ABSTRACT
Subseafloor sediment hosts a large, taxonomically rich, and metabolically diverse microbial ecosystem. However, the factors that control microbial diversity in subseafloor sediment have rarely been explored. Here, we show that bacterial richness varies with organic degradation rate and sediment age. At three open-ocean sites (in the Bering Sea and equatorial Pacific) and one continental margin site (Indian Ocean), richness decreases exponentially with increasing sediment depth. The rate of decrease in richness with increasing depth varies from site to site. The vertical succession of predominant terminal electron acceptors correlates with abundance-weighted community composition but does not drive the vertical decrease in richness. Vertical patterns of richness at the open-ocean sites closely match organic degradation rates; both properties are highest near the seafloor and decline together as sediment depth increases. This relationship suggests that (i) total catabolic activity and/or electron donor diversity exerts a primary influence on bacterial richness in marine sediment and (ii) many bacterial taxa that are poorly adapted for subseafloor sedimentary conditions are degraded in the geologically young sediment, where respiration rates are high. Richness consistently takes a few hundred thousand years to decline from near-seafloor values to much lower values in deep anoxic subseafloor sediment, regardless of sedimentation rate, predominant terminal electron acceptor, or oceanographic context.
IMPORTANCE Subseafloor sediment provides a wonderful opportunity to investigate the drivers of microbial diversity in communities that may have been isolated for millions of years. Our paper shows the impact of in situ conditions on bacterial community structure in subseafloor sediment. Specifically, it shows that bacterial richness in subseafloor sediment declines exponentially with sediment age, and in parallel with organic-fueled oxidation rate. This result suggests that subseafloor diversity ultimately depends on electron donor diversity and/or total community respiration. This work studied how and why biological richness changes over time in the extraordinary ecosystem of subseafloor sediment.
INTRODUCTION
Subseafloor sediment contains a diverse microbial ecosystem (1–3), with a total cell abundance comparable to that in terrestrial soil and in the world ocean (4). Subseafloor sedimentary communities push the boundaries of life as we know it; per-cell rates of respiration are often orders of magnitude lower than those in the surface world (5, 6), biomass turnover can take hundreds to thousands of years (7, 8), cell abundance can be as low as 10 cells per cm3 (9), and microbes in deep subseafloor sediment may be isolated from the surface world for millions of years (Ma) to tens of Ma. Subseafloor sediment, therefore, provides an unprecedented opportunity to investigate drivers of microbial diversity on a time scale of thousands to millions of years.
In the broadest context, distributions of microbial diversity result from combined effects of speciation, selection, dispersal, and ecological drift (10, 11). However, subseafloor conditions may severely impact the relative influence of these processes. For example, exceedingly low per-cell energy fluxes may place very high selection pressure on subseafloor populations, severely limit active dispersal (6) and cell abundance, and cause mean generation times to greatly exceed the already-long few-hundred-year to few-thousand-year time scale of biomass turnover (7) in subseafloor sediment (12). Generation times of hundreds to millions of years may in turn greatly lower the rates of speciation.
To document microbial diversity and its potential drivers in subseafloor sediment, we extracted and sequenced PCR amplicons for the V4 to V6 hypervariable region of the bacterial 16S rRNA gene from the sediment of four distinct locations: the Bering Sea (Integrated Ocean Drilling Program [IODP] expedition 323 site U1343) (13), the eastern equatorial Pacific (Knorr expedition 195-3 site EQP1), the central equatorial Pacific Ocean (Knorr 195-3 site EQP8), and the Bay of Bengal continental margin (Indian National Gas Hydrate Program [NGHP] site NGHP-1-14) (14) (Fig. 1).
FIG 1.
Sampling locations. (Map created with Generic Mapping Tools.)
MATERIALS AND METHODS
Sites.
The three open-ocean sites (Bering Sea site U1343 and Equatorial Pacific sites EQP1 and EQP8) have water depths of 1,953, 2,885, and 4,336 m below sea level (mbsl), respectively (see the supplemental material). The water depth at the Bay of Bengal continental margin site NGHP-1-14 is 895 mbsl (14). The Bering Sea and Bay of Bengal sites are characterized by high sea surface chlorophyll concentrations and very high sedimentation rates (0.34 and 1.04 mg/m3 and 250 m/Ma, and ca. 100 m/Ma at U1343 and NGHP-1-14, respectively). The equatorial Pacific sites are characterized by moderate sea surface chlorophyll concentrations and moderate mean sedimentation rates (0.16 and 0.32 mg/m3, and 75 and 4.8 m/Ma at EQP1 and EQP8, respectively). The total organic carbon (TOC) content of near-surface sediment is highest at the high sedimentation sites (0.6% and 1.7% at NGHP-1-14 and U1343, respectively) and lowest at the moderate sedimentation sites (0.1 and 0.02% at EQP1 and EQP8, respectively). The maximum sampled sediment depths range from 27 m below seafloor (mbsf) at EQP8 to 404 mbsf at U1343 (see Table S1 in the supplemental material). The concentration profiles of the dissolved metabolic products and substrates (dissolved inorganic carbon [DIC], methane, ammonium, oxygen, nitrate, and sulfate) indicate that microbial activity occurs throughout the sampled sequences (15).
Shipboard sampling and geochemistry.
Immediately after core recovery, we cleaned the cut face of the remaining core section with a sterile blade. For DNA analysis of NGHP-1-14, we cut 10-cm whole-core rounds from the core sections. For EQP1, EQP8, and U1343, we took samples for DNA analysis from the center of the cleaned core face using sterile 60-cm3 cutoff syringes. We froze the samples at −80°C for shore-based DNA analysis. Concentrations of DIC, sulfate, and, for site U1343, methane were measured during the expeditions, according to standard procedures (2, 13, 14, 16). We made TOC measurements, as previously described (17), on a Costech elemental analyzer. All geochemical and environmental data for site U1343 are deposited in the IODP database and accessible online in the IODP expedition 323 proceedings (13), except for the postcruise TOC (see the supplemental material). All dissolved geochemical measurements for sites EQP1, EQP8, and NGHP-1-14 are deposited at EarthChem (www.earthchem.org). The cell count data for EQP1, EQP8, and U1343 are available in a study by Kallmeyer et al. (4). The chlorophyll data are from a study by Gregg (18). We used the method of Sauvage et al. (19) to account for changes in DIC and alkalinity that resulted from the precipitation of carbonate during the recovery and processing of core samples from EQP1, EQP8, and U1343 (see the supplemental material).
Pyrosequencing, clustering, and diversity analyses.
We extracted DNA from the sediment samples using commercial kits (MoBio PowerSoil). We amplified the V4 to V6 hypervariable region of the 16S rRNA gene using the bacterial primer pair 518f-1064r and pyrosequenced the amplicons according to standard protocols on a 454 GS-FLX sequencer at the Josephine Bay Paul Center, Marine Biological Laboratory, Woods Hole, MA (20). To reduce error, we removed low-quality sequences (such as those with low average quality scores or deviations in read length) prior to analysis, as described in Huse et al. (21). Sequencing protocols, analyses, and initial results are accessible at the VAMPS website (https://vamps.mbl.edu/index.php) under the projects DCO_WAL_Bv6v4, KCK_EQP_Bv6v4, and JBK_IO_Bv6v4. For further analysis, we determined the taxonomy of each sample at the genus level using the SILVA database (22) on the VAMPS website. From EQP1 samples, we removed all sequences that correspond to Vibrio, because it was actively cultured in the laboratory used for EQP1 DNA extraction. No Vibrio DNA occurs in samples from the other sites, which were extracted in a different dedicated laboratory.
We used QIIME (23), as made available on N3phele (24), to cluster each sample into operational taxonomic units (OTUs) at the 3% similarity-level. To remove the effects of sampling intensity (number of reads) on downhole or intersite comparisons of richness estimates (25), we first randomly subsampled the number of reads in each sample to the lowest number found in any sample (n = 2,800). OTUs were picked from subsampled sequences using Uclust (26) with the furthest neighbor approach. Representative sequences for each OTU were assigned RDP taxonomy (27), aligned with PyNAST (28), and a distance matrix was calculated using UniFrac (29). Clustering average-neighbor OTUs with mothur's MiSeq standard operating procedures (SOP) (30, 31) identified more OTUs than with QIIME but exhibited similar trends of richness with sediment depth and age. To investigate patterns of diversity with changes in depth, we also clustered samples at multiple levels of similarity to generate comparisons between different similarity cutoff levels (see the supplemental material).
We used the distance matrix and OTU tables created with QIIME for statistical analyses of diversity. We calculated the Chao1 index (25) using QIIME. We compared these results to richness metrics calculated with CatchAll (32) (see the supplemental material). We performed Bray-Curtis similarity analyses, nonmetric multidimensional scaling, and Spearman rank correlation tests using the Primer 6 program (33).
Sediment age calculations.
We calculated sediment age estimates for U1343 using the sediment age model of Takahashi et al. (13). The U1343 age model is based on biostratigraphic and magnetostratigraphic data (13). No detailed chronostratigraphic data are available for EQP1 or EQP8; consequently, we estimated their sediment ages from their average sedimentation rates (sediment thickness [50] divided by basement age [41]). Because no published chronostratigraphic data are available for NGHP-1-14, our age model for that site is based on the biostratigraphically determined sedimentation rates of other NGHP sites in the same basin (ca. 110 m/Ma and ca. 125 m/Ma at NGHP-1-16 and NGHP-1-10, respectively [42]). The shallowest NGHP-1-14 samples may be younger than our age estimates, since relatively shallow vertical variation in its dissolved chemical profiles may have resulted from the deposition of ca. 13 m of sediment by a mass transport event about 1,400 years ago (34).
Reaction rate calculations.
To quantify the net rates of organic-fueled respiration from dissolved chemical data of EQP1, EQP8, and U1343, we used a modified version of the Matlab-based numerical procedures of Wang et al. (35). Similar calculations are not possible for continental margin site NGHP-1-14, because its dissolved chemical concentration profiles are not in diffusive steady state. We modified the approach of Wang et al. (35) by using an Akima spline, instead of a 5-point running mean, in order to generate a best-fit line to the chemical concentration data. We determined standard deviations through use of a Monte Carlo simulation (n = 30). For EQP1, EQP8, and U1343, we calculated organic-fueled respiration from DIC concentration profiles after first correcting DIC and alkalinity concentrations to account for carbonate precipitation during sediment recovery, processing, and storage (19). For U1343, we also calculated net organic-fueled respiration from the ammonium concentration profile to independently check the organic-fueled respiration rates calculated from DIC concentrations.
RESULTS
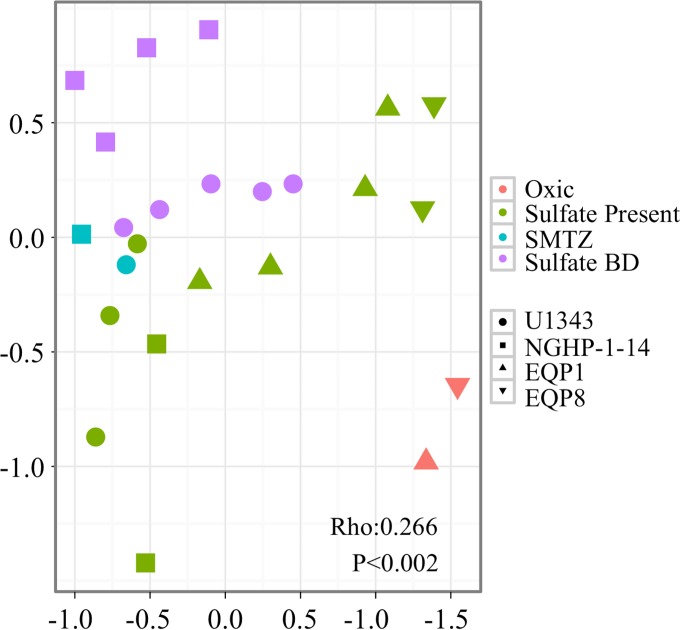
Abundance-weighted community composition broadly varies with the vertical succession of chemical redox zones (36, 37), with Bray-Curtis similarity scores exhibiting a clear gradient through (i) the oxygenated zone immediately beneath the seafloor, (ii) a deeper anoxic zone with abundant dissolved sulfate, (iii) a still-deeper sulfate/methane transition zone (SMTZ), and (iv) the deepest zone, with little or no sulfate but abundant dissolved methane (Fig. 2). This result expands on earlier discoveries that dominant microbial taxa in the SMTZ or in subseafloor hydrates differ from the dominant taxa in overlying or underlying sediment (38–40).
FIG 2.
Nonmetric multidimensional scaling (nMDS) plot. Bray-Curtis distances between samples represent the degree of community similarity (samples that contain similar communities are closer together in ordination space [stress = 0.01]). Symbol color indicates redox zone, and symbol shape indicates site location as shown in Fig. 1.
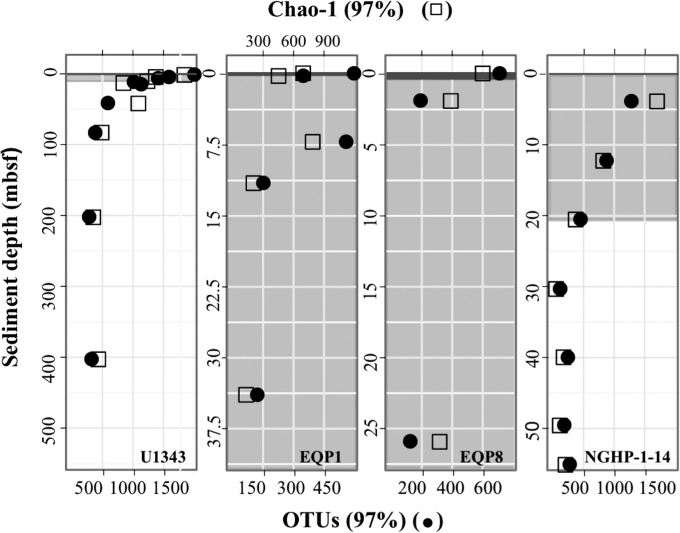
In contrast to abundance-weighted community composition, total bacterial diversity does not demonstrate a vertical succession of redox zones. Bacterial taxonomic richness, as measured by both Chao1 estimates and numbers of operational taxonomic units (OTUs) in samples normalized to equal numbers of reads, is highest near the seafloor and drops exponentially with increasing sediment depth at all four sites (Fig. 3). The trend is the same for parametric analyses (CatchAll [32]) (see the supplemental material). The rate of decrease in richness with depth varies greatly from site to site; OTU richness approaches low relatively stable values several tens of meters below the seafloor at site U1343 but within a couple of meters below seafloor at site EQP8. This exponential decline in richness occurs at every phylogenetic level: whether defined by similarity as high as 100% or as low as 85%, the number of operational taxonomic units in normalized samples declines with increasing sediment depth (see the supplemental material).
FIG 3.
Comparison of OTU richness to redox zones. Filled circles identify numbers of OTUs. Open squares show Chao1 richness values. Dark gray bar indicates oxygen penetration depth, light gray grid indicates the presence of sulfate, and white background indicates sulfate is below detection levels.
The rate of decrease in OTU richness is not clearly associated with any particular geochemical zone or transition between zones. The inflection from rapidly declining richness to relatively stable richness occurs within sulfate-replete sediment at EQP1 and EQP8, at or near the SMTZ at NGHP-1-14, and within the methane-rich sulfate-poor zone at site U1343.
The bacterial richness of our shallowest samples varied considerably from site to site, with richness of 97%-similar OTUs ranging from 1,951 at U1343 to 572 at EQP1 (Fig. 3). This variation is not surprising, because there are large environmental differences between sites (they underlie different oceanographic regimes and sample very different kinds of sediment), and because the shallowest samples differ greatly in sediment age (ranging from a few hundred years at U1343 to ∼10,000 years at EQP8). At greater depths, OTU richness at all sites decreases to values in a similar range (150 to 300 OTUs).
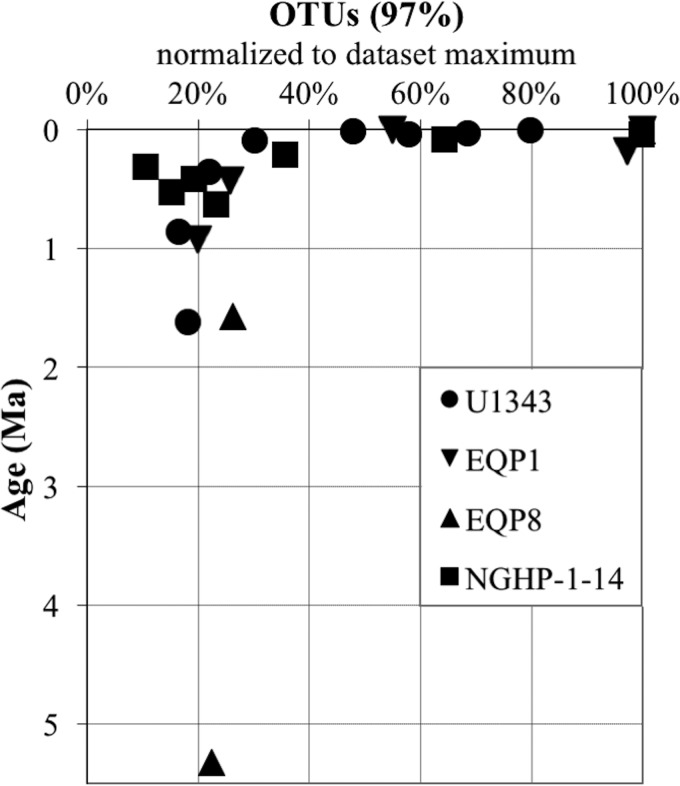
The transformation of sediment depth to sediment age shows that OTU richness decreases exponentially with age at all four sites (Fig. 4). It decreases rapidly in the youngest sediment and then stabilizes or decreases more slowly with greater age. Despite the large site-to-site differences in sedimentation rate, oceanographic context, and predominant electron acceptor regimes, OTU richness consistently takes a few hundred thousand years to decline from near-seafloor sediment levels to much lower values in deep-subseafloor sediment.
FIG 4.
OTU richness relative to sediment age. To normalize OTU counts between sites, we set the most OTU-rich sample for each site (always the shallowest sample, in the upper right corner) to 100% and calculated the richness of deeper samples calculated as percentages of that number. Symbol shape indicates site location.
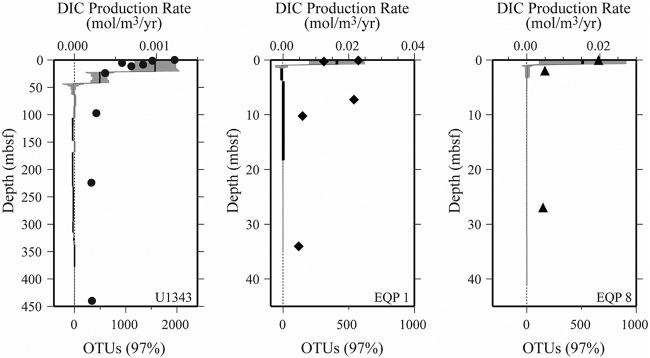
Vertical patterns of richness covary with DIC production rates at open-ocean sites U1343, EQP1, and EQP8 (Fig. 5). At EQP1 and EQP8, DIC production neatly represents gross organic-fueled respiration; the cored sequence is rich in external electron acceptors (nitrate and sulfate) and there is no evidence of major DIC sinks in the cored sediment (e.g., dissolved calcium or magnesium sinks indicating carbonate precipitation). The situation is slightly more complicated at U1343, where (i) the DIC profile is slightly modified by net DIC consumption (carbonate precipitation) in some intervals at depths greater than 100 mbsf (15) (Fig. 4) and (ii) external electron acceptors are scarce below the sulfate-methane interface at ∼8 mbsf (13). A comparison of the U1343 DIC production rates to net ammonium production rates demonstrates that these modifications are of secondary importance, because the calculated profile of DIC production broadly matches the vertical profile of organic-fueled degradation estimated from ammonium production (see the supplemental material).
FIG 5.
Relationship of OTU richness to organic respiration rate (as indicated by DIC production) at the open-ocean sites (U1343, EQP1, and EQP8). Black symbols indicate numbers of OTUs. Black lines and gray bars indicate reaction rates and two times the standard deviation, respectively. The data are plotted against sediment depth (mbsf) for each site.
At all three sites (U1343, EQP1, and EQP8), the rate of organic degradation indicated by net production of DIC and ammonium is highest near the seafloor, where OTU richness is also highest (Fig. 5). At U1343, organic degradation, like OTU richness, takes tens of meters to decline to extremely low values. At EQP1 and EQP8, the large declines in both organic degradation and OTU richness occur within the first few meters below the seafloor.
DISCUSSION
The relationships between abundance-weighted community composition and redox zones (Fig. 2) indicate that some taxa are influenced by the predominant terminal electron-accepting activity. In contrast, the lack of clear correspondence between bacterial richness and redox zonation suggests that the predominant terminal electron-accepting pathway does not exert primary control on OTU richness of subseafloor bacterial communities. Possible explanations of this lack of relationship between OTU richness and predominant terminal electron acceptors include the following: (i) most OTUs may represent taxa that are not involved in terminal electron acceptance and that operate similarly in successive redox zones (for example, fermentative taxa may be active in all of the anoxic zones), (ii) taxa directly involved in terminal electron acceptance may be capable of processing multiple kinds of electron acceptors, and (iii) terminal electron acceptance may not be limited to the predominant pathway, with terminal electron-accepting activity predominant in one redox zone also existing in other zones (for example, iron reducers may be present and active where sulfate reduction, methanogenesis, or sulfide oxidation predominate [51, 52, 53]).
The exponential decline in bacterial richness from seafloor to greater sediment depth is consistent with recent comparisons of bacterial OTUs in the ocean to OTUs in marine sediment (43). Based on the relative abundance of 16S V6 tags in the water column, shallow sediment (0 to 0.1 mbsf), and subseafloor sediment, these studies show that (i) marine sedimentary bacteria are dispersed via the ocean, and (ii) subseafloor sedimentary lineages are selected from the community present in shallow sediment (43).
The close match between the exponential decline in bacterial richness and the depth distribution of organic degradation rates at our open-ocean sites indicates that vertical variation in richness is closely tied to organic-fueled community activity. The pattern of organic degradation exponentially declining from seafloor to greater sediment depth was first observed decades ago. It is often explained with a “multi-G model,” in which organic matter is assumed to be composed of diverse organic compounds with different levels of reactivity (44). In such models, the most labile or biologically reactive organic substrates are respired at much higher rates than the least-labile substrates, leading the net rates of organic-fueled respiration to decrease exponentially with increasing sediment depth (44, 45).
A single EQP1 sample at 7.21 mbsf constitutes the only exception to the close correspondence between exponentially declining richness and exponentially declining organic degradation at these sites. This relatively OTU-rich sample contains an unusually high concentration of organic matter relative to adjacent sediment; its exceptionality is consistent with previous research that showed that discrete horizons of organic-rich sediment may sustain locally high respiration for millions of years (46).
Near continental shelves (such as the Indian Margin) and in upwelling regions (such as the Bering Sea and the equatorial Pacific), organic matter is the primary electron donor for subseafloor sedimentary communities (2). Consequently, the close correspondence between vertical patterns of richness and vertical patterns of organic degradation suggests that selection for organismal properties related to either total catabolic activity or electron donor diversity exerts the primary influence on bacterial OTU richness in anoxic subseafloor sediment. This correspondence also indicates that many bacterial taxa that are poorly adapted for subseafloor sedimentary conditions are degraded in the geologically young sediment where respiration rates are high.
This result sets a clear boundary for understanding bacterial OTU richness in anoxic subseafloor sediment. It also provides a potential basis for ultimately integrating OTU richness with other key properties that appear to be broadly related to total catabolic activity in subseafloor sedimentary communities, such as cell (4) or viral particle abundance (47) and activity (48). However, the exact traits that preferentially aid survival as catabolic activity and/or electron donor diversity decline remain to be determined; candidate traits include specialization to metabolize recalcitrant organic substrates, specific energy-conserving properties, such as membrane permeability (6), use of sodium ions for energy storage (6), spore formation (6), prophage modulation of metabolic activity (49), and/or a wide range of other properties (12).
Supplementary Material
ACKNOWLEDGMENTS
This research would not have been possible without the dedicated effort of the crews and scientific staff of the DV JOIDES Resolution and the RV Knorr. Samples were provided by Indian National Gas Hydrate Program expedition 01, expedition Knorr 195-3, and Integrated Ocean Drilling Program expedition 323. We especially thank Heather Schrum, Nils Risgaard-Petersen, and Laura Wehrmann for shipboard help on expedition 323 and Ann G. Dunlea for sharing ancillary geochemical measurements.
For postexpedition funding, we thank the U.S. National Science Foundation Biological Oceanography Program (grant NSF-OCE-0752336, and through the Center for Dark Energy Biosphere Investigations [C-DEBI], grant NSF-OCE-0939564), the U.S. Science Support Program associated with the U.S. IODP, and the Sloan Foundation (through the Deep Carbon Observatory Census of Deep Life). Part of this study is based on work supported while R. W. Murray was serving at the National Science Foundation.
Funding Statement
The work of E. A. Walsh, J. B. Kirkpatrick, R. Pockalny, and J. Sauvage was funded by the grants to S. D'Hondt.
Footnotes
This article is C-DEBI contribution 328.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00809-16.
REFERENCES
- 1.Inagaki F, Nunoura T, Nakagawa S, Teske AP, Lever MA, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, Nealson KH, Horikoshi K, D'Hondt S, Jørgensen BB. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci U S A 103:2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA, Cypionka H, Dickens GR, Ferdelman T, Hinrichs K-U, Holm NG, Mitterer R, Spivack AJ, Wang G, Bekins B, Engelen B, Ford K, Gettemy G, Rutherford SD, Sass H, Skilbeck CG, Aiello IW, Guèrin G, House CH, Inagaki F, Meister P, Naehr T, Niitsuma S, Parkes RJ, Schippers A, Smith DC, Teske AP, Wiegel J, Padilla CN, Acosta JLS. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216–2221. doi: 10.1126/science.1101155. [DOI] [PubMed] [Google Scholar]
- 3.Orsi WD, Edgcomb VP, Christman GD, Biddle JF. 2013. Gene expression in the deep biosphere. Nature 499:205–208. doi: 10.1038/nature12230. [DOI] [PubMed] [Google Scholar]
- 4.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Hondt S, Rutherford SD, Spivack AJ. 2002. Metabolic activity of subsurface life in deep-sea sediments. Science 295:2067–2070. doi: 10.1126/science.1064878. [DOI] [PubMed] [Google Scholar]
- 6.Hoehler TM, Jørgensen BB. 2013. Microbial life under extreme energy limitation. Nat Rev Microbiol 11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 7.Lomstein BA, Langerhuus AT, D'Hondt S, Jørgensen BB, Spivack AJ. 2012. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484:101–104. doi: 10.1038/nature10905. [DOI] [PubMed] [Google Scholar]
- 8.Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson RE, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske AP, House CH, Hinrichs K-U. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A 103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inagaki F, Hinrichs K-U, Kubo Y, Bowles MW, Heuer VB, Hong W-L, Hoshino T, Ijiri A, Imachi H, Ito M, Kaneko M, Lever MA, Lin Y-S, Methé BA, Morita S, Morono Y, Tanikawa W, Bihan M, Bowden SA, Elvert M, Glombitza C, Gross D, Harrington GJ, Hori T, Li K, Limmer D, Liu C-H, Murayama M, Ohkouchi N, Ono S, Park Y-S, Phillips SC, Prieto-Mollar X, Purkey M, Riedinger N, Sanada Y, Sauvage J, Snyder G, Susilawati R, Takano Y, Tasumi E, Terada T, Tomaru H, Trembath-Reichert E, Wang DT, Yamada Y. 2015. Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science 349:420–424. doi: 10.1126/science.aaa6882. [DOI] [PubMed] [Google Scholar]
- 10.Hamdan LJ, Coffin RB, Sikaroodi M, Greinert J, Treude T, Gillevet PM. 2013. Ocean currents shape the microbiome of Arctic marine sediments. ISME J 7:685–696. doi: 10.1038/ismej.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 12.D'Hondt S, Spivack AJ, Wang G. 2014. The underground economy (energetic constrains of subseafloor life), p 127–148. In Stein R, Blackman D, Inagaki F, Larsen H-C (ed), Earth and life processes discovered from subseafloor environments: a decade of science achieved by the Integrated Ocean Drilling Program (IODP). Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 13.Takahashi K, Ravelo AC, Alvarev Zarikian CA, Scientists E. 2011. Proceedings of the Integrated Ocean Drilling Program. Integrated Ocean Drilling Program Management International, Inc, Tokyo, Japan. [Google Scholar]
- 14.Collett TS, Riedel M, Cochran JR, Boswell R, Presley J, Kumar P, Sathe AV, Sethi A, Lall M, NGHP Expedition Scientists. 2015. Indian National Gas Hydrate Program Expedition 01 report. Scientific investigations report 2012-5054. US Geological Survey, Reston, VA. http://pubs.usgs.gov/sir/2012/5054/.
- 15.Wehrmann LM, Risgaard-Petersen N, Schrum HN, Walsh EA, Huh Y, Ikehara M, Pierre C, D'Hondt S, Ferdelman TG, Ravelo AC, Takahashi K, Zarikian CA, The Integrated Ocean Drilling Program Expedition 323 Scientific Party. 2011. Coupled organic and inorganic carbon cycling in the deep subseafloor sediment of the northeastern Bering Sea Slope (IODP exp. 323). Chem Geol 284:251–261. doi: 10.1016/j.chemgeo.2011.03.002. [DOI] [Google Scholar]
- 16.Gieskes JM, Gamo T, Brumsack H. 1991. Chemical methods for interstitial water analysis aboard JOIDES Resolution. Technical note 15. Ocean Drilling Program, Texas A&M University, College Station, TX: http://www-odp.tamu.edu/Publications/tnotes/tn15/technote15.pdf. [Google Scholar]
- 17.Verardo DJ, Froelich PN, McIntyre A. 1990. Determination of organic carbon and nitrogen in marine sediments using the Carlo Erba NA-1500 analyzer. Deep Sea Res A Oceanogr Res Pap 37:157–165. doi: 10.1016/0198-0149(90)90034-S. [DOI] [Google Scholar]
- 18.Gregg WW. 2005. Recent trends in global ocean chlorophyll. Geophys Res Lett 32:L03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauvage J, Spivack AJ, Murray RW, D'Hondt S. 2014. Determination of in situ dissolved inorganic carbon concentration and alkalinity for marine sedimentary porewater. Chem Geol 387:66–73. doi: 10.1016/j.chemgeo.2014.06.010. [DOI] [Google Scholar]
- 20.Marteinsson VT, Rúnarsson Á, Stefánsson A, Thorsteinsson T, Jóhannesson T, Magnússon SH, Reynisson E, Einarsson B, Wade N, Morrison HG, Gaidos E. 2013. Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J 7:427–437. doi: 10.1038/ismej.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook N, Milojicic D, Kaufmann R, Sevinsky J. 2012. N3phele: open science-as-a-service workbench for cloud-based scientific computing. Proc 2012 7th Open Cirrus Summit, 19 to 20 June 2012, Beijing, China. [Google Scholar]
- 25.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270. [Google Scholar]
- 26.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 27.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:294–296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunge J, Woodard L, Böhning D, Foster J, Connolly S, Allen H. 2012. Estimating population diversity with CatchAll. Bioinformatics 28:1045–1047. doi: 10.1093/bioinformatics/bts075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke KR, Gorley RN. 2006. PRIMER v6: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]
- 34.Hong W-L, Solomon EA, Torres ME. 2014. A kinetic-model approach to quantify the effect of mass transport deposits on pore water profiles in the Krishna-Godavari Basin, Bay of Bengal. Mar Pet Geol 58:223–232. [Google Scholar]
- 35.Wang G, Spivack AJ, Rutherford SD, Manor U, D'Hondt S. 2008. Quantification of co-occurring reaction rates in deep subseafloor sediments. Geochim Cosmochim Acta 72:3479–3488. doi: 10.1016/j.gca.2008.04.024. [DOI] [Google Scholar]
- 36.Froelich PN, Klinkhammer GP, Bender ML, Luedtke NA, Heath GR, Cullen D, Dauphin P, Hammond D, Hartman B, Maynard V. 1979. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim Cosmochim Acta 43:1075–1090. doi: 10.1016/0016-7037(79)90095-4. [DOI] [Google Scholar]
- 37.Jørgensen BB. 2006. Bacteria and marine biogeochemistry, p 169–206. In Schulz HD, Zabel M (ed), Marine geochemistry. Springer, Berlin, Germany. [Google Scholar]
- 38.Parkes RJ, Webster G, Cragg BA, Weightman AJ, Newberry CJ, Ferdelman TG, Kallmeyer J, Jørgensen BB, Aiello IW, Fry JC. 2005. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436:390–394. doi: 10.1038/nature03796. [DOI] [PubMed] [Google Scholar]
- 39.Fry JC, Webster G, Cragg BA, Weightman AJ, Parkes RJ. 2006. Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol Ecol 58:86–98. doi: 10.1111/j.1574-6941.2006.00144.x. [DOI] [PubMed] [Google Scholar]
- 40.Sørensen KB, Teske AP. 2006. Stratified communities of active Archaea in deep marine subsurface sediments. Appl Environ Microbiol 72:4596–4603. doi: 10.1128/AEM.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller RD, Sdrolias M, Gaina C, Roest WR. 2008. Age, spreading rates, and spreading asymmetry of the world's ocean crust. Geochem Geophys Geosyst 9:Q04006. doi: 10.1029/2007GC001743. [DOI] [Google Scholar]
- 42.Flores JA, Johnson JE, Mejía-Molina AE, Álvarez MC, Sierro FJ, Singh SD, Mahanti S, Giosan L. 2014. Sedimentation rates from calcareous nannofossil and planktonic foraminifera biostratigraphy in the Andaman Sea, northern Bay of Bengal, and eastern Arabian Sea. Mar Pet Geol 58:425–437. doi: 10.1016/j.marpetgeo.2014.08.011. [DOI] [Google Scholar]
- 43.Walsh EA, Kirkpatrick JB, Rutherford SD, Smith DC, Sogin M, D'Hondt S. 2015. Bacterial diversity and community composition from seasurface to subseafloor. ISME J 10:979–989. doi: 10.1038/ismej.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Middelburg JJ. 1989. A simple rate model for organic matter decomposition in marine sediments. Geochim Cosmochim Acta 53:1577–1581. doi: 10.1016/0016-7037(89)90239-1. [DOI] [Google Scholar]
- 45.Burdige DJ. 2007. Preservation of organic matter in marine sediments: controls, mechanisms, and an imbalance in sediment organic carbon budgets? Chem Rev 107:467–485. doi: 10.1021/cr050347q. [DOI] [PubMed] [Google Scholar]
- 46.Arndt S, Brumsack H-J, Wirtz KW. 2006. Cretaceous black shales as active bioreactors: a biogeochemical model for the deep biosphere encountered during ODP Leg 207 (Demerara Rise). Geochim Cosmochim Acta 70:408–425. doi: 10.1016/j.gca.2005.09.010. [DOI] [Google Scholar]
- 47.Engelhardt T, Kallmeyer J, Cypionka H, Engelen B. 2014. High virus-to-cell ratios indicate ongoing production of viruses in deep subsurface sediments. ISME J 8:1503–1509. doi: 10.1038/ismej.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelhardt T, Orsi WD, Jørgensen BB. 2015. Viral activities and life cycles in deep subseafloor sediments. Environ Microbiol Rep 7:868–873. doi: 10.1111/1758-2229.12316. [DOI] [PubMed] [Google Scholar]
- 49.Anderson RE, Brazelton WJ, Baross JA. 2013. The deep viriosphere: assessing the viral impact on microbial community dynamics in the deep subsurface. Rev Mineral Geochem 75:649–675. doi: 10.2138/rmg.2013.75.20. [DOI] [Google Scholar]
- 50.Divins DL. 2003. Total sediment thickness of the world's oceans and marginal seas. NOAA National Geophysical Data Center, Boulder, CO. [Google Scholar]
- 51.Wang G, Spivack AJ, D'Hondt S. 2010. Gibbs energies of reaction and microbial mutualism in anaerobic deep subseafloor sediments of ODP site 1226. Geochim Cosmochim Acta 74:3938–3947. doi: 10.1016/j.gca.2010.03.034. [DOI] [Google Scholar]
- 52.Holmkvist L, Ferdelman TG, Jørgensen BB. 2011. A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark). Geochim Cosmochim Acta 75:3581–3599. [Google Scholar]
- 53.Orsi WD, Jørgensen BB, Biddle JF. 8 April 2016. Transcriptional analysis of sulfate reducing and chemolithoautotrophic sulfur oxidizing bacteria in the deep subseafloor. Environ Microbiol Rep; doi: 10.1111/1758-2229.12387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.