ABSTRACT
Bacteriophage-encoded endolysins are highly diverse enzymes that cleave the bacterial peptidoglycan layer. Current research focuses on their potential applications in medicine, in food conservation, and as biotechnological tools. Despite the wealth of applications relying on the use of endolysin, little is known about the enzymatic properties of these enzymes, especially in the case of endolysins of bacteriophages infecting Gram-negative species. Automated genome annotations therefore remain to be confirmed. Here, we report the biochemical analysis and cleavage site determination of a novel Salmonella bacteriophage endolysin, Gp110, which comprises an uncharacterized domain of unknown function (DUF3380; pfam11860) in its C terminus and shows a higher specific activity (34,240 U/μM) than that of 14 previously characterized endolysins active against peptidoglycan from Gram-negative bacteria (corresponding to 1.7- to 364-fold higher activity). Gp110 is a modular endolysin with an optimal pH of enzymatic activity of pH 8 and elevated thermal resistance. Reverse-phase high-performance liquid chromatography (RP-HPLC) analysis coupled to mass spectrometry showed that DUF3380 has N-acetylmuramidase (lysozyme) activity cleaving the β-(1,4) glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine residues. Gp110 is active against directly cross-linked peptidoglycans with various peptide stem compositions, making it an attractive enzyme for developing novel antimicrobial agents.
IMPORTANCE We report the functional and biochemical characterization of the Salmonella phage endolysin Gp110. This endolysin has a modular structure with an enzymatically active domain and a cell wall binding domain. The enzymatic activity of this endolysin exceeds that of all other endolysins previously characterized using the same methods. A domain of unknown function (DUF3380) is responsible for this high enzymatic activity. We report that DUF3380 has N-acetylmuramidase activity against directly cross-linked peptidoglycans with various peptide stem compositions. This experimentally verified activity allows better classification and understanding of the enzymatic activities of endolysins, which mostly are inferred by sequence similarities. Three-dimensional structure predictions for Gp110 suggest a fold that is completely different from that of known structures of enzymes with the same peptidoglycan cleavage specificity, making this endolysin quite unique. All of these features, combined with increased thermal resistance, make Gp110 an attractive candidate for engineering novel endolysin-based antibacterials.
INTRODUCTION
Endolysins are bacteriophage (phage)-encoded proteins synthesized at the end of the lytic infection cycle, which degrade the peptidoglycan (PG) of the host bacterium to allow viral progeny release (1). The specific activity and structure of these proteins have boosted their study as new antimicrobials against pathogens including multidrug-resistant bacteria (2). Recently, engineered fusions of an endolysin and a selected outer membrane-permeabilizing peptide (Artilysins) were shown to display high activity against Gram-negative bacteria (3–5). In addition, the analysis of endolysins has also led to the development of new biotechnological tools for bacterial diagnostics and detection, among others (6).
The structure of endolysins varies depending on their origin. In general, most of the endolysins from phages infecting Gram-positive bacteria have a modular structure consisting of one or two N-terminal enzymatic active domains (EADs) and a C-terminal cell wall binding domain (CBD) separated by a short linker (7). In contrast, the vast majority of endolysins from phages infecting Gram-negative bacteria have a globular organization containing only an EAD, although a number of modular endolysins with different orientations of EADs and CBDs have also been described (4, 8). The CBDs are responsible for the recognition of the substrate and the high-affinity binding of these enzymes to the bacterial cell wall (9), whereas the EADs are responsible for the catalytic activity, i.e., the cleavage of specific bonds within the PG. The PG is a copolymer of alternating N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) residues linked by β-(1,4) glycosidic bonds. Lactyl groups of the MurNAc residues are substituted by a pentapeptide stem made of l- and d-amino acids, which is highly conserved in Gram-negative species (l-Ala-d-Glu-meso-diaminopimelic acid [mDAP]-d-Ala-d-Ala) but variable in Gram-positive species (10). The mDAP is substituted by l-lysine in most Gram-positive species, although it can still be found in Bacillus and Listeria and l-ornithine can be found in the third position of the stem peptides in the PG of Thermus thermophilus, spirochetes, and Bifidobacterium globosum (11).
Endolysins are classified according to enzymatic activity, in the following three groups: (i) glycosidases, which include glucosaminidases (EC 3.2.1.52), muramidases or lysozymes (EC 3.2.1.17), and lytic transglycosylases (EC 4.2.2.n1) targeting the β-(1,4) glycosidic bonds of the sugar backbone; (ii) amidases (EC 3.5.1.28), which target the amide bond between the sugar backbone and the peptide stems; and (iii) endopeptidases (EC 3.4.-.-), which hydrolyze the bond between two amino acids. Only a limited number of studies have analyzed the PG bond cleaved by endolysins (12–17), and most (automated) annotations of enzymatic specificity only rely on sequence similarity. As a result, available databases contain inaccurate descriptions of biochemical specificities. A major problem is associated with the fact that several endolysins were referred to as lysozymes despite the lack of a biochemical characterization. A typical example is the endolysin of the T7 bacteriophage originally named as “T7 lysozyme.” This erroneous designation still persists even though the T7 endolysin was experimentally demonstrated to be N-acetylmuramoyl-l-alanine amidase rather than N-acetylmuramidase (or lysozyme) (18). Another typical example is the bacteriophage lambda lysozyme, which has been shown to display lytic transglycosidase activity (19). Furthermore, sequence similarities used to assign a putative function to endolysins are sometimes very poor or limited, while Pfam designations are often not updated. This can lead to discrepancies between in silico and experimental results when the cleavage site determination is performed (20).
In this study, we report the functional and biochemical characterization of the modular Salmonella phage endolysin Gp110. Among the many endolysins we have reported before (21–23) and unpublished endolysins, Gp110 has outstanding enzymatic activity (between a 1.7- and 364-fold increase). In addition, the catalytic domain is encoded by a C-terminal domain of unknown function (DUF3380; pfam11860). These elements prompted us to characterize Gp110 in more detail, including the determination of its peptidoglycan cleavage specificity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type Pseudomonas aeruginosa PAO1 strain (ATCC 15692) was kindly provided by J. P. Pirnay (Lab MCT, Queen Astrid Military Hospital, Neder-Over-Heembeek, Belgium). The food isolate Salmonella enterica serovar Typhimurium LT2 (ATCC 700720) was provided by the Centre of Food and Microbial Technology of KU Leuven (Belgium). Chemically competent Escherichia coli TOP10 (Thermo Fisher Scientific, Waltham, MA, USA) and E. coli BL21(DE3)pLysS (Agilent Technologies, Santa Clara, CA, USA) cells were prepared for cloning and protein recombinant expression, respectively. All of these strains were grown at 37°C in lysogeny broth (LB) with shaking.
Cloning, large-scale expression, and purification.
The sequence for a putative endolysin encoded by the orf110 of the genome of the uncharacterized Salmonella phage 10 was kindly provided by K. Makhulatia (Eliava Institute, Georgia) and has been deposited to GenBank (accession no. KU705467). The orf110 was amplified using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and primers Gp110-F (5′-ATGGCCATTCTAAAACTTGGCAACC-3′) and Gp110-R (5′-GCAGAAACTCTTGTATGCTGCC-3′). The PCR product was cloned into the commercially available pEXP5-CT/TOPO expression vector (Thermo Fisher Scientific) according to the manufacturer's instructions and sequence verified using the BigDye Terminator v1.1 cycle sequencing kit and the ABI3130 genetic analyzer (Life Technologies, Carlsbad, CA, USA). The pEXP5-CT/TOPO vector provides a C-terminal 6×His tag for nickel-nitrilotriacetic acid (Ni-NTA) purification.
Recombinant expression of Gp110 was performed in 500 ml of LB at 37°C for 4 h, using E. coli BL21(DE3)pLysS cells after induction during mid-exponential growth of the culture (optical density at 600 nm [OD600nm] of 0.6) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After induction, the pellet was resuspended in lysis buffer (20 mM NaH2PO4-NaOH, 500 mM NaCl, 50 mM imidazole [pH 7.4]) and disrupted with a combination of three freeze-thawing cycles (−80°C/room temperature) and sonication (10 cycles of 30-s pulse and 30-s rest; Vibra-Cell Sonics and Materials, Newtown, CT, USA). Gp110 was purified using the His GraviTrap column kit (GE Healthcare Life Sciences, Buckinghamshire, UK), following the supplier's recommendations. Wash buffer and elution buffer were composed of 20 mM NaH2PO4-NaOH, 500 mM NaCl (pH 7.4) with 50 mM or 500 mM imidazole, respectively. Protein purity was estimated by SDS-PAGE. The Gp110 concentration was determined spectrophotometrically after the protein was dialyzed against phosphate-buffered saline (PBS) (pH 7.4) using Slide-A-Lyzer Mini dialysis units (Thermo Fisher Scientific, Waltham, MA, USA). The dialyzed protein was stored at 4°C without observed loss in activity.
Quantification and characterization of muralytic activity.
The hydrolytic activity of Gp110 was quantified on P. aeruginosa PAO1 cells with the outer membrane (OM) permeabilized by a chloroform-Tris-HCl treatment as described previously (24). Briefly, mid-exponentially growth phase (OD600nm of 0.6) P. aeruginosa PAO1 cells were incubated in a chloroform-saturated 0.05 M Tris-HCl buffer (pH 7.7) for 45 min. Afterwards, cells were washed in PBS (pH 7.4) and concentrated to an OD600 nm of 1.5 also in PBS. To determine the muralytic activity, 30 μl of Gp110 was added to 270 μl of OM-permeabilized P. aeruginosa PAO1 cells (final concentrations between 0.25 and 750 nM Gp110 for the dose-dependence curve), and the resulting decrease in optical density was measured spectrophotometrically (655 nm) in a microplate reader 680 (Bio-Rad, CA, USA). The muralytic activity of Gp110 was quantified in units per micromolar according to a standardized method described in reference 25.
The effects of pH and temperature on the lytic activity of the endolysin (final concentration, 2 nM) were assessed by the same method with some modifications. For the pH-dependent effect, OM-permeabilized P. aeruginosa PAO1 cells were resuspended in universal pH buffer (150 mM KCl, 10 mM KH2PO4, 10 mM sodium citrate, 10 mM H3BO4) adjusted to pH values between 3 and 12. To determine the effect of temperature on Gp110 activity, the endolysin (final concentration, 2 nM) was incubated for 10 min at either 30°C, 40°C, 50°C, 60°C, 70°C, 80°C, or 90°C, followed by a cooling step to room temperature. The P. aeruginosa PAO1 substrate was resuspended in universal pH buffer adjusted to the optimal pH for the endolysin activity, and the residual activity was tested at room temperature. For both experiments, the relative muralytic activity was calculated. All assays were performed in triplicate. Statistical analyses were performed using one-way analysis of variance (ANOVA) and the Tukey post hoc test.
In vitro antibacterial activity.
The antibacterial assay was performed similarly to that described previously (5). Mid-exponential growth phase P. aeruginosa PAO1 and S. Typhimurium LT2 cells (OD600nm of 0.6) were diluted in 5 mM HEPES-NaOH (pH 7.4) to a final density of 106 CFU/ml. Next, 100 μl of these cultures was mixed with 50 μl of Gp110 (2.5 μM final concentration, dialyzed against phosphate-buffered saline [pH 7.4]) and 50 μl of 5 mM HEPES-NaOH (pH 7.4) or EDTA (0.5 mM final concentration)-malate-lactate (both with a final concentration of 10 mM) dissolved in the same buffer. As a control, 100 μl of cells, 50 μl of 5 mM HEPES-NaOH (pH 7.4), and 50 μl of PBS (pH 7.4) were used. After incubation for 30 min at room temperature, mixtures were diluted in PBS (pH 7.4) and plated on LB agar plates. The antibacterial activity is quantified after 18 h of incubation at 37°C as the relative inactivation in logarithmic units [= log10(N0/Ni) with N0 representing the number of untreated cells and Ni the number of treated cells counted after incubation].
Analysis of PG fragments solubilized by Gp110.
The PG bond cleaved by Gp110 was determined using E. coli BW25113 Δlpp PG as a substrate. PG was extracted as previously described using boiling SDS (26). A total of 500 μg of pure PG was digested overnight at 37°C with 0.5 mg/ml Gp110 in a final volume of 200 μl. As a control, the same amount of PG was digested with 0.25 mg/ml Streptomyces globisporus mutanolysin (Sigma-Aldrich, MO, USA) in 25 mM phosphate buffer (pH 6). After centrifugation at 20,000 × g for 15 min, soluble muropeptides were reduced with sodium borohydride and separated by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Hypersil aQ C18 column (3 μm, 2.1 by 200 mm; Thermo Fisher Scientific, Waltham, MA, USA) coupled to an Agilent 6500 series quadrupole time of flight liquid chromatography-mass spectrometry (Q-TOF LC-MS) system. Muropeptides were eluted at a flow rate of 0.25 ml/min with a 0 to 15% gradient (buffer A: 0.1% [vol/vol] formic acid in water; buffer B: 0.1% [vol/vol] formic acid in acetonitrile) applied between 6 and 40 min. Bacillus subtilis 168 and Aerococcus viridans ATCC 10400 PG were also used as substrates. The hydrolysis conditions used for B. subtilis and A. viridans PG were similar to those described for E. coli PG.
Accession number(s).
The DNA sequence of orf110 was deposited in GenBank under accession number KU705467.
RESULTS
In silico analysis of Gp110.
Bioinformatic analysis of the Salmonella phage 10 genome revealed that orf110 encodes a 264-amino acid protein (deduced molecular mass, 28.9 kDa) predicted to be a putative endolysin by HHpred (27), with some similarities (21%) to Pseudomonas phiKZ phage endolysin (E-value, 3 × 10−20). Sequence similarity searches by BLASTP (28) indicate that Gp110 has 100% identity with the predicted peptidoglycan binding proteins froms three other Salmonella phages (PhiSH19, vB_SalM_SJ3, and Det7). Conserved domain analysis by Pfam (29) showed that Gp110 has a modular structure, with an N-terminal PG_binding_1 domain (pfam01471) and a C-terminal DUF3380 domain (pfam11860). PG_binding_1 from Gp110 has specific repeated motifs (DGIFGKAT and DGIAGPKT), a feature that seems to be common in proteins interacting with repetitive structures like peptidoglycan (30). These motifs match the consensus sequence [D-G-(Pho)2-G-K/N-G/N-T; Pho is hydrophobic amino acid] previously found in other endolysins with a Gram-negative background (21, 23). The DUF3380 domain is found in viruses and bacteria normally associated with PG_binding_1 and belongs to a family of functionally uncharacterized proteins.
Biochemical characterization of Gp110 peptidoglycan-degrading activity.
The predicted PG hydrolytic activity of Gp110 was confirmed and characterized using OM-permeabilized P. aeruginosa cells as a substrate (24) to allow the endolysin to reach the PG layer and exert its enzymatic activity, which is measured by a turbidity assay (25).
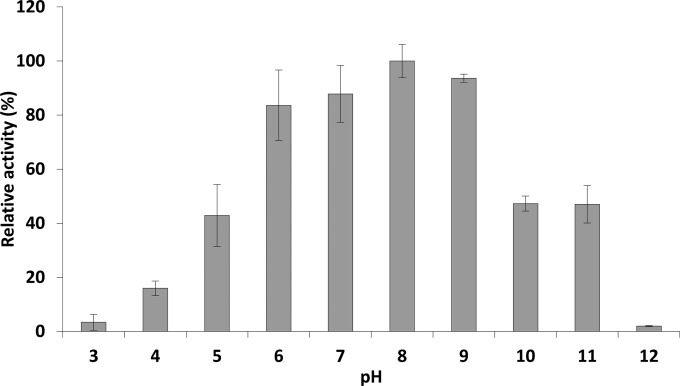
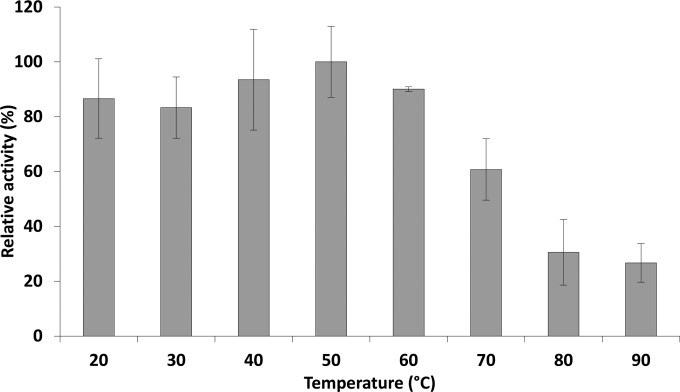
Incubation of Gp110 with the substrate in universal buffer adjusted to different pHs showed that this enzyme is active at pH values ranging from 4 to 11, maintaining between 16.0% ± 2.67% and 47.0% ± 6.9% of its maximal activity at pH 4 and 11, respectively. The highest activity is achieved in the pH range of 6 to 9 (between 83.6% ± 13.0% and 93.6% ± 1.4%, respectively) (Fig. 1). Of note is the significant decrease in activity below pH 6 and above pH 9, losing >50% of the activity (P < 0.05). Gp110 shows no significant loss in activity upon a 10-min heat exposure to temperatures between 20 and 60°C. Above 60°C, the activity gradually decreases with a remaining activity of 26.7% ± 7.0% after 10 min at 90°C (Fig. 2).
FIG 1.
pH dependence of Gp110 enzymatic activity. The muralytic activity of Gp110 is shown against OM-permeabilized P. aeruginosa PAO1 cells resuspended in a universal buffer adjusted to different pH values. Activity is expressed relative to the maximal muralytic activity at the optimal pH (pH 8). Each bar represents the mean of triplicate experiments, and error bars indicate the standard deviations. A one-way ANOVA and Tukey post hoc test indicated that there were no statistically significant differences between the activities at the pH range of 6 to 9 (all P > 0.05).
FIG 2.
Temperature resistance of Gp110. The residual enzymatic activity of Gp110 was analyzed after a 10-min incubation of the endolysin at different temperatures, ranging from 20°C to 90°C. PG hydrolase activity is expressed relative to the highest measured activity.
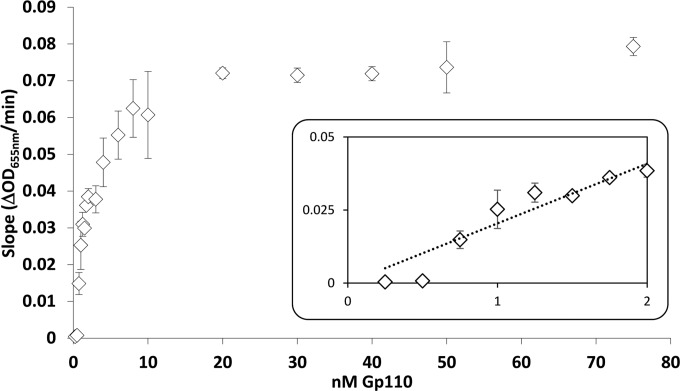
The specific activity of Gp110 was calculated at optimal pH under substrate-saturating conditions from the slope of the linear regression of the corresponding dose-dependent saturation curve as previously described (25) (Fig. 3). A linear dose response was observed between 0.25 and 1.25 nM Gp110. According to this method, the specific activity of Gp110 was calculated to be 34,240 U/μM, corresponding to complete clarification of turbid cell cultures in approximately 10 and 20 min with 10 and 1 nM Gp110, respectively.
FIG 3.
Saturation curve of Gp110 at optimal pH (pH 8). The muralytic activity was quantified against OM permeabilized P. aeruginosa PAO1 cells according to Briers et al. (25). The x and y axes display the amount of Gp110 (in nanomolar) added and the corresponding activity (OD655/min) measured, respectively. Each data point shows the average and error bars of three replicates. The insert zooms in on the region with a linear relationship between activity and enzyme concentration (substrate saturating conditions).
Gp110 antibacterial activity.
The in vitro antibacterial activity of the endolysin was tested against P. aeruginosa PAO1 and S. Typhimurium LT2 in the presence or absence of EDTA, malate, and lactate as OM permeabilizers (31). As expected, the activity of 2.5 μM Gp110 alone was insignificant against both strains. However, addition of 0.5 mM EDTA, together with the 2.5 μM Gp110, resulted in a reduction of 2.74 ± 0.11 log units in the case of P. aeruginosa PAO1, corresponding to nearly a 99.9% reduction in the number of viable cells and 0.38 ± 0.18 log unit in the case of S. Typhimurium LT2 cells. The addition of EDTA alone reduced the cell numbers with 0.62 ± 0.06 and 0.28 ± 0.29 log unit for P. aeruginosa PAO1 and S. Typhimurium LT2, respectively. Other OM permeabilizers such us malate and lactate did not significantly improve the antibacterial activity of the endolysin against S. Typhimurium LT2 (data not shown).
Determination of Gp110 PG cleavage specificity by LC-MS.
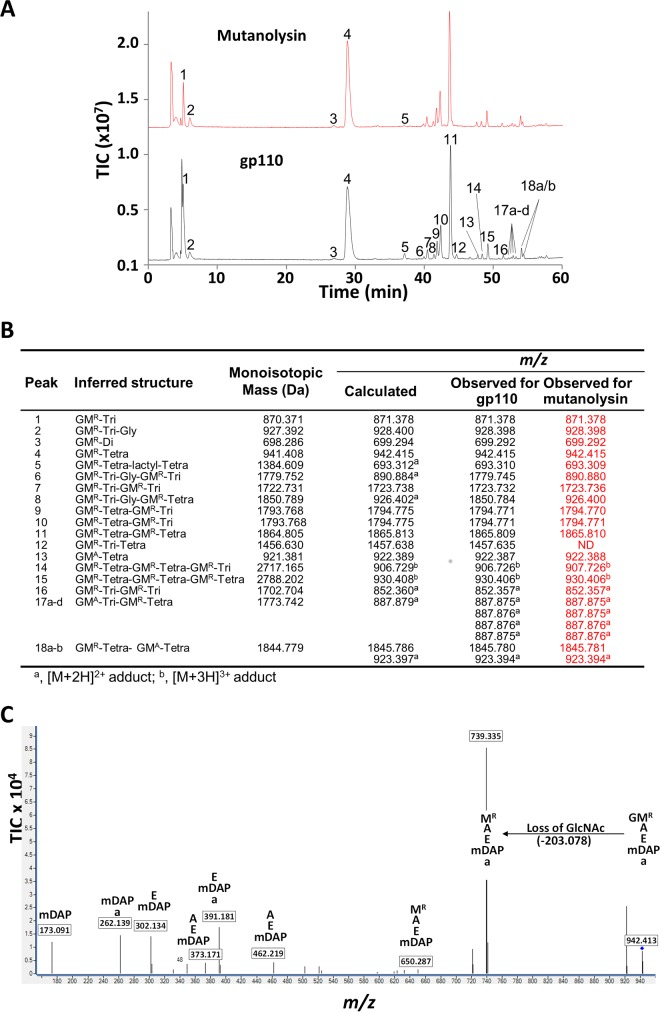
To determine the PG bond cleaved by the DUF3380 domain in Gp110, purified E. coli PG was incubated in the presence of recombinant Gp110, and soluble fragments were analyzed by RP-HPLC coupled to MS (Fig. 4). The muropeptide profile obtained for Gp110 is very similar to the one obtained for the N-acetylmuramidase mutanolysin (Fig. 4A), suggesting that Gp110 cleaves the β-1,4 glycosidic bonds of the PG sugar backbone. The major monomer (Fig. 4A, peak 4) generated after the digestion of the PG with Gp110 yielded a peak with an m/z at 942.415, matching the theoretical value expected for a disaccharide tetrapeptide (Fig. 4B), thus confirming the glycosidase activity. To determine whether DUF3380 possesses N-acetylmuramidase or N-acetylglucosaminidase activity, tandem mass spectrometry (MS/MS) was further performed on the major monomer (peak 4). The fragmentation event leading to the loss of a nonreduced GlcNAc residue (203.078 atomic mass units) indicated that Gp110 displays N-acetylmuramidase activity (Fig. 4C) instead of N-acetylglucosaminidase activity, which would involve the loss of 223.106 atomic mass units corresponding to reduced GlcNAc (32).
FIG 4.
Determination of Gp110 cleavage specificity. (A) LC-MS analysis of E. coli BW25113 Δlpp peptidoglycan digested by mutanolysin and recombinant Gp110. Soluble muropeptides were reduced and analyzed by RP-HPLC coupled to MS. Peaks corresponding to m/z values matching previously identified muropeptides are numbered. (B) Inferred structures, theoretical monoisotopic masses, and theoretical and observed m/z values of the peaks identified in panel A. Di, l-Ala-d-Glu; Tri, l-Ala-d-Glu-m-DAP; Tetra, l-Ala-d-Glu-m-DAP-d-Ala. (C) LC-MS/MS analysis of the major disaccharide peptide (peak 4) solubilized by Gp110. The fragmentation pattern of the [M + H]+ ion at m/z 942.414 was typical of a disaccharide tetrapeptide (DS-Tetra). The fragmentation event leading to the loss of a nonreduced GlcNAc residue (203.078) indicates that Gp110 displays N-acetylmuramidase (lysozyme) activity. The sequence of peptide fragments is indicated above their respective m/z values (boxed). A, l-Ala or d-Ala; a, C-terminal d-Ala; m-DAP, meso-diaminopimelic acid; E, γ-d-Glu; MR, reduced MurNAc; G, GlcNAc. TIC, total ion count.
Interestingly, Gp110 also displays enzymatic activity against B. subtilis and A. viridans PG (see Fig. S1 in the supplemental material). This indicates that this enzyme can cleave PG containing amidated mDAP or l-lysine residues at position 3 of the peptide stems. Unlike E. coli PG digestion products, which essentially contain tetrapeptide stems, both B. subtilis and A. viridans digestion products mostly contain tripeptide stems.
DISCUSSION
In this study, we describe a new endolysin from an uncharacterized phage infecting the Gram-negative pathogen S. Typhimurium. In silico analysis indicates that this endolysin has a modular structure harboring a DUF3380 EAD at the C terminus and a PG_binding_1 CBD at the N terminus. This modular structure is a common feature in endolysins with a Gram-positive background (7) but remains rare in endolysins with a Gram-negative background, which are mostly globular (22). Interestingly, the PG_binding_1 is also almost always restricted to Gram-positive-related endolysins (8), although there are a few exceptions (Salmonella phage PVP-SE1; Pseudomonas phages phiKZ, EL, 201phi21, and OBP; and phages infecting Burkholderia and Erwinia) (21, 23, 33). The DUF3380 domain has only been predicted in endolysins from phages infecting Burkholderia, Pseudomonas, and Erwinia (33), and it is classified as a pfam domain of unknown function. Noteworthy is the fact that there is another domain with unknown function (DUF3597, pfam12200) associated with an endolysin, but it has only been detected in the endolysin from the Listeria phage A118 (8, 33). According to our results, the biochemical analysis of DUF3380 using pure PG revealed that this domain has N-acetylmuramidase (EC 3.2.1.17) activity, cleaving the β-(1,4) bonds between N-acetylmuramic acid and N-acetylglucosamine in the sugar backbone of the PG. In addition, Gp110 can cleave PG with distinct PG compositions such as those in B. subtilis and A. viridans. Up to 690 proteins have a predicted DUF3380 domain, among which the large majority is present in bacterial proteins and only 10% are encoded by bacteriophages (InterPro database), which may indicate a horizontal transfer. According to the Pfam database, this domain is commonly found associated with a PG binding domain; however, it is also found in one-domain proteins (InterPro). Recently, using remote homology detection methods, such as FFAS03 (http://ffas.sanfordburnham.org) and data from publications collected by PubServer (http://pubserver.burnham.org), DUF3380 was designated a family of cell wall lytic enzymes (34), which is in accordance with our results, and we have confirmed this by determination of cleavage sites in the PG. Alignments of some sequences containing this DUF3380 domain show that the Gp110 E101 residue is conserved, suggesting that this is the catalytic residue. Moreover, this catalytic residue is followed by serine, which is a common feature in lysozymes. However, three-dimensional structure predictions of the DUF3380 domain show low homology with the tertiary structure of other lysozymes, which suggests a completely different fold. On the contrary, the CBD is strongly conserved, showing high homology with PG binding domains of other hydrolases.
In comparing Gp110 specific activity with that of other endolysins analyzed using the same method under optimal conditions (Table 1), this endolysin shows the highest enzymatic activity described to date: 1.7-fold more active than the second most active endolysin, OBPgp279, which also has a modular structure and predicted lysozyme-like muramidase activity (21). Compared to other Salmonella phage endolysins, Gp110 has 2.5- and 24.8-fold higher activities than PVP-SE1gp146 and PsP3gp10, respectively, and even 85.6-fold higher activity than Lys68 (Table 1). The optimal pH for Gp110 activity is pH 8 (Fig. 1), in contrast to that of other previously described endolysins with a Gram-negative background, which have optimal activities at neutral pH (21, 22, 23, 35). It is noteworthy that three other endolysins from phages infecting S. Typhimurium have been reported to show optimal activities at pH 9.5 (phage SPN1S endolysin) and pH 8.5 (phage SPN9CC endolysin and Lys394), respectively (36–38), whereas endolysins from phages infecting Salmonella enterica serovar Enteritidis (PVP-SE1gp146, Lys68, and PsP3gp10) have optimal activities at pH 7 (21, 22, 35). In terms of temperature resistance, Gp110 remains active after treatment at temperatures across the range tested (20 to 90°C), showing no significant activity loss with up to 10-min heat treatments at 60°C and only gradually decreasing activity at higher temperatures (Fig. 2). Whereas most endolysins irreversibly lose their enzymatic activities upon exposure to temperatures around 50°C (37, 39–43), some thermoresistant endolysins have also been described, including Lys68 from Salmonella phage phi68 (35), gp146 from Salmonella phage PVP-SE1 (21), the endolysins from bacteriophages Ph2119 and vB_Tsc2631 infecting the thermophile Thermus scotoductus (44, 45), and the lysin from deep-sea thermophilic bacteriophage GVE2 (46). Gp110 shows an intermediate profile with elevated temperature resistance compared to that of most mesophilic endolysins, which may correlate to increased shelf life of Gp110.
TABLE 1.
Comparison of Gp110 specific activity with other endolysins against P. aeruginosa PAO1a
| Endolysin | Activity (U/μM) | Structure | Reference or source |
|---|---|---|---|
| Gp110 | 34,240 | Modular | This work |
| OBPgp279 | 19,979 | Modular | 21 |
| LysEC8 | 17,103 | Globular | Unpublished results |
| PVP-SE1gp146 | 13,614 | Modular | 21 |
| EL188 | 4,735 | Modular | 23 |
| 201φ2-1gp229 | 4,469 | Modular | 21 |
| KZ144 | 2,058 | Modular | 23 |
| PsP3gp10 | 1,380 | Globular | 22 |
| P2gp09 | 829 | Globular | 22 |
| BcepC6Bgp22 | 786 | Globular | 22 |
| Lys68 | 400 | Globular | 35 |
| CR8gp3.5 | 315 | Globular | 3 |
| K11gp3.5 | 134 | Globular | 22 |
| KP32gp15 | 117 | Globular | 22 |
| LysAci7 | 94 | Globular | Unpublished results |
All specific activities were calculated following the method described in reference 25.
To verify if the biochemical activity of DUF3380 is translated into antibacterial activity, Gp110 (2.5 μM) was tested against P. aeruginosa PAO1 and S. Typhimurium LT2 cells in the presence or absence of 0.5 mM EDTA as an OM permeabilizer. As expected, the antibacterial activity of the protein without EDTA was very low against both strains due to the OM protective effect. However, in the presence of EDTA, the number of P. aeruginosa PAO1 viable cells was reduced by approximately 99.9%. The synergistic effect of EDTA in the antibacterial activity of endolysins with a Gram-negative background was first described with the endolysin EL188 and P. aeruginosa (47). The mild antibacterial activity of the endolysin-EDTA combination against S. Typhimurium LT2 cells was also observed for other endolysins (21, 36, 38) and can be explained by the lower degree of phosphorylation in Salmonella lipopolysaccharide (LPS) molecules than in Pseudomonas LPS molecules. As a consequence, S. Typhimurium LT2 has a significantly smaller amount of stabilizing divalent cations, resulting in lower susceptibility to EDTA permeabilization than P. aeruginosa.
Overall, we have characterized an endolysin that has, to our knowledge, the highest enzymatic activity against Gram-negative peptidoglycan reported to date. This high Gp110 activity can be explained by its enzymatically active domain, DUF3380, which was biochemically demonstrated to have N-acetylmuramidase activity and shows a low degree of homology with lysozymes. These features render Gp110 a novel attractive candidate for engineering to provide the enzyme with outer membrane-permeabilizing and consequently antibacterial properties.
Supplementary Material
Funding Statement
This work, including the efforts of Yves Briers, was funded by Research Foundation of Vlaanderen (FWO-Vlaanderen) (1517115N). This study was also funded by the ERANET Animal Health and Welfare project BLAAT (Rob Lavigne and Lorena Rodríguez-Rubio).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00446-16.
REFERENCES
- 1.Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol Rev 56:430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roach DR, Donovan DM. 2015. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 5:e1062590. doi: 10.1080/21597081.2015.1062590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5:e01379-14. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briers Y, Lavigne R. 2015. Breaking barriers: expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol 10:377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 5.Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay JP, Defraine V, Michiels J, Cenens W, Aertsen A, Miller S, Lavigne R. 2014. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:3774–3784. doi: 10.1128/AAC.02668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-Rubio L, Gutiérrez D, Donovan DM, Martínez B, Rodríguez A, García P. 2016. Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol 36:542−552. [DOI] [PubMed] [Google Scholar]
- 7.Nelson DC, Schmelcher M, Rodríguez-Rubio L, Klumpp J, Pritchard DG, Dong S, Donovan DM. 2012. Endolysins as antimicrobials. Adv Virus Res 83:299–365. doi: 10.1016/B978-0-12-394438-2.00007-4. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira H, Melo LD, Santos SB, Nobrega FL, Ferreira EC, Cerca N, Azeredo J, Kluskens LD. 2013. Molecular aspects and comparative genomics of bacteriophage endolysins. J Virol 87:4558–4570. doi: 10.1128/JVI.03277-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol 44:335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- 10.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 12.Navarre WW, Ton-That H, Faull KF, Schneewind O. 1999. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a d-alanyl-glycine endopeptidase activity. J Biol Chem 274:15847–15856. [DOI] [PubMed] [Google Scholar]
- 13.Paradis-Bleau C, Cloutier I, Lemieux L, Sanschagrin F, Laroche J, Auger M, Garnier A, Levesque RC. 2007. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage phiKZ gp144 lytic transglycosylase. FEMS Microbiol Lett 266:201–209. doi: 10.1111/j.1574-6968.2006.00523.x. [DOI] [PubMed] [Google Scholar]
- 14.Pritchard DG, Dong S, Baker JR, Engler JA. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079–2087. doi: 10.1099/mic.0.27063-0. [DOI] [PubMed] [Google Scholar]
- 15.Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG, Donovan DM. 2009. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol Lett 294:52–60. doi: 10.1111/j.1574-6968.2009.01541.x. [DOI] [PubMed] [Google Scholar]
- 16.Loessner MJ, Gaeng S, Wendlinger G, Maier SK, Scherer S. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett 162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- 17.Fischetti VA, Zabriskie JB, Gotschlich EC. 1974. Physical, chemical, and biological properties of type 6 M-protein extracted with purified streptococcal phage-associated lysin, p 26 In Haverkorn MJ. (ed), Streptococcal disease and the community. Excerpta Medica, Amsterdam, The Netherlands. [Google Scholar]
- 18.Inouye M, Arnheim N, Sternglanz R. 1973. Bacteriophage T7 lysozyme is an N-acetylmuramyl-l-alanine amidase. J Biol Chem 248:7247–7252. [PubMed] [Google Scholar]
- 19.Bienkowska-Szewczyk K, Lipinska B, Taylor A. 1981. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol Gen Genet 184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR. 2007. LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl Environ Microbiol 73:7150–7154. doi: 10.1128/AEM.01783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walmagh M, Briers Y, dos Santos SB, Azeredo J, Lavigne R. 2012. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201phi2-1 and PVP-SE1. PLoS One 7:e36991. doi: 10.1371/journal.pone.0036991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walmagh M, Boczkowska B, Grymonprez B, Briers Y, Drulis-Kawa Z, Lavigne R. 2013. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol 97:4369–4375. doi: 10.1007/s00253-012-4294-7. [DOI] [PubMed] [Google Scholar]
- 23.Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol Microbiol 65:1334–1344. doi: 10.1111/j.1365-2958.2007.05870.x. [DOI] [PubMed] [Google Scholar]
- 24.Lavigne R, Briers Y, Hertveldt K, Robben J, Volckaert G. 2004. Identification and characterization of a highly thermostable bacteriophage lysozyme. Cell Mol Life Sci 61:2753–2759. doi: 10.1007/s00018-004-4301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briers Y, Lavigne R, Volckaert G, Hertveldt K. 2007. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J Biochem Biophys Methods 70:531–533. doi: 10.1016/j.jbbm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172:451–464. doi: 10.1016/0003-2697(88)90468-X. [DOI] [PubMed] [Google Scholar]
- 27.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244−W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wren BW. 1991. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol 5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 31.Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckert C, Lecerf M, Dubost L, Arthur M, Mesnage S. 2006. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J Bacteriol 188:8513–8519. doi: 10.1128/JB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidová B, Šramková Z, Tišáková L, Oravkinová M, Godány A. 2014. Bioinformatics analysis of bacteriophage and prophage endolysin domains. Biologia 69:541–556. [Google Scholar]
- 34.Jaroszewski L, Koska L, Sedova M, Godzik A. 2014. PubServer: literature searches by homology. Nucleic Acids Res 42:W430–W435. doi: 10.1093/nar/gku450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveira H, Thiagarajan V, Walmagh M, Sillankorva S, Lavigne R, Neves-Petersen MT, Kluskens LD, Azeredo J. 2014. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PLoS One 9:e108376. doi: 10.1371/journal.pone.0108376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim JA, Shin H, Kang DH, Ryu S. 2012. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res Microbiol 163:233–241. doi: 10.1016/j.resmic.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Legotsky SA, Vlasova KY, Priyma AD, Shneider MM, Pugachev VG, Totmenina OD, Kabanov AV, Miroshnikov KA, Klyachko NL. 2014. Peptidoglycan degrading activity of the broad-range Salmonella bacteriophage S-394 recombinant endolysin. Biochimie 107(Pt B):293–299. doi: 10.1016/j.biochi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Lim JA, Shin H, Heu S, Ryu S. 2014. Exogenous lytic activity of SPN9CC endolysin against Gram-negative bacteria. J Microbiol Biotechnol 24:803–811. doi: 10.4014/jmb.1403.03035. [DOI] [PubMed] [Google Scholar]
- 39.Filatova LY, Becker SC, Donovan DM, Gladilin AK, Klyachko NL. 2010. LysK, the enzyme lysing Staphylococcus aureus cells: specific kinetic features and approaches towards stabilization. Biochimie 92:507–513. doi: 10.1016/j.biochi.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 40.Varea J, Monterroso B, Saiz JL, Lopez-Zumel C, Garcia JL, Laynez J, Garcia P, Menendez M. 2004. Structural and thermodynamic characterization of Pal, a phage natural chimeric lysin active against pneumococci. J Biol Chem 279:43697–43707. doi: 10.1074/jbc.M407067200. [DOI] [PubMed] [Google Scholar]
- 41.Sanz JM, Garcia JL, Laynez J, Usobiaga P, Menendez M. 1993. Thermal stability and cooperative domains of CPL1 lysozyme and its NH2- and COOH-terminal modules. Dependence on choline binding. J Biol Chem 268:6125–6130. [PubMed] [Google Scholar]
- 42.Obeso JM, Martinez B, Rodriguez A, Garcia P. 2008. Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int J Food Microbiol 128:212–218. doi: 10.1016/j.ijfoodmicro.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Heselpoth RD, Nelson DC. 2012. A new screening method for the directed evolution of thermostable bacteriolytic enzymes. J Vis Exp 69:4216. doi: 10.3791/4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plotka M, Kaczorowska AK, Stefanska A, Morzywolek A, Fridjonsson OH, Dunin-Horkawicz S, Kozlowski L, Hreggvidsson GO, Kristjansson JK, Dabrowski S, Bujnicki JM, Kaczorowski T. 2014. Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl Environ Microbiol 80:886–895. doi: 10.1128/AEM.03074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotka M, Kaczorowska AK, Morzywolek A, Makowska J, Kozlowski LP, Thorisdottir A, Skirnisdottir S, Hjorleifsdottir S, Fridjonsson OH, Hreggvidsson GO, Kristjansson JK, Dabrowski S, Bujnicki JM, Kaczorowski T. 2015. Biochemical characterization and validation of a catalytic site of a highly thermostable Ts2631 endolysin from the Thermus scotoductus phage vB_Tsc2631. PLoS One 10:e0137374. doi: 10.1371/journal.pone.0137374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye T, Zhang X. 2008. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl Microbiol Biotechnol 78:635–641. doi: 10.1007/s00253-008-1353-1. [DOI] [PubMed] [Google Scholar]
- 47.Briers Y, Walmagh M, Lavigne R. 2011. Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J Appl Microbiol 110:778–785. doi: 10.1111/j.1365-2672.2010.04931.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.