Abstract
The directional transport of auxin, known as polar auxin transport, allows asymmetric distribution of this hormone in different cells and tissues. This system creates local auxin maxima, minima and gradients that are instrumental in both organ initiation and shape determination. As such, polar auxin transport is crucial for all aspects of plant development but also for environmental interaction, notably in shaping plant architecture to its environment. Cell-to-cell auxin transport is mediated by a network of auxin carriers that are regulated at the transcriptional and post-translational levels. Here we review our current knowledge on some aspects of the ‘non-genomic’ regulation of auxin transport, putting an emphasis on how phosphorylation by protein and lipid kinases controls the polarity, intracellular trafficking, stability and activity of auxin carriers. We describe the role of several AGC kinases, including PINOID, D6PK and the blue light photoreceptor phot1, in phosphorylating auxin carriers from the PIN and ABCB families. We also highlight the function of some Receptor-Like Kinases (RLK) and two-component histidine kinase receptors in polar auxin transport, noticing that there are likely RLKs involved in coordinating auxin distribution yet to be discovered. In addition, we describe the emerging role of phospholipid phosphorylation in polarity establishment and intracellular trafficking of PIN proteins. We outline these various phosphorylation mechanisms in the context of primary and lateral root development, leaf cell shape acquisition as well as root gravitropism and shoot phototropism.
Keywords: kinase, auxin, intracellular trafficking, polarity, gravitropism, phototropism, root, endocytosis, Arabidopsis, receptor-like kinase, cytokinin, phosphoinositide
Introduction
Auxin (Indole 3-Acetic Acid, IAA) is a small molecule plant hormone derived from the amino acid tryptophan that controls virtually all aspects of the plant life. In particular, auxin is of paramount importance for both development and response to the environment and as such is a determining factor in the acquisition of the final shape of plants (Finet and Jaillais, 2012; Weijers and Wagner, 2016). A particularity of auxin is that it is actively transported across cells and tissues by specialized, plasma membrane localized, influx and efflux carriers. The combined activity of these transporters allows the generation of auxin gradients, as well as auxin maximums and minimums that are critical for organ patterning and differential growth during tropic responses (i.e. growth of the plants toward or against an environmental stimulus) (Finet and Jaillais, 2012). The combination of each auxin concentration in a given cell induces specific transcriptional programs that can have a wide range of outputs, including cell differentiation into various cell types and activation or inhibition of elongation growth. Auxin regulates transcription downstream of its perception by the TIR1 auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Weijers and Wagner, 2016). Briefly, TIR1 (and related family members called AUXIN SIGNALING F-BOX, AFBs) are F-BOX E3 ubiquitin ligases that interact with and trigger the degradation of AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional co-repressors in the presence of auxin (Weijers and Wagner, 2016). Aux/IAA degradation releases Aux/IAA-mediated inhibition of AUXIN RESPONSE FACTORs (ARFs), which are transcription factors that regulate the transcriptional output regulated by auxin (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Weijers and Wagner, 2016). Furthermore, this transcriptional profile is plugged into an intricate network of hormone, developmental and environmental responses, which together decide the output cellular responses (Finet and Jaillais, 2012; Jaillais and Chory, 2010). These downstream events in auxin signaling have been reviewed elsewhere and will not be further discussed in this review (Habets and Offringa, 2014; Hagen, 2015; Korasick et al., 2015; Larrieu and Vernoux, 2015).
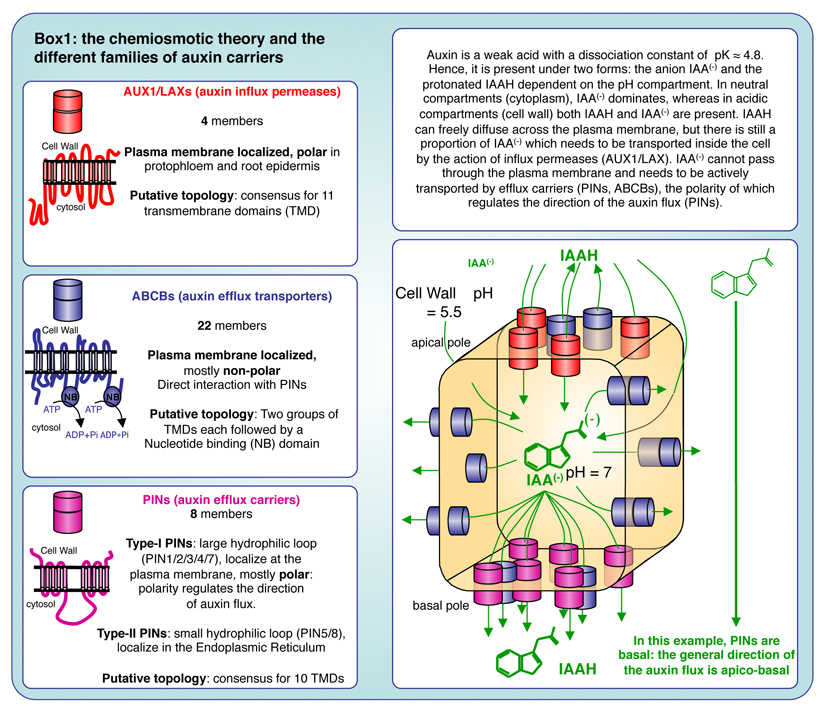
Upstream of gene regulation, a decisive and characteristic feature of auxin is the establishment of auxin gradients and/or maxima, which are established through cell-to-cell polar auxin transport. The chemiosmotic model of auxin transport that was formulated by Rubery and Sheldrake as well as by Mary Helen and Timothy Goldsmith, proposed that the protonation state of auxin, as a weak acid, is dictated by the pH of its environment (Goldsmith and Goldsmith, 1981; Goldsmith et al., 1981; Rubery and Sheldrake, 1973, 1974). The cell wall is acidic (pH ≈ 5.5), and therefore a significant proportion of auxin (≈ 20%) is in its protonated state (IAAH), which can freely diffuse across biological membranes. Inside the cell, the pH of the cytosol is neutral (pH ≈ 7) and most auxin is in its anionic form IAA-, which cannot diffuse across membrane and thereby, is trapped inside the cell (Box 1). As a consequence, the chemiosmotic hypothesizes postulated that plasma membrane auxin efflux carriers must exist to ensure the transport of auxin outside the cell. Indeed, it was latter demonstrated that there are three main families of transmembrane proteins that transport auxin across the plasma membrane: i) AUX1/LIKE AUX1 (AUX1/LAX), which are auxin influx permeases, ii) ATP-binding cassette subfamily B (ABCB) transporters, which are auxin efflux transporters and iii) PIN-FORMED proteins (PIN), which are auxin efflux carriers (Box 1) (Finet and Jaillais, 2012). All these subfamilies have been shown to independently transport auxin in planta and in heterologous systems (Barbez et al., 2013; Geisler et al., 2005; Grones and Friml, 2015; Petrasek et al., 2006; Yang and Murphy, 2009; Yang et al., 2006; Zourelidou et al., 2014). However, these studies also suggest that PINs and ABCBs interact and function both independently and interdependently to control polar auxin transport in planta (Bandyopadhyay et al., 2007; Blakeslee et al., 2007; Titapiwatanakun et al., 2009). In particular, PINs/ABCBs form protein complexes that are stabilized by ABCB proteins (Blakeslee et al., 2007; Titapiwatanakun et al., 2009). The PIN family is itself divided into several subfamilies based on the size of their intracellular loop: type-I PIN proteins (PIN1, PIN2, PIN3, PIN4 and PIN7) present a long hydrophilic loop (PINHL) and are mainly localized at the plasma membrane (box 1), PIN6 presents a partially reduced hydrophilic loop, while type-II PINs (PIN5 and PIN8), present a short hydrophilic loop and are localized in the Endoplasmic Reticulum (Cazzonelli et al., 2013; Ganguly et al., 2014; Krecek et al., 2009; Zazímalová et al., 2010)(Bennett, 2015; Finet and Jaillais, 2012; Habets and Offringa, 2014). In addition, the NRT1.1 nitrate sensor/transporter also facilitates auxin uptake at low NO3- concentration (Krouk et al., 2010).
Box1.
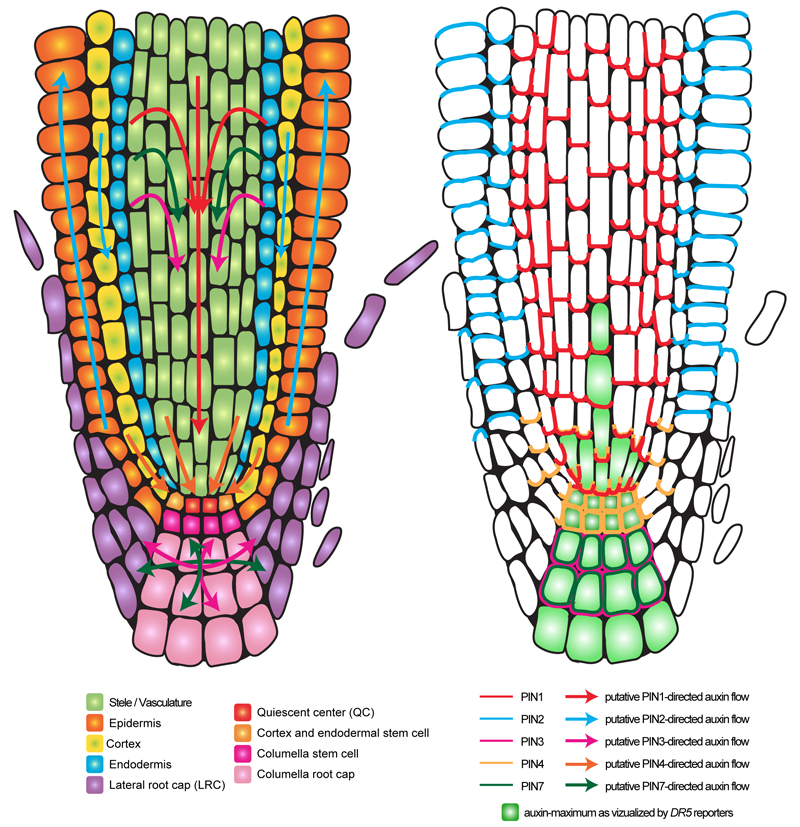
All these auxin carriers have been shown to be important for auxin fluxes in planta (Band et al., 2014). However, the auxin carrier family that has received the most attention is the PINs, because it confirmed another visionary prediction from the chemiosmotic theory: that a polar localization of an “auxin secreting system” could explain the directionality of auxin flow, providing that this polarity is coordinated at the tissue and organ level (Rubery and Sheldrake, 1974). In many cell types, type-I PIN proteins are indeed polarly localized and this polar localization was shown to directly correlate with the auxin flow direction (Adamowski and Friml, 2015; Band et al., 2014; Benkova et al., 2003; Blilou et al., 2005; Friml et al., 2002a; Friml et al., 2003; Galweiler et al., 1998; Habets and Offringa, 2014; Muller et al., 1998; Petrasek et al., 2006; Wisniewska et al., 2006). Although, to date, it is not possible to directly visualize the direction of auxin flow in plants, the polar localization of PIN proteins has been widely used as a proxy to deduce these fluxes (Wisniewska et al., 2006). The assumption that auxin transport route can be deduced from PIN polar localization is solidly backed-up by both experimental data and computer simulations (Band et al., 2014; Blilou et al., 2005; Grieneisen et al., 2007). PIN localization also correlates with expected auxin accumulation and depletion sites visualized by the synthetic output (transcriptional) reporters DR5, DR5rev and DR5v2, as well as auxin input reporters DII and R2DII (Benkova et al., 2003; Brunoud et al., 2012; Larrieu and Vernoux, 2015; Liao et al., 2015; Sabatini et al., 1999; Ulmasov et al., 1997; Vernoux et al., 2011). However, computational modeling also revealed the importance of both auxin efflux (by PINs and ABCBs) as well as influx (by AUX1/LAXs) carriers for the correct accumulation of auxin in the primary root tip (Band et al., 2014). Furthermore, PIN loss-of-function mutants (and higher order mutants, due to high functional redundancy between PIN family members and compensatory mechanisms) failed to form normal auxin maximum (Adamowski and Friml, 2015; Benkova et al., 2003; Blilou et al., 2005; Friml et al., 2002a; Friml et al., 2003; Vieten et al., 2005). Localizing the various PIN proteins therefore allowed drawing maps of auxin flow in various developmental contexts. As an example, Figure 1 shows putative auxin transport routes in the primary root, as inferred from PIN1, PIN2, PIN3, PIN4 and PIN7 localization. In the primary root, the most noticeable PIN localization are as follow: i) PIN1 (together with PIN3 and PIN7) in the stele, PIN2 in the cortex and PIN4 in three to four cells above the quiescent center (QC) localize at the basal end of the cell thereby directing auxin toward the tip of the root (acropetal auxin flow) (Band et al., 2014; Blilou et al., 2005; Friml et al., 2002a; Muller et al., 1998), ii) PIN4 in the QC and surrounding initials and PIN3/PIN7 in the columella are not polarly localized, redirecting auxin away from this region (basipetal auxin flow) (Band et al., 2014; Friml et al., 2002a; Friml et al., 2002b), iii) PIN2 in the epidermis is localized in the apical end of the cells, moving auxin back up the root and (Abas et al., 2006; Band et al., 2014; Blilou et al., 2005; Muller et al., 1998), iv) PIN1 in the endodermis and pericycle is not only localized at the basal pole of the cell, but also on the inner lateral plasma membrane domain (facing the stele), which allows recirculating auxin from the epidermis into the stele (Band et al., 2014; Blilou et al., 2005). This fountain-like pattern of auxin flow in the root creates auxin maximum in the QC, surrounding initials and to a lesser extent in the columella and protoxylem. Importantly, the differences in PIN localization are not only regulated by tissue specificity but are also dependent of the PIN proteins themselves. For example, when ectopically expressed in the epidermis, PIN1 is localized at the basal and apical end of the cell, while PIN2 is only apical (Marhavy et al., 2014; Wisniewska et al., 2006).
Figure 1. Polar auxin transport and PIN localization in the primary root tip.
Schematic representation of a longitudinal root section showing the different root tissues (left). The arrows represent putative auxin fluxes (left) as deduced from PIN protein localization (right). For clarity, PIN3 and PIN7 proteins have been omitted from the stele since their localization is redundant with that of PIN1.
While the map of auxin flows drawn in figure 1 appears relatively static, PIN protein localizations are in fact highly dynamic and they can be remodeled by both developmental and environmental cues. For example, a change of PIN protein localization reorients auxin toward the tip of young emerging primordia (e.g. lateral root), which is necessary for the emergence of the new organ (Benkova et al., 2003). In addition, PIN protein localizations, abundance, but also their activities, are altered during tropic responses (Rakusova et al., 2015). This high adaptability of the auxin transport machinery to various developmental and environmental responses has been shown over the years to not only rely on genomic changes, but also on post-translational control of the auxin carrier network. Among post-translational modifications, phosphorylation has been implicated at multiple stages in the control of auxin carriers’ localization and activity. Here, we will review how protein and lipid kinases dynamically set up and control polar auxin transport.
Phosphorylation and the establishment/maintenance of PIN polarity
Regulation of PIN polar targeting by AGCVIII-mediated PIN phosphorylation
In 1991, a recessive mutation in Arabidopsis was described to produce the so-called pin inflorescences, i.e: inflorescences that elongate but that do not produce flower primordia (Okada et al., 1991). The inflorescences of this mutant, named pin1, had reduced downward polar auxin transport. Moreover, wild-type plants grown in media containing high concentrations of auxin transport inhibitors, such as 1-N-Naphthylphthalamic acid (NPA) or 2,3,5-triiodobenzoic acid (TIBA), showed the same inflorescence phenotype, suggesting a role for PIN1 in polar auxin transport (Okada et al., 1991). PIN1 turned out to encode for a multipass membrane protein localized at the basal side of xylem cells in the stem (Galweiler et al., 1998) and later shown to directly transport auxin from the cytosol out of the cell (Petrasek et al., 2006).
In 1995, the pinoid (pid) mutant was described to have a very similar phenotype as pin1, with pin-like inflorescences harboring no or few flowers, which contained an aberrant number of flower organs, altered venation pattern and fused or multiple cotyledons (Benjamins et al., 2001; Bennett et al., 1995; Christensen et al., 2000). This phenotype suggests that PID might regulate polar auxin transport together with PIN1. Genetic analyses showed an enhanced phenotype of the double pin1pid mutant as compared to each single mutant, with a portion of double mutant seedlings having no cotyledon and fully fused first leaves (Furutani et al., 2004; Jaillais et al., 2007). This genetic analysis suggests that PIN1 and PID might act in two separate pathways. However, it was later revealed that this enhanced phenotype of the double mutant very likely reflects the ability of PID to regulate other PINs that act redundantly with PIN1 during embryogenesis (Friml et al., 2004). Alternatively, this phenomenon might be partially explained by the reported interaction between PID and ABCB1 (Henrichs et al., 2012).
The predicted PID protein is a serine-threonine protein kinase. In particular, PID protein falls into the AGC3 group of the plant specific AGCVIII protein kinase family (Christensen et al., 2000; Galvan-Ampudia and Offringa, 2007). PID is an active kinase in vitro (Christensen et al., 2000). Furthermore, overexpression of a kinase dead version of PID fails to induce any phenotype in transgenic Arabidopsis. By contrast, overexpression of wild type PID shows severe developmental defects, including small and dark green curled leaves, as well as reduced apical dominance and impaired elongation of the inflorescence (Christensen et al., 2000). Therefore, PID kinase activity is required for its function in planta. Moreover, 35S::PID plants are sterile or semifertile and, strong PID overexpression produced embryo lethality. At the seedling stage, both roots and hypocotyls are shorter and strongly agravitropic. In addition, emergence of lateral roots is delayed and only occurred upon collapse of the primary root meristem (Benjamins et al., 2001; Christensen et al., 2000). The 35S::PID seedling phenotypes are partially rescued by treatments with the polar auxin transport inhibitor NPA. In addition, the expression of the DR5::GUS auxin reporter in the root tip of 35S::PID plants is reduced, arguing that auxin content in the root tip might be lower due to enhancement of upwards auxin transport (i.e. towards the shoot apex). All these data suggested a role of PID kinase in the regulation of polar auxin transport (PAT), which turned out to be via direct phosphorylation of both PINs and ABCBs auxin efflux carriers (Henrichs et al., 2012; Michniewicz et al., 2007) (Fig. 2). Because of the intertwined relationships between PINs and ABCBs (Bandyopadhyay et al., 2007), it is still unclear to which extent the phenotypes reported above for PID loss- and gain-of-function can be directly attributed to phosphorylation on PINs and/or ABCBs.
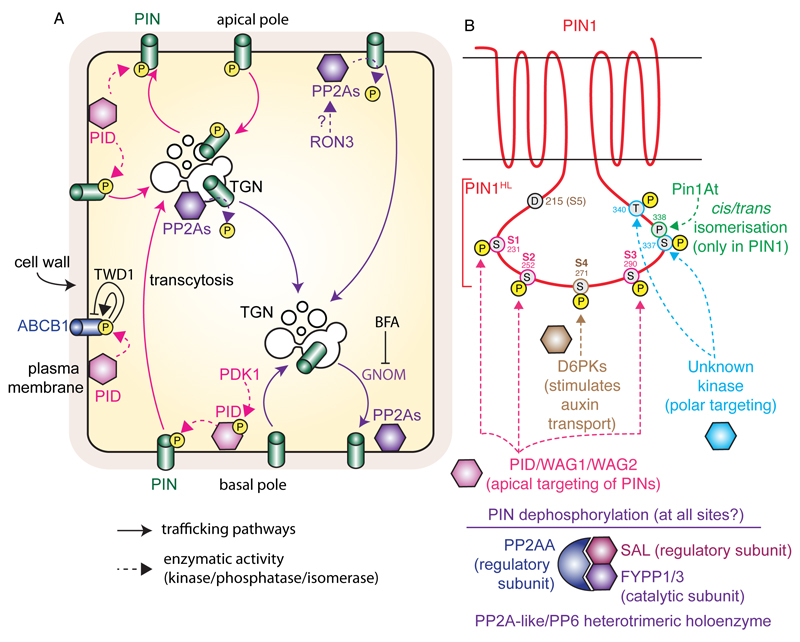
Figure 2. Model for PID and PP2As antagonistic activity on PIN phosphorylation, and the dual action of ABCB1 phosphorylation by PID.
A) Non-phosphorylated PINs undergo continuous endocytic recycling in a GNOM-dependent manner, which mediates their basal localization. The PID kinase phosphorylates PINs at the plasma membrane, which prohibits them from recycling via the GNOM-dependent route. Phosphorylated PINs instead undergo transcytosis to the apical pole of the cell. PINs are dephosphorylated by PP2As phosphatases, presumably at the plasma membrane and endosomes. PID also phosphorylates ABCB1, which promote ABCB1-mediated auxin export in the absence of TWD1, but inhibits it in its presence. B) Schematic representation of PIN1 topology and position of PIN1 phosphorylation site within PIN1 hydrophilic loop (PIN1HL). The action of the kinases/phosphatases/isomerase on specific residues is highlighted. Note that only D6PK main phosphorylation sites (S4 and S5) are highlighted, but that S1, S2 and S3 are also phosphorylated by this kinase, albeit less potently. Similarly, PID/WAG1/WAG2 are able to phosphorylate S4 and S5 but preferentially act on S1, S2 and S3. Dashed arrows represent direct phosphorylation, dephosphorylation or isomerisation events; arrows with chevron-shaped arrowhead represent trafficking pathways and blunt-ended lines represent inhibition. Yellow-filled circles with the letter ‘P’ represent phosphorylation, while grey-filled circles represent individual amino acid (each residue is numbered according to its position within the PIN1 protein).
PID phosphorylates ABCB1, likely on Ser634. Heterologous expression of PID and ABCB1 in tobacco suggests that PID enhances ABCB1 auxin transport activity (Henrichs et al., 2012) (Fig. 2). However, co-expression of PID and ABCB1 with the immunophilin-like TWISTED DARF1 (TWD1) inhibits ABCB1-mediated auxin efflux (Henrichs et al., 2012) (Fig. 2). Therefore, PID has a dual regulatory action on ABCB1 auxin transport, depending on the presence of TWD1. Interestingly, PID not only acts on ABCBs transporters, but also acts directly on PINs. In particular, Friml and colleagues (2004) reported that PID regulates auxin fluxes by determining the subcellular localization of PIN proteins (Fig. 2). PID activity is necessary to specify the delivery of these auxin transporters to the apical side of the cells. The authors showed that overexpression of PID induced apicalization of PINs (localization at the top of the cell or shootward) in root cells where they are normally localized in the basal membrane (at the bottom of the cell or rootward) (Friml et al., 2004). Basal localization of PIN proteins in subepidermal cell files ensures the auxin flux towards the root tip (Figure 1), which is necessary to maintain meristem activity. In 35S::PID plants, apical localization of PIN proteins enhances upwards auxin flux and, as a consequence, reduces the auxin maximum at the root tip and produces the collapse of the root meristem (Friml et al., 2004). By contrast, PIN2 localization in the apical membrane of root epidermal cells was not changed by PID overexpression, but was found in endomembrane compartment in the pid-9 loss-of-function (Friml et al., 2004; Sukumar et al., 2009). In addition, PID is mainly expressed in the epidermis of root meristem where PIN2 is apical and PID expression is absent in subepidermal cell files where PINs are basally localized (Friml et al., 2004; Kleine-Vehn et al., 2009; Michniewicz et al., 2007). PIN1 apicalization was also observed in embryo cells of 35S::PID plants and more clearly in transactivated RPS5A>>PID plants (RPS5a promoter being strongly expressed in dividing cells early during embryogenesis) (Friml et al., 2004). An apical-to-basal auxin flow established by basal PIN localization determines auxin maxima in the embryo hypophysis, which maintains primary root meristem formation during embryogenesis (Friml et al., 2003). Apicalization of PIN1 (and other PINs) in RPS5A>>PID embryo cells produces strong embryo patterning defects due to accumulation of auxin at the embryo proper to the detriment of the hypophysis (Friml et al., 2004). As a consequence, neither the root nor the cotyledons are properly specified in these plants. On the other hand, an apical-to-basal shift of PIN1 was observed in the epidermal cells of the pid inflorescences apex (Friml et al., 2004). PID expression at the flanks of the inflorescence apex nicely correlates with the PIN1 apical localization at these sites (Christensen et al., 2000). PIN1 mislocalization in pid plants disrupts the levels of auxin required for floral organ initiation leading to pin-like inflorescences similar to those of pin1 mutants (Friml et al., 2004).
Similar phenotypes to those of 35S::PID seedlings and RPS5A>>PID embryos were found in a set of loss-of-function mutants for multiple isoforms of the A subunits of protein phosphatase 2 (PP2AA1 (also known as ROOT CURLING ON NPA1, RCN1 (Garbers et al., 1996; Rashotte et al., 2001)), PP2AA2 and PP2AA3) (Michniewicz et al., 2007). PP2AAs are part of the PROTEIN PHOSPHATASE6 (PP6) holoenzyme and form a heterotrimeric complex with the catalytic subunit FYPP1/3 (for PHYTOCHROME-ASSOCIATED SERINE/THREONINE PROTEIN PHOSPHATASE1 and 3) and SAL (for SAL DOMAIN-LIKE which is the regulatory B subunit) (Fig. 2B) (Dai et al., 2012). The PP6 holoenzyme is a PP2A-like complex that incorporate the regulatory subunit PP2AA and SAL and will be referred with the generic name PP2A hereafter. Similar to PID overexpression, apicalization of PIN proteins was also observed in multiple mutants for pp2aa subunits (pp2a1;3 double mutants), fypp subunits (fypp1;3 double mutants) and the sal subunit (artificial microRNA targeting all four SAL genes) (Dai et al., 2012; Michniewicz et al., 2007). These observations indicated that PID and PP2A are part of the same pathway and that PID activity on PIN subcellular localization is counteracted by protein phosphatase PP2A (Michniewicz et al., 2007) (Fig. 2B). Mechanistically, PIN proteins where shown to be direct substrates of PID and PP2A (Ballesteros et al., 2013), suggesting that (de)phosphorylation is the signaling event that determines PIN polarity: apical when phosphorylated and basal when dephosphorylated (Dai et al., 2012; Michniewicz et al., 2007). In addition, the PP2A holoenzyme is regulated by ROTUNDA3 (RON3), which copurify with various PP2A components and controls PIN polar targeting (Karampelias et al., 2016).
PID phosphorylation of PIN proteins mainly occurs at the plasma membrane (Simon et al., 2016), where both type of proteins colocalize (Kleine-Vehn et al., 2009; Michniewicz et al., 2007) (Fig. 2A). Similar to 35S::PID and pp2a plants, PIN1 basal localization is also lost in gnom loss-of-function mutants or treatment with the fungal toxin brefeldine A (BFA) (Kleine-Vehn et al., 2009). PIN1 constitutively cycles from the plasma membrane to endosomal compartments and BFA blocks the recycling of PIN1 towards the plasma membrane, inducing its accumulation in intracellular compartments and, thus, the depletion from its polar membrane localization (Geldner et al., 2001). Basal PIN polar recycling is maintained by the activity of the GNOM protein, a GDP/GTP exchange factor for small G proteins of the ARF class (ARF-GEF) (Geldner et al., 2003; Steinmann et al., 1999). GNOM is targeted by BFA, which induces its ectopic localization in endosomes and as a result inhibits endocytic recycling (Geldner et al., 2003; Naramoto et al., 2014). Indeed, transgenic plants expressing BFA-resistant GNOM versions did not present changes in PIN1 polar localization upon BFA treatment (Geldner et al., 2003). Consistently with PIN apicalization in the root, gnom partial loss-of-function mutants (gnomR5) displayed reduced auxin maxima at the root tip and meristem collapse similar to 35S::PID plants (Geldner et al., 2004; Kleine-Vehn et al., 2009). Moreover, 35S::PID;gnomR5 plants showed strong developmental defects such as loss of embryonic apical-basal patterning which leads to aberrant root and shoot formation (Kleine-Vehn et al., 2009). The phenotypic similarities of the single 35S::PID and gnomR5 mutants, together with the enhancement of the developmental phenotypes in the double mutant plants and the apicalization of PIN1 observed in both mutants, suggested that PID and GNOM are antagonist modulators of PIN1’s subcellular localization. On the other hand, inhibition of GNOM by BFA treatment in 35S::PID plants or pp2a mutants (in which PIN1 is mislocalized to the apical side of root stele cells) did not induce accumulation of PIN1 in endosomes, suggesting that the apical localization of PIN proteins is GNOM-independent (Kleine-Vehn et al., 2009). It is likely that PIN phosphorylation by PID reduces the affinity of PIN proteins to undergo constitutive recycling at the basal membrane and drives its location to the apical membrane via the GNOM-independent pathway (Kleine-Vehn et al., 2009) (Fig. 2A). This model was confirmed by the observation that PIN2 transcytosis in root cortical cells induced by mild BFA treatments was enhanced in pp2a mutant and delayed in pid background (Kleine-Vehn et al., 2009). It was further supported by the fact that PIN1 carrying mutations that block its phosphorylation (phosphomutants) accumulated in intracellular compartments upon BFA treatment, while mutations that mimic PIN1 phosphorylation (phosphomimic) did not (see below).
PINOID phosphorylates PIN1 in its hydrophilic loop (HL) in the highly conserved PIN specific motif TPRXS(N/S) (Huang et al., 2010; Michniewicz et al., 2007; Zourelidou et al., 2014). Ser-231 (S1), Ser-252 (S2) and Ser-290 (S3) within the three TPRXS(N/S) motifs of PIN1HL were described as in vitro PID phosphorylation sites (Fig. 2B). The biological significance of PIN1 phosphorylation in planta was tested by generation of PIN1::PIN1-GFP transgenic plants that contain none, one, two or three of the phosphorylated Ser residues substituted by Glu (to mimic phosphorylation) or by Ala (a nonphosphorylatable residue) (Huang et al., 2010). The PIN1::PIN1S1A-GFP and PIN1::PIN1S1,3A-GFP lines showed developmental defects at the seedling and reproductive stages, which resembled those of pid loss-of-function mutants: formation of three cotyledons and aberrant number of floral organs. Expression of the triple PIN1S1,2,3A-GFP mutant caused severe defects leading to high frequency of embryo lethality or growth arrest upon germination (Huang et al., 2010). Moreover, expression of the PIN1S1,3A-GFP and PIN1S1,2,3E-GFP failed to complement the pin1 mutant phenotypes (aberrant number of cotyledons and inflorescence without few or no flowers) and, analysis of the subcellular localization of these PIN1 phosphomutants, revealed that the lack of complementation was at least in part due to mislocalization of PIN1. The WT version of PIN1 is localized at the apical membrane of the topmost cells of the inflorescence apex and the phosphomimic PIN1S1,2,3E-GFP protein showed the same apical localization, consistent with the notion that phosphorylated PIN1 induces its apical localization. By contrast, PIN1S1,3A-GFP and PIN1S1,2,3A-GFP proteins were targeted to the basal side, and were not shifted to the apical side of the cell when coexpressed with 35S::PID (Huang et al., 2010). In embryos, PIN1-GFP protein is mostly localized at the basal side of the membrane in provascular cells. This PIN1 localization is necessary to maintain auxin maxima and to define apico-basal axis during embryo development. On the other hand, PIN1 is localized at the apical side in epidermal cells of the embryo, and drives the accumulation of auxin at the tips of developing cotyledons. PIN1S1,2,3A-GFP protein was mainly accumulated in intracellular compartments in all cell types of the embryo or it was non-polarly localized when present at the membrane. This mislocalization contributes to the alteration of auxin response maxima establishment and consequently, to aberrant embryo development (Huang et al., 2010).
PID clusters together with PID2, WAG1 and WAG2 into the AGC3 group of plant AGCVIII kinases (Galvan-Ampudia and Offringa, 2007; Rademacher and Offringa, 2012; Santner and Watson, 2006). WAG1 and WAG2 were initially described as negative regulators of root waving, as wag1, wag2 and wag1wag2 mutants showed enhanced waving phenotype (Santner and Watson, 2006). These proteins were later shown to act redundantly with PID in the regulation of cotyledon development, root meristem size and root gravitropic response by modulating the subcellular localization of PIN proteins (Dhonukshe et al., 2010). The triple mutant plants pidwag1wag2 lack cotyledons. This phenotype might be explained by the observation of an aberrant basal (and sometimes lateral) localization of PIN1 in the embryo epidermal cells of these mutants (Dhonukshe et al., 2010). Basal PIN1 drives auxin to the bottom half of the embryo and thereby inhibits the formation of auxin maxima required for cotyledon initiation. This phenotype is absent or only partially observed in the pidwag1 and pidwag2 double mutants, respectively. pidwag1wag2 roots are agravitropic and shorter than wild-type roots, due to an increase on the auxin content at the root tip and the inability to create asymmetric auxin distribution upon gravistimulation as visualized using the DR5::GFP reporter. These defects are associated with the mislocalization of PIN2 at the basal side of epidermal cells where it is normally apical. Notably, and contrary to the case of PIN1 in the inflorescence apex, the basal PIN2 localization in root epidermal cells was not observed in pid single mutants, clearly demonstrating the redundant role of these three kinases in root development (Dhonukshe et al., 2010). Moreover, PID, WAG1 and WAG2 phosphorylate PIN2HL at the same Ser residues within the conserved TPRXS(N/S), ie: S1 (Ser-237), S2 (Ser-258) and S3 (Ser-310) and, PIN2::PIN2S1,2,3A-VENUS failed to complement the pin2 mutant (Dhonukshe et al., 2010). The basal localization of the non-phosphorylable PIN2 protein was not changed by overexpression of PID. Thus, phosphorylation of PIN2 by PID, WAG1 and WAG2 is necessary for its apical localization in root epidermis and for the correct establishment of auxin fluxes to modulate root meristem growth and response to gravitopic stimulus.
Although the main phosphorylation targets of PID/WAGs are the Ser residues S1, S2, S3 embedded in the TPRXS(N/S) motifs of PINHL, two additional Ser residues (named S4 and S5) within the hydrophilic loop have been recently shown to be phosphorylated by these kinases (Zourelidou et al., 2014) (Fig. 2B). These two phosphorylation sites are not as well conserved as residues S1 to S3. For example S4 (S271 in PIN1) is mutated to Asparagine (a non-phosphorylable residue) in PIN2 and S5 (position 215 in PIN1) is mutated to an aspartic acid (which might act as a natural phosphomimic or constitutive phosphorylation at this site) in PIN1 (Fig. 2B). S4 and S5 might therefore account for some PIN-specific regulation of their activity. These phosphorylation sites are not necessary to modulate the subcellular localization of PINs but to activate the efflux transport activity of these proteins (Fig. 2B). D6 PROTEIN KINASE (D6PK) phosphorylate PINs at the very same residues as PID/WAGs but with higher affinity for S4 and S5 residues (Zourelidou et al., 2014; Zourelidou et al., 2009). D6PK and its homologs D6PK-LIKE1 (D6PKL1), D6PK-LIKE2 (D6PKL2) and D6PK-LIKE3 (D6PKL3) belong to the AGC1 subgroup of protein kinases (Galvan-Ampudia and Offringa, 2007). D6PK localizes at the basal plasma membrane of the cells where it colocalizes with the PIN auxin efflux transporters. D6PK colocalizes with PIN1 in the main inflorescence stem, with PIN3 in the hypocotyl stem, PIN4 at the lateral root cap cells and PIN1 and PIN2 in the stele and cortex root cells respectively (Barbosa et al., 2014; Willige et al., 2013; Zourelidou et al., 2009). In addition, PIN1-type proteins are direct in vitro phosphorylation targets of D6PK and, PIN1 and PIN3 have been shown to be in vivo substrates as well. Plasma membrane localization of D6PK is essential to accomplish PIN phosphorylation (Barbosa et al., 2014; Willige et al., 2013; Zourelidou et al., 2014; Zourelidou et al., 2009). By contrast to PID and WAG1/WAG2, D6PK phosphorylation is only involved in the regulation of PIN activity (Zourelidou et al., 2014).
Finally, other Ser/Thr residues have been identified as in vivo phospho-residues within PINHL, but the kinase(s) responsible for this phosphorylation remains unknown. However, phosphorylation in Ser337/Thr340 of PIN1HL is necessary for PIN1 proper subcellular localization in embryo and inflorescence (Zhang et al., 2010b) (Fig. 2B). Phosphorylation of PIN1 at position S337, which is located within a Ser/Thr-Pro motif, a motif that is under the control of the peptidyl-prolyl cis/trans isomerase Pin1At (Xi et al., 2016) (Fig. 2B). This protein recognizes phosphorylated Ser/Thr residues in Ser/Thr-Pro motifs and catalyzes the cis/trans conformational change of the adjacent Proline residue. Pin1At binds to PIN1 and acts on several Ser/Thr-Pro motifs in the cytosolic loop. To date it is unknown what are the molecular impacts of these conformational changes on the PIN1 protein, but they are important for the PID/PP2A antagonistic effect on PIN1 polarity (Xi et al., 2016). The phosphosites that are recognized by Pin1At are different from the site S1 to S5 that are phosphorylated by PID/WAGs and D6PK and the kinase(s) that is(are) responsible for their phosphorylation is(are) currently unknown (Xi et al., 2016). This suggests that there are a series a kinases/phosphatases that (de)phosphorylate the cytosolic loop of PIN1 successively at different site to control its activity, polarity and trafficking. Interestingly, the Ser337 in PIN1 is conserved in other PINs but is not followed by a proline, suggesting that the Pin1At-mediated conformational change could be involved in PIN1-specific regulations.
The activity of PID/WAGs is itself regulated by phosphorylation, notably by the 3’- Phosphoinositide-dependent protein kinase 1 (PDK1). PDK1 is highly conserved among eukaryotes and is considered as a master regulator of AGC kinases in mammalian cells, due to its ability to activate other AGC kinases (Rademacher and Offringa, 2012). PDK1 possess a pleckstrin homology (PH) domain that is responsible of the interaction of PDK1 to regulatory phospholipids and of the localization of PDK1 at the plasma membrane. Interestingly, most of the AGC kinase substrates of PDK1 do not contain the PH domain and are localized either to the cytoplasm or to the plasma membrane by unknown mechanisms. PDK1 binding to its substrate proteins takes place through interaction between the PIF (PDK1-Interacting Fragment) sequence present at the C-terminus of the substrates and the N-terminal PIF-binding pocket of PDK1 (Rademacher and Offringa, 2012). PDK1 phosphorylates the catalytic domain of the target kinases within their activation loop, which stimulates their autophosphorylation and overall kinase activity.
In Arabidopsis, two homologs of the mammalian PDK1 have been found (PDK1–1 and PDK1–2) and define an independent subgroup of AGC kinases (Galvan-Ampudia and Offringa, 2007). Several members of the plant specific AGCVIII kinases have been shown to interact with and to be phosphorylated by PDK1 in vitro (Zegzouti et al., 2006a; Zegzouti et al., 2006b). Among them, the auxin polar transport regulators PID, WAG1 and WAG2. PDK1 phosphorylates PID on at least one conserved residue (S290) of the activation loop, which enhances PID kinase activity towards myelin basic protein (MBP), an artificial in vitro phosphorylation substrate of PID. The PIF domain of PID is essential for its interaction with PDK1 in vitro (Zegzouti et al., 2006a). On the other hand, WAG1 and WAG2, have been shown to interact with PDK1 in vitro, even if they do not contain a PIF domain within their sequence. However, PDK1 interaction with WAG1 and WAG2 do not produce an increase in the autophosphorylation and activity of these proteins in vitro (Zegzouti et al., 2006b). The requirement of PDK1 regulation of PID, WAG1 and WAG2 activity through its binding and phosphorylation in planta remains to be fully characterized. PID-GUS and PID-VENUS translational fusion lacking the PIF domain, are able to partially complement pid-loss of function mutant, arguing against the need of PDK1 modulation of PID activity to regulate auxin-dependent development (Benjamins et al., 2001; Michniewicz et al., 2007; Rademacher and Offringa, 2012). Alternatively to the control of kinase activity, PDK1 might modulate other aspects of the functionality of these proteins, such as their subcellular localization or interaction with downstream substrates.
Phospholipid phosphorylation by PIP Kinases and the control of PIN polarity
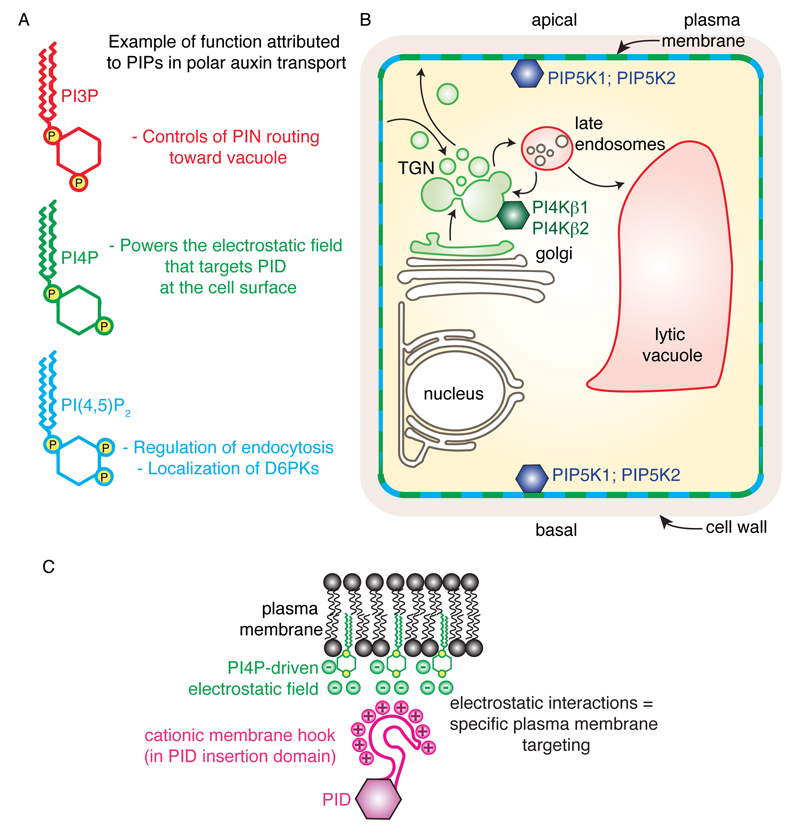
Phosphoinositides (also known as phosphatidylinositol phosphates or PIPs) are lipid molecules that are phosphorylated on their polar inositol head, at the inner leaflet of biological membranes (facing the cytosol) (Platre and Jaillais, 2016). They are minor phospholipids, accounting for less than one percent of total membrane lipids, yet they are critical for many membrane-associated events: i) they recruit proteins to membrane compartments through stereospecific interaction domains (e.g. Pleckstrin Homology domain (PH), Phox homology domain (PX), Fab1/YOTB/Vac1/EEA1 domain (FYVE)), ii) they are instrumental in cytoskeleton dynamics, iii) they can control enzyme and channel activity, iv) they are precursors of second messengers (e.g. Inositol-3-Phosphate, Diacylglycerol) and, v) they regulate virtually every aspect of membrane trafficking, including endocytosis and exocytosis (Holthuis and Levine, 2005; Jean and Kiger, 2012; Lemmon, 2008). Phosphoinositides form docking platforms at the cytosolic leaflet of membranes and are localized in specific pools in various organelles. Each compartment is composed of a unique combination of phosphoinositides providing a code for membrane identity (Kutateladze, 2010). Phosphoinositides can be phosphorylated on three different positions of the inositol ring, the 3rd, 4th and 5th position, either as single phosphorylation or in multiple combinations (Balla, 2013). Theoretically, there are seven possible PIP species, including three monophosphorylated species (PI3P, PI4P and PI5P), three bi-phosphorylated species (PI(3,4)P2, PI(3,5)P2 and PI(4,5)P2) and one tri-phosphorylated specie (PI(3,4,5)P3). Plant genomes lack type-I and type-II PI3-Kinases that phosphorylate PI4P and PI(4,5)P2 on the 3rd position of the inositol ring. As a result, plants do not produce any PI(3,4)P2 and PI(3,4,5)P3 and have only five different PIP species (Delage et al., 2013; Munnik and Nielsen, 2011). The three main phosphoinositides for which the subcellular localization is known in plants are PI3P, PI4P and PI(4,5)P2 (Simon et al., 2014; Simon et al., 2016; van Leeuwen et al., 2007; Vermeer et al., 2009; Vermeer et al., 2006) (Fig. 3). PI3P is localized in late endosomes, PI4P is mainly presents at the plasma membrane but there exist also a minor pool in early endosomes and PI(4,5)P2 is at the plasma membrane (Fig. 3) (Simon et al., 2014; Simon et al., 2016; van Leeuwen et al., 2007; Vermeer et al., 2009; Vermeer et al., 2006). Both PI4P and PI(4,5)P2 show a slight polar localization in root epidermal cells, being preferentially localized at the apical and basal end of the cell rather than on their lateral sides (Fig. 3) (Ischebeck et al., 2013; Tejos et al., 2014). PI4P is produced by PI4Ks (phosphatidylinositol 4-kinase) that phosphorylate phosphatidylinositol (PI) on the 4th position of the inositol ring (Balla, 2013). In turn, PIP5K (phosphatidylinositol 4-phosphate 5-kinase) phosphorylates PI4P on the 5th position of the inositol ring to produce PI(4,5)P2 (Balla, 2013). Two PIP5Ks (PIP5K1 and PIP5K2) out of 11 present in the Arabidopsis genome are localized in a polarized manner at the plasma membrane of root epidermis, with a preferential localization at the apical/basal pole of the cells (Ischebeck et al., 2013; Tejos et al., 2014) (Fig. 3). It is likely that the polar localization of PIP5K might account for the slight polarized localization of PI(4,5)P2. 12 PI4K genes are found in the Arabidopsis genome, but the three type-III PI4Ks (PI4Kα1, PI4Kβ1 and PI4Kβ2) account for almost the entire PI4K activity in planta (Delage et al., 2012). In roots, PI4Kβs are localized to the trans-Golgi Network (TGN), while PI4Kα1 localizes at the plasma membrane (Kang et al., 2011; Okazaki et al., 2015; Preuss et al., 2006).
Figure 3. Phosphoinositide localization in Arabidopsis root epidermis and function in regulating polar auxin transport.
A) Schematic representation of PI3P, PI4P and PI(4,5)P2 and example of their respective function in polar auxin transport. Note that they are likely involved in many more pathways and this represents only the studied examples. B) PIP subcellular localization in Arabidopsis root epidermis. The localization of the respective PIP kinases is indicated when known. Arrows with chevron-shaped arrowhead represent trafficking pathways. C) Model for the plasma membrane localization of PID by PI4P-driven electrostatics.
The polarly localized PIP5K1 and PIP5K2 kinases where shown to regulate polar auxin transport and in particular PIN protein polarity and trafficking (Ischebeck et al., 2013; Mei et al., 2012; Tejos et al., 2014). The single pip5k2 mutant produces less lateral root, is partially agravitropic, and has altered auxin maximum pattern as visualized by DR5::GUS and impaired polar auxin transport (Mei et al., 2012). The pip5k1pip5k2 double mutant has a stronger phenotype, suggesting functional redundancy between the two kinases (Ischebeck et al., 2013; Tejos et al., 2014). The phenotype of this double mutant is also reminiscent of polar auxin transport defects, including severely impaired primary root growth and altered gravitropic response, abnormal embryogenesis and disconnected vascular tissues in cotyledons (Ischebeck et al., 2013; Tejos et al., 2014). In this double mutant, the polarity of PIN1 is aberrant in the embryo, the primary root and in leafs, being either apolar or lacking a coordinated polarity from cell to cell (Tejos et al., 2014). This phenotype is reminiscent of the loss of PIN1 polarity in the gnom mutant, which is impaired in endocytic recycling (Geldner et al., 2004; Steinmann et al., 1999) and suggest that PIP5Ks might be involved in PIN trafficking. Indeed, the pip5k2 mutant has decelerated vesicular trafficking and altered response to BFA and the pip5k1pip5k2 double mutant shows decelerated endocytic recycling of both PIN1-GFP and PIN2-GFP (Ischebeck et al., 2013; Mei et al., 2012). PI(4,5)P2 is known to be involved in clathrin mediated endocytosis in metazoan, as it recruits many endocytic adaptors to the plasma membrane (Schmid and Mettlen, 2013). The pip5k1pip5k2 double mutant shows altered dynamics and spatial distribution of Clathrin Light Chain 2 (CLC2) at the plasma membrane (Ischebeck et al., 2013). This results suggest that PI(4,5)P2 might directly regulate the localization or function of some molecular actors of clathrin-mediated endocytosis, although this hypothesis has yet to be experimentally tested. It is possible that impaired endocytic trafficking in pip5ks mutants induces PIN polarity defects, which in turn might explain the polar auxin transport phenotype of the mutants. However, it is still not clear how the polar localization of PI(4,5)P2 contributes to this phenotype. Indeed, clathrin accumulates preferentially on the lateral side of the cells (Kleine-Vehn et al., 2011), where it presumably stimulates PIN endocytosis, which in turn is polarly recycled at the apical or basal poles of the cell. Therefore, PI(4,5)P2 accumulation does not directly correlate with the recruitment of clathrin (Kleine-Vehn et al., 2011). In addition, CLC2 dynamics in pip5k1pip5k2 double mutant was analyzed on the outer lateral face of epidermal cells (the side of the epidermis that is in contact with the microscope cover slide), while PIP5Ks are not or less localized in this polar domain. Analyzing PIN endocytosis and clathrin dynamics in apical/basal polar domains is technically challenging, as these poles are not in the plane of the microscope and represent a current limitation in the field.
It is possible that PI(4,5)P2 might also contribute to polarized exocytosis of PIN proteins, perhaps through the recruitment of the exocyst complex. Indeed, the EXO70 exocyst subunit is a known PI(4,5)P2 effector in animal and yeast and has been shown to regulate PIN trafficking in Arabidopsis (Drdova et al., 2013; Martin, 2015). Furthermore, PI4P and PI(4,5)P2 might control the localization of additional proteins involved in PIN protein trafficking, activity and/or polarity. For example, the AGC protein kinases PINOID, and D6PK interact in vitro with these lipids (Simon et al., 2016; Stanislas et al., 2015; Zegzouti et al., 2006a). In particular, PINOID is recruited to the plasma membrane by the electrostatic field generated at this membrane by PI4P (Simon et al., 2016) (Fig. 3). In addition, D6PK localization during root hair initiation is regulated by PIP5K3, another PIP5K expressed in trichoblasts (Stanislas et al., 2015). Finally, the apical-basal polarity of PI4P and PI(4,5)P2 seems to be a variable trait that can be seen only in weak expression lines and was not consistently reported in the literature (Simon et al., 2014; Simon et al., 2016; Tejos et al., 2014; van Leeuwen et al., 2007; Vermeer et al., 2009). The polarity indices (ratio of apical and basal fluorescent signal over lateral signal) of PI4P and PI(4,5)P2 reporters are only around 1.2 to 1.4, while it is around 3 for PIN2-GFP and PIP5Ks (Simon et al., 2016; Tejos et al., 2014). These ratios were shown to be statistically significant as compared to an overexpressed fluorescent aquaporin fusion protein (GFP-aqPIP2) by Tejos et al., (2014), but not by Simon et al., (2016). It is still unclear whether this apparent phosphoinositide polarity is indeed important for the observed phenotype of pipk mutants and PIN polarity establishment or whether it simply reflects the particular topology of root epidermal cells (Simon et al., 2016).
Interestingly, the expression of both PIP5K1 and PIP5K2 is induced by auxin, suggesting that auxin itself stimulates the phosphorylation of PI4P into PI(4,5)P2, and as such might induce PIN proteins endocytic trafficking and possibly feedback on their polarity (Mei et al., 2012; Tejos et al., 2014). Consistently with this hypothesis, the inducible overexpression of PIP5K1 or PIP5K2 induces PIN1 lateralization (Ischebeck et al., 2013). Such lateralization of PIN1 is reminiscent of the effect of auxin treatment on PIN1 polarity (Sauer et al., 2006). In the root, PIN1 lateralization following auxin treatment is dependent on ARF7 and ARF19, which are also required for PIP5K1 induction (Sauer et al., 2006; Tejos et al., 2014). As such, PIP5Ks and PI(4,5)P2 could be part of the machinery by which auxin controls the orientation of its own flux, a theory known as the canalization hypothesis. This model proposes a feedback effect of auxin on the directionality of intercellular auxin flow as a mean to polarize tissue and organ. This theory is at the base of many polar auxin transport computational models, although a clear understanding of the molecular mechanisms behind this auxin effect is lacking (Bennett et al., 2014). Furthermore, PIP5K1 or PIP5K2 gain-of-function induces the endocytosis and degradation of PIN2-GFP (Ischebeck et al., 2013), which is similarly triggered by either long-term auxin accumulation or depletion (Baster et al., 2013).
Altogether, PI phosphorylation into PI4P and then PI(4,5)P2 has a direct impact on PIN protein trafficking, polarity and auxin flow and this phosphorylation machinery is under transcriptional control of auxin.
Histidine Kinase Cytokinin Receptors and the regulation of PIN polarity and stability
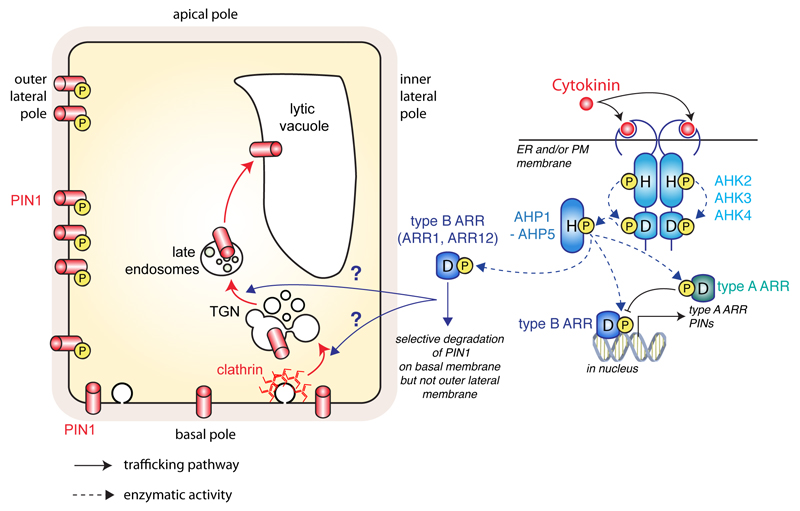
Auxin has long been shown to interact antagonistically with the phytohormone cytokinin (El-Showk et al., 2013). Cytokinin signaling is executed by a histidine-to-aspartate phosphorelay pathway (Hwang et al., 2012) (Fig. 4). This signaling cascade, also called two-components signaling system, is found in bacteria. In its simplest form, it involves a membrane histidine kinase, which acts as sensor or receptor, and response regulator proteins. Histidine kinase auto-phosphorylates on histidine residues, while response regulators are phosphorylated on conserved aspartate residues. Cytokinin signaling involves one intermediate between the histidine kinase and the response regulators in the form of a histidine phosphotranfer protein. In Arabidopsis, there are four cytokinin receptors (CYTOKININ-INDEPENDENT1, CKI1; and three ARABIDOPSIS HISTIDINE KINASE, AHK2 to AHK4), six histidine phosphotransfer proteins (ARABIDOPSIS HISTIDINE PHOSPHOTRANSFERT PROTEIN, AHP1 to AHP6) and two sub-families of response regulators (ARABIDOPSIS RESPONSE REGULATORs, ARRs), five type-A ARRs (ARR3, 9, 15-17) and eleven type-B ARRs (ARR1, 2, 10-14, 18-21) (Hwang et al., 2012).
Figure 4. Cytokinin phosphorelay and its role in PIN1 trafficking.
Schematic representation of the ‘classical’ cytokinin phosphorelay pathway from receptor to gene activation (right) and its ‘non-canonical’ role, independent of transcription but relying on AHKs and ARRs, in PIN1 trafficking during lateral root initiation (left). Dashed arrows represent direct phosphorylation events; arrows with chevron-shaped arrowhead represent trafficking pathways and arrows with triangle-shaped arrowhead represent signalling events; blunt-ended lines represent inhibition.
Binding of cytokinin in AHK’s extracellular ligand-binding domain triggers autophosphorylation on a conserved histidine residue in the N-terminal sensor kinase domain. The phosphoryl group is then transferred from this histidine to a conserved aspartate residue in the C-terminal receiver domain of the same AHK. The phosphoryl is then transferred from the aspartate in the AHK to a conserved histidine in AHPs, which are soluble proteins, unlike AHKs that are transmembrane receptors (Hwang et al., 2012). The phosphoryl on AHP’s histidine is then finally transferred to ARRs on conserved aspartate residues, hence the term “phosphorelay” that describes this signaling system. Type-B ARRs localize in the nucleus and are DNA binding transcription factors. They directly regulate the expression of primary cytokinin responsive genes, including type-A ARRs, which are negative feedback regulators. This canonical cytokinin signaling pathway regulates the expression of many genes and in particular it directly controls the expression of PINs (Simaskova et al., 2015). As such, the genomic response to cytokinin directly impact auxin fluxes (Fig. 4).
In addition, cytokinin also regulates polar auxin transport via non-genomic means, notably in the context of lateral root formation (Marhavy et al., 2011; Marhavy et al., 2014). Live imaging during lateral root outgrowth shows that cytokinin treatment induces rapid and pronounced PIN1 degradation (Marhavy et al., 2011). This effect is dependent on AHKs, in particular AHK4 as well as some type-B ARRs (ARR1 and ARR12). However, it is independent of protein synthesis and transcription as it still occurs following treatment with cycloheximide or cordycepin, which inhibit translation and transcription, respectively (Marhavy et al., 2011). PIN1 degradation following cytokinin treatment requires an intact endocytic pathway to route PIN1 from the plasma membrane to the lytic vacuole, where it is degraded. Interestingly, this cytokinin-induced degradation is not a general effect on membrane proteins: it seems to be specific for PIN1, PIN3 and PIN7, but has little effect on PIN2 or AUX1 localization in Arabidopsis root (Marhavy et al., 2011).
In addition to this effect on PIN1 stability, cytokinin also controls PIN1 polarity during lateral root emergence. During lateral root initiation, PIN1 polarity switches from a basal localization to a lateral localization, so that PIN1 is facing away from the stele and toward the tip of the new initium (Benkova et al., 2003). This redirects auxin flow and allows the formation of an auxin maximum at the tip of the new primordium, which is required for lateral root emergence. This lateralization of PIN1 is known to require endocytic trafficking (Geldner et al., 2004; Jaillais et al., 2007). Cytokinin treatment at low concentration specifically depletes PIN1-GFP from the basal end of the cell but not from the periclinal side (Marhavy et al., 2014). Again, this effect is dependent on an intact cytokinin phosphorelay pathway and is specific for PIN1. To show that cytokinin specifically depletes PIN1 at the basal end of the cell, Marhavý et al., missexpressed PIN1 in the PIN2 expression domain (epidermis and cortex). In this context, PIN1 is basaly localized in the cortex (similar as PIN2) but is localized at both basal and apical poles of the cell in the epidermis (Marhavy et al., 2014). Strikingly, cytokinin selectively depletes PIN1 at the basal pole but not the apical pole of epidermal cells. This effect was dependent on PIN1 phosphorylation status as cytokinin effect on PIN1 degradation was mitigated in pp2a loss-of-function mutants, PID overexpression lines or transgenic lines that constitutively express a PIN1 phosphomimic mutant (Marhavy et al., 2014). Overall, this study suggests that cytokinin not only induces PIN1 trafficking to the vacuole, but that this effect is specific for a specialized polar domain, very likely based on PIN1 phosphorylation status. This finding is highly relevant to lateral root formation, since the removal of PIN1 at the basal pole of the cell allows accumulation of PIN1 selectively at the periclinal plasma membrane and thereby redirection of the auxin flow toward the tip of the emerging primordium (Benkova et al., 2003). This suggests that cytokinin acts as a polarizing clue during organogenesis. However, the exact mechanisms by which cytokinin induces PIN1 degradation in the absence of transcription are currently unknown and one of the challenges for the coming years will be to understand these underlying mechanisms.
The TMK Receptor-Like Kinase (RLK) family in the control of ‘non-genomic’ auxin signaling and establishment of cell shape
Leaf pavement cells are highly polarized cells that present an interdigitated growth with the presence of lobes and necks. As such, they form an epidermal cell layer, which is characterized by its jigsaw-puzzle pavement. Exogenous auxin treatments promote lobe formation in a dose-dependent manner, while auxin biosynthesis mutants present a reduction of lobes that can be rescued by auxin treatments (Xu et al., 2014). In addition, PIN1 overexpression positively regulates the number of lobes in leafs and cotyledon pavement cells (Guo et al., 2015; Li et al., 2011). Moreover, the pin1 loss-of-function mutant also presents an altered PCs phenotype; the cells being long and narrow (Xu et al., 2014). By contrast to auxin biosynthesis mutants, exogenous auxin treatment does not rescue the pin1 cell shape phenotype, suggesting that PIN1-mediated polar auxin transport is essential for the formation of lobes and necks in PCs. However, plants grown on NPA have wild-type pavement cell morphology (Ringli et al., 2008; Xu et al., 2014). This result, together with the fact that auxin deficient mutants have relatively mild PC shape phenotype, suggests that polar auxin transport is a modulator, rather than a major regulatory pathway, of leaf cell morphogenesis.
PIN1 is slightly polarized at the tip of the lobes of pavement cells in young cotyledons (Guo et al., 2015; Xu et al., 2014). Evidence that this localization might be important for pavement cell morphogenesis came from a forward genetic screen for mutants with impaired PC shape. This screen isolated a loss-of-function mutant in FYPP1, the catalytic subunit of the PP2A holoenzyme (Fig. 2B), suggesting that the fppy1 mutants might have altered PIN1 phosphorylation and localization (Li et al., 2011). Indeed, PIN1 is delocalized to both necks and lobes of pavement cells in the fypp1 mutant (Li et al., 2011). In addition, overexpression of PINOID, which is known for its antagonistic action with PP2As on PIN1 phosphorylation, also induces a reduction of the number of lobes in PCs (Li et al., 2011). However, by contrast to the fypp1 loss-of-function mutant, PID gain-of-function delocalizes PIN1 from the lobe into the neck of pavements cells. Similar to the pin1 mutant, auxin treatment does not rescue pavement cell shape phenotype in fypp1 mutant and 35S::PID lines. Together, these results suggest that the PP2As and PID antagonistic effect on PIN1 phosphorylation status controls PIN1 targeting at the tip of the pavement cell lobes and that this polar localization is essential for the proper establishment of pavement cell morphogenesis (Li et al., 2011). TYPE-ONE PROTEIN PHOSPHATASE 4 (TOPP4), a catalytic subunit of PROTEIN PHOSPHATASE1 (PP1), is also required for pavement cell morphogenesis, PIN1 polar targeting in lobes and PIN1 dephosphorylation (Guo et al., 2015). Similar to FYPP1, TOPP4 directly interacts with PIN1 and antagonistically contributes to PIN1 dephosphorylation (Guo et al., 2015). Altogether, these results suggest that the phosphorylation switch observed in meristematic and embryo tissues, is active in pavement cells to control PIN1 polar targeting in lobes and cell morphogenesis.
Auxin activates ROPs [Rho-like guanosine triphosphatases (GTPase)], in particular, ROP2/ROP4 and ROP6 (Craddock et al., 2012; Yang and Lavagi, 2012). Interestingly, ROP2 and ROP6 have different subcellular localization, with ROP2 accumulating slightly more in the lobes than the neck regions and ROP6 slightly more in the neck than the lobes regions (Fu et al., 2005). Gain-of-function mutants of ROP2 and ROP6 have squared PCs that almost entirely lack lobes and necks (Fu et al., 2002; Lin et al., 2013; Poraty-Gavra et al., 2013). In addition, corresponding loss-of-function mutants also have a reduction in their number of lobes and necks, although these phenotypes are relatively weak (Fu et al., 2005; Fu et al., 2009; Lin et al., 2013). Rop2 and rop6 mutants present similar phenotypes, but they affect the cytoskeleton in different ways (Fu et al., 2005). On one hand, ROP2 and its effector ROP INTERACTIVE CRIB MOTIF-CONTAINING PROTEINS4 (RIC4) promote the assembly of filamentous actin in lobes, likely promoting targeted exocytosis and/or endocytosis events required for cellular outgrowth. On the other hand, ROP6 and its effector RIC1 promote the bundling of microtubules in the necks, which likely restrict growth notably through the thickening of cell wall by microtubule driven cellulose synthase activity (Fu et al., 2005; Fu et al., 2009; Lin et al., 2013; Nagawa et al., 2012; Sampathkumar et al., 2014). The downstream ROP6 effector RIC1 controls microtubule dynamics by directly binding to and controlling the activity of KATANIN (KAT), a microtubule-severing enzyme (Lin et al., 2013).
Both ROP2 and ROP6 are rapidly activated by auxin (i.e. auxin induces the production of GTP-loaded ROP2 and ROP6- the active form of these GTPases- that interact with their downstream effectors) (Wu et al., 2011; Xu et al., 2010). Auxin-mediated activation of ROP2 in the lobe region activates its downstream effector RIC4, which stabilizes actin in the lobe region and decreases PIN1 internalization (Nagawa et al., 2012). Reduction of PIN1 endocytosis in turn might increases PIN1 localization at the tip of the pavement cell’s lobes a localization that seems to be required for normal pavement cell shape establishment (Nagawa et al., 2012). Polarized PIN1 exports auxin preferentially at the lobe region, which activates ROP2 in this region, providing a positive feedback mechanism and self-organizing system for the polar PIN1 distribution in PCs. However, the effects of ROP/RIC signaling on PIN localization are relatively minor, suggesting that other signaling pathways might redundantly control PIN1 localization in PCs (Nagawa et al., 2012).
The receptor like kinases (RLK) from the TRANSMEMBRANE RECEPTOR KINASEs (TMKs) family (TMK1 to TMK4) have been identified upstream of ROP2 and ROP6 activation by auxin (Xu et al., 2014; Xu et al., 2010). This mutant presents squared pavement cells and this phenotype is not rescued by auxin. TMKs, which are receptor-like kinases are required for the auxin-mediated activation of ROP GTPases (Xu et al., 2014). Multiple mutants in TMK genes also present squared PCs and this phenotype is not rescued after auxin treatment (Xu et al., 2014). In addition, the ROP2 effector RIC4, which is localized at the plasma membrane in the tip of the lobe region and promotes the accumulation of actin microfilaments in wild-type plants, is mislocalized in the cytoplasm in the quadruple tmk1234 mutants (Xu et al., 2014; Xu et al., 2010). The delocalization of RIC4 in this mutant suggests that ROP2 is inactive in the absence of these proteins. In addition, RIC1 association with cortical microtubules was impaired in rop6-1 single mutant as well as in the tmk1234 quadruple mutant (Xu et al., 2014; Xu et al., 2010). Altogether, these results suggest that TMKs might be important for auxin perception upstream of ROP activation. Activated TMKs likely control downstream substrates by phosphorylation. These downstream effectors are currently unknown, but it could be ROP GTPases themselves. However, it is more likely that TMKs might activate ROP activators, such as ROP-GUANINE EXCHANGE FACTOR (ROP-GEF) (Miyawaki and Yang, 2014). Indeed, ROP-GEF have been shown to act downstream of other RLKs in plants, such as for example the FERONIA receptor (Duan et al., 2010; Miyawaki and Yang, 2014). Consequently, how exactly are ROPs activated by auxin and what is the role of TMK receptor kinases in this activation will require further investigations.
RLKs in the regulation of root gravitropism
Auxin and PIN dynamics during gravitropism
Root reorientation upon gravistimulation requires dynamic changes of auxin distribution at the tip of the root, which involve auxin transporters (Fig. 5). Using the synthetic auxin promoter DR5::GFP and DR5rev::GFP, it was detected that auxin accumulates in the lower part of the root (Ottenschlager et al., 2003; Paciorek et al., 2005). This auxin accumulation likely triggers a rapid growth arrest that is required for root bending. Using live imaging of the DII-VENUS auxin input sensor, the timing of this asymmetric auxin distribution was studied together with the dynamics of root bending (Band et al., 2012). In this study, Band et al., found that auxin accumulates at the lower side of the root within 30 minutes of a 90° gravistimulation. However, this auxin asymmetry is disappearing when the root tip is reaching a 40° angle to the horizontal. This phenomenon presumably corresponds to a tipping-point mechanism that allows to re-establish symmetric vertical growth (Band et al., 2012). Using DR5rev::3xVENUS-N7 (auxin output reporter), Baster et al., (2013) found an accumulation of the DR5 signal in the lower part of the root starting from 2 hours of gravistimulation up to 8h and then a decrease of the signal at 12h and 24h (Fig. 5). In parallel, they monitored a decrease of the auxin response at the upper side of the root notably at 4h and 8h post gravistimulation, which then came back up at 12h and 24h (Baster et al., 2013). Importantly, at 24h after gravistimulation, auxin maxima on both side of the root have a similar intensity and the root is growing straight down. This suggest that not only auxin accumulates at the lower side of the root but also is depleted at the upper side of the root, thereby increasing the auxin asymmetry and resulting elongation growth between the two sides (Fig. 5).
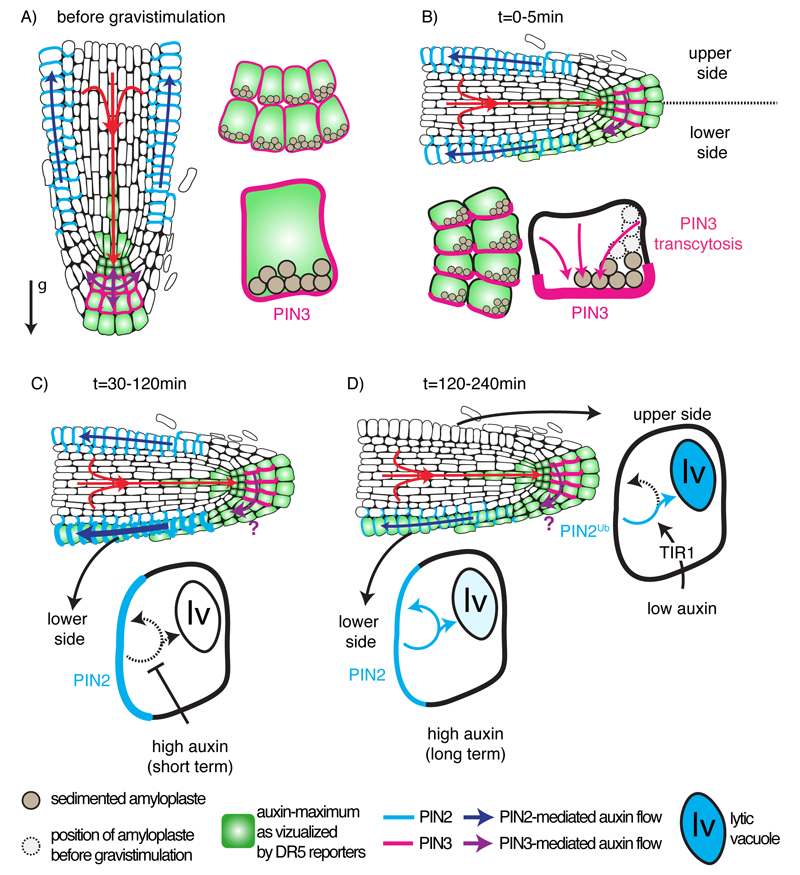
Figure 5. auxin fluxes and PIN trafficking during the gravitropic root response.
A) Auxin fluxes and PIN2/PIN3 localization in the primary root tip before gravistimulation. Note the position of sedimented amyloplasts in the collumela. The arrow with the ‘g’ letter indicates the gravity vector. B) Auxin fluxes and PIN2/PIN3 localization in the primary root tip from 0 to 30 min after gravistimulation. Note the movement of amyloplasts according to the new gravity vector, which triggers PIN3 transcytosis to the lower part of the cell and initiates the establishment of an asymmetric auxin maximum between the upper and lower part of the root. C) Auxin fluxes and PIN2/PIN3 localization in the primary root tip from 30 to 120 min after gravistimulation. High auxin in the lower part of the root inhibits PIN2 endocytosis, which promotes its localization at the plasma membrane and reinforce asymmetric auxin localization. Accumulation of auxin on the lower part of the root locally inhibits cell elongation, which triggers root bending. D) Auxin fluxes and PIN2/PIN3 localization in the primary root tip from 120 to 240 min after gravistimulation. Inhibition of endocytosis by auxin is transient and PIN2 endocytosis is re-established in the lower part of the root. In the meantime, low auxin on the upper part of the root triggers PIN2 degradation in a TIR1/AFBs-dependent manner. Note that the times indicated are compiled between those reported by Band et al., (2012) and Baster et al., (2013), but may vary according to the experimental setup and are only indicative. It is unknown at what point during the gravitropic response PIN3 localization is reset to apolar, hence the purple question mark in C and D.
Both auxin influx and efflux carriers are involved during the gravitropic response. The aux1 mutant is almost completely agravitropic, which highlight the importance of this auxin influx carrier in the root gravitropic response (Bennett et al., 1996). AUX1 is loading auxin that accumulates at the root tip into the lateral root cap, where it is then redistributed to the epidermis (Marchant et al., 1999; Swarup et al., 2005). Although aux1 is agravitropic, there is no evidence that AUX1 directly regulate the gravitropic responses. Rather, AUX1 is the driving force for redirection of auxin from the columella and, thus provides the source of auxin utilized in gravitropism. However, the first auxin transporter to dynamically change its localization following gravistimulation is PIN3 (Friml et al., 2002b). PIN3 is expressed in the root columella cells (Fig. 1), and it is localized in a non-polar fashion at the plasma membrane (Fig. 5A). Upon gravistimulation, PIN3 is rapidly (within minutes) relocalized by trancytosis (from one plasma membrane pole to another via endocytosis and recycling) at the lateral side facing the direction of the new gravity vector (Friml et al., 2002b; Kleine-Vehn et al., 2010) (Fig. 5B). This change of PIN3 localization triggers the redirection of auxin flow that initiate an asymmetric accumulation of auxin and the differential growth towards the new gravity vector. The auxin transporter PIN7 partially colocalizes with PIN3 in columella cells (Blilou et al., 2005) (Fig. 1). PIN7 also relocalizes according to the new gravity vector (Kleine-Vehn et al., 2010). Accordingly, PIN3 and PIN7 might act redundantly in redirecting the auxin fluxes during the gravitropic response (Kleine-Vehn et al., 2010). However, the agravitropic phenotype of pin3pin7 is relatively weak suggesting that other PINs might be involved as well or that some compensation mechanism exists. Gravity is mainly perceived in the columella by sedimenting statoliths, however the signal transduction leading to PIN3/PIN7 transcytosis and at what point during the gravitropic bending PIN3 localization is reset to apolar remains unknown (Fig. 5). The initial auxin asymmetry triggered by PIN3/PIN7 relocalization will then be amplified by differential trafficking of the PIN2 auxin carrier on the lower and upper side of the root depending on their respective auxin concentration (Fig. 5C). Two hours following gravistimulation and correlating with the change in auxin distribution, PIN2 plasma membrane localization is enhanced at the lower part of the root reinforcing auxin accumulation in this area (Baster et al., 2013; Paciorek et al., 2005) (Fig. 5). However, Peer and collaborators and Band and collaborators detected that auxin redirection occurs within 30 minutes (Band et al., 2012; Peer et al., 2014), therefore it is likely that all of the changes in PIN localization function in combination to amplify polar movements that follow cell to cell gradients. The accumulation of PIN2 at the lower part of the root is triggered by the initial increase in auxin, which inhibits endocytosis (Paciorek et al., 2005) (Fig. 5 and 6). Auxin inhibition of endocytosis is transient (Robert et al., 2010). As such, PIN2 plasma membrane accumulation is also transient and goes back to its original state 4h following gravistimulation (Baster et al., 2013) (Fig. 5D). Concomitantly, the level of PIN2 in the upper part of the root decreases (starting at 2h and culminating at 4h), before going back up to the original level 12h post gravistimulation (Abas et al., 2006; Baster et al., 2013; Jaillais et al., 2006) (Fig. 5D). On the upper part of the root, low auxin triggers PIN2 routing toward the vacuole, in a process that depends on PI3Kinase activity (Jaillais et al., 2006) (Fig. 3). PIN2 routing to the vacuole is regulated at the post-translational level by ubiquitination rather than phosphorylation but also requires genomic auxin signaling from the TIR1/AFBs auxin-signaling pathway (Abas et al., 2006; Baster et al., 2013; Korbei et al., 2013; Robert et al., 2010). The molecular steps between the perception of an auxin minimum by this TIR1/AFBs pathway and ubiquitination of PIN2 are so far unknown (Figure 6).
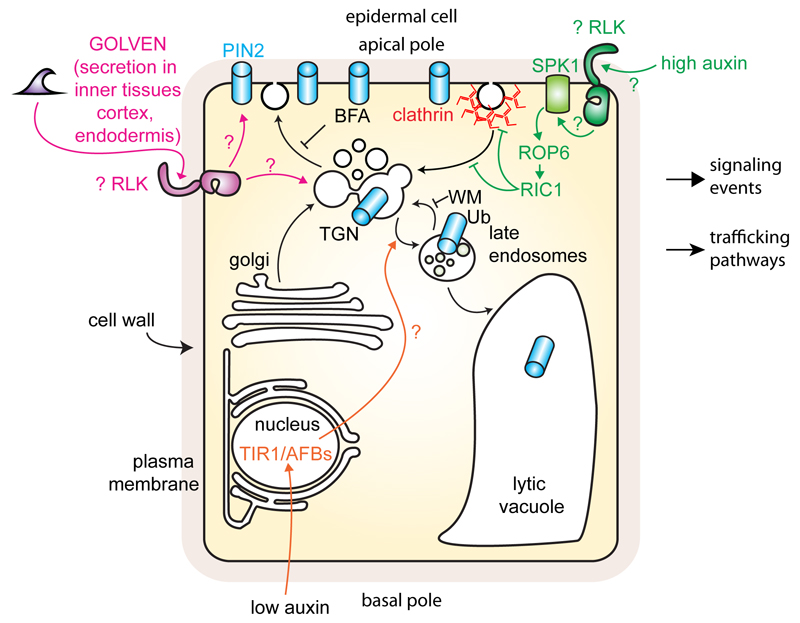
Figure 6. Missing RLKs in the regulation of PIN2 trafficking in epidermal root cells during gravitropism.
Schematic representation of PIN2 trafficking as regulated by the ‘non-genomic’ auxin pathway and GOLVEN peptides. Note that RLKs are likely involved in both pathways but have yet to be identified. Arrows with chevron-shaped arrowhead represent trafficking pathways and arrows with triangle-shaped arrowhead represent signalling events; blunt-ended lines represent inhibition.
‘Non-genomic’ auxin signaling and the inhibition of PIN2 endocytosis on the gravi-stimulated side - is TMK or another RLK upstream of ROP6?
Short-term auxin accumulation (or treatment) inhibits endocytosis, which is important to accumulate PIN2 at the lower side of the root and thereby amplify the asymmetric auxin accumulation between the two root sides (Paciorek et al., 2005; Robert et al., 2010). This pathway is independent of the canonical auxin ‘genomic’ perception that involves TIR1/AFB receptors but relies on a ‘non-genomic’ pathway that does not require de novo transcription (Robert et al., 2010). Similar to the ‘non-genomic’ pathway described above for pavement cell shape establishment, auxin inhibition of endocytosis involves ROP small GTPases (Chen et al., 2012; Lin et al., 2012). Auxin inhibits PIN internalization following BFA treatment, an inhibitor of protein recycling (Paciorek et al., 2005) (Fig. 5 and 6). Interestingly, loss-of-function mutants in rop6 or its downstream effector ric1 are insensitive to auxin inhibition of endocytosis and thereby show a general enhanced endocytosis (Chen et al., 2012; Lin et al., 2012). By contrast, ROP6 and RIC1 gain-of-function mutants have increased sensitivity to auxin inhibition of endocytosis and show lower endocytic rate. Consistently, rop6 and ric1 loss-of-function mutants respond more slowly to gravity, while their respective gain-of-function mutants had the opposite phenotype, their root bending faster than the wild type (Chen et al., 2012; Lin et al., 2012).
In addition, SPIKE1 (SPK1) was also shown to act directly upstream of ROP6 (Lin et al., 2012). SPK1 is a member of the dock homology region 2 (DHR2)-type Dock family of Rho guanine nucleotide exchange factors (ROP-GEFs) and acts as a GEF for ROPs in vitro (Basu et al., 2008). Similar to rop6 mutants, spk1 mutants present a reduced lobe formation in pavement cells and a slow gravitropic response (Lin et al., 2012). In addition, spk1 mutants present an increase in PIN2 endocytosis that was not recovered after auxin treatment (Lin et al., 2012). Importantly, pull-down assays, in vivo fluorescence resonance energy transfer (FRET) analysis and Co-IP show that SPK1 directly interacts with the GDP bound form of ROP6, and is required for ROP6 activation. While FRET analyses suggest that SPK1 might localize at the plasma membrane (PM) (Lin et al., 2012), an earlier report showed that SPK1 localizes in the endoplasmic reticulum (ER) (Zhang et al., 2010a). These results might point toward a role for ER-PM contact sites in ROP signaling, that should be explored further. Altogether, ROPs act in the ‘non-genomic’ auxin-signaling pathway to regulate PIN endocytosis in both roots and leaf pavement cells. However, they use different signaling components in these two cell types, ROP2-RIC4 in leaves and ROP6-RIC1 in roots (Chen et al., 2012; Lin et al., 2012; Nagawa et al., 2012; Robert et al., 2010; Xu et al., 2014; Xu et al., 2010).
It is currently unknown how auxin is perceived upstream of the SPK1-ROP6 module. However, the SPK1 ROP-GEF might be regulated by a receptor kinase upon auxin treatment. Indeed, FERONIA (FER) is a plasma membrane receptor kinase that regulates cell elongation and hormone crosstalks (Cheung and Wu, 2011; Kanaoka and Torii, 2010). This receptor is expressed in all the vegetative tissues and was discovered as a regulator of the communication between the pollen tube and the synergid cell (Escobar-Restrepo et al., 2007; Huck et al., 2003; Ngo et al., 2014; Rotman et al., 2008). However, fer mutant has a plethora of phenotypes including short root hairs that are often ruptured, bigger seeds, altered response to pathogens and abnormal response to mechanotransduction (Deslauriers and Larsen, 2010; Duan et al., 2010; Kessler et al., 2010; Mao et al., 2015; Shih et al., 2014; Yu et al., 2014; Yu et al., 2012). Using yeast two-hybrid, FER was shown to interact with 3 different ROP-GEF in Arabidopsis, GEF1, GEF4 and GEF10 (Duan et al., 2010; Huang et al., 2013). By analogy to the FER-GEF system, we can therefore hypothesize that a receptor kinase is likely to act upstream of the SPK1-ROP6 module to control ‘non-genomic’ auxin signaling (Miyawaki and Yang, 2014). One obvious candidate would by the TMK receptor kinases, which act upstream of ROP6 for ‘non-genomic’ auxin signaling during pavement cell shape establishment (Xu et al., 2014). TMK mutants have root phenotypes, but they need to be further examined, in particular for possible defects in endocytosis regulation (Dai et al., 2013). The question of how exactly these potential receptors are activated by auxin, will also require further studies.
GOLVEN peptides: an example of signaling ligands involved in the regulation of root gravitropism and PIN2 trafficking
Secreted peptides often act as signaling ligands that binds to RLKs and/or to a receptor-like proteins at the plasma membrane (Dievart and Clark, 2004). In Arabidopsis, GOLVENs belong to a small signaling peptide family encoded by 11 genes (Fernandez et al., 2013a; Fernandez et al., 2013b). The overexpression of all GLV peptides, with the exception of GLV4 and GLV8 causes root gravitropism defects (Fernandez et al., 2013a; Whitford et al., 2012). Furthermore, exogenous treatments with synthetic GLV peptides and GLV3 overexpression inhibit the formation of the lateral auxin gradient observed in the primary root after gravistimulation (Fernandez et al., 2013a). Exogenous treatment with synthetic GLV3 peptide stabilizes PIN2 both at the plasma membrane and endosomes, while PIN2 is degraded in glv3 loss-of-function mutant (Whitford et al., 2012) (Fig. 6). As such, GLV3 impacts differential PIN2 accumulation between the lower and upper part of the root following gravistimulation (see Fig. 5), which likely account for the agravitropic phenotype observed upon GLV3 treatment or in GLV3-overexpression lines. In addition, PIN2 internalization is triggered by exogenous treatment with other GLV peptides, including GLV3, 5, 6 and 11, but not with GLV1 or GLV4 (Fernandez et al., 2013a; Whitford et al., 2012). The molecular components that perceive GLV peptides at the plasma membrane are likely RLKs but they have not been identified yet.
Phosphorylation and the regulation of polar auxin transport during phototropism
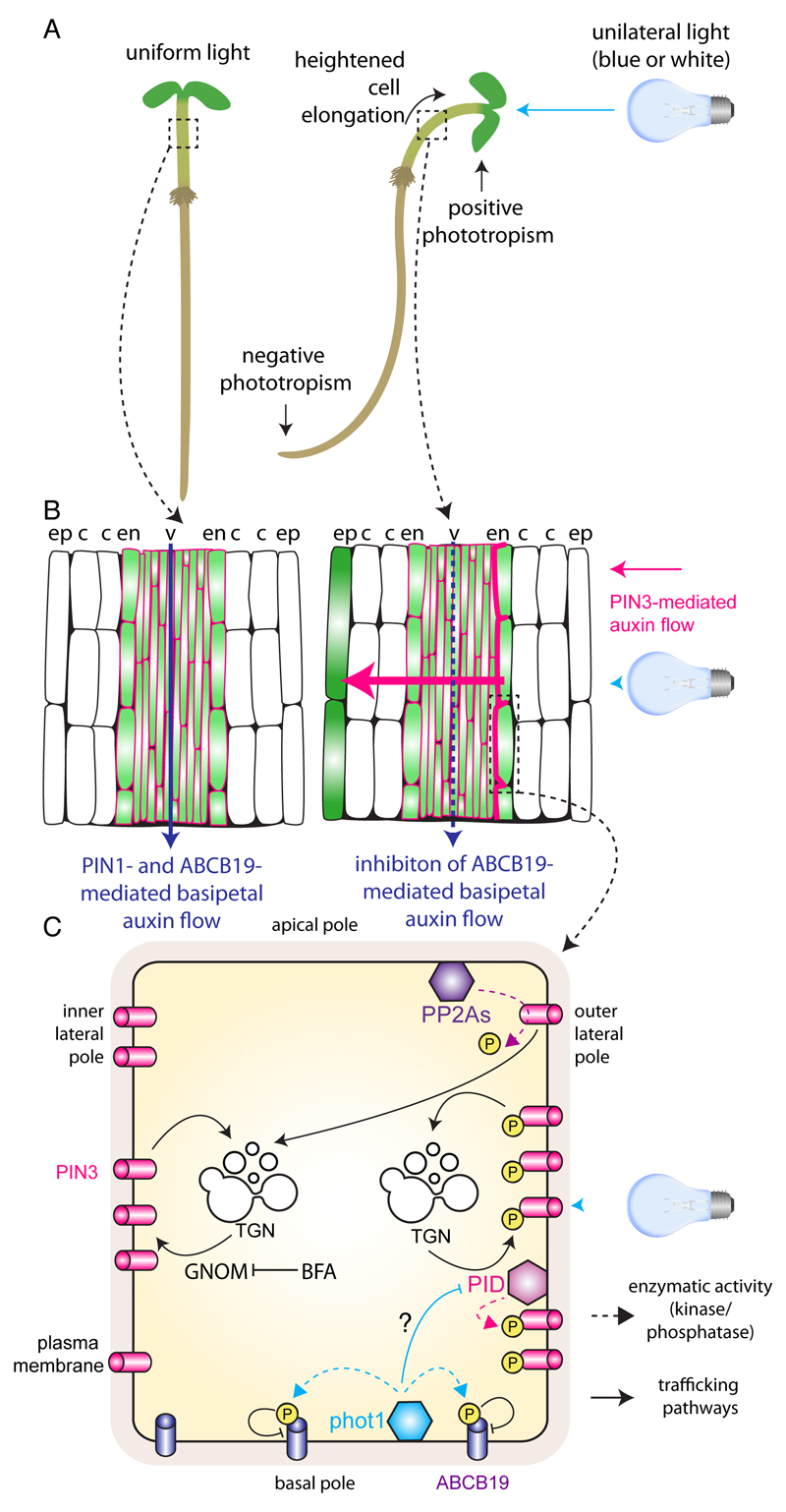
Phototropism, etymologically derived from the Greek words photo (=light) and tropos (to turn), can be defined as the tendency of growing plant organs to move or curve under the influence of light. Growth towards a light source is called positive phototropism, while growth away from the light source is called negative phototropism (Fig. 7). Plant stems generally exhibit positive phototropism, while roots exhibit negative phototropism. The adaptation of plant organogenesis and architecture to changing light conditions by means of tropic growth ensures the maintenance of plant fitness.
Figure 7. Protein kinases and auxin fluxes during the hypocotyl phototrophic response.
A) Seedling grown in uniform light (left) or under unilateral light (right). B) Schematic representation of a longitudinal section of hypocotyl showing putative auxin maximum (in green) and PIN3 localization (in pink) when seedling are grown in uniform light conditions or in darkness (left) and when they encounter unidirectional light (right). Arrows in dark blue represent PIN1- and ABCB19-mediated rootward auxin flow and the arrow in pink show PIN3-mediated lateralization of auxin flow toward the shaded side of then hypocotyl. C) Schematic representation of PIN3 trafficking in endodermis (illuminated side of the hypocotyl). Phot1 inhibits PID activity by unknown mechanisms, which limits PIN3 phosphorylation by PID and favours its routing by the GNOM-dependent pathway to the inner lateral pole of the cells. Phot1 also directly phosphorylates ABCB19, which inhibits its efflux activity and limits the downward auxin flow. ABCB19 inhibition and PIN3 lateralization stimulates auxin trafficking toward the shaded side of the hypocotyl, which promotes cell elongation and hypocotyl bending toward the light source. Note that phot1 action on ABCB19 and PIN3 trafficking has been represented in the same cell for convenience, but it is not known whether these events happen in the same place of the hypocotyl. Dashed arrows represent direct phosphorylation events; arrows with chevron-shaped arrowhead represent trafficking pathways and blunt-ended line represents inhibition.
The phototropic movement of plants attracted the interest of philosophers and scientists during the past centuries, giving rise to the basis of the actual knowledge and current models of phototropism (for a historical perspective of phototropic movement see (Whippo and Hangarter, 2006, 2009)). Notably, in the manuscript The Power of Movement in Plants (1880), Charles Darwin and his botanist son Francis Darwin established that the site of photoperception at the shoot tip and the site of curvature are physically separable in both monocots and dicots. This concept has been proven true to date for coleoptiles of monocotyledonous plants (Holland et al., 2009; Matsuda et al., 2011; Whippo and Hangarter, 2009). By contrast, in the dicot Arabidopsis, it has been reported that light perception and bending occurs at the same region of the hypocotyl and a role of the cotyledons in the phototropic response has been excluded (Christie et al., 2011; Preuten et al., 2013). Moreover, Darwin additionally established that a substance produced in the tip is transmitted to lower regions of the plant and is responsible of the induction of the phototropic curvature. This transmissible substance was posteriorly identified as the phytohormone auxin. The phototropic curvature follows the Cholodny–Went theory, which states that in response to tropic stimulus, the asymmetric auxin distribution causes differential growth of the two sides of the plant organ and, as a consequence, organ bending. In the case of shoot phototropism, unilateral light produces auxin accumulation at the shaded side of the hypocotyls, where it stimulates cell expansion (Fankhauser and Christie, 2015; Liscum et al., 2014; Sakai and Haga, 2012) (Fig. 7). Ultraviolet A (UV-A) and blue light (BL) (320–500 nm) are the most effective wavelengths to induce the phototropic bending. Analysis of Arabidopsis seedlings with altered phototropic responses lead to the identification of the blue-light (BL) photoreceptors responsible for phototropism, named phototropin 1 (phot1) and phototropin 2 (phot2) (Christie et al., 1998; Christie et al., 1999; Huala et al., 1997; Kagawa et al., 2001; Kinoshita et al., 2001; Liscum and Briggs, 1995; Sakai et al., 2001). Phot1 is involved in the phototropic response at both low and high BL intensities, while phot2 is only necessary for the phototropic response at high BL intensities. Phot1 (also called nph1) mutants show non-phototropic hypocotyl at BL intensities <10 µmol·m-2·s-1, whereas at higher fluence rates (10 and 100 µmol·m-2·s-1) the hypocotyls show a clear positive phototropic response, mediated by phot2 activity (Liscum et al., 2014; Sakai et al., 2001). By contrast, the phot2 (or npl1) single mutant shows normal phototropic responses at all BL intensities, due to phot1 activity. Only the phot1phot2 (or nph1npl1) double mutants show strong non-phototropic phenotype at both low and high BL (Sakai et al., 2001).
Analysis of the protein sequence of phototropins lead to their classification into the AGC4 group of AGC kinases and constitute the only members of this clade (Galvan-Ampudia and Offringa, 2007). Structurally, phototropins contain two functional domains: an N-terminal photosensor domain and a C-terminal serine/threonine kinase domain. The N-terminal photosensor domain contains two LOV (light, oxygen, or voltage) domains, LOV1 and LOV2, which bind flavin mononucleotid (FMN) as a cofactor, and function as the blue light-sensing moiety of the protein (Christie et al., 1998; Christie et al., 1999). In the dark, the LOV2 domain acts as a repressor of the C-terminal kinase domain and BL irradiation induces a conformational change that allows the activation of the phot kinase activity, triggering phot1 autophosphorylation and activation of a signaling cascade that ultimately leads to the establishment of an asymmetric auxin distribution between the illuminated and shaded sides of the hypocotyl (Fankhauser and Christie, 2015; Harper et al., 2003).
Among the downstream signaling components of the phot1-dependent phototropic response, NPH3 (for Non-phototropic hypocotyl 3) is required for the formation of the lateral auxin gradient upon directional light treatment (Haga et al., 2005). nph3 loss-of-function Arabidopsis mutants show defects in hypocotyl and root phototropism under a broad range of light intensities (Inada et al., 2004; Liscum and Briggs, 1995; Sakai et al., 2000). NPH3 is a BTB-containing protein belonging to the plant specific NPH3/RPT2 (or NRL) protein family (Motchoulski and Liscum, 1999; Pedmale et al., 2010; Pedmale and Liscum, 2007). Several members of this family act redundantly in auxin-mediated organogenesis and root gravitropism (which involves asymmetric auxin distribution), by modulating the subcellular localization of the polarly localized auxin-exporters PIN1 and PIN2 (Furutani et al., 2007; Furutani et al., 2011; Wan et al., 2012).
NPH3 and phot1 interact at the level of the plasma membrane where they both localize in dark conditions (Lariguet et al., 2006; Motchoulski and Liscum, 1999). NPH3 is phosphorylated in the dark but not as a direct substrate of phot1. Instead, NPH3 dephosphorylation occurs within minutes upon BL exposure and depends on phot1 activity (Pedmale and Liscum, 2007). NPH3 additionally interacts with Cullin3 (CUL3), forming a Cullin3 Ring E3 ubiquitin ligase that uses NPH3 as a substrate adaptor (CRL3NPH3) (Roberts et al., 2011). The CRL3NPH3 complex ubiquitinates phot1 upon irradiation with either low or high BL. Low BL induces mono- and multiubiquitination of phot1, and high BL induces mono-/multi- and polyubiquitination (Roberts et al., 2011). The biological significance of phot1 ubiquitination is poorly understood. It has been suggested that mono-/multiubiquitination induces phot1 internalization from the plasma membrane and might promote intracellular phot1 signaling by interaction/phosphorylation of cytoplasmic proteins (Liscum et al., 2014). On the other hand, polyubiquitination has been proposed to target phot1 for degradation via the 26S proteasome as a mechanism of receptor desensitization at high BL intensities (Roberts et al., 2011). However, a recent study using myristoylated and farnesylated (two lipid anchors that target and attach soluble proteins to the plasma membrane) phot1 proteins elegantly showed that the plasma membrane-bound fraction of phot1 represents the active form of the receptor and that internalization is not required for signaling (Preuten et al., 2015). These results would fit in a model in which phot1 modulates asymmetric auxin fluxes within the hypocotyl by phosphorylation of PIN proteins, but efforts to prove this direct interaction have failed (Ding et al., 2011). Nevertheless, phot1 binds in vivo and phosphorylates in vitro the auxin exporter ABCB19, a member of the MDR/PGP family of auxin transporters (Christie et al., 2011). ABCB19 phosphorylation by phot1 produces its inactivation (Christie et al., 2011) (Fig. 7). ABCB19 is a negative regulator of the phototropic response, as demonstrated by the enhanced hypocotyl bending of the abcb19 mutants in response to unilateral light (Noh et al., 2003). In addition to its intrinsic auxin transport activity, ABCB19 stabilizes PIN1 protein at the basal side of hypocotyl cells and promotes basipetal auxin flux (Noh et al., 2003). Disruption of ABCB19 activity by phot1 phosphorylation (or abcb19 mutation) reduces the vertical auxin flux (rootward), and produces a local accumulation of auxin at the bending zone of the hypocotyl. This available auxin might further be redistributed towards the shaded side of the hypocotyl by PIN-mediated lateral auxin transport (Christie et al., 2011) (Fig. 7).
The involvement of PIN proteins in the formation of lateral auxin gradients in response to unilateral light stimulus is still a matter of debate. Nonetheless, some PIN proteins, such as PIN3, have been proposed to function as modulators of lateral auxin transport in phototropism (Ding et al., 2011). PIN3 is mainly expressed in the endodermis of the hypocotyl (Friml et al., 2002b), but also found in the vasculature, cortex and epidermis. In etiolated Arabidopsis seedlings, PIN3 is apolarly localized at both the outer and inner lateral membranes of the endodermis cells (Fig. 7). Upon BL irradiation, PIN3 is depleted from the outer membrane and becomes more abundant at the inner side (facing the vasculature) of these cells. PIN3 polarization occurs only at the illuminated side of the hypocotyl, producing an overall polarization of PIN3 at the shaded side of the organ. Moreover, PIN3 polarization requires GNOM-dependent transcytosis (Ding et al., 2011) (Fig. 7). PIN3 re-localization might drive auxin back to the vasculature, and reduces the auxin content at the illuminated side of the hypocotyl. PIN3 polarization is abolished in phot1 mutants suggesting a role of phot1 in the modulation of lateral auxin flux through PIN3 phosphorylation. However, PIN3 is not a direct substrate of phot1. By contrast, PIN3 was found to be a direct substrate of PID, and PID activity is necessary for PIN3 lateralization (Ding et al., 2011). In both, 35S::PID plants and the triple mutant plants pidwag1wag2, PIN3 failed to re-localize upon unilateral light stimulus. In addition, PID expression is reduced upon light treatment. All these results lead the authors to propose a model in which, in the cells from the shaded side of the hypocotyl, PID activity is high and phosphorylates PIN3. The phosphorylated PIN3 is targeted to both sites of the endodermic cells (inner and outer lateral membranes). On the other hand, in the illuminated side of the hypocotyl, phot1 activation reduces PID activity, and non-phosphorylated PIN3 enters the polar trafficking pathway, with preferential localization at the inner cell side (Ding et al., 2011) (Fig. 7). This model proposes an indirect role of phot1 in the modulation of PIN3 lateralization, by regulating the level of expression and activity of PID through so far uncharacterized molecular mechanisms. A similar regulatory mechanism has been shown to involve PID (as well as PID2, WAG1 and WAG2) in the red-light enhancement of the phototropic curvature (Haga et al., 2014; Haga and Sakai, 2012, 2015). In this case, red-light treatment induces phytochrome-dependent down regulation of PID transcription and PID protein abundance, which increases the fraction of non-phosphorylated PINs (for instance, PIN3 and PIN7) delivered to the plasma membrane through the GNOM-dependent pathway (Haga et al., 2014).
Despite these evidences for a prominent role of PIN3 in phototropism, two main results suggest the existence of additional auxin transporters involved in lateral auxin fluxes: i) pin3 mutant plants only show a slightly reduced phototropic response (and the same is true for the 35S::PID and pidwag1wag2 mutant plants) and, ii) lateral auxin transport is not completely impaired in the pin3 mutant background. However the pin3pin4pin7 triple mutant have strong phototropic defect, suggesting that they redundantly mediate hypocotyl bending (Ding et al., 2011; Willige et al., 2013). On the other hand, using dark acclimated de-etiolated seedlings, Christie al., (2011) concluded that PIN3 is not responsible for lateral auxin redistribution, as pin3 mutants have the same pattern of auxin distribution as wild-type plants, as analyzed by the DR5rev::GFP reporter. Instead, these authors reported a reduction of PIN3 protein below the region of the hypocotyl curvature upon BL treatment, thus suggesting that PIN3 could promote auxin accumulation at the bending zone and indirectly contribute to lateral auxin distribution (Christie et al., 2011). Moreover, PIN3 lateralization in etiolated seedlings was detected after 3-4h of unilateral light treatment (Ding et al., 2011), while auxin gradient formation starts within less than one hour of illumination (for a timeline on early phototropic events we refer to the excellent recent review by Fankhauser and Christie (2015)). These observations might exclude PIN3 from the initial asymmetric auxin distribution and argue in favor of a role of PIN4, PIN7 or an unknown PIN-independent lateral auxin mechanism. However, it is also possible that either i) PIN3/PIN4/PIN7 localization happens within 1 hour but can be monitored by confocal microscopy only after 3-4h and/or ii) apparent polar localization is only part of the story and that auxin transport activity itself can also be polarized.
In parallel to the discovery of the strong non-phototropic phenotype of the pin3pin4pin7 triple mutant, the phenotypic characterization of the quadruple mutant d6pk0123 lead to the involvement of D6PK proteins in hypocotyl phototropism. In addition to the impaired hypocotyl phototropic response, the d6pk0123 mutant plants also show impaired gravitropic response in both hypocotyls and roots and, developmental defects such as reduced number of lateral roots (Willige et al., 2013; Zourelidou et al., 2009). All these phenotypes resembled those of auxin transport mutants. The hypocotyls of d6pk012 plants failed to establish the asymmetric auxin gradient necessary for hypocotyl bending upon unilateral light stimulation, although PIN3 lateralization occurred as in wild-type plants. However, d6pk0123 plants showed strong inhibition of the basipetal auxin transport, to the same extend as pin3pin4pin7 mutants. Together these results suggested that the non-phototropic hypocotyl phenotype of these plants was primary associated to defects on the main vertical auxin fluxes and suggested a genetic link between D6PK and PIN proteins (Willige et al., 2013). By contrast to PID/WAG-dependent PIN phosphorylation, D6PK-dependent PIN phosphorylation do not induce changes in the polar localization of PIN proteins (as observed by the proper lateralization of PIN3 in hypocotyls of d6pk012 mutants upon unilateral light stimulus), but instead it stimulates the auxin efflux activity of these transporters and ensures proper basipetal auxin transport in planta (Barbosa et al., 2014; Willige et al., 2013). Thus, the lack of phosphorylation of PIN3 in d6pk mutants results in a reduced auxin transport capacity of this protein, finally impairing the phototropic response of these plants. Hence, stimulation of PIN efflux activity by D6PKs is required for proper phototropism (Willige et al., 2013).
Conclusion
Throughout this review we highlighted the diverse mechanisms by which phosphorylation/dephosphorylation has been involved in ‘non-genomic’ regulation of polar auxin transport. Mainly, phosphorylation controls auxin carriers either by directly acting on their transport activity or their intracellular trafficking. The latter impacts the polarity, degradation and plasma membrane accumulation of PIN proteins. However, it is clear that much still need to be addressed to understand each pathway described in this review. Among the outstanding questions that remain unanswered are: How the differential phosphorylations of PIN proteins act either on their polarity or activity? How auxin and GOLVEN peptides are perceived to regulate PIN intracellular trafficking? What are the molecular steps, independent of transcription, between cytokinin perception and PIN polarity regulation? What are the proteins regulated by phosphoinositides that control PIN trafficking and polarity? How other environmental (e.g. halotropism (Galvan-Ampudia et al., 2013)) and developmental (e.g. GA signaling (Lofke et al., 2013; Willige et al., 2011)) regulatory cues coalesce in the regulation of PIN trafficking and activity? Addressing these questions will bring us closer to understanding how polar auxin transport is both a robust system in some contexts but highly dynamic and versatile in others. Integrating these different regulatory modules into one unifying model will also be a challenge for the coming years that will certainly require multidisciplinary approaches ranging from structural biology to quantitative biochemistry and cell imaging as well as mathematical modeling.
Acknowledgment
We thank Christine Miège, Matthieu Platre, John Christie, Antoine Larrieu, Carlos Galvan-Ampudia and Thomas Stanislas for discussions and comments on the manuscript. Y.J. is funded by ERC n°3363360-APPL and Marie Curie Action, n°PCIG-GA-2011-303601, under FP/2007-2013.
Footnotes
30 words statement: This review highlights our current knowledge on how phosphorylation by protein and phosphatidylinositol kinases controls the polarity, intracellular trafficking, stability and activity of auxin carriers.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nature Cell Biology. 2006;8:249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- Adamowski M, Friml J. PIN-dependent auxin transport: action, regulation, and evolution. The Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological Reviews. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros I, Dominguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartin M, Sanchez-Serrano JJ. Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. The Plant Journal. 2013;73:862–872. doi: 10.1111/tpj.12078. [DOI] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, Oh J, et al. Systems analysis of auxin transport in the Arabidopsis root apex. The Plant Cell. 2014;26:862–875. doi: 10.1105/tpc.113.119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Peret B, Oliva M, et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proceedings of the National Academy of Sciences, USA. 2012;109:4668–4673. doi: 10.1073/pnas.1201498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, Friml J, et al. Interactions of PIN and PGP auxin transport mechanisms. Biochemical Society Transactions. 2007;35:137–141. doi: 10.1042/BST0350137. [DOI] [PubMed] [Google Scholar]
- Barbez E, Lankova M, Parezova M, Maizel A, Zazimalova E, Petrasek J, Friml J, Kleine-Vehn J. Single-cell-based system to monitor carrier driven cellular auxin homeostasis. BMC Plant Biology. 2013;13:20. doi: 10.1186/1471-2229-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa IC, Zourelidou M, Willige BC, Weller B, Schwechheimer C. D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Developmental Cell. 2014;29:674–685. doi: 10.1016/j.devcel.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Baster P, Robert S, Kleine-Vehn J, Vanneste S, Kania U, Grunewald W, De Rybel B, Beeckman T, Friml J. SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. The EMBO Journal. 2013;32:260–274. doi: 10.1038/emboj.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB. A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proceedings of the National Academy of Sciences, USA. 2008;105:4044–4049. doi: 10.1073/pnas.0710294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development. 2001;128:4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR. Morphogenesis in pinoid mutants of Arabidopsis thaliana. The Plant Journal. 1995;8:505–520. [Google Scholar]
- Bennett T. PIN proteins and the evolution of plant development. Trends in Plant Science. 2015;20:498–507. doi: 10.1016/j.tplants.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Bennett T, Hines G, Leyser O. Canalization: what the flux? Trends in Genetics. 2014;30:41–48. doi: 10.1016/j.tig.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, Geisler M, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. The Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- Chen X, Naramoto S, Robert S, Tejos R, Lofke C, Lin D, Yang Z, Friml J. ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Current Biology. 2012;22:1326–1332. doi: 10.1016/j.cub.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Current Opinion in Plant Biology. 2011;14:632–641. doi: 10.1016/j.pbi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proceedings of the National Academy of Sciences, USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, Adamec J, et al. phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biology. 2011;9:e1001076. doi: 10.1371/journal.pbio.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C, Lavagi I, Yang Z. New insights into Rho signaling from plant ROP/Rac GTPases. Trends in Cell Biology. 2012;22:492–501. doi: 10.1016/j.tcb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, Wan J, et al. A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. The Plant Cell. 2012;24:2497–2514. doi: 10.1105/tpc.112.098905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Wang W, Patterson SE, Bleecker AB. The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS One. 2013;8:e60990. doi: 10.1371/journal.pone.0060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage E, Puyaubert J, Zachowski A, Ruelland E. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Progress in Lipid Research. 2013;52:1–14. doi: 10.1016/j.plipres.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Delage E, Ruelland E, Guillas I, Zachowski A, Puyaubert J. Arabidopsis type-III phosphatidylinositol 4-kinases beta1 and beta2 are upstream of the phospholipase C pathway triggered by cold exposure. Plant and Cell Physiology. 2012;53:565–576. doi: 10.1093/pcp/pcs011. [DOI] [PubMed] [Google Scholar]
- Deslauriers SD, Larsen PB. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Molecular Plant. 2010;3:626–640. doi: 10.1093/mp/ssq015. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137:3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE. LRR-containing receptors regulating plant development and defense. Development. 2004;131:251–261. doi: 10.1242/dev.00998. [DOI] [PubMed] [Google Scholar]
- Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, Friml J. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nature Cell Biology. 2011;13:447–452. doi: 10.1038/ncb2208. [DOI] [PubMed] [Google Scholar]
- Drdova EJ, Synek L, Pecenkova T, Hala M, Kulich I, Fowler JE, Murphy AS, Zarsky V. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. The Plant Journal. 2013;73:709–719. doi: 10.1111/tpj.12074. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences, USA. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S, Ruonala R, Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140:1373–1383. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Christie JM. Plant phototropic growth. Current Biology. 2015;25:R384–389. doi: 10.1016/j.cub.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiology. 2013a;161:954–970. doi: 10.1104/pp.112.206029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Hilson P, Beeckman T. GOLVEN peptides as important regulatory signalling molecules of plant development. Journal of Experimental Botany. 2013b;64:5263–5268. doi: 10.1093/jxb/ert248. [DOI] [PubMed] [Google Scholar]
- Finet C, Jaillais Y. Auxology: when auxin meets plant evo-devo. Developmental Biology. 2012;369:19–31. doi: 10.1016/j.ydbio.2012.05.039. [DOI] [PubMed] [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, Palme K. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002a;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002b;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell. 2005;120:687–700. doi: 10.1016/j.cell.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z. The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. The Plant Cell. 2002;14:777–794. doi: 10.1105/tpc.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Current Biology. 2009;19:1827–1832. doi: 10.1016/j.cub.2009.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, Torres-Ruiz RA, Tasaka M. The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development. 2007;134:3849–3859. doi: 10.1242/dev.009654. [DOI] [PubMed] [Google Scholar]
- Furutani M, Sakamoto N, Yoshida S, Kajiwara T, Robert HS, Friml J, Tasaka M. Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development. 2011;138:2069–2078. doi: 10.1242/dev.057745. [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Julkowska MM, Darwish E, Gandullo J, Korver RA, Brunoud G, Haring MA, Munnik T, Vernoux T, Testerink C. Halotropism is a response of plant roots to avoid a saline environment. Current Biology. 2013;23:2044–2050. doi: 10.1016/j.cub.2013.08.042. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends in Plant Science. 2007;12:541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruere J, Bernasconi P, Soll D. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. The EMBO Journal. 1996;15:2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, Ejendal KF, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. The Plant Journal. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jurgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, Jurgens G. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development. 2004;131:389–400. doi: 10.1242/dev.00926. [DOI] [PubMed] [Google Scholar]
- Goldsmith MH, Goldsmith TH. Quantitative predictions for the chemiosmotic uptake of auxin. Planta. 1981;153:25–33. doi: 10.1007/BF00385314. [DOI] [PubMed] [Google Scholar]
- Goldsmith MH, Goldsmith TH, Martin MH. Mathematical analysis of the chemosmotic polar diffusion of auxin through plant tissues. Proceedings of the National Academy of Sciences, USA. 1981;78:976–980. doi: 10.1073/pnas.78.2.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Grones P, Friml J. Auxin transporters and binding proteins at a glance. Journal of Cell Science. 2015;128:1–7. doi: 10.1242/jcs.159418. [DOI] [PubMed] [Google Scholar]
- Guo X, Qin Q, Yan J, Niu Y, Huang B, Guan L, Li Y, Ren D, Li J, Hou S. TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiology. 2015;167:1058–1075. doi: 10.1104/pp.114.249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets ME, Offringa R. PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytologist. 2014;203:362–377. doi: 10.1111/nph.12831. [DOI] [PubMed] [Google Scholar]
- Haga K, Hayashi K, Sakai T. PINOID AGC kinases are necessary for phytochrome-mediated enhancement of hypocotyl phototropism in Arabidopsis. Plant Physiology. 2014;166:1535–1545. doi: 10.1104/pp.114.244434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Sakai T. PIN auxin efflux carriers are necessary for pulse-induced but not continuous light-induced phototropism in Arabidopsis. Plant Physiology. 2012;160:763–776. doi: 10.1104/pp.112.202432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Sakai T. PINOID functions in root phototropism as a negative regulator. Plant Signaling & Behavior. 2015;10:e998545. doi: 10.1080/15592324.2014.998545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Takano M, Neumann R, Iino M. The Rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. The Plant Cell. 2005;17:103–115. doi: 10.1105/tpc.104.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G. Auxin signal transduction. Essays In Biochemistry. 2015;58:1–12. doi: 10.1042/bse0580001. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Henrichs S, Wang B, Fukao Y, Zhu J, Charrier L, Bailly A, Oehring SC, Linnert M, Weiwad M, Endler A, Nanni P, et al. Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. The EMBO Journal. 2012;31:2965–2980. doi: 10.1038/emboj.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, Roberts D, Liscum E. Understanding phototropism: from Darwin to today. Journal of Experimental Botany. 2009;60:1969–1978. doi: 10.1093/jxb/erp113. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nature Reviews Molecular Cell Biology. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. The Plant Cell. 2010;22:1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytologist. 2013;200:1089–1101. doi: 10.1111/nph.12432. [DOI] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130:2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Muller B. Cytokinin signaling networks. Annual Review of Plant Biology. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. The Plant Cell. 2004;16:887–896. doi: 10.1105/tpc.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Werner S, Krishnamoorthy P, Lerche J, Meijon M, Stenzel I, Lofke C, Wiessner T, Im YJ, Perera IY, Iven T, et al. Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. The Plant Cell. 2013;25:4894–4911. doi: 10.1105/tpc.113.116582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nature Structural and Molecular Biology. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miege C, Rollin C, Gaude T. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006;443:106–109. doi: 10.1038/nature05046. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miege C, Gaude T. The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell. 2007;130:1057–1070. doi: 10.1016/j.cell.2007.08.040. [DOI] [PubMed] [Google Scholar]
- Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nature Reviews Molecular Cell Biology. 2012;13:463–470. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Torii KU. FERONIA as an upstream receptor kinase for polar cell growth in plants. Proceedings of the National Academy of Sciences, USA. 2010;107:17461–17462. doi: 10.1073/pnas.1013090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA. Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans Golgi network compartments in Arabidopsis. Traffic. 2011;12:313–329. doi: 10.1111/j.1600-0854.2010.01146.x. [DOI] [PubMed] [Google Scholar]
- Karampelias M, Neyt P, De Groeve S, Aesaert S, Coussens G, Rolcik J, Bruno L, De Winne N, Van Minnebruggen A, Van Montagu M, Ponce MR, et al. ROTUNDA3 function in plant development by phosphatase 2A-mediated regulation of auxin transporter recycling. Proceedings of the National Academy of Sciences, USA. 2016;113:2768–2773. doi: 10.1073/pnas.1501343112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proceedings of the National Academy of Sciences, USA. 2010;107:22344–22349. doi: 10.1073/pnas.1013145107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. The Plant Cell. 2009;21:3839–3849. doi: 10.1105/tpc.109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martiniere A, Langowski L, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, Luschnig C, et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Molecular Systems Biology. 2011;7:540. doi: 10.1038/msb.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Jez JM, Strader LC. Refining the nuclear auxin response pathway through structural biology. Current Opinion in Plant Biology. 2015;27:22–28. doi: 10.1016/j.pbi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbei B, Moulinier-Anzola J, De-Araujo L, Lucyshyn D, Retzer K, Khan MA, Luschnig C. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Current Biology. 2013;23:2500–2505. doi: 10.1016/j.cub.2013.10.036. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nature Chemical Biology. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet P, Schepens I, Hodgson D, Pedmale UV, Trevisan M, Kami C, de Carbonnel M, Alonso JM, Ecker JR, Liscum E, Fankhauser C. PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proceedings of the National Academy of Sciences, USA. 2006;103:10134–10139. doi: 10.1073/pnas.0603799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu A, Vernoux T. Comparison of plant hormone signalling systems. Essays In Biochemistry. 2015;58:165–181. doi: 10.1042/bse0580165. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature Reviews Molecular Cell Biology. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Li H, Lin D, Dhonukshe P, Nagawa S, Chen D, Friml J, Scheres B, Guo H, Yang Z. Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Research. 2011;21:970–978. doi: 10.1038/cr.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nature Methods. 2015;12:207–210. doi: 10.1038/nmeth.3279. 202 p following 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Cao L, Zhou Z, Zhu L, Ehrhardt D, Yang Z, Fu Y. Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Current Biology. 2013;23:290–297. doi: 10.1016/j.cub.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, Scheres B, et al. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Current Biology. 2012;22:1319–1325. doi: 10.1016/j.cub.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Askinosie SK, Leuchtman DL, Morrow J, Willenburg KT, Coats DR. Phototropism: growing towards an understanding of plant movement. The Plant Cell. 2014;26:38–55. doi: 10.1105/tpc.113.119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. The Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofke C, Zwiewka M, Heilmann I, Van Montagu MC, Teichmann T, Friml J. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proceedings of the National Academy of Sciences, USA. 2013;110:3627–3632. doi: 10.1073/pnas.1300107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Yu F, Li J, Van de Poel B, Tan D, Li J, Liu Y, Li X, Dong M, Chen L, Li D, et al. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant, Cell and Environment. 2015;38:2566–2574. doi: 10.1111/pce.12570. [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. The EMBO Journal. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Parezova M, Petrasek J, Friml J, Kleine-Vehn J, Benkova E. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Developmental Cell. 2011;21:796–804. doi: 10.1016/j.devcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Marhavy P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benkova E. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Current Biology. 2014;24:1031–1037. doi: 10.1016/j.cub.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Martin TF. PI(4,5)P(2)-binding effector proteins for vesicle exocytosis. Biochimica et Biophysica Acta. 2015;1851:785–793. doi: 10.1016/j.bbalip.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Kajizuka T, Kadota A, Nishimura T, Koshiba T. NPH3- and PGP-like genes are exclusively expressed in the apical tip region essential for blue-light perception and lateral auxin transport in maize coleoptiles. Journal of Experimental Botany. 2011;62:3459–3466. doi: 10.1093/jxb/err019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Jia WJ, Chu YJ, Xue HW. Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Research. 2012;22:581–597. doi: 10.1038/cr.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Miyawaki KN, Yang Z. Extracellular signals and receptor-like kinases regulating ROP GTPases in plants. Frontiers in Plant Science. 2014;5:449. doi: 10.3389/fpls.2014.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. The EMBO Journal. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Current Opinion in Plant Biology. 2011;14:489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biology. 2012;10:e1001299. doi: 10.1371/journal.pbio.1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S, Otegui MS, Kutsuna N, de Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H, Nakano A, et al. Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. The Plant Cell. 2014;26:3062–3076. doi: 10.1105/tpc.114.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Developmental Cell. 2014;29:491–500. doi: 10.1016/j.devcel.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423:999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. The Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Miyagishima SY, Wada H. Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. The Plant Cell. 2015;27:663–674. doi: 10.1105/tpc.115.136234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proceedings of the National Academy of Sciences, USA. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jurgens G, Geldner N, Friml J. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Celaya RB, Liscum E. Phototropism: mechanism and outcomes. The Arabidopsis Book. 2010;8:e0125. doi: 10.1199/tab.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. The Journal of Biological Chemistry. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Platre MP, Jaillais Y. Guidelines for the use of protein domains in acidic phospholipid imaging. Methods in Molecular Biology. 2016;1376:175–194. doi: 10.1007/978-1-4939-3170-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poraty-Gavra L, Zimmermann P, Haigis S, Bednarek P, Hazak O, Stelmakh OR, Sadot E, Schulze-Lefert P, Gruissem W, Yalovsky S. The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiology. 2013;161:1172–1188. doi: 10.1104/pp.112.213165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. The Journal of Cell Biology. 2006;172:991–998. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Blackwood L, Christie JM, Fankhauser C. Lipid anchoring of Arabidopsis phototropin 1 to assess the functional significance of receptor internalization: should I stay or should I go? New Phytologist. 2015;206:1038–1050. doi: 10.1111/nph.13299. [DOI] [PubMed] [Google Scholar]
- Preuten T, Hohm T, Bergmann S, Fankhauser C. Defining the site of light perception and initiation of phototropism in Arabidopsis. Current Biology. 2013;23:1934–1938. doi: 10.1016/j.cub.2013.07.079. [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Offringa R. Evolutionary Adaptations of Plant AGC Kinases: From Light Signaling to Cell Polarity Regulation. Frontiers in Plant Science. 2012;3:250. doi: 10.3389/fpls.2012.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusova H, Fendrych M, Friml J. Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Current Opinion in Plant Biology. 2015;23:116–123. doi: 10.1016/j.pbi.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. The Plant Cell. 2001;13:1683–1697. doi: 10.1105/TPC.010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Bigler L, Kuhn BM, Leiber RM, Diet A, Santelia D, Frey B, Pollmann S, Klein M. The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. The Plant Cell. 2008;20:1470–1481. doi: 10.1105/tpc.107.053249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Covanova M, Hayashi K, et al. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143:111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E. Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin Ligase CRL3NPH3. The Plant Cell. 2011;23:3627–3640. doi: 10.1105/tpc.111.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman N, Gourgues M, Guitton AE, Faure JE, Berger F. A dialogue between the SIRENE pathway in synergids and the fertilization independent seed pathway in the central cell controls male gamete release during double fertilization in Arabidopsis. Molecular Plant. 2008;1:659–666. doi: 10.1093/mp/ssn023. [DOI] [PubMed] [Google Scholar]
- Rubery PH, Sheldrake AR. Effect of pH and surface charge on cell uptake of auxin. Nature: New Biology. 1973;244:285–288. doi: 10.1038/newbio244285a0. [DOI] [PubMed] [Google Scholar]
- Rubery PH, Sheldrake AR. Carrier-mediated auxin transport. Planta. 1974;118:101–121. doi: 10.1007/BF00388387. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sakai T, Haga K. Molecular genetic analysis of phototropism in Arabidopsis. Plant and Cell Physiology. 2012;53:1517–1534. doi: 10.1093/pcp/pcs111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proceedings of the National Academy of Sciences, USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K. RPT2. A signal transducer of the phototropic response in Arabidopsis. The Plant Cell. 2000;12:225–236. doi: 10.1105/tpc.12.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jonsson H, Meyerowitz EM. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. Elife. 2014;3:e01967. doi: 10.7554/eLife.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. The Plant Journal. 2006;45:752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wisniewska J, Reinohl V, Friml J, Benkova E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes and Development. 2006;20:2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Mettlen M. Cell biology: Lipid switches and traffic control. Nature. 2013;499:161–162. doi: 10.1038/nature12408. [DOI] [PubMed] [Google Scholar]
- Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Current Biology. 2014;24:1887–1892. doi: 10.1016/j.cub.2014.06.064. [DOI] [PubMed] [Google Scholar]
- Simaskova M, O'Brien JA, Khan M, Van Noorden G, Otvos K, Vieten A, De Clercq I, Van Haperen JM, Cuesta C, Hoyerova K, Vanneste S, et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nature Communications. 2015;6:8717. doi: 10.1038/ncomms9717. [DOI] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. The Plant Journal. 2014;77:322–337. doi: 10.1111/tpj.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Marques-Bueno MM, Armengot L, Stanislas T, Bayles V, Caillaud MC, Jaillais Y. A PI4P-driven electrostatic field controls cell membrane identity and signaling in plants. Nature Plants. 2016 doi: 10.1038/nplants.2016.89. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T, Hüser A, Barbosa IC, Kiefer P, Brackmann K, Pietra S, Gustavsson A, Zourelidou M, Schwechheimer C, Grebe M. Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nature Plants. 2015;1(11):15162. doi: 10.1038/nplants.2015.162. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiology. 2009;150:722–735. doi: 10.1104/pp.108.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nature Cell Biology. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- Tejos R, Sauer M, Vanneste S, Palacios-Gomez M, Li H, Heilmann M, van Wijk R, Vermeer JE, Heilmann I, Munnik T, Friml J. Bipolar plasma membrane distribution of phosphoinositides and their Requirement for auxin-mediated cell polarity and patterning in Arabidopsis. The Plant Cell. 2014;26:2114–2128. doi: 10.1105/tpc.114.126185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, Sakai T, et al. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. The Plant Journal. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JE, Gadella TW, Jr, Munnik T. Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. The Plant Journal. 2007;52:1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x. [DOI] [PubMed] [Google Scholar]
- Vermeer JE, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TW., Jr Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. The Plant Journal. 2009;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobena-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW, Jr, Munnik T. Visualization of PtdIns3P dynamics in living plant cells. The Plant Journal. 2006;47:687–700. doi: 10.1111/j.1365-313X.2006.02830.x. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- Wan Y, Jasik J, Wang L, Hao H, Volkmann D, Menzel D, Mancuso S, Baluska F, Lin J. The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2-based polar auxin transport in Arabidopsis root phototropism. The Plant Cell. 2012;24:551–565. doi: 10.1105/tpc.111.094284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D. Transcriptional responses to the auxin hormone. Annual Review of Plant Biology. 2016;67:539–574. doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP. Phototropism: bending towards enlightenment. The Plant Cell. 2006;18:1110–1119. doi: 10.1105/tpc.105.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP. The “sensational” power of movement in plants: A Darwinian system for studying the evolution of behavior. American Journal of Botany. 2009;96:2115–2127. doi: 10.3732/ajb.0900220. [DOI] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, Perez AC, Kleine-Vehn J, Vanneste S, Drozdzecki A, Leitner J, Abas L, Aerts M, Hoogewijs K, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Developmental Cell. 2012;22:678–685. doi: 10.1016/j.devcel.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Willige BC, Ahlers S, Zourelidou M, Barbosa IC, Demarsy E, Trevisan M, Davis PA, Roelfsema MR, Hangarter R, Fankhauser C, Schwechheimer C. D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. The Plant Cell. 2013;25:1674–1688. doi: 10.1105/tpc.113.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. The Plant Cell. 2011;23:2184–2195. doi: 10.1105/tpc.111.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Wu HM, Hazak O, Cheung AY, Yalovsky S. RAC/ROP GTPases and auxin signaling. The Plant Cell. 2011;23:1208–1218. doi: 10.1105/tpc.111.083907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Gong X, Yang Q, Yu H, Liou YC. Pin1At regulates PIN1 polar localization and root gravitropism. Nature Communications. 2016;7:10430. doi: 10.1038/ncomms10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusova H, Wang W, et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143:99–110. doi: 10.1016/j.cell.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. The Plant Journal. 2009;59:179–191. doi: 10.1111/j.1365-313X.2009.03856.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Current Biology. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Yang Z, Lavagi I. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Current Opinion in Plant Biology. 2012;15:601–607. doi: 10.1016/j.pbi.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Li J, Huang Y, Liu L, Li D, Chen L, Luan S. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Molecular Plant. 2014;7:920–922. doi: 10.1093/mp/ssu010. [DOI] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, Li L, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proceedings of the National Academy of Sciences, USA. 2012;109:14693–14698. doi: 10.1073/pnas.1212547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Anthony RG, Jahchan N, Bogre L, Christensen SK. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006a;103:6404–6409. doi: 10.1073/pnas.0510283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Li W, Lorenz TC, Xie M, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK. Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. The Journal of Biological Chemistry. 2006b;281:35520–35530. doi: 10.1074/jbc.M605167200. [DOI] [PubMed] [Google Scholar]
- Zhang C, Kotchoni SO, Samuels AL, Szymanski DB. SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Current Biology. 2010a;20:2144–2149. doi: 10.1016/j.cub.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencik A, Rolcik J, Friml J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proceedings of the National Academy of Sciences, USA. 2010b;107:918–922. doi: 10.1073/pnas.0909460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Absmanner B, Weller B, Barbosa IC, Willige BC, Fastner A, Streit V, Port SA, Colcombet J, de la Fuente van Bentem S, Hirt H, et al. Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. Elife. 2014;3:e02860. doi: 10.7554/eLife.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development. 2009;136:627–636. doi: 10.1242/dev.028365. [DOI] [PubMed] [Google Scholar]