Significance
Lipoxygenases are lipid-peroxidizing enzymes that have been classified according to their reaction specificity. ALOX15 (12/15-lipoxygenase) has been implicated in inflammatory resolution via biosynthesis of antiinflammatory and proresolving lipoxins. We found that lower mammals including lower primates express arachidonic acid 12-lipoxygenating ALOX15 orthologs, whereas higher primates express 15-lipoxygenating enzymes. Gibbons constitute the missing link interconnecting 12- and 15-lipoxygenating ALOX15 orthologs. To explore the evolutionary driving force for this specificity alteration, we quantified the lipoxin synthase activity of 12- and 15-lipoxygenating ALOX15 orthologs and observed that the lipoxin synthase activities of 15-lipoxygenating enzymes were significantly higher. These results suggest an evolution of ALOX15 specificity, which optimizes the biosynthetic capacity for antiinflammatory and proresolving lipoxins.
Keywords: eicosanoids, lipoxygenase, evolution, inflammation, protein design
Abstract
ALOX15 (12/15-lipoxygenase) orthologs have been implicated in maturational degradation of intracellular organelles and in the biosynthesis of antiinflammatory and proresolving eicosanoids. Here we hypothesized that lower mammals (mice, rats, pigs) express 12-lipoxygenating ALOX15 orthologs. In contrast, 15-lipoxygenating isoforms are found in higher primates (orangutans, men), and these results suggest an evolution of ALOX15 specificity. To test this hypothesis we first cloned and characterized ALOX15 orthologs of selected Catarrhini representing different stages of late primate evolution and found that higher primates (men, chimpanzees) express 15-lipoxygenating orthologs. In contrast, lower primates (baboons, rhesus monkeys) express 12-lipoxygenating enzymes. Gibbons, which are flanked in evolution by rhesus monkeys (12-lipoxygenating ALOX15) and orangutans (15-lipoxygenating ALOX15), express an ALOX15 ortholog with pronounced dual specificity. To explore the driving force for this evolutionary alterations, we quantified the lipoxin synthase activity of 12-lipoxygenating (rhesus monkey, mouse, rat, pig, humIle418Ala) and 15-lipoxygenating (man, chimpanzee, orangutan, rabbit, ratPhe353Ala) ALOX15 variants and found that, when normalized to their arachidonic acid oxygenase activities, the lipoxin synthase activities of 15-lipoxygenating ALOX15 variants were more than fivefold higher (P < 0.01). Comparative molecular dynamics simulations and quantum mechanics/molecular mechanics calculations indicated that, for the 15-lipoxygenating rabbit ALOX15, the energy barrier for C13-hydrogen abstraction (15-lipoxygenation) was 17 kJ/mol lower than for arachidonic acid 12-lipoxygenation. In contrast, for the 12-lipoxygenating Ile418Ala mutant, the energy barrier for 15-lipoxygenation was 10 kJ/mol higher than for 12-lipoxygenation. Taken together, our data suggest an evolution of ALOX15 specificity, which is aimed at optimizing the biosynthetic capacity for antiinflammatory and proresolving lipoxins.
Lipoxygenases (LOXs) form a diverse family of lipid-peroxidizing enzymes that catalyze the specific oxygenation of polyenoic fatty acids to their corresponding hydroperoxides (1, 2). Genomic LOX sequences occur in two domains (Bacteria, Eukarya) of terrestrial life (3). In mammals, LOXs have been implicated in the biosynthesis of lipid mediators (4, 5), but they may also play a role in cell differentiation (6, 7), apoptosis (8) and pathogenesis of inflammatory (9), hyperproliferative (10), and neurological (11) disorders. Targeted gene inactivation in mice provided useful information on the functionality of Alox12B (12R-lipoxygenase) (12) and Aloxe3 (epidermal lipoxygenase-3) (13), but Alox5 (5-lipoxygenase)- (14), Alox15- (15), and Alox12-deficient mice (16) do not show major defects unless challenged otherwise. In the human genome, six functional LOX genes have been identified, which encode for six distinct LOX isoforms (17).
Traditionally, mammalian LOXs have been classified according to their reaction specificity with arachidonic acid into 5-LOX, 12-LOX, and 15-LOX, but this classification has several problems (18). For those isoforms exhibiting their biological functions via the formation of bioactive mediators, the reaction specificity is of major biological importance. For instance, ALOX5 orthologs oxygenate arachidonic acid specifically to (6E,8Z,11Z,14Z)-5-hydroperoxy-6,8,11,14-eicosatetraenoic acid (5S-HpETE), which is further converted to various leukotrienes of the 5,6-series (4). In contrast, 15-lipoxygenating LOX isoforms are not capable of forming leukotrienes of the 5,6-series. Similarly, 12-lipoxygenating enzymes have been implicated in hepoxilin biosynthesis (5), but ALOX5 orthologs are not involved.
Although mammalian ALOX15 orthologs have been known for more than 40 y, their biological role is still a matter of discussion. The enzymes have been implicated in cell differentiation (6, 7) and in the pathogenesis of different diseases (1, 19), but there is no unifying concept for their functionality. Compared with other LOX-isoforms, mammalian ALOX15 orthologs are somewhat special. First, they are capable of oxygenating arachidonic acid to 12-HpETE and 15-HpETE in variable amounts (20–22). The molecular basis for this dual specificity has been explored (23), and three critical amino acids have been identified (i.e., “triad concept”). Alterations in the side-chain geometry of the triad determinants impacted the specificity of ALOX15 orthologs of men (24), extinct human subspecies (25), various nonhuman primates (22), rabbits (26), mice (27), rats (28), and pigs (29). Second, ALOX15 orthologs are capable of oxygenating phospholipids and cholesterol esters (30, 31), and thus, these LOX isoforms can modify biomembranes and lipoproteins. Third, highly developed primates such as modern and extinct humans (25, 32, 33), as well as orangutans (22, 34), express 15-lipoxygenating ALOX15 orthologs, whereas less developed mammals (21, 35–38), including lower primates, have 12-lipoxygenating enzymes. Although no functional data are currently available for most mammalian ALOX15 orthologs, multiple sequence alignments suggested that their reaction specificity was altered during late primate evolution from 12- to 15-lipoxygenation (3).
To test this “evolutionary concept of ALOX15 specificity” (3), we cloned the ALOX15 orthologs of selected primates and found that the evolutionary switch from 12- to 15-lipoxygenation occurred between Cercopithecidae and Hominidae. Moreover, we observed that 15-lipoxygenating ALOX15 orthologs exhibit a significantly higher biosynthetic capacity for antiinflammatory and proresolving lipoxins, suggesting that the targeted switch in enzyme specificity was aimed at optimizing the immune response.
Results
Prediction of Reaction Specificity of Mammalian ALOX15 Orthologs.
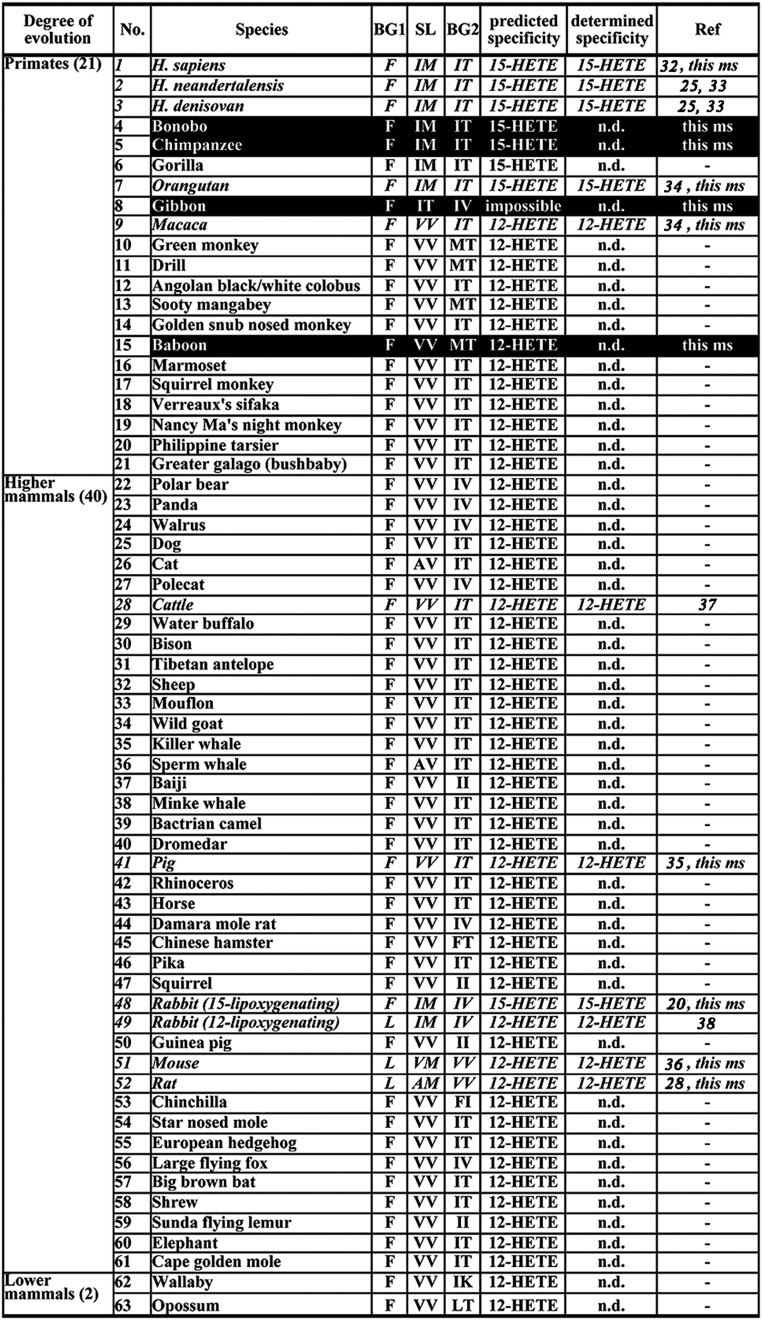
The evolutionary concept of alox15 specificity (3) suggests that mammals ranked low in evolution (mice, rats, pigs, cattle), including lower primates (rhesus monkeys), express 12-lipoxygenating ALOX15 orthologs. In contrast, higher primates (recent and extinct humans, orangutans) express 15-lipoxygenating isoforms (3). To test this concept, to characterize its mechanistic basis, and to explore the possible biological relevance of the evolutionary specificity alterations we used a research strategy involving multiple sequence alignments, cloning, and functional characterization of ALOX15 orthologs and in silico molecular dynamics (MD) simulations. First, we extracted cDNA sequences of ALOX15 orthologs from three major genomic databases. From the list of hits, incomplete sequences were eliminated and multiple amino acid alignments were carried out. A total of 63 complete ALOX15 sequences were retrieved for which all triad determinants have been sequenced (Fig. 1). The degree of amino acid conservation to human ALOX15 varied between 60% and 99% (SI Appendix, Table S1), and all sequences involved the iron liganding amino acids. Next, we compared the triad determinants [Sloane determinants (SL), Borngräber determinants 1 (BG1), Borngräber determinants 2 (BG2)] and found that, in lower mammals, at least one of the two major triad determinants (SL, BG1) is occupied by a small amino acid (Leu instead of Phe at the BG1 position, Val-Val, Ala-Val, Val-Met, Ala-Met instead of Ile-Met at the SL position). These data suggest that, according to the triad concept (23, 39), the ALOX15 orthologs of these mammals exhibit dominant 12-lipoxygenating activities. The only exception was the rabbit. For this species, two distinct ALOX15 cDNAs can be retrieved, and previous studies indicated pronounced differences in the reaction specificity of these ALOX15 isoforms (38).
Fig. 1.
Triad determinants of mammalian ALOX15 orthologs including predicted and measured reaction specificity. The triad determinants of ALOX15 orthologs were retrieved from the databases. Only those entries were considered for further evaluation, for which the triad determinants have completely been sequenced. ALOX15 orthologs, for which the reaction specificity has been determined experimentally, are indicated in italic letters. On black background, those ALOX15 orthologs are indicated for which the reaction specificity has been determined for the first time in this study to our knowledge.
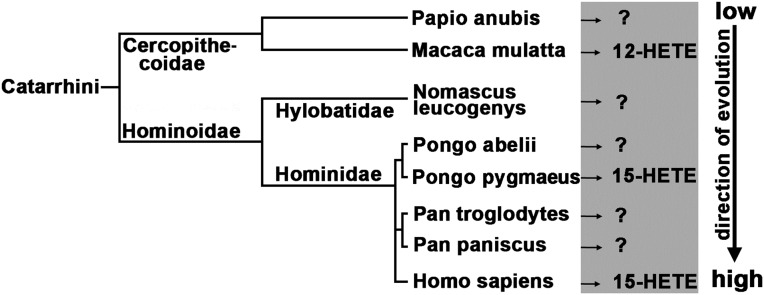
Summarizing the currently available data on the reaction specificity of mammalian ALOX15 orthologs (Fig. 1), one may conclude that the evolutionary change from 12- to 15-lipoxygenation has happened during later primate evolution (Fig. 2). Macaca mulatta is the most highly developed nonhuman primate for which a 12-lipoxygenating ALOX15 has been identified, and Pongo pygmaeus is the lowest nonhuman primate for which a 15-lipoxygenating ALOX15 has been described (22, 34). Unfortunately, the ALOX15 ortholog of the gibbon (Nomascus leucogenys), which is flanked in evolution by M. mulatta and P. pygmaeus, has not been specified. To test the evolutionary concept of ALOX15 specificity, we addressed the following problems: (i) What is the reaction specificity of the ALOX15 ortholog of Papio anubis, which precedes M. mulatta during primate evolution? The evolutionary concept of ALOX15 specificity suggests a 12-lipoxygenating ortholog. (ii) What is the reaction specificity of the ALOX15 ortholog of N. leucogenys? Such functional data are of particular interest (Fig. 2) because N. leucogenys is flanked in evolution by M. mulatta (12-lipoxygenating ALOX15) and P. pygmaeus (15-lipoxygenating ALOX15). (iii) The ALOX15 orthologs of Pongo abelii, Pan troglodytes, and Pan paniscus should constitute 15-lipoxygenating enzymes, but their reaction specificities have not yet been determined.
Fig. 2.
Simplified schematic view of late primate evolution. The main arachidonic acid oxygenation products are given in the gray shaded area. Unidentified product specificities are indicated by the question marks.
Reaction Specificity of ALOX15 Orthologs During Late Primate Evolution.
To characterize the later stages of mammalian ALOX15 evolution, we first compared the amino acid sequences of selected highly developed mammals with that of the human enzyme (Table 1). The corresponding sequence of the ancient human subspecies Homo neanderthalensis and Homo denisovan are 99.7% and 99.5% identical to that of modern humans (Homo sapiens), and mutagenesis data suggested that neither of the observed amino acid exchanges altered the reaction specificity (25). Thus, the two extinct human subspecies express a 15-lipoxygenating ALOX15 ortholog. Chimpanzees (Pan) are the closest living relatives of modern humans. When we compared the amino acid sequences of P. paniscus ALOX15 with that of P. troglodytes, we did not find any differences. Comparison with human ALOX15 indicated a 99.7% amino acid identity. Here only two different amino acids were detected (Table 1), and these exchanges were unlikely to be of functional relevance. The two orangutan subspecies (P. abelii, P. pygmaeus), which have identical amino acid sequences, share a 98.6% amino acid identity with the H. sapiens ortholog (eight different amino acids). For gibbons (N. leucogenys), rhesus monkeys (M. mulatta), and baboons (P. anubis), significantly lower degrees of amino acid conservation (Table 1) were calculated.
Table 1.
Degree of sequence conservation between ALOX15 orthologs of higher primates
| Species | Amino acid identity, % | Amino acid exchanges |
| H. sapiens | 100 | 0 |
| H. neanderthalensis | 99.7 | 2 |
| H. denisovan | 99.5 | 3 |
| P. paniscus | 99.7 | 2 |
| P. troglodytes | 99.7 | 2 |
| P. abelii | 98.6 | 8 |
| P. pygmaeus | 98.6 | 8 |
| N. leucogenys | 96.8 | 21 |
| M. mulatta | 94.6 | 35 |
| P. anubis | 95.0 | 35 |
The amino acid sequences of P. paniscus and P. troglodytes among Pan and of P. abelii and P. pygmaeus among Pongo are identical.
To predict the reaction specificities of the ALOX15 orthologs of these Catarrhini, we compared the amino acid sequences of the triad determinants (Fig. 3) and found that the sequences of the three amino acid clusters for H. sapiens, H. neanderthalensis, H. denisovan, P. troglodytes, P. paniscus, P. pygmaeus, and P. abelii were identical (Fig. 3). Thus, according to the triad concept, these enzymes should exhibit major 15-lipoxygenating activity, which is consistent with the functional data shown previously for the enzymes of H. sapiens and P. pygmaeus (22). The ALOX15 orthologs of M. mulatta and Mus musculus contain at least one small amino acid at the Sloane positions (i.e., SL), classifying these enzymes as 12-lipoxygenating ALOX15 variants. Moreover, in mice, the Borngräber 1 position (i.e., BG1) is occupied by a Leu, the side chain of which is less space-filling than the side chain of Phe present in 15-lipoxygenating ALOX15 orthologs. Baboons (P. anubis) have almost identical sequences around the triad determinants as rhesus monkeys (M. mulatta; Fig. 3), and thus, on the basis of sequence comparison, arachidonic acid 12-lipoxygenation could be predicted. The gibbon (N. leucogenys) ALOX15 ortholog shows interesting sequence differences at the triad determinants (Fig. 3), which made predictions of the reaction specificity risky. At the BG1 position, a bulky Phe is present, suggesting arachidonic acid 15-lipoxygenation. At the SL positions, Ile418 is conserved, but, instead of Met419 (van der Waals radius of 124 Å), a Thr (van der Waals radius of 93 Å) is found, suggesting a gain of space at the active site. At the BG2 position, the Ile593 is conserved, but a Val instead of a Thr occupies the adjacent position. This situation does not allow a precise prediction of the reaction specificity on the basis of the triad concept, and thus, direct experimental data are required.
Fig. 3.
Partial sequence alignment of ALOX15 orthologs of selected mammals. The sequence regions of the triad determinants (BG1, SL, BG2) are given, and the critical amino acids are indicated in bold. The amino acid differences with potential impact on the reaction specificity are labeled on gray background.
To obtain such data, we cloned the ALOX15 orthologs of P. anubis, P. abelii, and N. leucogenys, expressed them as recombinant proteins, and performed in vitro activity assays. As predicted, ALOX15 of P. anubis converted arachidonic acid mainly to 12-H(p)ETE (Fig. 4A), whereas the chimpanzee (P. troglodytes) ortholog produced mainly 15-H(p)ETE (Fig. 4B). Interestingly, the gibbon ALOX15 oxygenated arachidonic acid to a product mixture consisting of almost equal amounts of 12- and 15-H(p)ETE (Fig. 4C). Such pronounced dual reaction specificity of arachidonic acid oxygenation has never been described for any naturally occurring LOX isoform to our knowledge. For comparison (i.e., positive control), the specificity of the human ALOX15 was also tested (Fig. 4D), and our data confirmed dominant arachidonic acid 15-lipoxygenation. Because RP-HPLC does not reliably separate 12- and 8-hydroxy eicosatetraenoic acid (HETE) we carried out additional straight-phase (SP)-HPLC, which confirmed our conclusions. For more detailed quantification, for determination of the relative specific activities, and for statistical evaluation of the interspecies differences, we repeated expression and activity assays several times (Materials and Methods), and the results are summarized in Table 2. All enzyme preparations exhibit similar relative specific activities (the specific activity of the human enzyme was set at 100%). The ALOX15 orthologs of humans and chimpanzees are major 15-lipoxygenating enzymes. In contrast, gibbons express an ALOX15, which converts arachidonic acid to almost similar amounts of 12- and 15-H(p)ETE. To make sure that the products quantified were of enzymatic origin, we determined the enantiomer ratio and found that both 12- and 15-H(p)ETE were mainly the S isomer (Table 2).
Fig. 4.
Positional specificity of arachidonic acid oxygenation by ALOX15 orthologs representing late primate evolution. Arachidonate oxygenase activity assays and RP-HPLC analysis were carried out as described in Materials and Methods. Each chromatogram was scaled for the highest HETE peak. (A) Products formed by baboon ALOX15. (B) Products formed by chimpanzee ALOX15. (C) Products formed by gibbon ALOX15. (D) Products formed by human ALOX15.
Table 2.
Product pattern of primate ALOX15 orthologs
| Species | Relative specific activity, %* | Product share, % | |
| 15-HETE | 12-HETE | ||
| H. sapiens | 100 ± 41 | 78.7 ± 1.6 (99:1) | 21.3 ± 1.6 (98:2) |
| P. troglodytes | 135 ± 45 | 80.3 ± 2.1 (98:2) | 19.7 ± 2.1 (98:2) |
| N. leucogenys | 76 ± 14 | 50.5 ± 7.7 (98:2) | 50.0 ± 7.7 (99:1) |
| P. anubis | 37 ± 5 | 21.9 ± 2.5 (ND) | 78.1 ± 2.5 (ND) |
ALOX15 preparation and HPLC analysis of the reaction products were carried out as described in Materials and Methods. Four expression experiments were performed, and each sample was HPLC-quantified twice. Mean ± SD of the relative shares of the two major reaction products are given. Enantiomer compositions are given in parentheses. ND, not determined.
The relative specific activity was estimated by quantification of the product formation (RP-HPLC), correcting this value for the amount of LOX protein that was determined by immunoblotting.
Mutagenesis Studies on Gibbon ALOX15 Confirm Applicability of the Triad Concept.
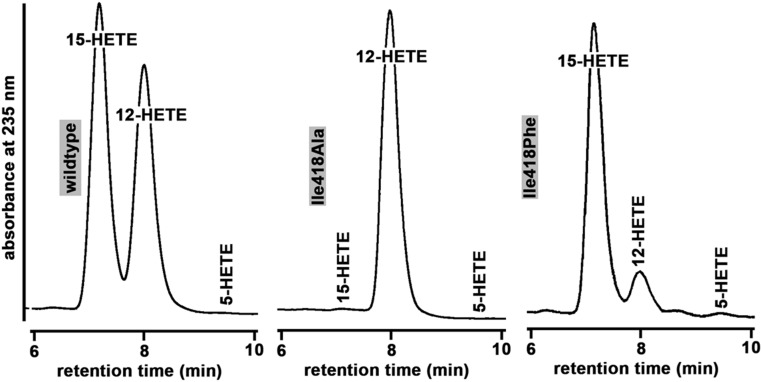
To test the applicability of the triad concept for gibbon ALOX15 and to quantify the relative contribution of the amino acid exchanges responsible for the high degree of dual positional specificity, we performed a number of site-directed mutagenesis studies. For this purpose, we first partly “gibbonized” the human ALOX15 to induce an increased share of 12-H(p)ETE formation (Met419Thr, Thr594Val). Here, we found that, as expected, the Met419Thr exchange increased the share of 12-H(p)ETE formation (Table 3). In fact, for this mutant, 12-H(p)ETE was the major arachidonic acid oxygenation product. In contrast, Thr594Val exchange did not lead to major alterations in the positional specificity (Table 3). These data suggest that the unusual positional specificity of the gibbon enzyme is mainly a result of the Met419Thr exchange. Next, we inserted at the major SL position a small Ala (Ile418Ala). An identical mutation in rabbit ALOX15 converted this enzyme almost completely to a 12-lipoxygenating variant (23). When we performed this mutation with the ALOX15 ortholog of gibbon (Fig. 5 and Table 3), orangutans, and chimpanzee (Table 4), we also observed dominant 12-lipoxygenation. Finally, we introduced a bulky Phe at this position (Ile418Phe). As expected, we found major 15-lipoxygenation (Fig. 5). Taken together, these mutagenesis data indicate that the triad concept is fully applicable for gibbon ALOX15.
Table 3.
Product pattern of ALOX15 mutants
| Species (mutant) | Relative product share, % | |
| 15-HETE | 12-HETE | |
| Human (WT) | 78.7 ± 1.6 | 21.3 ± 1.6 |
| Human (Met419Thr) | 34.7 ± 8.7 | 65.3 ± 8.7 |
| Human (Thr594Val) | 81.5 ± 1.2 | 18.6 ± 0.9 |
| Gibbon (WT) | 46.1 ± 5.2 | 53.9 ± 5.2 |
| Gibbon (Ile418Ala) | <1 | 99 ± 14 |
| Gibbon (Ile418Phe) | 83.6 ± 3.5 | 16.4 ± 3.5 |
Human and gibbon ALOX15 orthologs were expressed in E. coli. The product pattern was analyzed by consecutive RP-HPLC, SP-HPLC, and chiral-phase HPLC. Four independent expression experiments were performed, and each sample was quantified twice. Means ± SD of the relative shares of the two major reaction products are given.
Fig. 5.
Positional specificity of arachidonic acid oxygenation by gibbon ALOX15 mutants. Arachidonate oxygenase activity assay of gibbon ALOX15 and RP-HPLC analysis of the oxygenation products were carried out as described in Materials and Methods. Each chromatogram was scaled for the highest HETE peak.
Table 4.
Reaction specificity of mammalian ALOX15 orthologs and their Ile418Ala mutants
| Species/variant | Product share, % | |
| 12-H(p)ETE | 15-H(p)ETE | |
| Oryctolagus cuniculus (rabbits) | ||
| WT | 3 | 97 |
| Ile418Ala | 92 | 8 |
| H. sapiens (humans) | ||
| WT | 15 | 85 |
| Ile417Ala* | 94 | 6 |
| P. pygmaeus (orangutans) | ||
| WT | 14 | 86 |
| Ile417Ala* | 82 | 18 |
| P. troglodytes (chimpanzee) | ||
| WT | 20 | 80 |
| Ile419Ala | 99 | 1 |
ALOX15 expression and product analysis were carried out as described in Materials and Methods.
In the primary structure of human and orangutan ALOX15 orthologs, a Glu, which is present in the rabbit enzyme in the linker peptide interconnecting the N-terminal and the C-terminal domains, is lacking. Thus, amino acid numbering is shifted by one residue in comparison with the rabbit enzyme.
A Single Amino Acid Exchange Mimics the Evolutionary Alterations in Reaction Specificity.
Multiple mutagenesis studies (23, 24, 27, 29, 40) on a number of mammalian ALOX15 orthologs have previously confirmed the triad concept, and an Ala-scan of the triad determinants of rabbit ALOX15 indicated that a single Ile418Ala exchange is sufficient to completely convert the reaction specificity of this enzyme from 15- to 12-lipoxygenation (23). Thus, according to these data, a single point mutation is able to mimic the evolutionary switch in reaction specificity. To test whether similar dramatic changes in reaction specificity can be induced in other ALOX15 orthologs, we expressed human, orangutan, and chimpanzee ALOX15 and their corresponding Ile418Ala mutants. As indicated in Table 4, all WT enzymes were dominantly 15-lipoxygenating, whereas 12-H(p)ETE was the almost exclusive arachidonic acid oxygenation product of the Ile418Ala mutants. These data suggest that the Ile418Ala mutant of rabbit ALOX15 might be considered a suitable model for exploring the molecular basis for the evolutionary switch in reaction specificity of ALOX15 orthologs.
15-Lipoxygenating ALOX15 Orthologs Exhibit Significantly Higher Lipoxin-Synthesizing Capacities.
Our sequence and specificity data suggest an evolution-dependent alteration in reaction specificity, but the evolutionary driving forces for this process remained unclear. ALOX15 orthologs have been implicated in the biosynthesis of antiinflammatory and proresolving mediators (41–43), but it has not been explored whether the lipoxin synthase activities of 12- and 15-lipoxygenating ALOX15 orthologs are different. To address this point, we expressed five different 12-lipoxygenating (rhesus monkey, mouse, rat, pig, humIle418Ala) and five different 15-lipoxygenating (man, chimpanzee, orangutan, rabbit, ratPhe353Leu) ALOX15 variants and confirmed their reaction specificity (Table 5). Next, we tested the lipoxin synthase activity of these ALOX15 variants with two different substrates [5S-HETE, 5S,6R/S-dihydroxy eicosatetraenoic acid (DiHETE)]. Lipoxins can be biosynthesized via a number of different mechanisms, and these metabolic routes involve a concerted action of ALOX5, ALOX15, and/or ALOX12 (44, 45). One of the transcellular scenarios suggests that arachidonic acid is oxygenated by ALOX5, yielding 5S-H(p)ETE, which is subsequently further oxygenated by ALOX12 and/or ALOX15 isoforms to a mixture of LXA4 and LXB4 isomers. To compare the lipoxin-biosynthesizing activity of 12- and 15-lipoxygenating ALOX15 orthologs, we incubated our enzyme preparations with 5S-HETE (30 µM) for 10 min and quantified the formation of LXA4 and LXB4 isomers by RP-HPLC. The chemical identity of the lipoxin isomers was concluded from cochromatography with authentic standards and by comparison of their UV spectra (SI Appendix, Fig. S1). The experimental raw data were then normalized to the arachidonic acid oxygenase activity of the different enzyme preparations, and the relative lipoxin synthase activity of the human ALOX15 ortholog was set at 100%. From Table 5 it can be seen that 15-lipoxygenating ALOX15 orthologs exhibit a fivefold higher lipoxin synthase activity (P < 0.01) compared with the 12-lipoxygenating enzymes. The lower LX-synthase activity of the 12-lipoxygenating ALOX15 orthologs is consistent with the reaction mechanism. These ALOX15 isoforms convert 5-HETE mainly to 5-OH,12-OOH-ETE. This double oxygenation compound constitutes a dead-end product, which cannot be further transferred to lipoxin isomers. In contrast, the major product of 5-HETE oxygenation by 15-lipoxygenating ALOX15 orthologs was 5-OH,15-OOH-ETE, which can subsequently be further converted to lipoxin isomers.
Table 5.
Relative lipoxin synthase activity of mammalian ALOX15 orthologs
| Species | Relative lipoxin synthase activity, % | ||
| 15-/12-ratio | 5-HETE as substrate | 5,6-DiHETE as substrate | |
| 15-lipoxygenating | |||
| Human | 8.1 | 100.0 | 100 |
| Chimpanzee | 8.1 | 118.0 | 145.8 |
| Orangutan | 8.1 | 172.2 | 105.6 |
| Rabbit | 24.0 | 39.5 | 108.6 |
| ratI353F | 13.3 | 197.3 | 262.5 |
| Mean ± SD | 12.3 ± 6.9 | 125.4 ± 62.1* | 144.5 ± 68.4† |
| 12-lipoxygenating | |||
| Macaca | 0.01 | 25.7 | 19.9 |
| Mouse | 0.03 | 36.1 | 1.5 |
| Rat | 0.26 | 8.4 | 0.0 |
| Pig | 0.04 | 35.4 | 61.1 |
| humI418A | 0.11 | 29.2 | 2.1 |
| Mean ± SD | 0.09 ± 0.10 | 27.0 ± 11.2* | 17.1 ± 25.9† |
The relative lipoxin synthase activity of the ALOX15 orthologs was quantified as described in Materials and Methods. For 5S-HETE oxygenation, lipoxin A and lipoxin B isomers were quantified. During 5S,6(S/R)-DiHETE, only lipoxin A isomers were formed.
P = 0.008 by Student's t test.
P = 0.005 by Student's t test.
In parallel, we tested a distinct metabolic route of lipoxin biosynthesis. ALOX5 converts arachidonic acid to leukotriene A4 (5,6 epoxy arachidonic acid), which can further be transformed to leukotriene B4 or C4. Alternatively, it may undergo nonenzymatic epoxide hydrolysis to yield 5S,6(R/S)-DiHETE. The hydrolysis products can subsequently be oxygenated to lipoxin isomers by 12- and 15-lipoxygenating ALOX isoforms. When we incubated the recombinant ALOX15 orthologs with a 1:1 mixture of 5S,6S- and 5S,6R-DiHETE (30 µM) for 10 min and quantified the formation of LXA4 isomers, we found that, when normalized to identical arachidonic acid oxygenase activities, 15-lipoxygenating ALOX15 orthologs exhibit an almost 10-fold higher lipoxin A4 synthesizing capacity than 12-lipoxygenating orthologs (Table 5).
Taken together, these data indicate that 15-lipoxygenating ALOX15 variants exhibit a higher lipoxin synthase activity than the 12-lipoxygenating counterparts when normalized to an identical arachidonic acid oxygenase activity. It should be stressed at this point that our lipoxin synthase activity measurements do not involve detailed kinetic studies, and thus, direct comparison of basic kinetic parameters (KM, Vmax, catalytic efficiency) for the different ALOX15 orthologs was not possible. Moreover, our in vitro data might not adequately mirror the lipoxin synthase activity of the ALOX15 orthologs in vivo. To address this topic, experiments should be carried out with humanized knock-in mice, which express a 15-lipoxygenating mutant of murine Alox15. We are currently in the process of creating such animals.
MD Simulations and Quantum Mechanics/Molecular Mechanics Calculations.
As suggested by the triad concept, the evolutionary switch in reaction specificity might be related to a modified alignment of the substrate fatty acid at the active site of ALOX15 orthologs (2, 24, 46). For 15-lipoxygenating ALOX15 orthologs, the proS-hydrogen bound at carbon 13 (C13) was predicted to be located in close proximity to the iron-bound hydroxyl group functioning as hydrogen abstracting group. In contrast, for the 12-lipoxygenating ALOX15 orthologs, the fatty acid substrates may slide in deeper into the substrate-binding pocket approaching the proS-hydrogen at C10 to the enzyme-bound iron–hydroxyl complex (2). Because no X-ray structures are currently available for ALOX15–substrate complexes and because the geometry-based model of the triad concept does not consider energetic details, we performed MD simulations and quantum mechanics/molecular mechanics (QM/MM) calculations to explore the structure of enzyme–substrate complex for the 15-lipoxygenating rabbit ALOX15 and its 12-lipoxygenating Ile418Ala mutant.
Because of the structural flexibility of the fatty acid backbone, a large number of energetically similar arachidonic acid conformers are bound at the active site of WT rabbit ALOX15 (47), and similar results were obtained here for the 12-lipoxygenating Ile418Ala mutant. Next we screened the arachidonic acid conformers at the active site of the 12-lipoxygenating Ile418Ala mutant for catalytically productive structures using the following filtering criteria: (i) d(H13-OH) or d(H10-OH) ≤ 3.0 Å and (ii) d(C13-OH) > d(H13-OH) or d(C10-OH) > d(H10-OH). These criteria ensure that the C-H bond to be split during hydrogen abstraction is properly aligned in relation to the iron-bound hydroxyl. Applying this strategy, two sets of potentially productive conformers were extracted from our MD trajectories (SI Appendix, Table S5): (i) Structures suitable for proS-H abstraction from C13 (15-lipoxygenation) and (ii) structures suitable for proS-hydrogen abstraction from C10 (12-lipoxygenations). These data indicate that at the active site of the Ile418Ala mutant arachidonic acid conformers are present, which allow both 12- and 15-lipoxygenation. Similar results were previously reported for the 15-lipoxygenating WT enzyme (47). These data indicate that, on the basis of the original geometry-based triad concept, which stresses the binding distances, it may not be possible to reliably discriminate between enzyme–substrate complexes suitable for 12- or 15-lipoxygenation.
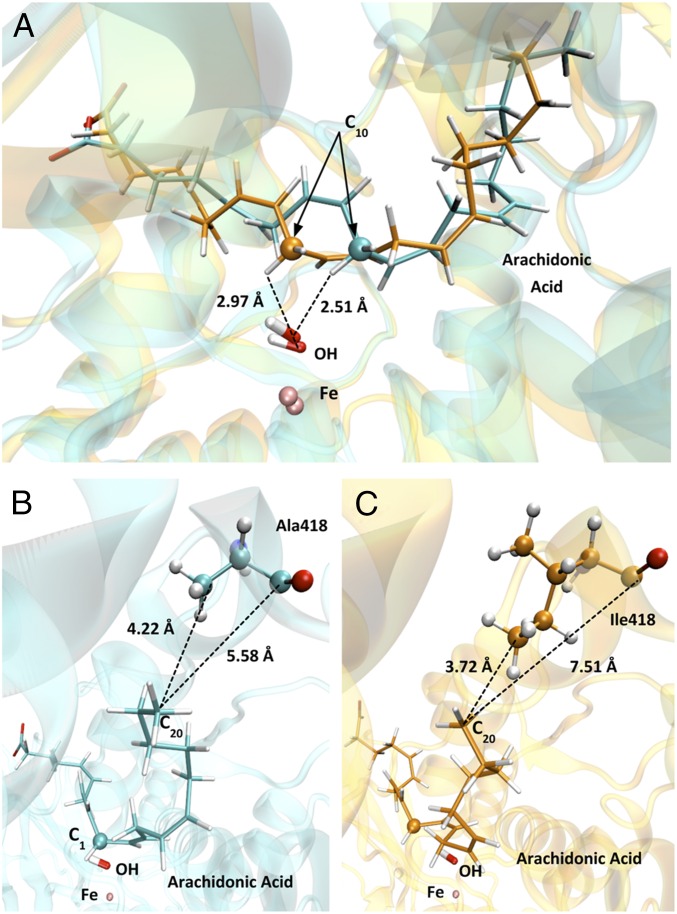
Next, we performed clustering analysis for the 12-lipoxygenating Ile418Ala mutant within the two sets of arachidonic acid conformers (12-lipoxygenating vs. 15-lipoxygenating), which was based on the rmsd of the heavy atoms of the substrate fatty acid. In Fig. 6A, a representative arachidonic acid conformer, which is suitably aligned at the active site of the 15-lipoxygenating WT enzyme, is shown. This complex is overlaid with a conformer, which is aligned for 12-lipoxygenation at the active site of the 12-lipoxygenating Ile418Ala mutant. Consistent with the triad concept, the arachidonic acid molecule has moved in deeper into the substrate-binding pocket to fill the vacancy, which is provided by the Ile418Ala exchange (Fig. 6 B and C and Movie S1).
Fig. 6.
Overlay (A) of WT ALOX15-AA (orange) and Ile418Ala ALOX15-AA (blue) active sites from two snapshots of the MD trajectories of both in silico models. The location of the arachidonic acid molecule with respect to the bottom of the cavity is shown in B and C. Arachidonic acid, the iron-bound hydroxyl ion (OH), and Fe are represented in sticks, and C10, Ala-418, and Ile418 are represented in balls and sticks.
The most populated arachidonic acid conformers observed at the active site of the 12-lipoxygenating Ile418Ala mutant were energetically optimized, and the corresponding reactant minima were located in the QM/MM potential energy surface. From those minima, potential energy profiles for the abstraction of the proS-hydrogen from C10 (H10-abstraction, structures I–X) and of the proS-hydrogen at C13 (H13-abstraction, structures XI–XX) were calculated. In the enzyme–substrate complexes suitable for 12-lipoxygenation (structures I–X), the distances between the proS-hydrogen at C10 and the iron-bound hydroxyl group [d(H10-OH)] range from 2.43 to 3.17 Å. Similarly, the distances between C10 and the iron bound hydroxyl [d(C10-OH)] vary between 3.28 to 3.67 Å (SI Appendix, Table S5). For structures XI to XX, which represent arachidonic acid conformers for 15-lipoxygenation, C13 is also located at reactive distances (SI Appendix, Table S5). These data indicate that, if one considers only the geometric distances, hydrogen abstraction from both C10 and C13 is possible.
Finally, we calculated the potential energy barriers for the transition states derived from the Michaelis complexes for structures I–X (suitable for 12-lipoxygenation) and XI–XX (suitable for 15-lipoxygenation). The energy barriers calculated for C13-hydrogen abstraction (15-lipoxygenation) ranged from 64 to 91 kJ/mol (SI Appendix, Table S6). In contrast, for C10-hydrogen abstraction (12-lipoxygenation), the energy barriers varied between 53 and 125 kJ/mol. To explore whether 12- or 15-lipoxygenation is preferentially catalyzed by the Ile418Ala mutant, the exponential average potential energy barriers (SI Appendix, Computational Methods) for H10- and H13-abstractions were calculated from the individual values given in SI Appendix, Table S6. Here, we found an exponential average potential energy barrier of 59 kJ/mol for H10-abstraction but 69 kJ/mol for H13-abstraction at 300 K. The 10-kJ/mol higher exponential average energy barrier for the transition states of 15-lipoxygenation is consistent with the experimental observation of preferential 12-lipoxygenation of the Ile418Ala mutant. For WT rabbit ALOX15, a 17-kJ/mol higher exponential average energy barrier was calculated for C10-hydrogen abstraction (47).
Summarizing the major finding of our in silico calculation, one may conclude that, for the rabbit ALOX15, the exponential average potential energy barrier for the 12-lipoxygenation is 17 kJ/mol higher than that for 15-lipoxygenation. On the contrary, for the Ile418Ala mutant, the exponential average potential energy barrier for 12-lipoxygenation is 10 kJ/mol lower than that for 15-lipoxygenation, and this difference explains preferential 12-lipoxygenation.
Discussion
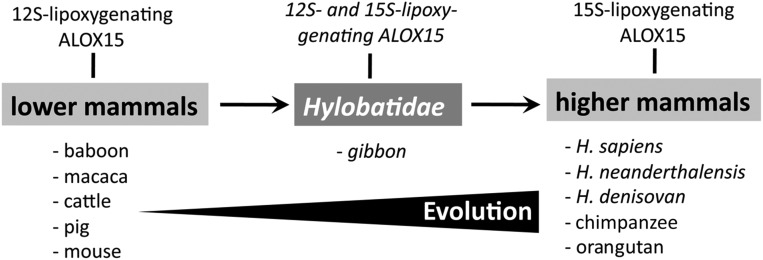
The conventional scenario of LOX classification, which stresses the positional specificity of the enzymes, is misleading because orthologous enzymes are frequently classified in different categories. This problem is particularly obvious for ALOX15 orthologs. Before this study was initiated, it was well established that some mammals (rhesus monkeys, mice, rats, pigs) express 12-lipoxygenating ALOX15 isoforms, whereas the orthologs of other mammals (H. sapiens, H. neanderthalensis, H. denisovan, P. pygmaeus) express 15-lipoxygenating enzymes. A systematic search of the currently available mammalian ALOX15 sequences (Fig. 1) suggested that lower mammals have small amino acids at either of the triad positions and thus, should function as 12-lipoxygenating enzymes (3, 22, 34). In contrast, highly developed mammals (H. sapiens, H. neanderthalensis, H. denisovan, P. pygmaeus) express 15-lipoxygenating enzymes as the triad positions are occupied by more space-filling amino acid residues. Unfortunately, the experimental basis for this evolutionary concept of ALOX15 specificity (3, 22, 34) was rather narrow because key ALOX15 orthologs have not been characterized. The aim of the present study was to express and characterize the ALOX15 orthologs of selected Catarrhini to confirm or disconfirm this concept and to define the evolutionary switching point at which 12-lipoxygenating enzymes have turned into 15-lipoxygenating orthologs. Because the reaction specificity of LOXs is important for LOX classification and their biological roles, answers to these questions appear to be of general interest. By applying a multiple research strategy involving detailed sequence comparison, experimental characterization of recombinant ALOX15 orthologs, multiple mutagenesis studies, and in silico simulations of the enzyme–substrate complex. we confirmed the evolutionary concept of ALOX15 specificity and identified Hylobatidae as the evolutionary switching point (Scheme 1).
Scheme 1.
Evolutionary concept of ALOX15 development. ALOX15 orthologs of lower mammals are 12-lipoxygenating enzymes, whereas higher mammals including humans express 15-lipoxygenating orthologs. Hylobatidae (gibbons) appear to be the evolutionary switching point because they express an ALOX15 ortholog with pronounced dual reaction specificity.
When we searched the major genomic databases for mammalian ALOX15 orthologs, we retrieved a total 63 complete entries (Fig. 1). Applying the triad concept of ALOX15 specificity (23, 39) to these sequences, one can conclude the reaction specificity of ALOX15 orthologs from their primary structures. Currently, specificity data are available for 15 mammalian ALOX15 orthologs (Fig. 1), and their primary structures indicate that all of them follow the triad concept. When we used the triad concept to predict the reaction specificity of the ALOX15 orthologs listed in Fig. 1, we found that, among the 63 primary structures, 62 follow the evolutionary concept of ALOX15 specificity, which suggests that higher primates express 15-lipoxygenating ALOX15 orthologs whereas lower primates and other mammals express 12-lipoxygenating enzymes. The only exception from this rule is the rabbit. In this species, a 12- and a 15-lipoxygenating ALOX15 isoform are expressed in a tissue-specific manner. Originally, the existence of two separate ALOX15 genes has been suggested (38), but completion of the rabbit genome did not confirm this suggestion. In fact, the major publically available genome databases [ENSEMBL (www.ensembl.org) and the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/)] suggest the existence of a single ALOX15 gene, which encodes for a 15-lipoxygenating ALOX15 ortholog. However, in rabbit monocytes, a mRNA encoding for a 12-lipoxygenating enzyme is expressed (38). This mRNA involves a TTA triplet encoding for Leu353, and mutagenesis studies indicated 12-lipoxygenation for this enzyme variant. The mechanistic details for the tissue-specific recoding process remains to be explored. It might well be that, in the future, additional exceptions from the evolutionary concept of ALOX15 specificity will be discovered, but, based on the currently available data, the likelihood of such discoveries is rather low (1.6%).
In addition to ALOX15, there is a dramatic difference in the reaction specificity of ALOX15B when mouse and human orthologs are compared. Human ALOX15B converts arachidonic acid to 15-H(p)ETE, whereas the mouse ortholog forms 8-H(p)ETE (48). Here, we explored whether this difference in reaction specificity of ALOX15B is also a consequence of an evolutionary process. Comparing the amino acid sequences of various mammalian ALOX15B orthologs, we found that there is no systematic development of ALOX15B reaction specificity. In fact, the retrieved sequence data suggest that most mammalian ALOX15B orthologs are arachidonic acid 15-lipoxygenating isoforms (SI Appendix, Table S2). The only exception we found was mouse Alox15b, which carries different amino acids at the critical positions (Jisaka determinants). Even the rat ALOX15B, which is most closely related to the mouse enzyme, should be a 15-lipoxygenating enzyme, as concluded from the sequence data.
To address the question about the driving force behind this evolutionary concept of ALOX15 specificity, we first compared the fatty acid specificity and membrane oxygenase activity of 12- and 15-lipoxygenating ALOX15 orthologs, but did not find conserved differences. However, when we tested the biosynthetic capacity of 12- and 15-lipoxygenating ALOX15 orthologs for antiinflammatory and proresolving lipoxins (Table 4), we found that 15-lipoxygenating ALOX15 orthologs exhibit a higher lipoxin synthase activity. Putting these results into a phylogenetic context, one may conclude that the evolutionary switch from 12- to 15-lipoxygenation was aimed at optimizing the lipoxin synthase activity. The 12-lipoxygenating ALOX15 orthologs of less highly developed mammals are capable of synthesizing proresolving lipoxins, but the 15-lipoxygenating orthologs of higher mammals can do so better. In other words, mammals expressing 15-lipoxygenating ALOX15 orthologs show a more efficient inflammatory resolution and this might confer them an evolutionary advantage. Because resolution actively concludes inflammation as immune response, the evolutionary switch in ALOX15 reaction specificity may be considered as part of a more complex developmental concept optimizing the immune system.
The initial version of the triad concept was based exclusively on geometric parameters (23, 39) stressing binding distances and alterations in the volume of the substrate-binding pocket. Previous MD simulations indicated that, at the active site of rabbit ALOX15, arachidonic acid conformers are present, which are suitable for both 12- and 15-lipoxygenation. In other words, binding distances and formal volume calculations cannot reliably discriminate between 12- or 15-lipoxygenation. However, calculations of the energy landscapes for the enzyme–substrate complexes indicated that, for WT rabbit ALOX15, the average energy barrier for the 15-lipoxygenating complexes is by 17 kJ/mol lower compared with the corresponding value for the 12-lipoxygenating complexes (47). These in silico results are consistent with experimental in vitro data (20). Here, we performed similar calculations for the 12-lipoxygenating Ile418Ala mutant of rabbit ALOX15 and found that the exponential average energy barrier of enzyme–substrate complexes suitable for 15-lipoxygenation was 10 kJ/mol higher than for 12-lipoxygenation, explaining the dominant 12-lipoxygenation of the mutant enzyme. Although these energy calculations do not prove the triad concept, they are consistent with the experimental data and move the concept a step forward. The data advance the simple distance-based hypothesis to an energy-based model, which broadens the mechanistic basis of the entire concept by including kinetic aspects.
Materials and Methods
Chemicals.
The sources of the chemicals used are specified in the SI Appendix.
Database Analysis.
The following genomic databases were used for searches of ALOX15 sequences: NCBI (www.ncbi.nlm.nih.gov/), ENSEMBL (www.ensembl.org/index.html), University of California, Santa Cruz, Genome Bioinformatics (https://genome.ucsc.edu/). The nucleotide sequences of the ALOX15 orthologs of different Catarrhini were extracted from the NCBI genome database with the following accession numbers: M. mulatta (XM_001094627), N. leucogenys (XM_012501351.1), P. abelii (NM_001133877), P. troglodytes (XM_003315313.2), P. paniscus (XM_008962574.1), and P. anubis (XM_009189398.1).
Cloning of the ALOX15 Ortholog from Pan troglodytes and Papio anubis.
The ALOX15 cDNAs from P. troglodytes and P. anubis were prepared by RT-PCR cloning from the blood of two individuals from each species. The methodological details of the cloning process are provided in SI Appendix. Because ALOX15 of P. paniscus has the identical amino acid sequence as P. troglodytes, separate cloning of the P. paniscus ALOX15 cDNA was not necessary.
Cloning of the ALOX15 Ortholog from N. leucogenys (Gibbon).
Because we were not able to obtain blood from this primate species, the complete ALOX15 cDNA was chemically synthesized (Biomatik), which includes a SalI restriction site immediately in front of the starting methionine and a HindIII site behind the stop codon. The synthesis product was inserted into a pME vector (Biomatik) for bacterial amplification. After SalI-HindIII digestion, the 2,000-bp restriction fragment was ligated into the linearized bacterial expression plasmid pET28b. This construct was completely sequenced.
Bacterial and Eukaryotic Expression of ALOX15 Isoforms.
WT and mutant ALOX15 isoforms of different mammalian species (man, chimpanzee, orangutan, gibbon, rhesus monkey, mouse, rat, pig) were expressed as N-terminal his-tag fusion proteins in Escherichia coli or in N2a cells, and the experimental details of expression are provided in the SI Appendix.
Site-Directed Mutagenesis.
Site-directed mutagenesis was carried out by using Pfu UltraII Hot Start 2× PCR Master Mix (Agilent Technologies), followed by DpnI digestion of the parent DNA. For each mutant, as many as five clones were screened for the LOX insert (digestion with a suitable combination of different restriction enzymes), and one clone was sequenced to confirm mutagenesis.
Fatty Acid Oxygenase Activity.
For the activity assay, the sequenced clone was replated, four well-separated colonies were picked, and protein expression was carried out as described earlier. For routine activity assay, different amounts of the stroma-free bacterial lysis supernatant (2–20 µL) were added to 250 µL PBS solution, and arachidonic acid (100 µM final concentration) was added. The mixture was incubated for different time periods (3–15 min at room temperature), the hydroperoxy compounds were reduced with SnCl2, the mixture was acidified to pH 3, and 0.25 mL of ice-cold methanol was added. The protein precipitate was spun down, and aliquots of the clear supernatant were injected directly to RP-HPLC.
Lipoxin Synthase Activity.
Lipoxin synthase activity was assayed by HPLC quantification of lipoxin A4 and B4 isomers formed during the incubation of the enzymes with different substrates. Bacterial lysate supernatants of mammalian ALOX15 orthologs were incubated in PBS solution with 5S-HETE or 5S,6(R/S)-DiHETE (30 µM) in the presence of 3 µM linoleic acid that served as enzyme activator (total assay volume, 0.1 mL). After 10 min of incubation at room temperature, the hydroperoxy derivatives were reduced (5 µL of an SnCl2 solution of 10 mg/mL). Then, 0.1 mL of ice cold MeOH was added, protein precipitate was spun down, and aliquots of the clear supernatant were injected to HPLC analysis. The lipoxin isomers formed were quantified, recording the absorbance at 300 nm by using a molar extinction coefficient of 53,000 (M cm)−1. The lipoxin synthase activity of the different enzyme preparations was first normalized for the arachidonic acid oxygenase activity of the enzymes, and then the corresponding value of the human ALOX15 was set 100% for each of the two substrates.
HPLC Analysis.
HPLC analysis of the LOX products was performed on a Shimadzu instrument equipped with a Hewlett-Packard diode array detector 1040 A by recording the absorbance at 235 nm. RP-HPLC was carried out on a Nucleosil C18 column (KS-system, 250 × 4 mm, 5-µm particle size; Marcherey-Nagel) coupled with a guard column (30 × 4 mm, 5-µm particle size). A solvent system of methanol/water/acetic acid (85/15/0.1 by volume) was used at a flow rate of 1 mL/min. SP-HPLC was performed on a Zorbax-SIL column (250 × 4 mm, 5-µm particle size) with the solvent system n-hexan/2-propanol/acetic acid (100/2/0.1 by volume) and a flow rate of 1 mL/min. Hydroxy fatty acid enantiomers were separated by chiral-phase HPLC. The 15-HETE enantiomers were separated as free fatty acids on a Chiralcel OD column (Daicel) by using a solvent system consisting of hexane/2-propanol/acetic acid (100/5/0.1 by vol.) and a flow rate of 1 mL/min. The 12-HETE enantiomers were separated as methyl ester on a Chiralcel OB column (Daicel) with a solvent system consisting of n-hexane/2-popanol/acetic acid (100/4/0.1 by volume) and a flow rate of 1 mL/min. Lipoxin isomers were analyzed by RP-HPLC using a Nucleosil C18 column (KS-system, 250 × 4 mm, 5-µm particle size; Marcherey-Nagel) coupled with a guard column (30 × 4 mm, 5-µm particle size) and the solvent system acetonitrile/water/acetic acid (38/62/0.1 by volume) at a flow rate of 1 mL/min.
Assay Repetitions and Statistical Evaluation.
For activity assays and for quantification of the product pattern, two to four different expression samples (50 mL each) were grown, and at least two activity assays were carried out per sample. Each activity assay was quantified twice by RP-HPLC, and the obtained raw data were statistically evaluated by using Microsoft Excel 2008 (12.8). Means ± SDs are given.
MD Simulations and QM/MM Calculations.
MD and QM/MM simulations were carried out for the Ile418Ala mutant of rabbit ALOX15 as described before (47). For this, we have generated molecular configurations of the Michaelis complex of the 12-lipoxygenating Ile418Ala mutant of rabbit ALOX15 by means of MD simulations. The system was fully solvated in an orthorhombic box of preequilibrated TIP3P water molecules, with dimensions of 125 Å × 79 Å × 85 Å. Seven sodium ions were added to neutralize the total charge of the system. The resulting model contains nearly 75,500 atoms. Three different trajectories of 5 ns of production at 300 K under periodic boundary conditions at constant pressure and temperature were run for the ALOX15 mutant. All MD simulations were run with CHARMM version c35-b1. More detailed information on methodological details, sequence comparison, and results obtained by MD simulations and QM/MM calculations are provided in the SI Appendix, Tables S3 and S4 and Figs. S2–S9.
Supplementary Material
Acknowledgments
We thank the German Primate Center (Göttingen, Germany) and the Max Planck Institute of Evolutionary Anthropology Leipzig for providing primate blood. This work was supported in part by Slovak Grant Agency (Bratislava, Slovakia) Research Grant 1/0392/14 (to M.P.), Deutsche Forschungsgemeinschaft Ku961/11-1, Spanish Ministerio de Economía y Competitividad Grant CTQ2014-53144-P, and “Programa Banco Santander-Universitat Autònoma de Barcelona.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.v.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604029113/-/DCSupplemental.
References
- 1.Haeggström JZ, Funk CD. Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chem Rev. 2011;111(10):5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov I, et al. Molecular enzymology of lipoxygenases. Arch Biochem Biophys. 2010;503(2):161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Horn T, et al. Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling. Prog Lipid Res. 2015;57:13–39. doi: 10.1016/j.plipres.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Gennaro A, Haeggström JZ. The leukotrienes: Immune-modulating lipid mediators of disease. Adv Immunol. 2012;116:51–92. doi: 10.1016/B978-0-12-394300-2.00002-8. [DOI] [PubMed] [Google Scholar]
- 5.Pace-Asciak CR. Pathophysiology of the hepoxilins. Biochim Biophys Acta. 2015;1851(4):383–396. doi: 10.1016/j.bbalip.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Rapoport SM, Schewe T. The maturational breakdown of mitochondria in reticulocytes. Biochim Biophys Acta. 1986;864(3-4):471–495. doi: 10.1016/0304-4157(86)90006-7. [DOI] [PubMed] [Google Scholar]
- 7.van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. A function for lipoxygenase in programmed organelle degradation. Nature. 1998;395(6700):392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- 8.Clària J. Regulation of cell proliferation and apoptosis by bioactive lipid mediators. Recent Patents Anticancer Drug Discov. 2006;1(3):369–382. doi: 10.2174/157489206778776961. [DOI] [PubMed] [Google Scholar]
- 9.Kühn H, O’Donnell VB. Inflammation and immune regulation by 12/15-lipoxygenases. Prog Lipid Res. 2006;45(4):334–356. doi: 10.1016/j.plipres.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Schneider C, Pozzi A. Cyclooxygenases and lipoxygenases in cancer. Cancer Metastasis Rev. 2011;30(3-4):277–294. doi: 10.1007/s10555-011-9310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palacios-Pelaez R, Lukiw WJ, Bazan NG. Omega-3 essential fatty acids modulate initiation and progression of neurodegenerative disease. Mol Neurobiol. 2010;41(2-3):367–374. doi: 10.1007/s12035-010-8139-z. [DOI] [PubMed] [Google Scholar]
- 12.Epp N, et al. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol. 2007;177(1):173–182. doi: 10.1083/jcb.200612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieg P, et al. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol. 2013;133(1):172–180. doi: 10.1038/jid.2012.250. [DOI] [PubMed] [Google Scholar]
- 14.Chen XS, Sheller JR, Johnson EN, Funk CD. Role of leukotrienes revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372(6502):179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- 15.Sun D, Funk CD. Disruption of 12/15-lipoxygenase expression in peritoneal macrophages. Enhanced utilization of the 5-lipoxygenase pathway and diminished oxidation of low density lipoprotein. J Biol Chem. 1996;271(39):24055–24062. [PubMed] [Google Scholar]
- 16.Johnson EN, Brass LF, Funk CD. Increased platelet sensitivity to ADP in mice lacking platelet-type 12-lipoxygenase. Proc Natl Acad Sci USA. 1998;95(6):3100–3105. doi: 10.1073/pnas.95.6.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funk CD, Chen XS, Johnson EN, Zhao L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 2002;68-69:303–312. doi: 10.1016/s0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn H, Thiele BJ. The diversity of the lipoxygenase family. Many sequence data but little information on biological significance. FEBS Lett. 1999;449(1):7–11. doi: 10.1016/s0014-5793(99)00396-8. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn H, Banthiya S, van Leyen K. Mammalian lipoxygenases and their biological relevance. Biochim Biophys Acta. 2015;1851(4):308–330. doi: 10.1016/j.bbalip.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant RW, Bailey JM, Schewe T, Rapoport SM. Positional specificity of a reticulocyte lipoxygenase. Conversion of arachidonic acid to 15-S-hydroperoxy-eicosatetraenoic acid. J Biol Chem. 1982;257(11):6050–6055. [PubMed] [Google Scholar]
- 21.Watanabe T, Medina JF, Haeggstrom JZ, Radmark O, Samuelsson B. Molecular cloning of a 12-lipoxygenase cDNA from rat brain. Eur J Biochem. 1993;212(2):605–612. doi: 10.1111/j.1432-1033.1993.tb17699.x. [DOI] [PubMed] [Google Scholar]
- 22.Vogel R, et al. Applicability of the triad concept for the positional specificity of mammalian lipoxygenases. J Biol Chem. 2010;285(8):5369–5376. doi: 10.1074/jbc.M109.057802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borngräber S, et al. Shape and specificity in mammalian 15-lipoxygenase active site. The functional interplay of sequence determinants for the reaction specificity. J Biol Chem. 1999;274(52):37345–37350. doi: 10.1074/jbc.274.52.37345. [DOI] [PubMed] [Google Scholar]
- 24.Sloane DL, Leung R, Barnett J, Craik CS, Sigal E. Conversion of human 15-lipoxygenase to an efficient 12-lipoxygenase: The side-chain geometry of amino acids 417 and 418 determine positional specificity. Protein Eng. 1995;8(3):275–282. doi: 10.1093/protein/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Adel S, et al. Leukotriene signaling in the extinct human subspecies Homo denisovan and Homo neanderthalensis. Structural and functional comparison with Homo sapiens. Arch Biochem Biophys. 2015;565:17–24. doi: 10.1016/j.abb.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn H, et al. Structural biology of mammalian lipoxygenases: Enzymatic consequences of targeted alterations of the protein structure. Biochem Biophys Res Commun. 2005;338(1):93–101. doi: 10.1016/j.bbrc.2005.08.238. [DOI] [PubMed] [Google Scholar]
- 27.Borngräber S, Kuban RJ, Anton M, Kühn H. Phenylalanine 353 is a primary determinant for the positional specificity of mammalian 15-lipoxygenases. J Mol Biol. 1996;264(5):1145–1153. doi: 10.1006/jmbi.1996.0702. [DOI] [PubMed] [Google Scholar]
- 28.Pekárová M, Kuhn H, Bezáková L, Ufer C, Heydeck D. Mutagenesis of triad determinants of rat Alox15 alters the specificity of fatty acid and phospholipid oxygenation. Arch Biochem Biophys. 2015;571:50–57. doi: 10.1016/j.abb.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H, et al. Site-directed mutagenesis studies on the iron-binding domain and the determinant for the substrate oxygenation site of porcine leukocyte arachidonate 12-lipoxygenase. Biochim Biophys Acta. 1994;1210(3):308–316. doi: 10.1016/0005-2760(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn H, Belkner J, Wiesner R, Brash AR. Oxygenation of biological membranes by the pure reticulocyte lipoxygenase. J Biol Chem. 1990;265(30):18351–18361. [PubMed] [Google Scholar]
- 31.Belkner J, Stender H, Kühn H. The rabbit 15-lipoxygenase preferentially oxygenates LDL cholesterol esters, and this reaction does not require vitamin E. J Biol Chem. 1998;273(36):23225–23232. doi: 10.1074/jbc.273.36.23225. [DOI] [PubMed] [Google Scholar]
- 32.Sloane DL, Dixon RA, Craik CS, Sigal E. Expression of cloned human 15-lipoxygenase in eukaryotic and prokaryotic systems. Adv Prostaglandin Thromboxane Leukot Res. 1991;21A:25–28. [PubMed] [Google Scholar]
- 33.Chaitidis P, et al. Lipoxygenase pathways in Homo neanderthalensis: Functional comparison with Homo sapiens isoforms. J Lipid Res. 2013;54(5):1397–1409. doi: 10.1194/jlr.M035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannesson M, Backman L, Claesson HE, Forsell PK. Cloning, purification and characterization of non-human primate 12/15-lipoxygenases. Prostaglandins Leukot Essent Fatty Acids. 2010;82(2-3):121–129. doi: 10.1016/j.plefa.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama C, et al. Arachidonate 12-lipoxygenase purified from porcine leukocytes by immunoaffinity chromatography and its reactivity with hydroperoxyeicosatetraenoic acids. J Biol Chem. 1986;261(35):16714–16721. [PubMed] [Google Scholar]
- 36.Freire-Moar J, Alavi-Nassab A, Ng M, Mulkins M, Sigal E. Cloning and characterization of a murine macrophage lipoxygenase. Biochim Biophys Acta. 1995;1254(1):112–116. doi: 10.1016/0005-2760(94)00199-9. [DOI] [PubMed] [Google Scholar]
- 37.De Marzo N, Sloane DL, Dicharry S, Highland E, Sigal E. Cloning and expression of an airway epithelial 12-lipoxygenase. Am J Physiol. 1992;262(2 pt 1):L198–L207. doi: 10.1152/ajplung.1992.262.2.L198. [DOI] [PubMed] [Google Scholar]
- 38.Berger M, et al. Simultaneous expression of leukocyte-type 12-lipoxygenase and reticulocyte-type 15-lipoxygenase in rabbits. J Mol Biol. 1998;278(5):935–948. doi: 10.1006/jmbi.1998.1737. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov I, Kuhn H, Heydeck D. Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15) Gene. 2015;573(1):1–32. doi: 10.1016/j.gene.2015.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sloane DL, Leung R, Craik CS, Sigal E. A primary determinant for lipoxygenase positional specificity. Nature. 1991;354(6349):149–152. doi: 10.1038/354149a0. [DOI] [PubMed] [Google Scholar]
- 41.Romano M. Lipoxin and aspirin-triggered lipoxins. Sci World J. 2010;10:1048–1064. doi: 10.1100/tsw.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HN, Surh YJ. Therapeutic potential of resolvins in the prevention and treatment of inflammatory disorders. Biochem Pharmacol. 2012;84(10):1340–1350. doi: 10.1016/j.bcp.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Serhan CN, Dalli J, Colas RA, Winkler JW, Chiang N. Protectins and maresins: New pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochim Biophys Acta. 2015;1851(4):397–413. doi: 10.1016/j.bbalip.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwab JM, Chiang N, Arita M, Serhan CN. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447(7146):869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serhan CN, et al. Reduced inflammation and tissue damage in transgenic rabbits overexpressing 15-lipoxygenase and endogenous anti-inflammatory lipid mediators. J Immunol. 2003;171(12):6856–6865. doi: 10.4049/jimmunol.171.12.6856. [DOI] [PubMed] [Google Scholar]
- 46.Kühn H, Schewe T, Rapoport SM. The stereochemistry of the reactions of lipoxygenases and their metabolites. Proposed nomenclature of lipoxygenases and related enzymes. Adv Enzymol Relat Areas Mol Biol. 1986;58:273–311. doi: 10.1002/9780470123041.ch7. [DOI] [PubMed] [Google Scholar]
- 47.Saura P, Suardiaz R, Masgrau L, Lluch JM, Gonzalez-Lafont A. Unraveling how enzymes can use bulky residues to drive site-selective C-H activation: The case of mammalian lipoxygenases catalyzing arachidonic acid oxidation. ACS Catal. 2014;4(12):4351–4363. [Google Scholar]
- 48.Jisaka M, Kim RB, Boeglin WE, Brash AR. Identification of amino acid determinants of the positional specificity of mouse 8S-lipoxygenase and human 15S-lipoxygenase-2. J Biol Chem. 2000;275(2):1287–1293. doi: 10.1074/jbc.275.2.1287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.