol.
Preparation of DmCry mutants
The DmCry W394F mutation was introduced by overlap extension polymerase chain reaction, and the obtained fragment was subsequently ligated into a pET28a vector (Merck Millipore, Darmstadt, Germany) via restriction cloning. The mutation was verified by sequencing (Eurofins Genomics, Ebersberg, Germany). The final plasmid was transformed into SoluBL21 competent Escherichia coli cells (Genlantis, San Diego, CA) for protein expression. Protein production was performed using the protocol for the WT (24).
Sample preparation for time-resolved spectroscopy
DmCry samples for trEPR measurements were prepared in a buffer containing 50 mM HEPES pH 7.0, 100 mM NaCl, 5 mM K3[Fe(CN)6], and 35% (v/v) glycerol (24), whereas CraCry samples were prepared in a buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl, 5 mM K3[Fe(CN)6], and 35% (v/v) glycerol. Sample concentrations were determined using ultraviolet-visible (UV-vis) spectroscopy (UV-2450, Shimadzu, Kyoto, Japan) and were adjusted to a final concentration of ∼1 mM. DmCry samples for transient absorption measurements were prepared in a buffer containing 50 mM HEPES pH 7.0, 100 mM NaCl, and 20% (v/v) glycerol (24), whereas CraCry samples were prepared in a buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl, and 20% (v/v) glycerol. Sample concentrations were determined using UV-vis spectroscopy (UV-2450, Shimadzu) and were adjusted to a final concentration of ∼0.05 mM.
Steady-state UV-vis and photoreduction assays
Absorption spectra were recorded using a V-660 spectrometer (Jasco, Easton, MD). The protein samples were measured in buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl, and 20% (v/v) glycerol. The protein solution was incubated for 5 min in the dark before the first spectrum was recorded. Spectra were measured at 10°C after different illumination times using a high-power LED emitting at 450 nm (9.7 mW cm−2, Roithner Lasertechnik GmbH, Vienna, Austria).
Fluorescence spectroscopy
The purified proteins were tested for correct folding and stability via a modified Thermofluor device optimized for flavoproteins (33). 20 μL of protein samples with ∼25 μM protein concentration were measured in a reverse transcription-polymerase chain reaction device (Rotor Gene-Q, QIAGEN, Venlo, The Netherlands). The experiment was performed using an excitation wavelength range between 470 and 480 nm and a green emission filter (510–515 nm). Melting curves were generated by a temperature gradient from 25–99°C, detecting fluorescence every 0.5°C. CraCry mutants were measured in buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl, and 35% (v/v) glycerol, and DmCry samples in a buffer containing 50 mM HEPES pH 7.0, 100 mM NaCl, and 35% (v/v) glycerol.
X-band trEPR spectroscopy
X-band trEPR spectra of CraCry and DmCry samples were recorded following pulsed laser excitation of the sample, which was performed using an OPO system (BBO-355-visible/IR, Opta, Bensheim, Germany) pumped by a Nd:YAG laser (GCR 190-10, Spectra Physics, Santa Clara, CA) at a wavelength of 460 nm and a pulse energy of (1.0 ± 0.2) mJ at a repetition rate of 1 Hz (pulse duration: 6 ns). A laboratory-build spectrometer was used in combination with a Bruker microwave bridge (ER046 MRT, Bruker, Billerica, MA). The sample was placed in a synthetic quartz (Suprasil) tube with an inner diameter of 1 mm, and irradiated in a dielectric resonator (ER 4118X-MD5, Bruker). The resonator was immersed into a laboratory-built nitrogen gas-flow cryostat and EPR data collected at a temperature of (270.0 ± 0.1) K stabilized by a temperature controller (ITC503, Oxford Instruments, Abingdon, UK). The microwave power was set to 2 mW and the microwave frequency controlled by a frequency counter (5352B, Keysight Technologies, Boeblingen, Germany). Signal acquisition was performed with a transient recorder (9354A, Teledyne LeCroy, Chestnut Ridge, NYA) at a bandwidth of 25 MHz. Distortions of the signal baseline introduced by the laser pulse were corrected by subtracting a signal collected at an off-resonant magnetic field position.
Spectral simulation of trEPR data
Simulations of trEPR spectra were performed using a self-written MATLAB (The MathWorks, Natick, MA) routine, based on the radical pair model and the FORTRAN program described previously (34). Additionally, the g and A tensors as well as the Drp tensor were included in the fitting process.
Transient absorption spectroscopy
Time-resolved optical absorption spectroscopy was performed at 277 K with a commercially available laser flash photolysis spectrometer (LP920K, Edinburgh Instruments Ltd., Kirkton Campus, UK) and data were recorded with a digital oscilloscope (TDS-3012C, Tektronix, Beaverton, OR). The protein sample was placed in a synthetic quartz (Suprasil) semimicro cell (108F-QS, Hellma GmbH & Co. KG, Muellheim, Germany). The temperature was regulated to (277.0 ± 0.1) K by a temperature controller (Alpha RA 8, Lauda, Lauda-Königshofen, Germany). Optical excitation was carried out using an OPO system (OPO PLUS, Continuum, San Jose, CA) pumped with a Nd:YAG laser (Surelite I, Continuum) at a wavelength of 460 nm, a pulse width of ∼6 ns, and a pulse energy of (4.0 ± 0.2) mJ. The repetition rate of the spectrometer was set to 6.67 mHz. To account for background signals, transients were measured alternately with and without optical excitation, and used for calculation of difference absorbance spectra with Beer-Lambert’s law.
Global analysis
To test the transient absorption data for a specific kinetic model, a target analysis was performed using the software Glotaran (Version 1.5.1 http://glotaran.org) (35). Errors were calculated by dividing two times the standard error given by Glotaran by the optical absorption of the sample at 448 nm.
Results
TrEPR spectroscopy
TrEPR allows time-resolved observation of short-lived radical pairs (RPs) and triplet states (36) generated by pulsed laser excitation. In contrast to conventional continuous wave EPR spectroscopy, which usually involves magnetic field modulation and lock-in signal amplification to improve the signal/noise ratio, trEPR data are recorded in a high-bandwidth direct-detection mode to maximize the time resolution of the experiment. Consequently, positive and negative signal amplitudes in trEPR correspond to enhance absorptive (A) and emissive (E) electron-spin polarization of the EPR transitions, respectively.
Various proteins from the Cry/Pl family have been investigated previously using time-resolved EPR methods (28, 37, 38, 39, 40); however, members of the animal (and animal-like) Cry subclade have not yet been examined. To close this gap, two proteins, CraCry and DmCry, were produced with their FAD cofactor being fully oxidized (FADox), because amino-acid-FAD RPs can only be generated starting from this redox state. Furthermore, it has been shown in DmCry protein samples and in intact cells that FADox is the physiological dark state of the photoreceptor, and thus, the intrinsic starting point of subsequent light-induced reactions (19, 41).
Upon illumination with blue light at ambient temperature, both CraCry and DmCry exhibit trEPR signatures with widths of ∼7 mT that can be attributed to spin-polarized paramagnetic species, and more specifically, to RPs on grounds of their spectral shape and narrow signal width (full contour plots are shown in the Supporting Material as Fig. S1, one-dimensional spectra obtained at maximum signal amplitudes are depicted in Fig. 1, A and C).
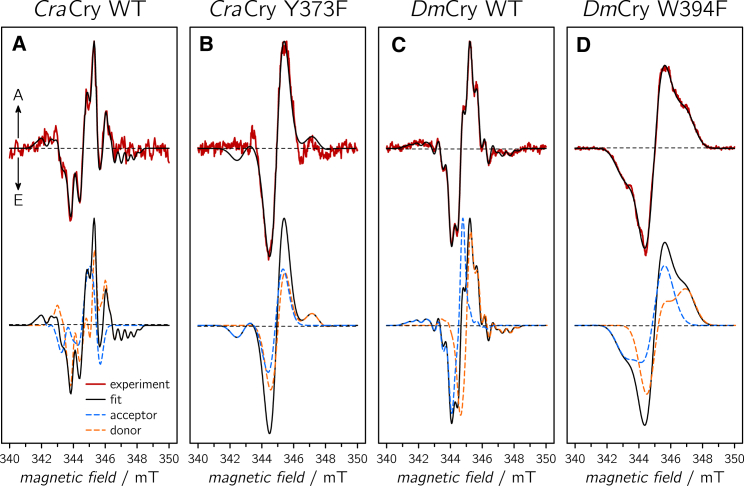
Figure 1.
trEPR experimental data (red curves) including their spectral simulation (black curves) of CraCry WT (A), CraCry Y373F (B), DmCry WT (C), and DmCry W394F (D). A and E arrows indicate enhanced absorptive and emissive signals, respectively. Lower panels depict contributions of the individual simulated radicals (donor depicted in orange, FAD as acceptor in blue) to the overall spectral simulation (drawn black curves). Spectra were recorded at microwave frequencies of 9.6650 GHz (CraCry WT), 9.6741 GHz (CraCry Y373F), 9.6650 GHz (DmCry WT), and 9.6756 GHz (DmCry W394F), respectively. For simulation details, see Table 1. To see this figure in color, go online.
Surprisingly, both WT proteins exhibit pronounced but different spectral shapes with features that have not yet been observed in other Crys. Specifically, the DmCry spectral shape is rather symmetric at around g = 2 and comprises two broad shoulders at 342.5 and 348.0 mT. CraCry, on the other hand, shows a more asymmetric pattern of approximately the same width but with a pronounced and well-resolved sequence of maxima and minima. To retrieve more information from the spectral shape, and to elucidate whether the signal patterns originate from a second overlapping RP species or from hyperfine couplings (hfcs), the signal kinetics were extracted from the two-dimensional trEPR data sets: Irrespective of the magnetic field position 1/e signal decay times of ∼9 μs (CraCry) and ∼4 μs (DmCry) have been obtained by exponential least squares fittings. It should be noted that these values represent relaxation times of the (light-generated) initial electron-spin polarization toward spin-state populations at thermal equilibrium in the presence of 5 mM K3[Fe(CN)6]; they should not be confused with the RP lifetimes. Additional overlapping RPs could be excluded for both proteins as the lifetime (and the spectral width) of a second RP is expected to be significantly different. Hence, the rather pronounced modulations of the overall E/A spectral shape must arise from hyperfine interactions of magnetic nuclei interacting with the unpaired electron spins on the individual radical pair halves.
Spectral simulations of the trEPR data are required for an unambiguous assignment of individual hyperfine patterns. For this purpose, our existing trEPR fitting routine (34) had to be extended to include anisotropic hfcs arising from various nuclei. As an initial starting point, published parameters for g-tensors, dipolar (D), and exchange (J) spin-spin couplings, angles between the two radicals (FAD•− and TrpC•(+), the terminal tryptophan residue of the Trp-triad) extracted from the DmCry structure (42, 43), and residual line-width parameters (37) were used, and additional hfcs were successively added to account for the complex hyperfine pattern. However, the rather large electron-electron coupling parameters (D and J) necessary for a distance of ∼19-Å between the proposed electron donor TrpC (W322 in CraCry and W342 in DmCry) and the electron acceptor FAD did not yield satisfactory simulations because the calculated spectra were too broad to reproduce the experimental data. Therefore, other amino acids at a larger distance to the FAD were taken into account as additional stepping stones in an extended electron-transfer pathway. Amino acid alignments of the animal Cry/(6,4) Pl subclade and first experimental data from Xenopus laevis (6,4) Pl (29) suggested a fourth residue (Y373 and W394 in CraCry and DmCry, respectively) that could in principle be involved in electron transfer of some Crys. Using a distance of ∼21.5 Å and the orientations of the corresponding amino-acid residues with respect to the flavin’s isoalloxazine moiety, both extracted from the DmCry crystal structure (Protein Data Bank: 4GU5; the orientation of Y373 in CraCry was assumed to be similar), the spectral shape could be fairly well reproduced using published hfcs for both radicals (albeit in different protein surroundings) as starting values (44). The best fits for both proteins are presented in the upper panels of Fig. 1, A and C, the contributions of the individual radicals are presented in the respective lower panels, and the respective parameters to calculate the trEPR spectra are summarized in Table 1.
Table 1.
Compilation of Parameters Used to Calculate trEPR Spectra of the Radical Pairs in CraCry and DmCry
| CraCry WT | CraCry Y373F | DmCry WT | DmCry W394F | |
|---|---|---|---|---|
| FAD | FAD | FAD | FAD | |
| g-tensor | ||||
| gx | 2.00435 | 2.00450 | 2.00413 | 2.00413 |
| gy | 2.00370 | 2.00350 | 2.00370 | 2.00370 |
| gz | 2.00220 | 2.00220 | 2.00220 | 2.00220 |
| giso | 2.00342 | 2.00340 | 2.00334 | 2.00334 |
| A(N(5)) / mT | ||||
| 1.876 | 2.199 | 1.933 | 1.840 | |
| 0.100 | 0.001 | 0.015 | 0.100 | |
| A(N(10)) / mT | ||||
| 0.825 | 0.997 | |||
| 0.100 | 0.001 | |||
| A(H(8α)) / mT | ||||
| Aiso | 0.380 | 0.442 | ||
| line broadening / mT | 0.500 | 1.067 | 0.376 | 1.498 |
| Y373 | W322 | W394 | W342 | |
| g-tensor | ||||
| gx | 2.01075 | 2.00381 | 2.00449 | 2.00349 |
| gy | 2.00450 | 2.00248 | 2.00258 | 2.00235 |
| gz | 2.00241 | 2.00230 | 2.00216 | 2.00211 |
| giso | 2.00589 | 2.00286 | 2.00308 | 2.00265 |
| A(H(1′)) / mT | ||||
| Ax | 2.158 | 0.050 | ||
| Ay | 1.992 | 1.200 | ||
| Az | 2.000 | |||
| A(N(1)) / mT | ||||
| 0.050 | ||||
| 1.200 | ||||
| A(H(5)) / mT | ||||
| Aiso | 0.383 | |||
| line broadening / mT | 0.303 | 1.020 | 0.391 | 1.286 |
| Radical Pair | ||||
| D / mT | –0.285 | –0.411 | –0.285 | –0.491 |
| J / mT | 0.002 | 0.030 | 0.005 | 0.015 |
From Fig. 1.
Briefly, only three hfcs (N(5), N(10), and H(8α)) were necessary to reproduce the FAD part, and only one (H(1′)) or two (N(1) and H(5)) for the tyrosine and tryptophan parts, respectively. Specifically, two axial nitrogen hfcs taken from EPR data of a stable FAD•− radical (44) and one isotropic hfc that corresponds to the unrestricted rotation of the 8α methyl group protons at ambient temperatures were required to yield an excellent agreement between experiment and simulation. The hfcs for the FAD part are quite similar for both proteins, thus supporting our general analysis concept (Table 1). Simulation of the tyrosine radical required one nearly axial hfc that accounts for one of the two H(1′) protons (45). As for all β-protons, their hfc values strongly depend on the orientation of the protons with respect to the π-plane of the aromatic ring (46). Here, an angle of 30° has been assumed (47). Finally, one axial and one isotropic hfc tensor that account for tryptophan N(1) and H(5) were included; their values have been taken from the literature (45).
To strengthen our assumption that in CraCry a tyrosine residue serves as terminal electron donor in FAD photoreduction, we repeated the trEPR experiments with a number of CraCry point mutants under otherwise identical experimental conditions. In all mutant proteins, redox-inactive phenylalanine residues have been chosen as replacements for tyrosine and/or tryptophan. All mutant proteins have been investigated for comparable stability and correct folding by temperature-dependent fluorescence spectroscopy (see Fig. S4). The resulting trEPR spectra are summarized in Fig. 1 C and Fig. 2.
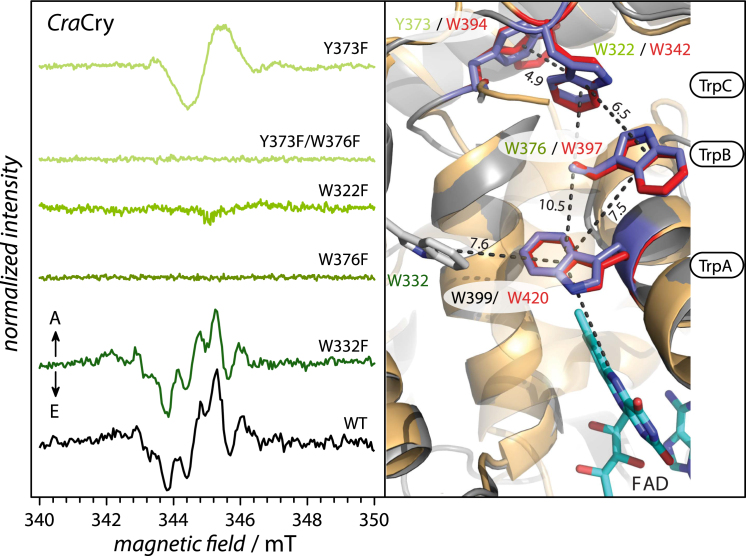
Figure 2.
TrEPR signals of CraCry mutants with A and E arrows indicating enhanced absorptive and emissive polarization of resonances, respectively. The experimental WT spectrum is shown in black, spectra of point mutants are shown as a green color gradient, where dark green resembles close and light green a long distance to the FAD cofactor. Spectra were recorded at microwave frequencies of 9.6650 GHz (CraCry WT), 9.6746 GHz (CraCry W332F), 9.6748 GHz (CraCry W376F), 9.6728 GHz (CraCry W322F), 9.6741 GHz (CraCry Y373F), and 9.6748 GHz (CraCry Y373F/W376F), respectively. On the right-hand side, an overlay of the DmCry structure (42, 43) and a CraCry model structure, including selected residues of the ET pathway, is shown. CraCry numbering is shown as a green color gradient, DmCry numbering is shown in red. To see this figure in color, go online.
Various CraCry mutants, namely W376F, W322F, and the double mutant W376F/Y373F, do not show any discernible trEPR signals under the present conditions, thus demonstrating that the bridging tryptophans in the Trp-triad are strictly essential for light-induced ET (Fig. 2). In contrast, substituting phenylalanine for W332, a tryptophan residue that is expected to be located close to W399 and W376 and that could in principle be involved in ET, exhibits a trEPR spectrum similar to that of the WT. Finally, the mutant Y373F exhibits a trEPR signal; however, the spectrum does not show any resolved hyperfine structure and is quite similar in its spectral shape to previously published trEPR data from the Cry-DASH subclade (28, 37, 38). Spectral simulation using values for W322 (TrpC) as RP partner in a flavin-based radical pair (due to larger D and J values that reflect the shorter distance between the two radicals, all tryptophan hfcs were treated as contributions to inhomogeneous line broadening) fit well with the experimental spectrum (Fig. 1 B; Table 1). Respective mutants from the DmCry protein, DmCry W394F and W342F, were also investigated under otherwise identical experimental conditions. Although the DmCry W342F sample did not show any trEPR signal (data not shown), a spectrum similar to the one of CraCry Y373F was obtained from the DmCry W394F mutant (Fig. 1 D). Again, spectral simulations using the parameters of W342 (TrpC) as electron donor led to very satisfactory agreement with the experiments (lower panel of Fig. 1 D).
As a first summary, trEPR spectra of WT CraCry and DmCry proteins can only be satisfactorily simulated if an aromatic amino acid different from the expected TrpC is taken into account and assumed to act as a terminal electron donor: Y373 in the case of CraCry and W394 in the case of DmCry. If these amino acids are replaced with phenylalanine residues, trEPR spectra can nevertheless be detected; however, spectral simulations indicate that TrpC serves as an electron donor in these mutants. On the other hand, disruption of the classical Trp-triad, e.g., by mutating TrpC, inhibits efficient ET in both proteins, and trEPR signals could not be detected.
Transient optical spectroscopy
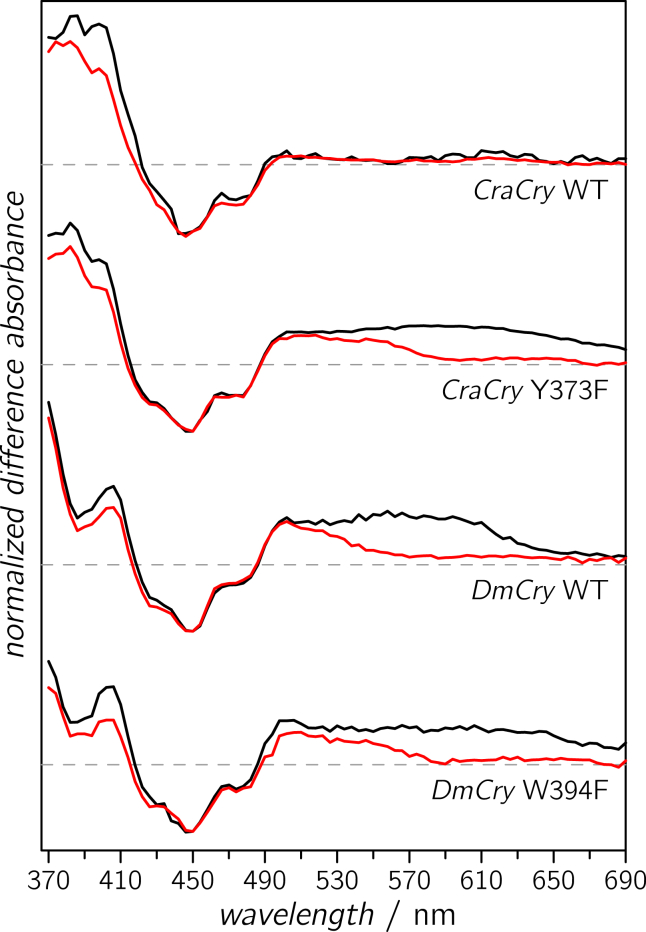
To yield further insights into the molecular processes upon illumination, we performed TA measurements on CraCry WT, CraCry Y373F, and the corresponding DmCry W394F protein sample; experimental data of DmCry WT were reproduced from the literature (24). All protein samples were photoexcited, and transient absorbance changes monitored in 4-nm steps over a wavelength range from 370 to 690 nm for either up to 10 μs or up to 10 ms after pulsed laser excitation (full data sets are shown in Fig. S2). Qualitatively, all to date published Cry/Pl difference spectra that were recorded with the flavin being initially fully oxidized, FADox, can be divided into different parts, namely a negative band at around 450 nm that has been assigned to FADox ground-state bleaching, and positive difference bands in the 375–415 nm and 500–650 nm regions. By global analyses and comparisons with reported spectra (21, 23, 24, 37, 48), the difference signals and their kinetics have been assigned to a two-step sequential reaction scheme: a deprotonation of the initially generated (protonated) tryptophanyl radical cation resembles the first species-associated spectrum (SAS) followed by a recombination of the RP2 consisting of FAD•− and (deprotonated) Trp• radicals.
To analyze the spectrum of CraCry WT, global analysis using an identical kinetic scheme was applied and two SAS (upper panel of Fig. 3) with lifetimes of 6 and 26 ms, respectively, were obtained (Table 2). However, both SAS are almost identical despite some minor discrepancies at 390 and 400 nm (see Fig. 3). There is no indication of any TrpH•+ deprotonation, which typically exhibits absorption differences at around 550 nm (49). This finding could either be rationalized if an amino acid other than tryptophan acts as terminal electron donor in CraCry WT, or (more unlikely) if tryptophan deprotonation is too fast for the time resolution of our TA setup (∼5 ns). Consequently, the two mutants CraCry Y373F and DmCry W394F were investigated and similar analysis procedures applied (Fig. 3, second and fourth panels). Again, two SAS, although with different spectra and different lifetimes of 0.48 and 8.46 μs for CraCry Y373F, and 0.62 and 6.76 μs for DmCry W394F, were detected. Both more short-lived components have identical broad absorption maxima between 550 and 600 nm, which coincides well with the absorption of a TrpH•+ radical species that deprotonates, thus forming RP2. These findings substantiate our notion that in both mutants a tryptophan-based radical is formed upon illumination. It is important to note that in both samples the final component in our model, RP2, decays three orders of magnitude faster as compared to CraCry WT and DmCry WT (Table 2).
Figure 3.
Global analyses of transient optical spectra of CraCry and DmCry. Species-associated difference spectrum components of RP1 (black) and RP2 (red) are shown for WT CraCry and CraCry Y373F. For comparison, the respective components of WT DmCry and DmCry W394F are depicted in the lower panels. To see this figure in color, go online.
Table 2.
Kinetic Data Extracted from TA Spectra
| Formation of RP2 | Decay of RP2 | |
|---|---|---|
| CraCry WT | ||
| Rate constant/μs−1 | (0.16 ±0.04) × 10−3 | (0.038 ± 0.009) × 10−3 |
| Lifetime/μs | (6.1 ± 1.4) × 103 | (26 ± 6) × 103 |
| CraCry Y373F | ||
| Rate constant/μs−1 | 2.08 ± 0.04 | 0.118 ± 0.001 |
| Lifetime/μs | 0.480 ± 0.008 | 8.46 ± 0.05 |
| DmCry WT (24) | ||
| Rate constant/μs−1 | 0.390 ± 0.009 | (0.147 ± 0.005) × 10−3 |
| Lifetime/ μs | 2.56 ± 0.06 | (6.8 ± 0.3) × 103 |
| DmCry W394F | ||
| Rate constant/μs−1 | 1.61 ± 0.05 | 0.148 ± 0.002 |
| Lifetime/μs | 0.620 ± 0.016 | 6.76 ± 0.09 |
Rate constants and respective lifetimes for formation and decays of RP2 were extracted from global analyses.
Steady-state photoreduction of CraCry
To receive further details of the ET properties of CraCry under steady-state conditions, photoreduction kinetics were recorded and analyzed (exemplary spectra and kinetics are depicted in Fig. 4, full data sets are depicted as Fig. S3).
Figure 4.
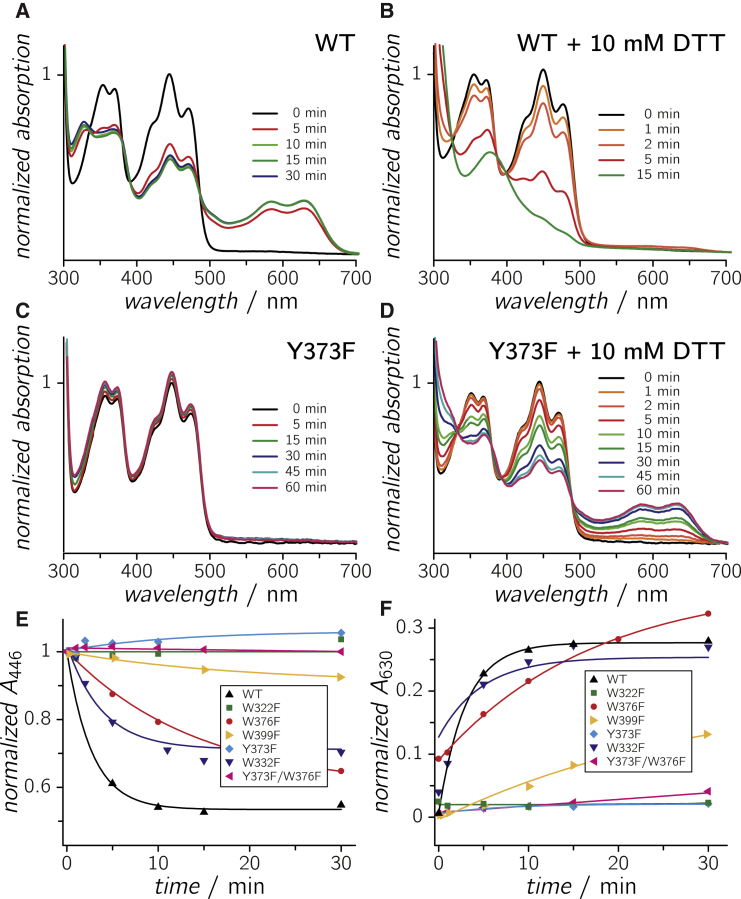
Blue light-induced spectral changes in the optical absorptions of WT and mutant CraCrys. Spectral changes in the optical absorptions of WT CraCry (with or without DTT as the external electron donor) after the indicated illumination times are shown in (A) and (B), respectively. Spectral changes in the optical absorptions of CraCry Y373F (with or without DTT) after the indicated illumination times are shown in (C) and (D), respectively. Absorption changes of all investigated CraCry samples without DTT, recorded at 446 and 630 nm, are depicted in (E) and (F). Monoexponential functions have been added as guide to the eye. To see this figure in color, go online.
By steady-state optical absorption spectroscopy it has been shown previously that CraCry WT is capable of forming all three biologically relevant FAD redox states, FADox, and after blue-light excitation FADH• and FADH– (32). In this contribution, we show that CraCry WT can be easily reduced to its semiquinone state (FADH•) without any reducing agent, such as dithiothreitol (DTT), within a few minutes of blue light illumination, and saturation is reached after ∼10 min (Fig. 4, A and F). Unlike other animal type-1 cryptochromes, CraCry can be converted to the fully reduced state, FADH–, without significant accumulation of an intermediate radical species by adding DTT (Fig. 4 B).
To obtain further information on the importance of the Trp-triad for photoreduction, several point mutants were investigated under identical experimental conditions. The distal tryptophan mutant, W322F, which is located close to the protein surface, shows no photoreduction activity even after extended periods of blue light illumination (Fig. 4, E and F). This lack of photoreduction that is not alleviated by addition of DTT coincides with expectations, as this mutation in the Trp-triad was found to abolish photoreduction completely in other cryptochromes (e.g., (19, 24, 26, 50)). Surprisingly, the medial Trp mutant, W376F, still undergoes photoreduction under the chosen steady-state conditions. Here, formation of the FADH• state proceeds ∼20% slower than in the WT (Fig. 4, E and F). In the presence of DTT, the medial W376F mutant still cannot perform the second reduction step toward FADH–, similar to the WT protein, but gets arrested in the FADH• state (Fig. S3). Interestingly, the proximal mutant W399F exhibits also a slight increase of its absorption between 500 and 700 nm upon illumination indicating radical state formation with low efficiency (Fig. 4, E and F).
Overall, our experiments lead to the assumption that ET in the photoreduction process of CraCry can occur in vitro via other residues than the conserved Trp-triad. Therefore, the two additional mutants of aromatic residues near the Trp-triad, Y373F and W332F, were also investigated. The photoreduction kinetic of the W332F mutant is fairly similar to that of the WT protein, whereas the tyrosine mutant, Y373F, lacks formation of any flavin radical species even after 1 h of blue light illumination (Fig. 4, E and F). Significant amounts of the neutral radical (including minute amounts of FADH–) can only be generated if 10 mM DTT are added (Fig. S3). Further distortion of the ET pathway by combining the Y373F mutant with the W376F mutant causes a nearly complete loss of photoreduction (Fig. 4, E and F). These findings show that Y373 and the integrity of the Trp triad are essential for efficient photoreduction, and thus, for any light-induced process in CraCry.
Discussion
Time-resolved spectroscopic characterization
trEPR spectroscopy has proven to be a valuable tool for the examination of ET pathways in a number of proteins because of its favorable time resolution and spectral sensitivity to molecular properties of the individual RP partner molecules. Hence, it is well suited to gain insight into intraprotein ET processes and their participating amino acids. In detail, trEPR spectra depend on the g-tensor, the hfcs, and the electron-electron interaction parameters of the paramagnetic species involved. As most of these parameters are orientation-dependent, and their anisotropy does not average out under the present conditions, also spatial information such as the orientation of the radicals with respect to each other, as well as distance estimations can be extracted from spectral simulations. In principle, the amino acids tyrosine and tryptophan exhibit sufficiently different g-tensor components to allow for radical discrimination even at X-band microwave frequencies (45), which has been demonstrated on a number of Xenopus laevis Cry-DASH mutants (37). However, the spectra of CraCry and DmCry presented here are the first ones, to our knowledge, that show pronounced hyperfine patterns, which prevented straightforward assignments to participating radicals exclusively on the basis of their g-tensor differences. Consequently, a minimum number of strong hfcs, namely N(5), N(10), and H(8α) for FAD•−, N(1) and H(5) for Trp•, and H(1′) for Tyr• had to be taken into account to achieve good agreements of experimental data and simulations (Fig. 1; Table 1). Dipolar couplings of –285 μT and exchange interactions of +5 μT for DmCry and +2 μT for CraCry, which are far smaller than published values for RPs with TrpC (34, 37) (W322 in CraCry and W342 in DmCry) and correspond to distances of about ∼21.5 Å, complete the necessary fitting parameters for an excellent agreement between experiment and theory. Although structural information is unavailable for CraCry to date, alignments predict that Y373 is at a similar position as W394 in DmCry. Nevertheless, this assumption leads to increased uncertainties in the distance and in particular in the orientation parameters of RPs in CraCry. In addition, it should be noted that the electron-electron interaction parameters that were optimized in the fitting process are just two out of a number of variables, and although many of them (e.g., g-tensors and hfcs) are known from independent experiments and had only narrow variability during fitting (see Results section for details), a straightforward error analysis for the derived distance between the radicals in the RP remains almost impossible. However, by manually varying D and J around their optimal values and compensating the resulting changes by spectral line width while keeping all other parameters constant, we are confident that our values are trustworthy within a distance uncertainty of ∼1 Å.
Bringing together the remarkably pronounced hyperfine structures and the weak electron-electron interactions led us to the conclusion that in both Crys not TrpC but more distant aromatic amino acids must serve as terminal electron donors. To corroborate this notion on the basis of our trEPR data on the WT proteins, additional experiments were carried out with a number of CraCry and DmCry point mutants. Whereas mutants of tryptophans within the conserved Trp-triad did not show any signal at all, the replacement of Y373/W394 lead to a trEPR signature that can be simulated using parameters of a flavin-based radical pair with TrpC (W322/W342, Fig. 1, B and D), as a terminal electron donor. Consistent results were obtained from transient optical absorption spectroscopy: in contrast to the CraCry WT protein, a tryptophan deprotonation process could be identified in CraCry Y373F and in DmCry W394F mutants (Fig. 3). Although the characteristic rates of this process in the two mutants are very similar, a difference of a factor of ∼1000 when compared to the respective WT proteins shows that both RP2s are much more short-lived (Table 2). This finding can be rationalized by the reduced surface exposure of residues W322 and W342: Formation of RP2 requires a proton acceptor close to the tryptophan, but neither an amino acid that could serve as proton acceptor nor pockets that could harbor water molecules can be found in the direct vicinity of TrpC. Therefore, direct recombination of RP1 back to the ground state is favored in these mutants.
Taken together, all experimental data point toward an aromatic amino acid that extends the conserved Trp-triad to a tetrad and is the terminal electron donor in CraCry and DmCry. There is one question which arises, and that is why there is no sign of a tyrosine radical observed in the difference TA data from the CraCry WT. In principle, a tyrosine radical should show absorptions at around 400 nm (51); however, only minute differences in absorption could be detected in this spectral region (upper panel of Fig. 3). One possible explanation is that overlapping difference absorptions from the FAD obscure positive absorptions that are expected for Tyr• formation, in particular as the absorption coefficient of a Tyr•, which is around 2 Lmmol–1cm–1 (51), is far smaller than that of FAD.
Similarities and differences of CraCry and DmCry and their implications for photoreceptor functions
Although the two cryptochromes investigated in this spectroscopic study belong to the same animal (and animal-like) Cry subclade, their proposed functions differ significantly. Therefore, it is surprising that both proteins use an identical tetrad of aromatic amino acids for RP formation and/or flavin photoreduction. Both proteins exhibit fast steady-state photoreduction properties. However, in contrast to DmCry (24), the FAD in CraCry can be easily converted into the fully reduced state via the neutral FADH• state (32). In addition, our steady-state photoreduction data show that CraCry exerts some light-driven electron transfer, when either the proximal or medial tryptophan has been mutated, although only to the radical FADH• state. In this way, CraCry contrasts DmCry, where corresponding mutations (W420F, W397F) cause a complete loss of the formation of its signaling active FAD•–state (42, 52). Complete loss of photoactive behavior can only be achieved in CraCry by mutation of the distal tryptophan of the Trp-triad, W322. This residue is supposed to be the terminal electron donor (TrpC) of the ET pathway in most other Crys and Pls. This result raises the question, whether there are other determinants for ET in CraCry apart from the different photochemistry, i.e., formation of FAD•– in DmCry and FADH• or FADH– in CraCry, which may allow partial bypassing of the Trp-triad. Among the candidate residues, W332 and Y373, we found that W332, a candidate for an ET side branch, caused only minute effects on the photochemistry of CraCry, whereas replacement of Y373 abolishes intramolecular ET in the absence of an exogenous reductant. In a model of the CraCry structure, this residue is only ∼4.9 Å distant from W322 and apparently elongates thereby the ET chain of the Trp triad (right panel of Fig. 2). The ability of the Y373F mutant to undergo photoreduction in the presence of DTT to the semiquinoid redox state indicates that the remaining Trp-triad is operational as long as the distal tryptophan can be reduced by a reductant during the μs-lifetime of the RP. However, one has to note that further reduction to the fully reduced FADH– state is not possible in the Y373F mutant. One possible explanation is that RP recombination after excitation of the FADH• cofactor outpaces ET from the exogenous reductant to the distal tryptophan W322. A similar inability to perform the second photoreduction step from FADH• to FADH– has been found for the medial tryptophan mutant W376F. In this mutant, excitation of FADox under steady-state conditions allows some electron transfer, presumably from the distal tryptophan and the nearby essential residue Y373, but again, no formation of the fully reduced FADH– state can be detected. In this context, it is noteworthy that in some members of the Cry/Pl family, the ET pathway can differ from the conserved Trp-triad (14, 28), or use other aromatic residues as side branches, as shown for the Methanosarcina mazei class II photolyase (53).
In summary, the length of the ET cascade of CraCry and DmCry is extended by ∼2 Å; however, steady-state reduction of the FAD cofactor seems to be equally efficient. It should also be mentioned at this point that appearance and kinetics of TA and/or trEPR spectra cannot be directly correlated to steady-state photoreduction kinetics as their experimental conditions are different. Yet, the terminal electron donor is different in both proteins: a tyrosine residue that forms a RP with a pronounced lifetime of 26 ms has been observed in CraCry. This finding shows that not only tryptophan residues are able to stabilize RP2 within a ms-lifetime, but also tyrosine residues may fulfill this function. Unfortunately, no information on the protonation state of the Tyr• radical can be obtained from our TA and trEPR data as g-values, electron-spin density distributions (and hence, hyperfine couplings) and optical absorptions are rather similar for Tyr• and Tyr•+ (45). Nevertheless, it can be summarized that both proteins show a surprisingly similar behavior after light excitation.
Are there any advantages for animal Cry function if ET occurs via a Trp/Tyr-tetrad and not via the normal Trp-triad? In principle, the Trp/Tyr-tetrad could be an evolutionary improvement because in animal Crys TrpC is not as surface-exposed as in other proteins of the Cry/Pl family. Here, structural information on CraCry would definitely help to support (or disprove) the argument. Furthermore, the direct consequences of the Trp/Tyr-tetrad for CraCry function remain vague. Because the protein is supposed to be involved in red-light photoreception (32), both the FADox state and the FAD•−/Tyr• RP may not be the primary light-absorbing states as none of these absorbs in the red-light region. Either the involvement of a second protein that absorbs in this range or the FADH• being the dark state of the photoreceptor seem conceivable for red-light photoreception.
On the other hand, significant consequences are expected for the proposed magnetoreceptor function of DmCry. In principle, the distance between the RP partners determines the spin-spin coupling parameters. Together with the strengths and anisotropies of hfcs they directly influence the singlet-to-triplet mixing rate, which is crucial for the ability of a molecule to sense magnetic fields (54, 55, 56, 57). Therefore, it is conspicuous that a fourth tryptophan with a different distance and orientation with respect to the flavin differentiates animal Crys from other proteins of the Pl/Cry family. This finding becomes even more important as most calculations of magnetic field effects in Crys have been performed using parameters of a flavin-TrpC radical pair derived from the classical Trp-triad (e.g., (58, 59)). Therefore, it will be intriguing to see how altered distance/orientation parameters of the fourth tryptophan—from now on named TrpD—affect the magnetic field effects of light-induced RPs in DmCry.
Conclusions
In this contribution, we describe how molecular spectroscopy corroborated by spectral simulations leads to a detailed picture of the ET pathways and photochemical processes in two animal (and animal-like) Crys upon blue-light excitation. Time-resolved spectroscopy with optical detection unravels aspects of the photochemistry, such as rate constants for the decay and transformations of involved radicals. Spectral simulations of trEPR data are well suited not only to distinguish between tryptophan-based and tyrosine-based RPs, but also to allow extracting structural information such as the distances between the RP partners. Therefore, the combination of the two techniques is crucial to obtain a clear picture of light-induced reactions in Crys.
Both Crys show ET along the conserved Trp-triad. However, a fourth aromatic amino acid residue extends this pathway. Whereas a tyrosine residue serves as a surface-exposed terminal electron donor in CraCry, a tryptophan is found for DmCry. When the respective fourth aromatic amino acids are exchanged to phenylalanines, RPs with similar TA/trEPR spectra as compared to published FAD-TrpC RPs are detected in both proteins (37); however, their lifetimes are drastically reduced as compared to those of the WT. On the other hand, if TrpC or TrpB are mutated, RP signatures could not be detected, which demonstrates the importance of the Trp-triad in these proteins for efficient ET.
Author Contributions
D.N., S.F., R.R., and B.P. gathered and analyzed the data. D.N. and B.P simulated the time-resolved spectroscopic data. S.F. and R.R. produced the samples. D.N., S.F., R.R., B.P., L.-O.E., S.W., and E.S. discussed the data, their interpretation and presentation. L.-O.E., S.W., and E.S. conceived and designed the study. E.S. coordinated the experiments. L.-O.E., S.W., and E.S. wrote the article.
Acknowledgments
E.S. and S.W. were supported by the Deutsche Forschungsgemeinschaft (DFG RTG1976, project P13), L.-O.E. was supported by the Deutsche Forschungsgemeinschaft (ES152/12-1) and the AFOSR (FA9550-14-1-0409). The authors thank Maria Mittag for providing the original CraCry plasmid.
Editor: David Cafiso.
Footnotes
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30442-8.
Contributor Information
Lars-Oliver Essen, Email: essen@chemie.uni-marburg.de.
Erik Schleicher, Email: erik.schleicher@physchem.uni-freiburg.de.
Supporting Material
References
- 1.Losi A., Gärtner W. The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu. Rev. Plant Biol. 2012;63:49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- 2.Chaves I., Pokorny R., Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 3.van der Horst G.T.J., Muijtjens M., Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 4.Stanewsky R., Kaneko M., Hall J.C. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 5.Maeda K., Robinson A.J., Hore P.J. Magnetically sensitive light-induced reactions in cryptochrome are consistent with its proposed role as a magnetoreceptor. Proc. Natl. Acad. Sci. USA. 2012;109:4774–4779. doi: 10.1073/pnas.1118959109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritz T., Adem S., Schulten K. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 2000;78:707–718. doi: 10.1016/S0006-3495(00)76629-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouritsen H., Ritz T. Magnetoreception and its use in bird navigation. Curr. Opin. Neurobiol. 2005;15:406–414. doi: 10.1016/j.conb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Gegear R.J., Casselman A., Reppert S.M. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454:1014–1018. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy M.J., Hughes R.M., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bugaj L.J., Choksi A.T., Schaffer D.V. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 11.Zoltowski B.D., Gardner K.H. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller M., Carell T. Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol. 2009;19:277–285. doi: 10.1016/j.sbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Paulus B., Bajzath C., Schleicher E. Flavoproteins and blue light reception in plants. In: Hille R., Miller S., Palfey B., editors. Handbook of Flavoproteins. Vol. 2: Complex Flavoproteins, Dehydrogenases and Physical Methods. Walter de Gruyter GmbH; Berlin/Boston: 2013. pp. 361–392. [Google Scholar]
- 14.Geisselbrecht Y., Frühwirth S., Essen L.-O. CryB from Rhodobacter sphaeroides: a unique class of cryptochromes with new cofactors. EMBO Rep. 2012;13:223–229. doi: 10.1038/embor.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei Q., Dvornyk V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS ONE. 2015;10:e0135940. doi: 10.1371/journal.pone.0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouly J.-P., Schleicher E., Ahmad M. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee R., Schleicher E., Batschauer A. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 18.Berndt A., Kottke T., Wolf E. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 2007;282:13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 19.Hoang N., Schleicher E., Ahmad M. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060160. e160.1559-e1160.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aubert C., Vos M.H., Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 21.Henbest K.B., Maeda K., Schleicher E. Magnetic-field effect on the photoactivation reaction of Escherichia coli DNA photolyase. Proc. Natl. Acad. Sci. USA. 2008;105:14395–14399. doi: 10.1073/pnas.0803620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y.F., Heelis P.F., Sancar A. Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry. 1991;30:6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- 23.Giovani B., Byrdin M., Brettel K. Light-induced electron transfer in a cryptochrome blue-light photoreceptor. Nat. Struct. Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- 24.Paulus B., Bajzath C., Schleicher E. Spectroscopic characterization of radicals and radical pairs in fruit fly cryptochrome - protonated and nonprotonated flavin radical-states. FEBS J. 2015;282:3175–3189. doi: 10.1111/febs.13299. [DOI] [PubMed] [Google Scholar]
- 25.Liu B., Liu H., Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 2010;13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oztürk N., Song S.-H., Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 27.Ozturk N., Selby C.P., Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc. Natl. Acad. Sci. USA. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biskup T., Hitomi K., Weber S. Unexpected electron transfer in cryptochrome identified by time-resolved EPR spectroscopy. Angew. Chem. Int. Ed. Engl. 2011;50:12647–12651. doi: 10.1002/anie.201104321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller P., Yamamoto J., Brettel K. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6-4) photolyases. Chem. Commun. (Camb.) 2015;51:15502–15505. doi: 10.1039/c5cc06276d. [DOI] [PubMed] [Google Scholar]
- 30.Collins B., Mazzoni E.O., Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Marley R., Giachello C.N.G., Jones A.R. Cryptochrome-dependent magnetic field effect on seizure response in Drosophila larvae. Sci. Rep. 2014;4:5799. doi: 10.1038/srep05799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beel B., Prager K., Mittag M. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24:2992–3008. doi: 10.1105/tpc.112.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forneris F., Orru R., Mattevi A. ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding. FEBS J. 2009;276:2833–2840. doi: 10.1111/j.1742-4658.2009.07006.x. [DOI] [PubMed] [Google Scholar]
- 34.Weber S., Biskup T., Norris J.R., Jr. Origin of light-induced spin-correlated radical pairs in cryptochrome. J. Phys. Chem. B. 2010;114:14745–14754. doi: 10.1021/jp103401u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snellenburg J.J., Laptenok S.P., van Stokkum I.H.M. Glotaran: a Java-based graphical user interface for the R package TIMP. J. Stat. Softw. 2012;49:1–22. [Google Scholar]
- 36.Bittl R., Weber S. Transient radical pairs studied by time-resolved EPR. Biochim. Biophys. Acta. 2005;1707:117–126. doi: 10.1016/j.bbabio.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Biskup T., Paulus B., Schleicher E. Variable electron transfer pathways in an amphibian cryptochrome: tryptophan versus tyrosine-based radical pairs. J. Biol. Chem. 2013;288:9249–9260. doi: 10.1074/jbc.M112.417725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biskup T., Schleicher E., Weber S. Direct observation of a photoinduced radical pair in a cryptochrome blue-light photoreceptor. Angew. Chem. Int. Ed. Engl. 2009;48:404–407. doi: 10.1002/anie.200803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber S., Kay C.W.M., Todo T. Photoactivation of the flavin cofactor in Xenopus laevis (6 - 4) photolyase: observation of a transient tyrosyl radical by time-resolved electron paramagnetic resonance. Proc. Natl. Acad. Sci. USA. 2002;99:1319–1322. doi: 10.1073/pnas.032469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelhard C., Wang X., Ahmad M. Cellular metabolites enhance the light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell. 2014;26:4519–4531. doi: 10.1105/tpc.114.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaidya A.T., Top D., Crane B.R. Flavin reduction activates Drosophila cryptochrome. Proc. Natl. Acad. Sci. USA. 2013;110:20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czarna A., Berndt A., Wolf E. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153:1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Levy C., Zoltowski B.D., Leys D. Updated structure of Drosophila cryptochrome. Nature. 2013;495:E3–E4. doi: 10.1038/nature11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okafuji A., Schnegg A., Weber S. G-tensors of the flavin adenine dinucleotide radicals in glucose oxidase: a comparative multifrequency electron paramagnetic resonance and electron-nuclear double resonance study. J. Phys. Chem. B. 2008;112:3568–3574. doi: 10.1021/jp077170j. [DOI] [PubMed] [Google Scholar]
- 45.Bleifuss G., Kolberg M., Lendzian F. Tryptophan and tyrosine radicals in ribonucleotide reductase: a comparative high-field EPR study at 94 GHz. Biochemistry. 2001;40:15362–15368. doi: 10.1021/bi010707d. [DOI] [PubMed] [Google Scholar]
- 46.McConnell H.M. Indirect hyperfine interaction in the paramagnetic resonance spectra of aromatic radicals. J. Chem. Phys. 1956;24:764–766. [Google Scholar]
- 47.Un S. The g-values and hyperfine coupling of amino acid radicals in proteins: comparison of experimental measurements with ab initio calculations. Magn. Reson. Chem. 2005;43:S229–S236. doi: 10.1002/mrc.1660. [DOI] [PubMed] [Google Scholar]
- 48.Langenbacher T., Immeln D., Kottke T. Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J. Am. Chem. Soc. 2009;131:14274–14280. doi: 10.1021/ja901628y. [DOI] [PubMed] [Google Scholar]
- 49.Bent D.V., Hayon E. Excited state chemistry of aromatic amino acids and related peptides. III. Tryptophan. J. Am. Chem. Soc. 1975;97:2612–2619. doi: 10.1021/ja00843a004. [DOI] [PubMed] [Google Scholar]
- 50.Zeugner A., Byrdin M., Ahmad M. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J. Biol. Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 51.Bent D.V., Hayon E. Excited state chemistry of aromatic amino acids and related peptides. I. Tyrosine. J. Am. Chem. Soc. 1975;97:2599–2606. doi: 10.1021/ja00843a002. [DOI] [PubMed] [Google Scholar]
- 52.Azarkh M., Groenen E.J.J. Simulation of multi-frequency EPR spectra for a distribution of the zero-field splitting. J. Magn. Reson. 2015;255:106–113. doi: 10.1016/j.jmr.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Kiontke S., Geisselbrecht Y., Essen L.-O. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 2011;30:4437–4449. doi: 10.1038/emboj.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulten K. Magnetic field effects in chemistry and biology. In: Treusch J., editor. Festkörperprobleme. Vieweg; Braunschweig: 1982. pp. 61–83. [Google Scholar]
- 55.Rodgers C.T., Hore P.J. Chemical magnetoreception in birds: the radical pair mechanism. Proc. Natl. Acad. Sci. USA. 2009;106:353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Efimova O., Hore P.J. Role of exchange and dipolar interactions in the radical pair model of the avian magnetic compass. Biophys. J. 2008;94:1565–1574. doi: 10.1529/biophysj.107.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., Berman G.P., Kais S. The radical pair mechanism and the avian chemical compass: quantum coherence and entanglement. Int. J. Quantum Chem. 2015;115:1327–1341. [Google Scholar]
- 58.Lee A.A., Lau J.C.S., Hore P.J. Alternative radical pairs for cryptochrome-based magnetoreception. J. R. Soc. Interface. 2014;11:20131063. doi: 10.1098/rsif.2013.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solov’yov I.A., Chandler D.E., Schulten K. Magnetic field effects in Arabidopsis thaliana cryptochrome-1. Biophys. J. 2007;92:2711–2726. doi: 10.1529/biophysj.106.097139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.