ABSTRACT
Deinococcus radiodurans and Escherichia coli expressing either PhoN, a periplasmic acid phosphatase, or PhoK, an extracellular alkaline phosphatase, were evaluated for uranium (U) bioprecipitation under two specific geochemical conditions (GCs): (i) a carbonate-deficient condition at near-neutral pH (GC1), and (ii) a carbonate-abundant condition at alkaline pH (GC2). Transmission electron microscopy revealed that recombinant cells expressing PhoN/PhoK formed cell-associated uranyl phosphate precipitate under GC1, whereas the same cells displayed extracellular precipitation under GC2. These results implied that the cell-bound or extracellular location of the precipitate was governed by the uranyl species prevalent at that particular GC, rather than the location of phosphatase. MINTEQ modeling predicted the formation of predominantly positively charged uranium hydroxide ions under GC1 and negatively charged uranyl carbonate-hydroxide complexes under GC2. Both microbes adsorbed 6- to 10-fold more U under GC1 than under GC2, suggesting that higher biosorption of U to the bacterial cell surface under GC1 may lead to cell-associated U precipitation. In contrast, at alkaline pH and in the presence of excess carbonate under GC2, poor biosorption of negatively charged uranyl carbonate complexes on the cell surface might have resulted in extracellular precipitation. The toxicity of U observed under GC1 being higher than that under GC2 could also be attributed to the preferential adsorption of U on cell surfaces under GC1. This work provides a vivid description of the interaction of U complexes with bacterial cells. The findings have implications for the toxicity of various U species and for developing biological aqueous effluent waste treatment strategies.
IMPORTANCE The present study provides illustrative insights into the interaction of uranium (U) complexes with recombinant bacterial cells overexpressing phosphatases. This work demonstrates the effects of aqueous speciation of U on the biosorption of U and the localization pattern of uranyl phosphate precipitated as a result of phosphatase action. Transmission electron microscopy revealed that location of uranyl phosphate (cell associated or extracellular) was primarily influenced by aqueous uranyl species present under the given geochemical conditions. The data would be useful for understanding the toxicity of U under different geochemical conditions. Since cell-associated precipitation of metal facilitates easy downstream processing by simple gravity-based settling down of metal-loaded cells, compared to cumbersome separation techniques, the results from this study are of considerable relevance to effluent treatment using such cells.
INTRODUCTION
Bioremediation strategies, such as bioreduction (1–3), biosorption (4–8), bioaccumulation (9, 10), and bioprecipitation (5, 11, 12, 13), have been studied for their potential to immobilize U from solutions. There is also a large body of work on microbial interactions with uranium relevant to environmental in situ bioremediation. The efficacy of U removal and fate of the metal at the end of the waste solution treatment are influenced by the chemical state of U prevalent under the given condition. U forms aqueous species as a result of complexation with ligands under different pH conditions (14). In open atmospheric systems, under oxygenic conditions, and with pH values lower than 3, U(VI) is present exclusively in the form of hexavalent uranyl cation, UO22+, which is the most bioavailable form of U (15, 16). Circumneutral pH favors the formation of positively charged uranyl hydroxide, [(UO2)3(OH)]5+, or [(UO2)4(OH)]+7complexes that are transformed to negatively charged ones at higher pH (pH 8 to 9). However, under strongly alkaline conditions, negatively charged uranyl-carbonate complexes, like [UO2(CO3)2]2− and [UO2(CO3)3]4−, predominate (17–19). It is important to understand how these different U species interact with bacterial cellular surfaces, especially for designing biological wastewater treatment systems. However, studies evaluating the effect of aqueous U speciation have been largely limited to biosorption, bioaccumulation, and bioreduction (9, 15, 16, 20).
Among the biological mechanisms involved in metal remediation, enzymatic bioprecipitation of heavy metals as metal phosphates is particularly attractive and is considered to be a promising approach for biological treatment of U effluents due to its high efficiency (14, 21). Bioprecipitation of metals as phosphates is mediated by phosphatases that cleave a phosphomonoester substrate (such as β-glycerophosphate) to release the phosphate moiety, which in turn precipitates heavy metals, such as U, Cd, Ni, Am, etc., from solutions (22, 23). Phosphatases are ubiquitous among prokaryotes that catalyze dephosphorylation of various substrates by hydrolysis of phosphoester or phosphoanhydride bonds (24, 25). Traditionally, phosphatases are broadly categorized as acid or alkaline phosphatases, based on the pH required for their optimum activity. Phosphatases play a crucial role in supporting microbial nutrition by releasing the assimilable phosphate from the organic source (24, 26). Phosphatases are either secreted outside the plasma membrane, where they are released in a soluble form, or retained as membrane-bound proteins. Phosphatases enable the release of inorganic phosphate (Pi) and organic by-products that can be transported across the membrane, thus providing cells with essential nutrients (25).
As uranyl phosphate precipitate is highly insoluble, it can serve as a long-term stable sink for U immobilization (27, 28), thus making phosphatase-mediated bioprecipitation a very attractive strategy for treating nuclear waste. In recent years, our laboratory has explored the ability of phosphatase-expressing recombinant bacterial cells to bioprecipitate U and other heavy metals from solutions (29). The PhoN enzyme from Salmonella enterica serovar Typhimurium (encoded by the phoN gene, GenBank accession no. X59036), a nonspecific acid phosphatase, was overexpressed in the Gram-positive radioresistant bacterium Deinococcus radiodurans [D. radiodurans(pPN1)], as well as in the Gram-negative bacterium Escherichia coli [E. coli(pPN1)] to achieve phosphatase-mediated bioprecipitation of U from acidic to neutral solutions (pH 6.8) (30, 31). In view of its phenomenal radioresistance, D. radiodurans was the organism of choice for its potential use in U bioprecipitation from nuclear effluents. Similarly, the phoK gene (GenBank accession no. EF143994), encoding a novel alkaline phosphatase from Sphingomonas sp. strain BSAR-1, was also introduced into both D. radiodurans [D. radiodurans(pK1)] and E. coli [E. coli(pK1)]. PhoN and PhoK show good activity between pH 5 and 7 and between pH 7 and 9, respectively. PhoK-expressing recombinants were shown to efficiently bioprecipitate U under carbonate-abundant conditions from alkaline solutions (11, 32). The crystalline precipitate formed by PhoN-/PhoK-expressing cells was identified as uranyl hydrogen phosphate by X-ray diffraction (XRD) analysis (11, 29, 32). Such crystalline aggregates of uranyl phosphate can be easily visualized by electron microscopy (11, 31).
The present study exploits the use of transmission electron microscopy to determine the effect of aqueous uranyl species predominant under two different geochemical conditions (GC) on the spatial distribution of the uranyl phosphate precipitate formed around D. radiodurans and E. coli cell surfaces. The information generated dissects the nature of interaction of U with bacterial cell surfaces under the given conditions and further indicates how these factors affect metal toxicity.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and expression of phosphatases.
The bacterial strains used in this study are listed in Table 1. Deinococcus radiodurans strain R1 was grown aerobically in TGY (1% Bacto tryptone, 0.1% glucose, and 0.5% yeast extract) liquid medium at 32°C ± 1°C with agitation (180 ± 5 rpm). E. coli DH5α cells were grown in Luria-Bertani (LB) medium at 37°C ± 1°C under agitation (180 ± 5 rpm). The antibiotic concentrations used for recombinant strains were 100 μg/ml of carbenicillin for E. coli and 3 μg/ml of chloramphenicol for D. radiodurans. Screening for the phosphatase-positive colonies was carried out on LB or TGY agar medium supplemented with 1 mg/ml phenolphthalein diphosphate (PDP) and methyl green (MG) at 50 μg/ml for E. coli or at 5 μg/ml for D. radiodurans transformants.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli DH5α | F− recA41 endA1 gyrA96 thi-1 hsdr17(rK− mK+) supE44 relA λlacU169 | Lab collection |
| D. radiodurans R1 | Wild-type strain | Lab collection |
| E. coli(pPN1) | E. coli DH5α containing plasmid pPN1 (phoN ORF, GenBank accession no. X59036, in plasmid pRAD1 under PgroESL promoter control)a | Appukutan et al. (30) |
| E. coli(pK1) | E. coli DH5α containing plasmid construct pK1 (phoK ORF, GenBank accession no. EF143994, in plasmid pRAD1 under PgroESL promoter control) | Nilgiriwala et al. (32); Kulkarni et al. (11) |
| E. coli(pRAD1) | E. coli DH5α containing shuttle vector plasmid pRAD1 | Kulkarni et al. (11) |
| D. radiodurans(pPN1) | D. radiodurans R1 containing plasmid construct pPN1 | Appukutan et al. (30) |
| D. radiodurans(pK1) | D. radiodurans R1 containing plasmid construct pK1 | Kulkarni et al. (11) |
| D. radiodurans(pRAD1) | D. radiodurans R1 containing shuttle vector plasmid pRAD1 | Appukutan et al. (30); Kulkarni et al. (11) |
ORF, open reading frame.
Determination of phosphatase activities.
PhoK and PhoN activities of the recombinant cells were determined in terms of Pi release from β-glycerophosphate (β-GP) substrate. Briefly, D. radiodurans(pK1), D. radiodurans(pPN1), and D. radiodurans(pRAD1) (Table 1) cells (optical density at 600 nm [OD600], ∼0.1) were suspended in 50 mM MOPS (morpholinepropanesulfonic acid) buffer (1 ml) either at pH 6.8 or 9, and 200 μl of 50 mM β-GP substrate was added, followed by incubation at 37°C for 30 min. Phosphate released in the supernatant was spectrophotometrically measured by the phosphomolybdic acid method (33). Protein concentration in equivalent cells was estimated by using Sigma total protein kit (Sigma-Aldrich, USA), based on Peterson's modification of the micro-Lowry method (34). Phosphatase activity was expressed as nanomoles of Pi released per minute per milligram of total cellular protein.
Uranium precipitation/biosorption assays.
Uranyl nitrate solution was prepared as a 100 mM stock solution by dissolving UO2(NO3)2·6H2O (Merck India Ltd.) in double-distilled water. To simulate a carbonate-dominated geochemical condition, a stock solution composed predominantly of uranyl carbonate complexes was prepared by the addition of a 1/10th volume of saturated ammonium carbonate to a 100 mM uranyl nitrate hexahydrate stock solution (final U concentration, 89 mM; carbonate concentration, 214 mM) (32, 35, 36). The formation of carbonate complexes with U was verified by monitoring the absorption spectra of solution in the visible light range between 400 and 500 nm (UV Unicam 300; Thermo Scientific) with specific peaks at 434 nm, 448 nm, and 464 nm (35, 36). U precipitation assays were carried out with PhoK/PhoN-expressing E. coli or D. radiodurans cells (OD600, ∼1) under the following two defined conditions: (i) geochemical condition 1 (GC 1) (carbonate-deficient condition; 10 mM MOPS [pH 7.0] with uranyl nitrate solution; final pH, 6.8), and (ii) geochemical condition 2 (GC 2) (carbonate-abundant condition; 10 mM MOPS [pH 9] and U solution prepared with ammonium carbonate, as explained above; final pH, 9.0). The concentration of carbonate in GC2 in all assays was always 2.4-fold higher than the U concentration used; i.e., the carbonate-to-U ratio of 2.4 was always maintained. The pH conditions (6.8 and 9) were optimized for the optimal phosphatase activity for U precipitation. The bioprecipitation assays were carried out at a U concentration of 1 mM and the sodium salt of β-glycerophosphate at a 5 mM concentration. The reaction mixture was subsequently kept at 30°C under static aerobic conditions for 18 h. The recombinant cells which carried the empty vector (i.e., pRAD1 lacking the phoN or phoK gene) were used as a control and were incubated with U under conditions similar to those mentioned above. The inability of these control cells to precipitate U confirmed the role of PhoK/PhoK in U precipitation and also ruled out spontaneous metal precipitation. An abiotic control (lacking cells) was also included. Aliquots (1.5 ml) were taken at different time intervals and subjected to centrifugation at 12,000 × g for 10 min. Residual U in the supernatant or U present in the pellet was estimated using Arsenazo III reagent, as described earlier (37). For biosorption experiments, the wild-type D. radiodurans or E. coli (DH5α) cells (OD600, ∼1) were suspended in 10 mM MOPS buffer for 10 min under conditions similar to those of GC1 and GC2 at U concentrations of 50, 100, 200, and 1,000 μM. In GC2, the carbonate concentration was 2.4-fold higher than that of U. The dry weight corresponding to the optical density of cells used in each experiment was determined and used to calculate the milligrams of U biosorbed or precipitated per gram (dry weight) of cells.
Transmission electron microscopy and X-ray diffraction analysis.
PhoN- or PhoK-expressing cells which had precipitated U or control cells were washed twice with 50 mM cacodylate buffer (pH 7.4) and fixed in a solution (2.5% glutaraldehyde and 0.5% para-formaldehyde) overnight at 4°C. Following three washes with cacodylate buffer, cells were embedded in 2% noble agar and dehydrated in a graded series of ethanol (30, 60, 75, 90, and 100%). After the removal of ethanol by treatment with propylene oxide, blocks were subsequently infiltrated with Spurr resin and propylene oxide (Sigma Aldrich) in ratios of 1:3, 1:1, and 3:1 (vol/vol) for 2 h in each case. The samples were finally infiltrated with Spurr resin for 16 h and then embedded by incubation at 60°C for 72 h. Thin sections of samples were prepared with an ultramicrotome (Leica, Germany), placed on 200-mesh Formvar-coated copper grids, and viewed with the Libra 120 Plus TEM (Carl Zeiss). Both stained (with 1.5% uranyl acetate) and unstained samples were viewed. Altogether, 121 fields were observed, and the results have been reported for observations seen in more than 85% of the fields visualized for each sample.
For X-ray diffraction analysis (XRD), the cells of D. radiodurans(pK1) and D. radiodurans(pPN1) after U precipitation were dried in an oven at 80°C for 4 h. The dried pellet was scraped and crushed into a fine powder. The powder was subjected to X-ray diffraction analysis using a high-precision Rigaku R-Axis D-max powder diffractometer using monochromatic Cu-Kα radiation (Solid State Physics Division [SSPD], Bhabha Atomic Research Centre [BARC], India). The diffraction pattern was recorded at 2θ, from 5° to 70°, with a step length of 0.02. The diffraction pattern obtained was compared to known standards in the International Centre for Diffraction Data (ICDD) database.
Uranium toxicity experiments.
E. coli or D. radiodurans was grown in LB or TGY liquid medium, respectively, until late-exponential phase of growth. Cells were washed twice with double-distilled water and suspended (OD600, ∼1) in 10 mM MOPS buffer, supplemented with U at 0 to 2 mM under GC1 or at 0 to 25 mM under GC2 for 4 h. Such U-exposed cells were washed free of uranium-containing medium, spotted (10 μl) on corresponding agar plates for the visual examination of growth or plated on agar plates to determine CFU, and growth after incubation of 24 h was recorded. In another set of experiments, cells (under resting conditions) were exposed to U, as described earlier, and such U-exposed cells (OD600, ∼0.5) were then inoculated into LB or TGY liquid broth medium with agitation (150 ± 5 rpm) or plated on LB or TGY agar plates. Growth was assessed by measuring the optical density at 600 nm (OD600) or by determining the CFU on agar plates (1.5% Bacto agar) after 24 h of incubation at 37°C for E. coli and after 48 h of incubation at 32°C for D. radiodurans.
Statistical analysis and thermodynamic modeling.
Each experiment was repeated three times. The reported values are the means of the results from three replicates from a representative experiment, and the error bars correspond to 95% confidence intervals. Aqueous U speciation under GC1 or GC2 was determined using Visual MINTEQ version 3.1 (http://vminteq.lwr.kth.se/download/) (9, 17).
RESULTS
PhoN is a cell-associated phosphatase, whereas PhoK is secreted extracellularly.
On histochemical (PDP-MG) plates (38) the recombinant cells expressing PhoN or PhoK appeared as intense green spots, whereas the control cells (carrying the empty vector pRAD1) showed no such color (Fig. 1A and B). Compared to the D. radiodurans cells expressing PhoN, PhoK-bearing strains formed darker green spots due to the higher specific activity of the phosphatase enzyme. In recombinants expressing PhoK (but not PhoN), a green halo extended outside the zone of growth into the medium, confirming the extracellular secretion of the PhoK enzyme reported earlier (Fig. 1A and B) (32). The spent medium from cultures was separated by centrifugation and spotted on PDP-MG plates. Unlike the spent medium of D. radiodurans(pPN1) or E. coli(pPN1) cells, the spent medium of D. radiodurans(pK1) (Fig. 1C) or E. coli(pK1) (Fig. 1D) cells showed intense green color, demonstrating extracellular secretion of the PhoK enzyme.
FIG 1.
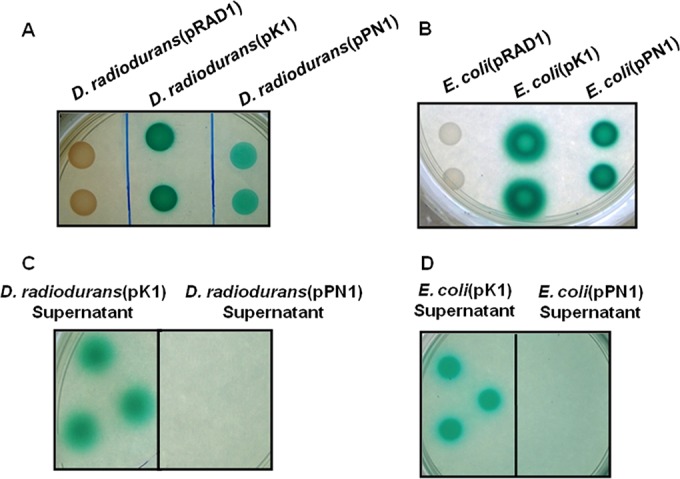
Phosphatase expression and secretion. Spotting of recombinant D. radiodurans (A) and E. coli (B) cells (10 μl; OD600, ∼1) to assess expression and activity of phosphatases, or of spent medium (10 μl) to identify extracellular secretion of PhoK in D. radiodurans (C) and E. coli (D) on modified histochemical plate containing PDP-MG (pH 7.0), was performed.
D. radiodurans(pPN1) cells show cell surface-associated precipitation under GC1, while D. radiodurans(pK1) cells exhibit extracellular precipitation under GC2.
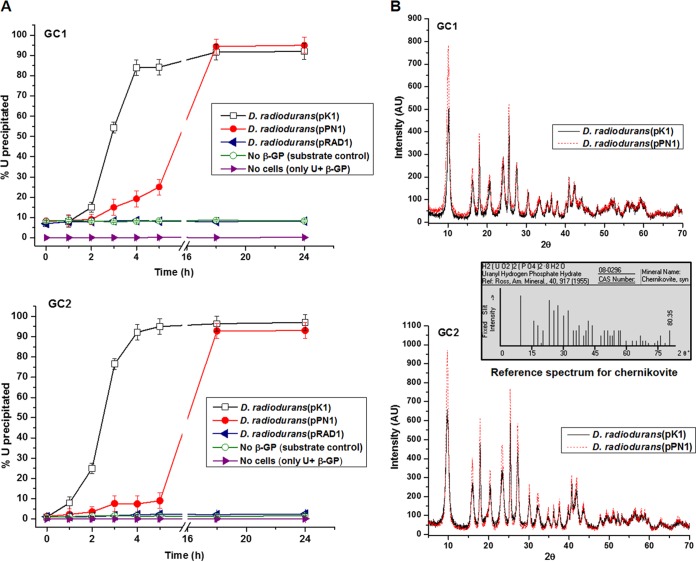
D. radiodurans(pPN1) and D. radiodurans(pK1) cells were used in U precipitation assays under GC1 and GC2, respectively. The GCs were selected for optimal U precipitation by the two enzymes employed, i.e., final pH 6.8 for the acid phosphatase PhoN, and pH 9.0 for the alkaline phosphatase PhoK. As reported earlier, the PhoK is a much higher-specific-activity enzyme than PhoN (at their respective pH optima) (11). D. radiodurans(pPN1) cells precipitated ∼25% U in 5 h under GC1 but only 15% under GC2 over the same time period (Fig. 2A), in accordance with pH dependence of its activity (Table 2). U precipitation by the D. radiodurans(pK1) cells was very rapid in both GCs, and more than 80% of U was precipitated by 5 h. However, when incubated for 18 h, both strains showed near-complete removal of U under both GC1 and GC2 (Fig. 2A). No detectable spontaneous precipitation of uranium occurred in the abiotic control (lacking cells) solution under either GC. Under both GCs, the D. radiodurans(pK1) cells showed a higher rate of phosphate hydrolysis than D. radiodurans(pPN1), commensurate with the higher specific activity of D. radiodurans(pPN1) (Table 2), thus accounting for the higher rate of U precipitation shown by the PhoK-expressing cells. In the D. radiodurans control cells (carrying pRAD1 alone), around 8.5% U was removed from the solution under GC1, and about 2.2% was removed under GC2 (Fig. 2A). XRD analysis identified that the U precipitate formed under all experimental conditions was chernikovite, a uranyl hydrogen phosphate, H2(UO2)2(PO4)2·8H2O (Fig. 2B).
FIG 2.
Uranium bioprecipitation using D. radiodurans and confirmation of precipitate as uranyl hydrogen phosphate under GC1 or GC2. (A) Cells (OD600, ∼1) of D. radiodurans(pRAD1), D. radiodurans(pPN1), or D. radiodurans(pK1) were used for U bioprecipitation. U removal kinetics was studied using cells incubated in 1 mM U with 5 mM β-glycerophosphate, and the percent metal removed from the supernatant is shown. (B) XRD spectra of cells incubated with 5 mM U and 10 mM β-glycerophosphate for 18 h. The reference spectrum for chernikovite is also included.
TABLE 2.
Hydrolysis of β-glycerophosphate by recombinant Deinococcus radiodurans strains
| Cells used | Pi release from β-GP (nmol/min/mg of total cellular protein)a |
|
|---|---|---|
| pH 6.8 | pH 9 | |
| D. radiodurans(pK1) | 270.6 ± 2.5 | 931.8 ± 5 |
| D. radiodurans(pPN1) | 32.1 ± 0.5 | 6.0 ± 0.5 |
| D. radiodurans(pRAD1) | 2.2 ± 0.1 | 0.49 ± 0.05 |
Pi release was measured in the buffer supernatant. Values reported are means with standard errors.
In the control cells [D. radiodurans(pRAD1)], where no precipitation was observed, the cell pellet displayed a pinkish color typical of D. radiodurans under both GCs (Fig. 3A and B). In D. radiodurans(pPN1) the cell pellet was a mix of pink (cells) and yellow (precipitate) (Fig. 3C). Interestingly, D. radiodurans(pK1) cells displayed a distinct yellowish precipitate of U trailing the pink cell pellet (Fig. 3D).
FIG 3.
Localization of U precipitate in recombinant D. radiodurans cells after U precipitation. D. radiodurans(pRAD1), D. radiodurans(pPN1), or D. radiodurans(pK1) cells were used for bioprecipitation assay of U (1 mM) under GC1 or GC2. After incubation for 18 h, the cells were either subjected to centrifugation to visualize the pellet or processed for TEM. In panels A, B, and D, electron micrographs from uranyl acetate-stained samples are shown. The U precipitate is indicated by arrows.
Both the geochemical results and the TEM analysis showed that U did not precipitate from solution in the control under either of the GCs, clearly establishing the phosphatase enzyme-mediated precipitation of U (Fig. 3A and B). D. radiodurans(pPN1) cells under GC1 formed cell surface-bound spicule-like precipitates (Fig. 3C). In contrast, D. radiodurans(pK1) cells under GC2 showed extracellular U precipitation exterior to the cell surface (Fig. 3D).
Uranyl phosphate precipitate is mostly cell bound under GC1, whereas under GC2, it is extracellular.
To ascertain if the aqueous uranyl species predominant under a particular geochemical condition indeed govern the precipitate location, the assay conditions were reversed. D. radiodurans(pPN1) cells were incubated with U under GC2, whereas D. radiodurans(pK1) cells were incubated with U under GC1. Visual examination of the cell pellet color after precipitation indicated that regardless of the phosphatase enzyme employed or its location, under GC1, the entire cell pellet turned yellowish (Fig. 3E), while under GC2, a yellow streak of uranyl phosphate trailed the pink cell pellet (Fig. 3D and F). TEM images of D. radiodurans(pK1) cells clearly showed cell surface-bound crystals under GC1, along with sparse presence of extracellular precipitate (Fig. 3E). In contrast, D. radiodurans(pPN1) cells displayed distinct extracellular uranyl phosphate crystals, with little cell surface-associated precipitate under GC2 (Fig. 3F).
The nature of the aqueous uranyl species predominant under a particular GC determines the eventual localization of U precipitate in E. coli.
Similar experiments performed with E. coli expressing PhoK or PhoN (Fig. 4) confirmed the relationship between aqueous U speciation and localization of U precipitate to be in accordance with the results obtained in D. radiodurans (Fig. 3). In the E. coli(pRAD1) control cells, no precipitate was observed (Fig. 4A and B). Regardless of the phosphatase employed for precipitation, under GC1, uranyl phosphate precipitate was cell associated (Fig. 4A), while under GC2, the precipitate was extracellular (Fig. 4B). In E. coli(pK1) cells under GC1, the precipitate was evenly bound all over the cell surface (Fig. 4A), whereas in E. coli(pPN1) cells, the precipitate was located in the periplasmic space at the polar ends (Fig. 4A).
FIG 4.
TEM analysis of recombinant E. coli cells after U precipitation. Cells (OD600, ∼1) of all three strains [E. coli(pRAD1), E. coli(pPN1), and E. coli(pK1)] were used in U (1 mM) precipitation assays for 18 h under GC1 (A) or GC2 (B) and processed for TEM. Arrows show the locations of uranyl phosphate precipitate.
Amount of uranium adsorbed by bacterial cells is higher under GC1 than GC2.
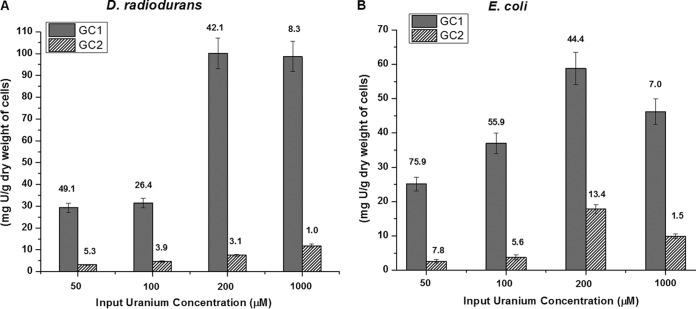
The amount of U adsorbed onto the wild-type bacterial cells lacking the PhoN/PhoK enzymes was monitored under either GC1 or GC2. At all the concentrations of U employed, both organisms showed higher biosorption of U under GC1 than under GC2. For instance, at a concentration of 1 mM input U under GC1, D. radiodurans adsorbed 98 mg of U/g (dry weight) of cells (i.e., 8.3% of the input U), whereas E. coli cells showed biosorption of 46 mg of U/g (dry weight) of cells (7% of the input U) (Fig. 5A and B). In contrast, both organisms adsorbed only 8 to 12 mg of U/g (dry weight) of cells (1 to 2% of the input U) under GC2 (Fig. 5A and B). Geochemical modeling studies using the MINTEQ software showed that under GC1, U largely formed positively charged uranium hydroxide complexes, while under GC2, the negatively charged uranyl carbonate-hydroxide complexes predominated (Table 3). Thus, greater biosorption of U occurs under GC1, where the positively charged hydroxide complexes predominate, rather than under GC2, where the negatively charged carbonate-hydroxide complexes of U are prevalent.
FIG 5.
Biosorption of U by D. radiodurans and E. coli. Wild-type E. coli or D. radiodurans cells (OD600, ∼1) were suspended in 10 mM MOPS buffer containing 50, 100, 200, or 1,000 μM U under either GC1 or GC2. Under GC2, the carbonate concentration was always 2.4 times higher than the U concentration used. The amount of U biosorbed per gram (dry weight) of cells is reported. The percent U removed is indicated by numbers above the respective bars.
TABLE 3.
U speciation in GC1 and GC2, as predicted by MINTEQ modelinga
| Component | GC1 |
GC2 |
||
|---|---|---|---|---|
| % of total concn (pH 6.8) | Species name | % of total concn (pH 9.0) | Species name | |
| UO22+ | 0.145 | UO2OH+ | 63.088 | UO2(CO3)34− |
| 0.035 | (UO2)2(OH)22+ | 26.529 | (UO2)3(OH)7− | |
| 51.899 | (UO2)2(OH)5+ | 4.252 | UO2(CO3)22− | |
| 47.742 | (UO2)4(OH)7+ | 1.741 | (UO2)4(OH)7+ | |
| 0.047 | (UO2)3(OH)7− | 1.249 | (UO2)3(OH)5+ | |
| 0.043 | (UO2)3(OH)42+ | 1.516 | (UO2)(OH)3− | |
| 0.109 | UO2(OH)2 (aq)b | 0.164 | (UO2)(OH)2 (aq) | |
| 0.032 | UO2(CO3) (aq) | |||
| NO31− | 100 | NO31− | ||
| CO32− | 1.169 | CO32− | ||
| 16.28 | HCO3− | |||
| 0.033 | H2(CO3) (aq) | |||
| 78.86 | UO2(CO3)34− | |||
| 0.013 | UO2(CO3)3 (aq) | |||
| 3.544 | UO2(CO3)32− | |||
| 0.062 | NaCO3− (aq) | |||
| NH41+ | 67.012 | NH41+ | ||
| 32.98 | NH3 (aq) | |||
| MOPS | 30.319 | MOPS | 98.652 | MOPS |
| 69.681 | H-MOPS (aq) | 1.348 | H-MOPS (aq) | |
| Na1+ | 99.974 | Na1+ | 99.974 | Na1+ |
| 0.022 | NaNO3 (aq) | 0.015 | NaHCO3(aq) | |
| Gly-2-phosphate | 66.77 | Gly-2-phosphate | 99.70 | Gly-2-phosphate |
| 33.23 | H-Gly-phosphate | 0.295 | H-Gly-phosphate | |
GC1, 1 mM uranyl nitrate hexahydrate, 5 mM β-glycerophosphate (sodium salt), and 10 mM MOPS (pH 6.8); GC2, 1 mM uranyl nitratehexahydrate, 5 mM β-glycerophosphate (sodium salt), 2.4 mM ammonium carbonate, and 10 mM MOPS (pH 9).
aq, aqueous.
Complexation of uranyl ions at alkaline pH decreases their toxicity to bacteria.
Exposure to 1.5 mM U under GC1 (pH 6.8) caused severe loss in cell viability in both D. radiodurans and E. coli cells, as visualized by a spot experiment (Fig. 6A). In contrast, the survival of cells remained largely unaffected even at 20 mM U under GC2 (Fig. 6A). In both D. radiodurans and E. coli, only 10% survival (determined by measuring CFU) was obtained compared to the control (cells not exposed to U) when incubated (on nutrient agar medium) under GC1 at 1.25 mM U, whereas under GC2, growth was unaffected even at 20 mM U (Fig. 6A). In a separate experiment, D. radiodurans and E. coli cells were exposed to 2 mM U under GC1 or 20 mM U under GC2 for 4 h and then inoculated (OD600, ∼0.5) in liquid nutrient medium without U. Growth of cells preincubated in U under GC2 remained unaltered compared to the control cells. On the other hand, cells that were preincubated in 2 mM U under GC1 did not show growth (Fig. 6B).
FIG 6.
Uranium sensitivity of D. radiodurans and E. coli. Pregrown wild-type D. radiodurans or E. coli cells were suspended in U under either GC1 or GC2 for 4 h under sterile conditions. In GC2, the carbonate concentration was 2.4 times higher than the U concentration used. The uranium-exposed cells were either spotted (10 μl) on LB or TGY agar plates for visual examination of growth or plated on agar plates to determine CFU (A) or inoculated into liquid broth medium (at OD600 of 0.5) and grown for 18 h (B). The CFU obtained after plating are indicated in parentheses.
DISCUSSION
Bacterial phosphatases can be efficiently expressed in appropriate host systems and effectively utilized to precipitate heavy metals, such as U, from effluents. Earlier, our laboratory expressed two different phosphatases (PhoK/PhoN) in E. coli and D. radiodurans and demonstrated the U removal potential of these engineered recombinant strains (11, 29–32) from solutions. PhoN-expressing cells (in GC1) formed cell-bound uranyl phosphate precipitates (Fig. 3), whereas PhoK-expressing cells (in GC2) generated extracellular uranyl phosphate precipitates, in conformity with the known cellular localization of the two enzymes (11, 29, 30, 32) (Fig. 3). This was substantiated by both TEM analysis and the nature of the cell pellet formed upon centrifugation of the assay mixture. The two-phase separation of the pellet in D. radiodurans(pK1) cells under GC2 resulted from extracellular U precipitation that was not associated with cells, while a more uniform mixture of cells (pink) and precipitate (yellow) was observed in D. radiodurans(pPN1) cells under GC1, indicating the presence of cell-associated precipitate.
A possible explanation for the differential localization of the U precipitate appeared to be the difference in the location of PhoN and PhoK enzymes. In addition to being cell associated, PhoK was secreted extracellularly by both D. radiodurans and E. coli, whereas PhoN remained solely contained within cells of both bacteria (Fig. 1). Alternatively, localization of the precipitate may be governed by uranium speciation. MINTEQ modeling studies showed that U mostly formed positively charged complexes under GC1, while the uranyl carbonate-hydroxide complexes formed under GC2 were predominantly negatively charged (Table 3). Therefore, the present study examined the possible effect of differentially charged aqueous U complexes on precipitate localization under the two GCs employed. The results showed that under GC1, the precipitate was largely cell associated, whereas under GC2, the precipitate was extracellular, with little cell surface association (Fig. 3 and 4). XRD results showed that the precipitated U phase was identical (chernikovite) under both GCs employed (Fig. 2B). Thus, the location of the enzymes had little role to play in determining whether the precipitate would be cell associated or extracellular.
Phosphatase assays, in the absence of uranium, show that the liberated phosphate is released into the supernatant buffer at pH 6.8 and 9 (Table 2). Bacterial cells usually carry a net negative surface charge at neutral pH (39, 40). With an increase in pH, the negative charge increases due to the increasing deprotonation of the functional groups found on the cell surface. In this study, the differential localization of the precipitate appears to be governed by the charge-dependent interaction of aqueous U species with the bacterial cell surface. The results indicate that the location of uranyl phosphate precipitate might be a consequence of the earlier event of the differential biosorption of U under the two different geochemical conditions. At pH 6.8 (GC1, in the absence of excess carbonate), U is adsorbed onto the bacterial cell surface, perhaps aided by the positive charge on the uranyl hydroxide aqueous species, which MINTEQ modeling suggests to be most prevalent. This initial complexation on the cell surface forms the nucleation sites, which are further consolidated by the codeposition of more incoming metal with the outgoing released inorganic phosphate (14, 40, 41), resulting in a build-up of polycrystalline metal phosphate precipitate on the cell surface (Fig. 3 and 4). At pH 9 (GC2, under carbonate-abundant conditions), MINTEQ modeling indicated U to be predominantly present as negatively charged uranyl carbonate-hydroxide aqueous complexes. It is difficult to distinguish the decreased biosorption and bioavailability of U due to (i) increasing carbonate concentration in the medium from the (ii) repulsion of negatively charged U species from the cell surfaces. However, the low level of U adsorbed onto the cell surface seems to be insufficient to consolidate the nucleation sites required for cell-associated precipitation. This probably leaves the negatively charged uranyl complexes free in the solution, resulting in extracellular precipitation of U upon encountering the phosphate released in the supernatant buffer either by PhoN or PhoK.
While the gross localization pattern of the precipitate seemed to be largely decided by the GC, subtle variations, apparently determined by the enzyme localization, were also observed. The periplasmic nonspecific acid phosphatases are known to concentrate at cell poles in Gram-negative bacteria (41–43). In accordance with this, the E. coli(pPN1) cells under GC1 displayed a periplasmic location of the precipitate at the poles (within the cells). The abundance of PhoN at the poles may result in localized sites of high phosphate concentration, leading to periplasmic accumulation of precipitate in E. coli (Fig. 4). This is unlike the exocellular (but still cell surface-associated) location of the precipitate in E. coli(pK1) under GC1, where the enzyme is released extracellularly and the precipitate is formed with the sorbed U on the external surface of the cell boundary. Deinococcus has a multilayered cell wall with a complex architecture (44), wherein the periplasm is not clearly defined. Further, the U hydroxyl species may not be able to permeate several layers of the cell envelope for uranyl hydrogen phosphate to be precipitated inside the cell wall. Thus, while geochemical conditions play a major role in determining the precipitate localization (i.e., cell associated and extracellular), finer variations may occur due to the different physiological conditions observed in the two bacteria.
The differential toxicity of U under the two geochemical conditions can also be attributed to the above-mentioned phenomenon. Binding of U to bacterial cell surfaces is evidently the first step toward obtaining cellular access, which subsequently causes the disruption of metabolic processes, eventually leading to lethality (9, 17). The predominantly negatively charged aqueous complexes of U formed under GC2 are repelled by the bacterial cell surface, making it difficult for U to acquire sufficient proximity to cells to cause significant damage. The presence of excess carbonate under such conditions may further decrease U availability to cells. On the other hand, the predominantly positively charged aqueous U species formed under GC1 would allow U to interact with cells, resulting in higher toxicity, as evidenced in this study (Fig. 6A and B). Also, increasing concentrations of uranium in GC1 lead to increased toxicity; however, there was no threshold concentration of uranium at which toxicity occurred in GC2 up to 25 mM U. Very likely, this was because under GC2, the carbonate concentration at each U concentration increased proportionally with the uranium concentration, thus continually limiting the amount of biosorption and, consequently, toxicity. These results also substantiate earlier reports that U biosorption decreases with increasing pH and increasing carbonate/bicarbonate concentration due to the higher degree of complexation of uranyl ion by hydroxides and carbonates (9, 18, 45). It has also been shown that increasing pH and higher bicarbonate/carbonate concentration exert lower toxicity (46–48), as in the presence of high carbonate concentration, bioaccumulation of U is reduced in bacteria, consequently leading to increased U tolerance (17, 18, 49).
The present study has provided illustrative insights into the interaction of U complexes with bacterial surfaces by imaging the location of the uranyl phosphate precipitates. Using recombinant bacterial strains expressing phosphatases, this work demonstrates the effect of aqueous speciation of U on its interaction with cell surfaces and on the eventual cellular/extracellular localization of the precipitated uranyl phosphate. The efficacy of recombinant strains expressing PhoN and PhoK in uranium removal from solutions has already been established, with the D. radiodurans(pK1) strain being much more efficient (11, 23, 29–32). This study shows that, unlike with biosorption (which has limited capacity and is often reversible) or bioaccumulation (which depends on metabolic activity and toxicity of metal), U species can be efficiently precipitated and removed from effluent solution by employing phosphatases. The cell-associated precipitation of metal has the advantage of easy downstream processing by simple gravity-based settling of metal-loaded cells, compared to cumbersome separation techniques (22, 41). With this notion and in light of the fact that localization of the uranium precipitate is determined primarily by aqueous uranium speciation, the results from this study are of relevance to effluent treatment using such cells. D. radiodurans(pK1) is an efficient strain, on account of the high specific activity of PhoK phosphatase over a wider pH range, and can be used under GC1 to obtain cell-associated precipitation. Under GC2, PhoK immobilized in a suitable matrix would be an appropriate choice for metal recovery.
ACKNOWLEDGMENTS
S.K.A. thanks the Department of Science and Technology, India, for the award of the Sir J. C. Bose National Fellowship and the Department of Atomic Energy for the award of the Raja Ramanna Fellowship. We gratefully acknowledge the XRD analysis of samples by Chitra Radhakrishnan and Rajul R. Choudhury, Solid State Physics Division, BARC, for the XRD analysis of samples and Namrata Waghmare for providing technical help in TEM analysis.
REFERENCES
- 1.Converse BJ, Wu T, Findlay RH, Roden EE. 2013. U(VI) reduction in sulfate-reducing subsurface sediments amended with ethanol or acetate. Appl Environ Microbiol 79:4173–4177. doi: 10.1128/AEM.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlett M, Moon HS, Peacock AA, Hedrick DB, Williams KH, Long PE, Lovley D, Jaffe PR. 2012. Uranium reduction and microbial community development in response to stimulation with different electron donors. Biodegradation 23:535–546. doi: 10.1007/s10532-011-9531-8. [DOI] [PubMed] [Google Scholar]
- 3.Boonchayaanant B, Nayak D, Du X, Criddle CS. 2009. Uranium reduction and resistance to reoxidation under iron-reducing and sulfate-reducing conditions. Water Res 43:4652–4664. doi: 10.1016/j.watres.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Merroun ML, Nedelkova M, Ojeda JJ, Reitz T, Fernández ML, Arias JM, Romero-González M, Selenska-Pobell S. 2011. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses. J Hazard Mater 197:1–10. doi: 10.1016/j.jhazmat.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary S, Sar P. 2011. Uranium biomineralization by a metal resistant Pseudomonas aeruginosa strain isolated from contaminated mine waste. J Hazard Mater 186:336–343. doi: 10.1016/j.jhazmat.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Acharya C, Chandwadkar P, Apte SK. 2012. Interaction of uranium with a filamentous, heterocystous, nitrogen-fixing cyanobacterium, Anabaena torulosa. Bioresour Technol 116:290–294. doi: 10.1016/j.biortech.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 7.Akhtar K, Waheed Akhtar M, Khalid AM. 2007. Removal and recovery of uranium from aqueous solutions by Trichoderma harzianum. Water Res 41:1366–1378. doi: 10.1016/j.watres.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Akhtar K, Khalid AM, Akhtar MW, Ghauri MA. 2009. Removal and recovery of uranium from aqueous solutions by Ca-alginate immobilized Trichoderma harzianum. Bioresour Technol 100:4551–4558. doi: 10.1016/j.biortech.2009.03.073. [DOI] [PubMed] [Google Scholar]
- 9.VanEngelen MR, Field EK, Gerlach R, Lee BD, Apel WA, Peyton BM. 2010. UO22+ speciation determines uranium toxicity and bioaccumulation in an environmental Pseudomonas sp. isolate. Environ Toxicol Chem 29:763–769. doi: 10.1002/etc.126. [DOI] [PubMed] [Google Scholar]
- 10.Malekzadeh F, Farazmand A, Ghafourian H, Shahamat M, Levin M, Colwell RR. 2002. Uranium accumulation by a bacterium isolated from electroplating effluent. World J Microbiol Biotechnol 18:295–302. doi: 10.1023/A:1015215718810. [DOI] [Google Scholar]
- 11.Kulkarni S, Ballal A, Apte SK. 2013. Bioprecipitation of uranium from alkaline waste solutions using recombinant Deinococcus radiodurans. J Hazard Mater 262:853–861. doi: 10.1016/j.jhazmat.2013.09.057. [DOI] [PubMed] [Google Scholar]
- 12.Nedelkova M, Merroun ML, Rossberg A, Hennig C, Selenska-Pobell S. 2007. Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. FEMS Microbiol Ecol 59:694–705. doi: 10.1111/j.1574-6941.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- 13.Liang X, Hillier S, Pendlowski H, Gray N, Ceci A, Gadd GM. 2015. Uranium phosphate biomineralization by fungi. Environ Microbiol 17:2064–2075. doi: 10.1111/1462-2920.12771. [DOI] [PubMed] [Google Scholar]
- 14.Newsome L, Morris K, Lloyd JR. 2014. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem Geol 363:164–184. doi: 10.1016/j.chemgeo.2013.10.034. [DOI] [Google Scholar]
- 15.Markich SJ. 2002. Uranium speciation and bioavailability in aquatic systems: an overview. ScientificWorldJournal 2:707–729. doi: 10.1100/tsw.2002.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belli KM, DiChristina TJ, Van Cappellen P, Taillefert M. 2015. Effects of aqueous uranyl speciation on the kinetics of microbial uranium reduction. Geochim Cosmochim Acta 157:109–124. doi: 10.1016/j.gca.2015.02.006. [DOI] [Google Scholar]
- 17.Carvajal DA, Katsenovich YP, Lagos LE. 2012. The effects of aqueous bicarbonate and calcium ions on uranium biosorption by Arthrobacter G975 strain. Chem Geol 330–331:51–59. [Google Scholar]
- 18.Gorman-Lewis D, Elias PE, Fein JB. 2005. Adsorption of aqueous uranyl complexes onto Bacillus subtilis cells. Environ Sci Technol 39:4906–4912. doi: 10.1021/es047957c. [DOI] [PubMed] [Google Scholar]
- 19.Krestou A, Panias D. 2004. Uranium (VI) speciation diagrams in the UO22+/CO32−/H2O system at 25°C. Eur J Miner Process Environ Prot 4:113–129. [Google Scholar]
- 20.Sheng L, Fein JB. 2014. Uranium reduction by Shewanella oneidensis MR-1 as a function of NaHCO3 concentration: surface complexation control of reduction kinetics. Environ Sci Technol 48:3768–3775. doi: 10.1021/es5003692. [DOI] [PubMed] [Google Scholar]
- 21.Macaskie LE, Empson RM, Cheetham AK, Grey CP, Skarnulis AJ. 1992. Uranium bioaccumulation by a Citrobacter sp. as a result of enzymically mediated growth of polycrystalline HUO2PO4. Science 257:782–784. doi: 10.1126/science.1496397. [DOI] [PubMed] [Google Scholar]
- 22.Macaskie LE, Jeong BC, Tolley MR. 1994. Enzymatically accelerated biomineralization of heavy metals: application to the removal of americium and plutonium from aqueous flows. FEMS Microbiol Rev 14:351–367. doi: 10.1111/j.1574-6976.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 23.Seetharam C, Soundarajan S, Udas AC, Rao AS, Apte SK. 2009. Lyophilized, non-viable, recombinant E. coli cells for cadmium bioprecipitation and recovery. Process Biochem 44:246–250. doi: 10.1016/j.procbio.2008.10.015. [DOI] [Google Scholar]
- 24.du Plessis EM, Theron J, Joubert L, Lotter T, Watson TG. 2002. Characterization of a phosphatase secreted by Staphylococcus aureus strain 154, a new member of the bacterial class C family of nonspecific acid phosphatases. Syst Appl Microbiol 25:21–30. doi: 10.1078/0723-2020-00098. [DOI] [PubMed] [Google Scholar]
- 25.Vincent JB, Crowder MW, Averill BA. 1992. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci 17:105–110. doi: 10.1016/0968-0004(92)90246-6. [DOI] [PubMed] [Google Scholar]
- 26.Gandhi NU, Chandra SB. 2012. A comparative analysis of three classes of bacterial non-specific acid phosphatases and archaeal phosphoesterases: evolutionary perspective. Acta Inform Med 20:167–173. doi: 10.5455/aim.2012.20.167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beazley MJ, Martinez RJ, Sobecky PA, Webb SM, Taillefert M. 2007. Uranium biomineralization as a result of bacterial phosphatase activity: insights from bacterial isolates from a contaminated subsurface. Environ Sci Technol 41:5701–5707. doi: 10.1021/es070567g. [DOI] [PubMed] [Google Scholar]
- 28.Benzerara K, Miot J, Morin G, Ona-Nguema G, Skouri-Panet F, Férard C. 2011. Significance, mechanisms and environmental implications of microbial biomineralization. C R Geosci 343:160–167. doi: 10.1016/j.crte.2010.09.002. [DOI] [Google Scholar]
- 29.Misra CS, Appukuttan D, Kantamreddi VSS, Rao AS, Apte SK. 2012. Recombinant D. radiodurans cells for bioremediation of heavy metals from acidic/neutral aqueous wastes. Bioeng Bugs 3:44–48. doi: 10.4161/bbug.3.1.18878. [DOI] [PubMed] [Google Scholar]
- 30.Appukuttan D, Rao AS, Apte SK. 2006. Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from dilute nuclear waste. Appl Environ Microbiol 72:7873–7878. doi: 10.1128/AEM.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appukuttan D, Seetharam C, Padma N, Rao AS, Apte SK. 2011. PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation. J Biotechnol 154:285–290. doi: 10.1016/j.jbiotec.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Nilgiriwala KS, Alahari A, Rao AS, Apte SK. 2008. Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl Environ Microbiol 74:5516–5523. doi: 10.1128/AEM.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames BN. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8:115–118. [Google Scholar]
- 34.Peterson GL. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 35.Francis A, Dodge C, Gillow J, Papenguth H. 2000. Biotransformation of uranium compounds in high ionic strength brine by a halophilic bacterium under denitrifying conditions. Environ Sci Technol 34:2311–2317. doi: 10.1021/es991251e. [DOI] [Google Scholar]
- 36.Acharya C, Joseph D, Apte S. 2009. Uranium sequestration by a marine cyanobacterium, Synechococcus elongatus strain BDU/75042. Bioresour Technol 100:2176–2181. doi: 10.1016/j.biortech.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 37.Fritz JS, Bradford EC. 1958. Detection of thorium and uranium. Anal Chem 30:1021–1022. doi: 10.1021/ac60138a002. [DOI] [Google Scholar]
- 38.Riccio ML, Rossolini GM, Lombardi G, Chiesurin A, Satta G. 1997. Expression cloning of different bacterial phosphatase-encoding genes by histochemical screening of genomic libraries onto an indicator medium containing phenolphthalein diphosphate and methyl green. J Appl Microbiol 82:177–185. doi: 10.1111/j.1365-2672.1997.tb02848.x. [DOI] [PubMed] [Google Scholar]
- 39.Van Loosdrecht MC, Lyklema J, Norde W, Schraa G, Zehnder AJ. 1987. Electrophoretic mobility and hydrophobicity as a measured to predict the initial steps of bacterial adhesion. Appl Environ Microbiol 53:1898–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.French S, Puddephatt D, Habash M, Glasauer S. 2013. The dynamic nature of bacterial surfaces: implications for metal-membrane interaction. Crit Rev Microbiol 39:196–217. doi: 10.3109/1040841X.2012.702098. [DOI] [PubMed] [Google Scholar]
- 41.Macaskie LE, Bonthrone KM, Yong P, Goddard DT. 2000. Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 146:1855–1867. doi: 10.1099/00221287-146-8-1855. [DOI] [PubMed] [Google Scholar]
- 42.Jeong BC, Hawes C, Bonthrone KM, Macaskie LE. 1997. Localization of enzymically enhanced heavy metal accumulation by Citrobacter sp. and metal accumulation in vitro by liposomes containing entrapped enzyme. Microbiology 143:2497–2507. doi: 10.1099/00221287-143-7-2497. [DOI] [PubMed] [Google Scholar]
- 43.Nesmeyanova MA, Tsfasman IM, Karamyshev AL, Suzina NE. 1991. Secretion of the overproduced periplasmic PhoA protein into the medium and accumulation of its precursor in phoA-transformed Escherichia coli strains: involvement of outer membrane vesicles. World J Microbiol Biotechnol 7:394–406. doi: 10.1007/BF00329408. [DOI] [PubMed] [Google Scholar]
- 44.Rothfuss H, Lara JC, Schmid AK, Lidstrom ME. 2006. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology 152:2779–2787. doi: 10.1099/mic.0.28971-0. [DOI] [PubMed] [Google Scholar]
- 45.Morcillo F, Gonźalez-Muñoz MT, Reitz T, Romero-Gonźalez ME, Arias JM, Merroun ML. 2014. Biosorption and biomineralization of U(VI) by the marine bacterium Idiomarina loihiensis MAH1: effect of background electrolyte and pH. PLoS One 9:e91305. doi: 10.1371/journal.pone.0091305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sani RK, Peyton BM, Dohnalkova A. 2006. Toxic effects of uranium on Desulfovibrio desulfuricans G20. Environ Toxicol Chem 25:1231–1238. doi: 10.1897/05-401R.1. [DOI] [PubMed] [Google Scholar]
- 47.Sepulveda-Medina PM, Katsenovich YP, Wellman DM, Lagos LE. 2015. The effect of bicarbonate on the microbial dissolution of autunite mineral in the presence of Gram-positive bacteria. J Environ Radioacst 144:77–85. doi: 10.1016/j.jenvrad.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 48.VanEngelen MR, Szilagyi RK, Gerlach R, Lee BD, Apel WA, Peyton BM. 2011. Uranium exerts acute toxicity by binding to pyrroloquinoline quinone cofactor. Environ Sci Technol 45:937–942. doi: 10.1021/es101754x. [DOI] [PubMed] [Google Scholar]
- 49.Fortin C, Dutels L, Garnier-Laplace J. 2004. Uranium complexation and uptake by a green alga in relation to chemical speciation: the importance of the free uranyl ion. Environ Toxicol Chem 23:974–981. doi: 10.1897/03-90. [DOI] [PubMed] [Google Scholar]