Abstract
Emerging evidence indicates that mitochondrial flashes (mitoflashes) are highly conserved elemental mitochondrial signaling events. However, which signal controls their ignition and how they are integrated with other mitochondrial signals and functions remain elusive. In this study, we aimed to further delineate the signal components of the mitoflash and determine the mitoflash trigger mechanism. Using multiple biosensors and chemical probes as well as label-free autofluorescence, we found that the mitoflash reflects chemical and electrical excitation at the single-organelle level, comprising bursting superoxide production, oxidative redox shift, and matrix alkalinization as well as transient membrane depolarization. Both electroneutral H+/K+ or H+/Na+ antiport and matrix proton uncaging elicited immediate and robust mitoflash responses over a broad dynamic range in cardiomyocytes and HeLa cells. However, charge-uncompensated proton transport, which depolarizes mitochondria, caused the opposite effect, and steady matrix acidification mildly inhibited mitoflashes. Based on a numerical simulation, we estimated a mean proton lifetime of 1.42 ns and diffusion distance of 2.06 nm in the matrix. We conclude that nanodomain protons act as a novel, to our knowledge, trigger of mitoflashes in energized mitochondria. This finding suggests that mitoflash genesis is functionally and mechanistically integrated with mitochondrial energy metabolism.
Introduction
The mitoflash is a recently discovered dynamic mitochondrial activity that is ubiquitously present across species ranging from worms to humans (1, 2, 3), as well as in experimental systems such as isolated mitochondria, intact cells, explanted organs, and even living animals (1, 3, 4, 5, 6, 7). Although the properties of individual mitoflashes are rather stereotypical, the rate of mitoflash occurrence is actively regulated by factors such as the metabolic state, oxidative stress, developmental stage, and ageing (3, 5, 6, 8, 9, 10, 11, 12). In addition, it has been shown that frequency-modulated mitoflash activity serves as a biomarker of pathological stresses as well as their amelioration by therapeutic interventions (11, 13, 14, 15). These recent findings strongly suggest that mitoflashes are elemental signaling events in eukaryotic cells.
Since the initial detection of mitoflashes with the biosensor mt-cpYFP, which dually senses superoxide and pH (1), the exact nature of the mitoflash has been intensely investigated, and individual groups have developed different methods and indicators to report mitoflashes or mitoflash-like events (8, 16, 17). It would be informative to systematically dissect the signal components of the mitoflash and obtain a more complete picture. Moreover, it remains to be determined how mitoflash activity is controlled by and coupled with other mitochondrial signals and functions. We and others have provided evidence that elevations of mitochondrial Ca2+ and reactive oxygen species (ROS) enhance the occurrence of mitoflashes (18, 19), particularly when the two regulators act synergistically (11). However, according to Mitchell’s chemiosmotic theory of ATP synthesis, protons and their vectorial movement across the inner mitochondrial membrane (IMM) are essential for ATP production (20, 21). This proton motive-force-centered bioenergetics is common to all forms of aerobic life, from single-cell organisms to humans. Thus, it is of great interest to determine whether the genesis of mitoflashes is linked to mitochondrial proton signaling and hence bioenergetics.
In this study, we first systematically characterized the constituent signals of mitoflashes with the aid of multiple recently developed indicators, as well as label-free imaging methods. More importantly, we investigated whether and how proton signaling controls mitoflash activity. We demonstrate that protons produced by electroneutral proton ionophores or by photolytic uncaging directly trigger mitoflashes. This result uncovers a heretofore unappreciated signaling role for mitochondrial protons, and sheds new, to our knowledge, light on how mitoflash activity is mechanistically and functionally integrated with energy metabolism.
Materials and Methods
Animal care
All procedures were carried out according to the rules of the American Association for the Accreditation of Laboratory Animal Care International and approved by the Animal Care Committee of Peking University (accredited by AAALAC International). This investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH publication No. 85-23, revised 1996).
Cardiomyocyte isolation and culture
Adult ventricular myocytes were enzymatically isolated from the hearts of adult male Sprague-Dawley rats as described previously (22) and in Supporting Materials and Methods in the Supporting Material. Cells were infected with adenovirus carrying mt-cpYFP, mt-EYFP, mt-grx-roGFP2, or mt-pHTomato at an m.o.i. of 20, and experiments were performed after 48–72 h in culture.
Confocal imaging of mitoflashes
Cells were suspended in Tyrode’s solution consisting of (in mM) 137 NaCl, 5.4 KCl, 1.2 MgCl2, 1.2 NaH2PO4, 1.8 CaCl2, 5.6 glucose, and 20 HEPES (pH 7.35, adjusted with NaOH). In rod-shaped cardiomyocytes or HeLa cells, mitoflashes were detected using an inverted confocal microscope (LSM 710; Zeiss, Jena, Germany) with a 40×, 1.3 NA oil-immersion objective. Time-lapse two-dimensional (xy) images were captured as described in Supporting Materials and Methods.
Proton uncaging from 2-nitrobenzaldehyde
Cells were loaded with 1 mM 2-nitrobenzaldehyde (NBA) as the proton donor, as previously described (23). Photolysis of NBA for proton uncaging was mediated by 405 nm laser illumination at various intensities (0.5–3% of full laser power of 15 mW) and applied to whole cells or predefined subcellular areas. The photolysis protocol was alternated with image acquisition at 488 nm excitation, in line-by-line (for whole-cell uncaging) or frame-by-frame fashion (for subcellular uncaging), and the entire protocol was repetitive at 1 Hz. Cytosolic pH was measured with SNARF-1, and mitochondrial pH was measuring using mt-EYFP or mt-pHTomato as described in Supporting Materials and Methods.
Image processing and mitoflash analysis
Confocal images were analyzed using custom-developed programs written in Interactive Data Language (Exelis Visual Information Solutions, Boulder, CO). Cell-motion artifacts and background fluorescence changes due to photoconversion and photobleaching were corrected by image processing, and individual mitoflashes were located and their parameters were quantified with the aid of FlashSniper (24).
Numerical simulation of proton spikes during uncaging
The dynamics between protons (H+) and pH buffers (B) in the matrix was depicted by the following ordinary differential equations:
where refers to the free proton concentration, is the proton-bound buffer concentration, is the total buffer concentration (25), is the association rate constant (26, 27), is the dissociation rate constant, and pKd = 7 is the dissociation constant (25). For initial conditions of uncaging, we set
For the numerical simulation, we used MATLAB ver. 2012a (The MathWorks, Natick, MA) running on a DELL desktop PC with an Intel i7-4790 CPU at 3.60 GHz and 8.0 GB of RAM. The mean lifetime (τ) of uncaged protons was estimated as the 63% decay time of the proton spike:
The mean distance of diffusion (l) was calculated as
where refers to the diffusion coefficient of protons (26). In this model, the buffer capacity can be defined as
Using the above parameters, we obtain a buffer power of 5.0 mM at pH 8.0, in agreement with the value measured experimentally (28).
Statistics
Data are expressed as mean ± SEM. When appropriate, Student’s t-test was applied to determine statistical significance.
Results
Multifaceted signal origin of mitoflashes
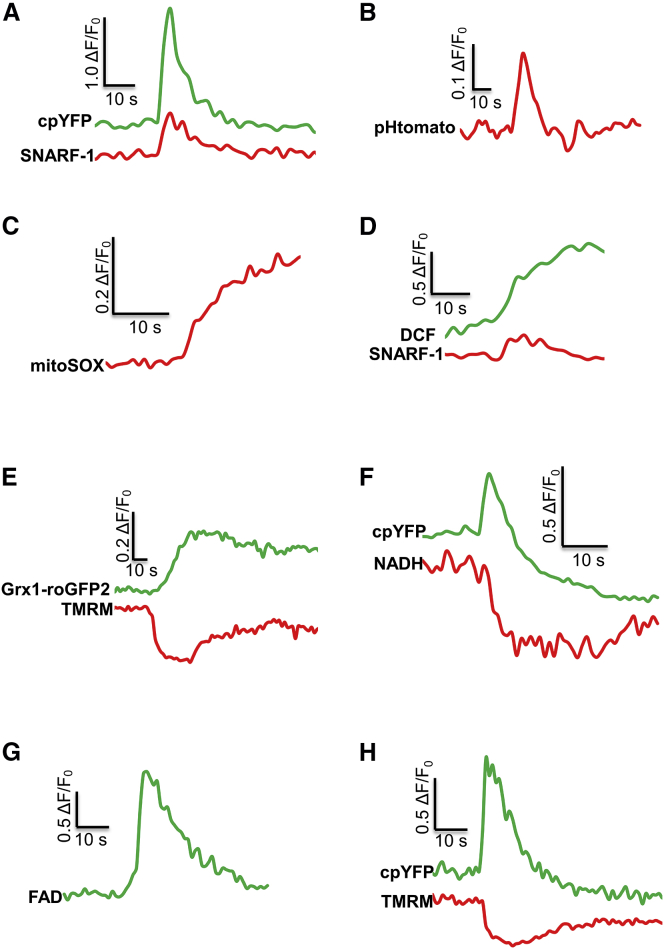
To further delineate the constituent signals of mitoflashes, we first performed multiparametric measurements of mitoflash activity in cardiomyocytes using a variety of genetically encoded or chemical fluorescent indicators. By imaging the mitochondria-targeted pH biosensor pHTomato (29) or the small-molecule dye SNARF-1, we found that the biosensor cpYFP-reported mitoflash signal (cpYFP flash) was matched by transient alkalinization of the mitochondrial matrix (Fig. 1, A and B), consistent with previous reports (2, 17, 30). The presence of a bursting production of mitochondrial ROS and an oxidative shift of the redox potential was demonstrated by using two chemical indicators and a genetically encoded, mitochondria-targeted probe. Specifically, ROS bursts were detected with mitoSOX (for superoxide) and 2, 7-dichlorodihydrofluorescein diacetate (DCF) (for total ROS) (4, 8, 30) (Fig. 1, C and D), and oxidative shifts of the redox potential were visualized with grx1-roGFP2 (31) (Fig. 1 E). Furthermore, label-free imaging of nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) autofluorescence revealed that a mitoflash also includes a transient oxidation of NADH (30) and FADH2, with the latter being manifested as an increase in FAD fluorescence (FAD-flash) (Fig. 1, F and G). Moreover, simultaneous measurements using tetramethylrhodamine methyl ester (TMRM) showed a rapid, partial loss of mitochondrial membrane potential (ΔΨm) followed by a gradual recovery (1) (Fig. 1, E and H), akin to an action potential in an excitable cell. Similar results were obtained in HeLa cells (Fig. S1). These findings confirm and extend previous reports and indicate that, in essence, a mitoflash reflects electrical and chemical excitation at the single-organelle level, with the latter comprising an ROS burst, a pH upstroke, and a redox shift toward oxidation.
Figure 1.
Multifaceted signals underlying mitoflashes. (A–H) Mitoflashes in adult cardiomyocytes were measured by multiple reporters, including mt-cpYFP (A, F, and H), SNARF-1 (A), pHTomato (B), mitoSOX (C), DCF (D), grx1-roGFP2 (E), and TMRM (E and H), as well as autofluorescence of NADH (F) and FAD (G). Increases in SNARF-1 and pHTomato fluorescence indicate alkalinization of the mitochondrial matrix. MitoSOX was used for the detection of superoxide production, and DCF was used to report changes in the total ROS level. The DCF signal was processed by subtracting the baseline increase. An increase in grx1-roGFP2 fluorescence reflects the shift of the mitochondrial redox potential toward oxidation, and a decrease in the TMRM signal reflects mitochondrial depolarization. NADH autofluorescence (420–470 nm) was measured at 720 nm two-photon excitation. FAD autofluorescence was measured between 500 and 650 nm at 488 nm excitation. An increase of FAD autofluorescence indicates a decrease of FADH2 content. All mitoflashes shown occurred spontaneously. To see this figure in color, go online.
It is noteworthy that the magnitude of mitoflashes reported by different indicators varied considerably, as did the mitoflash rise time (Fig. S2). This is not unexpected, because different indicators detect different signal components of the same events. Even for indicators that report the same modality (e.g., pHTomato and SNARF-1 for pH), the fold changes in fluorescence are also determined by the indicators’ affinity and dynamic range (e.g., Fmax/Fmin at extreme pH values). The large variability in rise times is mainly attributable to the indicators’ reversibility. DCF, mitoSOX, and grx1-roGFP2 are practically irreversible on the 10-s scale of a mitoflash, so their rise time reflects the entire duration of individual mitoflash events. Alternatively, it is also possible that different signal components of mitoflashes have differential kinetics (e.g., the pH signal appeared to peak sooner than other signals measured). Notably, we demonstrated a positive correlation between the amplitudes of cpYFP-reported mitoflashes and TMRM-reported ΔΨm oscillations (Fig. S3), indicating that these measured parameters are likely interlinked.
Triggering mitoflashes with ionophores
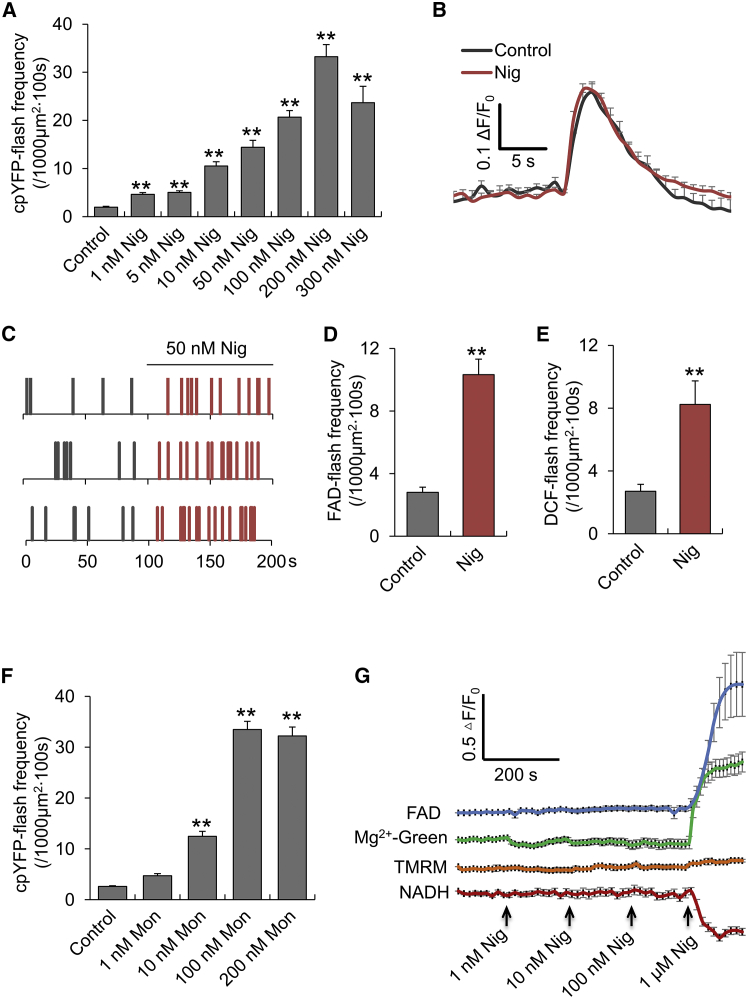
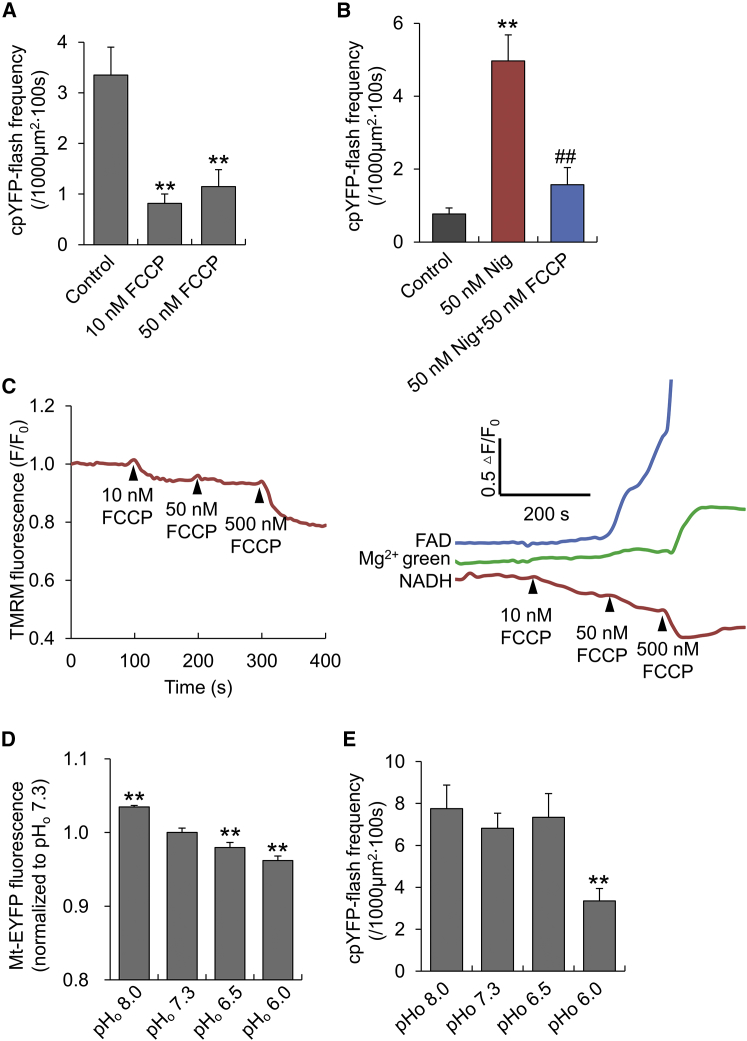
We hypothesized that mitoflash genesis and mitochondrial energy metabolism are functionally integrated, and as such, there might be a mechanistic link between mitochondrial proton signals and mitoflash ignition. To test this hypothesis, we used nigericin, an electroneutral H+/K+ antiporter, to induce proton influx. In cardiomyocytes expressing the biosensor cpYFP, nigericin (1–300 nM) potently stimulated mitoflash generation in a concentration-dependent manner without altering the properties of individual mitoflash events (Fig. 2, A and B). A time-course analysis showed that the mitoflash-stimulatory effect occurred immediately after nigericin application (Fig. 2 C). This concentration-dependent response is not specific to cardiomyocytes, because similar results were also found in HeLa cells expressing cpYFP (Fig. S4 A). Fig. 2, D and E, show that mitoflash activities, as reported by FAD autofluorescence or DCF, were all similarly increased by nigericin, indicating that spontaneous and nigericin-evoked mitoflashes have the same origin. Furthermore, we found that monensin, an electroneutral H+/Na+ antiporter, also increased mitoflash activity in a concentration-dependent manner (Fig. 2 F). This result indicates that the mitoflash-stimulatory effects of both nigericin and monensin are independent of their selectivity for the transported metal ions, but are dependent on the proton influx. In contrast, carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP), a proton ionophore, inhibited mitoflash activity at all concentrations tested (10–50 nM) (Fig. 3 A), and 50 nM FCCP mitigated the mitoflash activity induced by nigericin (Fig. 3 B).
Figure 2.
Mitoflashes induced by electroneutral ionophores. (A) Nigericin (Nig) concentration-dependently activated cpYFP-flashes in adult cardiomyocytes. n = 9–22 cells per group. ∗∗p < 0.01 versus control. (B) Averaged traces of cpYFP-flashes aligned by onset. Note that nigericin did not alter the properties of individual mitoflashes. n = 21 and n = 52 events from 10 cells for control and nigericin (Nig, 50–100 nM) groups, respectively. The mitoflashes in the control group occurred spontaneously. (C) Time course of the mitoflash response to nigericin in three representative cells. Individual mitoflash events are denoted by vertical ticks, and 50 nM nigericin was added at 100 s as indicated. (D and E) Nigericin (50 nM) stimulated mitoflashes reported with FAD (D) and DCF (E). n = 8–25 cells. ∗∗p < 0.01 versus control. (F) Concentration-dependent effect of monensin (Mon) on cpYFP-flash activity. n = 11–14 cells. ∗∗p < 0.01 versus control. (G) Concentration-dependent effects of nigericin on NADH, FAD, ΔΨm, and ATP in cardiomyocytes. Note that nigericin at 1–100 nM did not elicit discernible effects, but at 1 μM it caused a marked depletion of NADH, FADH2, and ATP. The NADH level was measured by detecting the autofluorescence (420–470 nm) at 720 nm two-photon excitation. The FADH2 level was indexed by monitoring FAD autofluorescence with excitation at 488 nm and emission at 500–650 nm. ΔΨm was measured using TMRM and the ATP content was measured with Mg2+-Green. An increase in Mg2+-Green fluorescence indicates a decrease in ATP content. n = 12–44 cells per trace. To see this figure in color, go online.
Figure 3.
Effects of FCCP and steady matrix pH on mitoflash activity. (A) Inhibition of mitoflash activity by FCCP. n = 9–10 cells per group. ∗∗p < 0.01 versus control. (B) FCCP reversed the nigericin (Nig)-induced mitoflash response. n = 6–8 cells. ∗∗p < 0.01 versus control; ##p < 0.01 versus Nig group. (C) Effects of FCCP on NADH, FADH2 (inversely related to FAD autofluorescence), ΔΨm (measured with TMRM), and ATP (measured with Mg2+-Green). n = 13–23 cells for each trace. Note that membrane depolarization and NADH oxidation started at 10 nM, followed by FADH2 oxidation at 50 nM and ATP depletion at 500 nM FCCP. (D) Mitochondrial pH changes in response to altered extracellular pH (pHo) in adult cardiomyocytes. Mitochondrial matrix pH was measured with mt-EYFP, and a decrease of its fluorescence indicates acidification of the matrix. n = 7 cells per group. ∗∗p < 0.01 versus pHo 7.3 group. (E) Effect of matrix pH changes (as shown in D) on mitoflash activity. n = 17–25 cells per group. ∗∗p < 0.01 versus pHo 7.3. To see this figure in color, go online.
Parallel measurements revealed that NADH and FADH2, measured by NADH and FAD autofluorescence, and cytosolic concentrations of ATP, indexed by Mg2+-Green fluorescence, remained unchanged at the concentrations of nigericin used to stimulate mitoflashes, but precipitously dropped at a higher concentration of nigericin (1 μM) (Fig. 2 G). As expected, FCCP decreased ΔΨm, oxidized NADH and FADH2, and caused significant ATP depletion (Fig. 3 C). Because mitochondrial energization is a prerequisite for mitoflash production (1), the mitoflash-inhibitory effect of FCCP is expected because the collapse of ΔΨm and de-energization directly undermine the mitochondrial electrical and chemical excitability. However, the mitoflash-stimulatory effect of electroneutral proton ionophores was surprising. Measurement with Rhod-2 and mitoSOX revealed no appreciable mitochondrial Ca2+ or basal ROS changes in response to 50 nM nigericin (Fig. S5), indicating that neither matrix Ca2+ nor ROS participate in ionophore induction of mitoflash activity. Thus far, the data strongly suggest that matrix protons trigger mitoflashes, provided that the mitochondria are energized and excitable.
Proton uncaging triggers mitoflashes
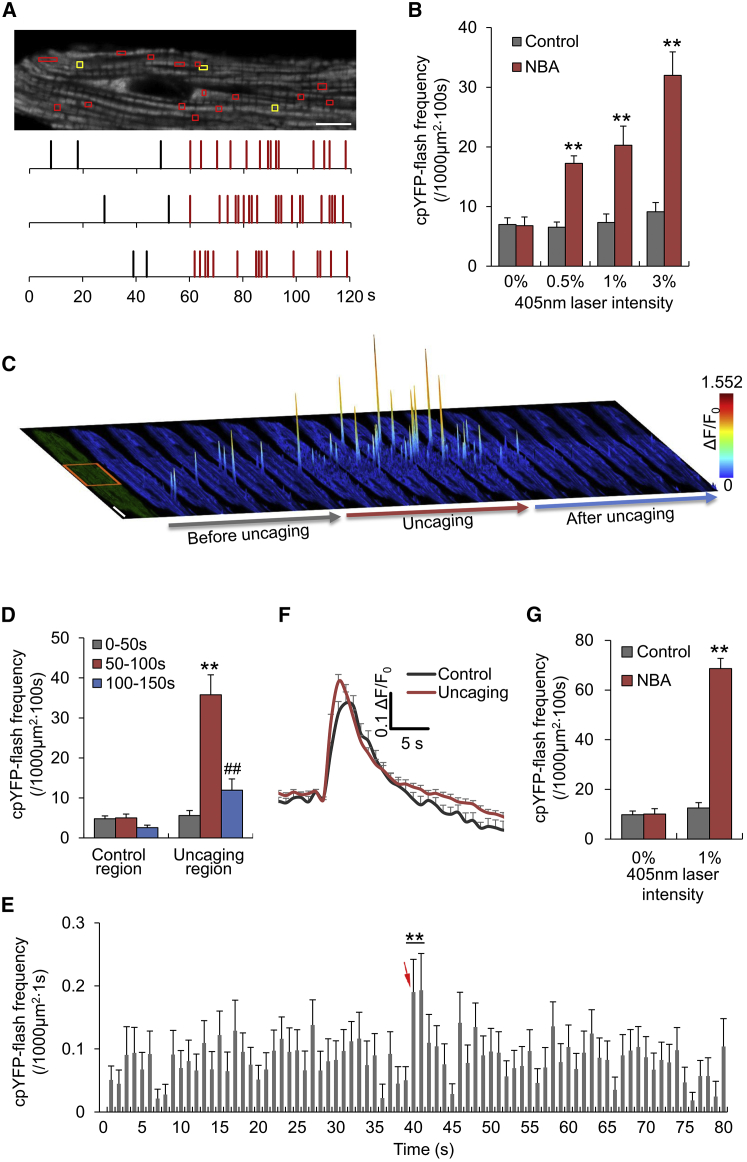
To directly test the hypothesis that protons trigger mitoflashes, we used proton uncaging with NBA, a membrane-permeable and photolabile proton donor (23). Photolytic illumination (405 nm laser at 0.5–3% of full power of 15 mW) was alternated with imaging illumination (488 nm laser) in the line-scan mode. Upon photolysis, the mitoflash frequency reported by mt-cpYFP in cardiomyocytes treated with 1 mM NBA rose abruptly (Fig. 4 A; Movie S1) in a laser-power-dependent manner (Fig. 4 B). The same illumination procedures, however, had no significant effect in the absence of NBA (Fig. 4 B), demonstrating that the increased mitoflash frequency was not due to phototoxicity. Similar results were obtained when mitoflashes were reported by pHTomato (Fig. S6). Likewise, proton uncaging greatly enhanced mitoflash activity in HeLa cells (Fig. S4 B). Despite the marked changes in the rate of occurrence, the evoked mitoflashes remained unaltered in terms of amplitude and duration (Fig. 4 F).
Figure 4.
Proton uncaging triggers mitoflashes. Adult cardiomyocytes expressing mt-cpYFP were loaded with 1 mM NBA, and proton uncaging at 405 nm illumination was alternated with imaging acquisition at 488 nm excitation (see Materials and Methods). (A) Mitoflash response to proton uncaging at 3% laser power. Top: intracellular mapping of mitoflash events before (yellow boxes, 0–60 s recording) and during (red boxes, 60–120 s recording) photolysis. Scale bar, 10 μm. Bottom: time course of mitoflash incidence (marked by vertical ticks) in three representative cells. The top row corresponds to the cell shown above. (B) Laser-power dependence of proton uncaging-induced mitoflashes. Cells without NBA treatment were used as controls. n = 5–15 cells per group. ∗∗p < 0.01 versus control. (C) Space-time confinement of mitoflashes responding to subcellular proton uncaging. Surface plots of mitoflashes registered in 10-s periods overlay the respective confocal images of the cell. A spike indicates a mitoflash event. The 405 nm laser (3% power) illumination was restricted to the central segment of the cell (red box) and applied between 50 and 100 s of the recording. Scale bar, 10 μm. (D) Statistics. n = 22 cells. ∗∗p < 0.01 versus control region; ##p < 0.01 versus photolysis. (E) Mitoflash response to short-duration NBA photolysis. The arrow denotes a 0.5-s 405-nm laser illumination at 3% intensity of full laser power (15 mW). Note that the increase in the rate of mitoflash occurrence was well confined within 2 s after proton uncaging. n = 62 cells. ∗∗p < 0.01 versus baseline or postphotolysis levels. (F) Averaged traces of cpYFP-flashes aligned by onset. n = 21 events from 11 cells for the control group; n = 52 events from 15 cells for the NBA photolysis group (uncaging). (G) Proton uncaging triggers mitoflashes in permeabilized adult cardiomyocytes bathed with 20 mM HEPES. n = 5–7 cells per group. ∗∗p < 0.01 versus control.
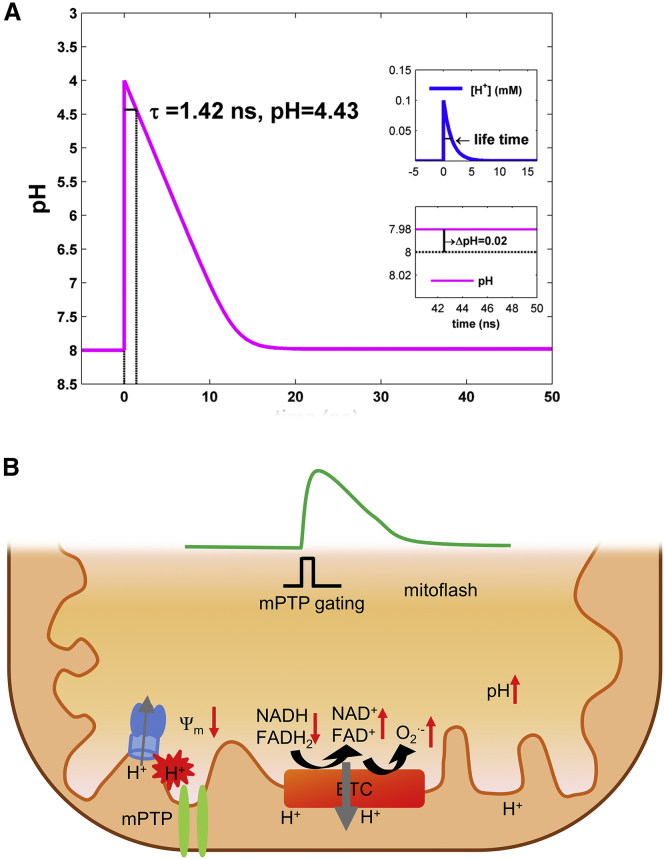
By controlling the space-time pattern of proton uncaging, we found that the increased mitoflash activity was sharply confined to subcellular regions of illumination. In the time domain, it quickly developed right at the onset of photolysis, but was readily reversible upon cessation of photolysis (Fig. 4, C and D). To better define the temporal localization of the mitoflash response, we applied a single 0.5-s photolysis protocol during image acquisition at 1 Hz. Upon photolysis, the rate of mitoflash occurrence abruptly increased by ∼1.4-fold in the first and second postphotolysis image frames, and then quickly returned to basal levels (Fig. 4 E), supporting the idea that the enhancement of mitoflash activity arises from a photolysis-elicited, short-lived surge of protons, or proton spike (Fig. 5 A).
Figure 5.
Simulation of a proton spike and schematic model for mitoflash genesis. (A) Matrix proton spike induced by proton uncaging. In this simulation, uncaging at time = 0 ns elicited a sudden drop in pH (from 8 to 4) or a surge of protons (upper inset) and a steady bulk matrix pH change of 0.02 unit (lower inset). The mean lifetime of the uncaged protons was 1.42 ns and the mean distance of diffusion was 2.06 nm. (B) A stochastic, flickering opening of the mPTP results in water and ionic fluxes to strain and depolarize the IMM. The mechanical strain causes dislocation of ETC molecules and disruption of the normal electron path, and thus greatly increases electron leakage to oxygen, resulting in a burst of superoxide formation. Meanwhile, depolarization accelerates electron transfer from the donor pool to ETC acceptors (e.g., from NADH to complex I, and from FADH2 to complex II) and shifts the redox potential toward oxidation. Accelerated ETC activity, driven dually by depolarization and oxygen deprivation of ETC electrons, stimulates coupled proton pumping across the IMM, giving rise to matrix alkalinization. Among others, nanodomain protons at the IMM may directly bind to a putative proton-binding site and thereby trigger transient opening of the mPTP to initiate a cascade of changes in a mitoflash. To see this figure in color, go online.
Next, we sought to discriminate possible actions of uncaged protons in the cytosolic and matrix compartments. Previous studies showed that mitoflash activity was preserved in cardiomyocytes after saponin permeabilization with mitochondrial respiration was supported by succinate, ADPm and Pi in the bathing solution (30). As shown in Fig. 4 G, photolysis elicited similar mitoflash responses in saponin-permeabilized cells in the presence of 20 mM HEPES as the pH buffer, indicating that the stimulation of mitoflashes by the uncaged protons occurred intramitochondrially (Fig. 4 G). Taken together, our results indicate that uncaged, short-lived protons trigger mitoflashes from the matrix of the mitochondria.
Mitoflash triggering is independent of steady matrix acidification
Notably, mitoflash triggering appeared to be independent of the bulk matrix pH changes produced by proton transport or proton uncaging. The mitoflash frequency responded abruptly to nigericin-mediated proton transport (Fig. 2 C), suggesting that the liberation of local high concentrations of protons at the inner surface of the IMM, which is rapidly established in the presence of ionophores, rather than gradual acidification of the bulk matrix, is the actual trigger. In the experimental setting of photolysis, we expect that a minuscule, steady decrease in matrix pH follows each proton spike, and bulk matrix acidification develops gradually during repetitive proton uncaging. We found that when the mitoflash response was fully developed, acidification was hardly measurable in either the cytosol (measured with SNARF-1) or the mitochondrial matrix (measured with mt-EYFP (32)) (Fig. S7). As photolysis continued, the matrix pH declined progressively, yet the frequency of mitoflashes remained steady at its plateau and was followed by a precipitous drop at the offset of photolysis when the pH decrease was maximal (Figs. 4, A–C, and S7). The sheer disparities in temporal kinetics indicate that mitoflash responses to electroneutral proton ionophores and NBA-mediated proton uncaging cannot be ascribed to bulk matrix pH acidification.
To directly appraise the effect of a steady matrix pH change, we manipulated the extracellular pH (pHo) to alter the mitochondrial pH. This maneuver failed to change the mitoflash frequency over a pHo range between 6.5 and 8, and mildly depressed it at pHo = 6.0 (Fig. 3, D and E). It is noteworthy that 50 nM nigericin induced a matrix pH change (measured by mt-pHTomato) similar to that elicited by pHo = 6.5 (Fig. S8). Proton uncaging at 3% laser power and 20 s after the onset of the photolysis protocol induced a matrix pH change comparable to that observed at pHo = 6.0 (Figs. 3 D and S7). These findings indicate that steady matrix acidification exerts a mild depressant effect on the genesis of mitoflashes, and provide compelling evidence that local and short-lived protons, rather than steady matrix acidification, act as an effective trigger of mitoflashes.
Discussion
To understand how mitoflashes are mechanistically and functionally integrated with mitochondrial biology, in this study we focused on the nature and physiological trigger of these discrete and quantal mitochondrial events. New lines of evidence indicate that mitoflashes reflect electrical and chemical excitation, manifested as action-potential-like membrane depolarization and sudden, dramatic, and transient changes in multifaceted and interrelated signals. In addition to bursting ROS production reported by mt-cpYFP, mitoSOX, and DCF, a transient redox shift toward oxidation was visualized with the use of grx1-roGFP2 in conjunction with label-free imaging of NADH and FAD autofluorescence, extending previous reports (1, 5, 8, 17, 30). The notion that a mitoflash comprises a pH component, i.e., transient matrix alkalinization, has been verified using both the chemical indicator SNARF-1 (30) and the genetically encoded probe pHTomato (29). In individual cardiomyocytes at any given moment, only a small fraction of mitochondria (0.1–5%) undergo stochastic electrochemical excitation (33). However, at the level of a single mitochondrion, a mitoflash engulfs the entire organelle, including all of its genomic, protein, and lipid contents. These results substantiate the emerging notion that mitoflashes play important signaling roles in eukaryotic cells.
Based on these and previous findings, we conceptualized a working model for mitoflash genesis in attempt to integrate diverse observations in a unifying framework (Fig. 5 B). As we proposed previously (33), a flickering opening of the mitochondrial permeability transition pore (mPTP) lets in water and ions, depolarizing and mechanically straining the mitochondrion. The decreased ΔΨm would facilitate proton pumping by the electron transport chain (ETC) complexes and accelerate electron transfer from the donors (NADH and FADH2) to the ETC acceptors (complexes I and II), oxidizing and alkalizing the matrix. Concomitantly, membrane strain may cause dislocation and disinsulation of the ETC at intermolecular junctures. As a result, more ETC electrons are captured by oxygen, giving rise to bursting production of superoxide and its ROS derivatives. Unlike the situation with persistent mPTP opening, transient proton influx through the flickering mPTP opening should be effectively buffered in the matrix and counteracted by the enhanced proton pumping mentioned above. This may explain the puzzling observation that despite the mPTP opening, only a low level of matrix acidosis developed. As to possible mechanisms of termination, the gradual decline of a mitoflash (reported by reversible indicators) would be collectively determined by the stochastic closure of the mPTP, repolarization of the IMM, depletion of the electron donor pool, and reinsulation of the ETC. Hence, the superoxide flash, oxidative redox shift, and matrix alkalinization simply reflect distinct but intertwined processes of one and the same event.
Our major finding, however, is that protons, which play a central role in energy metabolism, act as a direct trigger of mitoflashes, revealing a fundamental coupling of mitoflash genesis and bioenergetics. After matrix protons are taken up across the IMM or released from a photolabile donor, they immediately and reversibly increase the mitoflash frequency over a broad dynamic range. The fast kinetics of onset and offset, as demonstrated in the experimental settings with the space-time-patterned or single-pulse photolysis, provide strong support for the notion that short-lived uncaged or ionophore-liberated protons underlie the mitoflash-triggering effects. These features prompt us to speculate that in a respiring mitochondrion, a spike in protons in the matrix (or a local high-proton domain at the IMM) (Fig. 5 A) leads to protonation of putative proton-binding sites on mPTP or a closely related factor that initiates a cascade of changes in the form of a mitoflash. Once activated, individual mitoflashes display stereotypic properties that are independent of the strength and source of the proton trigger, which is characteristic of an excitable system (24). Since proton motive-force-centered bioenergetics is ubiquitous, we anticipate that this proton trigger mechanism may not be limited to the two cell types used in this study.
Another interesting finding is that protons trigger mitoflashes in a manner that is independent of bulk pH changes in the matrix. In the photolysis experiment, the full response of mitoflash triggering developed immediately upon photolytic illumination, before the matrix bulk pH displayed any appreciable changes. At the offset of proton uncaging, the mitoflash triggering response disappeared despite continued, steady changes in the matrix pH. By examining the effects of extracellular pH on mitochondrial pH and mitoflash genesis, we demonstrated that bulk matrix changes in pH have no or, in the case of acidification, even a depressant effect on mitoflash activity. The latter result is in general agreement with previous reports on acidification-mediated inhibition of classic mPTP gating (34, 35). To understand the disparity between steady pH changes and transient proton signals, we conducted a numerical analysis, which revealed that free protons are short-lived (lifetime ∼1.4 ns) and diffuse over a nanometer scale (∼2.1 nm) in the matrix environment (Fig. 5 A). Furthermore, free protons are extremely scarce in an alkaline mitochondrial matrix (∼0.4 protons in a mitochondrion 2 μm long and 200 nm in diameter at pH 8.0), and the matrix pH buffering capacity is estimated to be ∼5 mM per pH unit at pH 8.0 (i.e., at a 1:500,000 free/bound proton ratio) (28). As such, uncaging or ionophores would provide a dominant source of free protons for mitoflash triggering when present in the nanoscopic vicinity of the putative trigger sites.
In summary, we have shown that mitochondrial matrix protons act as a powerful trigger of mitoflashes, which are intimately interlinked with mitochondrial energization, ROS production, and pH oscillations. The proton trigger effect, as demonstrated in this study, is mediated by nanodomain, short-lived, free protons rather than changes in the matrix bulk pH. These findings not only unveil a novel, to our knowledge, proton signaling mechanism but also deepen our understanding of mitoflashes as elemental mitochondrial signaling events. Future investigations are warranted to determine the putative proton-sensing trigger site and define the specific functions of mitoflashes in proton-centered energy metabolism.
Author Contributions
H.C. and X.W. conceived and supervised the study, and wrote the manuscript. X.Z., Z.H., D.W., R.Z., R.Y., T.H., Y.Z., and C.J. performed mitoflash experiments and analyzed data. B.L. contributed conceptual insights through semiquantitative math modeling. F.G. made significant contributions to the experimental design and data interpretation. J.X. developed imaging processing programs and contributed to creating the figures. Y.W. prepared cardiomyocytes. All authors participated in data interpretation.
Acknowledgments
We thank Drs. P. Bernardi, V. Petronilli, S.S. Liu, W. Wang, M.Q. Dong, R.P. Xiao, Y.M. Wang, J.C. Luo, M. Ouyang, and C.Q. Song for valuable comments; C. Chen and T.P. Dick for grx1-roGFP2 plasmid; and I.C. Bruce and W.L. Yan for manuscript editing.
This work was supported by the National Key Basic Research Program of China (2013CB531200) and the National Science Foundation of China (31130067, 31470811, 31221002, 31327901, and 31500932).
Editor: Godfrey Smith.
Footnotes
Xianhua Wang, Xing Zhang, and Zhanglong Huang contributed equally to this work.
Supporting Materials and Methods, eight figures, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30459-3.
Contributor Information
Xianhua Wang, Email: xianhua@pku.edu.cn.
Heping Cheng, Email: chengp@pku.edu.cn.
Supporting Citations
Reference (36) appears in the Supporting Material.
Supporting Material
Arrows indicate the occurrence of mitoflashes. The basal fluorescence level is uniformly elevated upon 405 nm illumination due to the photoconversion effect of cpYFP. Scale bar, 10 μm.
References
- 1.Wang W., Fang H., Cheng H. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarzländer M., Logan D.C., Sweetlove L.J. Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell. 2012;24:1188–1201. doi: 10.1105/tpc.112.096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang H., Chen M., Cheng H. Imaging superoxide flash and metabolism-coupled mitochondrial permeability transition in living animals. Cell Res. 2011;21:1295–1304. doi: 10.1038/cr.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Huang Z., Wang X. Superoxide constitutes a major signal of mitochondrial superoxide flash. Life Sci. 2013;93:178–186. doi: 10.1016/j.lfs.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Wei L., Salahura G., Dirksen R.T. Mitochondrial superoxide flashes: metabolic biomarkers of skeletal muscle activity and disease. FASEB J. 2011;25:3068–3078. doi: 10.1096/fj.11-187252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen E.Z., Song C.Q., Dong M.Q. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature. 2014;508:128–132. doi: 10.1038/nature13012. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., Fang H., Cheng H. Imaging ROS signaling in cells and animals. J. Mol. Med. 2013;91:917–927. doi: 10.1007/s00109-013-1067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouvreau S. Superoxide flashes in mouse skeletal muscle are produced by discrete arrays of active mitochondria operating coherently. PLoS One. 2010;5:e13035. doi: 10.1371/journal.pone.0013035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou Y., Ouyang X., Cheng A. Mitochondrial superoxide production negatively regulates neural progenitor proliferation and cerebral cortical development. Stem Cells. 2012;30:2535–2547. doi: 10.1002/stem.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y., Mattson M.P., Cheng A. Permeability transition pore-mediated mitochondrial superoxide flashes regulate cortical neural progenitor differentiation. PLoS One. 2013;8:e76721. doi: 10.1371/journal.pone.0076721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou T., Zhang X., Cheng H. Synergistic triggering of superoxide flashes by mitochondrial Ca2+ uniport and basal reactive oxygen species elevation. J. Biol. Chem. 2013;288:4602–4612. doi: 10.1074/jbc.M112.398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Sun T., Xiong J.W. Remodeling of mitochondrial flashes in muscular development and dystrophy in zebrafish. PLoS One. 2015;10:e0132567. doi: 10.1371/journal.pone.0132567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Y., Fang H., Cheng H. Mitoflash altered by metabolic stress in insulin-resistant skeletal muscle. J. Mol. Med. 2015;93:1119–1130. doi: 10.1007/s00109-015-1278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Y., Ghosh P., Cheng A. Permeability transition pore-mediated mitochondrial superoxide flashes mediate an early inhibitory effect of amyloid beta1-42 on neural progenitor cell proliferation. Neurobiol. Aging. 2014;35:975–989. doi: 10.1016/j.neurobiolaging.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Zhang X., Cheng H. Proinflammatory cytokines stimulate mitochondrial superoxide flashes in articular chondrocytes in vitro and in situ. PLoS One. 2013;8:e66444. doi: 10.1371/journal.pone.0066444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santo-Domingo J., Giacomello M., Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 2013;32:1927–1940. doi: 10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breckwoldt M.O., Pfister F.M., Misgeld T. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat. Med. 2014;20:555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 18.Jian C., Hou T., Wang X. Regulation of superoxide flashes by transient and steady mitochondrial calcium elevations. Sci. China Life Sci. 2014;57:495–501. doi: 10.1007/s11427-014-4628-z. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W., Li K., Cheng H. Subsarcolemmal mitochondrial flashes induced by hypochlorite stimulation in cardiac myocytes. Free Radic. Res. 2014;48:1085–1094. doi: 10.3109/10715762.2014.932114. [DOI] [PubMed] [Google Scholar]
- 20.Nicholls D.G., Ferguson S.L. Academic Press; New York: 2002. Bioenergetics. [Google Scholar]
- 21.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 22.Cheng H., Lederer W.J., Cannell M.B. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 23.Swietach P., Spitzer K.W., Vaughan-Jones R.D. pH-Dependence of extrinsic and intrinsic H(+)-ion mobility in the rat ventricular myocyte, investigated using flash photolysis of a caged-H(+) compound. Biophys. J. 2007;92:641–653. doi: 10.1529/biophysj.106.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K., Zhang W., Cheng H. Superoxide flashes reveal novel properties of mitochondrial reactive oxygen species excitability in cardiomyocytes. Biophys. J. 2012;102:1011–1021. doi: 10.1016/j.bpj.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bazil J.N., Beard D.A., Vinnakota K.C. Catalytic coupling of oxidative phosphorylation, ATP demand, and reactive oxygen species generation. Biophys. J. 2016;110:962–971. doi: 10.1016/j.bpj.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgievskii Y., Medvedev E.S., Stuchebrukhov A.A. Proton transport via the membrane surface. Biophys. J. 2002;82:2833–2846. doi: 10.1016/S0006-3495(02)75626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zifarelli G., Soliani P., Pusch M. Buffered diffusion around a spherical proton pumping cell: a theoretical analysis. Biophys. J. 2008;94:53–62. doi: 10.1529/biophysj.107.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Tsien R.W. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 2012;15:1047–1053. doi: 10.1038/nn.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei-LaPierre L., Gong G., Wang W. Respective contribution of mitochondrial superoxide and pH to mitochondria-targeted circularly permuted yellow fluorescent protein (mt-cpYFP) flash activity. J. Biol. Chem. 2013;288:10567–10577. doi: 10.1074/jbc.M113.455709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutscher M., Pauleau A.L., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi A., Zhang Y., Herman B. Measurement of mitochondrial pH in situ. Biotechniques. 2001;30:804–808. doi: 10.2144/01304rv01. 810, 812 passim. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Jian C., Cheng H. Superoxide flashes: elemental events of mitochondrial ROS signaling in the heart. J. Mol. Cell. Cardiol. 2012;52:940–948. doi: 10.1016/j.yjmcc.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Bernardi P., Vassanelli S., Zoratti M. Modulation of the mitochondrial permeability transition pore. Effect of protons and divalent cations. J. Biol. Chem. 1992;267:2934–2939. [PubMed] [Google Scholar]
- 35.Szabó I., Bernardi P., Zoratti M. Modulation of the mitochondrial megachannel by divalent cations and protons. J. Biol. Chem. 1992;267:2940–2946. [PubMed] [Google Scholar]
- 36.Lukyanenko V., Gyorke S. Ca2+ sparks and Ca2+ waves in saponin-permeabilized rat ventricular myocytes. J. Physiol. 1999;521:575–585. doi: 10.1111/j.1469-7793.1999.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arrows indicate the occurrence of mitoflashes. The basal fluorescence level is uniformly elevated upon 405 nm illumination due to the photoconversion effect of cpYFP. Scale bar, 10 μm.