Abstract
Retinoid X receptors (RXRs) are transcription factors with important functions in embryonic development, metabolic processes, differentiation and apoptosis. A particular feature of RXRs is their ability to act as obligatory heterodimerisation partners of class II nuclear receptors. At the same time, these receptors are also able to form homodimers that bind to direct repeat (DR1) hormone response elements. Since the discovery of RXRs, most of the studies focused on its ligand binding and DNA-binding domains, while its N-terminal domain (NTD) harboring a ligand-independent activation function remained poorly characterized. Here, we investigated the solution properties of the NTD domain of RXRα alone and in the context of the full-length receptor using small-angle X-ray scattering (SAXS) and nuclear magnetic resonance (NMR) spectroscopy. We report the solution structure of the full-length homodimeric RXRα on DNA and show that the NTD remains highly flexible within this complex.
Introduction
Retinoid X receptors (RXRs) occupy a crucial place in nuclear receptor (NR) signaling network serving as obligatory dimerisation partners for several NRs including itself. RXRs are essential factors for many biological processes and consequently they play key roles in various diseases, including cancer (reviewed in 1, 2). RXRs comprise three isotypes, RXRα (NR2B1), RXRβ (NR2B2) and RXRγ (NR2B3) that are differentially expressed in tissues with RXRα being the predominant isotype 3. Numerous isoforms are generated from alternative splicing, the predominant one, RXRα1, encodes a 52 kDa protein. Two additional isoforms (α2 and α3) with 28 and 97 amino acid deletions in the N-terminal domain have been identified in mouse testis 4.
RXRs share a common domain organization with all other NRs comprising a variable N-terminal domain (NTD) harboring a ligand-independent activation function, the C-terminal ligand-binding domain (LBD), a key regulatory domain containing the ligand binding pocket and main dimerization surface, and the conserved DNA binding domain (DBD) which interacts with the core motif 5’-RGKTSA-3’ (R = A or G, K = G or T, S = C or G) 5, 6. RXR dimers recognize a repetition of the core motif with variable spacer size and relative orientation (i.e. Direct Repeat (DR), Inverted Repeat (IR) or Everted Repeat (ER)) that define the hormone response element (HRE). The specificity of the receptor binding results not only from the DNA sequence of the two half-sites, but also from the HRE geometry, since both spacing and relative orientation of the half-sites do influence the receptor binding affinity 7.
Our understanding of the NR structure-function relationship has considerably moved forward in the two last decades with the determination of the crystal structures of the isolated DBD and LBD, as well as the structures of the multi-domain complexes for some of them (reviewed in 8–10). However, little information about the NTD is known. The NTD of the NRs varies considerably in terms of length and amino acid sequence, and it harbors the weakest sequence conservation across the NR superfamily. Moreover, it was found that the majority of known NR isoforms differ mostly in their NTD domain 11. The NTD domain of most NRs possess an autonomous transcriptional ligand-independent activation function (AF-1) which, when linked to a heterologous DBD to form a chimeric fusion, can activate transcription in a constitutive manner. The NTD through its activation function AF-1 is involved in the modulation of the transcriptional activation of target genes in a cell-specific and promoter-dependent manner 12. For the majority of NRs, the NTD is also subjected to post-translational modifications, such as phosphorylation and sumoylation, that play essential roles in regulating the receptor transcriptional activities 12. Most of the biophysical and structural studies of NTDs of NRs have been focused on steroidal NRs 13–17. These studies have revealed that the NTDs are largely disordered and highly flexible (reviewed in 12). In the presence of osmolytes or upon interaction with protein partners, partial folding of the NTDs have been shown to occur and numerous proteins that interact with NRs NTDs have been identified (reviewed in 11). In addition, several studies have shown folding effects of the DNA binding on NTDs of steroidal NRs in the context of the intact protein 16, 18–20.
In the present study, we investigated the solution behavior of the NTD of RXRα and characterized its role in the DNA recognition. RXRs harbors NTDs that have been shown to be crucial for receptor transcriptional activities and subjected to post-translational modifications affecting the transactivation capability 21, 22. The NTD of RXRα is 130 amino acids long and is characterized by a large proportion of P and G residues, a usual feature for intrinsically disordered proteins. HDX experiments showed that RXRα NTD is highly accessible to solvent 23 and this domain was not observed in the crystal structure of multi-domain RXR heterodimer 24, 25. To further describe the NTD domain of RXRα and its potential role in DNA recognition, we used small-angle X-ray scattering (SAXS) and nuclear magnetic resonance (NMR) spectroscopy, two complementary biophysical methods that provide insights into intrinsically disordered regions of proteins.
Experimental Procedures
Constructs, expression and purification
The HsRXRαΔNTD (130-462) and HsRXRα NTD-DBD (1-220) were expressed as hexahistidine fusion proteins. The HsRXRα DBD (130-212) was expressed in fusion with Thioredoxine and hexahistidine tags in E. coli BL21DE3. The full-length HsRXRα (1-462) was expressed either in E. coli or in Sf9 insect cells as an N-terminal hexahistidine or Flag tagged fusion protein. All proteins were purified by affinity chromatography. Fusion proteins were removed by thrombin proteolysis. The cleaved protein was then gel filtrated. 9-cis retinoic acid was added in a 2-fold excess to saturate the receptor. The Ramp2 DR1 oligonucleotide strands (tgAGTTCAaGGGTCAat/acTCAAGTtCCCAGTta) were purchased from SIGMA and annealed as described previously 26 and added in a 1.2-fold excess to the dimers and the complex was gel-filtrated on a Superdex S200. Proteins were eluted in the final buffers (Tris 20 mM pH7.5, NaCl 100 mM, KCl 100 mM, Glycerol 5%, Chaps 2 mM, DTT 5 mM for the complexes without DNA and Tris 20 mM pH 7.5, NaCl 50 mM, KCl 50 mM, Glycerol 5%, Chaps 2 mM, DTT 5 mM for the complexes with DNA).
SAXS experiments
The synchrotron radiation X-ray scattering data were collected at the European Molecular Biology Laboratory (EMBL) at the X33 beamline (DESY, Hamburg) 27 using PILATUS detector at a sample-detector distance of 2.7 m, covering the range of momentum transfer 0.01 < q < 0.6 Å-1 (q = 4π sin(θ)/λ where 2θ is the scattering angle and λ = 0.15 nm is the X-ray wavelength) in 8 frames (15 seconds each) to check for possible radiation damage. All scattering measurements were carried out at 283 K using automated filling 28. The full-length RXR without DNA was measured on the SWING beamline at SOLEIL Synchrotron (Gif-sur-Yvette, France), using a 17×17 cm2 low-noise Aviex CCD detector positioned at a distance of 2.107m from the sample. Sample solutions were circulated in a thermostated Quartz capillary with a diameter of 1.5mm and 10 μm wall thickness, positioned within a vacuum chamber. Fifty frames of 2 s each were collected, normalized to the transmitted intensity, and subsequently averaged using the image analysis software Foxtrot (SWING beamline at SOLEIL Synchrotron). All complexes studied were measured at several solute concentrations in the range from 1 to 5 mg/ml. The data were processed following standard procedures using the program PRIMUS 29. The forward scattering I(0) and the radii of gyration Rg were evaluated using the Guinier approximation, assuming that at very small angles (q < 1.3/Rg), the intensity is represented as I(q) = I(0) exp(-(qRg)2/3). The maximum dimensions Dmax were computed using the indirect transform package GNOM 30 which also provides the distance distribution functions p(r). Low resolution models of the various complexes were constructed using the ab initio program DAMMIF 31 which represents the macromolecule by an assembly of densely packed beads and employs simulated annealing (SA) to build a compact interconnected configuration of beads that fits the experimental data. The most typical reconstructions were selected using the program DAMCLUST. The quaternary structure of the homodimer RXRαΔNTD in complex with Ramp2 was reconstructed at low resolution by rigid body modelling based on the atomic coordinates of its components using the crystal structures of the RXRα DBD-Ramp2 (PDB ID: 4CN2) and RXRα LBDs (PDB ID: 2ZY0) 32. The molecular modeling was performed by the program CORAL 33. The program employed a simulated annealing protocol performing random rigid body movements of LBDs restrained by proximity of the C-termini of DBDs and N-termini of LBDs for each chain in the homodimer whereby the hinge regions were sampled from the library of random loop conformations. For both ab initio and rigid body analysis, multiple runs were performed to verify the stability of the solution. The scattering from the atomic models was calculated using the program CRYSOL 34 which either predicts theoretical scattering patterns or fits the experimental data by adjusting the excluded volume and the contrast of the hydration layer. The conformational space of NTD moieties was explored using the Ensemble Optimization Method (EOM), which takes flexibility into account by allowing for the co-existence of multiple conformations in solution 35. EOM selects appropriate ensembles of configurations from large pools of random models of the protein (loops) and allows one to assess the range of different conformations, which the flexible protein can potentially adopt in solution. The models for the initial pool were created by random generation of the N-terminal loops connected to the DBD domains of RXRα. The theoretical scattering intensities of the randomized models were calculated using the program CRYSOL. The program GAJOE from the EOM package 35 employed a genetic algorithm to select from the pool of randomized structures the representative ensembles of 3D models whose averaged scattering profile fits the experimental data. Multiple runs of EOM enable one to compare the Rg and Dmax distributions of the selected structures versus the original random pool and to make a conclusion regarding degrees of protein compactness/extension and flexibility/rigidity.
Isothermal titration calorimetry experiments
ITC measurements were performed at 298 K on a MicroCal ITC200 (MicroCal). Purified proteins and DNA were dialyzed extensively against the buffer 20 mM Hepes pH 8.0, 100 mM sodium chloride and 1 mM TCEP. In the case of the NTD-DBD and full-length proteins the dialysis buffer additionally contained 2% glycerol. In a typical experiment 2 µl aliquots of DNA at 80 to 150 μM were injected into a 10 µM RXR dimer solution (200 μl sample cell). The delay between injections was 120 to 180 s to permit a complete relaxation of the system before the next injection. ITC titration curves were analyzed using the software Origin 7.0 (OriginLab). Standard free energies of binding and entropic contributions were obtained, respectively, as ΔG = −RT ln(Ka) and TΔS = ΔH – ΔG, from the Ka and ΔH values derived from ITC curve fitting using a single binding site model included in the Origin software.
NMR
NMR experiments were conducted at 279 K using a Bruker 700 MHz equipped with a TCI cryoprobe. Uniformly 15N, 13C labeled full-length RXR proteins were produced in E. coli using standard protocol for expression in minimal medium and were purified as described above. NMR samples were prepared as 150 μL aliquots of 40 μM protein in 50 mM phosphate buffer at pH 7.0, 50 mM of NaCl mixed with stoichiometric amounts of 9-cis retinoic acid in a 3 mm tube. 1H-15N HSQC spectra were recorded using standard pulse sequence and an acquisition time of 3 hours. Methyl correlations were obtained from 1H-13C SOFAST HMQC with a relaxation delay of 100 ms and a total acquisition time of 2 hours. Spectra of the protein in complex with the Ramp2 DR1 oligonucleotide were recorded after addition of stoichiometric amount of the DNA duplex to the protein sample. The interaction between the RXR homodimer and the DNA was monitored in the NMR tube by observing the imino-protons in the 12-14 ppm region of the proton spectrum.
Results
RXRα binding to DR1
Homodimeric RXRs bind direct repeats of the hexanucleotide half-site separated by 1 nucleotide (DR1) 3. We have previously characterized the effects of genomic variation in natural DR1 sequences on RXR binding and their structural impact on the RXR homodimer DBD structure 36. The largest cooperative binding and dimerization surface was observed for the RXR homodimer complex with Ramp2 DR1 36. Such DNA-driven regulation of DBD positioning may propagate to other domains of the receptor, including the NTD. To further investigate the underlying principles of this allosteric control, we determined the binding affinities and thermodynamic parameters of DNA binding by the full-length RXRα homodimer and by its truncated form lacking the NTD (RXRαΔNTD). Figure 1A-C show representative isotherms obtained from ITC measurements, and a detailed analysis of thermodynamic parameters corresponding to the averaged values of at least three independent experiments is presented in Table 1. RXRα binds to the Ramp2 DR1 with an affinity in the nM range. The presence of the NTD leads to a slight but consistent loss of affinity by a factor of two that results from both slightly less favorable enthalpic and entropic contributions to the DR1 binding energy.
Figure 1. Quantification of the interaction between RXR and Ramp2 DR1 by ITC.
Representative ITC isotherms for the binding of the Ramp2 DR1 duplex to the (A) RXRα NTD-DBD, (B) to the RXRα ΔNTD and (C) to the full-length RXRα. The top panels show the raw ITC data expressed as the change in thermal power with respect to time over the period of titration. Lower panels: change in molar heat is expressed as a function of molar ratio of corresponding DR1 to dimer-equivalent RXR. The solid lines in the lower panels represent the fit of data to a one-site model using the ORIGIN software.
Table 1. Thermodynamic parameters obtained from ITC measurements for the binding of RXRα homodimer to DR1 response elements.
| Dimer | Kd (nM) | N | ΔHobs kcal.mol−1 | ΔSobs cal x mol-1 x deg-1 |
|---|---|---|---|---|
| RXRα DBD | 38 ± 15 | 1.1 | -39 ± 3 | -98 |
| Ramp2 DR1 | ||||
| RXRα NTD-DBD | 93 ± 15 | 1.01 | -28 ± 0.7 | -63 |
| Ramp2 DR1 | ||||
| RXRα ΔNTD | 47 ± 4 | 1.08 | -28 ± 1 | -58 |
| Ramp2 DR1 | ||||
| RXRα full-length | 79 ± 15 | 1.0 | -24 ± 2 | -49 |
| Ramp2 DR1 |
Errors were calculated from 3 independent measurements performed at 298 K. All uncertainties are given to one standard deviation. N corresponds to the number of moles of dimer per mole of DNA. Concentrations were measured by UV spectrophometry.
Overall architecture of RXRα-DNA complexes in solution
To characterize the architecture of the RXRα homodimer in solution, we used Small Angle X-ray Scattering (SAXS). Scattering data were obtained for the isolated domains either in the DNA-free or DNA-bound form (LBD dimer, DBD-DNA) and for multi-domain complexes (ΔNTD-DNA, NTD-DBD-DNA), as well as for the full-length RXRα protein DNA-free and in complex with Ramp2 DR1 (Figure 2 and Table 2). All studied samples were homogeneous with linear Guinier plots. Several independent scattering data sets provided consistent results and clearly established the stoichiometry of the dimers. A reasonable fit was found between the experimental SAXS data for the folded domains (LBD, DBD-DNA) and the scattering curves calculated from the X-ray crystal structures (Table 2). These complexes showed a symmetrical distance distribution function (Figure 2B). In contrast, for the multi-domain RXRα complexes the radii of gyration (Rg) and the maximum size (Dmax) values suggested an extended shape (Table 2). Low resolution models were reconstructed ab initio from the corresponding experimental scattering patterns using the program DAMMIF 31. A series of reconstructions yielded superimposable results. The typical ab initio model of RXRαΔNTD-DR1 (presented in Figure 2C) revealed an elongated shape and provided a good fit to the experimental data. A more detailed modeling was performed using the available high resolution models of the individual domains and adjustment of the relative position of the domains by rigid-body refinement against the scattering data using CORAL 33. The model obtained by rigid body refinement agrees well with the ab initio models as seen from the superposition in Figure 2C. Importantly, the RXRα LBD dimer is asymmetrically positioned with respect to the two-half-sites of the DR1, (Figure 2D), an asymmetry that has already been observed for heterodimers harboring RXR 37–39. The refined model of RXRαΔNTD-Ramp2 DR1 also suggests a potential interaction between the DBD of the monomer bound at the 5' side of the DR1 and the hinge and or the LBD of the second monomer. For the constructs containing the NTD, (NTD-DBD-DNA, full-length, full-length-DNA), the e measured SAXS parameters (Table 2) indicate that, the NTD is rather extended as shown by a significant increase of the Dmax parameter, consistent with the disordered state of this domain.
Figure 2. SAXS of RXRα homodimeric complexes.
(A) Scattering profiles of RXRα DBD-Ramp2 DR1 (1, red), RXRα LBD (2, green), RXRαΔNTD-Ramp2 DR1 (3, blue), RXRα NTD-DBD-Ramp2 DR1 (4, pink), and RXRα-Ramp2 DR1 (5, cyan). Experimental data are denoted by black dots, the corresponding fits are given as solid lines. Fits are computed from the crystal structures of RXRα LBD (PDB: 2ZY0) and of RXRα DBD-Ramp2 DR1 (PDB: 4CN2) or from refined SAXS refined models. The profiles are arbitrary displaced in logarithmic scale for better visualization. (B) Distance distribution functions computed from the X-ray scattering patterns using the program GNOM. (C) Molecular envelope of the complex RXRαΔNTD-Ramp2 shown as grey spheres together with the pseudo-atomic refined structure of RXRαΔNTD-Ramp2 DR1. (D) Pseudo-atomic model RXRαΔNTD-Ramp2 DR1 is asymmetric with the LBD dimer positioned at the 5’end of the DNA. The blue dotted lines indicate the hinge domain connecting the DBD to LBD.
Table 2. Small Angle X-ray parameters.
| Complexes | Rg, Å | Dmax, Å | χ |
|---|---|---|---|
| RXRα DBD-Ramp2 DR1 | 18.5 ± 0.7 | 67 ± 5 | 1.18 |
| RXRα LBD | 24.5 ± 0.5 | 80 ± 5 | 1.41 |
| RXRα NTD-DBD-Ramp2 DR1 | 43 ± 2 | 160 ± 20 | 1.50 |
| RXRα ΔNTD-Ramp2 DR1 | 36 ± 0.5 | 125 ± 7 | 1.08 |
| RXRα full-length | 41 ± 3 | 150 ± 20 | 0.90 |
| RXRα full-length-Ramp2 DR1 | 44 ± 2 | 160 ± 20 | 0.93 |
Rg, and Dmax, are the radius of gyration and maximum size, calculated from the scattering data. χ is the discrepancy between the experimental data and the predicted scattering curves. The predicted scattering curves were obtained from the appropriate crystal structures in case of RXR LBD and RXR DBD-Ramp2 DR1, from the rigid body model for RXR ΔNTD-Ramp2 DR1 and from EOM ensembles for the two constructs containing NTD.
Characterization of the N-terminal domain of RXRα
The analysis of the primary sequence of the NTD of RXRα is presented in Figure S1. A consensus of the prediction methods indicates that the NTD is disordered. The overall charge and hydrophobicity values calculated with PONDR (www.pondr.com) are <R> = 0.004 and <H> = 0.466, respectively, and consensus prediction of secondary structure indicates a disordered random coil. The NTD of RXRα is characterized by a large proportion of P and G residues, a usual feature for intrinsically disordered proteins. However some amino acids, less frequently found in disordered regions, such as F, H and M are also present all along the NTD, indicating that some structuring may occur, either transiently or upon interaction with partners.
Experimental evidence of the disordered state of the RXRα NTD was provided by liquid state NMR. The full-length RXRα was produced in E. coli and labeled with 13C and 15N. 1H-15N and 1H-13C HSQC spectra were recorded at 279K. At this temperature, no signal from the folded parts of the protein is expected due to very fast transverse relaxation times resulting from the slow tumbling correlation time of the large RXRα homodimer. Indeed, the 1H-15N HSQC correlation spectrum displays about 90 sharp peaks with amide proton frequencies characteristic of unfolded state (between 7.8 and 8.3 ppm) (Figure 3). The number of correlation peaks (111) is rather close to the number of correlations expected from the sequence composition of the RXRα NTD (133). The missing resonances may be due to cis-trans isomerization of the numerous proline residues (21) that are present in the RXRα NTD. Furthermore, 13 high intensity peaks could be identified in the region of the 1H-15N HSQC that corresponds to glycine 15N chemical shifts (107-112 ppm), a number that correlates well with the 14 glycines present in the RXRα NTD. These observations suggest that RXRα NTD is disordered in solution. Upon addition of the Ramp2 DR1, most of the amide resonances are unaffected with only 7 correlation peaks displaying slight chemical shift perturbations. Overall, these experiments indicate that the NTD of RXRα in context of the full-length protein is mostly disordered, and its state is only marginally affected upon binding to DNA.
Figure 3. Superimposition of the 15N HSQC spectrum of full length RXRα at 279K before (in blue) and after (in red) addition of Ramp2 DR1.
The 15N HSQC spectrum of RXRα full-length (in blue) displays 91 sharp peaks. About 111 peaks were expected from the 133 residues of the NTD (that contains 21 prolines). Upon addition of Ramp2 DR1 (in red), most of the resonances are unaffected with only 7 peaks displaying slight chemical shift perturbations. The missing resonances may be due to cis-trans proline isomerization. The region corresponding to Glycine residues is shown in a box.
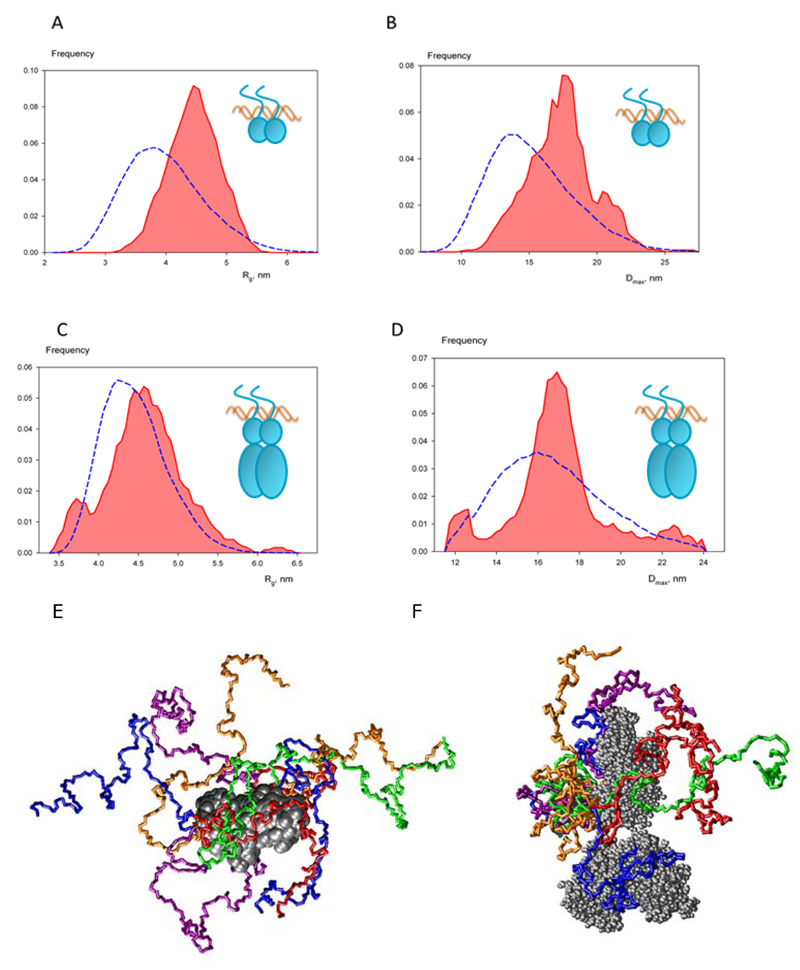
To further characterize the RXRα NTD, we analyzed the ensemble of conformations populated by the RXRα NTD in the context of the full-length RXRα and compared it to the ensemble explored by the NTD-DBD construct using the Ensemble Optimization Method (EOM) 35. In the EOM approach, a genetic algorithm is used to select a representative ensemble of NTD conformations from a large pool of randomly generated models to minimize the discrepancy between the experimental SAXS profile and the ensemble-averaged scattering profile.
For the NTD-DBD-DNA complex, the representative ensemble shown in Figure 4E is in agreement with NMR data indicating a highly disordered and flexible domain. The selected ensemble neatly fits the corresponding scattering profile (Figure 4A and Table 2) and suggests rather extended NTD conformations in both monomers. The extended NTD conformation is also supported by the distribution of Dmax and especially Rg values which is noticeably larger than the average of the random pool (Figure 4A and 4B). EOM analysis performed on the complex with the full-length RXRα produced in insect cells yields a rather good fit to the SAXS profile (Figure 2A and Table 2). Surprisingly, EOM results for the full-length RXRα-DNA complex clearly indicate a more compact conformational ensemble compared to the NTD-DBD/DNA construct (Figure 4F). This finding is also supported by the similar Rg values observed for the full length RXRα and RXRα NTD-DBD complexes, as defined by Guinier approximation (Table 2, and Rg and Dmax distributions in Figure 4C and 4D). The EOM results obtained for the RXRfull-length without DNA (data not shown) could not be compared to the ensemble of conformers of the NTD in context of the DNA bound complex because of the flexibility of the DBDs and the absence of dimerization of the DBDs in absence of DNA. Overall, these data demonstrate a high degree of flexibility of the NTDs that is restrained by the space occupied by the hinge and LBD dimer in the context of the full-length DNA bound protein. The lower number of extended conformations of the NTD in context of the full-length protein may also be explained by possible transient interactions between the NTD and the folded domains as was previously shown for steroidal NRs 40, 41.
Figure 4. Ensemble of conformations of the intrinsically unfolded N-terminal domain of RXRα.
(A-D) Size and shape distributions: Rg (A and C) and Dmax (B and D) for RXRα NTD-DBD-Ramp2 DR1 (A and B) and RXRα-Ramp2 DR1 (C and D). Distributions are computed from selected structures (colored red) and compared to random pools generated by EOM (blue dashed lines). (E) Ensemble of conformers of the NTDs of RXRα in RXRα NTD-DBD-Ramp2 DR1. The DNA is not represented for sake of clarity. (F) Ensemble of conformers of the NTDs of RXRα in the full-length complex.
Discussion
The integrity of the nuclear receptor protein is required for its full transcriptional activity in the cell. In particular, the NTD of NR is essential for the modulation of the transactivation potential of the receptor in a cell-, promoter- and/or ligand-dependent manner. Structural studies reported up to now have mainly focused on the two isolated globular receptor domains, the DBD and the LBD, preventing a complete understanding of the molecular mechanisms underlying the allosteric regulations achieved by NR.
In the present study, we have characterized the solution behavior of the RXRα NTD in context of the intact RXR homodimer either free or bound to the Ramp2 DR1 DNA. The Ramp2 DR1 has been shown to modulate the conformation of the RXR DBD-DNA allowing a strong homodimerization and increased binding affinity 36. SAXS studies of the functional RXRα homodimer bound to Ramp2 DR1 revealed the asymmetry of the NR dimer that is induced by the non-symmetric DNA target, positioning both LBDs at the 5’end of the target DNA. This observation is reminiscent to the solution and cryo-EM structures of heterodimeric NRs, such as RXR-RAR or RXR-VDR, bound to their cognate direct repeat HREs 37, 38. It highlights the critical role played by the DNA together with the hinge region linking the DBD to the LBD in promoting proper inter-domain interactions that stabilize a dominant receptor topology. Indeed, RXR heterodimers are able to recognize highly diverse HREs (DR0-8 and IR) through adapting different conformations. While the in vivo relevance of RXR homodimer remains to be proven, deep sequencing data have revealed that regulatory DNA binding sites are very often pre-bound by RXR in absence of ligand stimulation suggesting a possible role of RXR homodimer as a pre-existing DNA-bound inhibitor of transcription that readily exchanges its dimeric partner in a ligand-driven way 42, 43. A role of RXR homodimer in transcriptional regulation in macrophages has also been recently described 44, 45.
Using SAXS and NMR, we were able to provide insights on the hydrodynamic behavior of the RXR NTD region in the context of the full-length protein. NMR indicates that the NTD is mostly disordered and remains flexible upon binding to Ramp2 DR1. The slight decrease of binding affinity induced by the presence of the disordered NTD observed by ITC experiments may be explained by local interactions between NTD residues and the DNA as suggested by the few chemical shift perturbations observed upon DNA binding. SAXS data provided further evidence about the disordered state of NTD in DNA-bound proteins, in agreement with HDX experiments 23. Similarly to the RXRα NTD, the NTDs of steroidal NRs and of the ecdysone receptor have been shown to be intrinsically disordered, adopting a premolten globule-like state 46, 47. However, the NTDs of steroidal NRs are known to undergo folding into secondary/tertiary structure upon interaction with multiple proteins and DNA 48. Such DNA-induced folding of the steroidal NRs NTDs contrasts with the weak perturbations of chemical shifts observed for the RXRα NTD upon DNA binding. The difference between the displayed properties of the NTDs of RXRα and other steroidal NRs in the DNA-bound form may be related to their distinct primary sequence organization. In steroidal NRs, the NTD itself is often highly modular and it comprises multiple regions contributing to receptor-dependent gene regulation 12 while the NTD of RXRα does not display such distinct regulatory areas.
The NTD of RXR, like the NTDs of steroidal NRs containing the major transactivation function AF1, is naturally intrinsically disordered, and this structural plasticity is functionally important for NR activity. Folding of the AF1/NTD domain upon interaction with protein partners is likely to contribute to cell-specific receptor actions. The RXR-NTD exists as an ensemble of conformers with a different degree of local order, which prime this region of the receptor to rapidly respond to changes in the intracellular environment through hormone binding and post-translation modifications 49, 50. For RXRα, several proteins have been shown to potentiate the transcriptional activity of the AF1 domain of RXR, such as Bcl3 51. It would be of high interest to investigate how the conformational ensemble of the RXRα NTD is modulated upon interactions with protein partners.
Associated Content
The Supporting Information is available free of charge on the ACS Publications website at xxx.
One figure describes the domain organization of hRXRα and the in silico analysis of the RXRα NTD sequence composition and disorder predictions.
Acknowledgements
We thank Isabelle Kolb-Cheynel (IGBMC Baculovirus Facility) for productions in insect cells and the Structural Biology and Genomics platform of the IGBMC.
Funding
The project was supported by the Centre National pour la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Agence Nationale de Recherche (ANR-11-BSV8-023), the Association pour la Recherche sur le Cancer (ARC) (SFI20121205585) and Alsace contre le Cancer, the Fondation pour la Recherche Médicale (FRM), the WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, www.wenmr.eu) and the BMBF research grant BioSCAT (Contract no: 05K2012) and French Infrastructure for Integrated Structural Biology (FRISBI) (ANR-10-INSB-05-01), and INSTRUCT as part of the European Strategy Forum on Research Infra-structures (ESFRI). D.I.S. and M.V.P. were supported by the EU FP7 infrastructures grant Biostruct-X, Contract No 283570. A.Y.B was supported by a FRM doctoral fellowship (FDT20140930978).
Abbreviations
- NR
nuclear receptor
- RXR
retinoid X receptor
- DBD
DNA binding domain
- LBD
ligand binding domain
- NTD
N-terminal domain
- AF
activation function
- DR
direct repeat
- DR1
direct repeat separated by one nucleotide
- IR
inverted repeat
- ER
everted repeat
- HRE
hormone response element
- ITC
isothermal titration calorimetry
- SAXS
small angle X-ray scattering
- NMR
nuclear magnetic resonance
- Cryo-EM
cryo electron microscopy
- HDX
hydrogen-deuterium exchange
- Kd
dissociation constant
- Rg
gyration radius
- Dmax
maximal dimension
Footnotes
Notes
The authors declare no competing financial interest.
References
- [1].Lefebvre P, Benomar Y, Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- [2].Gilardi F, Desvergne B. RXRs: collegial partners. Sub-cellular biochemistry. 2014;70:75–102. doi: 10.1007/978-94-017-9050-5_5. [DOI] [PubMed] [Google Scholar]
- [3].Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58:712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- [4].Brocard J, Kastner P, Chambon P. Two novel RXR alpha isoforms from mouse testis. Biochem Biophys Res Commun. 1996;229:211–218. doi: 10.1006/bbrc.1996.1782. [DOI] [PubMed] [Google Scholar]
- [5].Mader S, Leroy P, Chen JY, Chambon P. Multiple parameters control the selectivity of nuclear receptors for their response elements. Selectivity and promiscuity in response element recognition by retinoic acid receptors and retinoid X receptors. J Biol Chem. 1993;268:591–600. [PubMed] [Google Scholar]
- [6].Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes & development. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- [7].Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- [8].Jin L, Li Y. Structural and functional insights into nuclear receptor signaling. Adv Drug Deliv Rev. 2010;62:1218–1226. doi: 10.1016/j.addr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Molecular and cellular endocrinology. 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- [10].Gallastegui N, Mackinnon JA, Fletterick RJ, Estebanez-Perpina E. Advances in our structural understanding of orphan nuclear receptors. Trends in biochemical sciences. 2015;40:25–35. doi: 10.1016/j.tibs.2014.11.002. [DOI] [PubMed] [Google Scholar]
- [11].Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kumar R, McEwan IJ. Allosteric modulators of steroid hormone receptors: structural dynamics and gene regulation. Endocrine reviews. 2012;33:271–299. doi: 10.1210/er.2011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Warnmark A, Wikstrom A, Wright AP, Gustafsson JA, Hard T. The N-terminal regions of estrogen receptor alpha and beta are unstructured in vitro and show different TBP binding properties. J Biol Chem. 2001;276:45939–45944. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- [14].Wardell SE, Kwok SC, Sherman L, Hodges RS, Edwards DP. Regulation of the amino-terminal transcription activation domain of progesterone receptor by a cofactor-induced protein folding mechanism. Mol Cell Biol. 2005;25:8792–8808. doi: 10.1128/MCB.25.20.8792-8808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry-Us. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- [16].Kumar R, Baskakov IV, Srinivasan G, Bolen DW, Lee JC, Thompson EB. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem. 1999;274:24737–24741. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- [17].Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002;277:20079–20086. doi: 10.1074/jbc.M201003200. [DOI] [PubMed] [Google Scholar]
- [18].Brodie J, McEwan IJ. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. Journal of molecular endocrinology. 2005;34:603–615. doi: 10.1677/jme.1.01723. [DOI] [PubMed] [Google Scholar]
- [19].Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of human progesterone B-receptors: biophysical and biochemical comparison to A-receptors. J Biol Chem. 2001;276:23825–23831. doi: 10.1074/jbc.M102611200. [DOI] [PubMed] [Google Scholar]
- [20].Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- [21].Harish S, Ashok MS, Khanam T, Rangarajan PN. Serine 27, a human retinoid X receptor alpha residue, phosphorylated by protein kinase A is essential for cyclicAMP-mediated downregulation of RXRalpha function. Biochem Biophys Res Commun. 2000;279:853–857. doi: 10.1006/bbrc.2000.4043. [DOI] [PubMed] [Google Scholar]
- [22].Mascrez B, Mark M, Krezel W, Dupe V, LeMeur M, Ghyselinck NB, Chambon P. Differential contributions of AF-1 and AF-2 activities to the developmental functions of RXR alpha. Development. 2001;128:2049–2062. doi: 10.1242/dev.128.11.2049. [DOI] [PubMed] [Google Scholar]
- [23].Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson JA. Structure of the retinoid X receptor alpha-liver X receptor beta (RXRalpha-LXRbeta) heterodimer on DNA. Nat Struct Mol Biol. 2014;21:277–281. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- [25].Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Juntunen K, Rochel N, Moras D, Vihko P. Large-scale expression and purification of the human vitamin D receptor and its ligand-binding domain for structural studies. Biochem J. 1999;344(Pt 2):297–303. [PMC free article] [PubMed] [Google Scholar]
- [27].Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, Svergun D. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Crystallogr. 2007;40:S190–S194. [Google Scholar]
- [28].Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI, Roessle M. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J Appl Crystallogr. 2008;41:913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- [30].Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- [31].Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lippert WP, Burschka C, Gotz K, Kaupp M, Ivanova D, Gaudon C, Sato Y, Antony P, Rochel N, Moras D, Gronemeyer H, et al. Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology. ChemMedChem. 2009;4:1143–1152. doi: 10.1002/cmdc.200900090. [DOI] [PubMed] [Google Scholar]
- [33].Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- [35].Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- [36].Osz J, McEwen AG, Poussin-Courmontagne P, Moutier E, Birck C, Davidson I, Moras D, Rochel N. Structural basis of natural promoter recognition by the retinoid X nuclear receptor. Scientific reports. 2015;5:8216. doi: 10.1038/srep08216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, Svergun DI, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- [38].Orlov I, Rochel N, Moras D, Klaholz BP. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 2012;31:291–300. doi: 10.1038/emboj.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Maletta M, Orlov I, Roblin P, Beck Y, Moras D, Billas IM, Klaholz BP. The palindromic DNA-bound USP/EcR nuclear receptor adopts an asymmetric organization with allosteric domain positioning. Nat Commun. 2014;5:4139. doi: 10.1038/ncomms5139. [DOI] [PubMed] [Google Scholar]
- [40].Metivier R, Penot G, Flouriot G, Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–1970. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- [41].He B, Wilson EM. The NH(2)-terminal and carboxyl-terminal interaction in the human androgen receptor. Mol Genet Metab. 2002;75:293–298. doi: 10.1016/S1096-7192(02)00009-4. [DOI] [PubMed] [Google Scholar]
- [42].Chatagnon A, Veber P, Morin V, Bedo J, Triqueneaux G, Semon M, Laudet V, d'Alche-Buc F, Benoit G. RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nunez V, Alameda D, Rico D, Mota R, Gonzalo P, Cedenilla M, Fischer T, Bosca L, Glass CK, Arroyo AG, Ricote M. Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc Natl Acad Sci U S A. 2010;107:10626–10631. doi: 10.1073/pnas.0913545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Menendez-Gutierrez MP, Roszer T, Fuentes L, Nunez V, Escolano A, Redondo JM, De Clerck N, Metzger D, Valledor AF, Ricote M. Retinoid X receptors orchestrate osteoclast differentiation and postnatal bone remodeling. J Clin Invest. 2015;125:809–823. doi: 10.1172/JCI77186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lavery DN, McEwan IJ. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry-Us. 2008;47:3360–3369. doi: 10.1021/bi702221e. [DOI] [PubMed] [Google Scholar]
- [47].Nocula-Lugowska M, Rymarczyk G, Lisowski M, Ozyhar A. Isoform-specific variation in the intrinsic disorder of the ecdysteroid receptor N-terminal domain. Proteins. 2009;76:291–308. doi: 10.1002/prot.22342. [DOI] [PubMed] [Google Scholar]
- [48].Hilser VJ, Thompson EB. Structural dynamics, intrinsic disorder, and allostery in nuclear receptors as transcription factors. J Biol Chem. 2011;286:39675–39682. doi: 10.1074/jbc.R111.278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Current opinion in structural biology. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- [50].Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chemical reviews. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- [51].Na SY, Choi HS, Kim JW, Na DS, Lee JW. Bcl3, an IkappaB protein, as a novel transcription coactivator of the retinoid X receptor. J Biol Chem. 1998;273:30933–30938. doi: 10.1074/jbc.273.47.30933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.