Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most malignant tumors and is difficult to diagnose in the early phase. This study was aimed at obtaining the metabolic profiles and characteristic metabolites of pancreatic intraepithelial neoplasia (PanIN) and PDAC tissues from Sprague–Dawley (SD) rats to establish metabonomic methods used in the early diagnosis of PDAC. In the present study, the animal models were established by embedding 7,12‐dimethylbenzanthracene (DMBA) in the pancreas of SD rats to obtain PanIN and PDAC tissues. After the preprocessing of tissues, 1H nuclear magnetic resonance (NMR) spectroscopy combined with multivariate and univariate statistical analysis was applied to identify the potential metabolic signatures and the corresponding metabolic pathways. Pattern recognition models were successfully established and differential metabolites, including glucose, amino acids, carboxylic acids and coenzymes, were screened out. Compared with the control, the trends in the variation of several metabolites were similar in both PanIN and PDAC. Kynurenate and methionine levels were elevated in PanIN but decreased in PDAC, thus, could served as biomarkers to distinguish PanIN from PDAC. Our results suggest that NMR‐based techniques combined with multivariate statistical analysis can distinguish the metabolic differences among PanIN, PDAC and normal tissues, and, therefore, present a promising approach for physiopathologic metabolism investigations and early diagnoses of PDAC.
Keywords: Biomarker, metabonomics, nuclear magnetic resonance, pancreatic cancer, pancreatic intraepithelial neoplasia
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant tumor with an extremely poor prognosis. Patients often have locally advanced unresectable PDAC or metastases when diagnosed. Only 10–20% of cases are resectable and the rate of 5‐year survival is under 5.5%,1 making PDAC the fourth most common cause of death among tumors.2 Given the rapid progress of these tumors, early diagnosis of PDAC could significantly improve the prognosis. Previous study shows that patients with tumors <2 cm in diameter could have a 5‐year survival rate as high as 30–60% while those with tumors <1 cm in diameter could have a 5‐year survival rate of 75%, indicating that the therapeutic effect for patients with early PDAC is far better than for those with advanced lesions.3 Unfortunately, current diagnostic methods lack sensitivity and specificity to detect PDAC in the early stages. CA‐199 is the most commonly used tumor marker in serum and displays a direct correlation with the occurrence and process of the disease.4 However, the sensitivity and specificity of diagnosis is only 59–64% to 69–82% when the dosage is 40–100 μ/mL, respectively.5 Image examinations also have some difficulties in PDAC diagnosis. Endoscopic ultrasonography‐guided fine needle aspiration is an invasive examination that may lead to serious complications, while computed tomography and magnetic resonance imaging often fail to detect tumors under 2 cm in diameter. Therefore, there is a pressing need to develop a new diagnostic approach for early PDAC detection.
Nuclear magnetic resonance (NMR) spectroscopy is a highly sensitive and sophisticated technique used to analyze biochemical samples that can provide a wealth of metabolic information. The metabolic information is helpful in understanding the underlying metabolic mechanisms of diseases and establishing early diagnoses. NMR spectroscopy has been used in detecting the metabolic alterations in various types of cancers, including brain tumors, breast cancer and prostate cancer.6, 7, 8, 9, 10 Due to the fact that magnetic resonance spectroscopy (MRS) has been recognized as a useful diagnostic method for detecting and identifying brain, breast and prostate cancers,11 increasing numbers of researchers have been looking at NMR‐based metabonomic methods as promising in the clinical diagnosis of pancreatic cancer.12, 13, 14 Tesiram et al.15 report that total choline, taurine and glucose plus triglycerides were significantly increased in cancerous serum compared to controls. OuYang et al.16 observe remarkably lower levels of 3‐hydroxybutyrate, 3‐hydroxyisovalerate, lactate and trimethylamine‐N‐oxide and significant higher levels of isoleucine, triglyceride, leucine and creatinine in serum from pancreatic cancer patients compared with controls. Fang et al.17 compare the NMR spectra of pancreatic tissues from rat models of PDAC and controls, and show elevated levels of leucine, isoleucine, valine, alanine, lactate and taurine with decreased levels of betaine in pancreatic tissues of PDAC. These studies suggest the feasibility of using NMR spectroscopy to differentiate cancer from normal or other benign lesions.
Metabonomic analyses of both serum and pancreatic tissues from pancreatic intraepithelial neoplasia (PanIN) and PDAC are important for developing earlier diagnostic methods; however, the metabonomic information from tissues will be more favorable for the establishment of in vivo detection methods, such as 1H MRS, which will target at tumor tissue. Therefore, in this study, we aim to establish the metabolic profiles and characteristic metabolites of PDAC and PanIN, and evaluate the feasibility of NMR spectroscopy in early diagnosis of pancreatic cancer by comparing the NMR‐based metabolic profiling of PanIN, PDAC and normal tissues in rat models.
Materials and Methods
Animal models
The animal experiments were conducted in the Experimental Animals Center of Fujian Medical University. The study protocol was in accordance with the principles of the National Institutes of Health guide for the care and use of Laboratory animals and approved by the Ethics Committee of the Fujian Medical University Union Hospital. Seventy‐five male Sprague–Dawley (SD) rats (4–6 weeks old, weighing 150–170 g, obtained from the experimental animals center of Fujian Medical University, Fuzhou, China) were fed on standard laboratory conditions, and maintained at a temperature of 24 ± 2°C and a relative humidity of 50 ± 10%. SD rats were divided into three groups: experimental group I (n = 30), experimental group II (n = 30) and a control group (n = 15). The rats in the experimental groups were fasted for 12 h prior to the surgery. The modeling started with a 2‐cm transverse incision inferior to the xiphoid. Then, 7 mg of 7,12‐Dimethylbenzanthracene (DMBA; Sigma‐Aldrich, St Louis, MO, USA) was implanted under the capsule of the body and the tail of the pancreas. A purse string suture surrounding the embedding position of DMBA on pancreas was made to prevent leakage of the chemical powder. The incision was sutured and hemorrhages were staunched. Except that the DMBA was replaced by 7 mg of NaCl2, the identical operation was carried out for the control group.
Tissue samples
Three weeks after embedding DMBA, the experimental group was killed while experimental group II and the control group were killed 12 weeks after surgery. During these procedures, all rats were on continuous airway anesthesia (Matrx VMR, MIDMARK, USA) using isoflurane (Jiupai Pharmaceutical, Hubei, China, 2.5% gas concentration, 0.5 L/min). After the rats' death, the pancreatic mass tissues were collected. For PanIN and PDAC tissues, the normal regions (which, visually, were not in accordance with the characteristic of tumorous lesions), were cleared off and confirmed by histological observation. The tissues collected were divided equally into two specimens: one for histological analysis and another for 1H NMR analyses. Then those samples for NMR analysis were quick‐frozen in liquid nitrogen and stored at −80°C.
Histology of pancreatic samples
The samples for histology were fixed in 10% formalin solution for 12 h immediately after death. Paraffin embedding and standard procedures followed, and 4‐μm sections were made and stained with HE. All slices were randomly double‐blind confirmed histologically by experienced pathologists. All PanIN cases (PanIN‐1, 2 and 3) that occurred in experimental I were assigned to the PanIN group and all PDAC cases in experimental II were assigned to the PDAC group.
Sample preprocessing
Tissue samples (300 mg each) were defrosted on ice. The tissue was mixed with 0.6 mL ultrapure water and 1.2mL methanol, and then homogenized for 3 min (MiniBeadbeater‐16; BIO SPEC, Bartlesville, OK, USA) in 7‐mL lap tubes. After the addition of 1.2 mL chloroform and 1.2 mL ultrapure water, the mixture was vortexed for 60 s. After 15 min standing, each sample was centrifuged at 10 397g for 10 min and the supernate was collected. Then, the supernate was lyophilized in vacuum freeze‐drying equipment (LGJ‐10C; Four‐ring Science Instrument Plant, Beijing, China) for 24 h to eliminate water and methanol. For NMR analysis, the lyophilizate was dissolved in 550 μL of 150‐mM deuterated phosphate buffer (NaH2PO4 and K2HPO4, pH 7.4, including 0.1% sodium 3‐[trimethylsilyl] propionate‐2,2,3,3‐d4 [TSP]) followed with a 10‐min centrifugation at 10 000 rpm. The 500‐μL upper layer liquid was transferred into a 5‐mm NMR tube for NMR analysis.
Nuclear magnetic resonance spectroscopy
1H NMR spectroscopy was performed on aqueous extracts in the Varian NMR System (Agilent Technologies, Palo Alto, CA, USA) operating at 500.13 MHz proton frequency at 298 K. For each sample, spectra were acquired using the 1H NOESYPR1D pulse sequence with water suppression: RD‐90°‐t 1‐90°‐t m‐90°‐ACQ. A total of 128 scans with a spectral width of 20 ppm, accompanied with a data point of 32 K, were collected for all spectra. The acquisition time was 1.36 s with a relaxation delay (RD) of 2 s, a fixed interval t 1 of 4 μs and the mixing time of 100 ms.
All free induction decays (FID) were multiplied by an exponential weighting function equivalent to a line‐broadening of 1 Hz to increase the signal‐to‐noise ratio, followed by a Fourier transformation. All spectra were manually corrected for phase and baseline using MestReNova (V9.0; Mestrelab Research, Santiago de Compostela, Galicia, Spain). The chemical shift in spectra was referenced to TSP at δ0.0. The spectral regions δ9.7–0.7 were integrally segmented into discrete regions of 0.002 ppm, and the spectral regions of δ4.74–4.92 and δ3.33–3.39 were removed to eliminate the interference of water and methanol signals for analysis. Then, the integrated data were normalized to a constant sum before multivariate statistical analysis.
Multivariate statistical analysis
Multivariate statistic analysis was performed using the software SIMCA‐P+ (V11.0; Umetrics AB, Umea, Sweden). Principal component analysis (PCA) was implemented using the approach of mean center scaling to simplify the multivariate data into a few principal components, which can highlight intrinsic trends and the existence of outliers.
Partial least squares discriminant analysis (PLS‐DA) and orthogonal partial least squares discriminant analysis (OPLS‐DA) were included in the supervised pattern method to maximize the distinction between different groups in samples. Sequentially, these methods were carried out using Pareto scaling to discover the statistically significant metabolite variations in PDAC and PanIN groups. The results are visualized by using score plots and loading plots to highlight the group clusters and variates, respectively. Then the model coefficients were back‐calculated from the coefficients incorporating the weight of the variables and plotted with color‐coded coefficients to enhance the interpretability of the models. Thus, the metabolic components that contributed to the variation between groups could be displayed and extracted from the color‐coded loading plots. The coefficient plots were color‐coded according to the absolute value of coefficients, with a hot color corresponding to a significant difference between classes and a cool color corresponding to no significant differences. Based on significance at P < 0.05 and degree of freedom = 10, the correlation coefficient |r| > 0.576 was set to the cut‐off value, which was determined by the test for the significance of Pearson's correlation coefficient. In addition, through 10‐fold cross‐validation and the response permutation testing (RPT, permutation number = 200), the quality and validity of PLS‐DA pattern recognition models could be fully assessed. The cross‐validation parameters R 2 and Q 2 in the permutated plot represent the degree of model fitting and the predictive ability of the derived models, respectively. If the maximum value of Q 2 from the permutation test is not more than the Q 2 of the real model, the model could serve as a predictable model.
Univariate statistical analysis
The relative concentrations of those metabolites with discriminatory significance derived from multivariate statistical analysis were calculated by integrating their characteristic signals in the NMR spectra. A one‐way ANOVA was performed by using Students' t‐test (SPSS, version 19.0; IBM SPSS, Chicago, IL, USA) to statistically confirm the differences in metabolite levels among different groups. The relative concentration was expressed as means ± SD, and a P‐value < 0.05 was considered as statistically significant.
Results
Animal models and the corresponding 1H nuclear magnetic resonance spectra from pancreatic tissues
To investigate the metabolic alteration in each phase of PDAC, we used the rats embedded with DMAB to induce PanIN and PDAC. Confirmed by histological observation (Fig. 1), PanIN (PanIN‐2, n = 11), PDAC (n = 15) and control (n = 14) rats were included in NMR detections. Typical 500‐MHz 1H NMR spectra of aqueous pancreas extracts from each group are presented in Figure 2. The resonance assignment and metabolite identification for these extracts were conducted based on the previous literature and public databases.18, 19 These one‐dimensional NMR spectra, including 64 metabolites (Fig. 2), could provide a rough overview of all metabolites. A certain degree of metabolic differences on the spectra could be noticed between control, PanIN and PDAC (e.g. lactate, vitamin K and betaine). Due to the high similarity of the NMR spectra on visual inspection, the acquisition of differential metabolites information is limited. Thus, to obtain more precise and detailed information, multivariate statistical analysis was implemented on NMR spectral data.
Figure 1.
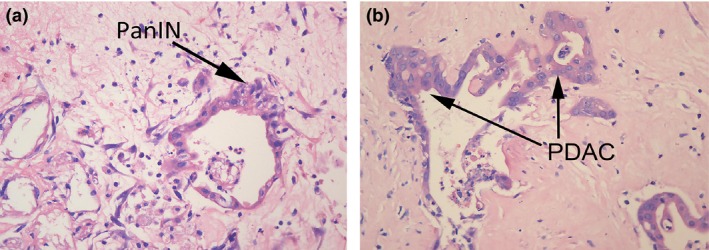
(HE ×400) Representative histology of pancreatic tissues from DMBA‐treated Sprague–Dawley rats. (a) Histological section of the pancreas with the pancreatic intraepithelial neoplasia (PanIN)‐II lesion (arrow) demonstrates small papillae of lesion and mild‐to‐moderate nucleic abnormalities, including some loss of polarity, enlarged nuclei and pseudostratification. (b) Histological section of the pancreas with the PDAC lesion (arrow).
Figure 2.
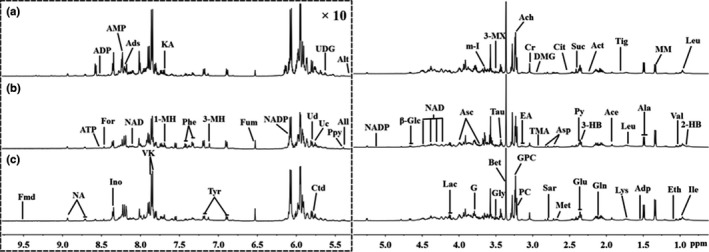
Representative 500 MHz 1H NOESYPR1D nuclear magnetic resonance (NMR) spectra (δ0.7–5.3 and δ5.3–9.7) from the aqueous extracts of (a) control, (b) pancreatic intraepithelial neoplasia and (c) pancreatic ductal adenocarcinoma pancreatic tissues. The spectral regions of δ5.3–9.7 (in the dashed box) were magnified 10 times compared with the regions of δ0.7–5.3 for the purpose of clarity. The assignments of peaks were noted. 3‐HB, 3‐Hydroxybutyrate; 2‐HB, 2‐Hydroxybutyrate; 3‐MH, 3‐Methylhistidene; 1‐MH, 1‐Methylhistidene; 3‐MX, 3‐Methylxanthine; Ace, acetate; Ach, acetylcholine; Act, acetone; Ads, adenosine; ADP, adenosine 3′,5′‐diphosphate; AMP, adenosine monophosphate; ATP, adenosine triphosphate; Adp, adipate; Ala, alanine; Alt, allantoate; All, allantoin; α‐Glc, α‐Glucose; Asc, ascorbate; Asp, aspartate; β‐Glc, β‐Glucose; Bet, betaine; Cho, choline; CL, cholate; Cit, citrate; Cr, creatine; Ctd, cytidine; DMG, dimethylglycine; Eth, ethanol; EA, ethanolamine; Fmd, formaldehyde; For, formate; Fum, fumarate; G, glycerol; Gln, glutamine; Glu, glutamate; GPC, glycerolphosphocholine; Gly, glycine; Ino, inosine; Ile, isoleucine; KA, kynurenate; Lac, lactate; Leu, leucine; Lys, lysine; Met, methoinine; MM, methylmalonate; m‐I, myo‐Inositol; NA, nicotinamide; NAD, nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; PC, phosphocholine; Phe, phenylalanine; Ppy, phosphoenolpyruvate; Py, pyruvate; Sar, sarcosine; Suc, succinate; Tau, taurine; Tig, tiglinate; TMA, trimethylamine; Tyr, tyrosine; Uc, uracil; Ud, uridine; UDG, uridine diphosphate glucose; Val, valine; VK, vitamin K.
Metabonomic profile of normal, PanIN and PDAC rats
Principal component analysis was performed to provide an overview of the 1H NMR data collected from aqueous extracts of the control, PanIN and PDAC pancreatic tissues. As the score plots reveal (Fig. 3a), the first two principal components (PC1 and PC2) could explain 67.4% of the total variance and no outlier was found. A certain degree of separation existed between the control group and PanIN and PDAC groups in the PCA score plot; however, PanIN and PDAC groups were mixed with each other due to their similar metabolic profiles. Therefore, PLS‐DA was performed on these three groups to further maximize the systematic variance and to eliminate the interference information. On visual inspection, the separation in the PLS‐DA score plot (Fig. 3b) was much more obvious than for PCA. However, a moderate overlap can still be observed between PanIN and PDAC groups, indicating that the metabolic alterations that occurred in the lesions were similar in a certain degree.
Figure 3.
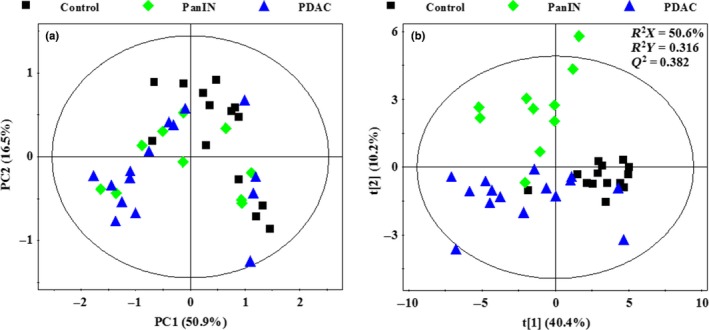
(a) Principal component analysis scores plot based on 1H NOESYPR1D nuclear magnetic resonance (NMR) spectra of control (■), pancreatic intraepithelial neoplasia (PanIN) ( ), PDAC (
), PDAC ( ). (b) PLS‐DA scores plot based on 1H NOESYPR1D NMR spectra of control (■), PanIN (
). (b) PLS‐DA scores plot based on 1H NOESYPR1D NMR spectra of control (■), PanIN ( ), PDAC (
), PDAC ( ).
).
Partial least squares discriminant analysis was also performed on groups in pairs (control vs PanIN, control vs PDAC, PanIN vs PDAC). Pronounced separations can be observed in all three plots (Fig. 4a), which represent the discriminating metabolic profile between pair‐wise groups. According to the cross‐validation parameters, R 2 (the total explained variation) and Q 2 (the predictability of the models) (Fig. 4, left panels), for all three models demonstrate strong predictability and reliable subsequent identification of the characteristic metabolites. The validation plots from permutation tests (Fig. 4, right panels) and 10‐fold cross‐validation suggested that the models based on the NMR data from aqueous extracts were effective and authentic. In addition, the smaller R 2 and Q 2 values in the PanIN versus PDAC model than in others and a little overfitting in this model (Fig. 4c) also indicated similar metabolic profiles and the continuity of the physiological changes from PanIN to PDAC.
Figure 4.
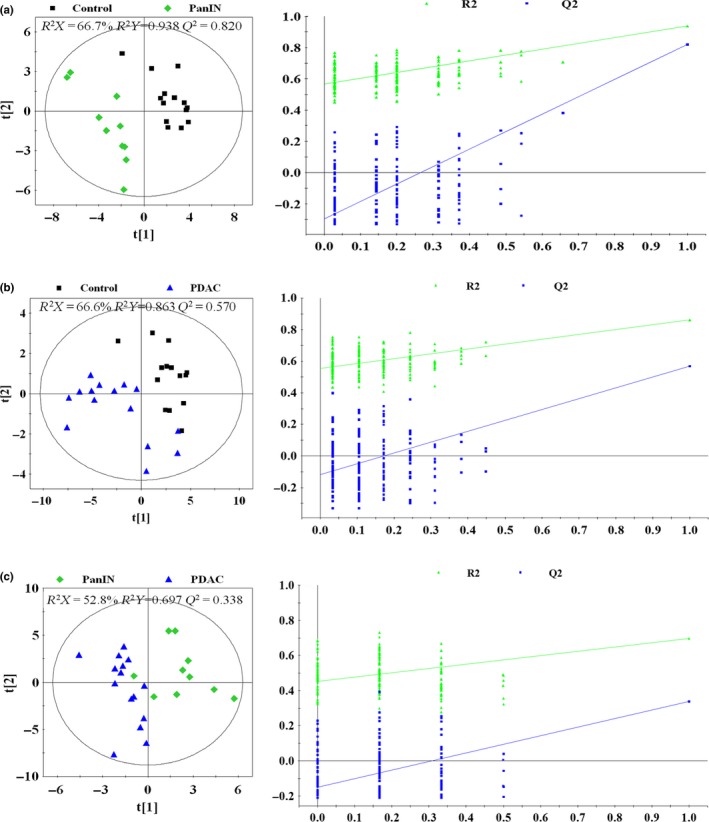
PLS‐DA scores plots (left panels) and plots of permutation tests (n = 200) of PLS‐DA (right panels) based on 1H NOESYPR1D nuclear magnetic resonance (NMR) spectra of aqueous extracts from normal Sprague–Dawley (SD) rat pancreas (control, black box ■), pancreatic intraepithelial neoplasia (pancreatic intraepithelial neoplasia [PanIN], green diamond  and SD rat pancreatic ductal adenocarcinoma (pancreatic ductal adenocarcinoma [PDAC], blue triangle
and SD rat pancreatic ductal adenocarcinoma (pancreatic ductal adenocarcinoma [PDAC], blue triangle  ). (a) Control versus PanIN; (b) control versus PDAC; (c) PanIN versus PDAC.
). (a) Control versus PanIN; (b) control versus PDAC; (c) PanIN versus PDAC.
To understand the detailed metabolic information following the progress of the disease and to identify the metabolites responsible for the metabonomic differences, OPLS‐DA was also conducted on the NMR data among normal, PanIN and PDAC groups for one predictive component and one orthogonal component (Fig. 5). The score plots demonstrate the obvious distinctions between pair‐wise groups (Fig. 5, left panel), while the loading plots with color‐coded correlation coefficients could clearly display the metabolites contributed to metabonomic difference in each pair‐comparison. (Fig. 5, middle and right panels). Finally, 37 of the characteristic metabolites and their corresponding variations in PanIN and PDAC groups were identified and their differences in relative concentrations among different groups were confirmed by univariate statistical analysis (Table 1). Only a few results from univariate statistical analysis were different from those from OPLS‐DA, including acetylcholine, aspartate, lysine, NADP+ and nicotinamide, which could be due to the very low level of metabolites, the statistical difference between multivariate and univariate analysis, and the possible overlap between signals from different metabolites in NMR spectra, thus demonstrating a high reliability of multivariate statistical analysis. Metabonomic pathways analysis was conducted using the KEGG database. The primary pathways correlate to characteristic metabolites of PanIN and PDAC, and the numbers of common tendency metabolites involved in corresponding pathways are listed in Table 2.
Figure 5.
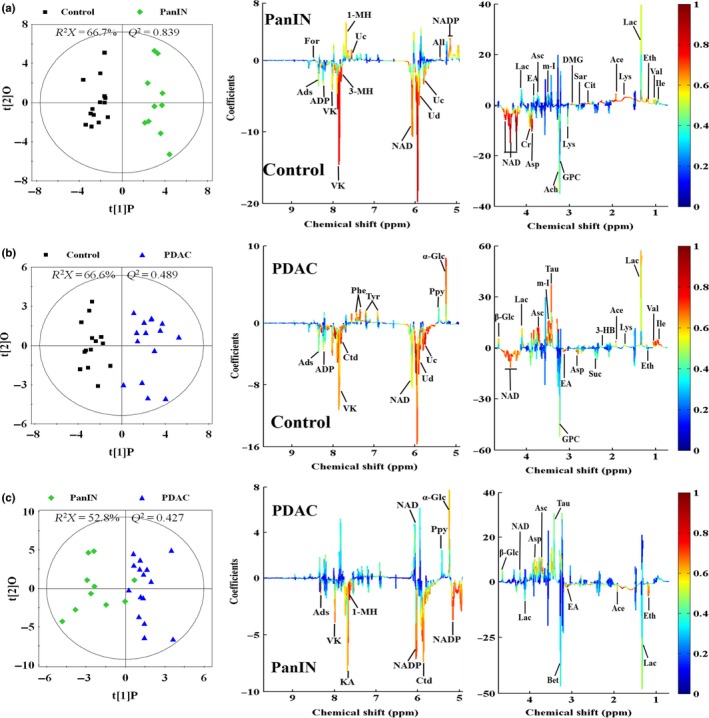
OPLS‐DA scores plots (left panels) derived from 1H nuclear magnetic resonance (NMR) spectra of aqueous extracts and corresponding coefficient loading plots (middle and right panels) from normal Sprague–Dawley rat pancreas (control, black box ■), pancreatic intraepithelial neoplasia (pancreatic intraepithelial neoplasia [PanIN], green diamond  ) and SD rat pancreatic ductal adenocarcinoma (pancreatic ductal adenocarcinoma [PDAC], blue triangle
) and SD rat pancreatic ductal adenocarcinoma (pancreatic ductal adenocarcinoma [PDAC], blue triangle  ). (a) Control versus PanIN; (b) control versus PDAC; (c) PanIN versus PDAC. The color map shows the significance of metabolites variations between the two classes. Keys of the assignments were shown in Figure 2.
). (a) Control versus PanIN; (b) control versus PDAC; (c) PanIN versus PDAC. The color map shows the significance of metabolites variations between the two classes. Keys of the assignments were shown in Figure 2.
Table 1.
OPLS‐DA coefficients and relative concentrations of metabolites derived from the nuclear magnetic resonance data in different groups
| Metabolites | r † | Relative concentration‡ | ||||
|---|---|---|---|---|---|---|
| C‐PanIN | C‐PDAC | PanIN‐PDAC | C | PanIN | PDAC | |
| 1‐Methylhistidine | 0.718 | — | −0.771 | 0.48 ± 0.30 | 1.25 ± 0.85** | 0.57 ± 0.40# |
| 2‐Hydroxybutyrate | 0.750 | 0.701 | — | 1.18 ± 0.35 | 2.07 ± 1.78* | 2.07 ± 0.71*** |
| 3‐Hydroxybutyrate | 0.912 | 0.612 | −0.728 | 0.42 ± 0.10 | 0.80 ± 0.30*** | 0.61 ± 0.20**,# |
| 3‐Methylhistidine | −0.775 | −0.753 | — | 0.42 ± 0.09 | 0.25 ± 0.12** | 0.26 ± 0.15** |
| Acetate | 0.793 | 0.657 | −0.730 | 0.77 ± 0.08 | 1.23 ± 0.18*** | 1.04 ± 0.19***,# |
| Acetylcholine | −0.673 | — | 0.643 | 3.24 ± 0.51 | 2.20 ± 0.76*** | 2.86 ± 1.03 |
| Adenosine | −0.578 | −0.651 | −0.712 | 1.71 ± 0.33 | 1.17 ± 0.75* | 1.07 ± 0.60** |
| Adipate | 0.711 | — | −0.698 | 1.01 ± 0.38 | 2.56 ± 1.62** | 1.28 ± 0.61# # |
| Ascorbate | — | 0.764 | 0.708 | 26.11 ± 2.63 | 26.74 ± 4.31 | 29.37 ± 3.32*,# |
| Aspartate | −0.900 | −0.745 | 0.796 | 21.10 ± 1.96 | 16.32 ± 2.34*** | 20.24 ± 2.51##, # |
| Betaine | −0.916 | −0.654 | — | 12.72 ± 0.89 | 9.89 ± 1.96*** | 10.90 ± 1.66*** |
| Citrate | 0.861 | — | −0.713 | 0.89 ± 0.13 | 1.64 ± 0.65*** | 0.94 ± 0.63# |
| Creatine | −0.936 | — | 0.844 | 2.90 ± 0.28 | 2.45 ± 0.37** | 2.79 ± 0.42# |
| Cytidine | −0.859 | −0.791 | −0.688 | 3.26 ± 0.53 | 1.78 ± 0.70*** | 1.88 ± 0.82*** |
| Dimethylglycine | 0.797 | — | −0.710 | 0.03 ± 0.02 | 0.09 ± 0.03*** | 0.03 ± 0.05# # |
| Glutamate | 0.726 | 0.586 | — | 11.43 ± 1.19 | 12.55 ± 1.91* | 12.33 ± 1.68* |
| Glycerol | — | 0.715 | 0.607 | 39.15 ± 2.70 | 37.56 ± 5.26 | 42.70 ± 5.30*,# |
| Inosine | −0.780 | −0.591 | — | 1.75 ± 0.24 | 0.85 ± 0.36*** | 1.21 ± 0.42*** |
| Isoleucine | 0.730 | 0.772 | — | 0.47 ± 0.10 | 0.72 ± 0.20*** | 0.79 ± 0.24*** |
| Kynurenate | 0.629 | −0.719 | −0.725 | 1.88 ± 0.51 | 2.26 ± 0.78* | 1.53 ± 0.66*,# |
| Lactate | 0.725 | 0.686 | −0.582 | 8.42 ± 1.38 | 15.43 ± 3.62*** | 14.41 ± 5.68** |
| Leucine | 0.780 | 0.761 | — | 1.55 ± 0.17 | 1.73 ± 0.25* | 2.03 ± 0.41*** |
| Lysine | 0.852 | — | −0.665 | 5.45 ± 0.60 | 7.25 ± 1.91*** | 5.69 ± 1.24**,# |
| Methionine | 0.874 | −0.620 | −0.645 | 1.03 ± 0.15 | 1.33 ± 0.33* | 0.92 ± 0.22*,# |
| myo‐Inositol | 0.591 | 0.730 | — | 7.67 ± 0.45 | 8.36 ± 0.93* | 8.67 ± 0.95*** |
| Nicotinamide | −0.851 | −0.825 | −0.740 | 0.30 ± 0.03 | 0.26 ± 0.05* | 0.32 ± 0.06# |
| NAD+ | −0.941 | −0.766 | — | 0.05 ± 0.05 | 0.02 ± 0.02* | 0.02 ± 0.02* |
| NADP+ | −0.779 | −0.804 | −0.750 | 0.02 ± 0.01 | 0.01 ± 0.01** | 0.02 ± 0.02 |
| Phenylalanine | 0.622 | 0.777 | — | 0.35 ± 0.10 | 0.55 ± 0.23** | 0.64 ± 0.32** |
| Sarcosine | 0.746 | — | −0.761 | 0.10 ± 0.02 | 0.21 ± 0.15* | 0.06 ± 0.05# |
| Taurine | — | 0.768 | 0.706 | 9.64 ± 1.59 | 11.47 ± 2.82 | 15.87 ± 4.40***,## |
| Uracil | −0.793 | −0.743 | −0.810 | 1.36 ± 0.13 | 0.98 ± 0.33** | 0.85 ± 0.39** |
| Uridine | −0.907 | −0.739 | −0.749 | 5.77 ± 0.97 | 3.38 ± 1.18*** | 3.77 ± 1.44*** |
| Uridine diphosphate | −0.817 | −0.810 | — | 4.01 ± 0.55 | 2.16 ± 0.96*** | 2.08 ± 1.18*** |
| Valine | 0.734 | 0.753 | — | 0.85 ± 0.17 | 1.24 ± 0.32*** | 1.27 ± 0.34*** |
| Vitamin K | −0.904 | −0.850 | — | 5.28 ± 1.03 | 2.26 ± 0.84*** | 3.01 ± 1.70*** |
| α‐Glucose | — | 0.732 | 0.659 | 2.31 ± 0.63 | 3.13 ± 1.49 | 4.85 ± 2.16***,# |
†Correlation coefficients: positive and negative signs indicate positive and negative correlation in the concentrations. |r| > 0.576 is the cut‐off value for significance based on significance of P = 0.05 and degrees of freedom = 10. “—” means |r| < 0.576. &Relative concentration derived by integrating the characteristic signals of each metabolite in the nuclear magnetic resonance spectra, and the concentrations are expressed as means ± SD *P < 0.05, **P < 0.01, ***P < 0.001 versus control group; # P < 0.05, ## P < 0.01, ### P < 0.001 versus PanIN group. C, control group. PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma.
Table 2.
Involved pathways corresponding to metabolic differences in sample tissues between pairwise groups
| Pathways | Control‐PanIN† | Control‐PDAC† | Common tendency‡ |
|---|---|---|---|
| Biosynthesis of antibiotics | 10/32 | 9/31 | 8/10 |
| ABC transporters | 9/32 | 11/31 | 8/12 |
| Central carbon metabolism in cancer | 8/32 | 8/31 | 7/9 |
| 2‐Oxocarboxylic acid metabolism | 8/32 | 6/31 | 6/8 |
| Biosynthesis of amino acids | 8/32 | 7/31 | 5/9 |
| Protein digestion and absorption | 8/32 | 8/31 | 6/10 |
| Aminoacyl‐tRNA biosynthesis | 7/32 | 7/31 | 6/7 |
| Pyrimidine metabolism | 4/32 | 4/31 | 4/4 |
| Nicotinate and nicotinamide metabolism | 4/32 | 4/31 | 4/4 |
| Other specific amino acid | 11/32 | 9/31 | 8/11 |
†For each pathway, the ratio of metabolites involved in any metabolic pathway to all significant metabolites corresponding to the pairwise group difference. ‡For each pathway, the ratio of metabolites with common variation tendency in all significant metabolites which are involved in corresponding pathways in pairwise groups of control‐ pancreatic intraepithelial neoplasia (PanIN) and control‐pancreatic ductal adenocarcinoma (PDAC).
Discussion
To meet the need for early diagnosis of PDAC, various methods have been used to find corresponding biomarkers. NMR spectroscopy demonstrates advantages over other detection methods as a noninvasive and effective tool for the identification of characteristic metabolites. In this study, DMBA was implanted in the pancreas of SD rats to establish animal PanIN and PDAC models.20 This chemical could induce a series of pathological changes from the healthy state to PDAC with different induction times, thus simulating the progress of PDAC in humans. Because the tissue of PanIN is often too small to dissect from non‐tumor regions, metabonomic information on non‐tumor regions could interfere the identification of metabolite biomarkers, which may exert an influence on the sensitivity of NMR spectroscopy analysis. Nonetheless, a rigorous sampling process and histological diagnoses were conducted to guarantee accurate sampling, to limit the diluted effects of the non‐tumor regions on the PanIN tissue, and recognition pattern models demonstrated clear separations in pair‐comparisons in our study, indirectly validating the availability of sample collection. PanIN, a precancerous lesion of PDAC with a high possibility of progressing to cancer,21, 22 is often generated in the small intralobular pancreatic ducts. Typically, these are characterized by flat or papillary lesions and different degrees of cytologic abnormalities. The metabonomic profile of PanIN and metabolic differences between PanIN and PDAC can provide a host of beneficial information for studying pathophysiological alterations as well as searching for biomarkers in the early phase of PDAC.23 In the present study, all rats from experimental group I were diagnosed with PanIN‐2 but without PanIN‐1 or PanIN‐3. This is predictable because the identification of PanIN‐1, a very early lesion with low risk, is very difficult and PanIN‐3 is often observed in PDAC groups only24 due to the high invasiveness, especially with other promoters.25 Therefore, PanIN‐2 could be treated as the beginning of invasive progression leading to highly invasive PDAC.26 Thus, the metabonomic profiles can be highly representative for that of PDAC at early stage. In terms of preprocessing, with the advantages of a neutral environment, simplicity and convenience, the extraction method using methanol–chloroform27, 28 can ensure effective separation and purification of metabolites without external impurities entering the aqueous layer. In our case, we focused on the water‐soluble metabolites rather than the fat‐soluble metabolites because the former are because the former are more conductive to screen out the characteristic metabolites served to clinical practice.
Metabonomic difference in PanIN compared with the normal tissue
To date, few metabonomic studies have been conducted to investigate the metabolic variations relevant to PanIN. In the present study, obvious metabonomic alterations in both PanIN and PDAC compared to control groups were observed, including in glucose, amino acids, carboxylic acids and coenzymes (Table 1). With the KEGG pathway analysis, the pathway of central carbon metabolism in cancer, which involved abnormal concentration changes of glutamate, aspartate, methionine, phenylalanine, leucine, citrate, lactate and isoleucine in PanIN, was seductive due to tumor metabolic reprogram related with abnormal functions of tumor suppressor genes of P53, SIRT3, SIRT6 and the oncogenes of Ras, PI3K, Akt and c‐Myc. A higher concentration of lactate was observed in PanIN, which can be attributed to the oxygen deficit from rapid growth and aerobic glycolysis without the ability to remove lactate from the tumor microenvironment and can contribute to metastases of tumors.29 In addition, a high concentration of glutamate in tissue was a significant phenomenon associated with reconstructed glutamine metabolism, which is one of the most significant mechanisms to promote the aerobic glycolysis in cancer cells. Because of the hypoxia in cancer tissues, the cytosolic acetyl‐CoA cannot be produced through tricarboxylic acid circle and the glutamate plays an important role to contribute carbon to lipogenic acetyl‐CoA through the reductive citric acid pathway.30 The high concentration of glutamate can also activate receptors of N‐methyl‐d‐aspartate and alpha‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionic acid, as a result, to be a switch to increase the chance of malignant transformation of PanIN and enhance the invasion and migration of PDAC.31 Meanwhile, the metabolic changes of the branched chain amino acids in PanIN, including leucine, isoleucine and valine, may be tightly connected with cellular transformation, which caused the inactivation of the tumor necrosis factor‐alpha converting enzyme and resulted an in elevated possibility of PDAC genesis.32 The concentration differences of the metabolites in ABC transporters metabolism were also noticeable in comparison (e.g. lysine, aspartate, myo‐inositol and betaine). Given the high relevance with progression of PDAC,33 the discrepancy of ABC transporters metabolism may enhance the transport of nutritions into cells and promote intracellular anabolism. However, due to the complicated network connections among metabolites, numerous differential metabolites could be affected and identified in PanIN besides the metabolites mentioned above. Obviously, the specific mechanism and bioinformation involved have yet to be fully understood and further analyses are required for deeper insight. In general, the metabolic changes of PanIN are in accordance with the tumor metabolism reprogramming in cancer and the corresponding metabolites could be valuable for the metabonomic‐based detection and diagnosis of PanIN.
Metabonomic difference in PanIN compared with pancreatic ductal adenocarcinoma
In previous reports, higher concentrations of taurine, glutamate, lactate, leucine, isoleucine and valine and lower concentration of betaine were identified as biomarkers in PDAC tissue,17, 34 which were also observed in the present study. It is meaningful to find those with the same trends in the variation of PanIN, which may serve as biomarkers not only for PDAC but also for pancreatic precancerous lesions. Furthermore, acknowledging the metabonomic difference between PanIN and PDAC tissue is beneficial for a better understanding of the development mechanism and the establishment of diagnostic strategies targeting the early stage of PDAC. Through comparison to the control, we noticed that several differential metabolites in PanIN and PDAC shared the same trends in variation and relevant pathways (Table 2), such as the metabolism of glycolysis, amino acids, nucleic acid and nicotinamide, which are important components in tumor metabolism.35, 36, 37 This discovery can be interpreted as the intercommunity and continuity of metabolic abnormality from PanIN to PDAC and regarded as indirect evidence of PDAC's genealogical theory,21, 22 thus implying the feasibility to diagnose PanIN and PDAC based on similar mutual metabolite clusters. Only kynurenate and methionine showed a difference in the trends in variation, which were increased in PanIN but decreased in the PDAC. Considering being a crucial participator in tryptophan metabolism, the content variation of kynurenate can be associated with the disorder of tryptophan metabolism which can further affect the nicotinamide metabolism and glycolysis. In addition, its abnormal concentration was reported to be associated with inflammation that contributed to tumorgenesis.38, 39 Methionine is on the central position in the methionine cycle and the most important methyl donor for synthesis of nitrogen substances. Because the methionine and tryptophan are transported into the cancer cells by the l‐type amino‐acid transporter 1 (LAT1), which has been proved to be overexpressed in the PDAC and is highly related to patients' prognosis,40 it is reasonable to speculate that molecular events along with the progression from PanIN to PDAC may impact on LAT1 and cause these differential metabolic variations in the two lesions. In addition, the high level of dimethylglycine and sarcosine in PanIN can be correlated with the enhanced biosynthesis of serine, glycine and cysteine, which are significant for growth and proliferation of cancer cells. Moreover, the high level of taurine in PDAC was in accordance with the previous report23 but different in PanIN. Taurine is an organic acid highly collected in immune cells41, 42 and proved to be associated with apoptosis,43, 44 reflecting the body's immune response toward the tumorgenesis. Because modeling and preprocessing procedures are different, it is still debatable to determine the content alterations of taurine in PanIN.
To sum up, we utilized an NMR‐based metabonomic technique to demonstrate that significant metabolic changes occur in glucose, amino acids and carboxylic acids in PanIN and PDAC tissues, which were involved in the tumor metabolism reprogramming. Although trends in the variation of metabolites in PanIN and PDAC were similar compared with normal pancreas tissues, the metabolic differences were obvious enough to distinguish PanIN from not only the control but also PDAC. The specific metabolic biomarkers of PanIN were identified and recognition models of pair‐comparisons were established with considerable sensitivity and specificity, which can aid in establishing noninvasive metabonomic diagnostic methods to distinguish PDAC and PanIN from normal pancreas tissues in the future. NMR‐based metabonomic strategies present a promising approach for metabolism investigations and early diagnosis of PanIN and PDAC.
Disclosure Statement
All the authors declare no conflict of interest and all the authors have not financial or personal relationships with other people or organization that could inappropriately influence this work.
Acknowledgments
This work is sponsored by the National Natural Science Foundation of China (No. 81272581), the United Fujian Provincial Health and Education Project for Tackling the Key Research (No. WKJ‐FJ‐10) and the National Key Clinical Specialty Discipline Construction Program of China and Key Clinical Specialty Discipline Construction Program of Fujian.
Cancer Sci 107 (2016) 836–845
Funding Information
United Fujian Provincial Health and Education Project for Tackling the Key Research (Grant/Award Number: “WKJ‐FJ‐10″), National Key Clinical Specialty Discipline Construction Program of China, Key Clinical Specialty Discipline Construction Program of Fujian, National Natural Science Foundation of China (Grant/Award Number: “81272581”).
References
- 1. Matsuda T, Marugame T, Kamo K et al Cancer incidence and incidence rates in Japan in 2005: based on data from 12 population‐based cancer registries in the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J Clin Oncol 2011; 41: 139–47. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62: 10–29. [DOI] [PubMed] [Google Scholar]
- 3. Chari ST. Detecting early pancreatic cancer: problems and prospects. Semin Oncol 2007; 34: 284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kokhanenko N, Ignashov AM, Varga EV et al Role of the tumor markers CA 19‐9 and carcinoembryonic antigen (CEA) in diagnosis, treatment and prognosis of pancreatic cancer. Vopr Onkol 2001; 47: 294–7. [PubMed] [Google Scholar]
- 5. Singh S, Tang SJ, Sreenarasimhaiah J, Lara LF, Siddiqui A. The clinical utility and limitations of serum carbohydrate antigen (CA19‐9) as a diagnostic tool for pancreatic cancer and cholangiocarcinoma. Dig Dis Sci 2011; 56: 2491–6. [DOI] [PubMed] [Google Scholar]
- 6. Rehman L, Rehman UL, Azmat SK, Mohammad Hashim AS. Magnetic resonance spectroscopy: novel non‐invasive technique for diagnosing brain tumors. J Coll Physicians Surg Pak 2015; 25: 863–6. [PubMed] [Google Scholar]
- 7. Bartella L, Morris EA, Dershaw DD et al Proton MR spectroscopy with choline peak as malignancy marker improves positive predictive value for breast cancer diagnosis: preliminary study. Radiology 2006; 239: 686–92. [DOI] [PubMed] [Google Scholar]
- 8. Thomas MA, Wyckoff N, Yue K et al Two‐dimensional MR spectroscopic characterization of breast cancer in vivo. Technol Cancer Res Treat 2005; 4: 99–106. [DOI] [PubMed] [Google Scholar]
- 9. Huzjan R, Sala E, Hricak H. Magnetic resonance imaging and magnetic resonance spectroscopic imaging of prostate cancer. Nat Clin Pract Urol 2005; 2: 434–42. [DOI] [PubMed] [Google Scholar]
- 10. Squillaci E, Manenti G, Mancino S et al MR spectroscopy of prostate cancer. Initial clinical experience. J Exp Clin Cancer Res 2005; 24: 523–30. [PubMed] [Google Scholar]
- 11. Kwock L, Smith JK, Castillo M et al Clinical role of proton magnetic resonance spectroscopy in oncology: brain, breast, and prostate cancer. Lancet Oncol 2006; 7: 859–68. [DOI] [PubMed] [Google Scholar]
- 12. Khan SA, Cox IJ, Thillainayagam AV, Bansi DS, Thomas HC, Taylor‐Robinson SD. Proton and phosphorus‐31 nuclear magnetic resonance spectroscopy of human bile in hepatopancreaticobiliary cancer. Eur J Gastroenterol Hepatol 2005; 17: 733–8. [DOI] [PubMed] [Google Scholar]
- 13. Lin Z, Jin H, Guo X et al Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by H‐1 nuclear magnetic resonance‐based metabonomic profiles. Clin Biochem 2012; 45: 1064–9. [DOI] [PubMed] [Google Scholar]
- 14. Napoli C, Sperandio N, Lawlor RT, Scarpa A, Molinari H, Assfalg M. Urine metabolic signature of pancreatic ductal adenocarcinoma by 1H nuclear magnetic resonance: identification, mapping, and evolution. J Proteome Res 2011; 11: 1274–83. [DOI] [PubMed] [Google Scholar]
- 15. Tesiram YA, Lerner M, Stewart C, Njoku C, Brackett DJ. Utility of nuclear magnetic resonance spectroscopy for pancreatic cancer studies. Pancreas 2012; 41: 474–80. [DOI] [PubMed] [Google Scholar]
- 16. OuYang D, Xu J, Huang H, Chen Z. Metabolomic profiling of serum from human pancreatic cancer patients using 1H NMR spectroscopy and principal component analysis. Appl Biochem Biotechnol 2011; 165: 148–54. [DOI] [PubMed] [Google Scholar]
- 17. Fang F, He X, Deng H et al Discrimination of metabolic profiles of pancreatic cancer from chronic pancreatitis by high‐resolution magic angle spinning 1H nuclear magnetic resonance and principal components analysis. Cancer Sci 2007; 98: 1678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cui Q, Lewis IA, Hegeman AD et al Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol 2008; 26: 162–4. [DOI] [PubMed] [Google Scholar]
- 19. Wishart DS, Knox C, Guo AC et al HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 2009; 37: D603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rivera JA, Graeme‐Cook F, Werner J et al A rat model of pancreatic ductal adenocarcinoma: targeting chemical carcinogens. Surgery 1997; 122: 82–90. [DOI] [PubMed] [Google Scholar]
- 21. Hruban RH, Adsay NV, Albores‐Saavedra J et al Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001; 25: 579–86. [DOI] [PubMed] [Google Scholar]
- 22. Hruban RH, Takaori K, Klimstra DS et al An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol 2004; 28: 977–87. [DOI] [PubMed] [Google Scholar]
- 23. Wang AS, Lodi A, Rivera LB et al HR‐MAS MRS of the pancreas reveals reduced lipid and elevated lactate and taurine associated with early pancreatic cancer. NMR Biomed 2014; 27: 1361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Esposito I, Seiler C, Bergmann F, Kleeff J, Friess H, Schirmacher P. Hypothetical progression model of pancreatic cancer with origin in the centroacinar–acinar compartment. Pancreas 2007; 35: 212–7. [DOI] [PubMed] [Google Scholar]
- 25. Hwang IK, Kim H, Lee YS et al Presence of pancreatic intraepithelial neoplasia‐3 in a background of chronic pancreatitis in pancreatic cancer patients. Cancer Sci 2015; 106: 1408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sipos B, Frank S, Gress T, Hahn S, Kloppel G. Pancreatic intraepithelial neoplasia revisited and updated. Pancreatology 2009; 9: 45–54. [DOI] [PubMed] [Google Scholar]
- 27. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959; 37: 911–7. [DOI] [PubMed] [Google Scholar]
- 28. Jensen SK. Improved Bligh and Dyer extraction procedure. Lipid Technol 2008; 20: 280–1. [Google Scholar]
- 29. Cheng LL, Lean CL, Bogdanova A et al Enhanced resolution of proton NMR spectra of malignant lymph nodes using magic‐angle spinning. Magn Reson Med 1996; 36: 653–8. [DOI] [PubMed] [Google Scholar]
- 30. Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci 2013; 104: 275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herner A, Sauliunaite D, Michalski CW et al Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras‐MAPK signaling. Int J Cancer 2011; 129: 2349–59. [DOI] [PubMed] [Google Scholar]
- 32. Perez L, Kerrigan JE, Li X, Fan H. Substitution of methionine 435 with leucine, isoleucine, and serine in tumor necrosis factor alpha converting enzyme inactivates ectodomain shedding activity. Biochem Cell Biol 2007; 85: 141–9. [DOI] [PubMed] [Google Scholar]
- 33. Mohelnikova‐Duchonova B, Brynychova V, Oliverius M et al Differences in transcript levels of ABC transporters between pancreatic adenocarcinoma and nonneoplastic tissues. Pancreas 2013; 42: 707–16. [DOI] [PubMed] [Google Scholar]
- 34. He XH, Li WT, Gu YJ et al Metabonomic studies of pancreatic cancer response to radiotherapy in a mouse xenograft model using magnetic resonance spectroscopy and principal components analysis. World J Gastroenterol 2013; 19: 4200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He Y, Chu SH, Walker WA. Nucleotide supplements alter proliferation and differentiation of cultured human (Caco‐2) and rat (IEC‐6) intestinal epithelial cells. J Nutr 1993; 123: 1017–27. [DOI] [PubMed] [Google Scholar]
- 36. Zhang JG, Zhao G, Qin Q et al Nicotinamide prohibits proliferation and enhances chemosensitivity of pancreatic cancer cells through deregulating SIRT1 and Ras/Akt pathways. Pancreatology 2013; 13: 140–6. [DOI] [PubMed] [Google Scholar]
- 37. Chini CC, Guerrico AM, Nin V et al Targeting of NAD metabolism in pancreatic cancer cells: potential novel therapy for pancreatic tumors. Clin Cancer Res 2014; 20: 120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor MW, Feng GS. Relationship between interferon‐gamma, indoleamine 2,3‐dioxygenase, and tryptophan catabolism. FASEB J 1991; 5: 2516–22. [PubMed] [Google Scholar]
- 39. Wirleitner B, Rudzite V, Neurauter G et al Immune activation and degradation of tryptophan in coronary heart disease. Eur J Clin Invest 2003; 33: 550–4. [DOI] [PubMed] [Google Scholar]
- 40. Kaira K, Sunose Y, Arakawa K et al Prognostic significance of l‐type amino‐acid transporter 1 expression in surgically resected pancreatic cancer. Br J Cancer 2012; 107: 632–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dingley AJ, King NJ, King GF. An NMR investigation of the changes in plasma membrane triglyceride and phospholipid precursors during the activation of T‐lymphocytes. Biochemistry 1992; 31: 9098–106. [DOI] [PubMed] [Google Scholar]
- 42. Wright LC, Obbink KL, Delikatny EJ, Santangelo RT, Sorrell TC. The origin of 1H NMR‐visible triacylglycerol in human neutrophils. Highfatty acid environments result in preferential sequestration of palmitic acid into plasma membrane triacylglycerol. Eur J Biochem 2000; 267: 68–78. [DOI] [PubMed] [Google Scholar]
- 43. Opstad KS, Bell BA, Griffiths JR, Howe FA. Taurine: a potential marker of apoptosis in gliomas. Br J Cancer 2009; 100: 789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X, Tu S, Wang Y, Xu B, Wan F. Mechanism of taurine‐induced apoptosis in human colon cancer cells. Acta Biochim Biophys Sin 2014; 46: 261–72. [DOI] [PubMed] [Google Scholar]


