Significance
The DNA in mitochondria is essential for efficient energy production. Critical for mitochondrial DNA replication and expression are sequences concentrated in the so-called control region. We report that many mitochondrial DNAs contain a triple-stranded region whose third strand is RNA and maps to the control region. These R-loops contribute to DNA architecture and replication in the mitochondria, and aberrant R-loop processing causes disease.
Keywords: RNase H1, R-loop, mitochondrial DNA, DNA segregation, mitochondrial disease
Abstract
The genetic information in mammalian mitochondrial DNA is densely packed; there are no introns and only one sizeable noncoding, or control, region containing key cis-elements for its replication and expression. Many molecules of mitochondrial DNA bear a third strand of DNA, known as “7S DNA,” which forms a displacement (D-) loop in the control region. Here we show that many other molecules contain RNA as a third strand. The RNA of these R-loops maps to the control region of the mitochondrial DNA and is complementary to 7S DNA. Ribonuclease H1 is essential for mitochondrial DNA replication; it degrades RNA hybridized to DNA, so the R-loop is a potential substrate. In cells with a pathological variant of ribonuclease H1 associated with mitochondrial disease, R-loops are of low abundance, and there is mitochondrial DNA aggregation. These findings implicate ribonuclease H1 and RNA in the physical segregation of mitochondrial DNA, perturbation of which represents a previously unidentified disease mechanism.
Mammalian mtDNA contributes 13 critical proteins of the oxidative phosphorylation system that produces much of the cell’s energy. Consequently, aberrant or insufficient mtDNA causes cell and tissue dysfunction that manifests in a range of human diseases (1). In most cells and tissues mtDNA is formed of circles of dsDNA. The two strands are denoted heavy (H) and light (L) owing to their different base compositions. Important cis-elements that function as origins of replication, the replication terminus, and transcriptional promoters are concentrated in the control region (CR) (2–5). Many molecules of mtDNA bear a third strand of DNA, known as “7S DNA,” which forms a displacement (D) loop covering much of the CR. The D-loop spans approximately half a kilobase of the 16.5 kb of mammalian mtDNA (6–8) and is present on 1–65% of mtDNA molecules (9). The frequent synthesis of 7S DNA across such a pivotal region of the mitochondrial genome implies an important role for the D-loop in mtDNA metabolism, and it has been implicated in protein recruitment and mtDNA organization (10, 11).
Ribonuclease H1 (RNase H1) degrades RNA hybridized to DNA (12) and is essential for mtDNA maintenance in mice (13). Recently, pathological mutations in RNase H1 (RNASEH1), including RNASEH1, c.424G > A; p.Val142Ile (V142I), have been reported to cause adult-onset neuromuscular disease (14). We have studied fibroblasts with V142I RNase H1 and find no evidence of the primer retention associated with loss of the gene in murine cells (15). However, the human cells carrying mutant RNase H1 have markedly reduced levels of a newly identified RNA that forms an R-loop on the mtDNA. The RNA is similar in length and location to the D-loop but is complementary to 7S DNA. A low level of the mitochondrial R-loop is associated with aggregation of mtDNA, suggesting a role for the R-loop in mtDNA organization and segregation.
Results
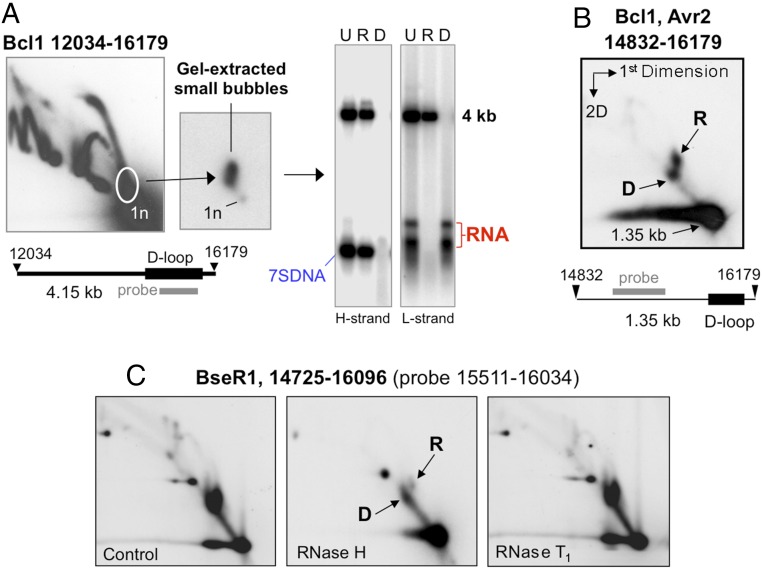
Analysis of RNA hybridized to mtDNA must contend with the ready degradation of the RNA during extraction (16). Previous analysis of fragments of mtDNA encompassing the CR demonstrated that they included molecules with 7S DNA, as expected for triple-stranded D-loops (10), but no RNA (SI Appendix, Fig. S1A). More recently, however, interstrand cross-linking was found to preserve RNA/DNA hybrids of mtDNA and to enhance markedly the signal associated with D-loop–like structures (SI Appendix, Fig. S1B and ref. 17). Therefore, we sought to determine whether the increased signal reflected RNA preservation or solely increased D-loop stability. Because intact RNA/DNA hybrids are easier to isolate from solid tissues than from aneuploid cultured cells (17), we refined our isolation procedure for mouse liver mtDNA. This protocol employs a protease treatment on ice (SI Appendix, Materials and Methods), which improves the quality of the mtDNA, as evidenced by the replication intermediates resolved by 2D agarose gel-electrophoresis (2D-AGE) (SI Appendix, Fig. S2). The revised procedure was used for all subsequent analysis of murine mtDNA. To enrich D-loops and small bubble structures and to determine their nucleic acid composition, we gel-extracted material from the base of an initiation arc and treated it with RNase HI, DNase, or no enzyme before denaturation and 1D-AGE. Blot hybridization to strand-specific probes revealed L-strand RNA complementary to 7S DNA and of similar mobility and abundance (Fig. 1A). Analysis of smaller fragments containing the CR resolved the small bubble structures into two discrete species, D and R, of similar abundance (Fig. 1B). Ban2, which cuts 7S DNA bound to mtDNA (i.e., the D-loop) (SI Appendix, Fig. S3A), cleaved species D but not species R (SI Appendix, Fig. S3B), indicating the latter were refractive to digestion at nucleotide position (np) 15,749 (in the CR), possibly owing to an RNA component, because restriction enzymes are unable to cleave RNA/DNA hybrids (e.g., ref. 18). Species R proved sensitive to in vitro RNase H treatment but was largely insensitive to single-stranded RNase (RNase T1) (Fig. 1C), indicating it contains an RNA hybridized to DNA for much of its length. Therefore, we conclude that species D contains the well-recognized D-loop, whereas species R is a previously unknown form of mtDNA containing an R-loop whose RNA is approximately the complement of the D-loop.
Fig. 1.
Many molecules of murine mtDNA contain a mitochondrial R-loop complementary to the 7S DNA of the mitochondrial D-loop. (A) Purified mouse mtDNA (SI Appendix, Fig. S2) was digested with Bcl1 and fractionated by 2D-AGE before gel extraction of the material close to the base of the initiation arc (circled on a representative blot, hybridized to probe np 15,551–16,034). Repeated 2D-AGE of the gel-extracted nucleic acids confirmed that the majority were more massive than the linear restriction fragment, and heat denaturation and repeated 1D-AGE of samples, untreated (U), treated with RNase HI (R), or DNase (D) as previously (18), revealed an L-strand RNA complementary to 7S DNA. (B) A double digestion of mouse mtDNA to create a smaller CR-containing fragment reveals two discrete species, D and R, well separated from the linear 1.35-kb fragment. (C) 2D-AGE of BseR1-digested mouse mtDNA and enzyme treatments indicate species R is more sensitive to Eco-RNase HI than species D, although both are largely resistant to RNase T1.
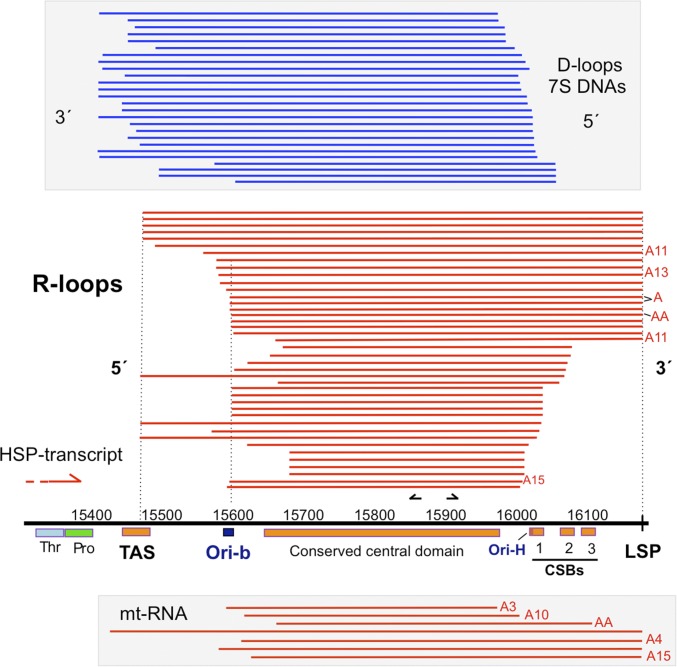
Further mtDNA preparations and gel extractions were performed to generate material for the conversion of the R-loop RNA to DNA via reverse transcription (SI Appendix, Materials and Methods), and sequencing analysis confirmed that the R-loop maps to the CR (Fig. 2). Seven of 41 RNAs sequenced were oligo-adenylated (Fig. 2 and SI Appendix, Table S1), all at the terminus mapping closest to light-strand promoter (LSP). Oligo or polyadenylation occurs at the 3′ end of mitochondrial and other messenger RNAs and thus indicates that these RNAs are L-strand, concordant with the results of the strand-specific probes (Fig. 1A). The 3′ ends most frequently mapped precisely to LSP (20 of 41 sequences), and none extended beyond this point (Fig. 2 and SI Appendix, Table S1), suggesting that the LSP is the ultimate terminus for R-loop synthesis. Of the 41 R-loops sequenced, the eight longest (with respect to the inferred 5′ end) began at np 15,476 ± 2 nt, within a region (np 15,423–15,483) (19) predicted to form a secondary structure (20) known as the “termination-associated sequence” (TAS) because of its proximity to the 3′ end of the 7S DNAs (8). Seventeen of the other 5′ ends were clustered around np 15,600, within 25 nt of the second origin of replication in the CR, Ori-b (15). We propose to name the RNA component of the abundant mitochondrial R-loop “LC-RNA” [for “light- (or lagging-) strand, CR RNA”), because the term “7S RNA” has been applied to the transient primer sequence spanning LSP to origin of replication H (Ori-H) on the H-strand (15, 21).
Fig. 2.
Mapping of the mouse mitochondrial R-loop. RNAs recovered from Bcl1-digested mouse liver mtDNA fractionated by 2D-AGE (Fig. 1A and main text) or from undigested purified mouse liver mtDNA were DNase-treated, circularized, converted to DNA by RT-PCR, cloned, and sequenced. The lengths of the R-loops (red lines) are inferred from the junctions (SI Appendix, Table S1) and are aligned to the CR of murine mtDNA. Circularized RT-PCR applied to purified mouse mitochondrial RNA (mt-RNA) yielded a small number of clones (red lines at the bottom of the figure). Circularized PCR, cloning, and sequencing were used to map the ends of gel-extracted 7S DNAs (blue lines). Short black arrows mark the approximate position of the primers used for RT-PCR. A, adenine residues added posttranscriptionally to the 3′ end of the RNAs; CSB, conserved sequence block; LSP, light strand promoter; Ori-b and Ori-H, origins of replication; Pro, tRNA proline gene; TAS, termination-associated sequence; Thr, tRNA threonine gene.
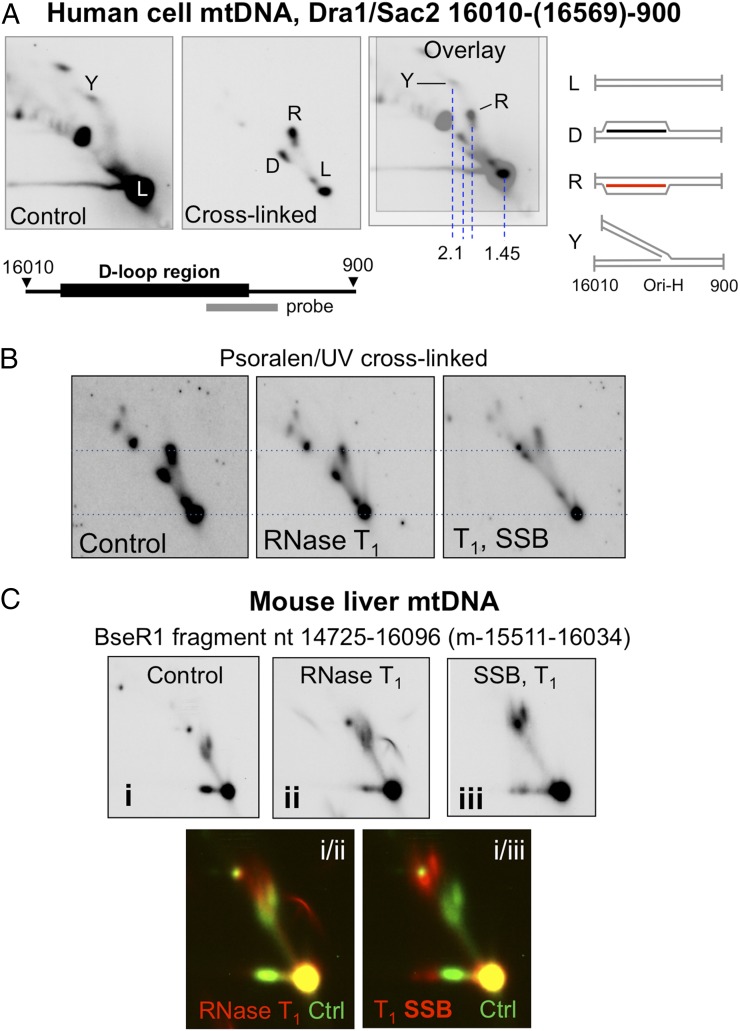
Although in our experience the preservation of RNA/DNA hybrids is more difficult in aneuploid cells than in solid tissues (17), a similar pair of small bubble-like structures was evident in human mtDNA of 143B osteosarcoma cells subjected to interstrand cross-linking (Fig. 3A), and species refractive to Ban2 digestion at np 15,749 of murine mtDNA (i.e., equivalent to species R in SI Appendix, Fig. S3B) were detected in heart, brain, and kidney (SI Appendix, Fig. S4A). Hence, R-loops as well as D-loops appear to be a ubiquitous feature of mammalian mtDNA. Because the gel-extracted mouse mtDNA material contained both LC-RNA and 7S DNA (Fig. 1A), it was not clear whether the two species resided on different molecules; however, the greater resolution achieved with the smaller 1.35-kb restriction fragment indicated that the two small bubble-like structures (species D and R) had similar mobility in the first-dimension electrophoresis step (Fig. 1B), suggesting that their masses were substantially the same, as would be expected for species containing either 7S DNA or LC-RNA but not both. This inference was corroborated by the analysis of human mtDNA, because a specific replication (Y) fork structure marks the position where a species of ∼2.1 kb of duplex DNA resolves, similar to that predicted for molecules containing both LC-RNA and 7S DNA, whereas species D and R ran considerably closer to the linear duplex fragments of 1.45 kb (see overlay in Fig. 3A). Moreover, the single-stranded DNA content is approximately the same in D and R, as evidenced by the similar mobility shift when preincubated with single-stranded DNA-binding protein (SSB) before gel fractionation (Fig. 3 B and C). Nevertheless, a short initiation-type arc of a length appropriate for a combined D/R loop was detectable in cross-linked rat liver mtDNA samples (SI Appendix, Fig. S4B), suggesting that some molecules contain both LC-RNA and 7S DNA.
Fig. 3.
Interstrand nucleic acid cross-linking stabilizes mitochondrial D-loops and R-loops in cultured human cells, and both are mobility shifted by SSB. (A) Human 143B osteosarcoma cells were psoralen/UV cross-linked or left untreated (control), and the isolated DNA was digested with Sac2 and Dra1 before 2D-AGE and blot hybridization to a probe spanning np 16,341–151 that detects the D-loop region (np 16,107–191). An overlay shows the mobility of species D and R relative to a recognized species on the replication fork arc (Y). Vertical dashed blue lines indicate the mobility of various species in the first-dimension electrophoresis step that is indicative of their mass. Interpretations of the structures of the nucleic acids appear to the right of the gel images: L or 1n, linear duplex mtDNA fragment; Y, replication fork; D-loop, 1n + 7S DNA; R-loop, 1n + LC-RNA (see text for details). The slower mobility of the R-loop in the second dimension compared with the D-loop might reflect a general feature that distinguishes the two types of triple-stranded molecule (i.e., the different properties of molecules with a third strand of RNA as opposed to DNA), or it might indicate a more complex arrangement of LC-RNA that includes segments of RNA–RNA pairing as well as RNA/DNA hybrid. (B and C) Species D and R of human (B) and mouse (C) mtDNA are mobility shifted by incubation with SSB. RNase T1 was applied before SSB incubation to remove any RNA tails that might have accentuated the mobility shift. The probe for mouse mtDNA spanned np 15,511–16,034. Merged false-color images [green (C, i) merged with red (C, ii or C, iii)] at the foot of the panel C provide a direct comparison of the effect of SSB on the mobility of D- and R-loops.
The existence of the mitochondrial R-loop raises questions about its formation and processing and its roles in mtDNA metabolism. RNase H1 is a strong candidate for the enzyme involved in R-loop processing, because it is known to target RNA/DNA hybrids in the CR (15). The identification of a homozygous missense mutation in RNASEH1, V142I, in the affected members of two unrelated families with neuromuscular disease (SI Appendix, Fig. S5A and Table S2) offered an opportunity to explore the enzyme’s impact on mtDNA and the mitochondrial R-loop using patient-derived fibroblasts and muscle of affected individuals.
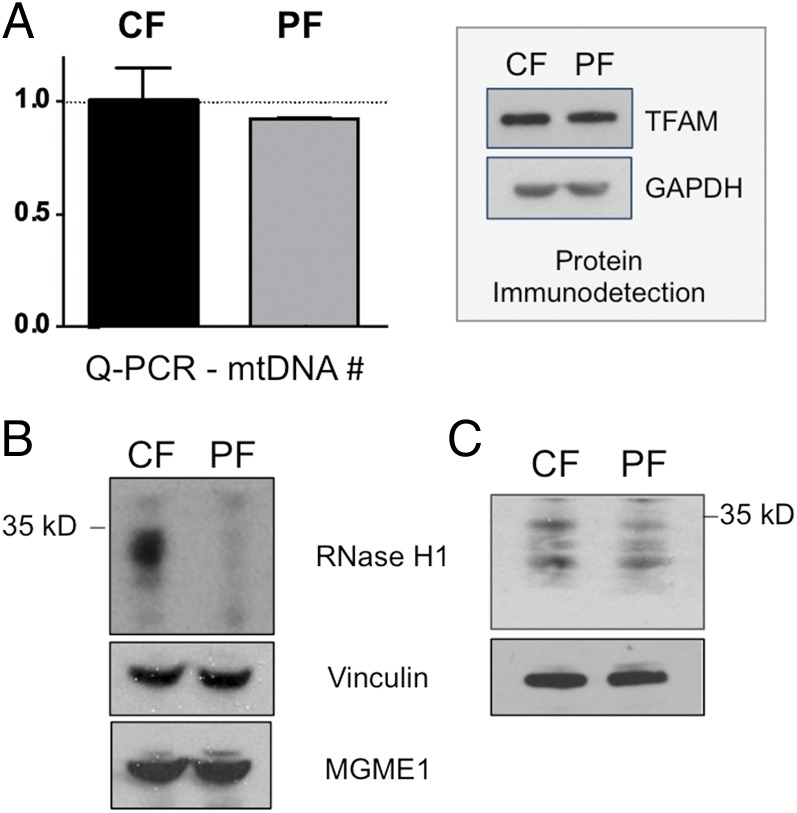
Recently we showed that murine cells lacking the enzyme are unable to maintain mtDNA (15). In contrast, fibroblasts carrying V142I RNase H1 had 92% of the average mtDNA copy number of three controls and normal levels of the mtDNA packaging protein transcription factor A, mitochondrial precursor (TFAM) (Fig. 4A). Concordant with the mtDNA copy number data, the RNase H1 variant V142I produces milder pathology in humans than seen with gene ablation in the mouse: adult-onset neuromuscular disease (encephalomyopathy) (this report and ref. 14), as opposed to embryonic lethality (13). However, analysis of the steady-state level of the protein using detergent-solubilized cell lysates, stored frozen, implied that the V142I substitution resulted in almost complete loss of RNase H1, because the protein was barely detectable (Fig. 4B). The apparent absence of protein could indicate either a much greater redundancy for RNase H1 in humans than in mice or that the immunoblotting results were misleading. In favor of the latter possibility, electrophoresis yielded a markedly higher signal for V142I RNase H1 in samples freshly prepared immediately after cell lysis than in samples stored frozen (Fig. 4C). Hence, we infer that the mutant protein is less stable than wild-type RNase H1 and that the presence of an appreciable amount of RNase H1 protein in the patient-derived fibroblasts correlates with the less severe phenotype of the human disease compared with the results of Rnaseh1 excision in the mouse.
Fig. 4.
V142I RNase H1 protein is unstable, but the mutant protein does not appreciably alter mtDNA copy number. (A) Quantitative PCR to a fragment of a nuclear and a mitochondrial gene (SI Appendix) was used to determine the relative abundance of mtDNA in patient-derived (PF) V142I RNase H1 fibroblasts (V142I) compared with three control human fibroblast lines (CF); data are shown as the mean ± SEM from three independent experiments. (Inset) A representative immunoblot for TFAM and the reference protein GAPDH. (B and C) Immunoblots of proteins from control and patient-derived fibroblasts stored frozen (B) or freshly prepared (C): RNase H1, MGME1 [a mitochondrial endonuclease (Atlas Antibodies)], and vinculin (a reference protein). Proteins were fractionated by 4–12% (wt/vol) SDS/PAGE.
A prominent feature of the loss of RNase H1 in murine cells is the retention of primers on the mtDNA in the CR (15). It has long been proposed that 7S DNA arises from a primer initiating at LSP and that the 5′ ends of the 7S DNAs mark the RNA–DNA transition point (22). Fractionation of 7S DNA from mouse embryonic fibroblasts (MEFs) without Rnaseh1 indicates it is contiguous with a segment of RNA, detected by a probe spanning LSP to Ori-H, whereas in cells retaining Rnaseh1 the RNA extension is not detectable (Fig. 5A). Therefore, Rnaseh1 ablation leads to (LSP–Ori-H) primer retention on 7S DNA molecules as well as on productive nascent strands (15). Because 7S DNA is highly abundant, it was chosen to determine whether the human mutant RNase H1 compromises primer processing in the CR of human mtDNA.
Fig. 5.
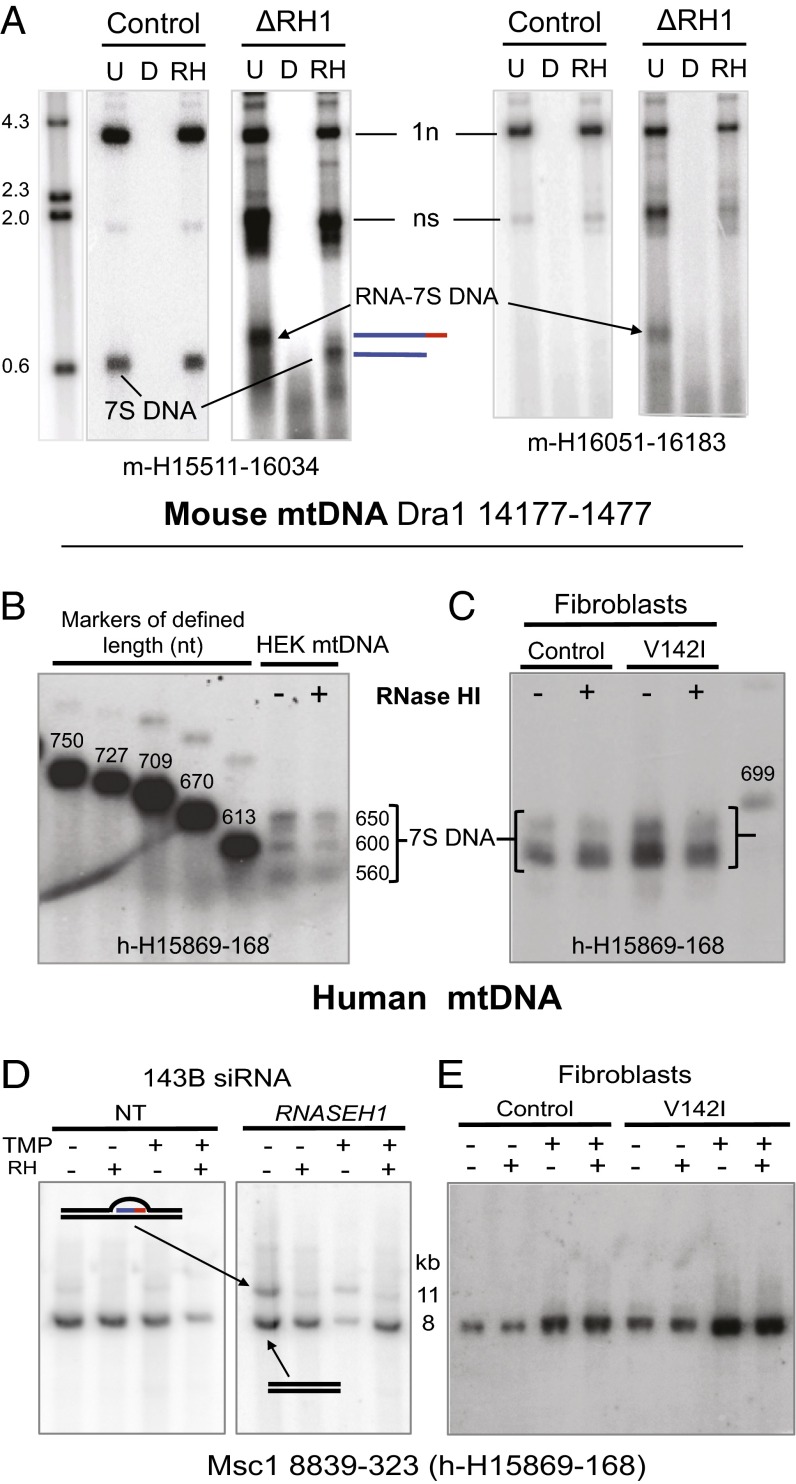
7S DNA molecules are persistently attached to their primer RNA in murine cells lacking RNase H1, but there is no evidence of primer retention in human cells with V142I RNase H1. (A) MEF mtDNA from cells treated for 8 d without (control) or with 4HT to excise the Rnaseh1 gene (ΔRH1) was digested with Dra1 and left untreated (U) or treated with DNase (D) or Eco-RNase HI (RH); denatured in 80% formamide, 15 min at 85 °C; and separated by 1D-AGE (1% Tris-borate). The λ-Hind3 DNA ladder was run in parallel to provide size markers. Southern hybridization with a riboprobe to the 5′ end of the H-strand (H15,511–16,034) detected the full-length H-strand of 3.6 kb (1n) and nascent strands (ns) (described in detail in ref. 15) and 7S DNA. 7S DNA without a primer is not detected by m-H16,051–16,183 but is detected by m-H15,511–16,034 (albeit only in ΔRH1 samples). Line drawings appear to the right of the gel image to denote 7S DNA with and without an RNA primer; red line, RNA; blue lines, DNA. (B) 7S DNAs of the human mitochondrial D-loop range from ∼560–650 nt in HEK cells and lack a RNA primer in human cells with wild-type (control) or mutant (V142I) RNase H1. (C) Sucrose gradient-purified mtDNAs of HEK cells and fibroblasts were separated by 1D-AGE [2% agarose, sodium borate (46)] after treatment with (+) or without (−) Eco-RNase HI and denaturation in 80% formamide, 15 min at 85 °C and were hybridized with a riboprobe complementary to nt 15,869–168 of human H-strand mtDNA. The mtDNA was run in parallel with a series of markers of defined length (SI Appendix). (D and E) DNA was extracted from whole 143B cells after 6 d of siRNA [nontargeting (NT) or targeting RNASEH1] (D) and from control or patient-derived (V1421) fibroblasts (E). DNA was digested with Msc1, fractionated by 1D-AGE (1% Tris-acetate-EDTA), and blot hybridized to the indicated probe. Cells were subjected to UV/psoralen cross-linking before isolation of nucleic acid and Msc1 and RNase HI (RH) digestion, as indicated. The compositions of the 8- and 11-kb species are interpreted in the line drawings. The 11-kb band has a blocked site at np 323 resulting from the R-loop. Red lines indicate RNA, and black lines indicate DNA.
Before assessing 7S DNA primer retention in V142I RNase H1 fibroblasts, we first analyzed mitochondrial D-loop length and properties in HEK cells. Fractionation by sodium-borate gel-electrophoresis resolved four 7S DNA species of ∼560–650 nt, which are concordant with those defined previously (8) with free 5′ ends mapping to np 111, 150, 168, and 191 (23). Treatment with RNase HI did not alter their lengths (Fig. 5B), suggesting that none of the 7S DNAs had a retained primer (of 10 or more ribonucleotides). The lengths of the 7S DNAs of human fibroblasts with wild-type or mutant (V142I) RNase H1 were similar to those of HEK cells, irrespective of in vitro RNase HI treatment (Fig. 5C). Therefore, in the human mutant fibroblasts there was no evidence of retention of a primer from LSP to Ori-H, which would have extended the 7S DNAs by 200 nt, one third of their usual length.
Because RNA/DNA hybrids of mtDNA can be lost readily during isolation (16, 24), we also carried out a blocked-site assay to screen for retained primers. In MEFs lacking Rnaseh1, the failure to remove the LSP–Ori-H primer creates a stretch of RNA/DNA hybrid in the mtDNA that is refractive to restriction digestion at a site (np 16,179), immediately downstream of LSP (np 16,190) (SI Appendix, Fig. S6A and ref. 15). Msc1 was selected for the equivalent analysis of human mtDNA, because its cleavage site, at np 323, lies between LSP (np 407) and Ori-H (multiple proposed sites at or downstream of np 300) (SI Appendix, Fig. S6B). First we applied the procedure to mtDNA of human 143B osteosarcoma cells, after silencing RNase H1 for 144 h (as previously described in ref. 25). The experiment potentially served two purposes, providing a test for the hypothesis that RNase H1 performs the same role in human and murine cells, and if so, providing a positive control for RNA retention in human mtDNA. RNASEH1 gene silencing in 143B cells markedly increased the amount of Msc1-digested mtDNA that was uncut at np 323 (but not at np 8,939), particularly in cells subjected to psoralen/UV cross-linking that improves the recovery of RNA/DNA hybrids (17). Moreover, site blockage was largely relieved when the samples were coincubated with Eco-RNase HI for the final hour of the restriction digestion (Fig. 5D). These findings suggest that an acute shortage of RNase H1 in human (143B) cells leads to the retention of the primer from LSP to the DNA transition site, as in murine fibroblasts (15). In contrast, the V142I RNase H1 was not associated with any detectable site blockage at np 323 of human mtDNA (Fig. 5E), again indicating there is no persistent primer from LSP to Ori-H in the patient-derived fibroblasts. Thus, human mitochondria containing the V142I RNase H1 are able to remove the primers associated with 7S DNA. Either the residual activity of the mutant enzyme (14) is sufficient for this task, or there is redundancy for primer processing in some human cell types but not in others.
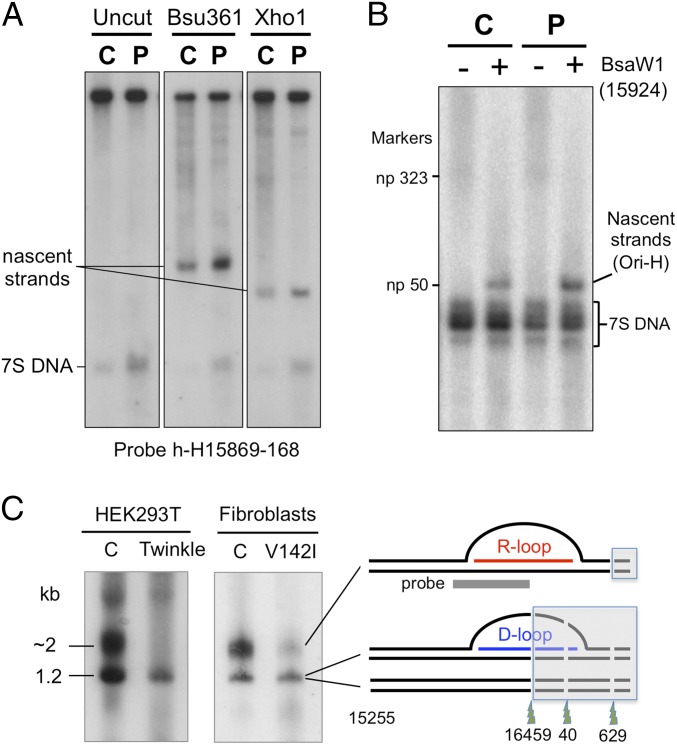
The V142I mutation was associated with an approximately eightfold increase in the abundance of 7S DNAs relative to control fibroblasts (Fig. 6A), as reported previously (14). The earlier report also measured a two- to fourfold increase in mitochondrial replication intermediates in the mutant fibroblasts detected by 2D gel-electrophoresis. However, very few products of strand-asynchronous/bootlace replication were detected (14), although they form the majority of mitochondrial replication intermediates in other cell types and in solid tissues (3, 18); therefore their disintegration during extraction might have confounded the analysis. In a different approach, we intentionally separated the strands of mtDNA after cleaving at np 14,955 (Xho1) or 14,445 (Bsu361), sites respectively 1.7 and 2.2 kb downstream of Ori-H. Fragments of these lengths, i.e., nascent strands of DNA with a 5′ end mapping to Ori-H, were approximately three times more abundant in RNase H1 mutant cells than in control fibroblasts (Fig. 6A). Thus, this finding establishes that mitochondrial replication intermediates are present at a higher steady-state level in cells with V142I RNase H1, and the increase in nascent strands suggests that this level is attributable to slow mtDNA replication rather than to slow turnover of the mtDNA. As with the 7S DNAs (Fig. 5C), there was no evidence of a retained primer associated with the nascent strands of mtDNA in V142I RNase H1 fibroblasts (Fig. 6B and SI Appendix, Fig. S6C), in contrast to murine cells lacking RNase H1 (15).
Fig. 6.
V142I RNase H1 has opposite effects on the abundance of mitochondrial D-loops and R-loops. (A) Whole-cell DNA from control (C) and V142I RNase H1 (P) fibroblasts was denatured in 80% (vol/vol) formamide for 15 min at 85 °C and fractionated by 1D-AGE. Where indicated, samples were digested with Bsu361 or Xho1. After blot transfer, nascent H-strands and 7S DNA were detected by hybridization to probe h-H15,869–168. (B) Equivalent DNA samples were denatured after digestion with BsaW1 to shorten the nascent strands and allow higher-resolution mapping on 2% (wt/vol) agarose, sodium borate gels (15, 46); under these conditions nascent strands with retained primers would resolve above the np 323 marker (SI Appendix, Fig. S6C). (C) Ban2- and Acc1-digested whole-cell DNAs of control HEK cells (C) or cells expressing Twinkle DNA helicase (Twinkle) and from control (C) or V142I RNase H1 (V142I) fibroblasts were subjected to 1% agarose, 1D-AGE and blot hybridized to h-H15,869–168. HEK cell DNA was psoralen/UV cross-linked before extraction (SI Appendix).
In terms of our prior knowledge of mtDNA replication and maintenance, it was difficult to explain the marked increase in 7S DNA caused by mutant RNase H1 (Fig. 6A), because the only candidate substrate, the so-called “7S RNA” (primer), appears to be processed normally in cells with V142I RNase H1 (Figs. 5 and 6B and SI Appendix, Fig. S6C). If RNase H1 acts exclusively on RNA/DNA hybrids and does not have an unrecognized function, 7S DNA levels must be influenced by RNA other than 7S RNA (the primer spanning LSP to Ori-H). The mitochondrial R-loop is a clear alternative candidate. As a test of the potential impact of V142I RNase H1 on the mitochondrial R-loop, we again used a blocked-site assay by digesting human fibroblast DNA with Acc1 and Ban2 (Fig. 6C). Human mtDNA has an Acc1 site at np 15,255, ∼850 nt downstream of the 3′ end of the D-loop. Ban2 has two sites in the D-loop region of human mtDNA [and, extrapolating from the mouse data (Fig. 2), also in the R-loop] at np 16,459 and 40 and a third site outside the CR at np 629. Hence, the R-loop should prevent cleavage by Ban2 at np 16,459 and np 40, generating a fragment of 1.95 kb. [D-loops, on the other hand, are susceptible to Ban2 digestion (SI Appendix Fig. S3)]. A species commensurate with a fragment containing blocked sites at np 16,459 and np 40 was readily detectable in HEK cells and in control fibroblasts, but this species was markedly less abundant in the V142I RNase H1 fibroblasts (and in cells expressing high levels of recombinant Twinkle DNA helicase) (Fig. 6C). The mitochondrial R-loop, resolved by 2D-AGE analysis of mtDNA after interstrand cross-linking, was also less abundant in fibroblasts containing V142I RNase H1 than in controls (SI Appendix, Fig. S7). Hence, there were fewer R-loops in the fibroblasts of the patient with mutant RNase H1 than in control fibroblasts.
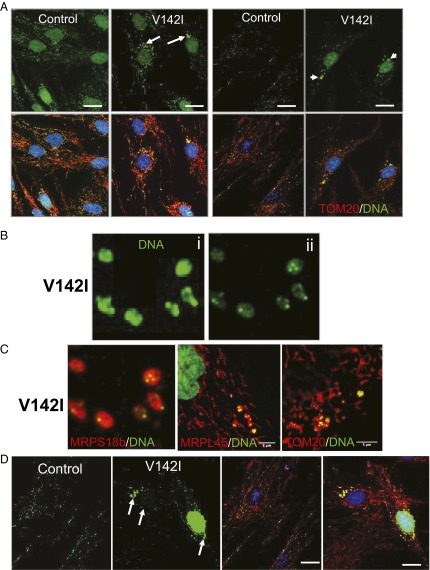
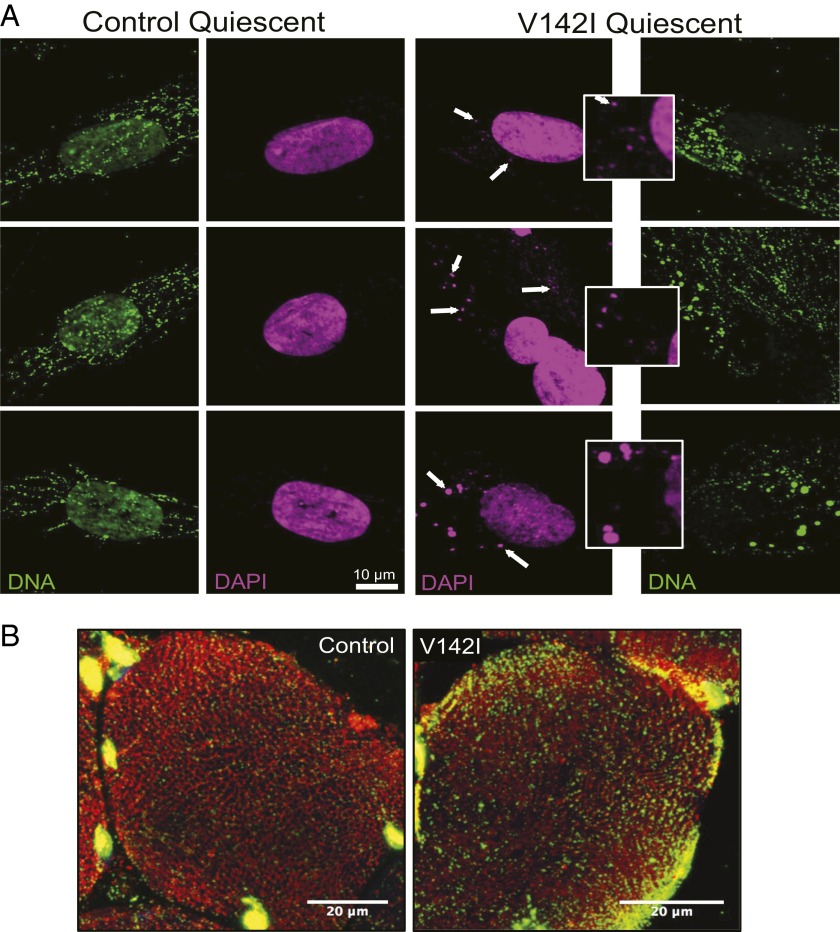
The high abundance of mitochondrial R-loops in normal cells and tissues and the maintenance of nearly normal mtDNA copy numbers when R-loops are scarce (Figs. 1, 4A, and 6C and SI Appendix, Fig. S7) suggested that many R-loops are not directly involved in replication. The portion of the mtDNA including the CR is implicated in the association of the mtDNA with the inner mitochondrial membrane (26), and we have described multicopy fragments of mtDNA encompassing the CR, held together by protein, that are putative intermediates of the segregation process. These findings led us to propose that the D-loop is involved in the organization and segregation of mtDNA (10, 11); the same inferences apply to the mitochondrial R-loop. Therefore, we determined the distribution and organization of mtDNA in fibroblasts with mutant and wild-type RNase H1. Immunocytochemistry showed that more than one-third of the mutant primary fibroblasts have abnormally large mtDNA foci (nucleoids) (Fig. 7A and SI Appendix, Fig. S8) occupying as much as 200 times the volume of the typical (modal) mtDNA foci of control fibroblasts. Deconvolution analysis of the microscope images revealed closely packed, smaller, discrete foci ∼0.3 µm in diameter (Fig. 7B), similar to those in the control fibroblasts analyzed here and previously (27), leading us to infer that the enlarged mtDNA foci comprise clusters of single copies of mtDNA. The multipartite nature of the enlarged mitochondrial nucleoids suggests that RNase H1 V142I impedes the physical segregation of mtDNA molecules. Attribution of the enlarged nucleoid phenotype to the pathological variant of RNase H1 is supported by observations that the loss of RNase H1 in murine fibroblasts or its depletion in human 143B cells also results in the formation of abnormally large mtDNA foci (SI Appendix, Fig. S9). The inability to distribute mtDNA normally is associated with perturbation of the mitochondrial network, because the mitochondria themselves form dense bodies around the aggregates of mtDNA (Fig. 7C). Hence, the distribution of mtDNA and mitochondria might be coupled. Despite the aggregation of nucleoids and mitochondria, the clusters of mtDNAs remained replication competent, because labeling of cellular DNA for a period of 10 h with the nucleotide analog BrdU and immunocytochemistry to the incorporated BrdU produced a staining profile similar to that of anti-DNA labeling (Fig. 7D). Mitochondrial disease manifests in nondividing cells, so quiescent cells potentially offered a context more closely resembling that of the affected tissues (muscle and brain). After cell growth was stopped via serum starvation, mtDNA foci were larger than in the corresponding proliferating cells; in the patient-derived cells the clusters of mtDNA reached a size and DNA concentration that was visible with DAPI staining, in contrast to control cells (Fig. 8A). Next, we extended the analysis of mtDNA organization to muscle sections and found that clustering was also evident in an affected tissue, muscle and that the distribution of mtDNA foci was not as uniform as in control fibers (Fig. 8B and SI Appendix, Fig. S10). MtDNA clustering and disorganization were evident in a majority of the patient’s muscle fibers, whereas only a small percentage displayed mitochondrial proliferation based on succinate dehydrogenase staining (SI Appendix, Fig. S5 B, ii). Therefore the enlarged mtDNA foci are attributable to impaired segregation and are not a secondary consequence of mitochondrial biogenesis and proliferation.
Fig. 7.
V142I RNase H1 causes mtDNA aggregation and disruption of mitochondrial morphology. (A) Confocal single optical images of primary human fibroblasts from a control and from the index case with V142I RNase H1 labeled with antibodies to DNA stained green; the outer mitochondrial membrane protein, translocase of outer membrane 20 (TOM20), and the mitochondrial ribosomal proteins MRPS18b and MRPL45, stained red. DNA in the mitochondria produces colocalization of green and red (yellow). In some cases nuclear DNA was stained with DAPI (blue). White arrows indicate the mtDNA clusters observed in the patient-derived cells. (B, i) A raw confocal optical section of a region of a V142I patient fibroblast depicting antibody staining against mtDNA (green). (ii) The reconstruction of i treated with a custom deconvolution algorithm to extend the resolution and showing that the large mtDNA foci comprise multiple smaller units of similar-sized particles. (C) Such particles colocalize (yellow) with staining for antibodies against the mitochondrial ribosomal subunits MRPS18b and MRPL45 and the outer mitochondrial membrane protein TOM20. (D) Single confocal optical section of V142I RNaseH1 and control fibroblasts treated with 20 µM BrdU. Replicating DNA is visualized with antibodies against BrdU (green), and mitochondria are visualized with anti-Tom20 (red); cell nuclei are stained with DAPI (blue). Images show clustering of the BrdU signal in V142I RNaseH1 fibroblasts cells to the DNA clustering in other panels and in SI Appendix, Fig. S3. (Scale bars: 20 µm in A and D; 5 µm in C.)
Fig. 8.
V142I RNase H1 quiescent cells and muscle display mtDNA aggregation. Confocal optical images of quiescent human fibroblasts (A) and a 12-μm transverse section of muscle (B) from a control and from the index case with V142I RNase H1. Samples were labeled with antibodies to DNA (green) and the outer mitochondrial membrane protein TOM20 (red) and were stained with DAPI (magenta). White arrows and zoomed images highlight some of the mtDNA clusters observed in the patient-derived cells stained with DAPI. (B) Tom20 and DNA merged images; for individual components, DAPI staining, and additional muscle fibers see SI Appendix, Fig. S10.
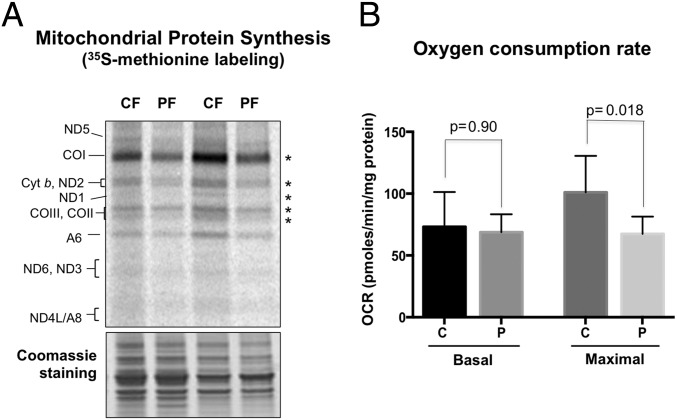
In the cultured cells with V142I RNase H1 there was no evident perturbation of transcription based on the steady-state levels of four mature mitochondrial transcripts, nor was there an increase in precursor RNAs, suggesting that RNA processing is not compromised in the mutant fibroblasts (SI Appendix, Fig. S11). However, as evidenced by [35S]methionine labeling of mitochondrial translation products (Fig. 9A), the mitochondria of V142I RNase H1 fibroblasts displayed rates of protein synthesis one-third lower than that of controls (with the exception of ATP synthase subunit A6, which gave inconsistent results), and their respiratory capacity was compromised (Fig. 9B). These defects might relate to the perturbation of the intimate connections between the translation machinery and the mtDNA (28), perhaps because the mitochondrial R-loop includes a putative ribosome-binding sequence (21).
Fig. 9.
V142I RNase H1 impairs mitochondrial translation and respiration. (A) One-hour [35S]methionine labeling of nascent mitochondrial polypeptides in control (CF) or V142I RNase H1 patient (PF) fibroblasts, fractionated by 12% SDS/PAGE. Tentative polypeptide assignments are identified to the left of the gel: ND1-6, cyt b, COX1-3 and A6, A8 are subunits of respiratory chain complexes I, III, and IV and ATP synthase, respectively. The panel shows two of four experiments. The pair of samples to the right was derived from cells grown to full confluence. V142I cells displayed a 34% decrease in mitochondrial translation compared with the two control cell lines analyzed, based on quantitation of the indicated bands (*). (B) Mitochondrial oxygen consumption rate (OCR) was measured using a Seahorse flux analyzer before (basal) and after the addition of the uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, Sigma; maximal), having subtracted the nonmitochondrial (rotenone-insensitive) OCR. The data represent the mean ± SEM of four independent experiments, each performed in duplicate. Statistical analysis was performed using an unpaired two-tailed Student's t test. Maximal respiration of the V142I (P) cells was significantly lower than that of the control (C) cells (P = 0.018).
Discussion
The discovery of pathological mutations in human RNase H1 advances our understanding of mitochondrial disorders and highlights the fundamental role of the enzyme and RNA in mtDNA metabolism. The pathologies associated with the human mutation and the loss of RNase H1 activity in the mouse are strikingly different. Rnaseh1 ablation is incompatible with development beyond early embryogenesis (13), whereas the missense mutation in RNASEH1 results in an adult-onset neuromuscular disease. The molecular defects also differ: An absence of murine RNase H1 results in mtDNA depletion and primer retention (15), but neither occurs with the human mutant RNase H1 (Figs. 4A and 5). Two striking changes in the mtDNA of cells harboring V142I RNase H1 are the increase in D-loops and the decrease in the newly recognized R-loop.
The detection of the mitochondrial R-loop comes almost half a century after the first descriptions of the mitochondrial D-loop (6, 7). Both are triple-stranded species with the third strand located in the major noncoding region, which frequently is described as the least conserved part of the mitochondrial genome; however, apart from three short hypervariable regions (29), most of the CR is well conserved, and the portion known as the “conserved central domain” (8, 20) defines much of the overlap between the D-loop and the R-loop (Fig. 2). The high abundance and location of the triple-stranded species and the perturbations described in this report, combined with earlier studies of the D-loop, suggest they have important roles in mtDNA maintenance and expression.
The V142I substitution results in lower RNase H activity than the wild-type enzyme (14) but is associated with a marked decrease in R-loops in the fibroblasts of the patient studied here (Fig. 6C and SI Appendix, Fig. S7). Human cells might activate a back-up mechanism for processing RNA/DNA hybrids in response to RNase H1 deficiency but with poorer powers of discrimination, causing the mitochondrial R-loop to be dislodged and degraded prematurely. However, the V142I RNase H1 protein appears to be unstable (Fig. 4 B and C), potentially because of its susceptibility to oxidation (30), raising the intriguing possibility that the mutant protein is modified in vivo to accelerate its turnover or render it inactive (30) to prevent a dominant negative phenotype from manifesting. Inactivation by oxidation would be desirable if the V142I substitution disrupted RNase H1’s interaction with a protein partner(s) that restricted its actions. Even at 40% of full activity (14), unregulated V142I RNase H1 would be expected to degrade RNA/DNA hybrids inappropriately and thereby could increase the rate of turnover of mitochondrial R-loops. All that said, a definitive demonstration that R-loop depletion is the result of V142I RNase H1 awaits the creation of a cellular or animal model with this mutation.
The increase in the abundance of D-loops in cells with mutant RNase H1 can be explained in one of two ways. The D-loop is the precursor of the mitochondrial R-loop. Thus, more D-loops are made to increase R-loop synthesis, enabling some R-loops to survive long enough to perform their (critical) function(s), i.e., to compensate for unfettered RNase H1 (V142I) degrading the mitochondrial R-loop rapidly or accelerated R-loop turnover caused by an imperfect substitute for the mutant enzyme. Alternatively, the reciprocal relationship between the D-loop and the R-loop in the fibroblasts carrying mutant RNase H1 could result from overlapping functions: D-loops might not be optimal for mtDNA segregation but serviceable when R-loops are scarce, although the increase in D-loops appears disproportionate. Further study of the D-loop could help resolve its relationship to the R-loop; if D-loops beget R-loops, then decreasing the level of D-loops in the fibroblasts with mutant RNase H1 would decrease R-loop levels still further, and situations that result in a decrease in D-loops would be accompanied by a decrease in R-loops or increased R-loop stability.
The impact of RNase H1 on mtDNA segregation strongly implies RNA involvement. Such a role might have been anticipated earlier, because mitochondria retain features of their prokaryotic ancestors and RNA has long been proposed to play an important role in nucleoid organization in bacteria (31). Furthermore, RNA polymerase has been linked to bacterial chromosomal segregation (32). Thus, the mtDNA aggregates of V142I RNase H1 cells, formed when the abundance of mitochondrial R-loops is low, potentially point to an analogous arrangement and process in the mitochondria, in which RNase H1 is required to process products of the mitochondrial RNA polymerase (POLRMT) to facilitate mtDNA segregation.
There are two ways in which POLRMT could generate the mitochondrial R-loop. To produce all the mature RNAs required for gene expression, the heavy-strand promoter-derived (HSP) polycistronic transcript must cover almost the entire mitochondrial genome (33); the tRNA gene furthest from the promoter, tRNAThr, is separated from the CR by the anti-sense sequence of tRNAPro. Thus, POLRMT needs to extend the synthesis of the minimal H-strand polycistronic RNA only a short distance beyond tRNAThr to enter the CR and create an RNA corresponding to the mitochondrial R-loop. The other means by which POLRMT could readily generate the mitochondrial R-loop is via promoter-independent synthesis on the single-stranded template formed by the D-loop. The site in the CR known as “Ori-b” is a good candidate: It functions as a starting point for DNA synthesis on both strands (3, 15), presumably preceded by a short primer in the case of the L-strand, and more than 40% of the sequenced R-loops have a 5′ end within 25 nt of Ori-b (Fig. 2). Any triplex regions of DNA in the D-loop would be disrupted in the process of LC-RNA synthesis, reducing the stability of the 7S DNA and thus facilitating its removal, leaving the mitochondrial R-loop. The removal of 7S DNA can readily be achieved, given that RNA/DNA hybrids have higher melting temperatures than equivalent duplex DNAs and that a number of DNA helicases are poor at unwinding RNA/DNA duplexes (34). Both methods of generating R-loops might exist for different purposes. Polycistronic HSP transcripts extending to LSP can enable transcript-dependent (also known as “bootlace”) replication (TDR) (17), because, according to the model, RNA is hybridized to the lagging strand template from its inception. That is, TDR requires an RNA covering much of the CR (LC-RNA). If the LC-RNA for TDR derives from HSP transcripts, then there is a tight coupling of transcription and replication, and they are consecutive, not concurrent, events [viz. transcription initiates at HSP and proceeds around the mtDNA terminating at LSP, with the final portion of the transcript serving as the (lagging strand) initiator RNA for TDR]. Synthesizing R-loops on D-loops, rather than deriving them from HSP transcripts, would separate R-loop production from mitochondrial gene expression, and these R-loops could be dedicated to DNA organization and segregation.
In both schemes for POLRMT-mediated synthesis of R-loops, the putative cloverleaf structure TAS could play a key regulatory role. Binding of a protein to TAS could arrest transcription [in the manner of mitochondrial transcription termination factor (mTERF) at another site in the mitochondrial genome (35)] and thereby prevent POLRMT progression into the CR, denying the mtDNA the first RNA needed for TDR. In the case of R-loop synthesis via the D-loop, TAS could facilitate recruitment of POLRMT or other accessory factors to the appropriate start site. POLG2 is an excellent candidate for binding to TAS, based on its D-loop–binding properties (10) and homology to amino acyl tRNA synthetases (36). Furthermore the mitochondrial R-loops have a displaced DNA strand that binds SSB (Fig. 3B), suggesting an additional role for mitochondrial SSB in mtDNA maintenance.
The discovery of the mitochondrial R-loop may necessitate a reassessment of almost all aspects of mtDNA metabolism. Because R-loop numbers are low in the cells of a patient with mutant RNase H1 and mtDNA abnormalities, investigation of the abundance and behavior of the mitochondrial R-loop in the context of pathological mutants of Twinkle DNA helicase (37), mitochondrial genome maintenance exonuclease 1 (MGME1) (38), FARS2 (39), FBXL4 (40), and POLG (41) could prove informative. Human cultured cells are an appropriate model system for the study of this and many other aspects of mtDNA replication and maintenance. For a protracted period it was thought that cultured cells did not recapitulate the prominent mtDNA abnormalities (depletion or multiple deletions) seen in the tissues of patients. However, although proliferating cultured cells of patients with defects in deoxynucleotide metabolism display normal mtDNA levels, in most cases mtDNA depletion occurs when these cells exit the cell cycle (e.g., refs. 42 and 43), and a revaluation of the phenomenon of multiple deletions suggests these abnormalities are also present in cultured cells. The existing data from muscle samples are consistent with the view that multiple deletions are broken replication intermediates. It is possible, even likely, that most replication intermediates are intact before extraction, because linear DNA molecules are unlikely to persist for long, given the dangers of strand invasion and the aberrant recombination products they could generate. Thus, the accumulated replication intermediates seen in the mutant RNase H1 fibroblasts (Fig. 6A) could be directly related to the multiple deletions detected in the muscle of the patient; however, their contribution to the pathology in this and the allied disorders remains to be established.
Materials and Methods
Methods are fully described in SI Appendix. MtDNA from tissue was prepared as described previously (17) but without a 50 °C incubation step. Digestion and fractionation of native or denatured mtDNA was as performed previously (15, 17). Mitochondrial translation was via [35S]methionine labeling in the presence of emetine (44), and oxygen consumption was measured using a XF flux analyzer. CR RNA was sequenced after reverse transcription and cloning. MEF mtDNAs were prepared as described previously (15). Cell culture, RNAi, mtDNA isolation, nucleic acid digestion, modification, and analysis were as described previously (15, 17, 25, 45). This study was performed under the ethical guidelines issued by University College London for clinical studies. Written informed consent was obtained from all subjects before genetic testing.
Supplementary Material
Acknowledgments
J.B.H. was an NIH-CamGrad Scholar (2006–2010). M.M. is supported by the European Commission, MEET Project Grant 317433. The study was funded by a Medical Research Council (MRC) Intramural Award (to I.J.H.) and MRC Senior Non-Clinical Fellowship MC_ PC_ 13029 (to A.S.). R.D.S.P., H.H., and M.G.H. are supported by MRC Centre for Neuromuscular Diseases Grant G0601943, the UK National Health Service Specialised Service for Rare Mitochondrial Diseases of Adults and Children, and the National Institute for Health Research University College London Hospitals/University College London Biomedical Research Centre. R.J.C. is supported by the NIH Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600537113/-/DCSupplemental.
References
- 1.Area-Gomez E, Schon EA. Mitochondrial genetics and disease. J Child Neurol. 2014;29(9):1208–1215. doi: 10.1177/0883073814539561. [DOI] [PubMed] [Google Scholar]
- 2.Kasamatsu H, Vinograd J. Unidirectionality of replication in mouse mitochondrial DNA. Nat New Biol. 1973;241(108):103–105. doi: 10.1038/newbio241103a0. [DOI] [PubMed] [Google Scholar]
- 3.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18(6):651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Fish J, Raule N, Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306(5704):2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 5.Chang DD, Clayton DA. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: A functional promoter consists of multiple upstream domains. Mol Cell Biol. 1986;6(9):3253–3261. doi: 10.1128/mcb.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnberg A, van Bruggen EF, Borst P. The presence of DNA molecules with a displacement loop in standard mitochondrial DNA preparations. Biochim Biophys Acta. 1971;246(2):353–357. doi: 10.1016/0005-2787(71)90147-x. [DOI] [PubMed] [Google Scholar]
- 7.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc Natl Acad Sci USA. 1971;68(9):2252–2257. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walberg MW, Clayton DA. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annex BH, Williams RS. Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle. Mol Cell Biol. 1990;10(11):5671–5678. doi: 10.1128/mcb.10.11.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Re M, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37(17):5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176(2):141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller W, Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci USA. 1972;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerritelli SM, et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11(3):807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 14.Reyes A, et al. RNASEH1 Mutations Impair mtDNA Replication and Cause Adult-Onset Mitochondrial Encephalomyopathy. Am J Hum Genet. 2015;97(1):186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes JB, et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci USA. 2015;112(30):9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pohjoismäki JL, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010;397(5):1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyes A, et al. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013;41(11):5837–5850. doi: 10.1093/nar/gkt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasukawa T, et al. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25(22):5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayona-Bafaluy MP, et al. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31(18):5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown GG, Gadaleta G, Pepe G, Saccone C, Sbisà E. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J Mol Biol. 1986;192(3):503–511. doi: 10.1016/0022-2836(86)90272-x. [DOI] [PubMed] [Google Scholar]
- 21.Ojala D, Crews S, Montoya J, Gelfand R, Attardi G. A small polyadenylated RNA (7 S RNA), containing a putative ribosome attachment site, maps near the origin of human mitochondrial DNA replication. J Mol Biol. 1981;150(2):303–314. doi: 10.1016/0022-2836(81)90454-x. [DOI] [PubMed] [Google Scholar]
- 22.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 23.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1997;272(24):15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 24.Yang MY, et al. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111(4):495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 25.Ruhanen H, Ushakov K, Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim Biophys Acta. 2011;1813(12):2000–2007. doi: 10.1016/j.bbamcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci USA. 1977;74(4):1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, et al. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40(13):6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz S, Weisser HJ, Heizmann J, Pollak S. A third hypervariable region in the human mitochondrial D-loop. Hum Genet. 1997;101(3):384. [PubMed] [Google Scholar]
- 30.Lima WF, et al. Human RNase H1 activity is regulated by a unique redox switch formed between adjacent cysteines. J Biol Chem. 2003;278(17):14906–14912. doi: 10.1074/jbc.M211279200. [DOI] [PubMed] [Google Scholar]
- 31.Pettijohn DE, Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Kruse T, et al. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 2006;20(1):113–124. doi: 10.1101/gad.366606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bestwick ML, Shadel GS. Accessorizing the human mitochondrial transcription machinery. Trends Biochem Sci. 2013;38(6):283–291. doi: 10.1016/j.tibs.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santamaría D, et al. DnaB helicase is unable to dissociate RNA-DNA hybrids. Its implication in the polar pausing of replication forks at ColE1 origins. J Biol Chem. 1998;273(50):33386–33396. doi: 10.1074/jbc.273.50.33386. [DOI] [PubMed] [Google Scholar]
- 35.Terzioglu M, et al. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013;17(4):618–626. doi: 10.1016/j.cmet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Fan L, Sanschagrin PC, Kaguni LS, Kuhn LA. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: Implications for a dual role as a primer recognition factor and processivity clamp. Proc Natl Acad Sci USA. 1999;96(17):9527–9532. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spelbrink JN, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28(3):223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 38.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45(2):214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almalki A, et al. Mutation of the human mitochondrial phenylalanine-tRNA synthetase causes infantile-onset epilepsy and cytochrome c oxidase deficiency. Biochim Biophys Acta. 2014;1842(1):56–64. doi: 10.1016/j.bbadis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnen PE, et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93(3):471–481. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Goethem G, Dermaut B, Löfgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28(3):211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 42.Dalla Rosa I, et al. MPV17 Loss Causes Deoxynucleotide Insufficiency and Slow DNA Replication in Mitochondria. PLoS Genet. 2016;12(1):e1005779. doi: 10.1371/journal.pgen.1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saada A. Deoxyribonucleotides and disorders of mitochondrial DNA integrity. DNA Cell Biol. 2004;23(12):797–806. doi: 10.1089/dna.2004.23.797. [DOI] [PubMed] [Google Scholar]
- 44.Dalla Rosa I, et al. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014;42(13):8500–8515. doi: 10.1093/nar/gku513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyes A, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39(12):5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brody JR, Kern SE. Sodium boric acid: A Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques. 2004;36(2):214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.