Abstract
Infection by the opportunistic pathogen Pseudomonas aeruginosa is a leading cause of morbidity and mortality seen in cystic fibrosis (CF) patients. This is mainly due to the genotypic and phenotypic changes of the bacteria that cause conversion from a typical nonmucoid to a mucoid form in the CF lung. Mucoid conversion is indicative of overproduction of a capsule-like polysaccharide called alginate. The alginate-overproducing (Alg+) mucoid phenotype seen in the CF isolates is extremely unstable. Low oxygen tension growth of mucoid variants readily selects for nonmucoid variants. The switching off mechanism has been mapped to the algT/U locus, and the molecular basis for this conversion was partially attributed to mutations in the algT/U gene itself. To further characterize molecular changes resulting in the unstable phenotype, an isogenic PAO1 derivative that is constitutively Alg+ due to the replacement of the mucA with mucA22 (PDO300) was used. The mucA22 allele is common in mucoid CF isolates. Thirty-four spontaneous nonmucoid variants, or sap (suppressor of alginate production) mutants, of PDO300 were isolated under low oxygen tension. About forty percent of the sap mutants were rescued by a plasmid carrying algT/U (Group A). The remaining sap mutants were not (Group B). The members of Group B fall into two subsets: one similar to PAO1, and another comparable to PDO300. Sequence analysis of the algT/U and mucA genes in Group A shows that mucA22 is intact, whereas algT/U contains mutations. Genetic complementation and sequencing of one Group B sap mutant, sap22, revealed that the nonmucoid phenotype was due to the presence of a mutation in PA3257. PA3257 encodes a putative periplasmic protease. Mutation of PA3257 resulted in decreased algT/U expression. Thus, inhibition of algT/U is a primary mechanism for alginate synthesis suppression.
Keywords: exopolysaccharide, alginate, cystic fibrosis, PalgT/algU, biosynthetic genes
1. Introduction
Infection by opportunistic pathogens such as Staphylococcus aureus, Haemophilus influenzae, Burkholderia cepacia and Pseudomonas aeruginosa is a leading cause of morbidity and mortality seen in patients with cystic fibrosis (CF), an autosomal recessive genetic disorder. Despite an improved understanding of the basic genetic defect responsible for CF, P. aeruginosa continues to be the number one killer in these patients (Govan and Harris, 1986; Pedersen, 1992). This is mainly due to the ability of the bacteria to undergo genotypic and phenotypic changes from the typical nonmucoid (Alg−) form to a mucoid (Alg+) phenotype. This mucoid conversion, readily observed in the P. aeruginosa colony morphology, is indicative of the overproduction of a capsule-like polysaccharide called alginate (Evans and Linker, 1973). Alginate consists of repeating units of mannuronic and guluronic acid, which may be O-acetylated (reviewed in (Remminghorst and Rehm, 2006)). The potential roles of alginate in pathogenesis include a mechanism for bacterial adherence, a barrier to phagocytosis and a mechanism to neutralize oxygen radicals (for review, see (Govan and Deretic, 1996)). Alginate also affects leukocyte functions, such as the oxidative burst and interference with opsonization, and plays an immunomodulatory role via induction of proinflammatory cytokines and suppression of lymphocyte transformation (Bayer et al., 1991; Pedersen, 1992; Song et al., 2003).
The alginate biosynthetic genes are located in a large operon at 3.96 Megabase pairs (Mbp) of the chromosome (reviewed in (Mathee et al., 2002; Remminghorst and Rehm, 2006)). Overexpression of the first gene in this operon, algD, which encodes guanosine diphosphate (GDP)-mannose dehydrogenase, is believed to commit the metabolic sugar intermediates to alginate production. The algD operon is transcriptionally controlled by genes from several loci (Fig. 1). A number of these genes are located around 5.9 Mbp, and encode two response regulators, AlgR (also called AlgR1) and AlgB (Mathee et al., 2002; Remminghorst and Rehm, 2006). Additionally, an operon at 0.83 Mbp of the chromosome, consisting of the algT/U-mucA-mucB-mucC-mucD genes, plays a pivotal role in converting a normally nonmucoid cell to mucoid form (Fyfe and Govan, 1980; Ohman and Chakrabarty, 1981; Martin et al., 1993a; Martin et al., 1993b). The first gene, algT (also known as algU; henceforth referred to as algT/U), encodes a sigma factor essential for activation of alginate genes, including its own (Deretic et al., 1994; DeVries and Ohman, 1994). AlgT/U regulates the algB (Goldberg and Dahnke, 1992; Wozniak and Ohman, 1993), algR (Wozniak and Ohman, 1994), algD (Wozniak and Ohman, 1994), and amrZ (Wozniak et al., 2003) operons. Together, these data suggest that these alginate genes form a cascade with the algT/U gene at the top. The 22-kDa AlgT/U sigma factor (σ22) has similarity to alternative bacterial sigma factors with high homology to SigE (σE) from Escherichia coli and Salmonella typhimurium (DeVries and Ohman, 1994; Martin et al., 1994). σE is required for transcription of a stress regulon that responds to an extracytoplasmic signal (Hiratsu et al., 1995; Raina et al., 1995; Rouviere et al., 1995).
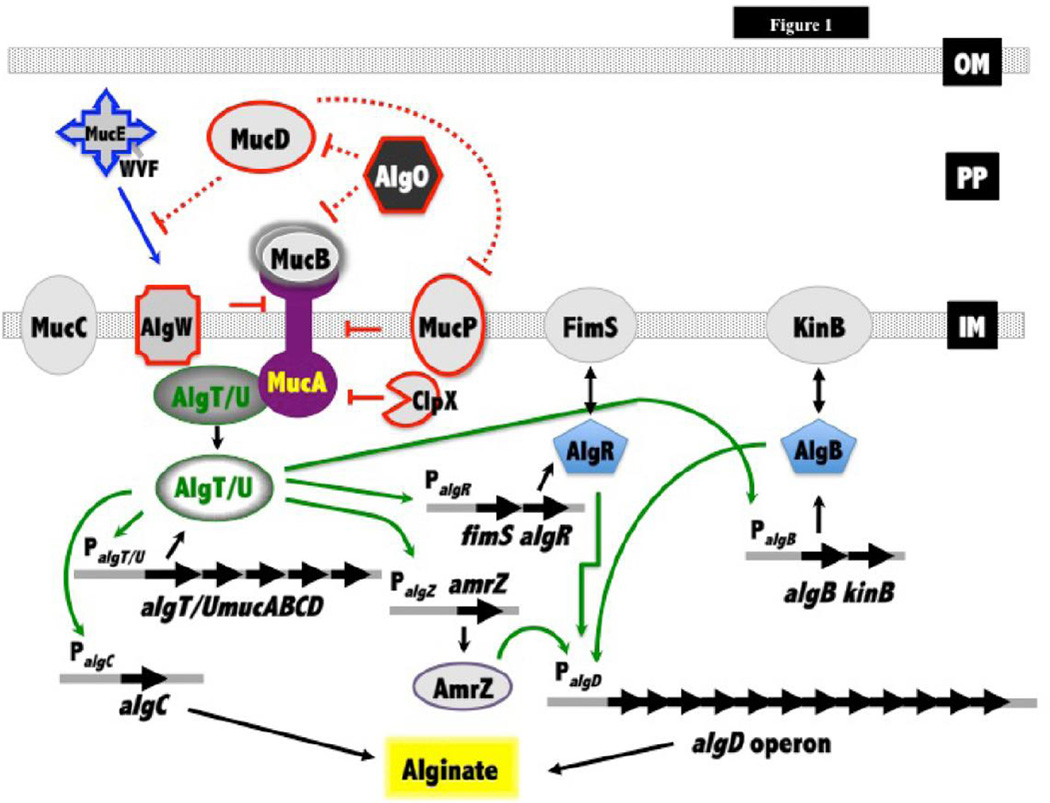
Fig. 1.
Summary of alginate production regulation. Alginate production is controlled by the alginate biosynthetic operon (algD operon). Expression of algD is regulated by the response regulators AlgR and AlgB, the ribbon-helix-helix regulator AmrZ, and the ECF sigma factor AlgU. The sensor kinase partners of AlgR and AlgB are FimS and KinB, respectively. AlgT/U activity is inhibited by MucB, MucD and the anti-sigma factor MucA. MucA activity is regulated by the AlgW, MucP and MucD proteases. The WVF C-terminal amino acid triad of MucE has been shown to prime the AlgW protease to cleave MucA. In a mucA strain such as PDO300 (PAOmucA22), AlgT/U inhibition by MucA, MucB, and MucD is removed. MucC has been shown to both positively and negatively regulate alginate production. Free AlgT/U activates all the genes in the alg regulon: fimS-algR, algB-kinB, amrZ/algZ and algD operons, as well as itself. This study demonstrated that in a mucA background, loss of the periplasmic protease AlgO results in loss of alginate production (see text for details).
MucA is the AlgT/U anti-sigma factor and thus functions as a negative regulator of alginate production in clinical settings (Mathee et al., 2002). Inactivation of the mucA gene often results in the conversion of the nonmucoid strain to its mucoid form (Martin et al., 1993b). Direct interaction between σ22 and MucA has been previously demonstrated (Schurr et al., 1996; Xie et al., 1996).
Downstream of the algT/U and mucA genes is the mucB (also known as algN) gene, which encodes a periplasmic protein (Schurr et al., 1996; Mathee et al., 1997) that interacts with MucA and negatively regulates alginate production (reviewed in (Mathee et al., 2002)). Alginate production appears to be regulated by sequential proteolysis of the transmembrane protein MucA similar to the E.coli σE regulatory pathway (Qiu et al., 2007). This begins with cleavage of MucA by the periplasmic AlgW (Qiu et al., 2007), followed by secondary cleavage at the inner membrane by MucP (Qiu et al., 2007) and subsequent degradation of the remaining peptide by ClpXP (Qiu et al., 2008). Binding of MucB to MucA inhibits AlgW from cleaving MucA (Cezairliyan and Sauer, 2009). AlgW is activated by an amino acid triad signal in MucE, a DegS homologue (Qiu et al., 2007). MucE activation of AlgW is suppressed by the HtrA/DegP homolog, MucD (previously known as AlgY) (Qiu et al., 2007). Inactivation of mucD results in an Alg+ phenotype (Boucher et al., 1996; Ohman et al., 1996). The mucC (also known as algM) gene product seems to have a controversial modulatory role (Ohman et al., 1996; Boucher et al., 1997).
In the early stages of infection, aggressive antibiotic therapy is able to eradicate initial and intermittent colonization of the CF lungs by P. aeruginosa (Frederiksen et al., 1997). However, when the colony morphology of bacteria isolated from sputum samples is of the Alg+ form, the organisms can no longer be eliminated from the lungs despite aggressive antibiotic therapy (Frederiksen et al., 1997). The selection pressure for mucoid conversion common to P. aeruginosa strains that thrive in the complex CF respiratory environment is not well understood. However, using an in vitro system, Mathee et al. (1999) established that repeated exposure of a P. aeruginosa biofilm to activated polymorphonuclear leukocytes (PMNs), or low levels of hydrogen peroxide, can give rise to mucoid variants with defective mucA (Mathee et al., 1999). Yet, the Alg+ phenotype seen in the CF isolates is extremely unstable (Ohman and Chakrabarty, 1981; MacGeorge et al., 1986; Schurr et al., 1994). Low oxygen tension growth of mucoid variants readily selects for nonmucoid variants (Ohman and Chakrabarty, 1981). The locus responsible for the switching off mechanism in the CF isolate FRD had been previously mapped to the algT/U locus (Flynn and Ohman, 1988a). Subsequently, the molecular basis for this conversion was partially attributed to mutations in the algT/U gene itself (Deretic et al., 1994; DeVries and Ohman, 1994). Expression of algT/U has been correlated with down regulation of flagella synthesis (Garrett et al., 1999). In addition, algT/U expression is repressed by AmpR, a LysR-type transcriptional regulator involved in β-lactam resistance and virulence factor expression (Balasubramanian et al., 2011). This further suggests a high level of coordinated regulation with respect to virulence, with AlgT/U serving a major central role.
This study was undertaken to elucidate other mechanisms involved in the conversion from the clinical mucoid to the nonmucoid form. This was done using a genetically well-defined mucoid strain PDO300. PDO300 is a derivative of the prototypic nonmucoid PAO1 that contains a mucA22 mutation (Mathee et al., 1999). Thirty-four spontaneous nonmucoid variants termed sap mutants for suppressor of alginate production were isolated from the mucoid PDO300. Of these, 14 sap strains were complemented by the algT/U gene (Group A mutants). Sequence analysis of the algT/U and mucA genes of the Group A sap mutants revealed that the mucA22 allele is intact, whereas the algT/U gene is altered. The remaining mutants (Group B) were subdivided into two subgroups based upon expression of a 62-kDa outer membrane protein observed in PDO300, but not in PAO1. The mutation in one of the Group B strains was mapped to the E. coli prc homolog, PA3257, called algO.
2. Materials and Methods
2.1. Bacterial strains, plasmids, media and primers
The P. aeruginosa and E. coli strains and plasmids used are listed in Table 1. E. coli was grown on Luria-Bertani (LB) medium supplemented with tetracycline (Tc, 10 µg ml−1), chloramphenicol (Cm, 30 µg ml−1), ampicillin (Ap, 50 µg ml−1) and kanamycin (Km, 25 µg ml−1) when required. P. aeruginosa was grown on LB or on LB/PIA agar, which is a 1:1 mixture of LB agar and Pseudomonas isolation agar (PIA) (Mathee et al., 1997). Antibiotic supplements for P. aeruginosa were 100 µg ml−1 tetracycline and 300 µg ml−1 carbenicillin (Cb). Cultures were all grown at 37°C unless stated otherwise. All primers used were synthesized by Integrated DNA Technologies (Coralville, IA).
Table 1.
Bacterial strains, plasmids, cosmids and primers used in this study.
| Strains | Relevant Genotype and/or Phenotype | Reference |
|---|---|---|
| E. coli | ||
| DH5α | Φ80 lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF) U169 |
Invitrogen (Bethesda Research Laboratories) |
| HB101 |
proA2 leuB6 thi-1 lacY1 hsdR hsdM recA13 supE44 rpsL20 |
(Boyer and Roulland-Dussoix, 1969; Bolivar and Backman, 1979) |
| MTP194 | Tcr, PAO1 fragment 3628056–3652056 (pMO011713) |
(Huang et al., 2000) |
| MTP227 | Tcr, PAO1 fragment 4284570–4307322: pepA | (Huang et al., 2000) |
| MTP264 | Tcr, PAO1 fragment 4965548–4993321: algW | (Huang et al., 2000) |
| MTP315 | Tcr, PAO1 fragment 5904709–5927122: algP algQ algR fimS |
(Huang et al., 2000) |
| MTP264 | Tcr, PAO1 fragment 4965548–4993321: algW | (Huang et al., 2000) |
| MTP46 | Tcr, PAO1 fragment 832401–853309: mucD | (Huang et al., 2000) |
| MTP227 | Tcr, PAO1 fragment 4284570–4307322: pepA | (Huang et al., 2000) |
| MTP280 | Tcr, PAO1 fragment 5290517–5306250: dksA | (Huang et al., 2000) |
| MTP236 | Tcr, PAO1 fragment 4462016–4484172: rlpA | (Huang et al., 2000) |
| P. aeruginosaa | ||
| FRD406 | algT::Tn501-250 (Alg+) | (Flynn and Ohman, unpublished data) |
| PAO1 | Prototypic strain, nonmucoid | (Holloway and Matsumoto, 1984) |
| PDO300 | PAOmucA22, constitutively mucoid | (Mathee et al., 1999) |
| PKM800 | PAOmucA22 algO96 (sap22; Alg−) | This study |
| PKM801 | PAOalgT15-1 mucA22 (sap28; Alg−) | This study |
| PKM802 | PAOalgT15-2 mucA22 (sap29; Alg−) | This study |
| PKM803 | PAOalgT15-3 mucA22 (sap33; Alg−) | This study |
| PKM804 | PAOalgT15-4 mucA22 (sap34; Alg−) | This study |
| PKM805 | PAOalgT17-1 mucA22 (sap50; Alg−) | This study |
| PKM806 | PAOalgT17-2 mucA22 (sap51; Alg−) | This study |
| PKM807 | PAOalgT24-1 mucA22 (sap37; Alg−) | This study |
| PKM808 | PAOalgT43-1 mucA22 (sap44; Alg−) | This study |
| PKM809 | PAOalgT43-2 mucA22 (sap45; Alg−) | This study |
| PKM810 | PAOalgT80::A1 mucA22 (sap40; Alg−) | This study |
| PKM811 | PAOalgT80::A2 mucA22 (sap41; Alg−) | This study |
| PKM812 | PAOalgT118-1 mucA22 (sap18; Alg−) | This study |
| PKM813 | PAOalgT122-1 mucA22 (sap38; Alg−) | This study |
| pKM814 | PAOalgT122-2 mucA22 (sap39; Alg−) | This study |
| Plasmids | ||
| pRK600 | Cmr; Nms; pRK2013 Nmr::Tn9 | (Finan et al., 1986) |
| pRK2013 | Kmr; ori ColE1, RK2-Tra+ | (Figurski and Helinski, 1979) |
| pCD100 | algT/U Tc Hg | This study |
| pJG293 | algT/U Mob Tc Km | This study |
| pRTS4500 | TOPO pCR2.1 containing algO and promoter with HindIII ends (PAO1 fragment 3642833–3645080) |
pTOPO-AlgO; This study |
| pRTS6000 |
algO fragment from pRTS4500 cloned into pME6030 using HindIII |
pAlgO; This study |
| Primers (5′ - 3′ sequence) | ||
| DR1FmucA | CTG CGC GAG TTC GAA GGT TTG A | This study |
| DR2RmucA | GCT GCC ATT GCG CTC GTA GAC | This study |
| DR3FalgT | CTT GGC AAG ACG ATT CGC TGG GAC | This study |
| DR4RalgT | CCT GCA GGG CTT CAC GAC TC | This study |
| cos-1 | CGCCCTCTGGTAAGGTTG | (Huang et al., 2000) |
| KAN-2 FP-1 | ACCTACAACAAAGCTCTCATGAACC | Epicentre |
| prc_F_HindIII | ATTAATAAGCTTCGCTGCCTCCATAGTGGG | This study |
| prc_R_HindIII | AATTATAAGCTTTTATGACGCTCCCGCTGA ACT |
This study |
| prc_F2 | CGAGAGCCTGCTGATCGA | This study |
| prc_F3 | AACGACCAGACCAGCAAG | |
| prc_F4 | AACTGAAGCTGACCCTGG | This study |
| GC_prc_F2 | TCGATCAGCAGGCTCTCG | This study |
| GC_prc_F3 | CTTGCTGGTCTGGTCGTT | This study |
| GC_prc_F4 | CCAGGGTCAGCTTCAGTT | This study |
| GC_prc_F5 | TTATGACGCTCCCGCTGA | This study |
sap strains whose mutations have not been mapped are not listed in the strain table
2.2. Isolation of sap mutants
Previously, we constructed an alginate-producing variant of PAO1 by replacing the mucA gene with the mucA22 allele that is frequently found in CF isolates (Mathee et al., 1999). In order to study the switch of P. aeruginosa from mucoid to nonmucoid form under defined conditions, we used PDO300 as the mucoid parent strain for isolation of spontaneous nonmucoid variants. Ten tubes containing two mL LB medium were inoculated with a single colony of PDO300 and incubated at 37°C without agitation. After 48 hours the cultures were diluted and plated on LB plates and the percentage of mucoid colonies was determined. Thirty-six nonmucoid isolates were chosen for further analysis by taking ten colonies from each of the first two tubes and two colonies from each of the last eight tubes. The nonmucoid phenotype of two of these was unstable and so only thirty-four were further analyzed.
2.3. Construction of pCD100 and pJG293
The plasmid pCD100 was constructed by digesting genomic DNA of the alginate producing strain FRD406 (DeVries and Ohman, unpublished data). FRD406 contains a chromosomal transposon Tn501-250 insertion located 3 kilobases (kb) upstream of algT/U which could be used as an adjacent selectable marker. Genomic DNA from FRD406 was digested with BamHI, and the fragments were ligated into the cosmid vector pEMR2 (Flynn and Ohman, 1988b). The bank of clones was then screened for Tn501, which confers mercury resistance and is not restricted by BamHI. One positive clone was digested with BamHI; the products were ligated into a broad host-range vector pRK404, transformed into E. coli HB101, and tested for mercury resistance. One clone chosen for further study, pCD100, was found to contain a single 17-kb BamHI fragment of P. aeruginosa DNA plus the additional 8.2-kb Tn501-250 insertion. Plasmid pCD100 was then transferred to PAO1 by triparental mating. The plasmid pJG293 was constructed by XhoI digestion of pJG102 (Goldberg et al., 1993). The XhoI fragment containing the algT/U gene was ligated into XhoI digested pCP13 (Darzins and Chakrabarty, 1984) resulting in pJG293.
2.4. DNA manipulations
All molecular techniques were performed according to standard protocols (Sambrook et al., 1989). DNA sequencing was done using the Big Dye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA) and analyzed on an Applied Biosystems 3100 Genetic Analyzer. To ensure high fidelity, the top strand was sequenced twice, while the bottom strand was done once (standard 2+1 sequencing). Sequencing primers are listed in Table 1.
2.5. Triparental mating
Plasmids and cosmids were conjugated into P. aeruginosa via triparental mating using helper strains pRK600 and pRK2013 (Figurski and Helinski, 1979; Finan et al., 1986). Conjugants were selected on LB:PIA plates (1:1) containing Tc (100 µg ml−1).
2.6. En masse complementation
En masse complementation was performed using the minimal tiling path PAO1 library that contains 336 clones ((Huang et al., 2000) available from Paul Phibbs, East Carolina University). The four plates were individually pooled, and the pooled cultures used as donors for triparental matings. To score for mucoidy, LB:PIA (1:1) plates with and without glycerol were used to reduce false positives. After one-two days of growth at 37°C, colonies were restreaked to confirm the mucoid phenotype.
2.7. Amplification of the algT/U, mucA, and algO loci
Genomic DNA was prepared from PAO1, PDO300 and the sap strains and the polymerase chain reaction (PCR) was used to amplify the mucA, algT/U or algO loci using the Expand High Fidelity PCR kit (Roche Applied Science, Indianapolis, IN). Primer sequences are listed in Table 1. The mucA, algT/U, and algO loci were PCR amplified using primer pairs DR1FmucA/DR2RmucA, DR3FalgT/DR4RalgT and prc_F_HindIII and prc_R_HindIII, respectively.
2.8. Membrane fractionation
The outer membrane fraction of P. aeruginosa strains was obtained by sarkosyl treatment of sonicated extracts as described (Filip et al., 1973). Fractions were separated by SDS polyacrylamide gel electrophoresis (PAGE) and visualized by Coomassie Brilliant Blue staining.
2.9. Measurement of β-lactamase, protease and β-galactosidase activities
For β-lactamase activity, strains were grown in LB with induction by benzylpenicillin (500 mg/L) for 2.5 hours and cells lysed by sonication. For the LasA protease and LasB elastase assays, strains were incubated at 37°C in LB with rapid aeration (shaking at 250 rpm) for 18 hours under standardized conditions. For β-galactosidase assays, O/N cultures of P. aeruginosa strains containing plasmid with an algT/U-lacZ transcriptional fusion (pKMG37) were diluted 1/100 into fresh LB supplemented with the appropriate antibiotic. Cells were harvested when an OD600 of 0.6–0.8 was reached. In all cases, enzyme activity was measured as described (Kong et al., 2005).
2.10. Antibiotic resistance assay
Antibiotic resistance profiles of the sap mutants were generated using the E-test antibiotic kit according to the manufacturer’s instructions (bioMérieux, l’Etoile, France).
2.11. Alginate assay
Alginate concentrations in culture supernatants were measured after extensive dialysis using a colorimetric assay for uronic acids (Jain and Ohman, 1998). To determine the alginate concentration, a set of standards was made with sodium alginate (Sigma, St. Louis, MO). The alginate concentration was expressed in mg per ml supernatant.
2.12. Cosmid DNA identification
Cosmid DNA was extracted from P. aeruginosa and transformed into E. coli (Sambrook et al., 1989). Cosmid DNA was then purified from E. coli and identified by sequencing the insert junction using the cos-1 primer.
2.13. Tn mutagenesis and characterization
To identify the ORF of interest, the complementing cosmid was mutagenized using the EZ::TN transposon kit (Epicentre, Madison, WI) according to the manufacturer’s protocol except that the incubation time was doubled from two to four hours. The mutagenized cosmids were transformed into E. coli TOP10 (Invitrogen, Carlsbad, CA). Transformants were plated on selective media (Tc 20 µg ml−1 and Km 30 µg ml−1). More than 500 transformants were pooled and used as donors in triparental matings with sap22. Conjugants were screened for loss of the mucoid phenotype, indicating disruption of a vital ORF. From these strains, the cosmid DNA was extracted as described above. After transforming the Tn-containing cosmid into E. coli, sequencing was performed using the KAN-2 FP-1 primer (Table 1).
2.14. Construction of pAlgO
The algO gene was PCR amplified with the primers prc_F_HindIII and prc_R_HindIII (Table 1), both carrying HindIII restriction sites. The resulting amplicon that contained the algO open reading frame in addition to approximately 120 bp of upstream and 20 bp of downstream sequences, was cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) to form pTOPO-AlgO. The HindIII-fragment from pTOPO-AlgO was subcloned into the HindIII site of pME6030 (Heeb et al., 2000) to form pRTS6000. The complementing construct is referred to as pAlgO.
2.15. AlgT/U structural modeling
The sequence of PAO1 AlgT/U from the Pseudomonas genome database (Stover et al., 2000) was used as input to the Swiss Model Server (Peitsch, 1995; Guex and Peitsch, 1997; Schwede et al., 2003; Kopp and Schwede, 2004; Arnold et al., 2006). Using the automated mode, the sequence was modeled based upon the structure of RpoE (chain 1or7A) (Campbell et al., 2003). ANOLEA and What Check were used to determine the quality of the model. Similar methods were used to model the point mutations in algT/U.
2.16. Statistics
The significance testing for the alginate assay and lacZ promoter fusions was carried out using ANOVA and the Bonferroni correction in the MS Excel plugin Analyse-It v1.71 (http://www.analyse-it.com/).
3. Results and Discussion
3.1. Isolation of suppressor of alginate production (sap) mutants
A set of nonmucoid revertants, termed sap strains, was generated by growing a defined mucA22 mutant (Alg+ PDO300) without aeration. Approximately 90 percent of the colonies (2059 of 2300 from 10 independent colonies) had reverted to the nonmucoid phenotype. Thirty-four sap isolates were chosen for further analysis. Previously, spontaneous nonmucoid variants were isolated and characterized to better understand the alginate pathway, however, these studies were not performed in a sequenced strain background (DeVries and Ohman, 1994; Schurr et al., 1994). DeVries and colleagues used ethylmethane sulfonate to isolate a mucoid histidine auxotroph, FRD39, from FRD1, a prototrophic CF isolate that is mucoid due to the presence of a mucA22 allele (Ohman and Chakrabarty, 1981). Nonmucoid variants were then isolated by incubating FRD39 under low oxygen tension (without aeration) (DeVries and Ohman, 1994). The second study by Schurr et al., (Schurr et al., 1994) used a mucoid derivative of PA0381, PA0578, that was isolated upon treatment with carbenicillin and selection on Pseudomonas isolation agar (Govan and Fyfe, 1978). This strain is mucoid due to the presence of the mucA22 allele (Fyfe and Govan, 1980). Spontaneous nonmucoid derivatives of PA0578 were isolated by repeated passage on PIA (Schurr et al., 1994). The advantage of using PDO300 is that it is a defined derivative of PAO1 in which the wild type mucA was replaced with the mucA22 allele (Mathee et al., 1999). PAO1 is well characterized and its genome has been sequenced (Stover et al., 2000).
3.2. Analysis of sap mutants
The growth rates in LB of PAO1, PDO300 and of all 34 sap mutants were virtually identical (data not shown). Pulse-field gel electrophoresis analysis of chromosomal DNA revealed no obvious genetic rearrangements in any of the strains (data not shown). PDO300 produced approximately 271 µg of alginate per ml whereas all sap mutants and PAO1 produced virtually none (data not shown). The antibiotic resistance profiles of PDO300 and of each of the sap strains were identical to that observed with PAO1 (antibiotics tested: piperacillin, aztreonam, ceftazidime, imipenem, meropenem, colistin, ciprofloxacin, ofloxacin, tobramycin and netilmycin; data not shown).
3.3. sap alleles fall into two different complementation groups
In order to investigate which of the 34 sap mutants could be complemented back to the mucoid phenotype by introducing the algT/U locus in trans, the plasmid pCD100 carrying algT/U-mucA22-mucB-mucC-mucD was introduced by triparental mating into each of the 34 variants. When pCD100 was introduced, 14 (41%) of the sap mutants turned mucoid whereas 20 (59%) remained nonmucoid. Introduction of the plasmid pJG293, containing only the algT/U gene, into each of the 14 variants complemented by pCD100, restored the mucoid phenotype. This suggests that the mutation was located in the algT/U gene. Introduction of pCD100 or pJG293 into PAO1 resulted in an Alg+ phenotype (data not shown).
It was possible that the 20 isolates not complemented by pCD100 had mutations in regulatory genes such as algR and algB, in the biosynthetic algD operon or in other known or unknown loci. Plasmids containing the algR, algP, algQ loci in a 20-kb fragment or the entire algD operon, pAL (Darzins and Chakrabarty, 1984) and pAlg2 (Chitnis and Ohman, 1990), respectively, were introduced into each of the 20 strains. None of the strains became mucoid suggesting that these have mutations in genes not previously associated with alginate production (C. Ceballos, L. Florez, R. Smiddy, R. Sautter, L. Schneper, and K. Mathee, personal communication). Thus, the sap alleles were classified into two groups, those in which the mucoid phenotype is restored by algT/U complementation (Group A) and those which are not (Group B).
3.4. Outer membrane profile
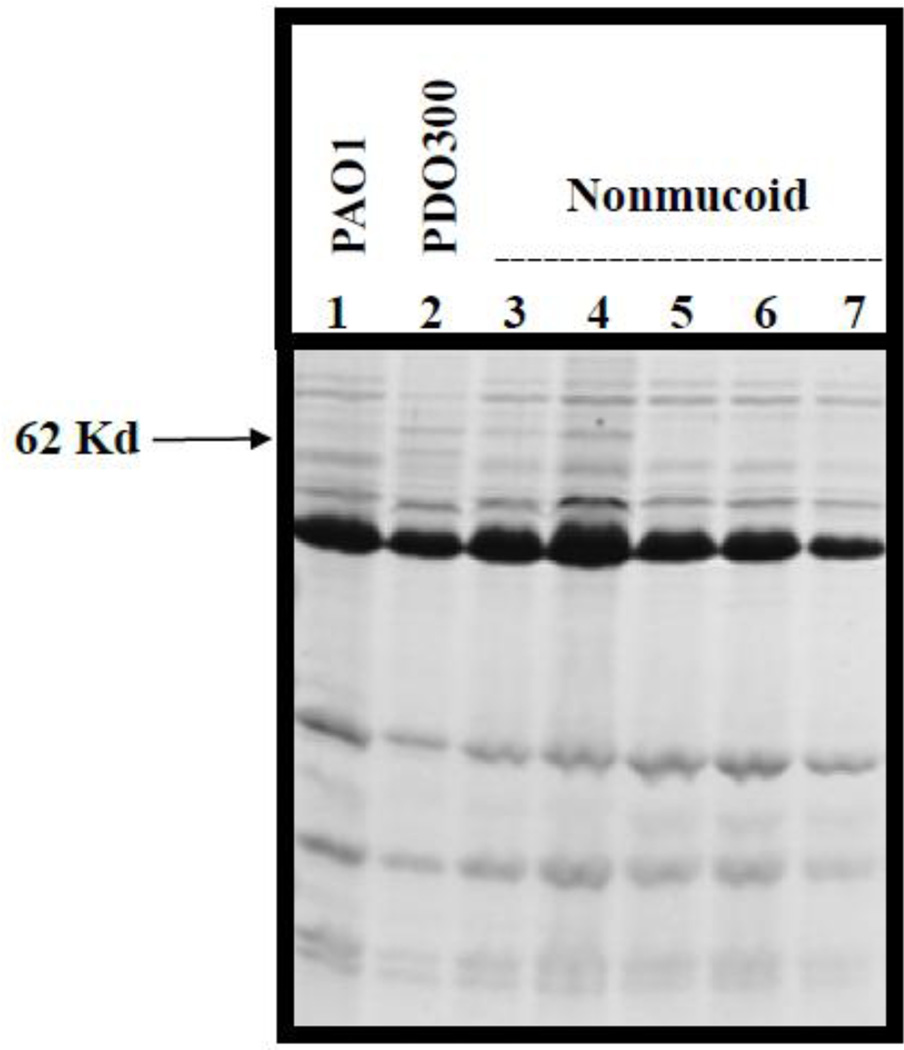
Alginate overproducing strains have outer membrane protein profiles with a prominent 54-kDa protein that is not observed in nonmucoid strains (Goldberg and Ohman, 1987; Grabert et al., 1990). The identity of the 54 kDa protein has been shown to be the product of the algE gene (Mathee et al., 1999), which is transcribed as part of an 18-kb operon of biosynthetic genes and appears to encode the porin component of a secretory complex involved in polymer export to the bacterial surface (Chu et al., 1991; Rehm et al., 1994a; Rehm et al., 1994b; Hay et al., 2010). The mobility of the AlgE protein in SDS-PAGE is variable (Grabert et al., 1990). Several proteins of unknown identity were differentially expressed in the 34 sap mutants. One of these proteins has an apparent molecular weight of 62-kDa and was expressed in PDO300 but not in PAO1 (Fig. 2, compare Lanes 1 and 2). Several attempts to determine the N-terminal amino acid sequence of the 62-kDa protein failed. However, based on the presence and absence of this protein, the nonmucoid revertants that are not complemented by algT/U can be divided into two subgroups, B1 (e.g. Fig. 2, lane 7) and B2, respectively (e.g. Fig. 2, Lanes 3 and 4). Those that are complemented by algT/U were placed into subgroup A regardless of the presence or absence of the 62-kDa protein (e.g. Fig. 2, Lanes 5 and 6).
Fig. 2.
Outer membrane protein profiles of parental strains and representative nonmucoid variants. Outer membrane proteins were isolated from PAO1 (lane 1), PDO300 (lane 2) and the nonmucoid variants (Lanes 3 – 7), fractionated by SDS-PAGE and visualized with Coomassie Blue. Lanes 3 and 4 contain nonmucoid variants that are not complemented by a plasmid containing algT/U but have a 62 kDa protein similar to PDO300. These have been grouped into subgroup B2. Lanes 5 and 6 contain sap strains representative of Group A. Lane 7 contains a nonmucoid variant that is not complemented by algT/U and has an OMP profile similar to PAO1, representative of subgroup B1.
3.5. Phenotypic determinants of the sap mutant groups
The sap strains were analyzed for phenotypes previously shown to correlate with alginate production, antibiotic resistance, and pathogenicity. Specifically, basal and inducible β-lactamase activities, as well as the ability to produce the LasA protease and LasB elastase were quantified in the sap mutants (Fig. 3).
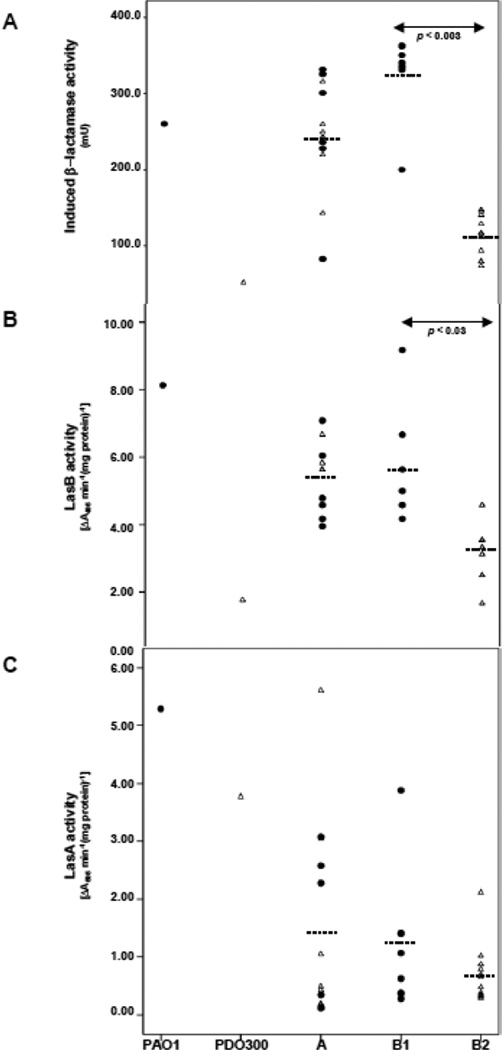
Fig. 3.
Scatter plots of PAO1, PDO300, and the sap strains’ (A) induced beta-lactamase (B) LasB elastase and (C) LasA protease activities. Closed circles represent strains that do not express detectable Omp62 and open triangles represent strains that express Omp62. Dashed lines represent average enzymatic activities of the subgroup. Significant p-values are noted.
3.5.1. The amount of inducible β-lactamase varies in nonmucoid variants
P. aeruginosa isolates from CF patients typically produce high levels of chromosomal β-lactamase, a group 1 cephalosporinase, encoded by ampC (Giwercman et al., 1991; Campbell et al., 1997). PDO300, like PAO1, produces a low basal level of β-lactamase. However, the inducible level of β-lactamase (following treatment with 500 µg ml−1 benzyl-penicillin) is reduced by approximately 30% in PDO300 compared to PAO1 (Mathee et al., 1999). Consequently, the resistance to beta-lactam antibiotics of typical CF strains is not associated with the mucA22 mutation. The 34 nonmucoid variants in the present study all had basal levels of β-lactamase similar to PAO1 and PDO300 (data not shown). The inducible levels of β-lactamase, however, varied considerably between the 34 variants with several exhibiting levels greater than that of PAO1 (Fig. 3A).
3.5.2 Protease production is altered in the alginate-producing variants
P. aeruginosa secretes a number of proteases including LasB protease (elastase) and LasA protease (staphylolytic protease) and alkaline protease which aid in its pathogenesis by compromising host barriers (Kharazmi, 1989). Alginate-producing strains, including PDO300, exhibited reduced levels of LasA and LasB activities (Mohr et al., 1990; Mathee et al., 1999a). It was possible the protease activities were restored in the sap mutants. As expected, PDO300 exhibited less LasB (Fig. 3B) and LasA (Fig. 3C) activities than PAO1. The LasB activity was completely restored in one sap strain, similar to PDO300 in another, and at intermediate levels in the rest. In the case of LasA, activity was completely restored in one sap strain and in one remained the same as in PDO300. The Group B1 sap strain that had restored LasB activity, also exhibited the highest LasA activity of Group B1 members. However, in most of the sap strains (65%) the LasA protease levels were less than 25% of that of PDO300. These results clearly indicate the complex multi-tiered relationship between alginate and protease production. At least in one case, LasB has been shown to facilitate cleavage of NDK to its active form to generate GTP necessary for alginate production (Kamath et al., 1998). Identification of the sap mutations is critical to better understand the role of these genes in regulating these two phenotypes important for virulence.
3.5.3 Comparison of sap groups
The phenotypic properties of the three groups of sap strains were compared using the non-parametric Mann-Whitney test (Mann and Whitney, 1947). Group A strains, which were complemented by pCD100, had higher LasB activities than Group B (p-value < 0.0008). The 62-kDa outer membrane protein containing (PDO300-like) B2 subgroup had higher induced β-lactamase (p-value < 0.0003) and LasB levels (p-value < 0.03) compared with the B1 (PAO1-like) subgroup (Fig. 3). LasA activities (p-value 0.54) did not reflect this grouping. LasB elastase has been demonstrated to have a positive correlation with algD expression and the mucoid phenotype (Storey et al., 1997; Kamath et al., 1998). Although the results presented here are more supportive of the early studies, further characterization of the sap strains is warranted.
3.6. Molecular analysis of the sap alleles
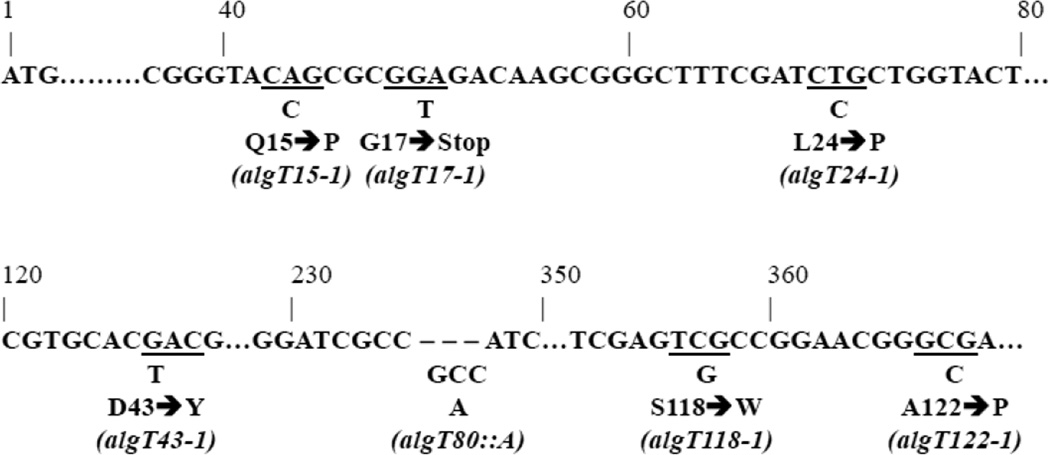
The 14 sap mutants that could be restored to Alg+ by trans-complementation with algT/U contained the original mucA22 allele. Thus, these strains are likely to have a mutation in algT/U. The algT/U coding sequence in each of the 14 Group A variants was sequenced (see Fig. 3). All had mutations in algT/U and five alleles were isolated multiple times. Identical alleles were from the same culture and thus are likely to be siblings. Four of the sap alleles (algT15-1, algT15-2, algT15-3, and algT15-4) contained the same CAG (Gln) to CCG (Pro) transversion in codon 15 substituting a polar amino acid for a nonpolar one. Two sap alleles (algT43-1 and algT43-2) altered codon 43 from GAC (Asp) to TAC (Tyr); two (algT80-1 and algT80-2) contained an insertion of GCC after codon 79 resulting in the addition of an Ala residue at position 80; two (algT122-1 and algT122-2) contained a GCG (Ala) to CCG (Pro) transversion at codon 122; and two (algT17-1 and algT17-2) encode a truncated protein, containing a GGA (Gly) to TGA (stop) mutation. The remaining sap mutants were unique substitutions. Codon 24 of algT24-1 contained a CTG (Leu) to CCG (Pro) transition. This nonmucoid revertant was isolated from a unique mucoid colony. The remaining Group A sap strain was isolated from the same mucoid parent as the algT15 alleles. This sap strain resulted from a single transversion from TCG (Ser) to TGG (Trp) at codon 118. These results are summarized in Figure 4. Analysis of the DNA sequences of these nonmucoid revertant strains demonstrated that most algT/U mutations (70% in this study) were transversions (Fig. 4), although transitions are favored (Jukes, 1987).
Fig. 4.
algT/U sequence analysis of the nonmucoid variants. Partial nucleotide sequence of the algT/U gene from PAO1 and the nonmucoid variants. Dots indicate the sequences in between that are not shown. Underlined triplets represent codons found to have mutations in algT/U; and the mutations are given below. Alteration in amino acid is represented below each mutated codon (e.g. amino acid 15 mutated from Q to P). The allelic name for the mutation is given in parentheses.
In the aforementioned previous studies of nonmucoid revertants, the Alg- phenotype was also linked to the algT/U locus. Out of 28 isolates, nine had missense mutations in codons 18 and 29, in five and four of the isolates, respectively (DeVries and Ohman, 1994). In the study by Schurr et al. (Schurr et al., 1994), two out of three nonmucoid variants had mutations in algT/U. One had a nonsense mutation in codon 74 and the other had a frameshift mutation at codon 160. The third isolate had an intact algT/U gene. No further analysis was done with the third isolate. In this study, we found seven novel algT/U mutations (Fig. 4).
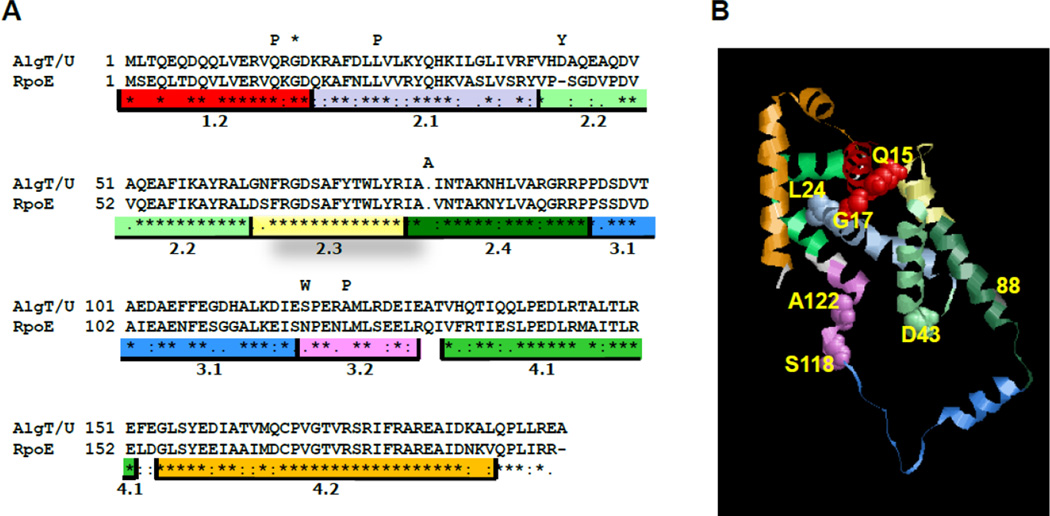
AlgT/U is approximately 64% identical and 79% homologous to E. coli RpoE. Both are classified as members of the σ70 family that have four conserved regions (1–4) that are further subdivided (e.g. 2.1 – 2.4) (Fig. 5A) (Lonetto et al., 1992). To better understand the effects of the individual point mutations, the protein sequence of AlgT/U was aligned with E. coli RpoE (Fig. 4A, (Larkin et al., 2007)) and the tertiary structure of AlgT/U was modeled using Swiss Model based upon the RpoE crystal structure (Fig. 5B) (Guex and Peitsch, 1997; Campbell et al., 2003; Schwede et al., 2003; Arnold et al., 2006). The atomic empirical mean force potential ANOLEA values (Melo et al., 1997) and GROMOS (van Gunsteren et al., 1996) suggested favorable energy environments of most amino acids except for residues 106 to 120. This is not surprising given that residues 112 through 119 of RpoE are disordered (Campbell et al., 2003). Residues 106 to 120 were not further analyzed.
Fig. 5.
Mutations found in the AlgT/U protein. (A) Alignment of P. aeruginosa PAO1 AlgT/U with E. coli RpoE. Symbols below the alignment (*, :, and .) represent identical, highly conserved, and conserved residues, respectively. The sigma factor regions are separated in nine colored boxes based on homology analysis (Lonetto et al, 1992). Mutations identified in this study are indicated above. Note the stop codon mutant was included in this figure as it resulted in a truncated protein. (B) Modeled structure of AlgT/U based upon alignment with RpoE. Residues are colored according to (A). Residues that were identified to be mutant in this study are labeled.
Conserved region 1.2 is important in ensuring that promoter-bound sigma factors are associated with RNA polymerase. It is required for open complex formation and the transition from abortive transcription to elongation (Baldwin and Dombroski, 2001). Three nonmucoid revertants (amino acids 15 and 17, Fig. 4 and amino acid 18 (DeVries and Ohman, 1994)) were isolated in this region. This suggests these three amino acids are critical for the σ factor function (DeVries and Ohman, 1994). The model suggests a structure consisting of nine alpha helices. Located at the end of the first helix, Gln15 potentially hydrogen bonds with Val11, Leu62 and Phe65. Substitution of Gln15 with Pro could potentially not only affect the end of the first helix, perhaps by loosening the helix, but also affect hydrogen bonding to Leu62 and Phe65.
The algT24, algT43 and algT80 alleles contain mutations in regions 2.1, 2.2, and 2.4 respectively. Region 2.1 is important in binding to core RNA polymerase, region 2.2 is involved in promoter melting and region 2.4 has been implicated in binding to the −10 promoter region (Helmann and Chamberlin, 1988). The mutation in region 2.1 at amino acid 24 (Leu to Pro) is in the primary core binding “RpoD Box” (Lesley and Burgess, 1989; Nagai and Shimamoto, 1997). De Vries and Ohman also isolated a second mutation in the region at amino acid 29. Mutation in this region will prevent holoenzyme formation. In the model, substitution of Leu24 with Pro does not appear to affect hydrogen bonding with Arg20 and Ala21. As expected intuitively and shown by modeling, Pro is not a favorable amino acid at that position. Similarly, mutation of Asp43 to Tyr, does not seem to affect hydrogen bond formation, but is not an energetically favorable substitution.
The alg118 and alg122 alleles both contain mutations in region 3.2, which is involved in binding to core RNA polymerase (Lonetto et al., 1992). Mutation in this region will prevent the sigma factor from binding to the core enzyme efficiently (Nagai and Shimamoto, 1997). This region was disordered and not represented in the model. In this study, no mutations mapped to regions 2.3 or 4.2, as previously reported by Schurr et al. (Schurr et al., 1994).
3.7. En masse mating yields a novel gene involved in alginate production
The remaining 20 sap mutants that remained nonmucoid even when algT/U was expressed in trans, were mated with a minimal tiling path (MTP) PAO1 genomic cosmid library (Huang et al., 2000). As expected, the mucoid phenotype was restored in several of the sap mutants. One of the sap mutants (sap22) that was complemented back to mucoidy was further characterized. This particular sap strain belongs to the PAO1-related subgroup of Group B, meaning it did not express the 62 kDa protein and had relatively high levels of elastase.
3.8. Identification of the sap22 complementing cosmid
The identity of the sap22 mutant complementing cosmid was determined by sequencing the junction of the insert. This cosmid, pMO011713, contains PAO1 genomic sequences from coordinates 3628056 to 3652056 (Table 1, (Huang et al., 2000)), encompassing a total of 25 ORFs. This cosmid did not contain any known alg genes, suggesting the presence of novel ORFs involved in the production of alginate. To ensure that the cosmid pMO011713 indeed complemented the nonmucoid revertant sap22 to a mucoid phenotype, the sap mutant and PAO1 were remated with E. coli containing cosmid clones from the original MTP library (Table 1). PAO1 and sap22 yielded a nonmucoid and mucoid phenotype, respectively with pMO011713.
3.9. Transposon mutagenesis of pMO011713
Since pMO011713 contains 25 genes, the cosmid was subjected to transposon mutagenesis to map the novel gene involved in the alginate production. The mutant library was introduced into sap22. Loss of the mucoid phenotype in transconjugants indicated the disruption of the complementing gene on the cosmid by the Tn insertion. Of the 150–200 transconjugants screened, six nonmucoid clones were recovered, and the phenotype confirmed by restreaking on fresh media. Once the loss of phenotype was verified, the cosmids were moved into E. coli and the cosmid DNA purified and sequenced using a primer that hybridized within the transposon insertion. All six clones had a Tn insertion in the same ORF, PA3257, which encodes a predicted polypeptide of 698 amino acids. The Tn insertions occurred at the following amino acid positions: 160, 370, 139, 587, 179 and 364. These six mutants strongly suggested that PA3257 is involved in alginate production. Henceforth this ORF is referred to as algO.
3.10. Analysis of PA3257
According to the genome annotation, PA3257 is a 2096-bp ORF, encoding a putative 78-kDa protein with a calculated pI of 6.22 (using ExPasy, http://www.expasy.ch/tools/pi_tool.html). This putative protein is 42 % identical to the E. coli periplasmic protease Prc that plays a role in antibiotic resistance and stress response (Hara et al., 1991). The E. coli Prc protease was also shown to process carboxy-termini of its target proteins, and consequently is also referred to as the tail-specific protease (Tsp; (Silber et al., 1992)). As expected, AlgO has a typical signal peptide with a hydrophobic uncharged amino acid residue followed by a cleavage site for signal peptidases (Izard et al., 1996). The signal sequence, as identified by SignalP 3.0, is predicted to be NH2-MRHYSAISRISMKRFLPR TALLLLLGASSLPLFA↓S (Bendtsen et al., 2004). This ORF also has a strong ribosome-binding site seven to ten bases immediately upstream from the putative initial methionine codon. Sequence analysis shows the presence of a potential stem-and-loop structure followed by a T string approximately 40 bp downstream from the proposed translation termination codon, indicating the presence of a ρ-independent terminator (Yang and Roberts, 1989).
Analysis of the 698 amino acid long P. aeruginosa AlgO sequence reveals the presence of two conserved domains. The region between amino acids 252–337, corresponds to a PDZ binding domain, which is implicated in protein-protein interactions and C-terminus processing (Beebe et al., 2000). Amino acids 340 to 545 represent the Tsp domain, which is involved in the degradation of C-termini based on the carboxy-tail amino acid sequence (Silber et al., 1992).
3.11. The algO gene alone is sufficient to restore the mucoid phenotype
To demonstrate that the phenotypic change is due to a single gene, a low copy plasmid containing PA3257 under the control of its own regulatory sequences was introduced into the nonmucoid sap22. Twenty-four hours after plating the mating on selective media, colonies showing a strong mucoid phenotype appeared (data not shown). These colonies were restreaked and showed significant alginate production after 24 hours of incubation. The pAlgO plasmid was also introduced into PAO1 as a negative control. PAO1 (pAlgO) remained strictly nonmucoid and pigmented, suggesting that overexpression of the algO gene alone is not capable of inducing the mucoid phenotype (data not shown). To ensure that alginate was secreted, the quantity of alginate produced by various strains was determined using the carbazole assay ((Jain and Ohman, 1998); Table 2). As expected, PAO1, sap22 and sap22 (pME6030) produced negligible amounts of alginate. PAO1 in the presence of pAlgO exhibited basal but relatively insignificant alginate production (Table 2). The two strains, Alg+ PDO300 and Alg+ sap22 (pAlgO), both generated significant quantities of alginate (291 ± 77 and 227 ± 77 µg ml−1, respectively) (Table 2). Thus, sap22 likely harbors a mutation in the algO gene and alginate production can be restored in this strain to levels comparable to the parent by algO expression in trans.
Table 2.
Transcomplementation of sap22 by pAlgO restores high levels of secreted alginate
| Strain | Relevant genotype | Alginate Phenotype |
Alginate (± SD)a |
|---|---|---|---|
| PAO1 | Wild-type | Alg− | BD |
| RTS2700 | PAO1 (pAlgO) | Alg− | 25 ± 35 |
| PDO300 | PAO mucA22 | Alg+ | 291 ± 77 |
| RTS22 | PAO mucA22 sap22 | Alg− | BD |
| RTS2702 | PAO mucA22 sap22 (pME6030) | Alg− | 25 ± 33 |
| RTS226 | PAO mucA22 sap22 (pAlgO) | Alg+ | 227 ± 77 |
Mean secreted alginate is expressed in µg ml−1 (± standard deviation) from at least three independent cultures determined as described in the Materials and Methods section. BD, below detection.
3.12. Sequencing of the sap22 allele reveals a mutation in the algO gene
To confirm that the sap22 strain contained a mutation within algO, the gene and promoter were sequenced in their entirety from both sap22 and the cloned algO gene from PAO1. The sequences of algO from the PAO1 genome database and the cloned gene in pAlgO were identical. The sequence from the algO gene in sap22 showed an insertion of a single nucleotide, thymidine (T), at position 3,643,250 on the genome. This mutation corresponds to codon 96, and results in a frameshift leading to premature termination at codon 309, which is converted from glutamic acid to UGA, the Opal stop codon. The mutant allele in sap22 is referred to as algO96.
3.13. Loss of algO leads to increased algT/U expression
Since P. aeruginosa AlgO is involved in alginate production, it was postulated that the expression of algT/U may be compromised in the sap22 mutant. To address this, a PalgT/U-lacZ promoter fusion plasmid was introduced into PAO1, PDO300 and sap22. As predicted, PAO1 activity levels were minimal, whereas PDO300 showed a nearly three-fold increase in PalgT/U expression (Table 3). Interestingly, the loss of algO in the Alg− sap22 strain led to increased algT/U expression by 60 % (p < 0.01). However, the increased AlgT/U still failed to confer a mucoid phenotype.
Table 3.
A mutation in algO/prc increases activity of the algU promoter
| Strain | Relevant Genotype | Alginate Phenotype |
β-galactosidasea (Miller Units) |
|---|---|---|---|
| DH5α | E. coli (PalgT/U-lacZ) | Alg− | 11 ± 12 |
| PAO1 | Wild-type (PalgT/U-lacZ) | Alg− | 179 ± 45 |
| PDO300 | PAO mucA22 (PalgT/U-lacZ) | Alg+ | 520 ± 62 |
| RTS22 | PAO mucA22 sap22 (PalgT/U-lacZ) | Alg− | 855 ± 65 |
Mean β-galactosidase activity (± standard deviation) expressed in Miller units (Miller, 1972) from at least three independent cultures.
3.14. BlastP and ClustalW analysis of AlgO
Homologs of P. aeruginosa algO were found using BlastP (Altschul et al., 1990; Altschul and Gish, 1996; Altschul et al., 1997). P. aeruginosa algO shows strong homology to the E. coli gene prc (Reiling et al., 2005; Winsor et al., 2005). The E. coli Prc protein contains 682 amino acids, with a mass of ∼80 kDa, whereas P. aeruginosa AlgO has a calculated molecular weight of 78 kDa with 42 % identity (Silber et al., 1992). Prc homologs from E. coli, S. typhimurium and Haemophilus influenze were aligned with AlgO using ClustalW because of their high identity with AlgO (data not shown; (Thompson et al., 1994)). Like E. coli Prc, P. aeruginosa AlgO has the conserved PDZ and Tsp domains, with 74 % and 80 % sequence similarity, respectively.
Prc cleaves the nonpolar C-termini of its targets (Nagasawa et al., 1989; Silber et al., 1992). The catalytic triad in E. coli Prc is conserved in AlgO (Ser-479, Asp-490, Lys-504) as are many residues flanking the triad. E. coli Prc residues (Gly-424/425, Glu-481, Thr-501) that contribute to its stability as measured by shifts in the circular dichroism (CD) spectra (Keiler et al., 1995), are also conserved in AlgO. The overall conservation of residues 470–500 in AlgO with Prc and other Prc homologs, suggests that AlgO may indeed function as a serine-protease.
3.15. P. aeruginosa algO does not share phenotypes similar to E. coli prc
The E. coli prc mutant was discovered due to its inability to properly process penicillin binding protein, PBP3 (Hara et al., 1989). Hara et al demonstrated that the expression of prc was essential for growth under osmotic or thermal stress (Hara et al., 1991). The loss of prc was linked to a weakened heat-shock response due to the loss of two crucial heat-shock proteins, DnaK and GroEL (Hara et al., 1991). These same mutants failed to grow in a salt-free medium (Hara et al., 1989; Hara et al., 1991). Based upon these data, we speculated that similar effects may be seen in the P. aeruginosa algO mutant strain sap22. However, in P. aeruginosa the effect was not significant (data not shown). Similar results were obtained in a mucA22 algO::Tcr strain (Reiling et al., 2005). These data suggest that P. aeruginosa AlgO does not influence the response to heat or osmotic stress in P. aeruginosa. The contribution of AlgO to antibiotic resistance is also negligible (data not shown) compared with the contribution of Prc in E. coli. However, the antibiotic resistance mechanisms present in P. aeruginosa are more complicated, and thus the organism may adapt with greater ease.
In E. coli, the loss of prc was complemented by an array of genes (Bass et al., 1996). Two of these genes, hhoA and hhoB, are HtrA homologs (serine proteases) that restored growth at 41°C (Bass et al., 1996). This same study also identified dksA (dnaK suppressor) as a prc suppressor, and may do so via heat-shock protein inhibition or by promoting stress-response factors (Bass et al., 1996). Another second-site suppressor was mapped to the rlpA gene encoding a periplasmic lipoprotein involved in cell wall synthesis (Takase et al., 1987; Bass et al., 1996). Introduction of cosmids containing the P. aeruginosa homologs of phpA, dksA and rlpA into the sap22 strain did not restore the mucoid phenotype (data not shown). This implies that AlgO plays a specific role in alginate production. However, this study used sap22, which has a mutation in two genes, mucA and algO. Thus, one cannot rule out that loss of algO in the prototypic strain PAO1 can be complemented by these second site suppressors. An alternative explanation is that these E. coli suppressors need to be overexpressed to suppress the algO− phenotype.
4. Conclusion
Alginate-producing P. aeruginosa has been attributed to be the leading cause of morbidity and mortality seen in CF patients. Thus, elucidation of the molecular mechanism responsible for the conversion from nonmucoid to the intractable mucoid form is of potential therapeutic interest. Conversion from the nonmucoid to mucoid form involves mutations in the mucA gene that encodes the anti-sigma factor for the extracytoplasmic function sigma factor AlgT/U (Mathee et al., 1997). The mucoid phenotype is unstable and it appears that there may be multiple pathways involved in the conversion from mucoid to nonmucoid form. This study using Alg+ PAOmucA22 showed that 90 % of progenies converted to a nonmucoid phenotype under low oxygen tension at 37°C. The molecular basis for this conversion has partially been attributed to mutations in the algT/U gene itself (DeVries and Ohman, 1994; Schurr et al., 1994). Of the 34 nonmucoid variants analyzed, 14 had previously unidentified mutations in algT/U (Group A; Fig. 4). Biochemical characterization of the mutants isolated in this study will reveal the contribution of each mutated residue to its function as a sigma factor. During the nonmucoid conversion, presumably these algT/U mutations alter the protein’s conformation disabling its interaction with the RNA polymerase. The modus operandi is to slow down alginate production by mutating essential genes required for alginate production, and in this case the function of σ22.
Clearly, acquired mutation in the algT/U gene is not the sole method involved in the conversion from mucoid to nonmucoid form. The remaining sap mutants (Group B) were subdivided based upon the absence (subgroup B1) or presence (subgroup B2) of a 62-kDa OMP protein. Strains lacking functional AlgT/U show consistently higher virulence factor production (Fig. 3). This is expected, as alginate production is metabolically taxing on the cells. It is not known whether AlgT/U may be repressing virulence gene expression indirectly. Groups A and B show subtle, yet distinguishable, differences in β-lactamase and elastase levels. The algT/U strains exhibited higher virulence protein levels, but were far below PAO1 levels. In subgroup B2, the revertants show a marked decrease in virulence, as measured by LasA and LasB activity, but still express OMP62. This deviates from PAO1, in which decreased OMP62 was correlated with increased virulence (higher levels of LasA and LasB activity).
Although the sap mutants were classified into three broad groups in this study, within each subgroup, there is variation. Together, these data suggest the bacteria may be regulating the mucoid phenotype at multiple loci, with direct effects on virulence gene products. Mapping of the mutation in one of these sap strains identified a role for a putative protease, AlgO, in positively regulating alginate production. Further studies are needed to determine how AlgO regulates alginate production; whether it acts on truncated MucA proteins as has been proposed (Reiling et al., 2005), or whether it affects other members of the pathway, perhaps by inhibiting MucB or activating one of the other proteases (Fig. 1). Regardless, AlgO affects alginate production at least partly by affecting algT/U expression. Thus, under conditions where alginate production is not beneficial, the bacteria evolve to downregulate AlgT/U function, either by directly mutating algT/U or by affecting its expression. Further characterization of the additional sap mutants not complemented by algT/U as well as determination of the exact mechanism of AlgO action and its substrates should reveal the complex mechanisms involved in the control of alginate production and may provide leads to potential therapeutic targets.
Highlights.
We screened for genes involved in alginate production
Multiple pathways are involved in mucoid to nonmucoid reversion
Reversion is in part attributed to loss of AlgT/U and AlgO function
AlgT/U and AlgO positively regulate alginate production
Acknowledgments
We thank members of the Mathee Laboratory for insightful discussions. We thank Dr. Paul Phibbs at East Carolina University for the minimal tiling path cosmid library. This work was supported in part by Public Health Service grant AI-19146 from the National Institute of Allergy and Infectious Diseases (D.O.), Veterans Administration Medical Research Funds (D.O.), grants from the Danish Biotechnology Program (A.K., M.G. and S.M.), and by a grant from the Danish Health Council (K.M., S.M., and N.H.), NIH/NIGMS grant R25 GM61347 (DR and RTS). The authors would like to thank all members of the Mathee laboratory for their support and thoughtful discussions, but would also especially like to acknowledge the assistance of Dr. Kok-Fai Kong, Robert J. Smiddy and Camila Ceballos for thoughtful review of the manuscript prior to submission.
Abbreviations
- A
adenosine
- Ala
alanine
- Alg−
nonmucoid
- Alg+
alginate overproducing mucoid
- ANOLEA
atomic non-local environment assessment
- Ap
ampicillin
- Asp
aspartic acid
- bp
base pair
- C
cytidine
- Cb
carbenicillin
- CF
cystic fibrosis
- DNA
deoxyribonucleic acid
- F
phenylalanine
- G
guanosine
- Gln
glutamine
- Gly
glycine
- Kb
kilobase(s)
- KDa
kilodalton(s)
- Km
kanamycin
- LB
Luria-Bertani broth
- Leu
leucine
- Mb
megabase
- O/N
overnight
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PIA
Pseudomonas isolation agar
- Pro
proline
- Ser
serine
- SDS
sodium dodecyl sulfate
- T
thymidine
- Tc
tetracycline
- Tn
transposon
- Trp or W
tryptophan
- Tyr
tyrosine
- V
valine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Balasubramanian D, Kong KF, Jayawardena SR, Leal SM, Sautter RT, Mathee K. Co-regulation of {beta}-lactam resistance, alginate production and quorum sensing in Pseudomonas aeruginosa. J Med Microbiol. 2011;60:147–156. doi: 10.1099/jmm.0.021600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin NE, Dombroski AJ. Isolation and characterization of mutations in region 1.2 of Escherichia coli sigma70. Mol Microbiol. 2001;42:427–437. doi: 10.1046/j.1365-2958.2001.02642.x. [DOI] [PubMed] [Google Scholar]
- Bass S, Gu Q, Christen A. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer AS, Speert DP, Park S, Tu J, Witt M, Nast CC, Norman DC. Functional role of mucoid exopolysaccharide (alginate) in antibiotic-induced and polymorphonuclear leukocyte-mediated killing of Pseudomonas aeruginosa. Infect Immun. 1991;59:302–308. doi: 10.1128/iai.59.1.302-308.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bolivar F, Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Schurr MJ, Yu H, Rowen DW, Deretic V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 1997;143:3473–3480. doi: 10.1099/00221287-143-11-3473. [DOI] [PubMed] [Google Scholar]
- Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- Campbell JI, Ciofu O, Hoiby N. Pseudomonas aeruginosa isolates from patients with cystic fibrosis have different beta-lactamase expression phenotypes but are homogeneous in the ampC,ampR genetic region. Antimicrob Agents Chemother. 1997;41:1380–1384. doi: 10.1128/aac.41.6.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Ohman DE. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990;172:2894–2900. doi: 10.1128/jb.172.6.2894-2900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, May TB, Chakrabarty AM, Misra TK. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene. 1991;107:1–10. doi: 10.1016/0378-1119(91)90290-r. [DOI] [PubMed] [Google Scholar]
- Darzins A, Chakrabarty AM. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Schurr MJ, Boucher JC, Martin DW. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filip C, Fletcher G, Wulff JL, Earhart CF. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Kunkel B, De Vos GF, Signer ER. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Ohman DE. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988a;170:1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Ohman DE. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988b;170:3228–3236. doi: 10.1128/jb.170.7.3228-3236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen B, Koch C, Hoiby N. Antibiotic treatment of initial colonization with Pseudomonas aeruginosa postpones chronic infection and prevents deterioration of pulmonary function in cystic fibrosis. Pediatr Pulmonol. 1997;23:330–335. doi: 10.1002/(sici)1099-0496(199705)23:5<330::aid-ppul4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Fyfe JA, Govan JR. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol. 1980;119:443–450. doi: 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- Garrett ES, Perlegas D, Wozniak DJ. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giwercman B, Jensen ET, Hoiby N, Kharazmi A, Costerton JW. Induction of beta-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother. 1991;35:1008–1010. doi: 10.1128/aac.35.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JB, Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992;6:59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Goldberg JB, Gorman WL, Flynn JL, Ohman DE. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JB, Ohman DE. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J Bacteriol. 1987;169:1593–1602. doi: 10.1128/jb.169.4.1593-1602.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978;4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- Govan JR, Harris GS. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. [PubMed] [Google Scholar]
- Grabert E, Wingender J, Winkler UK. An outer membrane protein characteristic of mucoid strains of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1990;56:83–87. doi: 10.1111/j.1574-6968.1990.tb04127.x. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hara H, Nishimura Y, Kato J, Suzuki H, Nagasawa H, Suzuki A, Hirota Y. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5882–5889. doi: 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ID, Rehman ZU, Rehm BH. Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl Environ Microbiol. 2010;76:1806–1812. doi: 10.1128/AEM.02945-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O’Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW, Matsumoto M. Pseudomonas aeruginosa PAO. In: O’Brien SJ, editor. Genetic Maps. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1984. pp. 194–197. [Google Scholar]
- Huang B, Whitchurch CB, Croft L, Beatson SA, Mattick JS. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb Comp Genomics. 2000;5:189–203. doi: 10.1089/omi.1.2000.5.189. [DOI] [PubMed] [Google Scholar]
- Izard JW, Rusch SL, Kendall DA. The amino-terminal charge and core region hydrophobicity interdependently contribute to the function of signal sequences. J Biol Chem. 1996;271:21579–21582. doi: 10.1074/jbc.271.35.21579. [DOI] [PubMed] [Google Scholar]
- Jain S, Ohman DE. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH. Transitions, transversions, and the molecular evolutionary clock. J Mol Evol. 1987;26:87–98. doi: 10.1007/BF02111284. [DOI] [PubMed] [Google Scholar]
- Kamath S, Kapatral V, Chakrabarty AM. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol. 1998;30:933–941. doi: 10.1046/j.1365-2958.1998.01121.x. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Silber KR, Downard KM, Papayannopoulos IA, Biemann K, Sauer RT. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci. 1995;4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazmi A. Interactions of Pseudomonas aeruginosa proteases with the cells of the immune system. Antibiot Chemother. 1989;42:42–49. doi: 10.1159/000417602. [DOI] [PubMed] [Google Scholar]
- Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lesley SA, Burgess RR. Characterization of the Escherichia coli transcription factor sigma 70: localization of a region involved in the interaction with core RNA polymerase. Biochemistry. 1989;28:7728–7734. doi: 10.1021/bi00445a031. [DOI] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGeorge J, Korolik V, Morgan AF, Asche V, Holloway BW. Transfer of a chromosomal locus responsible for mucoid colony morphology in Pseudomonas aeruginosa isolated from cystic fibrosis patients to P. aeruginosa PAO. J Med Microbiol. 1986;21:331–336. doi: 10.1099/00222615-21-4-331. [DOI] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Annals of Mathematical Statistics. 1947;18:50–60. [Google Scholar]
- Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993a;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993b;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- Mathee K, Kharazmi A, Hoiby N. Role of exopolysaccharide in biofilm matrix formation: The alginate paradigm. Norwich, U.K: Horizon Scientific Press; 2002. [Google Scholar]
- Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. Proc Int Conf Intell Syst Mol Biol. 1997;5:187–190. [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Nagai H, Shimamoto N. Regions of the Escherichia coli primary sigma factor sigma70 that are involved in interaction with RNA polymerase core enzyme. Genes Cells. 1997;2:725–734. doi: 10.1046/j.1365-2443.1997.1600357.x. [DOI] [PubMed] [Google Scholar]
- Nagasawa H, Sakagami Y, Suzuki A, Suzuki H, Hara H, Hirota Y. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J Bacteriol. 1989;171:5890–5893. doi: 10.1128/jb.171.11.5890-5893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Mathee K, McPherson CJ, DeVries CA, Ma S, Wozniak DJ, Franklin M. ASM Press. 1996. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa; pp. 472–483. [Google Scholar]
- Pedersen SS. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 1992;28:1–79. [PubMed] [Google Scholar]
- Peitsch MC. Protein modeling by E-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BH, Boheim G, Tommassen J, Winkler UK. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J Bacteriol. 1994a;176:5639–5647. doi: 10.1128/jb.176.18.5639-5647.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BH, Grabert E, Hein J, Winkler UK. Antibody response of rabbits and cystic fibrosis patients to an alginate-specific outer membrane protein of a mucoid strain of Pseudomonas aeruginosa. Microb Pathog. 1994b;16:43–51. doi: 10.1006/mpat.1994.1004. [DOI] [PubMed] [Google Scholar]
- Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology. 2005;151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. Bacterial alginates: from biosynthesis to applications. Biotechnol Lett. 2006;28:1701–1712. doi: 10.1007/s10529-006-9156-x. [DOI] [PubMed] [Google Scholar]
- Rouviere PE, De Las Penas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Schurr MJ, Martin DW, Mudd MH, Deretic V. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J Bacteriol. 1994;176:3375–3382. doi: 10.1128/jb.176.11.3375-3382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber KR, Keiler KC, Sauer RT. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C-termini. Proc Natl Acad Sci U S A. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Wu H, Ciofu O, Kong KF, Hoiby N, Rygaard J, Kharazmi A, Mathee K. Pseudomonas aeruginosa alginate is refractory to Th1 immune response and impedes host immune clearance in a mouse model of acute lung infection. J Med Microbiol. 2003;52:731–740. doi: 10.1099/jmm.0.05122-0. [DOI] [PubMed] [Google Scholar]
- Storey DG, Ujack EE, Mitchell I, Rabin HR. Positive correlation of algD transcription to lasB and lasA transcription by populations of Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Infect Immun. 1997;65:4061–4067. doi: 10.1128/iai.65.10.4061-4067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Takase I, Ishino F, Wachi M, Kamata H, Doi M, Asoh S, Matsuzawa H, Ohta T, Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987;169:5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren WF, Billeter SR, Eising AA, Hunenberger PH, Kruger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 Manual and User Guide. 1996 [Google Scholar]
- Winsor GL, Lo R, Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock RE, Brinkman FS. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Involvement of the alginate algT gene and integration host factor in the regulation of the Pseudomonas aeruginosa algB gene. J Bacteriol. 1993;175:4145–4153. doi: 10.1128/jb.175.13.4145-4153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Sprinkle AB, Baynham PJ. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol. 2003;185:7297–7300. doi: 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Roberts JW. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc Natl Acad Sci U S A. 1989;86:5301–5305. doi: 10.1073/pnas.86.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]