Abstract
Nrf2 is a bZIP transcription factor regulating the expression of antioxidant and detoxification genes. We have found that Nrf2 knockout mice have an increased infarction size in response to regional ischemic reperfusion and have a reduced degree of cardiac protection by means of ischemic preconditioning. With cycles of brief ischemia and reperfusion (5′I/5′R) that induce cardiac protection in wild type mice, an elevated Nrf2 protein was observed without prior increases of Nrf2 mRNA. When an mRNA species is being translated into a protein, it is occupied by multiple ribosomes. The level of ribosome-associated Nrf2 mRNA increased following cycles of 5′I/5′R, supporting de novo Nrf2 protein translation. A dicistronic reporter assay indicated a role of the 5′ untranslated region (5′ UTR) of Nrf2 mRNA in oxidative stress induced Nrf2 protein translation in isolated cardiomyocytes. Western blot analyses after isolation of proteins binding to biotinylated Nrf2 5′ UTR from the myocardium or cultured cardiomyocytes demonstrated that cycles of 5′I/5′R or oxidants caused an increased association of La protein with Nrf2 5′ UTR. Ribonucleoprotein complex immunoprecipitation assays confirmed such association indeed occurring in vivo. Knocking down La using siRNA was able to prevent Nrf2 protein elevation by oxidants in cultured cardiomyocytes and by cycles of 5′I/5′R in the myocardium. Our data point out a novel mechanism of cardiac protection by de novo Nrf2 protein translation involving interaction of La protein with 5′ UTR of Nrf2 mRNA in cardiomyocytes.
Keywords: Ischemic stress, Protein translation, RNA binding, Cardiac protection
1. Introduction
Chemical stress is known to result in an inhibition of protein synthesis in general [1–3]. How certain genes bypass the general control of protein synthesis and be translated selectively remains unclear. Under normal physiological conditions, protein synthesis is initiated following the recognition of 7-methyl guanine cap at the 5′ end of mRNA species by eukaryotic initiation factor-4E (eIF4E), the binding of poly (A) tail binding proteins (PABPs), and the recruitment of 43S preinitiation complex onto the mRNA species [1,2]. After scanning of 5′ untranslated region (5′ UTR) for the start codon, eIF2 bound GTP is hydrolyzed, followed by joining of tRNA and the 60S subunit of the ribosome, marking the final step for translation initiation. Under stress conditions, 5′ methyl guanine cap dependent translation is inhibited.
Certain mRNA species can bypass 5′ methyl guanine cap dependent translation and undergo protein translation via an internal ribosome entry site (IRES) [2–5]. IRES trans-acting factors (ITAFs) play an important role in recognizing the mRNA species and for coordinating stress induced protein translation [5,6]. Stress induced protein translation can occur rapidly, often within 1 h or sooner following the insult [7–9]. A number of viral proteins have been shown to undergo IRES dependent translation in host cells during viral infection. Yeast is another popular model for studying the impact of stress on protein translation. Whether cellular proteins undergo stress induced translation has not been studied in an animal model of cardiac injury.
Ischemic preconditioning is a well documented phenomenon where cycles of brief ischemic episodes induce protection against myocardial injury. Despite the fact that this phenomenon has been known for over 25 years [10], the mechanism underlying such protection remains not fully understood. With ischemia, an increased amount of H2O2 is detectable in the heart [11–13]. Reperfusion enhances oxidant generation through abnormal mitochondrial function and activated xanthine oxidase [14]. Patients with myocardial ischemia undergoing percutaneous transluminal coronary angioplasty to restore the blood flow show elevation of biomarkers of oxidative stress in the blood [15,16]. Despite the overwhelming evidence suggesting an association of oxidative stress with ischemia and reperfusion, clinical trials with antioxidants have not yielded a clear protective effect against cardiac injury [17–21]. High doses of antioxidant vitamins may even be harmful [22]. These lines of evidence are consistent with our hypothesis that an initial exposure to low or mild doses of oxidants may serve to activate endogenous defense mechanisms.
A list of genes found upregulated during ischemic preconditioning suggests activation of NF-E2 related factor-2 (Nrf2) transcription factor. These genes, for example, heme oxygenase-1 (HO-1), glutamate-cysteine ligase catalytic subunit (GCLC), NAD(P)H: quinone oxidoreductase-1, thioredoxin, cytochrome b and aldose reductase, contain the antioxidant response cis-element (ARE) in the promoters [23–25]. ARE is a consensus sequence of TGACnnnGC and a binding site of Nrf2 transcription factor [26,27]. Genome-wide profiling studies indicate that Nrf2 participates in regulation of a long list of genes, including growth factors, signaling molecules and transcription factors [28,29]. These studies suggest that Nrf2 not only is a controller for cellular defense but also mediates cellular repair and proliferation or regeneration.
With isolated cardiomyocytes, oxidants induce elevation of Nrf2 protein rapidly, within 10 min [30]. Measurements of Nrf2 mRNA and pharmacological approaches with actinomycin D have eliminated transcription as a cause of Nrf2 protein increase. Lack of Nrf2 protein stabilization, evidence of [35S]-methionine incorporation, and an inhibitory effect of cycloheximide point to de novo translation contributing to Nrf2 protein increase [30]. With HeLa cells, we have identified La autoantigen as a protein binding to Nrf2 mRNA using LC–MS/MS based proteomics [31]. Here we address whether de novo Nrf2 protein translation occurs in an animal model of ischemic reperfusion, and whether La binding to Nrf2 mRNA mediates de novo Nrf2 protein translation in cardiomyocytes in vitro and in vivo.
2. Material and methods
2.1. Ischemia and reperfusion in mice
Male C57BL/6J mice (8–10 weeks old, Harlan) were cared for in accordance with NIH guideline for laboratory animals. The surgical protocol was approved by the Institutional Animal Care and Use Committee. After induction of deep anesthesia with Avertin (2.5%), a tracheotomy was performed to ventilate animals. Upon exposure of the pericardium through a left lateral thoracotomy at the third intercostal space, an 8–0 sterile suture was placed underneath the left anterior descending (LAD) artery 1–3 mm from the tip of the left atrium. Two ends of the suture were passed through a 1–2 mm PE50 hollow tube to form a cross so that by pulling two ends of the suture, the tube was placed perpendicular to the LAD. Ischemia was produced by clamping the suture against the tube tightly and was evidenced by a visible blanched area distal to the ligation site. For reperfusion, the suture was released from the clamps to allow restoration of blood flow. For ischemic preconditioning, two cycles of 5 min ischemia and 5 min reperfusion (2× 5′I/5′R) were performed. The protective effect of preconditioning was determined by permanent LAD occlusion to induce myocardial infarction following 2× 5′I/5′R.
To quantify the area-at-risk (AAR), hearts were perfused with 1 ml of 1% trypan blue via the abdominal aorta catheter following retrograde perfusion with saline. Upon excision, the hearts were sliced into 1 mm thick transverse sections. The sections were incubated in 1% 2,3,5-triphenyl-tetrazolium chloride (TTC) in phosphate buffered saline solution (pH 7.4) at 37 °C for 30 min. The sections were then fixed in 10% formalin overnight at 4 °C. Total ventricle area, AAR and infarct area were quantified from the sections using ImageJ (NIH) planimetry. When the infarct area was quantified without comparing to AAR, the left ventricle was sectioned into 5 even transverse slices for incubation in 1% TTC for 20 min at 37 °C followed by fixation in 10% formalin overnight at 4 °C. Each TTC-stained tissue slice was photographed on both sides for measurements of infarct areas by computerized planimetry using NIH image J software [32]. To measure cardiac troponin I (cTnI), the blood was collected via the abdominal vena cava and subsequently centrifuged for serum collection, which was used for cTnI measurements using an ELISA kit (Life Diagnostics).
2.2. Western blot analyses
Heart tissues were quickly frozen and later ground into powder in a liquid nitrogen bath. Tissue lysates in extraction buffer (Cell Signaling Technology) were centrifuged at 14,000 ×g for collection of supernatants. The 5× Laemmli sample buffer [65 mM Tris, pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, with 5% fresh β-mercaptoethanol] was added to the extracts and boiled for 10-min. After SDS-PAGE, Western blot was performed using primary antibodies against Nrf2 (ab62352, Abcam) and secondary antibodies conjugated with horseradish peroxidase for an enhanced chemiluminescence reaction.
2.3. Isolation of RNA associated with ribosomes
The quick frozen left ventricular tissue (~20 mg) was ground in a liquid nitrogen bath into powder to render it soluble in 1 ml of lysis buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1% Nonidet-P40, 40 mM dithiothreitol, 500 U/ml RNAsin, 1% deoxycholate). After centrifugation (12,000 g, 10 s, at 4 °C) to remove nuclei and insoluble debris, the supernatants were supplemented with 0.5 ml of 2× extraction buffer (0.2 M Tris–HCl, pH 7.5, 0.3 M NaCl, 150 µg/ml cycloheximide, 650 µg/ml heparin, and 10 mM phenyl-methyl-sulfonyl fluoride) for further centrifugation (14,000 g, 10 min, at 4 °C) to remove mitochondria and membranous debris. The supernatant was layered onto a 5 ml linear sucrose gradient (10%–35% sucrose, supplemented with 10 mM Tris–HCl at pH 7.5, 140 mM NaCl, 1.5 mM MgCl2, 10 mM dithiothreitol, 100 µg/ml cycloheximide, 0.5 mg/ml heparin) and centrifuged in a SW41Ti rotor (Beckman) at 38,000 rpm, 4 °C for 180 min [33]. RNAs were extracted from ribosomal fractions using Trizol and ethanol precipitation.
2.4. Real time RT-PCR
Total RNA or ribosome-associated RNA (1 µg) was converted to cDNA with a Revertaid kit (Fermentas). An equal amount of cDNA was mixed with SYBR Green Master Mix for real-time PCR in a CFX96 thermocycler (Bio-Rad) for detection of Nrf2 mRNA using the primer pair of TCCATTTCCGAGTCACTGAACCCA (forward) and TGACTCTGACTCCGGCATTTCACT (reverse) with 50 °C for 30 min followed by 95 °C for 15 min to activate the Taq polymerase, and then PCR for 39 cycles at 95 °C for 15 s and 60 °C for 60 s. After PCR, melting curves were acquired by temperature shift from 55 °C to 95 °C to ensure that a single product was amplified during PCR. The 18S rRNA was measured in parallel with the primer pair of TCAACTTTCGATGGTAGTCGCCGT (forward) and TCCTTGGATGTGGT AGCCGTTTCT (reverse) to demonstrate an equal amount of RNA templates between samples.
2.5. Cell culture, H2O2 treatment and transfection
Neonatal rat cardiomyocytes were prepared as described [30]. After seeding at 0.3 × 106 cells per well of 6-well plates or 2.5 × 106 cells per 100-mm dish, myocytes were cultured in low glucose Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) for 3 days before 24-h culture in 0.5% FBS/DMEM and then treatment with 100 µM H2O2 for 10 min. The cells were placed in fresh 0.5% FBS/DMEM after H2O2 treatment. Forty eight hours before H2O2 treatment, cells were transfected using Fugene 6 with a dicistronic luciferase reporter plasmid, which was constructed with SV40 promoter driven transcription and 5′ methyl guanine cap driven Renilla luciferase in front of human Nrf2 5′ UTR and Firefly luciferase [34]. Following H2O2 treatment, Firefly versus Renilla luciferase was measured using a dual luciferase kit (Promega).
2.6. Protein pull down by biotinylated RNA
Human Nrf2 5′ UTR (555 nucleotides) was cloned into pJET 1.2 vector for in vitro transcription using T7 polymerase in the presence of biotin-11-UTP. Gel purified biotinylated RNA of Nrf2 5′ UTR (3 µg) was incubated with tissue lysates (500 µg proteins) for 1 h at 25 °C. Bound proteins were isolated with Streptavidin Sepharose beads (Amersham Biosciences) for analysis by SDS-PAGE and Western blots.
2.7. Ribonucleoprotein immunoprecipitation (RIP)
Endogenous RNA–protein complex was isolated by immunoprecipitation using antibodies against La protein [35]. Proteins were extracted from cardiomyocytes (10 × 106 cells) or left ventricular tissues (10 mg) with swelling buffer (5 mM HEPES, pH 8.0, 85 mM KCl, 0.5% Nonidet P-40) for removal of the nuclei. Cytoplasmic extracts were incubated with Protein A/G plus agarose beads precoated with 3 µg of antibodies against La protein, or rabbit IgG for controlling nonspecific binding. “Inputs” represent cytoplasmic extracts without antibody or IgG incubation for Western blot to show equal loading. After protein binding, the beads were washed 5 times with NT2 buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM NgCl2, 0.05% Nonidet P-40), then incubated at 30 °C with 20 units of RNase-free DNase I in 100 µl NT2 buffer for 15 min. Following washes, proteins in the immunoprecipitates were digested in 0.5 mg/ml proteinase K at 55 °C for 15 min. RNAs were extracted from the immunoprecipitates using Trizol with isopropyl ethanol precipitation for RT-PCR.
2.8. siRNA design and synthesis
The siRNA (TGCTAGAGACAAGTAGTTTATTTAGTA) against mouse and rat La mRNA was designed with IDT online tools (http://www.idtdna.com). An oligonucleotide containing T7 promoter sequence was used for in vitro transcription to produce the siRNA. For transfecting cardiomyocytes, siRNA (300 pmol for final 300 nM) was mixed with oligofectamine (Invitrogen, Carlsbad, CA) in the Opti-MEM media for 6 h incubation with cells. At 48 h after transfection, cells were treated with 100 µM H2O2. To deliver siRNA in vivo, mice were cannulated with a PE10 polyethylene tube in the jugular vein and 15 µg of siRNA in 300 µl PBS was injected. Ischemic reperfusion surgery was performed 24 h after siRNA injection.
2.9. Statistical analyses
The statistical difference was determined by 2-tailed Student's t test when two samples were compared or by one way ANOVA when multiple groups of data were compared.
3. Results
3.1. Nrf2 knockout mice are more sensitive to ischemic reperfusion injury
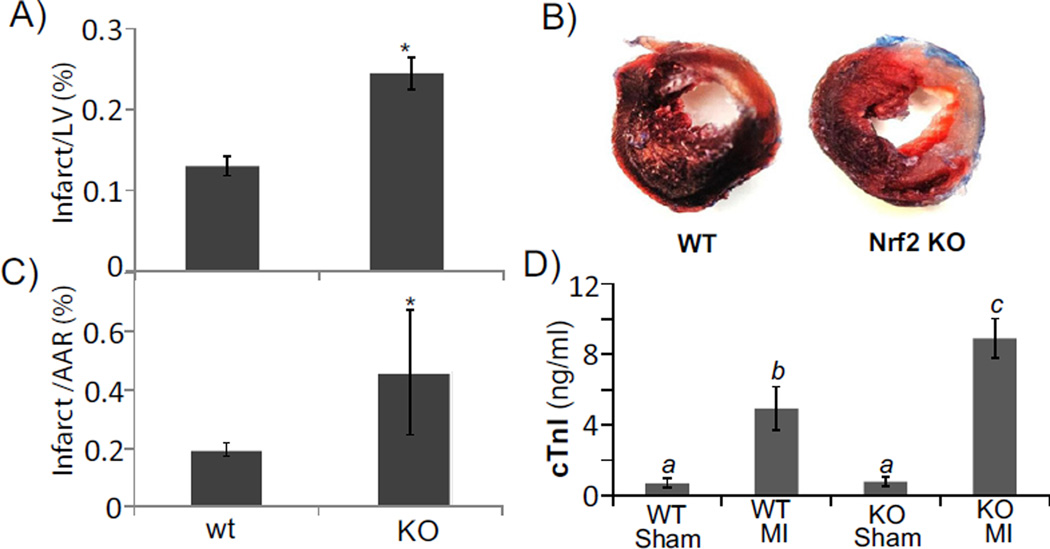
Nrf2 has been reported as a cytoprotective gene in multiple organs and tissues. Whether it plays a role in myocardial infarction has not been determined. We performed LAD coronary artery occlusion surgery in wild type (WT) littermates or Nrf2 knockout (KO) mice to determine whether lack of Nrf2 results in an increased myocardial injury. Infarction size is about 15% of the left ventricle if LAD coronary artery is ligated for 30 min followed by 24 h of reperfusion [36]. By not inducing the maximal level of infarct, we can address whether knocking out Nrf2 sensitizes the animals to ischemic reperfusion injury. In our hands, 30 min ischemia followed by 24 h of reperfusion caused an average of 13% infarction in the left ventricle of wild type mice. The same protocol caused Nrf2 KO mice to have much bigger infarct size (Fig. 1A, B). In addition to an increase in infarct size or infarct size over the area-at-the risk (AAR, Fig. 1C), serum cardiac troponin I (cTnI) concentration was measured for quantitative comparison of cardiac injury. The data show that Nrf2 KO mice had much higher level of cTnI in the blood (Fig. 1D), indicating worse cardiac injury.
Fig. 1.
Nrf2 KO mice are more sensitive to ischemic reperfusion injury. Nrf2 KO mice and wild type littermates were subjected to 30 min ischemia by LAD coronary artery occlusion followed by 24 h reperfusion before quantification of infarct size or infarct over area-at-risk (AAR) following trypan blue perfusion and TTC staining. The data represent average ± standard deviations from 4 animals in each group. When two groups of means are compared, a statistical difference (p < 0.05) was determined by Student's t test and is labeled with *. When multiple groups of means were compared, ANOVA was used to determine statistical differences (p < 0.05). The means labeled “a” are significantly different from the means labeled “b” or “c”, while the difference between means labeled with “b” and “c” is statistically significant.
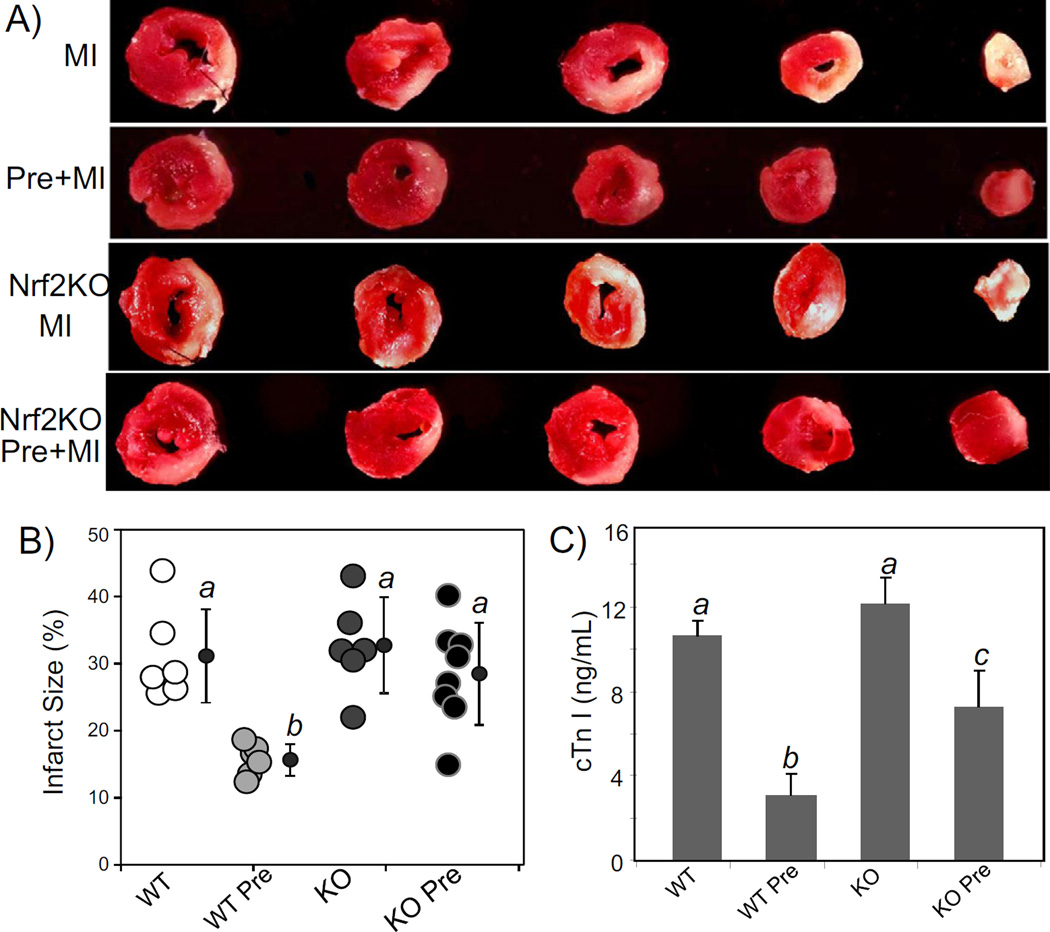
Permanent occlusion of LAD causes about 30% infarction of the left ventricle in wild type animals [32]. Since this is the maximal level of cardiac injury for an animal to survive the surgery [32,36,37], this model allows us to study cardiac protection by ischemic preconditioning and test whether Nrf2 KO mice have lost cardiac protection. WT littermates with 2 cycles of 5 min ischemia and 5 min of reperfusion (2× 5′I/5′R) showed a marked protection against myocardial infarction by permanent LAD occlusion, resulting in an approximate 50% reduction in infarct size (15.6 ± 2.4% versus 31.2 ± 7.0%; p < 0.001, Fig. 2A, B). Significant reduction in blood cTnI levels provides additional measurements of cardiac protection by 2× 5′I/5′R in WT mice (Fig. 2C). In contrast, 2× 5′I/5′R did not elicit as much protective effect in Nrf2 KO mice, as measured by infarct size and blood cTnI, compared to WT mice (Fig. 2). The average infarct size of Nrf2 KO mice with preconditioning was not significantly different from that without preconditioning (28.5 ± 7.6% versus 31.2 ± 7.0%; p > 0.5). These data support that Nrf2 mediates cardiac protection as measured by ischemic preconditioning.
Fig. 2.
Nrf2 KO mice lose preconditioning induced cardiac protection. Wild type (WT) and Nrf2 KO mice at 8–10 weeks old were used for LAD coronary artery occlusion surgery. For preconditioning, mouse hearts were subjected to 5 min occlusion and 5 min release of the LAD coronary artery for 2 cycles (2× 5′I/5′R). For control group not having 2× 5′I/5′R or preconditioning group with 2× 5′I/5′R, the coronary artery was permanently occluded. At 24 h after permanent occlusion, the hearts and blood were collected for TTC staining to measure areas of infarction (A, B) and for cTnI concentration assay (C) respectively. A series of transverse sections of representative hearts downstream of the ligature are shown (A). Total myocardial areas or areas of infarct were quantified by using NIH Image J software. Each open circle represents the measurement from one animal and average means with standard deviations from 6 to 8 animals are shown with filled-in circles (B). ANOVA was used to determine the significant difference (p < 0.05) between the means. The means labeled “a” are significantly different from that labeled “b” or “c”, whereas the label “b” indicates significant difference from the mean labeled “c”.
3.2. Brief cycles of I/R caused elevation of Nrf2 protein
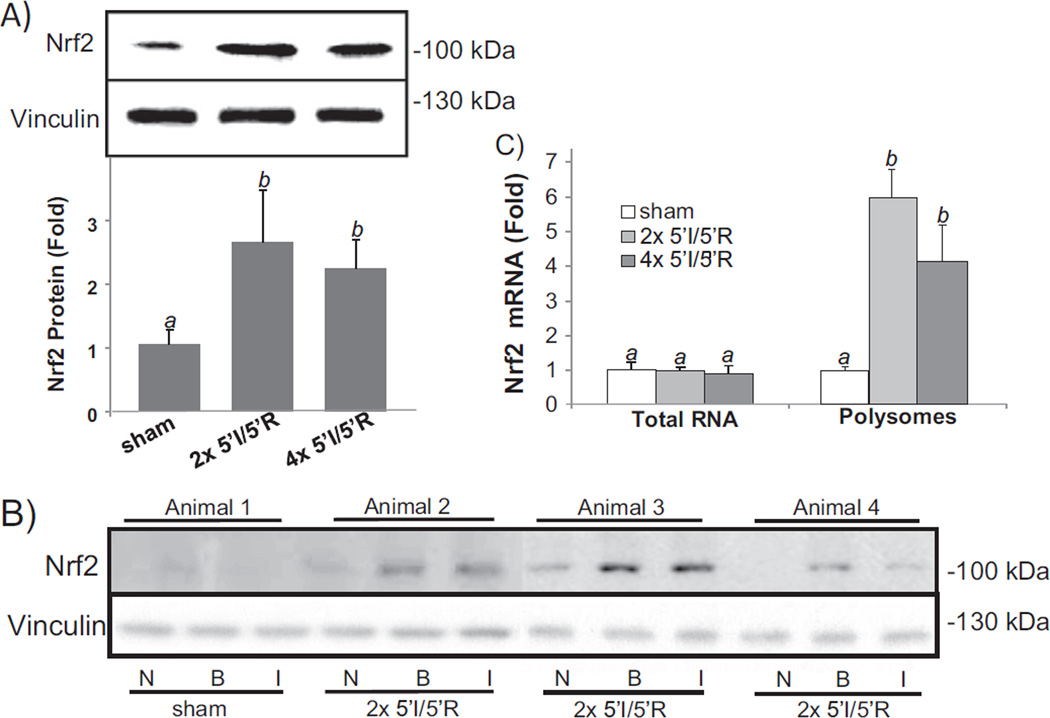
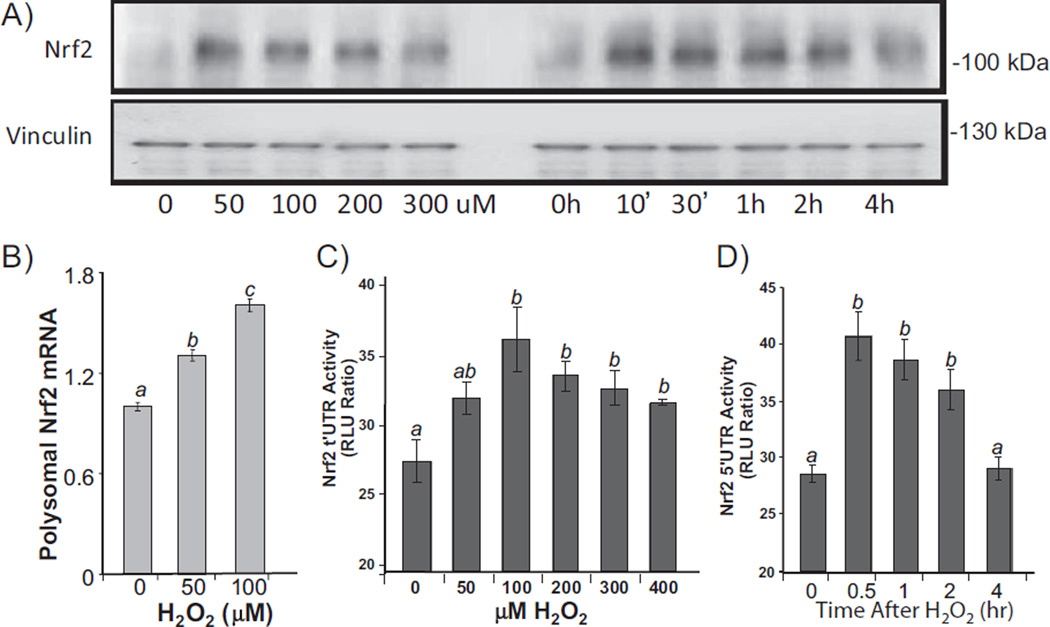
Previous works from our laboratory found that oxidative stress causes de novo Nrf2 protein translation at the cellular level [30,31]. To address whether such stress induced Nrf2 protein translation occurs in vivo, we measured Nrf2 protein levels in the myocardium following ischemic reperfusion, which is known to induce oxidative stress. Biological relevant Nrf2 is detected at 95–110 kDa by Western blot following denaturing SDS-gel electrophoresis [38]. With 2× or 4× 5′I/5′R, rapid elevation of Nrf2 protein was detectable with whole heart tissue lysates (Fig. 3A). When the ischemic region was dissected from the non-ischemic region after Evan's blue perfusion, elevated Nrf2 was observed in the ischemic region and the border zone (Fig. 3B). Severe ischemia and reperfusion stress, such as 30′I/30′R, 30′I/60′R, and 60′I/90′R did not cause Nrf2 protein elevation (data not shown).
Fig. 3.
Brief cycles of I/R cause elevation of Nrf2 protein. Mouse hearts were subjected to 2× or 4× 5′I/5′R. Tissue extracts from the whole hearts were used for Western blots (80 µg protein/lane) with vinculin as a loading control (A). The bar graph represents the relative density of the Nrf2 band over vinculin in the same sample as means ± standard deviations from 13 animals (A). Ischemic (I) versus non-ischemic (N) areas and 1–2 mm border zone (B) were rapidly dissected following Evan's blue dye perfusion for Western blot (B). A sham operated control was used for collecting areas corresponding to I, N or B (B). Total RNA or polysomal RNA was extracted from the whole heart tissue lysates for real time RT-PCR (C). The data represent means ± standard deviations from 3 (total RNA) or 4 animals (polysomal RNA, C). ANOVA was used to determine the significant difference (p < 0.05) between the means. The means labeled “a” are significantly different from that labeled “b”.
To demonstrate that rapid increase of Nrf2 protein resulted from de novo protein translation in the myocardium, we measured the association of Nrf2 mRNA with ribosomes. When an mRNA strand is being translated, it is occupied with multiple ribosomes. Ribosomes were isolated by sucrose gradient ultracentrifugation from the heart tissue after sham operation, and 2× or 4× 5′I/5′R for measurements of Nrf2 mRNA. With total heart tissue lysates, 2× or 4× 5′I/5′R did not cause an increase in Nrf2 mRNA, eliminating transcription as a cause of Nrf2 protein increase (Fig. 3C). Increased Nrf2 mRNA was found in ribosomal fractions from hearts with 2× or 4× 5′I/5′R (Fig. 3C), supporting de novo Nrf2 protein translation occurred in the ischemic area.
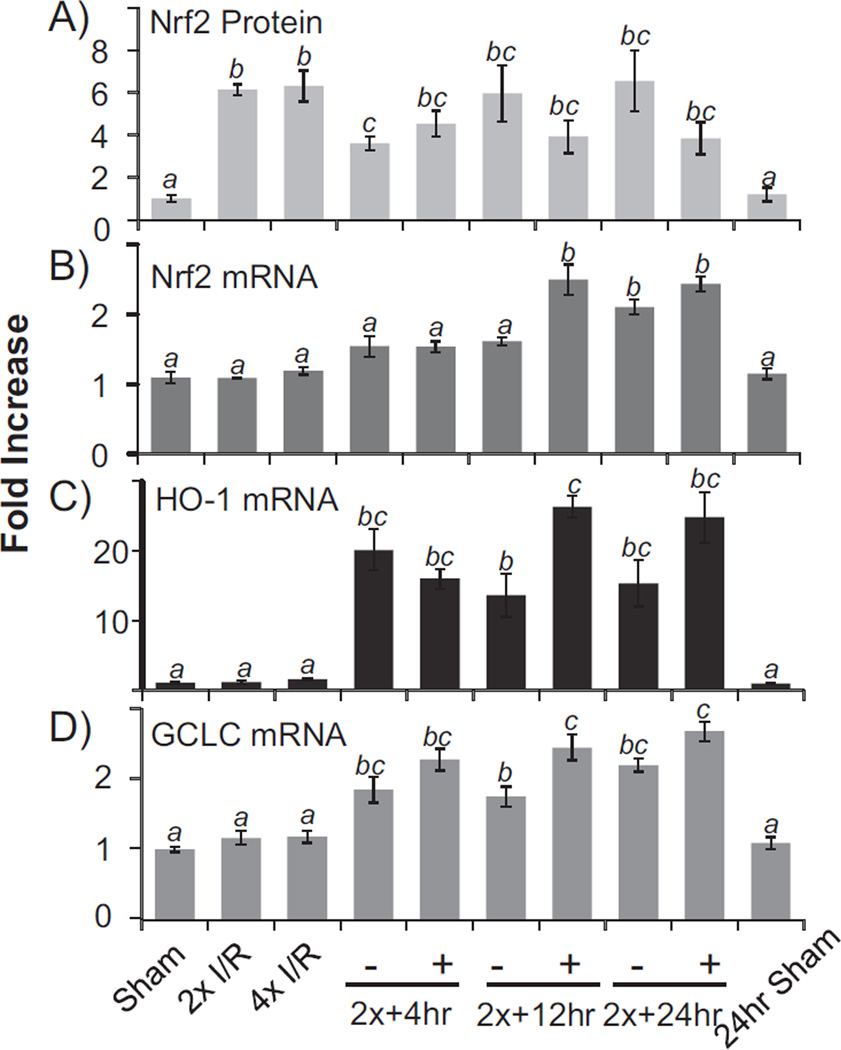
Nrf2 is a transcription factor regulating the expression of genes containing ARE in the promoters [27,39]. To address whether elevated Nrf2 protein is functional, we measured the transcripts of Nrf2 downstream genes. Nrf2 gene contains an ARE in the promoter and has been shown to undergo self regulated transcription [40]. Increases of Nrf2 mRNA were observed at 4, 12 and 24 h after 2 cycles of 5′I/5′R (Fig. 4B). In the early time point when Nrf2 protein was first elevated, Nrf2 mRNA remained unchanged (Fig. 4A, B), again supporting de novo Nrf2 protein translation in the absence of transcription in the early time points. Two additional ARE containing genes were measured: HO-1 and GCLC, both of which showed elevated transcripts in later time points (Fig. 4C, D). Therefore elevated Nrf2 protein appears to be functional as a transcription factor.
Fig. 4.
Transcription of Nrf2 downstream genes following elevation of Nrf2 protein. Mouse hearts were subjected to 2× or 4× 5′I/5′R. Immediately following 2× 5′I/5′R, the LAD coronary artery was permanently occluded for 4, 12, or 24 h for the + groups, whereas the – groups serve as sham operated controls, which had 2× 5′I/5′R but not permanent coronary artery occlusion. Tissue extracts from the whole hearts were used for Western blot (A) or harvesting total RNA for real time RT-PCR (B–D). The data represent means ± standard deviations from 3 animals. ANOVA was used to compare means and statistical differences were indicated by different letters. The means labeled “a” are significantly different from the means labeled “b”, “c” or “bc”, whereas “b” indicates significant difference of the mean from that labeled “c”. The mean labeled “bc” is not significantly different from that of “b” or “c”.
3.3. La protein binds to Nrf2 mRNA in cardiomyocytes
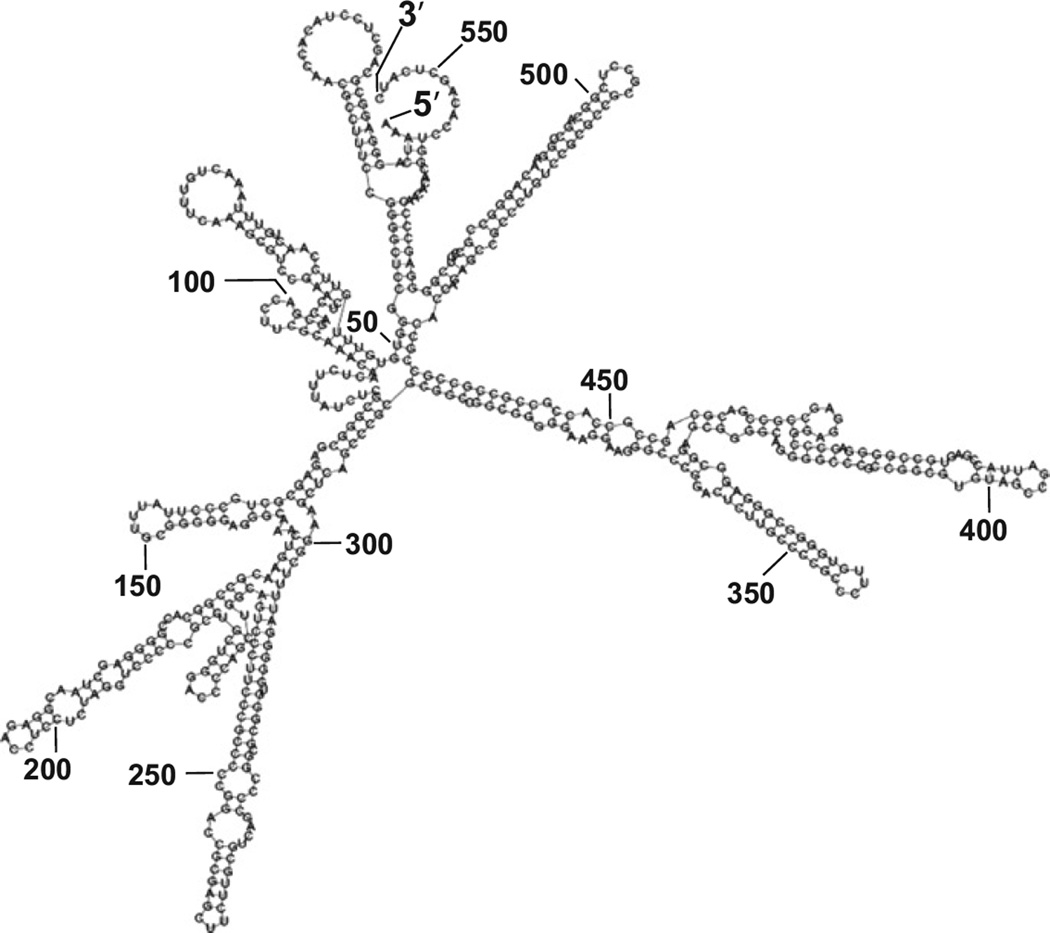
Genes that undergo stress induced protein translation often harbor IRES in 5′ UTR [2–5]. Human Nrf2 gene encodes an mRNA species containing 555 nucleotides of 5′ UTR with 70% sequence being G or C, which contributes to formation of a “stems and loops” secondary structure typical of IRES based on the prediction by Zucker's MFold algorithm (Fig. 5). An IRES interacts with its trans-acting factors (ITAFs) to regulate protein translation under stress conditions [5,6]. In order to test that Nrf2 5′ UTR plays a role in stress induced Nrf2 protein translation, we cloned the sequence of Nrf2 5′ UTR into a dicistronic vector with two luciferases: SV40 promoter and 5′ m7GpppN driven Renilla luciferase in front of Nrf2 5′ UTR regulated Firefly luciferase [34]. The Renilla luciferase corrects for transcription rate and general translation, whereas Firefly luciferase activity reflects Nrf2 5′ UTR driven translation. We used primary cultured cardiomyocytes to test the activation of Nrf2 5′ UTR by oxidative stress, since it is possible to transfect the cells with the reporter construct. Treatment of 100 µM H2O2 caused rapid elevation of Nrf2 protein in cultured cardiomyocytes (Fig. 6A). Measurements of Nrf2 mRNA in ribosomal fractions indicate that H2O2 caused an increased association of Nrf2 mRNA with ribosomes (Fig. 6B). When the dicistronic reporter construct was transfected into cardiomyocytes, a dose and time dependent elevation of Firefly luciferase was observed (Fig. 6C, D), indicating a role of Nrf2 5′ UTR in Nrf2 protein translation.
Fig. 5.
Nrf2 5′ UTR secondary structure predicted by Zucker's MFOLD algorithm.
Fig. 6.
Nrf2 5′ UTR mediated protein translation. Primary cultured rat cardiomyocytes were treated with H2O2 at indicated doses for 10 min before harvesting at 1 h (A, B, C) or with 100 µM H2O2 for 10 min before harvesting at indicated time (A, D). Levels of Nrf2 protein were measured by Western blot (30 µg protein/lane) with vinculin as a loading control (A). Ribosome-associated RNA was isolated for real time RT-PCR (B). Cells in 6-well culture dishes were transfected with 1 µg DNA of pRL–luc–Nrf2 5′ UTR-FL-luc dicistronic vector. At 48 h after transfection and 24 h culture in 0.5% FBS/DMEM, cells were treated with various doses of H2O2 for 10 min and harvested at 1 h later (C), or with 100 µM H2O2 for 10 min and harvested at indicated time (D) for dual luciferase assays. ANOVA was used to compare means and statistically significant differences (p < 0.05) are indicated by different letters. The mean labeled “a” is significantly different from the means labeled “b” or “c”, while “b” indicates statistical difference from that labeled “c”. The mean labeled “ab” is not significantly different from “a” or “b”.
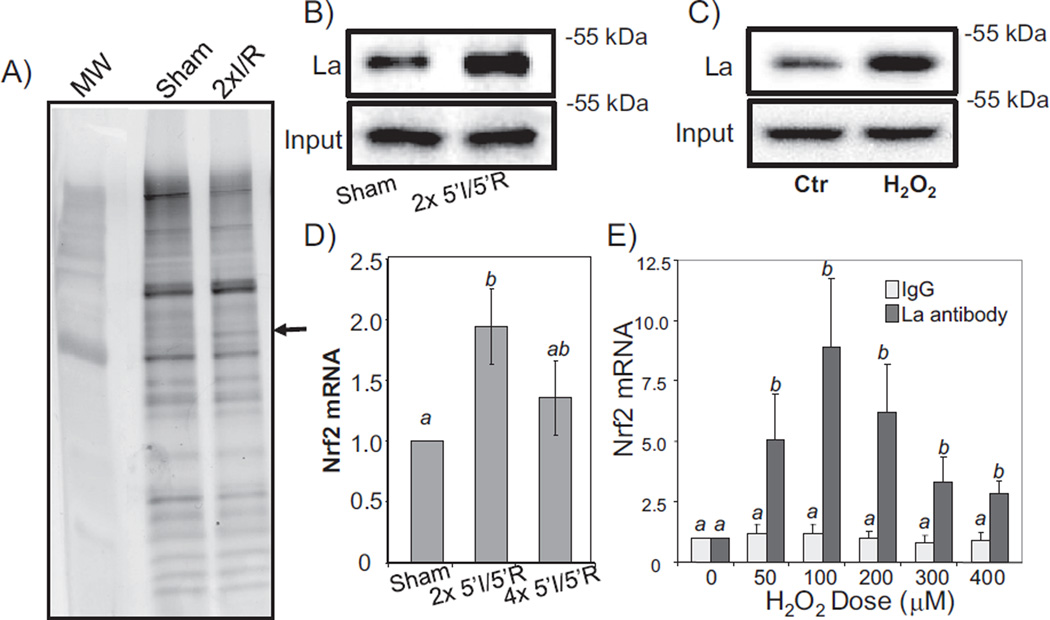
Using LC–MS/MS based proteomics, we have found La autoantigen as a protein binding to Nrf2 mRNA [31]. With myocardial tissue lysates, a protein bound to Nrf2 5′ UTR at the molecular weight equivalent to La autoantigen was found following 2× 5′I/5′R (Fig. 7A). In vitro biotinylated Nrf2 5′ UTR binding assay using tissue or cell lysates demonstrated that La indeed increased binding to Nrf2 5′ UTR in response to 2× 5′I/5′R in the myocardium (Fig. 7B) or to H2O2 treatment in cardiomyocytes (Fig. 7C). Ribonucleoprotein complex immunoprecipitation (RIP) assays allowed us to demonstrate protein–RNA interaction indeed occurring at cellular or organismic levels. This assay utilizes antibodies against La autoantigen for immunoprecipitation using cell lysates or myocardial tissue lysates, and measures Nrf2 mRNA by RT-PCR using the immunocomplex of La protein. This approach has led to the finding that 2× and 4× 5′I/5′R in the myocardium caused an increased association of La with Nrf2 mRNA (Fig. 7D). With H2O2 treatment in cardiomyocytes, a dose dependent La association with Nrf2 mRNA was observed (Fig. 7E).
Fig. 7.
La autoantigen interacts with Nrf2 5′ UTR. Biotinylated Nrf2 5′ UTR generated via in vitro transcription was used as a bait to isolate bound proteins from myocardial tissues (A, B) or cardiomyocytes (C). Bound proteins were eluted for SDS gel electrophoresis (A) or Western blot to detect La autoantigen (B, C). The lysates from the whole hearts with 2× or 4× 5′I/5′R, or from primary cultured cardiomyocytes harvested at 1 h after various doses of H2O2 treatment for 10 min were used for immunoprecipitation with an antibody specific to La autoantigen (D, E). The immunoprecipitates were used for real time RT-PCR to detect Nrf2 mRNA (D, E). The data are means ± standard deviations from triplicates of one experiment representative of three. ANOVA was used to compare means and statistically significant differences (p < 0.05) are indicated by different letters. The mean labeled “a” is significantly different from that labeled “b”, while “ab” indicates no significant difference from that labeled “a” or “b”.
3.4. La protein mediates de novo Nrf2 protein translation
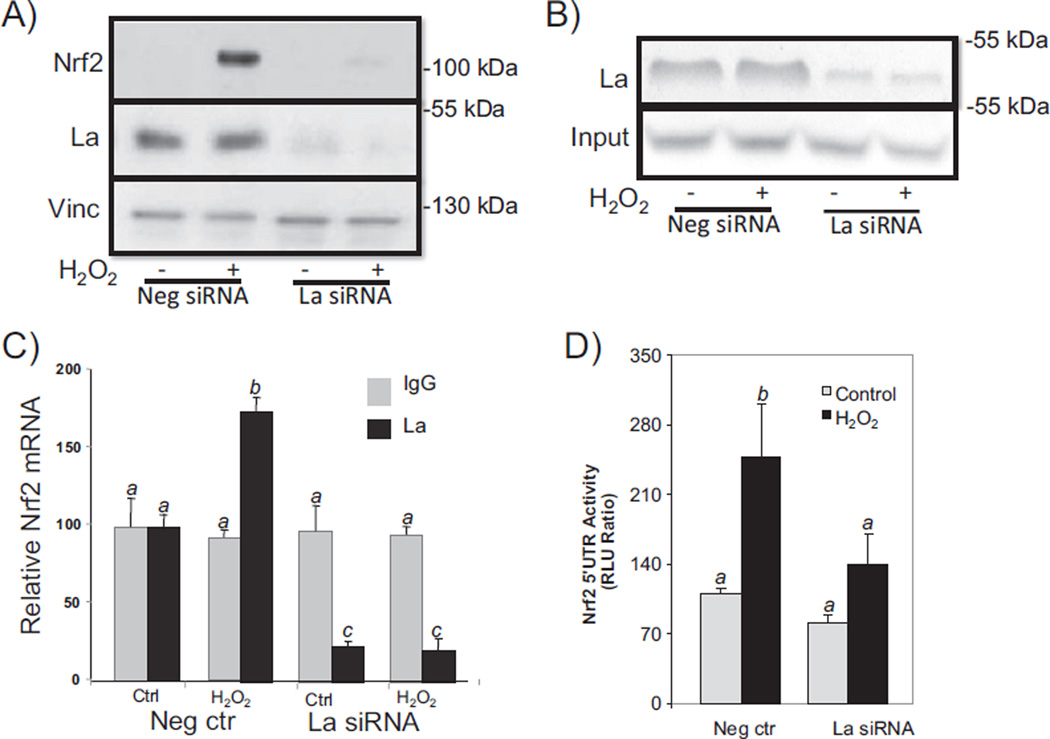
To demonstrate that La autoantigen indeed regulates Nrf2 protein translation, we used an RNA interference strategy to reduce La protein expression. With cardiomyocytes in culture, Nrf2 protein was induced with H2O2 treatment in cells transfected with the negative control, i.e. a scrambled RNA sequence (Fig. 8A). With siRNA against La, transfected cardiomyocytes showed reduction of La protein and corresponding inhibition of H2O2 induced Nrf2 protein (Fig. 8A). The siRNA also blocked La interaction with Nrf2 mRNA as measured by in vitro binding to biotinylated Nrf2 5′ UTR and RIP assays (Fig. 8B & C). Measurements of Nrf2 5′ UTR activity using cardiomyocytes transfected with the dicistronic reporter construct also indicated an inhibitory effect of La siRNA against H2O2 induced activation (Fig. 8D).
Fig. 8.
La autoantigen regulates H2O2 induced Nrf2 protein translation in cardiomyocytes. Cardiomyocytes were transfected with 300 nM siRNA sequence against La autoantigen or scrambled sequence (negative siRNA). At 48 h after transfection, cells were treated with 100 µM H2O2 for 10 min before harvesting 1 h later for Western blot (A), in vitro biotinylated Nrf2 5′ UTR binding then Western blot (B), or immunoprecipitation of La complex then real time RT-PCR to measure Nrf2 mRNA (C). La siRNA was cotransfected with pRL–luc–Nrf2 5′ UTR-FL-luc dicistronic vector for measurement of Nrf2 5′ UTR driven Firefly luciferase over Renilla luciferase 1 h after 10 min treatment of 100 µM H2O2 (D). ANOVA was used to compare means and the statistically significant differences (p < 0.05) are indicated by different letters. The means labeled “a” are significantly different from the means labeled “b” or “c”, while“b” indicates significant difference from the means labeled “c”.
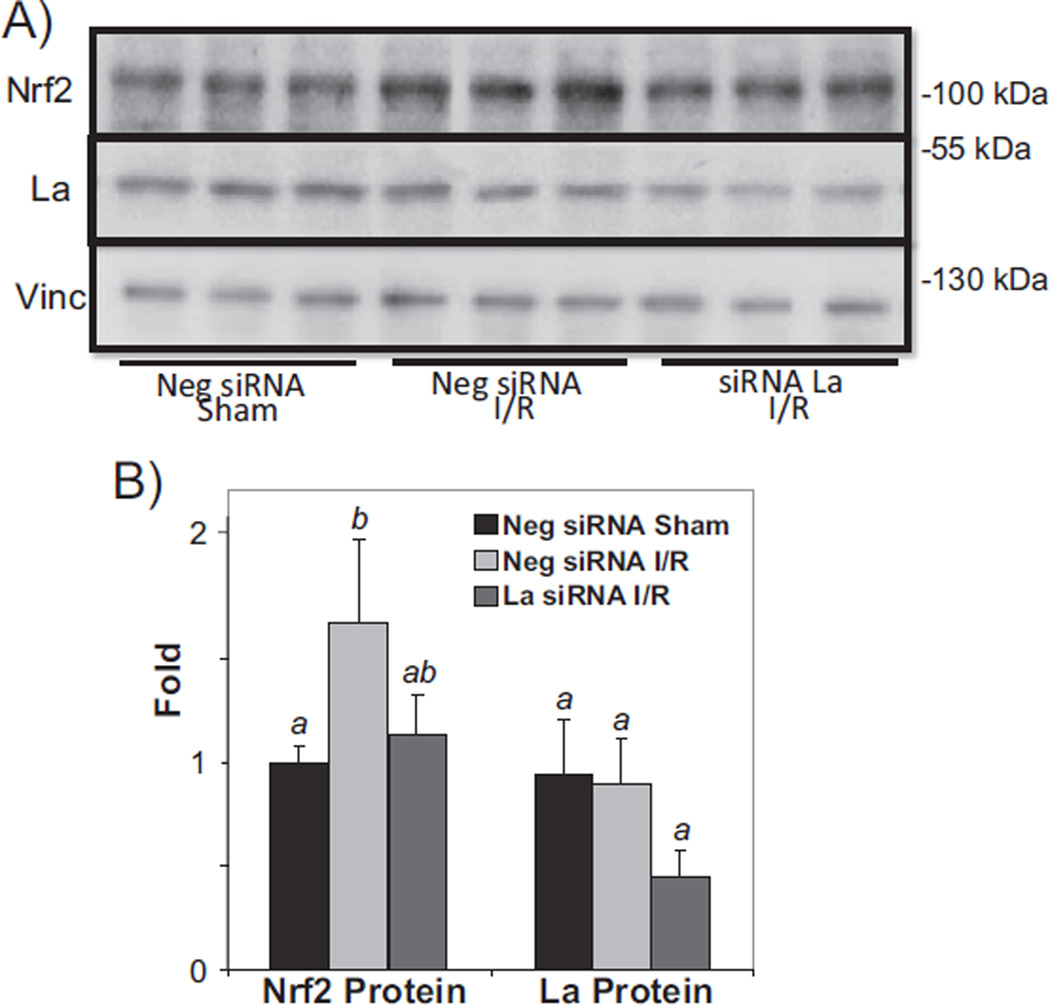
When siRNA was delivered in vivo, we found an approximate 60% reduction of La protein in the myocardium after a bolus injection of siRNA via the jugular vein (15 µg, ~500 µg/kg, Fig. 9A). Compared to the effect of 2× 5′I/5′R on elevation of Nrf2 protein, mice with La siRNA failed to elevate Nrf2 protein in the myocardium (Fig. 9B). These results demonstrate that La autoantigen was a key factor for regulating Nrf2 protein translation.
Fig. 9.
La autoantigen regulates Nrf2 protein induction in vivo. siRNA or negative control siRNA (15 µg) was injected via the jugular vein 24 h before surgery to induce 2× 5′I/5′R (C). Tissue lysates from the whole heart were used for Western blot (80 µg protein/lane), with each lane representing one sample from one animal (A). The intensities of the bands were quantified by NIH image J software and presented as means ± SD from 3 animals (B). ANOVA was used to compare means and the statistically significant differences (p < 0.05) are indicated by different letters. The means labeled “a” are significantly different from the means labeled “b”, while “ab” indicates no significant difference from that labeled “a” or “b”.
4. Discussion
This study reveals that Nrf2 KO mice have an increased sensitivity to ischemic injury. Nrf2 protein elevation correlates with cardiac protection by ischemic preconditioning. We found that 2× or 4× 5′I/5′R caused elevation of Nrf2 protein likely through de novo translation in the myocardium. An increase of La protein binding to Nrf2 5′ UTR appeared to mediate oxidant or I/R induced de novo Nrf2 protein translation in cardiomyocytes or the myocardium. Since Nrf2 controls the transcription of a cluster of antioxidant and detoxification genes, de novo translation of Nrf2 protein constitutes an effective measure for rapid activation of endogenous defense.
Infarct size quantification and blood cTnI measurements indicate that Nrf2 KO mice have reduced but not completely abolished protection by ischemic preconditioning. Our data are consistent with the literature suggesting the cardiac protective function of Nrf2 [37, 41–44]. How Nrf2 protects the myocardium from tissue injury remains to be addressed. The protective effect of ischemic preconditioning is observed in two phases: early phase, immediately after cycles of brief ischemic reperfusion, and late phase, typically 24 h after the initial episode of brief ischemia [45]. The time course studies of Nrf2 and its downstream genes suggest that Nrf2 may participate in both early and late phases of preconditioning induced cardiac protection.
Induction of Nrf2 protein is unlikely the sole mechanism of cardiac protection by ischemic preconditioning. In addition to elevated oxidant generation, ischemic reperfusion causes increases in reactive nitrogen species, profound changes in energy metabolism and altered ionic homeostasis. These biochemical events may trigger cellular defense mechanisms in addition to Nrf2. The classic view of the mechanism of cardiac protection by preconditioning includes release of adenosine, bradykinin, endothelin and endorphins. Some of these factors activate survival signal pathways following binding to G-protein coupled receptors on cell surface, including phosphoinositide 3 kinase (PI3K)/Akt, ERK1/2, and protein kinase C [46–48]. PI3K/Akt, p38 MAPK and protein kinase C have been suggested to promote Nrf2 expression and nuclear translocation [43,49]. Activation of HIF-1 and NF-kB transcription factors, and elevated expression of inducible nitric oxide synthase (iNOS), manganese superoxide dismutase, heat shock proteins and cyclooxygenase-2 also mediate the cardiac protective effect of preconditioning [50–53]. Among the genes under the control of Nrf2, HO-1 is also a downstream target of the redox sensing transcription factor HIF-1 [54,55]. Overexpression of HO-1 alone in the myocardium via a genetic approach is sufficient for cardiac protection [56]. In parallel with protective roles of individual genes, preserving mitochondrial integrity and function appears to be a key event in preconditioning induced cardiac protection [57]. Therefore Nrf2 governed pathway is one among several parallel or cross-talking pathways contributing to cardiac protection.
Most importantly, our data here suggest that stress induced protein translation occurs in experimental animals. Yeast and cell lines have been typically used to demonstrate selective protein translation under stress. IRES in 5′ UTR enables translation initiation of selective proteins, bypassing the global decline of protein translation by 5′ m7GpppN cap dependent mechanism [2–5]. IRES mediated protein translation was first discovered with viral proteins, and is heavily studied in cell lines during viral infection. About 50 cellular genes have now been reported to undergo IRES mediated translation. Examples of these genes include c-myc, c-Jun, HIF-1α, p27/Kip1, Apaf1, bcl-2, XIAP and GRP78 (http://ifr31w3.toulouse.inserm.fr/iresdatabase). It is commonly thought that IRES sequences are GC rich, a feature essential for formation of secondary structures containing “stems and loops”. The sequence of Nrf2 5′ UTR fits this characteristics (Fig. 5). Transfection experiments with the dicistronic reporter construct support Nrf2 5′ UTR indeed containing an IRES. Since over 50% of cellular ATP is utilized for general protein translation and ATP synthesis is impaired during stress, selective protein translation serves to conserve energy [2]. Evolutionarily, selective protein translation has the advantage of increasing the ratio of the proteins necessary for dealing with stress.
An IRES recruits ITAFs, which promote the binding of eIFs and ribosomes for translation initiation. With Nrf2 5′ UTR, we have found that La protein acts as an ITAF responding to oxidants or brief cycles of I/R by turning on Nrf2 protein translation. The La protein was first discovered as an autoantigen in 20–30% of lupus patients or 60% of patients with Sjögren's syndrome, and is therefore named Sjögren's syndrome antigen B [58]. This protein is encoded by a gene well conserved among eukaryotes, and contains two RNA recognition motifs. Isolation of ribonucleoprotein complex with antibodies specific to La revealed the binding of La to a large variety of RNA species [58]. Newly synthesized transcripts from RNA polymerase III, including ribosomal RNA and tRNA, appear to bind to La at their common UUU-3′ OH end in the nuclei [59]. Such interaction contributes to nuclear retention and protection against nuclease digestion of the nascent transcripts. Cytoplasmic La protein binds to 5′-UTR terminal oligopyrimidine track (5′-TOP) and activates translation of mRNAs containing 5′-TOP, which encode ribosomal proteins and translation elongation factors [60]. In the cytoplasm, La regulates translation of viral proteins and a few cellular proteins through binding to IRES [58,61, 62]. La protein has been reported to be phosphorylated, acetylated or sumoylated. Immunoassays including 2-D Western blot have not revealed alteration of these posttranslational modifications with H2O2 treatment (data not shown). Therefore although we have found a role of La in oxidative stress induced Nrf2 protein translation, how oxidative stress causes an increased binding of La to Nrf2 5′ UTR remains to be investigated.
Acknowledgments
Source of funding
Work from our laboratory has been supported by NIH R01 HL 076530, R01 HL089958, R21ES017473, and T32 ES007091, Arizona Biomedical Research Commission (QMC), and Mark and Mary Anne Fay Investigator Award (BX), and Marjorie Hornbeck Estate Research Award (JZ) from University of Arizona Sarver Heart Center. Proteomic facility core of Southwest Environmental Health Sciences Center is supported by NIH P30 ES006694 grant.
We would like to thank technical assistance and contributions of Drs. Elena Sheveleva and Thai Nho Dinh.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 6.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 7.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K–mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J. Biol. Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damiano F, Alemanno S, Gnoni GV, Siculella L. Translational control of the sterol-regulatory transcription factor SREBP-1 mRNA in response to serum starvation or ER stress is mediated by an internal ribosome entry site. Biochem. J. 2010;429:603–612. doi: 10.1042/BJ20091827. [DOI] [PubMed] [Google Scholar]
- 10.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 11.Shlafer M, Myers CL, Adkins S. Mitochondrial hydrogen peroxide generation and activities of glutathione peroxidase and superoxide dismutase following global ischemia. J. Mol. Cell. Cardiol. 1987;19:1195–1206. doi: 10.1016/s0022-2828(87)80530-8. [DOI] [PubMed] [Google Scholar]
- 12.Vandeplassche G, Hermans C, Thone F, Borgers M. Mitochondrial hydrogen peroxide generation by NADH-oxidase activity following regional myocardial ischemia in the dog. J. Mol. Cell. Cardiol. 1989;21:383–392. doi: 10.1016/0022-2828(89)90649-4. [DOI] [PubMed] [Google Scholar]
- 13.Grill HP, Zweier JL, Kuppusamy P, Weisfeldt ML, Flaherty JT. Direct measurement of myocardial free radical generation in an in vivo model: effects of postischemic reperfusion and treatment with human recombinant superoxide dismutase. J. Am. Coll. Cardiol. 1992;20:1604–1611. doi: 10.1016/0735-1097(92)90457-x. [DOI] [PubMed] [Google Scholar]
- 14.Zweier JL, Talukder MA. The role of oxidants and free radicals in reperfusion injury. Cardiovasc. Res. 2006;70:181–190. doi: 10.1016/j.cardiores.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Araya G, Nettle D, Castro P, Miranda F, Greig D, Campos X, Chiong M, Nazzal C, Corbalan R, Lavandero S. Oxidative stress after reperfusion with primary coronary angioplasty: lack of effect of glucose–insulin–potassium infusion. Crit. Care Med. 2002;30:417–421. doi: 10.1097/00003246-200202000-00025. [DOI] [PubMed] [Google Scholar]
- 16.Iuliano L, Pratico D, Greco C, Mangieri E, Scibilia G, FitzGerald GA, Violi F. Angioplasty increases coronary sinus F2-isoprostane formation: evidence for in vivo oxidative stress during PTCA. J. Am. Coll. Cardiol. 2001;37:76–80. doi: 10.1016/s0735-1097(00)01040-8. [DOI] [PubMed] [Google Scholar]
- 17.Stephens NG, Parsons A, Schofield PM, Kelly F, Cheeseman K, Mitchinson MJ. Randomised controlled trial of vitamin E in patients with coronary disease: Cambridge Heart Antioxidant Study (CHAOS) Lancet. 1996;347:781–786. doi: 10.1016/s0140-6736(96)90866-1. [DOI] [PubMed] [Google Scholar]
- 18.Yusuf S, Dagenais G, Pogue J, Bosch J, Sleight P. Vitamin E supplementation and cardiovascular events in high-risk patients. The Heart Outcomes Prevention Evaluation Study Investigators. N. Engl. J. Med. 2000;342:154–160. doi: 10.1056/NEJM200001203420302. [DOI] [PubMed] [Google Scholar]
- 19.Keith ME, Jeejeebhoy KN, Langer A, Kurian R, Barr A, O'Kelly B, Sole MJ. A controlled clinical trial of vitamin E supplementation in patients with congestive heart failure. Am. J. Clin. Nutr. 2001;73:219–224. doi: 10.1093/ajcn/73.2.219. [DOI] [PubMed] [Google Scholar]
- 20.G. Heart Protection Study Collaborative. MRC/BHF Heart Protection Study of antioxidant vitamin supplementation in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 2002;360:23–33. doi: 10.1016/S0140-6736(02)09328-5. [DOI] [PubMed] [Google Scholar]
- 21.Arad Y, Spadaro LA, Roth M, Newstein D, Guerci AD. Treatment of asymptomatic adults with elevated coronary calcium scores with atorvastatin, vitamin C, and vitamin E: the St. Francis Heart Study randomized clinical trial. J. Am. Coll. Cardiol. 2005;46:166–172. doi: 10.1016/j.jacc.2005.02.089. [DOI] [PubMed] [Google Scholar]
- 22.Miller ER, Pastor-Barriuso R, Dalal D, Riemersma RA, Appel LJ, Guallar G. Meta-analysis: high-dosage vitamin E supplementation may increase all-cause mortality. Ann. Intern. Med. 2005;142:37–46. doi: 10.7326/0003-4819-142-1-200501040-00110. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Hamid T, Keith RJ, Zhou G, Partridge CR, Xiang X, Kingery JR, Lewis RK, Li Q, Rokosh DG, Ford R, Spinale FG, Riggs DW, Srivastava S, Bhatnagar A, Bolli R, Prabhu SD. Cardioprotective and antiapoptotic effects of heme oxygenase-1 in the failing heart. Circulation. 2010;121:1912–1925. doi: 10.1161/CIRCULATIONAHA.109.905471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das DK. Thioredoxin regulation of ischemic preconditioning. Antioxid. Redox Signal. 2004;6:405–412. doi: 10.1089/152308604322899477. [DOI] [PubMed] [Google Scholar]
- 25.Das DK, Maulik N. Cardiac genomic response following preconditioning stimulus. Cardiovasc. Res. 2006;70:254–263. doi: 10.1016/j.cardiores.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1–Nrf2–ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 28.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 29.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of Nrf2 protein in activation of antioxidant response element by oxidants. Mol. Pharm. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Dinh T, Kapperler K, Chen Q. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.015032. (M111.015032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael LH, Entman ML, Hartley CJ, Youker KA, Zhu J, Hall SR, Hawkins HK, Berens K, Ballantyne CM. Myocardial ischemia and reperfusion: a murine model. Am. J. Physiol. 1995;269:H2147–H2154. doi: 10.1152/ajpheart.1995.269.6.H2147. [DOI] [PubMed] [Google Scholar]
- 33.del Prete MJ, Vernal R, Dolznig H, Mullner EW, Garcia-Sanz JA. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han B, Zhang JT. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell. Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael LH, Ballantyne CM, Zachariah JP, Gould KE, Pocius JS, Taffet GE, Hartley CJ, Pham TT, Daniel SL, Funk E, Entman ML. Myocardial infarction and remodeling in mice: effect of reperfusion. Am. J. Physiol. 1999;277:H660–H668. doi: 10.1152/ajpheart.1999.277.2.H660. [DOI] [PubMed] [Google Scholar]
- 37.Ashrafian H, Czibik G, Bellahcene M, Aksentijevic D, Smith AC, Mitchell SJ, Dodd MS, Kirwan J, Byrne JJ, Ludwig C, Isackson H, Yavari A, Stottrup NB, Contractor H, Cahill TJ, Sahgal N, Ball DR, Birkler RI, Hargreaves I, Tennant DA, Land J, Lygate CA, Johannsen M, Kharbanda RK, Neubauer S, Redwood C, de Cabo R, Ahmet I, Talan M, Gunther UL, Robinson AJ, Viant MR, Pollard PJ, Tyler DJ, Watkins H. Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway. Cell Metab. 2012;15:361–371. doi: 10.1016/j.cmet.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau A, Wang T, Whitman S, Zhang D. The predicted molecular weight of Nrf2: it is what it is not. Antioxid. Redox Signal. 2012 doi: 10.1089/ars.2012.4754. http://dx.doi.org/10.1089/ars.2012.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 40.Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Sano M, Shinmura K, Tamaki K, Katsumata Y, Matsuhashi T, Morizane S, Ito H, Hishiki T, Endo J, Zhou H, Yuasa S, Kaneda R, Suematsu M, Fukuda K. 4-Hydroxy-2-nonenal protects against cardiac ischemia–reperfusion injury via the Nrf2-dependent pathway. J. Mol. Cell. Cardiol. 2010;49:576–586. doi: 10.1016/j.yjmcc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Stein AB, Bolli R, Dawn B, Sanganalmath SK, Zhu Y, Wang OL, Guo Y, Motterlini R, Xuan YT. Carbon monoxide induces a late preconditioning-mimetic cardioprotective and antiapoptotic milieu in the myocardium. J. Mol. Cell. Cardiol. 2012;52:228–236. doi: 10.1016/j.yjmcc.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng C, Sun Z, Tong G, Yi W, Ma L, Zhao B, Cheng L, Zhang J, Cao F, Yi D. Alphalipoic acid reduces infarct size and preserves cardiac function in rat myocardial ischemia/reperfusion injury through activation of PI3K/Akt/Nrf2 pathway. PLoS One. 2013;8:e58371. doi: 10.1371/journal.pone.0058371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai Y, Cui W, Xin Y, Miao X, Barati MT, Zhang C, Chen Q, Tan Y, Cui T, Zheng Y, Cai L. Prevention by sulforaphane of diabetic cardiomyopathy is associated with up-regulation of Nrf2 expression and transcription activation. J. Mol. Cell. Cardiol. 2013;57:82–95. doi: 10.1016/j.yjmcc.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Napoli C, Pinto A, Cirino G. Pharmacological modulation, preclinical studies, and new clinical features of myocardial ischemic preconditioning. Pharmacol. Ther. 2000;88:311–331. doi: 10.1016/s0163-7258(00)00093-0. [DOI] [PubMed] [Google Scholar]
- 46.Clerk A, Cullingford TE, Fuller SJ, Giraldo A, Markou T, Pikkarainen S, Sugden PH. Signaling pathways mediating cardiac myocyte gene expression in physiological and stress responses. J. Cell. Physiol. 2007;212:311–322. doi: 10.1002/jcp.21094. [DOI] [PubMed] [Google Scholar]
- 47.Downey JM, Davis AM, Cohen MV. Signaling pathways in ischemic preconditioning. Heart Fail. Rev. 2007;12:181–188. doi: 10.1007/s10741-007-9025-2. [DOI] [PubMed] [Google Scholar]
- 48.Burgoyne JR, Mongue-Din H, Eaton P, Shah AM. Redox signaling in cardiac physiology and pathology. Circ. Res. 2012;111:1091–1106. doi: 10.1161/CIRCRESAHA.111.255216. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Xiao Z, Yao J, Zhao G, Fa X, Niu J. Participation of protein kinase C in the activation of Nrf2 signaling by ischemic preconditioning in the isolated rabbit heart. Mol. Cell. Biochem. 2013;372:169–179. doi: 10.1007/s11010-012-1458-9. [DOI] [PubMed] [Google Scholar]
- 50.Bolli R. Preconditioning: a paradigm shift in the biology of myocardial ischemia. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H19–H27. doi: 10.1152/ajpheart.00712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otani H. Ischemic preconditioning: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2008;10:207–247. doi: 10.1089/ars.2007.1679. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Cohen MV, Downey JM. Mechanism of cardioprotection by early ischemic preconditioning. Cardiovasc. Drugs Ther. 2010;24:225–234. doi: 10.1007/s10557-010-6236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanada S, Komuro I, Kitakaze M. Pathophysiology of myocardial reperfusion injury: preconditioning, postconditioning, and translational aspects of protective measures. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1723–H1741. doi: 10.1152/ajpheart.00553.2011. [DOI] [PubMed] [Google Scholar]
- 54.Ockaili R, Natarajan R, Salloum F, Fisher BJ, Jones D, Fowler III AA, Kukreja RC. HIF-1 activation attenuates postischemic myocardial injury: role for heme oxygenase-1 in modulating microvascular chemokine generation. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H542–H548. doi: 10.1152/ajpheart.00089.2005. [DOI] [PubMed] [Google Scholar]
- 55.Dawn B, Bolli R. HO-1 induction by HIF-1: a new mechanism for delayed cardioprotection? Am. J. Physiol. Heart Circ. Physiol. 2005;289:H522–H524. doi: 10.1152/ajpheart.00274.2005. [DOI] [PubMed] [Google Scholar]
- 56.Yet SF, Tian R, Layne MD, Wang ZY, Maemura K, Solovyeva M, Ith B, Melo LG, Zhang L, Ingwall JS, Dzau VJ, Lee ME, Perrella MA. Cardiac-specific expression of heme oxygenase-1 protects against ischemia and reperfusion injury in transgenic mice. Circ. Res. 2001;89:168–173. doi: 10.1161/hh1401.093314. [DOI] [PubMed] [Google Scholar]
- 57.Hausenloy DJ, Ong SB, Yellon DM. The mitochondrial permeability transition pore as a target for preconditioning and postconditioning. Basic Res. Cardiol. 2009;104:189–202. doi: 10.1007/s00395-009-0010-x. [DOI] [PubMed] [Google Scholar]
- 58.Wolin SL, Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 59.Maraia RJ, Intine RV. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 2001;21:367–379. doi: 10.1128/MCB.21.2.367-379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, Conte MR. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 61.Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]