Abstract
Pseudomonas aeruginosa is well known for its antibiotic resistance and intricate regulatory network, contributing to its success as an opportunistic pathogen. This study is an extension of our transcriptomic analyses (microarray and RNA-Seq) to understand the global changes in PAO1 upon deleting a gene encoding a transcriptional regulator AmpR, in the presence and absence of β-lactam antibiotic. This study was performed under identical conditions to explore the proteome profile of the ampR deletion mutant (PAOΔampR) using GeLC-MS analysis. The proteomic data identified ~53% of total PAO1 proteins and expanded the master regulatory role of AmpR in determining antibiotic resistance and multiple virulence phenotypes in P. aeruginosa. AmpR proteome analysis identified 853 AmpR-dependent proteins, which include 102 transcriptional regulators and 21 two-component system proteins. AmpR also regulates cyclic di-GMP phosphodiesterases (PA4367, PA4969, PA4781) possibly affecting major virulence systems. Phosphoproteome analysis also suggests a significant role for AmpR in Ser, Thr and Tyr phosphorylation. These novel mechanisms of gene regulation were previously not associated with AmpR. The proteome analysis also identified many unannotated and misannotated ORFs in the P. aeruginosa genome. Thus, our data sheds light on important virulence regulatory pathways that can potentially be exploited to deal with P. aeruginosa infections.
Keywords: Pseudomonas aeruginosa, AmpR, Proteome, Phosphoproteome, cyclic di-GMP
Graphical abstract
INTRODUCTION
Pseudomonas aeruginosa, a Gram-negative opportunistic pathogen, frequently causes life-threatening infections in cystic fibrosis (CF) patients, and several other hospitalized immunocompromised individuals such as those with burn wounds, medical implants, cancer and AIDs [1-4]. The extensive range of infections caused by P. aeruginosa is due, in part, to the multitude of virulence factors coded for by its genome. The genome of P. aeruginosa strain PAO1 encodes a predicted 5569 open reading frames (ORFs) and remains one of the largest sequenced bacterial genomes on a single chromosome with 36% of hypothetical proteins [5]. While most bacterial pathogens adopt a strategy of genome reduction [6, 7], the plasticity of the P. aeruginosa genome that allows incorporation of acquired DNA, has enabled the bacterium to thrive in a diverse range of habitats [8]. The genome also aids in clinical setting by encoding numerous virulence factors to establish and maintain an infection, as well as for different antibiotic resistance mechanisms [9, 10].
P. aeruginosa is intrinsically primed to evade antibiotics [11]. The membrane impermeability [12, 13] and ability to modify drug targets [14], compounded by the expression of multiple efflux pumps [15] and β-lactamases (AmpC and PoxB) [16, 17] makes it a formidable pathogen. The major β-lactamase, AmpC, is a clinically-important, chromosomally-encoded enzyme that mediates resistance to most cephalosporins [16]. In P. aeruginosa and Enterobacteriaceae members, AmpR, a LysR-type transcriptional regulator (LTTR) [18, 19], induces the expression of ampC in the presence of β-lactams [20-23]. In addition to ampC, AmpR was previously shown to regulate genes involved in P. aeruginosa virulence [24, 25]. Recent transcriptome analyses have shown that the P. aeruginosa AmpR regulon is quite extensive [26, 27]. AmpR microarray studies showed AmpR-regulation of non-β-lactam resistance through MexEF-OprN efflux system, as well as several virulence determinants under quorum sensing control, secretion systems and biofilm formation [26]. Deep sequencing of RNA further revealed the role of AmpR in other processes such as oxidative stress, iron acquisition and heat shock, most of which are mediated by regulation of small RNAs [27]. Importantly, the transcriptome studies reveal that AmpR activates expression of genes associated with acute infection establishment and represses those that control chronic infection phenotypes [26, 27]. Thus, it is important to know whether AmpR-mediated differential regulation of genes is also evident at the protein level. The dynamics of the P. aeruginosa proteome during exposure to β-lactams and the role of AmpR in the process are yet to be elucidated.
This study describes the shotgun proteomic analysis of the wild type P. aeruginosa PAO1 and its isogenic ampR deletion mutant, PAOΔampR in the presence and absence of β-lactam stress, similar to previous transcriptome studies [26, 27]. Proteins were identified using two proteomic database search tools namely, Crux [28] and InsPecT [29]. The proteome data further expanded the AmpR regulon to include novel virulence mechanisms. The data also revealed a role for AmpR in protein phosphorylation, and identified several previously unannotated or misannotated ORFs in P. aeruginosa genome.
MATERIALS AND METHODS
Bacterial cell culture and primers
P. aeruginosa strains PAO1 [5] and PAOΔampR [26] were grown and harvested essentially identical to the two transcriptome (array and deep-sequencing of RNA) studies [26, 27]. Briefly, the cells were grown to an OD600 of 0.8 in LB medium and divided into two pools. To one pool, added 100 μg/mL of benzyl penicillin (induced sample) and a second pool was kept without antibiotic as control (uninduced). The cells were further grown for two hours before harvesting. The culture OD600 at that point was ~ 4.0. The numbers of replicates for each of the conditions were: two each for uninduced and induced PAO1 and three each for uniduced and induced PAOΔampR, making a total of 10 samples. The sub-inhibitory β-lactam exposure did not have a significant effect on the growth of PAO1 and PAOΔampR.
GeLC-MS analysis of PAO1 and PAOΔampR samples
For each samples, 1 gram of cell pellet was resuspended in 5 mL of guanidium chloride solution (8 M guanidium chloride, 5% n-propanol, 10 mM NH4HCO3 pH 8.6 and 10 mM DTT added fresh) and sonicated at room temperature. The sonicated sample was aliquoted into four Lysing Matrix B tubes (MP Biomedical; 1 mL per tube) and vortexed for 30 seconds. The extracts were processed and analyzed as described [30] at the Proteomics facility at the Harvard-Partners Center for Genetics and Genomics. Briefly, each sample was run on a NuPAGE and 10 slices were obtained from each. The protein fragments from each gel slice were characterized using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on a LTQ XL mass spectrometer (Thermo Fisher Scientific Inc, USA).
Data analysis
The binary RAW files were converted to the generic mzXML format [31] using MakeMS2 (http://proteome.gs.washington.edu/software/makems2/). P. aeruginosa PAO1 protein sequences were obtained from the Pseudomonas Community Annotation Project (PseudoCAP) [32] and combined with the commonly encountered contaminant protein list available from ftp://ftp.thegpm.org/fasta/cRAP. All pre-and post-processing of text files were carried out using sed, awk and perl scripts [33].
Database searching and protein identification
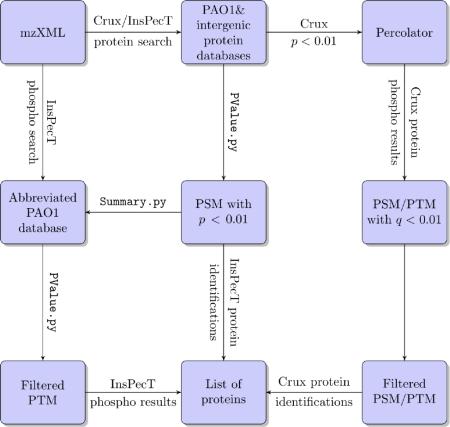
An overview of the work-flow is shown in Fig. 1. FASTA files containing the PAO1 protein sequences (5569 entries) and common contaminant sequences (112 entries) was used to generate the forward and shuffled database using PrepDB.py and ShuffleDB.py for InsPecT (Version 20100804) [34] and using create-index command for Crux (Version 1.31) [28]. Iodoacetic acid derivative of cysteine was specified as a fixed modification, while oxidation of methionine, phosphorylation of serine, threonine and tyrosine were specified as variable modifications in both Crux and InsPecT searches.
Fig. 1.
Overview of the work-flow for GeLC-MS analysis using Crux and Inspect.
For Crux, the maximum number of modified methionine was set to two and phosphorylated serine, threonine and tyrosine residues was set to three. For InsPecT, the maximum number of variable modifications was set to three. The instrument type was set to ESI-ION-TRAP for InsPecT, all other parameters were default for both Crux and InsPecT database searches. The output files from Crux search-for-matches were filtered using Percolator [35] to determine the peptide spectrum matches (PSM) having minimal false discovery rate. Only those PSM that fulfilled the criteria of q < 0.01 [35] were chosen. Search results from InsPecT were filtered using PValue.py and only PSM with p < 0.01 were chosen. In InsPecT Summary.py was used to generate a subset of identified protein database (1574 entries) to search against, for identifying phosphorylation modifications. The results were further analyzed using PhosphateLocalization.py script to filter post-translational modifications (PTM) based on phosphate localization score (PLS) and only those PTM having PLS > 20 were considered.
For determining novel proteins, the PseudoCAP [32] annotated intergenic regions of the PAO1 genome were translated in all six frames using the EMBOSS [36] command transeq, combined with the common contaminants FASTA file and was used for searching the tandem mass spectra using Crux and InsPecT as described above. Data analysis using a third program, BioWorks (Ver 3.3.2) generated data similar to Crux (data not shown) and so was not used further.
Distribution and functional enrichment analysis
Functional categorization of all the identified proteins followed that of the PseudoCAP [32]. Gene distribution under individual functional categories was plotted as percentage of query genes vs. percentage of PAO1 genes in each functional category.
Enrichment analysis
Functional enrichment analysis was performed for both the total proteome dataset and AmpR-regulated proteins. Enrichment of a particular functional category in the total proteome data was assessed by comparing the percentage of proteins in that category in the proteome dataset with its percentage distribution in PAO1. A higher or lower percentage compared to PAO1 signified enrichment (E) or underrepresentation (U), respectively. The significance of E or U was determined by p-values, computed by hypergeometric distribution. A conservative-threshold of 0.05/N (N=26, number of functional categories) was chosen to account for multiple hypotheses setting. Thus, a p-value ≤ 0.002 was considered significant.
Similarly, the protein datasets in regulated by AmpR in the absence and presence of β-lactam were also tested for E/U of specific functional categories by comparing percentage of proteins in a particular functional category in each AmpR-dependent dataset against percentage of that category in the total proteome dataset.
RNA isolation, cDNA synthesis and quantitative real-time PCR (qPCR)
Total RNA was isolated from PAO1 and PAOΔampR following the same culture condition as for the proteome assay. The antibiotic was added to the cells grown to mid-log phase and the cells with or without antibiotic exposure was further incubated for one hour before harvesting in the stationary phase. We had previously demonstrated that the gene expression trend is the same, whether we exposed the cells for 40 minutes or for two hours [26]. RNA isolation, cDNA synthesis and qPCR assays were performed as described earlier [26]. Ten nanograms of cDNA were used per reaction well in the qPCR assays. The clpX gene (PA1802) was used as an internal control to ensure equal amounts of RNA were used in all samples. Assays were performed at least in biological triplicates, each with technical triplicates, for every gene analyzed. Melt curves were determined to ensure primer specificity. Gene expression in PAOΔampR was normalized to the corresponding PAO1 values, for both the β-lactam uninduced and induced conditions and is presented as fold-expression in PAOΔampR. All data were analyzed for statistical significance using t-test on GraphPad statistical analysis software. Primers used for the qPCR analysis are listed in Supplementary Table 1.
Verification of unannotated ORFs
The basic alignment search tool [37, 38] was used to map identified peptides to ORFs not previously annotated in the PseudoCAP [32]. To verify that the putative ORF was expressed, reverse transcriptase PCR (RT-PCR) was performed on RNA isolated from PAO1. Briefly, PAO1 RNA was isolated and cDNA was synthesized as described earlier [26]. As a control, the cDNA synthesis was also performed in the absence of reverse transcriptase enzyme and the sample was processed along with the cDNA. Using cDNA as template, putative ORFs were amplified using primers listed in Supplementary Table 1. The amplification products were analyzed using standard DNA gel electrophoresis.
RESULTS AND DISCUSSION
GeLC-MS analysis of P. aeruginosa proteome
It has been well-established that the β-lactam antibiotic exposure results in the AmpR-induced expression of ampC encoding a β-lactamase in P. aeruginosa and many Enterobacteriaceae members [23, 24]. Further, transcriptome studies and complementary assays demonstrated that the AmpR regulon is extensive and includes additional genes involved in antibiotic resistance, virulence and metabolism [24-27]. However, the transcriptional profile does not necessarily reflect proteomic profile due to post-transcriptional regulation. In addition, transcriptome analyses [26, 27] could have missed key AmpR-regulated targets. In order to complement the previous transcriptomic studies [26, 27] to determine the global regulatory role of AmpR, proteomic analyses of PAO1 and PAOΔampR were performed using GeLC-MS.
PAO1 and PAOΔampR cells were grown under identical conditions as in our previous transcriptome (microarray and RNA-Seq) studies in the presence or absence of β-lactam [26, 27]. The total cell lysate was separated by SDS-PAGE and the MS-MS spectra for each gel slice were obtained as described under materials and methods. Analysis of the GeLC-MS data was performed using two different tools, Crux [28] and InsPect [29]. Crux uses a modified SEQUEST algorithm [39], whereas InsPecT uses a combination of database searching and de novo peptide tag-based filtering algorithms [29]. It should also be noted that the mere absence of a protein in a group does not mean that the protein is not expressed, but that it was not detectable in that condition using our proteomic methodology.
The list of genes encoding the non-redundant peptides found in the GeLC-MS analysis by Crux and InPecT is given in Supplementary Table 2. These analyses led to the identification of 2965 non-redundant proteins in all of the samples analyzed (Supplementary Table 2). A total of 1302 proteins were detected by both the analysis tools, whereas 15 and 1648 proteins were exclusively identified in Crux and InsPecT, respectively.
Our proteome data thus identified 53% of the total PAO1 encoded proteins. Previous proteome studies in P. aeruginosa identified between 1% and 30% of total proteins [40-44]. The number of proteins identified in the current analysis is significantly higher than the previous studies. The widely varying results between the studies can potentially be explained by the differential sensitivities of the various analytical techniques (iTraq, MudPit, 2-DE), the strains (PAO1 and isogenic mutants, PA14, AES-1R) and the media (LB, PIA, CF media) used.
Overall classification of identified proteins based on their functional categories
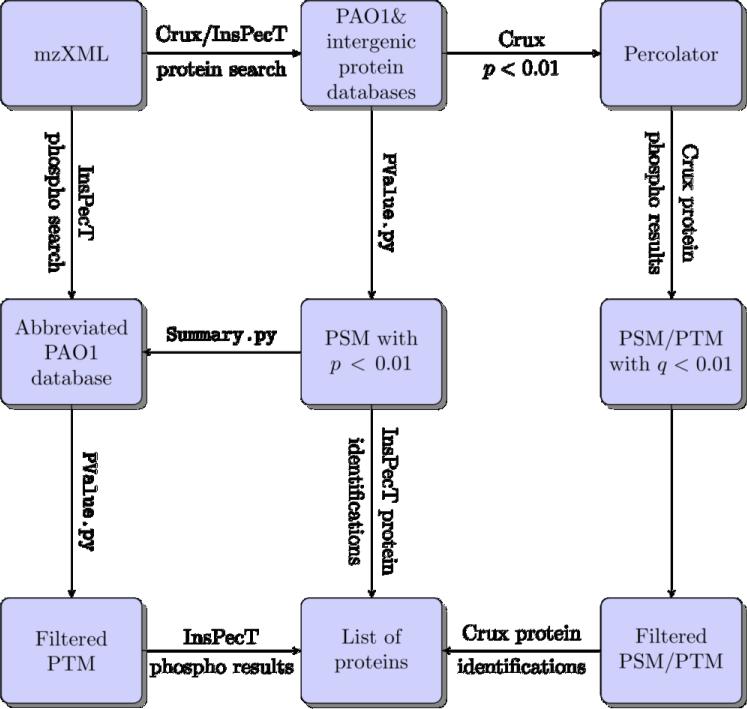
In order to find out if the proteins identified by GeLC-MS analysis are uniformly distributed or skewed in some specific categories, functional categorization of the 2965 non-redundant proteins was followed as that of PseudoCAP [32].
For each functional category (Categories a to z), the percentage of ORFs in the PAO1 database was compared to the percentage of gene products in that category identified in the proteome data (Fig. 2; Supplementary Table 3). There was an overall good representation of the genes under various functional categories in our proteome data, compared to the distribution in PAO1 (Fig. 2). The largest number of proteins identified in our analysis (31%) belonged to the hypothetical, unclassified or unknown class (Category c), which makes up 36% of the PAO1 genome. Thus, the proteins identified in Category c in our proteome data are no longer hypothetical. The next highest number of proteins identified belonged to putative enzymes (Category o), membrane proteins (Category d) and transcriptional regulators (Category l) in both PAO1 (8.1, 11.5 and 7.8%, respectively) and proteome data (8.4, 7.0 and 6.8%, respectively; Fig. 2; Supplementary Table 3).
Fig. 2. Functional categorization of the total proteins identified in GeLC-MS analysis by PseudoCAP (winsor 2011).
All the non-redundant proteins (2965) identified by crux and Inspect were functionally categorized and plotted as percentage distribution of each category in PAO1 (purple) vs. proteome data (blue). Hypergeometric analysis was performed to determine significantly enriched (*) or under-represented (#) categories (p-values of ≤ 0.002). The functional categories are (a) DNA replication, recombination, modification and repair; (b) fatty acid and phospholipid metabolism; (c) hypothetical; (d) membrane proteins; (e) amino acid biosynthesis, metabolism; (f) translation, post-translational modification, degradation; (g) cell wall/LPS/capsule; (h) transport of small molecules; (i) energy metabolism; (j) biosynthesis of cofactors, prosthetic groups, carriers; (k) adaptation, protection; (l) transcriptional regulators; (m) two-component regulatory systems; (n) secreted factors- toxins, enzymes, alginate; (o) putative enzymes; (p) chaperones, heat-shock proteins; (q) central intermediary metabolism; (r) nucleotide biosynthesis and metabolism; (s) carbon compound catabolism; (t) motility and attachment; (u) chemotaxis; (v) related to phage, transposon, plasmid; (w) protein secretion, export apparatus; (x) cell division; (z) transcription, RNA processing, degradation.
The percentage of ORFs in each functional category in proteome data and PAO1 genome were compared to determine if there is under/over representation of a particular category. The significance of enrichment (E) or under-representation (U) was determined using hypergeometric distribution p-value (Supplementary Table 3). Significant enrichment was seen for proteins involved in vital cellular functions (* in Fig. 2; Supplementary Table 3) such as DNA replication, recombination, modification and repair (Category a), cell division (Category x), transcription, RNA processing and degradation (Category z), amino acid biosynthesis and metabolism (Category e), translation (Category f), nucleotide biosynthesis and metabolism (Category r), central intermediary metabolism (Category q), biosynthesis of cofactors (Category j), energy metabolism (Category i) and cell-wall/LPS (Category g). In addition, it was not surprising that the proteins in the categories of adaptation and protection (Category k) and chaperones and heat shock proteins (Category p) were over-represented because exposure of cells to β–lactam antibiotic results in stress [45].
The significantly under-represented proteins (# in Fig. 2; Supplementary Table 3) belonged to the functional classes of hypothetical (Category c), membrane proteins (Category d) and transcriptional regulators (Category l). Although not statistically significant, substantially reduced number of proteins involved in protein secretion apparatus (Category w), and carbon compound metabolism (Category s) were detected (Supplementary Table 3). Expression of many of these proteins are condition-specific, thus low representation of these functional groups is expected as they may not be expressed under the experimental condition used in the study.
Identification of AmpR- and AmpR-β-lactam-dependent proteins
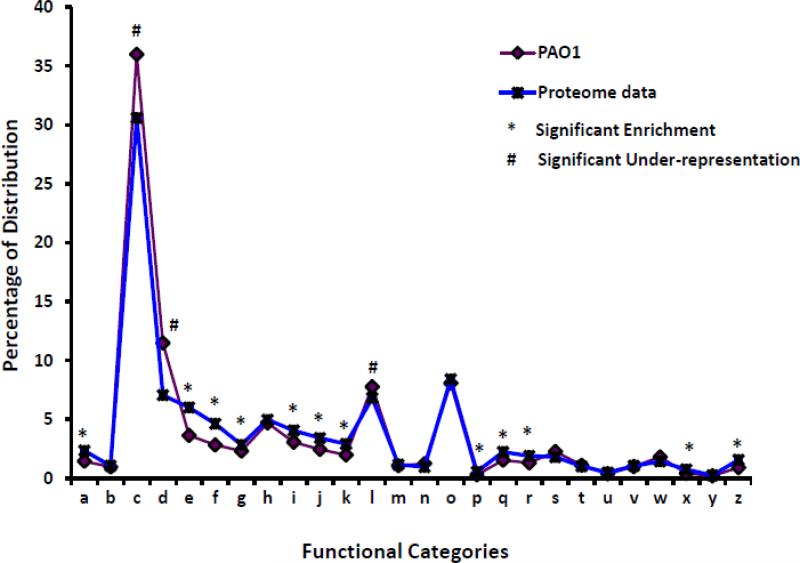
The GeLC-MS data identified numerous proteins expressed under individual conditions. In the absence of β-lactam exposure, the analyses identified 2139 and 2267 proteins in PAO1 and PAOΔampR, respectively. In the presence of β-lactam the PAO1 and PAOΔampR expressed 2052 and 2150 proteins, respectively (Table 1). However, there can be potential overlaps in the proteins that are expressed under different conditions. Hence, these numbers do not reflect the true difference between β-lactam or AmpR-dependent proteins. In order to determine the exclusively AmpR-dependent proteins in the presence and absence of β-lactam antibiotics, a four-way Venn diagram was plotted (Fig. 3). The complete list of identified proteins in each of the Venn groups is given in Supplementary Table 4.
Table 1.
Summary of proteins identified in each sample condition by GeLC-MS analysis.
| Sample | β-lactam | Crux | InsPecT | Combined |
|---|---|---|---|---|
| PAO1 | − | 848 | 2109 | 2139 |
| + | 990 | 2029 | 2052 | |
| PAOAampR | − | 1003 | 2246 | 2267 |
| + | 552 | 2011 | 2150 | |
| Total | 1317 | 2950 | 2965 | |
The number of proteins present in each condition tested identified by either the Crux or InsPecT algorithms is indicated. The number of unique proteins identified with either algorithm is summarized in the combined column.
Fig. 3. Venn diagram of proteins identified in different samples.
Distribution of non-redundant proteins identified in PAO1 and PAOΔampR without (uninduced) and with (induced) sub-MIC β-lactam. Distribution of phosphoproteins identified in each group is shown in the inset (grey squares).
The proteins expressed under all conditions are found in Group O and, most likely, are part of the core proteome (discussed in the following section). The 585 proteins in Groups G, H, K, L, M and N were eliminated from further analysis, as they could not be assigned to any one class unequivocally. Thus, of the remaining 947 proteins, we identified a total of 853 AmpR-dependent proteins (Groups A, E, C and J, Fig. 3) and 94 β-lactam dependent proteins (Groups F and I, Fig. 3).
The 207 proteins found in Groups A and E are positively regulated by AmpR, since they are produced only in the presence of AmpR and not in any other condition. Similarly, AmpR negatively regulates the proteins present in Groups C and J (284 proteins), since they are produced only in the absence of ampR. A similar logic was applied to identify 106 positively regulated and 256 negatively regulated proteins that are AmpR-dependent only under β-lactam stress (Groups B and D, Fig. 3), and 94 proteins that are differentially expressed in response to β-lactam exposure and independent of AmpR (Groups F and I, Fig. 3).
Proteins expressed under all conditions: Core proteome
Of the 2965 proteins identified, 1433 proteins (26% of PAO1 genome) were detected in all four conditions irrespective of the presence of AmpR or antibiotic (Group O; Fig. 3). Many of the previously identified AmpR-regulated ORFs such as AmpC, LasR, RhlR, the MexAB-OprM efflux pump, and some Psl proteins were present in Group O. This is probably because in addition to the housekeeping proteins that are expected to be expressed under all test conditions, Group O is also likely to contain AmpR-regulated proteins that have a basal-level of expression (higher or lower expression in ampR mutants compared to PAO1). The reason could be that our current proteome analysis, unlike the previous transcriptome analyses, is not quantitative and will not detect changes in protein levels between the conditions. Thus, AmpR potentially regulates subsets of the proteins that are detected in Group O. Another possibility is that the basal level expression of the proteins ensures a rapid response to the different stress conditions. As one would expect, many genes of Group O are involved in cellular metabolism and ribosome biosynthesis (Supplementary Table 4). Accordingly, functional categorization and hypergeometric distribution analysis (Supplementary Table 5) revealed a significant enrichment in categories e (amino acid biosynthesis and metabolism), f (translation, post translation and modification), k (adaptation, protection), q (central intermediary metabolism), r (nucleotide biosynthesis and metabolism), x (cell division) and z (transcription, RNA processing and degradation). As expected many of the proteins involved in murein biosynthesis (MurA, MurC, MurD, MurE, MurF, MurG, Mur I and MraW), the peptidoglycan formation (PBP1A, PBP1B, PBP2, PBP3, PBP5, PBP6), the cell wall shape (EnvA/LpxC, EnvB, and LpxD) and others (AmpDh3, Mp1, MltA, AmiB and LdcA) were detected in Group O (Supplementary Table 4).
The characterized proteins in the adaptation and protection category (k), which was significantly enriched, include OstA, Ohr, AmpC, HtpX, SodB, KatA, LasR, RhlR, Lon, PpkA, CheZ, and several chemotactic transducers. In addition, proteins of the major RND efflux pumps MexAB-OprM, MexEF-OprN and its regulator MexT, and TriABC were also detected under all conditions (Supplementary Table 4).
Among the functional categories significantly under-represented in Group O were categories c (hypothetical), d (membrane proteins), l (transcriptional regulators) and m (two-component regulatory systems). A low representation of these in the core set is probably due to their condition specific expression. The outer membrane protein category is under-represented and the proteome analysis detected the following characterized members: OprD, OprH, OprF, OprQ, OprI, Opr86, OprC, OprG, OpdO, OprL, OstA and IcmP. The analysis also detected the sigma factors, RpoS, RpoD, RpoN, AlgU/T and FliA and global regulators, Anr, Vfr, Dnr, Crc, GacA and Hfq (Supplementary Table 4).
A major adaptive phenotype of P. aeruginosa during chronic infection is the production of alginate. This study detected the following alginate-specific regulatory proteins: AlgU, MucA, MucB, MucC, MucD, AlgO/Prc, AlgW, AlgP, AlgR, AlgC, AlgB, and Ndk [46-49]. However, none of the proteins from algD operon involved in alginate biosynthesis [50] were detected in this category (Supplementary Table 4). Previous studies have demonstrated that genes of the AlgT/U regulon are expressed to deal with cell envelope homeostasis [51-53]. However, our proteome data suggests that the regulators of this system may have additional roles, since the proteins are detected in all four conditions.
Analysis of AmpR-dependent proteins (independent of β-lactam)
The functional categorization and enrichment analysis of AmpR-dependent proteins in the absence of β-lactam antibiotics was done as described earlier. In the protein set positively regulated by AmpR (Groups A and E, Fig. 3; Supplementary Table 6), significant enrichment was seen only in Category c (the hypothetical, unclassified, unknown), which accounts for 46% of the proteins in this group. Only Category f proteins (translation, post-translational modification and degradation) were found to be significantly under-represented (Supplementary Table 6).
The proteins that are positively regulated by AmpR (Supplementary Table 7) include major virulence determinants, such as the LasA protease (PA1871), the alkaline protease secretion protein AprD (PA1246), and the phospholipase PlcB (PA0026). The proteins LasA, PlcB and AprD are under QS regulation [54, 55]. The QS process is positively regulated by AmpR [26, 27], supporting the proteome findings. The proteome data also identified modulators of cyclic di-GMP (c-di-GMP) levels such as BifA (PA4367), CdpA (PA4969) TpbB (PA1120) to be under AmpR-regulation (Supplementary Table 7). The role of AmpR in c-di-GMP signaling is discussed in a later section. Two other proteins in the AmpR-dependent group are the sensor kinases of two TCSs, RoxS (PA4494) and RocS2 (PA3044). The RoxSR TCS plays a crucial role in attachment of P. aeruginosa to the epithelial cell surface to initiate the infection process [56]. The RocS2A2 TCS is a major regulator of fimbrial gene expression, affecting the attachment process to host cell surfaces [57]. AmpR-dependent expression of these two proteins suggests a role for AmpR in the establishment of infection and warrants further investigation.
Among the proteins negatively regulated by AmpR (detected in the absence of ampR; Groups C and J, Fig. 3; Supplementary Table 6), proteins in Categories m and n corresponding to two-component regulatory systems and secreted factors, respectively, were significantly enriched. Proteins in Categories e (amino acid biosynthesis and metabolism) and f (translation) were under-represented.
AmpR was previously shown to negatively regulate chronic infection phenotypes, such as biofilm formation [26]. The type VI secretion system (T6SS) is one of the attributes of chronic infection [58]. In accordance with this data, the proteome analysis shows negative regulation of proteins of the T6SS Tse3 (PA3484), TssJ1 (PA0080) and TssG1 (PA0089; Supplementary Table 7). Similarly, AmpR negatively regulates BfiS (PA4197) and BfiR (PA4196), which play critical roles in biofilm formation [59], and PelG (PA3058), a biofilm matrix protein involved in pellicle formation [60]. These findings add further support to our previous data [26, 27].
AmpR regulates some proteins only under β-lactam exposure
The proteins that were expressed only in the presence of AmpR and β-lactam (Group B, Fig 3; Supplementary Table 6) were not enriched for any functional category. However, there was a significant under-representation of proteins involved amino acid biosynthesis and metabolism (Category e). Although not statistically significant (p-value ≤ 0.002), this group had a large number of transcriptional regulators (13%; p-value of 0.007). This group included an ECF sigma factor VreR (PA0676), a positive regulator of P. aeruginosa virulence [61], and KynR (PA2082), an activator of the kynurenine pathway for anthralinate (a PQS precursor) biosynthesis [62]. These findings agree with previous studies demonstrating that AmpR is a positive regulator of many acute virulence factors, including those regulated by the PQS system [26, 27]. AmpR also positively regulates two metabolic regulators - PcaQ (PA0152), a homolog of a phenolic compound catabolism regulator in Agrobacterium tumefaciens [63], and BkdR (PA2246) that is involved in amino acid metabolism in P. putida [64]. BkdR is also regulated by Crc [65]. Since both AmpR and Crc have previously been demonstrated to regulate metabolism [26, 66], there is a potential interplay between these regulators and needs further investigation.
The proteins that are expressed only in the absence of AmpR and β-lactam (Group D, Fig. 3) were significantly enriched in Categories d (membrane proteins), and l (transcriptional regulators), whereas under-represented in Categories e (amino acid biosynthesis and metabolism) and q (central intermediary metabolism; Supplementary Table 6). Proteins whose synthesis was negatively regulated by AmpR in the presence of β-lactam (Supplementary Table 7) encompassed those involved in cofactor biosynthesis (CobC, CobD, CobV) and proteins involved in antibiotic resistance (MexXY, ArnT, Aph, MexD). The cob genes are part of an 11-gene operon that is involved in biosynthesis of the cofactor cobalamine [32]. Cobalamine has been demonstrated to enhance growth under anaerobic conditions and suppress biofilm formation in P. aeruginosa [67]. The MexXY RND efflux system confers resistance to aminoglycosides and macrolides [68, 69]. Aph (PA4119) is an aminoglycoside phosphotransferase [70], which is encoded as part of a two-gene operon along with a transcriptional regulator PA4120 [32]. It is interesting to note that AmpR negatively regulates expression of aminoglycoside resistance proteins in response to β-lactam stress. This is not very surprising since one would expect up-regulation of resistance to β-lactam and not to aminoglycoside in response to β-lactam exposure. However, there was no differential regulation of the mexXY or aph in our transcriptome data [26, 27]. The current proteome analysis suggests potential AmpR-mediated regulation of aminoglycoside resistance, further expanding the AmpR regulon. This was further confirmed by a differential susceptibility profile for aminoglycosides, amikacin and tobramycin, observed between PAO1 and PAOΔampR (data not shown).
Furthermore, upon β-lactam exposure, AmpR negatively regulates additional proteins involved in chronic infection phenotype such as those involved in chaperone-usher pathway, CupB2 (PA4085), CupB3(PA4084), alginate regulation, AlgZ/FimS (PA5262), biofilm formation, MifS (PA5512), PslG (PA2237) and PslJ (PA2240), and the T6SS proteins, TssE1 (PA0087) and Tse2 (PA2702; Supplementary Table 7). The negative regulation of cupB2 by AmpR in the presence of β-lactam was confirmed by qPCR (2 fold, p-value 0.004). The proteome data thus, supports the role of AmpR as a positive and negative regulator of acute and chronic infection phenotypes, respectively.
An interesting observation is downregulation of HacB (PA0305), an acyl homoserine lactone acylase by AmpR. HacB degrades AHL molecules and serves to quench QS signaling [71]. Thus, AmpR not only activates QS genes by controlling expression of major QS regulators [27] but also downregulates QS quenchers.
AmpR regulates c-di-GMP signaling
C-di-GMP-mediated signaling plays a critical role in determining P. aeruginosa pathogenesis [72, 73]. Intracellular c-di-GMP levels are modulated by diguanylate cyclases (DGCs) containing GGDEF domains, and phosphodiesterases (PDEs) containing EAL domains [54] that synthesize and degrade c-di-GMP, respectively. The P. aeruginosa genome encodes 39 proteins that are capable of modulating c-di-GMP levels [32, 74]. The proteome data suggests that two of these PDEs, BifA (PA4367) and CpdA (PA4969) were positively regulated by AmpR. qPCR analyses concurred with the proteome data: BifA (−2.7 fold, p-value 0.0003), CpdA (−2.5 fold, p-value 0.0001). Another PDE, PA4781 was determined to be under negative AmpR-regulation in the proteome data. However, PA4781 expression was positively regulated by AmpR, as seen in qPCR analysis (−2.0 fold, p-value 0.0003). Elevated c-di-GMP levels in the cell positively regulate chronic infection phenotypes, such as biofilm formation [75]. P. aeruginosa AmpR negatively regulates biofilm formation [26] and one possible mode is by regulating PDE gene expression.
AmpR regulon includes transcriptional regulators
P. aeruginosa AmpR has an extensive regulon. We hypothesized that AmpR-mediated gene regulation is mediated by intermediate transcriptional regulators. Accordingly, previous transcriptome analyses identified 22 [26] and 66 [27] transcriptional regulators under AmpR-mediated regulation. The proteome data identified that the presence of 102 putative transcriptional regulators (~24% of transcriptional regulators encoded by PAO1) was found to be AmpR-dependent under different categories (Supplementary Table 6 and 7). However, the specific role for most of these is yet to be elucidated [32, 76]. Eighteen of the AmpR-regulated transcriptional regulators identified by proteome analysis overlapped with either of the transcriptome studies (Table 2). Many of these regulators have no assigned role. The known regulators include PrtN (PA0610) and PrtR (PA0611), which are part of the regions of genome plasticity in P. aeruginosa [8] and regulate pyocin production to confer a survival advantage [77]. ParR, a transcriptional regulator of the ParRS TCS, contributes to adaptive resistance to polymyxin B and colistin by activating the arn operon involved in LPS modification [78]. AmpR-mediated regulation of ParR, identified in the RNA-Seq and proteome studies, expands the role of AmpR in non-β-lactam resistance [26].
Table 2.
AmpR regulated transcriptional regulators overlapping with Array and RNA-Seq analysis.
| PA# | Gene Name | Product Name | Array | RNA-Seq | Proteome |
|---|---|---|---|---|---|
| PA0601 | probable two-component response regulator | − | + | + | |
| PA0610 | prtN | transcriptional regulator PrtN | + | − | + |
| PA0611 | prtR | transcriptional regulator PrtR | + | − | + |
| PA1142 | probable transcriptional regulator | − | + | + | |
| PA1359 | probable transcriptional regulator | − | + | + | |
| PA1799 | parR | two-component response regulator, ParR | − | + | + |
| PA1961 | probable transcriptional regulator | − | + | + | |
| PA2588 | probable transcriptional regulator | + | + | + | |
| PA2718 | probable transcriptional regulator | − | + | + | |
| PA2877 | probable transcriptional regulator | − | + | + | |
| PA3027 | probable transcriptional regulator | − | + | + | |
| PA3034 | probable transcriptional regulator | − | + | + | |
| PA3077 | probable two-component response regulator | − | + | + | |
| PA3630 | probable transcriptional regulator | − | + | + | |
| PA4145 | probable transcriptional regulator | − | + | + | |
| PA4781 | cyclic di-GMP phosphodiesterase | + | − | + | |
| PA4983 | dmsR | probable two-component response regulator | − | + | + |
| PA4987 | probable transcriptional regulator | − | + | + |
Presence and absence of a AmpR dependent transcriptional regulator is detected by (+) and (−), respectively.
One transcriptional regulator that was found to be AmpR- regulated in all three studies is PA2588 (Table 2). PA2588 is flanked by PqsH (PA2587), which is involved in synthesis of the PQS signaling molecule [79], and a two-gene operon (PA2589-PA2590) that is potentially involved in iron uptake [32]. AmpR is known to positively regulate both the iron uptake process PQS-mediated QS regulation, affecting diverse virulence phenotypes [27]. The role of PA2588 in P. aeruginosa is not yet known, but given the genomic location and the potential role of AmpR in its regulation, it warrants further study.
Phosphoproteome analysis
Post translational modifications (PTM) are beginning to be widely accepted in microbial systems not only as means of regulation [80] but also in the control of protein-protein interactions [81]. Recent evidence suggest that apart from the previously known histidine/aspartate phosphorylation, the phosphorylation of serine, threonine and tyrosine residues are common in bacteria and are no longer exclusive to eukaryotes [82, 83]. In P. aeruginosa, Ser/Thr/Tyr phophorylations have been shown to regulate virulence factor production [84, 85]. Analysis of bacterial phosphoproteomes has traditionally been less common than proteome analysis since the identification of PTM tremendously increases the time required for database searching.
We used InsPecT to determine if there are phosphorylation modifications in any of the identified proteins from P. aeruginosa (Table 3). InsPecT identified total 51 proteins with 52 unique phosphorylation modifications (PLS >20, p-value <0.05) (Table 3). The cell division protein ZipA (PA1528) was phosphorylated on serine at two different positions (Table 3). Out of the 52 phosphorylation sites, 24 (46.2%) were on the Ser, 17 (32.7%) were on the Thr and 11 (21.2%) were on Tyr.
Table 3.
List of identified phosphopeptides from PAO1 and PAOAampR samples in the presence and absence of β-lactam antibiotic.
| PA # | Protein description | Phosphopeptide |
|---|---|---|
| PA0389 | hypothetical protein | R.HGWASphosRLWPNLLGEIGIYR.V |
| PA0427 | Major intrinsic multiple antibiotic resistance efflux outer membrane protein OprM precursor | R.AAFFPSISLTANAGTMSphosR.Q |
| PA0437 | cytosine deaminase | K.ALLSphosHEDVKQRAWQTLK.W |
| PA0620 | probable bacteriophage protein | K.GRVTAGMALAATDIPGLDASphosK.L |
| PA0943 | hypothetical protein | K.QMPISphosGNASR.S |
| PA1174 | periplasmic nitrate reductase protein NapA | K.GKTLYphosDVLFRNGQVDR.F |
| PA1206 | hypothetical protein | K.QAYIAMDVETphosIATIR.D |
| PA1528 | cell division protein ZipA | R.DESphosGFKGPALLQNILESGLR.F |
| PA1528 | cell division protein ZipA | K.LKFKLDRSFANLPDDDGDSphosAELLGPAR.V |
| PA1544 | transcriptional regulator Anr | R.FRARGFSAQQFRLAMSphosR.N |
| PA1588 | succinyl-CoA synthetase beta chain | K.ILVESCTDIDKELYphosLGAVVDRSSRR.I |
| PA1589 | succinyl-CoA synthetase alpha chain | R.SLADIGKALAELTphosGWEVK.K |
| PA1805 | peptidyl-prolyl cis-trans isomerase D | K.GEDFAALAKEFSphosQDIGSAATGGDLGYAGR.G |
| PA2015 | putative isovaleryl-CoA dehydrogenase | R.AYLYphosAVAAACDRGETTRK.D |
| PA2229 | conserved hypothetical protein | R.CHPDWSLLRLSphosEVLFDR.R |
| PA2291 | probable glucose-sensitive porin | K.MSphosGSGTKGALLPVELIWQPK.V |
| PA2304 | AmbC | R.NYRAGLGLSphosWREAFQTDSR.A |
| PA2462 | hypothetical protein | K.GQTDETphosVRQSQIVAQGNLAIK.A |
| PA2492 | transcriptional regulator MexT | R.TphosLFDDPLFVRTGR.S |
| PA2735 | probable restriction-modification system protein | K.YphosRDVILPFTVLR.R |
| PA2744 | threonyl-tRNA synthetase | K.KEAADFIKLTLQVYphosR.D |
| PA2950 | hypothetical protein | R.ADYphosKELQPEVQSRVEELWDK.V |
| PA3040 | conserved hypothetical protein | R.GKIHDSLKRARDTphosLR.D |
| PA3168 | DNA gyrase subunit A | K.GQQLISphosMLIPESGAQILTASER.G |
| PA3227 | peptidyl-prolyl cis-trans isomerase A | R.NGFADVPSphosDDVVILSAKR.L |
| PA3313 | hypothetical protein | K.ELKVSphosAIPDEAPTELLR.K |
| PA3552 | ArnB | K.NLTphosCAEGAMFVSDDSALAERVR.R |
| PA3567 | probable oxidoreductase | R.DLLVEVRAISVNPVDTphosK.V |
| PA3659 | probable aminotransferase | R.CQILFLCSphosPGNPTGALVPLETLK.K |
| PA3700 | lysyl-tRNA synthetase | R.YphosPFEVSPLARR.N |
| PA3728 | hypothetical protein | R.HRFSVNTQELDLTphosLMPR.G |
| PA3796 | hypothetical protein | K.LTphosPDGQAPQGDLDIGSLLAR.F |
| PA3817 | probable methyltransferase | R.LYGRSAISphosKLEMNILR.G |
| PA4251 | 50S ribosomal protein L5 | K.ITphosGQKPVVTYARK.S |
| PA4256 | 50S ribosomal protein L16 | R.GSphosKVSFGEYALKATSRGR.L |
| PA4414 | UDP-N-acetylmuramoylalanine--D-glutamate ligase | R.YphosLARRGLPFAVVDTR.E |
| PA4448 | histidinol dehydrogenase | R.SYHEKQKQGSphosWR.Y |
| PA4496 | probable binding protein component of ABC transporter | K.AKIVTYEWGEYphosIKR.A |
| PA4576 | probable ATP-dependent protease | R.AFNQRYDRALDSphosVERR.A |
| PA4700 | penicillin-binding protein 1B | R.SRNSKARPAPGLNKWLSphosWALK.L |
| PA5022 | conserved hypothetical protein | K.MTphosPTLLKNQLTEIPWGSGVR.E |
| PA5044 | type 4 fimbrial biogenesis protein PilM | R.RYphosGLSVEEAGLAKK.Q |
| PA5050 | primosomal protein N' | R.LALPSPLRRLFDYphosRAPR.G |
| PA5171 | arginine deiminase | R.KAGVEVITphosISASELGR.G |
| PA5194 | hypothetical protein | K.ASGWLVQVTphosEPLFR.L |
| PA5232 | conserved hypothetical protein | K.TVETphosANEREKLMFR.V |
| PA5240 | thioredoxin | K.DGNVEATKVGALSphosK.S |
| PA5345 | ATP-dependent DNA helicase RecG | R.RRSphosLLVRLQDGSGTLSLR.F |
| PA5372 | choline dehydrogenase | R.GRPNLTIVTphosHALSDR.I |
| PA5492 | conserved hypothetical protein | R.HPLTphosDFDRLMLDWAQASQLPIHVLMTK.A |
| PA5497 | class II (cobalamin-dependent) ribonucleotide-diphosphate reductase subunit, NrdJa | R.IRGSphosVLHAKYSRYMQR.V |
| PA5556 | ATP synthase alpha chain | R.NEGTphosIVSVSDGIVR.I |
Interestingly the distribution of phosphorylated proteins varied greatly in PAOΔampR samples. While PAO1 samples showed only five (uninduced) and one (induced) phosphorylated peptides, 28 (uninduced) and 29 (induced) phosphopeptides were detected in PAOΔampR samples. Owing to the potential overlap between the peptides detected in the four conditions, a four way Venn-diagram was constructed to identify AmpR-dependent phosphorylations (inset, Fig. 3). Using the same logic as for the AmpR-dependent proteins (Fig. 3), phosphorylation of two proteins were positively regulated (Fig. 3, A and E), and 26 were negatively regulated by AmpR (Fig. 3, C and J). AmpR negatively regulated phosphorylation of another 19 in the presence of β-lactam (Fig. 3, D). The list of AmpR-dependent phosphorylated proteins is given in Supplementary Table 8. Phosphorylations under negative AmpR-regulation included proteins involved in energy utilization such as SucC (PA1588), SucD (PA1589), a transcription regulator Anr (PA1544), outer membrane protein OprM (PA0427) involved in the antibiotic efflux, proteins involved in translation including PpiA (PA3227), RplE (PA4251), and RplP (PA4256). Anr is an important regulator of anaerobic growth and biofilm formation in P. aeruginosa [86, 87]. AmpR-mediated negative phosphorylation in the presence of β-lactam stress included proteins involved in antibiotic resistance such as a transcriptional regulator MexT (PA2492), and proteins involved in cell wall synthesis such as a penicillin binding protein MrcB (PA4700) and MurD (PA4414; Supplementary Table 8). AmpR has been previously demonstrated to negatively regulate expression of mexT using qPCR, and consequently, the functioning of the MexEF-OprN efflux pump [26]. This could potentially be mediated by differential phosphorylation of MexT. However, at this point, it is not clear how phosphorylation affects the activity of these proteins.
Unannotated gene analysis
High-throughput proteomic data have been successfully employed to aid microbial genome annotation [88-91]. Having identified proteins that are expressed under different conditions such as antibiotic stress and the presence or absence of AmpR, we then asked the question if our tandem mass spectra could provide clues to correct potentially misannotated ORFs in the PAO1 genome. The tandem mass spectra were searched against a database containing protein sequences resulting from all possible six-frame translation of all the intergenic regions in the PAO1 genome as described in materials and methods. The hypothesis is that if the so-called intergenic regions contain protein-encoding genes, peptide sequences corresponding to these proteins should be identified in the database search. Crux identified 30 peptides belonging to 13 intergenic regions, while InsPecT identified 218 peptides belonging to 57 intergenic regions.
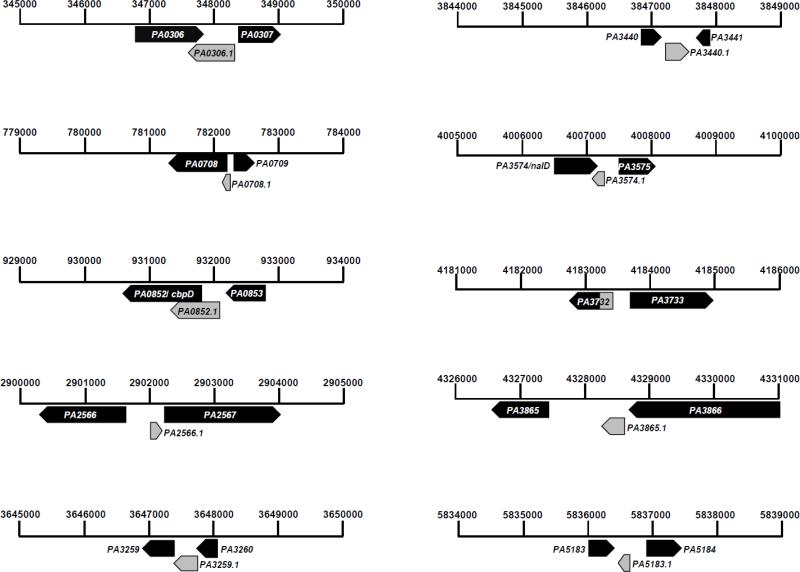
Based on sequence analysis with PAO1 and other sequenced P. aeruginosa strains (PA7, PA14, LESB58), primers were designed to test nine intergenic regions by reverse transcriptase polymerase chain reaction (RT-PCR; Supplementary Table 1). Our results suggest that at least nine novel, unannotated conserved ORFs exist in the PAO1 genomes that are misannotated as intergenic regions (Table 4, Fig. 4). All of previously unannotated novel ORFs were tested by RT-PCR (Supplementary Fig. 1). Additionally, eleven peptides were mapped to intergenic regions that were in frame with the downstream annotated ORF (Table 5), suggesting that the putative start codon was erroneously annotated in the Pseudomonas database. One of these, PA3732, was tested by RT-PCR (Supplementary Fig. 1). The new proposed coordinates for these misannotated ORFs based upon the peptides are listed in Table 5. All the proposed new ORFs in PAO1 genome that were confirmed by RT-PCR are depicted on PAO1 co-ordinates in Fig. 4. The confirmation of new ORFs reflects the limitations of existing software to predict all the ORFs in the genome accurately.
Table 4.
List of Unannotated ORFs in PAO1 genome verified by reverse transcription PCR.
| Proposed ORF | Proposed PAO1 coordinates | Orthologs |
|---|---|---|
| PA0306.1 | 348473 - 347835 | PSPA7 0398, PA14 04010, PALES 31051, PA14 36010, PACG_03535 |
| PA0708.1 | 782205 - 782113 | none |
| PA0852.1 | 932105 - 931395 | unannotated orthologs in PA14, PA7 and LESB58 |
| PA2566.1 | 2902048 - 2902155 | unannotated orthologs in LESB58 and PA14 |
| PA3259.1 | 3647746 - 3647399 | PSPA7 1865, unannotated orthologs in PA14 and LESB58 |
| PA3440.1 | 3847538 - 3847200 | PALES 16201, PA14 19600, PSPA7 1687 |
| PA3574.1 | 4007324 - 4007130 | PA14 18070, PSPA7 1570, unannotated ortholog in LESB58 |
| PA3732.1 | 4183405 - 4182785 | PaerPA01004315, PSPA7 1387, PaerPAb 06389, PA39016 001680026 |
| PA3865.1 | 4327697 - 4327362 | PA14 13950 |
| PA5183.1 | 5836685 - 5836470 | PALES 55771, PA14 68470, PSPA7 5926 |
Fig. 4.
Proposed new ORFs shown on the PAO1 genome co-ordinates.
Table 5.
List of Misannotated ORFs in PAO1 genome.
| Proposed ORF | Proposed PAO1 coordinates | Orthologs |
|---|---|---|
| PA0369 | 413840 - 413364 | PA14, C3719, PACS2, 39016, LESB58, 2192, PA7 |
| PA0459 | 517837 - 520635 | PA14, 39016, 2192, PACS2, LESB58, C3719, PA7 |
| PA0799 | 878818 - 876617 | 39016, PA14, LESB58, C3719, PACS2, 2192, PA7 |
| PA1926 | 2104177-2106447 | 2192, LESB58, PACS2, 39016, PA14, C3719 |
| PA2731 | 3090231 - 3089644 | |
| PA3248 | 3635589 - 3634924 | C3719, PACS2, LESB58, 2192, 39016, PA14, PA7 |
| PA3732 | 4183405 - 4182785 | LESB58, PACS2, C3719, 39016, 2192, PA14, PA7 |
| PA3861 | 4321104 - 4322627 | C3719, 39016, LESB58, PA14, 2192, PACS2, PA7 |
| PA4985 | 5601131 - 5599884 | PACS2, C3719, 39016, PA14, LESB58, 2192, PA7 |
| PA5117 | 5764611 - 5762659 | LESB58,PACS2, C3719, 39016, 2192, PA14 |
| PA5322 | 5991168 - 5993774 | PACS2, C3719, LESB58, 2192, PA14, PA7 |
Comparison of AmpR transcriptome and proteome
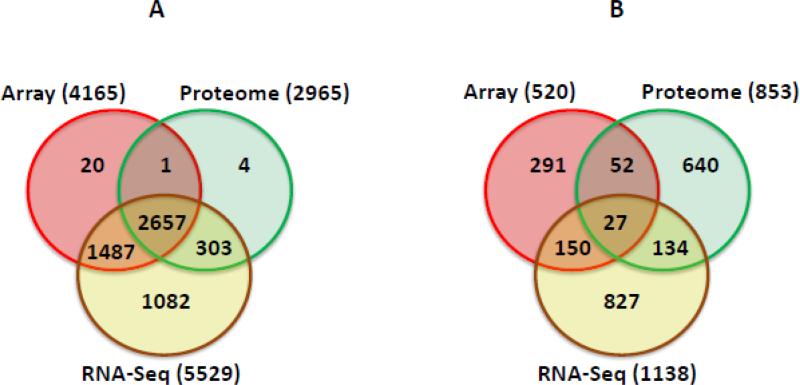
We had previously analyzed the transcriptome of AmpR using both DNA microarrays [26] and RNA deep sequencing [27]. Comparing the normalized data of all four conditions from the two-transcriptome studies with the proteome data revealed an extensive overlap (Fig 5A). As expected, the transcriptome data had much more reads that were not identified in the proteome studies, and this can be attributed to short half-lives of the mRNA, tRNA reads and various kinds of post-transcriptional regulation. Moreover, almost all the proteins detected in the proteome analysis were detected in one of the two-transcriptome studies (Fig 5A). Various technical (sample preparation, instrumentation etc.), functional (posttranscriptional or post-translational modifications) or analysis software variations can account for the differences observed in the three approaches. The findings also highlight the fact that in order to obtain a comprehensive picture, studying gene expression on a global scale should involve a combination of approaches.
Fig. 5. Comparative analyses of AmpR microarray, RNA-Seq and proteome datasets.
Venn diagram was constructed using the datasets from the microarray, RNA-Seq and Proteome analysis (A) Comparison of total identified datasets (B) Comparison of the AmpR-dependent datasets.
The transcriptome data and complementary assays established the global regulatory nature of AmpR [26], which also includes regulatory RNAs [27]. Comparison of the transcriptome (Array and RNA-Seq) and proteome datasets revealed that 27 ORFs are AmpR-regulated in all the three experiments (Fig. 5B; Supplementary Table 9). As expected, many of the genes/proteins that were detected in all three assays contribute to P. aeruginosa virulence. These include the Psl exopolysaccharide biosynthetic proteins, PslF, PslG and PslH (PA2236-PA2238) that play a role in biofilm formation [60], the PQS response protein PqsE [92], a component of the MexGHI-OpmD efflux pump MexI [93], and the phenazine-modifying enzyme PhzS ([94]; Supplementary Table 9) The findings support previous phenotypic assays and qPCR studies that determined a role for AmpR in regulating biofilm formation, QS and antibiotic resistance [26, 27].
Even though we found 2121 non-redundant AmpR dependent genes combining all the three analysis, 363 genes overlapped between any two analyses (Fig. 5B). Thus, conservatively speaking, actual AmpR regulon can be anywhere between >363 < 2121 genes. The 363 AmpR dependent genes identified in any two analyses included a long list of β-lactam and non-β-lactam resistance genes, QS-regulated genes, genes of the phenazines biosynthetic operon, the hydrogen cyanide biosynthetic genes, T6SS and those involved in biofilm and alginate production, etc. (Supplementary Table 9). These sufficiently paint the global picture of AmpR regulation in P. aeruginosa.
Further, there were many AmpR regulated proteins that were unique to in any one of the analysis (Fig. 5; Supplementary Table 9). Proteome, array and RNA-Seq identified 640, 291, 827 AmpR dependent genes, respectively, that did overlap with any other list. The RNA-Seq gene set also included the AmpR regulated small RNAs [27].
The AmpR-dependent proteins detected uniquely in the proteome analysis further identified many other virulence genes such as the alkaline protease AprD (PA1246), the BfiSR (PA4196-4197) TCS and proteins of the chaperone-usher pathway (CupB2, B3 and C3) (Supplementary Table 9). In addition, regulation of few of the c-di-GMP PDEs by AmpR was identified by proteomic data. While one (PA4781) overlapped with array, two unique PDEs CpdA (PA4969) and BifA (PA4367) were identified by AmpR proteome analysis (Supplementary Table 9). The role of AmpR in modulating c-di-GMP levels needs further investigation. Thus, both the previous transcriptome and the current proteome data not only attest to its role as global regulator of virulence in P. aeruginosa, but also revealed new modes of gene regulation by AmpR.
CONCLUSION
Our study is the first report to look at the proteomic response of ampR mutant of P. aeruginosa in the presence and absence of β-lactam antibiotic. The data presented here not only supports the previous transcriptomic studies [26, 27], but also strengthens the master regulatory role of P. aeruginosa AmpR in regulating antibiotic resistance, virulence factors as well as protein phosphorylation. In addition, we also describe the Ser/Thr/Tyr phosphorylations in PAO1 and PAOΔampR in the presence and absence of β-lactam antibiotics that were not previously reported. The huge difference between the phosphoproteomes of PAO1 and PAOΔampR under the same conditions is interesting and needs to be explored further. Finally, we identified and confirmed several unannotated and misannotated ORFs in the P. aeruginosa genome. Since the function of over ~36% of the ORFs in the PAO1 genome is yet to be determined, it will take a considerable amount of research to decipher the specific role of these newly identified proteins. Thus, the information provided in this analysis opens up new areas of research in understanding complex strategies that contribute to the success of P. aeruginosa as a pathogen. Global approaches like the current study will not only aid in connecting previously isolated areas of research but also in finding therapeutic strategies to combat this formidable pathogen.
Supplementary Material
SIGNIFICANCE.
The AmpR proteome data not only confirmed the role of AmpR in virulence and resistance to multiple antibiotics, but expanded the perimeter of AmpR regulon. The data presented here points to the role of AmpR in regulating cyclic di-GMP levels and phosphorylation of Ser, Thr and Tyr, adding another dimension to the regulatory functions of AmpR. We also identify some previously unannotated/misannotated ORFs in the P. aeruginosa genome, indicating the limitations of existing ORF analyses software. This study will contribute towards understanding complex genetic organization of P. aeruginosa. Whole genome proteomic picture of regulators at higher nodal positions in the regulatory network will not only help us link various virulence phenotypes but also design novel therapeutic strategies.
HIGHLIGHTS OF THE STUDY.
■ Proteome data complements previous transcriptome studies with PAOΔampR.
■ Previously unidentified AmpR-regulated virulence mechanisms expand AmpR regulon.
■ Proteome analysis detected 53% of total proteins, 15% of which were AmpR-regulated.
■ AmpR regulates phosphodiesterases, and Ser, Thr, Tyr protein phosphorylation.
■ Proteome data identified novel and previously misannotated open reading frames.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health-Minority Biomedical Research Support SCORE grants (grant numbers S06 GM08205, 5SC1AI081376 to K.M.) and Florida International University (FIU) Research Assistantship (Herbert Wertheim College of Medicine, to D.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–51. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 2.Alp E, Coruh A, Gunay GK, Yontar Y, Doganay M. Risk factors for nosocomial infection and mortality in burn patients: 10 years of experience at a university hospital. J Burn Care Res. 2012;33:379–85. doi: 10.1097/BCR.0b013e318234966c. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita K, Ohara M, Kojima T, Nishimura R, Ogawa T, Hino T, Okada M, Toratani S, Kamata N, Sugai M, Sugiyama M. Prevalence of drug-resistant opportunistic microorganisms in oral cavity after treatment for oral cancer. J Oral Sci. 2013;55:145–55. doi: 10.2334/josnusd.55.145. [DOI] [PubMed] [Google Scholar]
- 4.Gomes MZ, Machado CR, da Conceicao Mde S, Ortega JA, Neves SM, Lourenco MC, Asensi MD. Outbreaks, persistence, and high mortality rates of multiresistant Pseudomonas aeruginosa infections in a hospital with AIDS-predominant admissions. Braz J Infect Dis. 2011;15:312–22. doi: 10.1016/s1413-8670(11)70198-2. [DOI] [PubMed] [Google Scholar]
- 5.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–64. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 6.Georgiades K. Genomics of epidemic pathogens. Clin Microbiol Infect. 2012;18:213–7. doi: 10.1111/j.1469-0691.2012.03781.x. [DOI] [PubMed] [Google Scholar]
- 7.Kelkar YD, Ochman H. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics. 2013;193:303–7. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:3100–5. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73:338–44. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg JB. Why is Pseudomonas aeruginosa a pathogen? F1000 Biol Rep. 2010:2. doi: 10.3410/B2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock RE, Martinez JL. The intrinsic resistome of Pseudomonas aeruginosa to beta-lactams. Virulence. 2011;2:144–6. doi: 10.4161/viru.2.2.15014. [DOI] [PubMed] [Google Scholar]
- 12.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zavascki AP, Carvalhaes CG, Picao RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 15.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol. 2009;12:512–9. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–82. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong KF, Jayawardena SR, Del Puerto A, Wiehlmann L, Laabs U, Tummler B, Mathee K. Characterization of poxB, a chromosomal-encoded Pseudomonas aeruginosa oxacillinase. Gene. 2005;358:82–92. doi: 10.1016/j.gene.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 18.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–23. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 19.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Ann Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc Natl Acad Sci U S A. 1985;82:4620–4. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg F, Normark S. Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J Bacteriol. 1987;169:758–63. doi: 10.1128/jb.169.2.758-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindberg F, Lindquist S, Normark S. Genetic basis of induction and overproduction of chromosomal class I beta-lactamase in nonfastidious Gram-negative bacilli. Rev Infect Dis. 1988;10:782–5. doi: 10.1093/clinids/10.4.782. [DOI] [PubMed] [Google Scholar]
- 23.Lodge J, Busby S, Piddock L. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC beta-lactamase promoter. FEMS Microbiol Lett. 1993;111:315–20. doi: 10.1111/j.1574-6968.1993.tb06404.x. [DOI] [PubMed] [Google Scholar]
- 24.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother. 2005;49:4567–75. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian D, Kong KF, Jayawardena SR, Leal SM, Sautter RT, Mathee K. Co-regulation of beta-lactam resistance, alginate production and quorum sensing in Pseudomonas aeruginosa. J Med Microbiol. 2011;60:147–56. doi: 10.1099/jmm.0.021600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. The regulatory repertoire of Pseudomonas aeruginosa AmpC beta-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian D, Kumari H, Jaric M, Fernandez M, Turner K, Dove S, Narasimhan G, Lory S, Mathee K. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat-shock and the oxidative stress response. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt942. 10.1093/nar/gkt942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park CY, Klammer AA, Kall L, MacCoss MJ, Noble WS. Rapid and accurate peptide identification from tandem mass spectra. J Proteome Res. 2008;7:3022–7. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner S, Shu HJ, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: Identification of posttransiationally modified peptides from tandem mass spectra. Analytical Chemistry. 2005;77:4626–39. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 30.Kudva IT, Krastins B, Sheng H, Griffin RW, Sarracino DA, Tarr PI, Hovde CJ, Calderwood SB, John M. Proteomics-based expression library screening (PELS): a novel method for rapidly defining microbial immunoproteomes. Mol Cell Proteomics. 2006;5:1514–9. doi: 10.1074/mcp.T600013-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 32.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noble WS. A quick guide to organizing computational biology projects. PLoS Comput Biol. 2009;5:e1000424. doi: 10.1371/journal.pcbi.1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–39. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 35.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–5. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 36.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino acid sequences in apProtein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Blonder J, Goshe MB, Xiao W, Camp DG, 2nd, Wingerd M, Davis RW, Smith RD. Global analysis of the membrane subproteome of Pseudomonas aeruginosa using liquid chromatography-tandem mass spectrometry. J Proteome Res. 2004;3:434–44. doi: 10.1021/pr034074w. [DOI] [PubMed] [Google Scholar]
- 41.Hare NJ, Solis N, Harmer C, Marzook NB, Rose B, Harbour C, Crossett B, Manos J, Cordwell SJ. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol. 2012;12:16. doi: 10.1186/1471-2180-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hare NJ, Soe CZ, Rose B, Harbour C, Codd R, Manos J, Cordwell SJ. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J Proteome Res. 2012;11:776–95. doi: 10.1021/pr200659h. [DOI] [PubMed] [Google Scholar]
- 43.Damron FH, Davis MR, Jr., Withers TR, Ernst RK, Goldberg JB, Yu G, Yu HD. Vanadate and triclosan synergistically induce alginate production by Pseudomonas aeruginosa strain PAO1. Mol Microbiol. 2011;81:554–70. doi: 10.1111/j.1365-2958.2011.07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damron FH, Owings JP, Okkotsu Y, Varga JJ, Schurr JR, Goldberg JB, Schurr MJ, Yu HD. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J Bacteriol. 2012;194:1317–30. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–57. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 46.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene. 2012;498:242–53. doi: 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–86. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 49.Ohman DE, Mathee K, McPherson CJ, DeVries CA, Ma S, Wozniak DJ, Franklin M. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular Biology of Pseudomonads. American Society for Microbiology; Washington D.C.: 2002. pp. 472–83. [Google Scholar]
- 50.Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood LF, Ohman DE. Identification of genes in the sigma 22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. MBio. 2012:3. doi: 10.1128/mBio.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–26. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 53.Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 54.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hurley BP, Goodman AL, Mumy KL, Murphy P, Lory S, McCormick BA. The two-component sensor response regulator RoxS/RoxR plays a role in Pseudomonas aeruginosa interactions with airway epithelial cells. Microbes Infect. 2010;12:190–8. doi: 10.1016/j.micinf.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol Microbiol. 2011;79:1353–66. doi: 10.1111/j.1365-2958.2010.07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286:12317–27. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiol. 2005;151:985–97. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 61.Llamas MA, van der Sar A, Chu BC, Sparrius M, Vogel HJ, Bitter W. A Novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 2009;5:e1000572. doi: 10.1371/journal.ppat.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knoten CA, Hudson LL, Coleman JP, Farrow JM, 3rd, Pesci EC. KynR, a Lrp/AsnC-type transcriptional regulator, directly controls the kynurenine pathway in Pseudomonas aeruginosa. J Bacteriol. 2011;193:6567–75. doi: 10.1128/JB.05803-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parke D. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J Bacteriol. 1996;178:266–72. doi: 10.1128/jb.178.1.266-272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madhusudhan KT, Lorenz D, Sokatch JR. The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli. J Bacteriol. 1993;175:3934–40. doi: 10.1128/jb.175.13.3934-3940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hester KL, Lehman J, Najar F, Song L, Roe BA, MacGregor CH, Hager PW, Phibbs PV, Jr., Sokatch JR. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J Bacteriol. 2000;182:1144–9. doi: 10.1128/jb.182.4.1144-1149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonnleitner E, Valentini M, Wenner N, Haichar FZ, Haas D, Lapouge K. Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One. 2012;7:e44637. doi: 10.1371/journal.pone.0044637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee KM, Go J, Yoon MY, Park Y, Kim SC, Yong DE, Yoon SS. Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect Immun. 2012;80:1639–49. doi: 10.1128/IAI.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morita Y, Tomida J, Kawamura Y. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol. 2012;3:408. doi: 10.3389/fmicb.2012.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–70. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hachler H, Santanam P, Kayser FH. Sequence and characterization of a novel chromosomal aminoglycoside phosphotransferase gene, aph (3')-IIb, in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:1254–6. doi: 10.1128/aac.40.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahjudi M, Papaioannou E, Hendrawati O, van Assen AH, van Merkerk R, Cool RH, Poelarends GJ, Quax WJ. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology. 2011;157:2042–55. doi: 10.1099/mic.0.043935-0. [DOI] [PubMed] [Google Scholar]
- 72.An S, Wu J, Zhang LH. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol. 2010;76:8160–73. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frangipani E, Visaggio D, Heeb S, Kaever V, Camara M, Visca P, Imperi F. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ Microbiol. 2013 doi: 10.1111/1462-2920.12164. doi: 10.1111/462-2920.12164. [DOI] [PubMed] [Google Scholar]
- 74.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–44. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balasubramanian D, S.K M, E S-H, L S, X Y, G T, G N, K M . Transcriptional regulatory network in Pseudomonas aeruginosa. In: M.M. B, editor. Bacterial gene regulation and transcriptional networks. Caiser Academic Press; United Kingdom: 2013. [Google Scholar]
- 77.Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 78.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother. 2010;54:3372–82. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–80. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–94. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–82. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 82.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 83.Ravichandran A, Sugiyama N, Tomita M, Swarup S, Ishihama Y. Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics. 2009;9:2764–75. doi: 10.1002/pmic.200800655. [DOI] [PubMed] [Google Scholar]
- 84.Kelly-Wintenberg K, South SL, Montie TC. Tyrosine phosphate in a- and b-type flagellins of Pseudomonas aeruginosa. J Bacteriol. 1993;175:2458–61. doi: 10.1128/jb.175.8.2458-2461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 86.Zimmermann A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–90. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 87.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol. 2010;12:1719–33. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 88.Payne SH, Huang ST, Pieper R. A proteogenomic update to Yersinia: enhancing genome annotation. BMC Genomics. 2010;11:460. doi: 10.1186/1471-2164-11-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ansong C, Tolic N, Purvine SO, Porwollik S, Jones M, Yoon H, Payne SH, Martin JL, Burnet MC, Monroe ME, Venepally P, Smith RD, Peterson SN, Heffron F, McClelland M, Adkins JN. Experimental annotation of post-translational features and translated coding regions in the pathogen Salmonella Typhimurium. BMC Genomics. 2011;12:433. doi: 10.1186/1471-2164-12-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim W, Silby MW, Purvine SO, Nicoll JS, Hixson KK, Monroe M, Nicora CD, Lipton MS, Levy SB. Proteomic detection of non-annotated protein-coding genes in Pseudomonas fluorescens Pf0-1. PLoS ONE. 2009;4:e8455. doi: 10.1371/journal.pone.0008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okamoto A, Yamada K. Proteome driven re-evaluation and functional annotation of the Streptococcus pyogenes SF370 genome. BMC Microbiol. 2011;11:249. doi: 10.1186/1471-2180-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farrow JM, 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–51. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aendekerk S, Diggle SP, Song Z, Hoiby N, Cornelis P, Williams P, Camara M. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151:1113–25. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 94.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–65. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.