Significance
G-quadruplex (GQ) is a four-stranded DNA structure that forms in the human genome and influences gene expression and DNA replication. Stably formed GQ structures can act as a physical blockade to disrupt genomic processes. To counteract such an effect, cells possess special helicases dedicated to unfolding GQ structures. In our study, we examined three such GQ-resolving helicases, using single-molecule imaging, that show distinct binding preference to different conformations (folding directions) of GQ DNA. Nevertheless, all three GQ resolvases use the same mechanism, which involves repetitive cycles of GQ unfolding in successive runs. Such activity leads to efficient GQ hybridization with a complementary DNA and dislodging chemical GQ ligand. Our findings have implications for designing new GQ ligands for therapeutic purposes.
Keywords: G-quadruplex, resolving activity, RHAU, BLM, WRN
Abstract
G-quadruplex (GQ) is a four stranded DNA secondary structure that arises from a guanine rich sequence. Stable formation of GQ in genomic DNA can be counteracted by the resolving activity of specialized helicases including RNA helicase AU (associated with AU rich elements) (RHAU) (G4 resolvase 1), Bloom helicase (BLM), and Werner helicase (WRN). However, their substrate specificity and the mechanism involved in GQ unfolding remain uncertain. Here, we report that RHAU, BLM, and WRN exhibit distinct GQ conformation specificity, but use a common mechanism of repetitive unfolding that leads to disrupting GQ structure multiple times in succession. Such unfolding activity of RHAU leads to efficient annealing exclusively within the same DNA molecule. The same resolving activity is sufficient to dislodge a stably bound GQ ligand, including BRACO-19, NMM, and Phen-DC3. Our study demonstrates a plausible biological scheme where different helicases are delegated to resolve specific GQ structures by using a common repetitive unfolding mechanism that provides a robust resolving power.
G-quadruplex (GQ) is a noncanonical secondary structure of DNA in which two or more sets of four guanines form into stacks of tetrad rings stabilized by monovalent cations, such as potassium or sodium (1). GQ DNA can exhibit different conformations dictated by the arrangement of the loop sequences that intervene between the consecutive runs of guanines. GQ is classified as “parallel” when all guanine bases point in the same direction and “nonparallel” when they do not (2). Recent studies reported evidence that the GQ structures form in human cells and that the distribution and abundance change throughout the cell cycle (3, 4). Bioinformatic studies have revealed that the human genome is highly enriched with potential GQ-forming sequences and that many are associated with genes implicated in human diseases (5).
The formation of GQ DNA and GQ RNA can modulate essential cellular processes, including replication, transcription, and translation (6–8). Stable GQ structures can act as physical barriers to down-regulate gene expression or block replication, for example. Such inhibitory effects can be counteracted by helicases, such as Bloom helicase (BLM), Werner helicase (WRN), FANCJ, PIF1, and RNA helicase AU (associated with AU rich elements) (RHAU) (G4 resolvase 1), all of which can bind and unfold GQ structures (9–12). Ablation of RHAU in germ cells caused an accumulation of G4 structures (13). Many GQ-resolving helicases have a higher affinity toward GQ DNA than to other forms of single-strand (ss) or double strand (ds) DNA. Although they differ in directionality and substrate specificity, it is not clear whether these helicases have an overlapping or complementary role in resolving GQ DNA structures. A recent study reported the structure of the GQ recognition domain of RHAU, which revealed an exquisite architecture specifically designed to fit into the parallel GQ structure (14). In agreement, our earlier study also demonstrated by EMSA that RHAU exhibits specific binding to parallel GQ DNA (15). Such distinct structural features seen in this protein suggest that different helicases may be delegated to unfold a subset of specific structures of GQ DNAs in cells.
We sought to investigate several outstanding questions regarding the GQ-resolving activity: (i) the potential role of GQ-unfolding helicases in targeting specific conformers of GQ DNA, (ii) the mechanism by which helicases resolve the GQ structures, (iii) the outcome of the resolving activity in the context of duplex DNA, and (iv) the ability of resolvases to compete with GQ binding ligand. We tested three GQ-resolving helicases (RHAU, BLM, and WRN) on a set of parallel and nonparallel GQ DNAs. We show that the three helicases exhibit distinct GQ-conformation specificity, yet they all use a common mechanism of repetitive unfolding to destabilize the GQ structure in successive runs. Such activity is sufficient to enable annealing, in a cis-specific manner, and removing a GQ ligand bound to GQ DNA. Our findings point to a plausible orchestration of different types of helicases assigned to unfold specific subsets of GQ conformations.
Results
RHAU Binding and Activity Is Specific to Parallel GQ Structure.
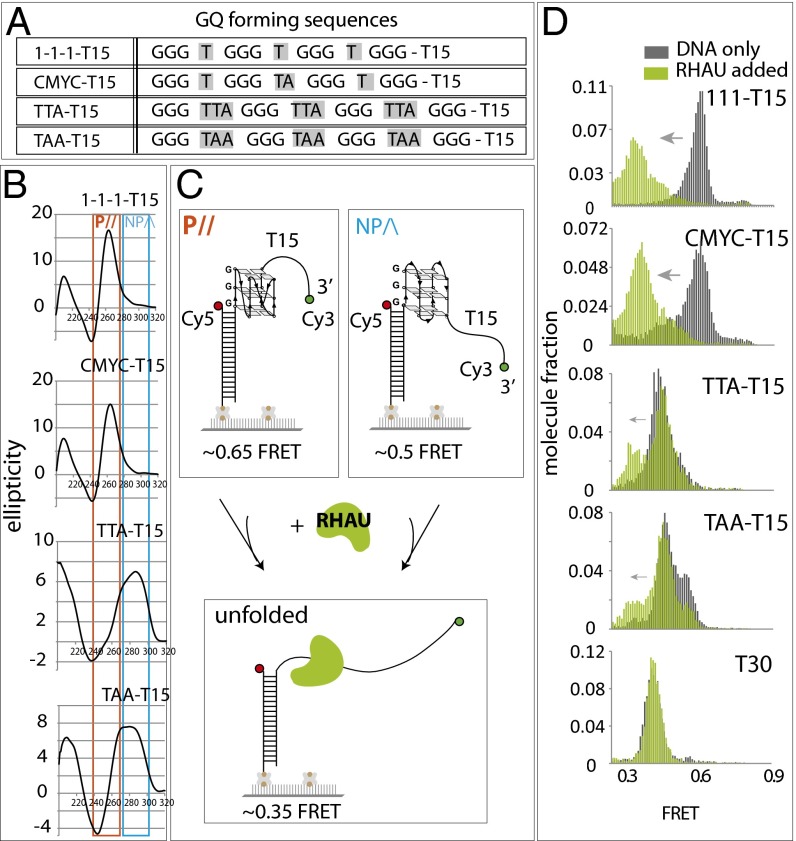
We reported previously that RHAU (G4R1, DDX36) interacts preferentially with parallel GQ conformation (15), and the structural basis of the parallel GQ specificity was provided by a recent study (14). To investigate the GQ-resolving activity of RHAU, we prepared FRET-DNA substrates, including 111 and CMYC, which represent primarily parallel conformation, and TTA and TAA, which exhibit a primarily antiparallel (nonparallel) signature of GQ folding (15). Each DNA is named after the composition of the loop sequence as shown (Fig. 1A). The T15 was added for optimal loading of RHAU (11, 15, 16). The circular dichroism (CD) analysis on four DNAs confirmed that both 111-T15 and CMYC-T15 still fold into highly parallel GQs and that the TTA-T15 and TAA-T15 exhibit nonparallel folding as reported previously (15, 17–20) (Fig. 1B). Each GQ-DNA substrate is prepared as a partially duplexed DNA with Cy3 (green) and Cy5 (red) dyes located near either end of the GQ-forming region to measure the folding (formation) and unfolding (resolution) of GQ that occurs within the FRET-sensitive distance range (Fig. 1C). The biotin at one end of the dsDNA enables tethering individual DNA molecules to a NeutrAvidin-coated surface (21).
Fig. 1.
RHAU binding and activity is specific to parallel GQ. (A) GQ DNA sequences used in the study. (B) CD spectrum of 111-T15, CMYC-T15, TTA-T15, and TAA-T15. Orange and blue boxes indicate signature of parallel GQ and nonparallel GQ, respectively. (C) Schematic diagram of single-molecule FRET experiment depicting parallel and nonparallel GQ unfolded by RHAU. (D) FRET histogram of above GQ substrates before (gray) and after (green) addition of RHAU.
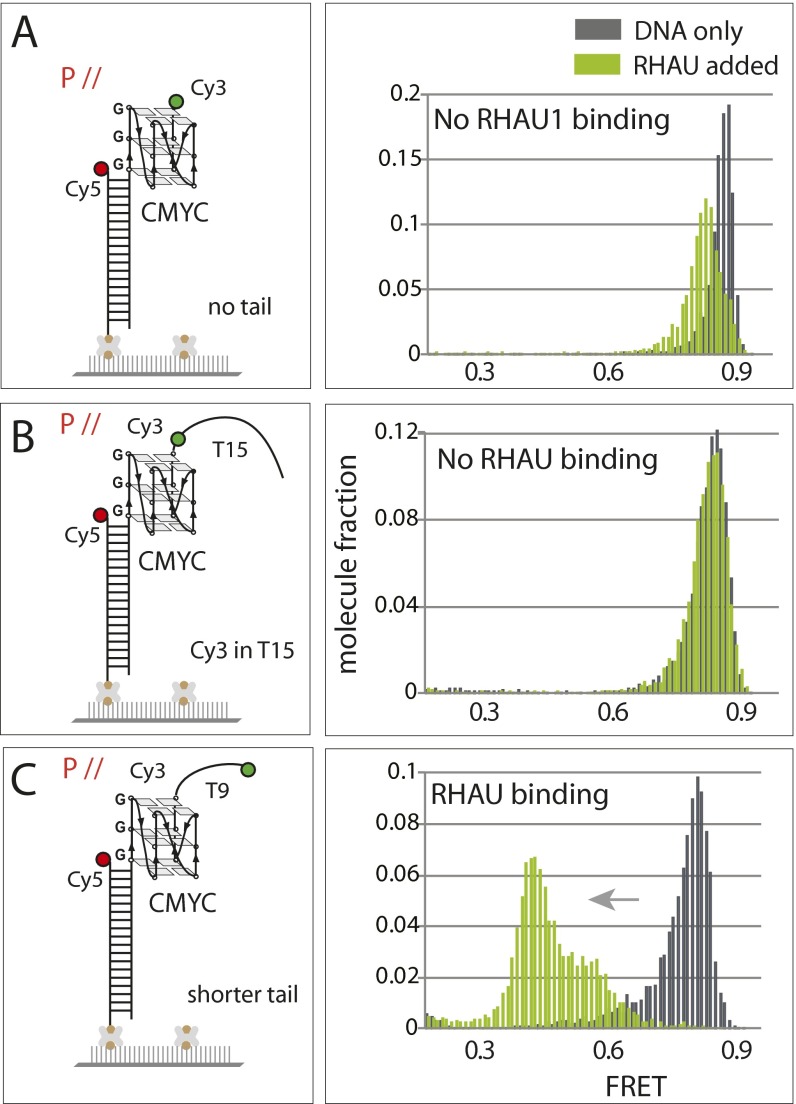
The FRET values that are collected from over 4,000 single molecules obtained from approximately 20 fields of view are built into a FRET histogram. The 111-T15 and CMYC-T15 yielded FRET peaks at 0.65–0.7 whereas the TTA-T15 and TAA-T15 displayed a lower FRET peak at 0.5 (Fig. 1D, in gray). The difference in the FRET peaks likely arises from the different conformations of GQ as depicted in Fig. 1C. Upon addition of RHAU (7.5 nM), the histograms shifted to lower FRET values in 111-T15 and CMYC-T15, but not significantly in the TTA-T15 and TAA-T15 substrates, suggesting that RHAU binds specifically to the parallel conformers of GQ (Fig. 1D, in green). In addition, the FRET decrease exhibited in the two parallel GQ substrates suggests that the RHAU is likely disrupting the folded structure of GQ. The FRET change was not detected when the GQ DNA did not possess an ssDNA tail or when the Cy3 dye was located adjacent to the GQ structure (Fig. S1 A and B). We note that 9 nt of ssDNA were sufficient for RHAU binding (Fig. S1C). Based on the substrates tested above, RHAU likely makes an intimate contact with both the GQ (parallel) and the ssDNA.
Fig. S1.
RHAU binding requires parallel GQ with uninterrupted ssDNA overhang. (A) FRET construct for GQ DNA without ssDNA tail and the FRET histogram collected before and after addition of RHAU. RHAU does not bind parallel GQ without an ssDNA overhang. (B) RHAU does not bind if the ssDNA overhang contains a fluorophore internally (3 nt away from GQ). (C) The 9 nt of ssDNA is sufficient to allow RHAU binding.
RHAU Resolves GQ Structure by Repetitive Unfolding Activity.
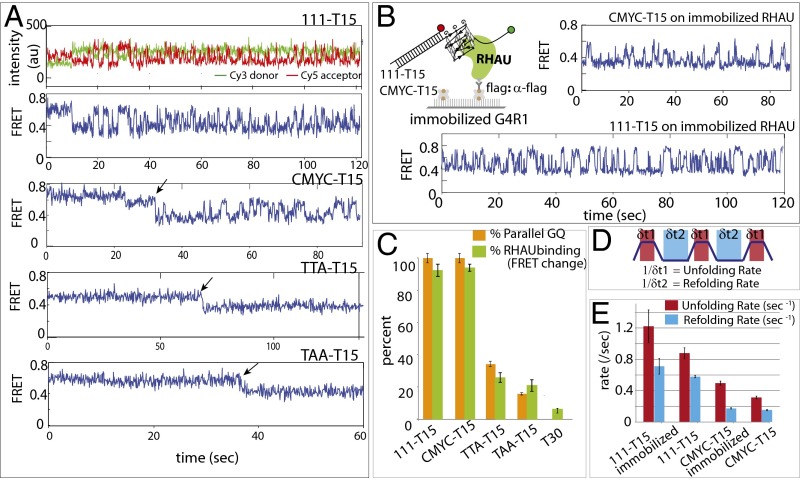
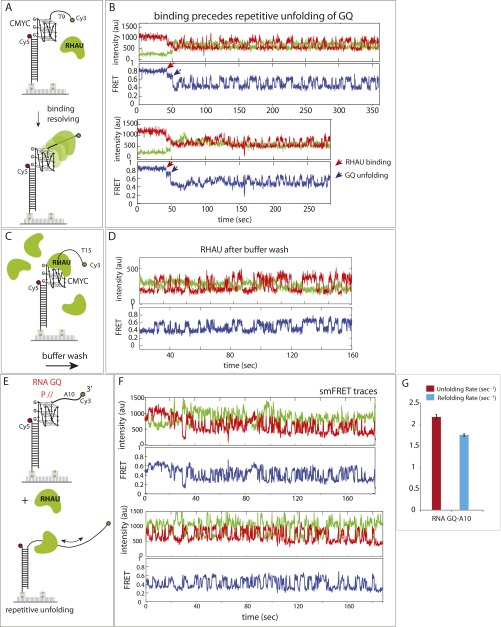
To study the mechanism responsible for the RHAU activity seen above, we examined single molecule FRET traces. Binding of RHAU to both 111- and CMYC-T15 induced FRET fluctuation (anticorrelated change between Cy3 and Cy5 signals), reporting on a periodic distance change between the two FRET dyes, likely resulting from successive GQ-unfolding activity (Fig. 2A, Top). This activity, which does not depend on ATP, is consistent with the broad FRET peak shown in Fig. 1D. When RHAU was applied to a substrate with a shorter ssDNA tail (CMYC-T9), the initial protein binding and subsequent unfolding repetition were well-distinguished as a stepwise decrease from 0.8 to 0.7, followed by FRET fluctuation that oscillates between 0.35 and 0.6 (Fig. S2 A and B). These data suggest that the FRET fluctuation likely arises from an activity of a single RHAU rather than successive binding and rebinding of multiple units. Further, the FRET fluctuation continued after a buffer wash of excess proteins in reaction, also suggesting that a single RHAU is responsible for the repetitive cycle of unfolding and refolding of GQ (Fig. S2C). For TTA- and TAA-T15 DNAs, a majority of traces showed no change in FRET, with a small fraction of molecules displaying a one step FRET decrease (Fig. 2A, Bottom). To test more definitively whether the repetitive FRET fluctuation seen in 111- and CMYC-T15 arises from a single unit of RHAU or successive binding of many units, we immobilized flag-tagged RHAU on a single-molecule imaging surface coated with anti-flag antibody (22, 23). CMYC-T15 DNA applied to immobilized RHAU produced a highly similar repetitive FRET pattern, suggesting that the FRET fluctuation is indeed due to a single protein activity rather than successive binding of multiple RHAU (Fig. 2B). Therefore, we interpret the fluctuating signal as repetitive GQ unfolding–refolding activity by a single RHAU.
Fig. 2.
RHAU unfolds parallel GQ repetitively. (A) Representative smFRET traces reporting on RHAU activity on 111-T15, CMYC-T15, TTA-T15, and TAA-T15. (B) A reciprocal experiment where RHAU was immobilized to a single-molecule surface to which GQ-FRET substrate was added. (C) Bar graph comparison of parallel GQ vs. RHAU-bound GQ for all substrates tested. (D) Schematic diagram of dwell time measurement of GQ spent in folded (red) and unfolded (blue) state. (E) Unfolding and refolding rates measured for 111-T15, CMYC-T15, TTA-T15, and TAA-T15 in the presence of RHAU for both substrate-tethered and protein-immobilized configurations.
Fig. S2.
(A and B) FRET constructs and representative smFRET traces. RHAU binding and repetitive unfolding are clearly distinguished, denoted as red and blue arrows, respectively, on a shorter ssDNA overhang (9 nt) substrate. (C and D) Repetitive unfolding persists even after buffer removal of excess RHAU proteins, suggesting a monomer-based activity. (E and F) RHAU exhibits the same type of repetitive unfolding on an RNA-GQ substrate. (G) The unfolding and refolding kinetic on RNA-GQ far exceeds that on DNA-GQ.
To quantify the GQ binding by RHAU, we counted the fraction of molecules that display FRET change (i.e., the FRET fluctuation for 111- and CMYC-T15 and one step FRET decrease for TTA- and TAA-T15) and plotted this result alongside the percentage of parallel GQ conformation (15) (Fig. 2C). The result suggests that RHAU’s binding is highly specific to parallel GQ. Although we detected FRET change, we did not observe any FRET fluctuation in TTA- or TAA-T15 substrates, indicating that the parallel conformers of these DNA do not stimulate repetitive resolving activity of RHAU. We also show that the dye-to-dye distance does not interfere with detecting RHAU activity on nonparallel GQs (Fig. S3). RHAU exhibited similar activity on RNA-GQ, likely because of the RNA-GQ adopting parallel conformation (24), which indicates that RHAU likely employs the same mechanism in unfolding both the RNA- and DNA-GQs.
Fig. S3.
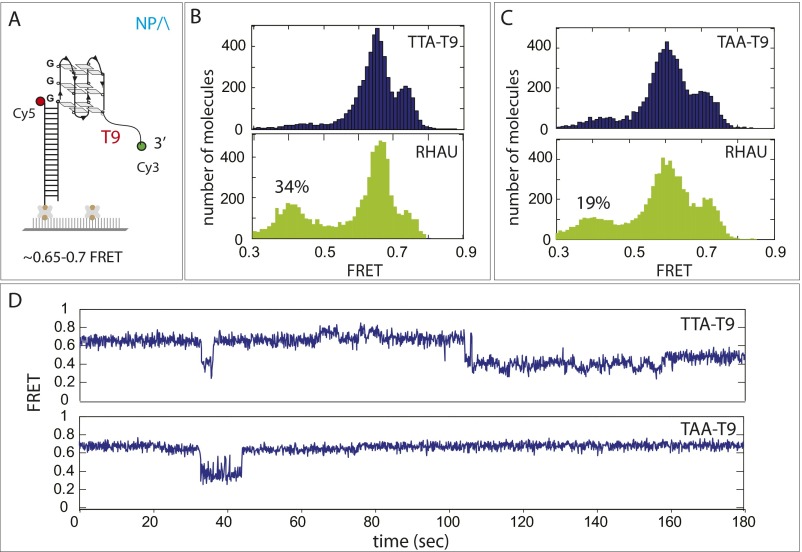
RHAU activity on TTA-T9. (A) Schematic of TTA-T9 FRET construct DNA was designed to match the dye-to-dye distance of parallel GQ by reducing 6 nucleotides from T15. (B and C) FRET histograms before (Upper) and after (Lower) RHAU display a partial shift corresponding to 34% and 19% for TTA-T9 and TAA-T9, respectively, consistent with the result obtained in Fig. 2C. (D) Single-molecule FRET traces from TTA-T9 and TAA-T9 showing stepwise FRET change induced by RHAU binding.
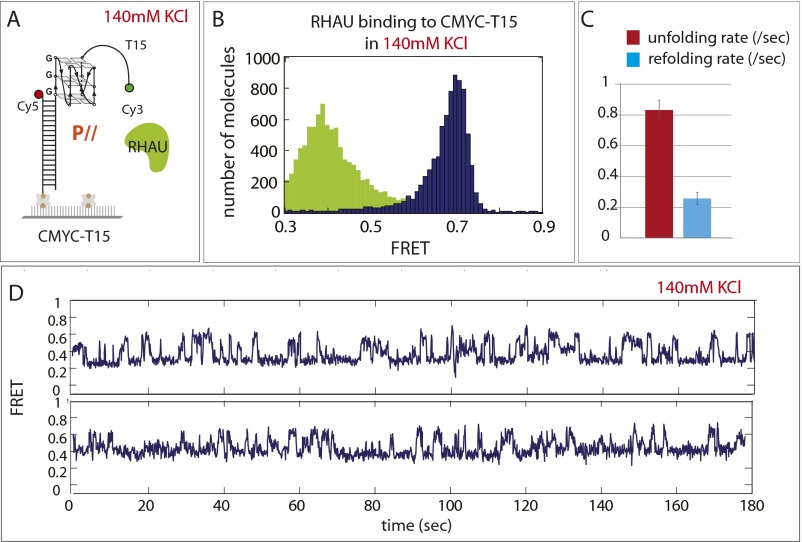
For kinetic analysis of the GQ-unfolding activity in 111- and CMYC-T15, we measured the dwell times that represent the unfolding time (Fig. 2D, δ1, red) and refolding time (Fig. 2D, δ2, light blue). Based on the dwell times collected from more than 500 cycles of events, we obtained the unfolding rate (Fig. 2E, red bar) and refolding rate (Fig. 2E, light blue bar) for 111- and CMYC-T15. Interestingly, both rates were approximately threefold higher for 111-T15 than in CMYC-T15, likely governed by thermal stability of the GQ (25): that is, more stable GQ leads to faster refolding (26). Consistently, RNA-GQ tested above showed three- to sixfold higher rate than the case of 111- and CMYC-T15, respectively (Fig. S2G). Furthermore, when RHAU was applied to CMYC-T15 in 140 mM KCl, where the GQ folding is expected to be more stable, RHAU displayed similar activity with higher unfolding rate than at 50 mM KCl (Fig. S4). Taken together, our results indicate that RHAU activity is specific to parallel conformation of GQ, whether it is composed of DNA or RNA, and that the unfolding and refolding rate scales with GQ-folding strength.
Fig. S4.
RHAU-unfolding activity of CMYC-T15 in 140 mM KCl. (A) Schematic diagram of RHAU applied to CMYC-T15 (parallel) FRET DNA. (B) FRET histogram before (navy) and after (green) addition of RHAU shows similar unfolding signature seen in 50 mM KCl. (C) Unfolding and refolding rate of RHAU measured in 140 mM KCl. (D) Representative single-molecule FRET traces.
RHAU Activity Enables Hybridization with a Complementary Strand in cis.
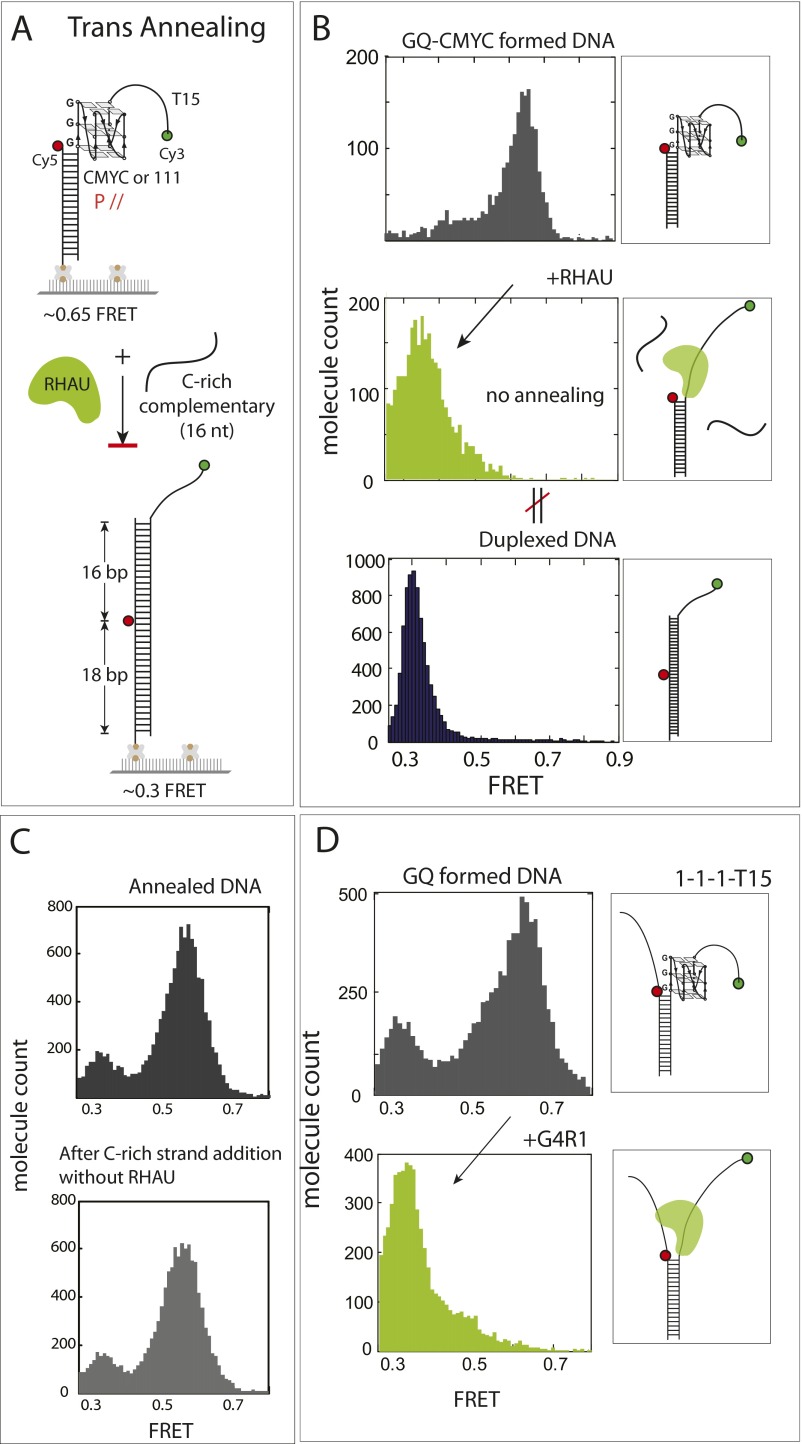
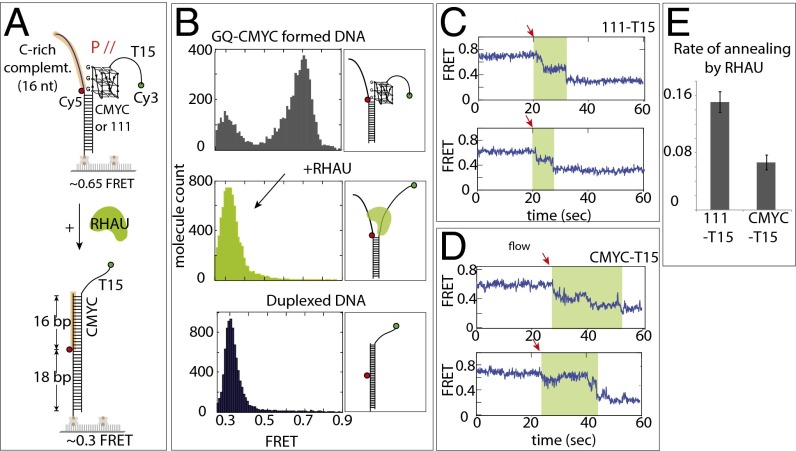
We asked whether the RHAU activity is sufficient to resolve the GQ to hybridize with a cDNA. To test this question, we applied to GQ-DNA 10 nM unlabeled oligonucleotide, which bears a complementary sequence of the GQ-folding sequence (Fig. S5A). However, we observed the same FRET change as when RHAU was added alone (Fig. S5B), suggesting that the RHAU’s unfolding activity does not lead to annealing with the cDNA added in trans. It is likely that RHAU occupies and occludes the site of DNA required for hybridization. Next, we tested annealing reaction in the format of partially unwound duplex DNA, or in cis, which is more relevant to genomic processes such as DNA replication and transcription (Fig. 3A). To form this DNA, we preformed the GQ strand by heating and gradually cooling the G-rich strand independently, followed by mixing with the complementary strand at room temperature. The two FRET dyes were situated such that the GQ formation and annealing would result in high and low FRET, respectively (Fig. 3A). As expected, the GQ-folded CMYC-T15 DNA exhibited a major peak at high FRET (0.7). When RHAU was added, a sharp low FRET (0.3) peak emerged, reflecting an efficient annealing by the GQ-resolving activity of RHAU (Fig. 3B). We confirmed that the low FRET of 0.3 arises from the fully duplexed DNA (Fig. 3B, Bottom). We confirmed that lack of G4R1 does not lead to annealing, which reflects stability of GQ in this splayed arm context (Fig. S5C). RHAU applied to 111-T15 also showed the same annealing activity as CMYC-T15 (Fig. S5D).
Fig. S5.
RHAU does not stimulate trans annealing. (A) Schematic diagram of trans annealing assay. (B) FRET histogram of DNA only (Top), RHAU added (Middle), and DNA annealed without preforming GQ (Bottom). RHAU does not promote trans annealing: that is, when excess concentration of the cDNA-to-GQ strand was applied in solution. (C) FRET histogram of GQ-formed DNA before and after addition of cDNA. (D) FRET histogram of preformed GQ DNA construct before (Upper) and after RHAU addition (Lower).
Fig. 3.
RHAU induces annealing only in cis configuration. (A) Schematic diagram of RHAU-promoted cis annealing of CMYC-T15. (B) smFRET histograms of DNA only (Top), RHAU applied (Middle), and DNA preannealed (i.e., duplexed DNA) (Bottom). (C and D) Single-molecule traces from RHAU-assisted stepwise annealing of complementary strand on 111-T15 and CMYC-T15. The red arrow indicates the moment at which annealing is initiated. The green shaded area represents the annealing duration. (E) Annealing rate measured for 111-T15 and CMYC-T15.
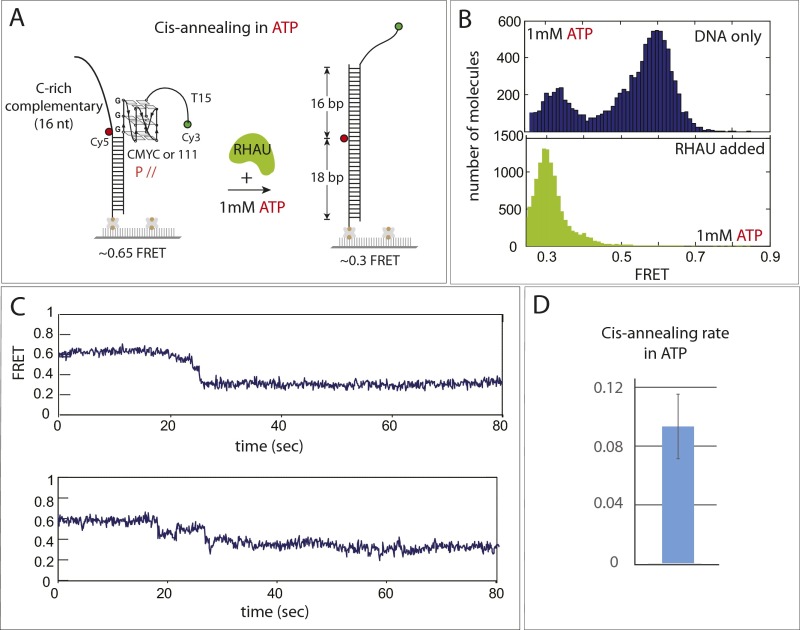
The representative FRET traces show that the high FRET (0.7) transitions to low FRET (0.3) in two to three steps (Fig. 3C), likely suggesting a sequential unfolding of the GQ structure and concomitant stepwise annealing with the complementary strand. The FRET fluctuation was no longer present, indicating that the repetitive activity is not exhibited during or after the annealing reaction, suggesting that the RHAU’s repetitive activity occurs only in the context where the complementary C-rich strand is not readily accessible: for example, when bound to proteins. The annealing rate between the 111- and CMYC-T15 substrates indicates that annealing is about threefold faster in 111-T15 than in CMYC-T15 (Fig. 3 C–E), reflecting the difference seen in the unfolding rate between the two substrates (Fig. 2E). The cis annealing experiment performed in the presence of ATP yielded the same result, indicating that the annealing activity is not affected by ATP (Fig. S6).
Fig. S6.
Cis annealing in ATP. (A) Schematic diagram of cis annealing performed in the presence of ATP. (B) FRET histogram before and after annealing reaction indicates that annealing happened upon addition of RHAU, in the same way as shown in Fig. 3 in the absence of ATP. (C) Representative smFRET traces revealing stepwise annealing. (D) The rate of cis-annealing reaction.
RHAU Acts to Remove GQ-Bound Ligand.
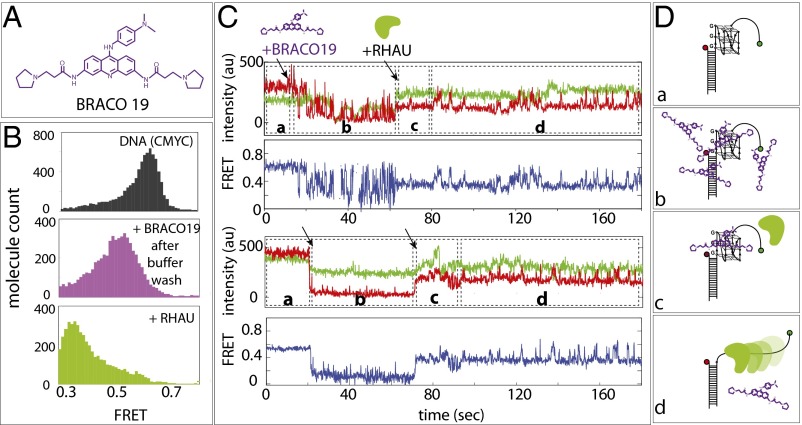
We sought to test whether RHAU activity is sufficient to dislodge GQ ligand bound to GQ DNA. We chose BRACO-19 (Fig. 4A), which binds to parallel GQ and displayed the most distinct signature of binding in the smFRET platform. The addition of BRACO-19 followed by buffer wash shifted the FRET peak from 0.6 to 0.5 (Fig. 4B, purple). Subsequently, the addition of RHAU shifted the FRET peak to low FRET (Fig. 4B, lime-green), which signifies RHAU repetitive unfolding, suggesting that the ligand is removed by the RHAU activity (Fig. 4B).
Fig. 4.
RHAU-resolving activity dislodges GQ ligand. (A) Chemical structure of BRACO19. (B) smFRET histograms of CMYC-T15 (black, Top), with Braco19 after excess buffer wash (purple, Middle), and after adding RHAU (yellow, Bottom). (C) smFRET flow traces of CMYCT15 -Braco19-RHAU interactions. Braco19 and RHAU were added at 20 s and 60 s to the CMYC-T15, respectively, as denoted by black arrows. (D) Schematic diagrams of (a) CMYC-T15, (b) excess Braco19, (c) RHAU added, and (d) RHAU displacing BRACO19 and resolving GQ.
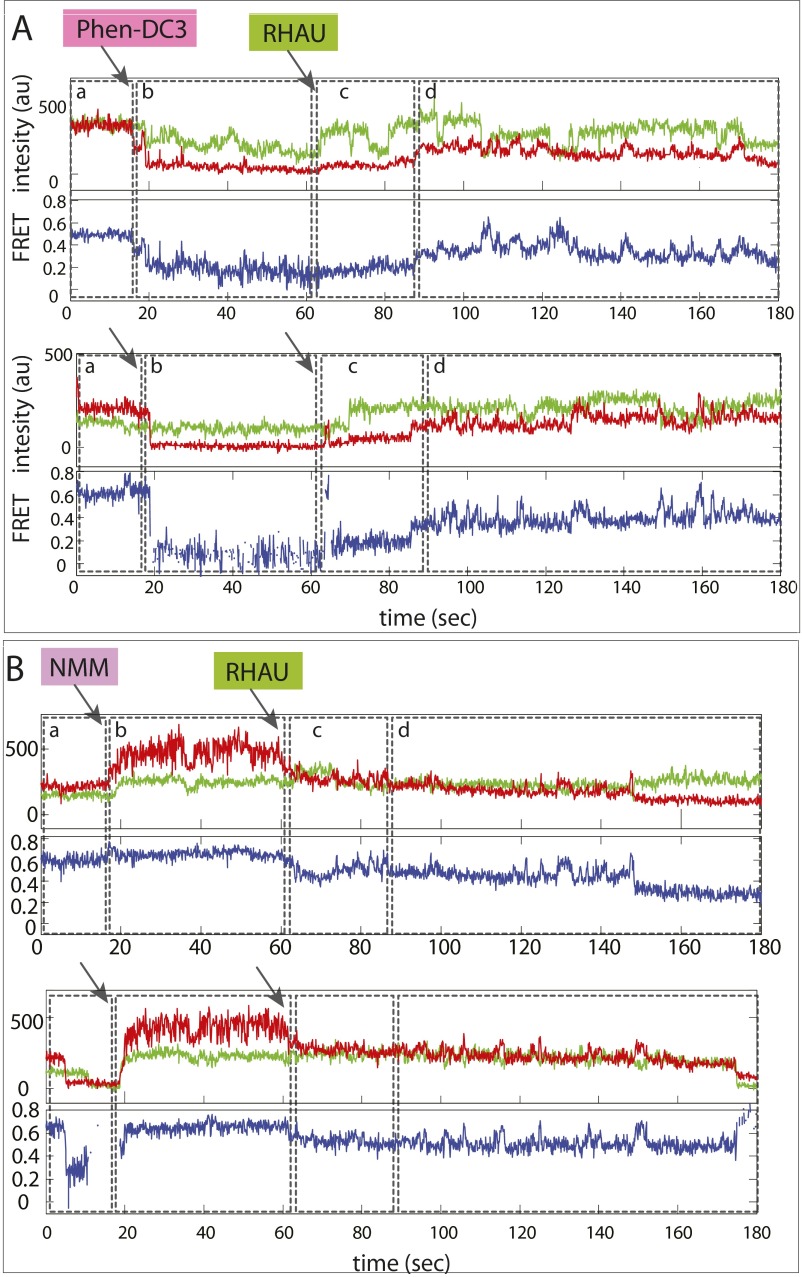
The real-time flow measurement revealed that BRACO-19 binding induced rapid fluctuation or quenching of the Cy5 signal (Fig. 4 C and D, stage b). When RHAU was applied to this platform, we initially obtained a steady FRET at around 0.5, which represents the ligand-bound state of CMYC-T15 GQ (Fig. 4C, stage c). Subsequently, the FRET fluctuation ensued, indicating that RHAU’s repetitive GQ-resolving activity is initiated after clearing BRACO-19 (Fig. 4C, stage d). We tested two more GQ ligands, Phen-DC3 and NMM. In both cases, addition of RHAU halted the Cy5 signal change and induced the same FRET fluctuation pattern (Fig. S7), indicating that RHAU activity is sufficient to dislodge GQ ligand from GQ DNA (Fig. 4D). Although many earlier studies used the 1–5 µM range, we used a 50–100 nM concentration based on our Kd determination (15). We limited the ligand concentration because 1–5 µM ligand gets adsorbed to the surface and quenches all fluorescence, preventing us from conducting single molecule detection. Therefore, what we demonstrate here is that a single GQ ligand bound to GQ can be successfully removed by a single RHAU’s resolving activity.
Fig. S7.
RHAU activity leads to dislodging GQ ligand. Representative smFRET traces obtained while adding Phen-DC3 (A) and NMM (B). Both show evidence of ligand binding, followed by RHAU-unfolding activity (FRET fluctuation).
RHAU Activity Is Independent of ATP.
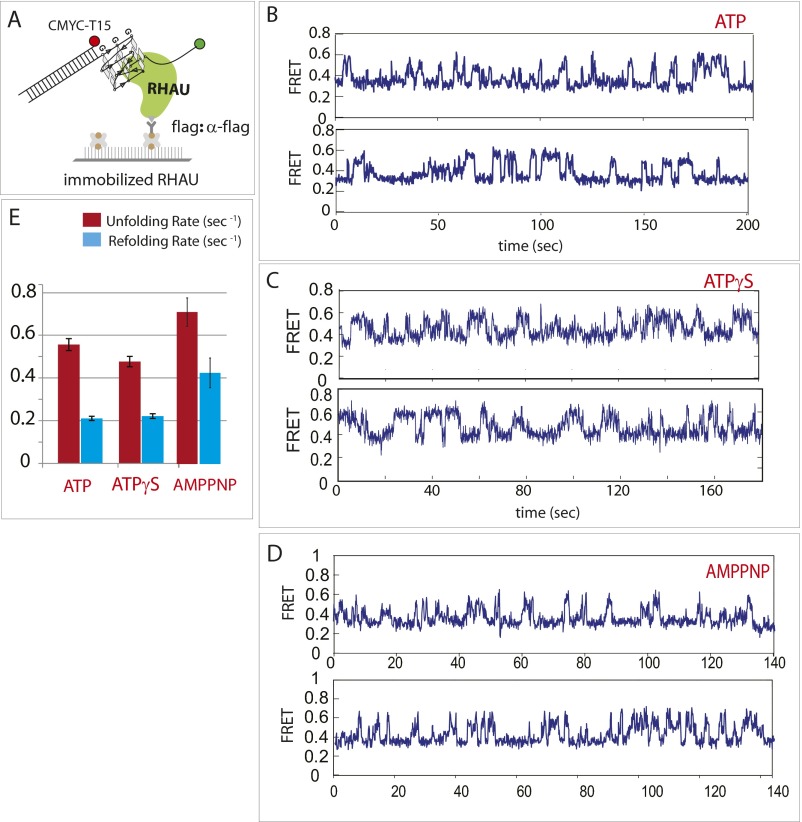
To study the role of ATP, we used an immobilized protein assay, which provides a flexible platform to change buffer conditions while DNA molecules remain bound to RHAU on the surface (Fig. S8A). As shown in Fig. 2B, we observed rapid FRET fluctuation in the absence of ATP. When ATP and ATPγS and AMPPNP were applied to the RHAU–DNA complex independently, we obtained fluctuating FRET signals that resembled the ones detected in the absence of ATP (Fig. S8) The long-lived FRET signal obtained in a single-molecule spot indicates continuous interaction between the single DNA molecule and RHAU in the presence or absence of ATP. The rate of unfolding and refolding of GQ in the presence of ATP, ATPγS, and AMPPNP was comparable with the rates obtained in the absence of ATP (Fig. S8D), suggesting that the repetitive unfolding activity is independent of ATP.
Fig. S8.
Repetitive GQ-unfolding activity of RHAU is ATP-independent. (A) CMYC-T15 binding on flag tag-immobilized RHAU on a single-molecule surface. (B) smFRET trace of RHAU on CMYC-T15 in the presence of ATP. (C) Same as B, in the presence of ATPγS. (D) Same as B, in the presence of AMPPNP. (E) CMYC refolding (blue) and unfolding (red) rates in the presence of ATP, ATPγS, and AMPPNP.
BLM Displays Repetitive Unfolding of GQ Regardless of Conformation.
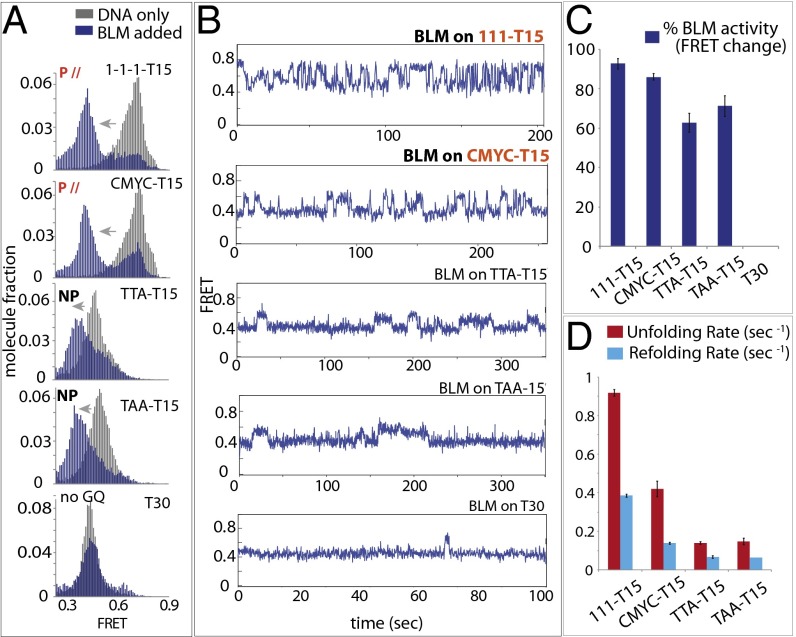
Recent single-molecule studies have reported that truncated BLM interacts with telomeric GQ (9, 27, 28). We used the same set of substrates used above to test the BLM activity. In all four cases, the high FRET that corresponds to GQ-DNA (Fig. 5A, in gray) transitioned to lower FRET (Fig. 5A, in navy blue), indicating that 200 nM full-length BLM interacts with all GQ structures tested here except for the T30, the non-GQ control DNA. This result suggests that, unlike RHAU, BLM interacts with GQ-DNA regardless of the conformation.
Fig. 5.
BLM binds both parallel and nonparallel GQ and exhibits repetitive unfolding activity. (A) FRET histogram of BLM bound (blue) and unbound (gray) to 111-T15, CMYC-T15, TTA-T15, and TAA-T15. (B) smFRET traces of BLM bound to all substrates. (C) Percentages of BLM-bound GQ-DNA molecules in a bar graph. (D) Refolding and unfolding rates of all GQ-DNAs by BLM.
The representative smFRET traces for individual GQ-DNA displayed FRET fluctuations for 111- and CMYC-T15 that resembled the pattern obtained for RHAU (Fig. 5B, Top). In the case of TTA- and TAA-T15, the FRET fluctuation was significantly slowed down. Although the majority (∼90%) of 111- and CMYC-T15 exhibited FRET fluctuation, a substantial fraction (60–70%) of TTA- and TAA-T15 also displayed FRET changes, again reflecting that BLM acts on GQ in a conformation-independent manner (Fig. 5C). The BLM binding and GQ-unfolding activity was independent of ATP, similar to the case of RHAU, which is in agreement with previous studies (9, 27, 28). We also tested two substrates that contain ssDNA on the 5′ side of GQ (TTA). BLM exhibited a similar FRET fluctuation, in agreement with previous studies (28, 29), which indicates that a similar mechanism is at work (Fig. S9). Similar to RHAU, the unfolding rate of BLM was significantly higher than the refolding rate for all substrates. Both rates obtained for 111- and CMYC-T15 were highly similar to the ones measured for RHAU, suggesting that the two proteins may use a similar unfolding mechanism.
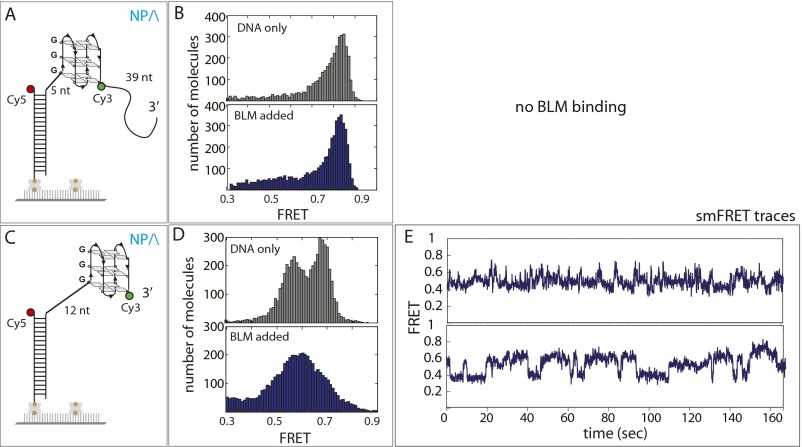
Fig. S9.
BLM binding and activity on DNA with 5′ ssDNA. (A and C) GQ (TTAGGG, telomeric) containing FRET DNA constructs that possess ssDNA on the 5′ side of GQ. (B) FRET histogram remains the same after BLM is added, indicating no binding of BLM to this GQ DNA, which is likely due to the Cy3 dye on the 3′ side of GQ and the short length of ssDNA on the 5′ side. (D) FRET histogram before and after BLM addition changes, indicating protein loading to this DNA, from the 5′ side of 12 nt ssDNA. (E) Representative smFRET traces display fluctuating FRET, indicating a repetitive unfolding initiated from the 5′ side.
WRN Displays Selective Binding and Repetitive Unfolding on Telomeric GQ.
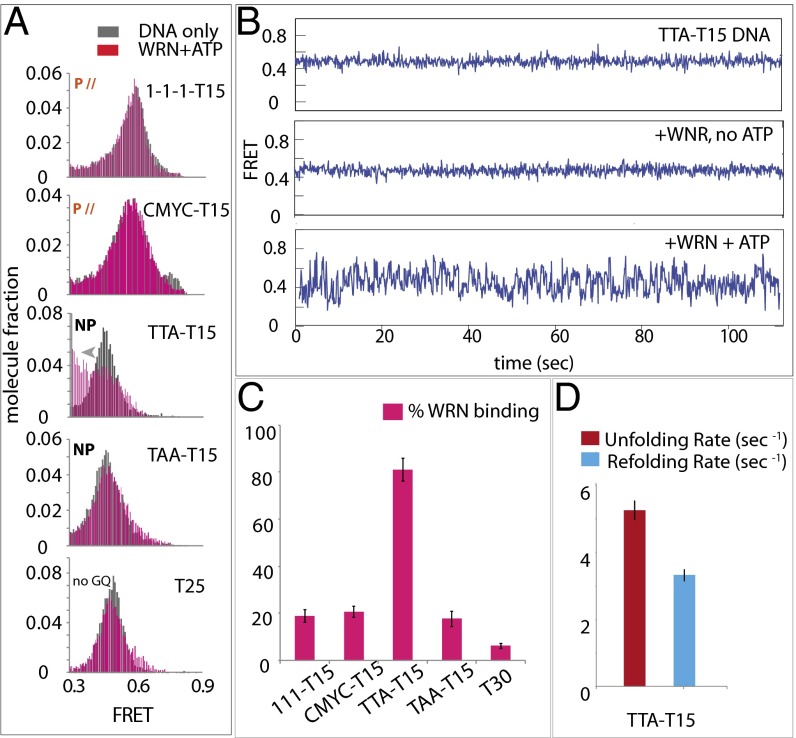
We set out to test another RecQ family helicase, WRN, implicated in GQ unwinding. We used all four GQs and the non-GQ control DNA used above. Unlike RHAU, WRN induced FRET shift only on telomeric GQ, TTA-T15 applied with ATP, suggesting a highly specific binding of WRN toward the telomeric GQ and requirement for ATP (Fig. 6A) (25).
Fig. 6.
WRN binding is specific to telomeric GQ in the presence of ATP. (A) FRET histograms of 111-T15, CMYC-T15, TTA-T15,TAA-T15, and T30 (non-GQ control) (in gray) with WRN plus ATP (pink). (B) smFRET traces of all GQ-DNAs with WRN and ATP. (C) Percentage of WRN-bound 111-T15, CMYC-T15, TTA-T15, TAA-T15, and T25 in the presence of ATP in a bar graph. (D) Unfolding (red) and refolding (blue) rates of TTA-T15 in the presence of WRN and ATP.
Unlike a steady FRET signal for all substrates (Fig. 6B), WRN displayed rapid FRET fluctuation on telomeric GQ, likely representing a repetitive unfolding and refolding of telomeric GQ. The majority (>80%) of telomeric GQs induce binding of WRN that entails repetitive FRET fluctuation whereas less than 20% of traces showed a FRET change in the other substrates. Interestingly, telomeric GQ unfolding and refolding was 5–10 times faster than that measured for RHAU and BLM (Fig. 6D). WRN is unique in that its binding and GQ-resolving activity is highly specific to telomeric GQ and is ATP-dependent.
Discussion
Formation and Resolution of GQ.
Genomic DNA is compacted into nucleosome units that are tightly organized into chromatin. Therefore, DNA cannot be rearranged into any other secondary structures under these conditions. In every cell cycle, however, the entire genomic DNA undergoes a round of replication in which the entire molecule of dsDNA is unraveled into single-stranded DNA. Transcription also entails local melting of DNA at active transcription sites. It is thought that these processes facilitate certain sequences of ssDNA to form noncanonical secondary structures, such as the G-quadruplex and trinucleotide repeat (TNR) hairpin (30). Stably folded GQ structures can be long-lasting, leading to alterations in the subsequent replication or transcription activity (31, 32). Counteracting the formation of such GQ structures is a special class of helicases evolved to specifically target and resolve stably formed GQ structures (33). Highly stable DNA-GQ structures often adopt parallel conformations (26). In agreement, our previous in vitro study demonstrated that only parallel GQ folding can be sustained in the context of dsDNA (34). Nevertheless, it is possible that even weak GQ structures may be stabilized by GQ-binding proteins (35). It is expected that the GQ formation and deformation will be maintained and regulated by these proteins. Here, we examined three GQ-resolving helicases, RHAU, BLM, and WRN, which displayed distinct substrate specificity, yet a very similar unfolding mechanism.
Conformation-Specific Unfolding.
Many previous studies focused on unwinding of telomeric GQ substrates (10, 27, 29, 36, 37). The highly selective binding of RHAU to parallel GQ was anticipated from the previous structure (14). Based on this structural feature, we envision that the N-terminal RHAU makes a specific contact on the top of parallel GQ whereas the helicase domain uses its two RecA-like domains to pry open the folded GQ structure. Our data also reveal a requirement of a segment of ssDNA, likely reflecting that RHAU uses the adjacent ssDNA as an anchor to engage in the GQ-unwinding activity. The robust unwinding activity of RHAU on RNA-GQ confirms that the substrate specificity of RHAU arises from its recognition of the parallel configuration of GQ regardless of DNA or RNA. Although RHAU uses the same mechanism to repetitively unfold parallel structures of CMYC-T15, 111-T15, and RNA-GQ, we do observe different unfolding and refolding rates depending on the folding strength of the GQ. We further tested this effect by applying a 140-mM KCl condition in which GQ folding is tighter (Fig. S4), which reflects that the unfolding activity by RHAU may not be equivalent on all parallel GQs and that the proficiency may depend on the GQ-folding stability. Unlike RHAU, BLM binds all GQ constructs, displaying an indiscriminate binding to both parallel and nonparallel GQ structures. The most selective substrate specificity was seen in WRN, which interacted only with the telomeric GQ in the presence of ATP. This finding is consistent with the high telomeric specificity of WRN displayed in DNA deletion suppression (25, 38) and unwinding activity (39).
Repetitive Unfolding Mechanism.
Although unwinding of nucleic acid is inherently a reversible process, the repetitive activity can prevent the single-stranded segments from rewinding and interacting with unwanted proteins. Our results demonstrate that all three GQ resolvases use the same mechanism of repetitive GQ unfolding albeit with different substrate specificity. Our previous studies demonstrated the repetitive translocation activity of DNA motors (22, 40, 41) and an RNA motor (42), involved in the DNA repair pathway and antiviral signaling, respectively. Recent single-molecule studies on BLM (truncated version) (29, 43) and on PifI (37) also showed repetitive unfolding activity on telomeric GQs. Although the ATP-independent unfolding activity by RHAU and BLM that we present here is consistent with some previous studies that reported ATP independent GQ unwinding activity by BLM (9, 28), others have presented ATP-dependent unwinding by RHAU and BLM. Our results cannot be directly compared with these previous studies for the following reasons. First, we have monitored the protein activity on intramolecular GQs whereas others have used bimolecular or tetramolecular GQs (16) (12) as unwinding substrates. Second, we have used full-length BLM unlike many other studies that resorted to a truncated form of BLM or the core-BLM fragments (27–29). Third, the DNA substrates designed for the study had a different composition: for example in the ssDNA length, polarity, and composition. Taken together, the repetitive motion is likely a conserved mechanism among a diverse family of helicases as an efficient means of preserving the ssDNA or RNA from unintended processes. Although the repetitive nature of unfolding bears resemblance to the previous ATP-dependent DNA and RNA motor proteins, the ATP-independent GQ unfolding by RHAU and BLM observed here likely employs a different mechanism.
DNA Annealing and Ligand Displacement.
RHAU induced DNA annealing only in the cis configuration where the two complementary strands were located in the same DNA molecule (Fig. 3). In our previous study, RNA helicase A exhibited repeated cycles of unwinding, which stimulated unwinding in trans (44). The cis-specific annealing by RHAU suggests that the protein does not unfold the GQ completely, but likely only partially disrupts the structure. This suggestion is also in agreement with the narrow range of FRET fluctuation between 0.35 and 0.6 (Fig. 2A), which is much lower than the expected distance change from unfolding of 15–16 nucleotides. The emerging model is that RHAU latches onto the top of the parallel GQ by its N-terminal domain (14) and that the helicase grabs onto the propeller arms of the GQ structure to spring back and forth, stretching and relaxing in repetition. Interestingly, the repetitive unwinding is abolished in the presence of the complementary strand in cis. Instead, there is a stepwise pairing between the GQ and complementary strand (Fig. 3C). These data suggests that RHAU disrupts the GQ enough to allow for partial annealing, which may trigger further unwinding and subsequent annealing. It is probable that the G triplets are released one at a time, gradually allowing the stepwise annealing to occur. Based on this result, it is likely that the repetitive unwinding is needed only when the complementary strand is absent or when it is not accessible due to being occupied by another protein, for example. Again, such activity will preserve and sequester the unwound GQ DNA until the complementary strand becomes accessible. We also demonstrated that the unwinding activity by RHAU dislodged the chemical ligands BRACO-19, NMM, and Phen-DC3 bound to GQ DNA (Fig. 4 and Fig. S4). Our results clearly indicate that the GQ-bound ligand is easily displaced when RHAU initiates unwinding. Many previous studies reported that GQ ligands such as BRACO-19 stabilize the GQ structure and interfere with downstream processes, such as telomerase extension and gene expression (45). Although these results seem to contradict our findings, the concentrations of ligand used in these studies are likely high enough (1–100 µM) to outcompete the resolvase enzyme, resulting in more pronounced stabilization of GQ, which leads to the corresponding inhibitory effect.
Materials and Methods
DNA-Protein Measurement.
WRN protein was purified as described previously (46). Reactions were performed in 10 mM Tris, pH 7.5, 25 mM KCl, 1 mM MgCl2, and 2 mM ATP. BLM was purified as described previously (47), and single-molecule reactions were performed with 100 nM BLM in the presence of 20 mM Tris, pH 7.6, 50 mM KCl, 3 mM MgCl2, and 2 mM ATP. RHAU was purified as described previously (15), and single-molecule reactions were performed with 7.5 nM RHAU in the presence of 50 mM Tris-Acetate, pH 7.6, 50 mM KCl, 50 mM NaCl, 0.5 mM Mg(Ac)2, and 1 mM ATP, ATPγS, and AMPPNP. See Table 1 for DNA sequences.
Table 1.
Extended materials and methods: DNA sequences
| Sequence | 5′ to 3′ |
| CMYC-T15/3Cy3/ | TGG CGA CGG CAG CGA GGC TT GGGTGGGTAGGGTGGG TTT TTT TTT TTT TTT/3Cy3/ |
| 111-T15/3Cy3/ | TGG CGA CGG CAG CGA GGC TT GGGTGGGTGGGTGGG TTT TTT TTT TTT TTT/3Cy3/ |
| TTA-T15/3Cy3/ | TGG CGA CGG CAG CGA GGC TT GGGTTAGGGTTAGGGTTAGGG TTT TTT TTT TTT TTT/3Cy3/ |
| TAA-T15/3Cy3/ | TGG CGA CGG CAG CGA GGC TT GGGTAAGGGTAAGGGTAAGGG TTT TTT TTT TTT TTT/3Cy3/ |
| T15/3Cy3/ | TTT TTT TTT TTT TTT/3Cy3/ |
| T25/3Cy3/ | TTT TTT TTT TTT TTT TTT TTT TTT T |
| CMYC-T9/3Cy3/ | TGG CGA CGG CAG CGA GGC TT GGGTGGGTAGGGTGGG TTT TTT TTT/3Cy3/ |
| C4 | CCC A CCC TA CCC A CCC |
| CMYC complementary /iCy5/ | CCC A CCC TA CCC A CCC AA G/iCy5/CC TCG CTG CCGTCG CCA /3Bio/-3 |
| 111 complementary/iCy5/ | CCC A CCC A CCC A CCC AA G/iCy5/CC TCG CTG CCGTCG CCA /3Bio/-3 |
| /5Cy5/18merBio | /5′Cy5/GCC TCG CTG CCG TCG CCA/3′Bio/ |
| /5Cy5/18mer | /5′Cy5/GCC TCG CTG CCG TCG CCA |
| Biotin CMYC 51 mer | /5BiodT/-GGC CGC TTA TGG GGA GGG TGG GGA GGG TGG GGA AGG TGG GGA GGA GAC TCA |
DNA sequences used in this study. The underline portion corresponds to G-quadruplex forming sequence.
smFRET Data Analysis.
Software for analyzing single-molecule FRET data is available for download from https://cplc.illinois.edu/software/.
SI Materials and Methods
DNA Sample Preparation.
Oligonucleotides required to make the partial DNA duplex substrates were purchased from IDT with either Cy3 or Cy5 dyes (Table 1). For internal labeling purpose, oligonucleotides were purchased with an amino modifier C6 dT, reacted with NHS-ester–conjugated Cy3 or Cy5 (GE Healthcare). Labeling protocol was as follows: 10 mM dye was incubated with 0.1 mM DNA in 100 mM sodium bicarbonate, pH 8.5, buffer overnight at 4 °C in a dark room. Excess dye was removed by the ethanol precipitation method. The final pellet was dissolved in 50 μL of ddH20, and the ethanol precipitation was performed again to remove any residual dyes. The dried DNA pellet was dissolved in 20 μL of 10 mM Tris⋅HCl, pH 8.0, and stored sample at −20 °C.
All GQ DNA constructs were prepared by mixing a 3′ Cy3 or internal labeled Cy3 sequence with the complementary 5′ Cy5 sequence with or without 3′ biotin at a molar ratio of 1.5:1 in T50 (10 mM Tris⋅HCl, pH 7.5, 50 mM NaCl, or 100 mM KCl), incubated for 2 min at 95 °C on heat block, and then slowly cooled down to room temperature in a time frame of 2–3 h. For single-molecule imaging, 0.8 mg/mL glucose oxidase, 0.625% glucose, ∼1–3 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic (Trolox), and 0.03 mg/mL catalase were added to the reaction buffers.
DNA-Protein Studies.
WRN protein was purified as described previously (46). Reactions were performed in 10 mM Tris, pH 7.5, 25 mM KCl, 1 mM MgCl2, and 2 mM ATP. BLM was purified as described previously (47), and single-molecule reactions were performed with 100 nM BLM in the presence of 20 mM Tris, pH 7.6, 50 mM KCl, 3 mM MgCl2, and 2 mM ATP. RHAU was purified as described previously (15), and single-molecule reactions were performed with 7.5 nM RHAU in the presence of 50 mM Tris-acetate, pH 7.6, 50 mM KCl, 50 mM NaCl, 0.5 mM Mg(Ac) 2, and 1 mM ATP or ATPγS.
DNA Preparation for the cis Annealing Experiment.
GQ-bearing ssDNA with 3Cy3 (Table 1) was heated up to 95 °C for 2 min on a heat block. Thereafter, the temperature was gradually decreased 2 °C/min until it reached 60 °C, followed by 5 °C /min decrease until it reached 22 °C on a thermocycler. Subsequently, the internally Cy5-labeled complementary strand was (Table 1) added to GQ-containing strand and incubated at room temperature for 5 min. The annealed mixture was immediately diluted to 100 pM in 10 mM Tris⋅HCl, pH 7.5, and 100 mM KCl to immobilize it on a quartz surface.
CD Measurements.
All circular dichroism (CD) spectras were recorded at room temperature (23 °C ± 1 °C) on a JASCO J-715 spectropolarimeter over the range of 200–320 nm using a 1-mm path length quartz cuvette with a reaction volume of 100 or 200 μL. The GQ oligonucleotide concentration was 10 μM. Each sample was subjected to an average of two scans for 3 min, before it was subtracted from the buffer and tailed duplex spectrum. All CD measurements were carried out in the buffer containing 10 mM Tris⋅HCl, pH 7.5, 50 mM KCl, and 50 mM NaCl at room temperature (23 °C ± 1 °C).
Single-Molecule Fluorescence Data Acquisition.
Single-molecule fluorescence experiments were carried out on quartz slides (Finkenbeiner). To minimize surface interactions with the DNA, quartz slides and coverslips were coated with polyethylene glycol (PEG) (21). Briefly, the slides and coverslips were cleaned and treated with methanol, acetone, and potassium hydroxide, burned, treated with aminosilane, and coated with a mixture of 97.5% mPEG (m-PEG-5000; Laysan Bio) and 2.5% biotin PEG (biotin-PEG-5000; Laysan Bio).
Partial duplex DNA molecules were annealed and immobilized on the PEG-passivated surface via biotin–neutravidin interaction at the concentration range of 50–100 pM. Excess donor molecules were washed away with T 50 buffer. For measurement, the reaction buffer was supplemented with an oxygen scavenging system [0.8 mg/mL glucose oxidase, 0.625% glucose, ∼3 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic (Trolox), and 0.03 mg/mL catalase]. All experiments and measurements were carried out at room temperature (23 °C ± 1 °C).
Prism-type total internal reflection microscopy was used to acquire single-molecule FRET. A 532-nm Nd:YAG laser was guided through a prism to generate an evanescent field of illumination. A water-immersion objective was used to collect the signal, and a 550-nm long pass filter was used to remove the scattered light. Cy3 signals were collected using a 630-nm dichroic mirror and sent to a charge-coupled device camera. Data were recorded with a time resolution of 100–200 ms as a stream of imaging frames and analyzed with scripts written in interactive data language to give fluorescence intensity time trajectories of individual molecules.
smFRET Data Analysis.
Basic data analysis was carried out by scripts written in Matlab, with FRET efficiency, E, calculated as the intensity of the acceptor channel divided by the sum of the donor and acceptor intensities. Histograms were generated using over 6,000 molecules collected and were fit to Gaussian distributions using Origin 8.0, with the peak position left unrestrained. Dwell times were collected by measuring the time the molecule spent in a particular FRET state. The means and the SEs were plotted. Software for analyzing single-molecule FRET data is available for download from https://cplc.illinois.edu/software/.
Acknowledgments
This work was supported by American Cancer Society Grant RSG-12-066-01-DMC; NIH Grant 1DP2GM105453; National Science Foundation Physics Frontiers Center Program 0822613 through the Center for the Physics of Living Cells (to R.T, H.H., and S.M.); and funds from the Intramural Program of the National Institute on Aging, National Institutes of Health (V.A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603724113/-/DCSupplemental.
References
- 1.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 2.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35(22):7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem. 2014;6(1):75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5(3):182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16(8):910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3(4):218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1(3):167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budhathoki JB, et al. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014;42(18):11528–11545. doi: 10.1093/nar/gku856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giri B, et al. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39(16):7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MC, Murat P, Abecassis K, Ferré-D’Amaré AR, Balasubramanian S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015;43(4):2223–2231. doi: 10.1093/nar/gkv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, et al. A G-quadruplex DNA structure resolvase, RHAU, is essential for spermatogonia differentiation. Cell Death Dis. 2015;6:e1610. doi: 10.1038/cddis.2014.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heddi B, Cheong VV, Martadinata H, Phan AT. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc Natl Acad Sci USA. 2015;112(31):9608–9613. doi: 10.1073/pnas.1422605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tippana R, Xiao W, Myong S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014;42(12):8106–8114. doi: 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creacy SD, et al. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J Biol Chem. 2008;283(50):34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31(4):1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J Am Chem Soc. 2011;133(44):17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathad RI, Hatzakis E, Dai J, Yang D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011;39(20):9023–9033. doi: 10.1093/nar/gkr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrus A, et al. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu Y, et al. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang H, et al. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22(6):842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey S, Agarwala P, Maiti S. Effect of loops and G-quartets on the stability of RNA G-quadruplexes. J Phys Chem B. 2013;117(23):6896–6905. doi: 10.1021/jp401739m. [DOI] [PubMed] [Google Scholar]
- 25.Damerla RR, et al. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11(16):3036–3044. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126(50):16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 27.Budhathoki JB, Stafford EJ, Yodh JG, Balci H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015;43(12):5961–5970. doi: 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee S, et al. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun. 2014;5:5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu WQ, Hou XM, Li M, Dou SX, Xi XG. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43(9):4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11(11):786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemmens B, van Schendel R, Tijsterman M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat Commun. 2015;6:8909. doi: 10.1038/ncomms9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piazza A, et al. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34(12):1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bharti SK, et al. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem. 2013;288(39):28217–28229. doi: 10.1074/jbc.M113.496463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreig A, et al. G-quadruplex formation in double strand DNA probed by NMM and CV fluorescence. Nucleic Acids Res. 2015;43(16):7961–7970. doi: 10.1093/nar/gkv749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brázda V, Hároníková L, Liao JC, Fojta M. DNA and RNA quadruplex-binding proteins. Int J Mol Sci. 2014;15(10):17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210(2):191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy S, Li B, Comai L. Processing of human telomeres by the Werner syndrome protein. Cell Cycle. 2010;9(16):3137–3138. doi: 10.4161/cc.9.16.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards DN, Machwe A, Chen L, Bohr VA, Orren DK. The DNA structure and sequence preferences of WRN underlie its function in telomeric recombination events. Nat Commun. 2015;6:8331. doi: 10.1038/ncomms9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437(7063):1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 41.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142(4):544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323(5917):1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28(4):405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh HR, Xing L, Kleiman L, Myong S. Repetitive RNA unwinding by RNA helicase A facilitates RNA annealing. Nucleic Acids Res. 2014;42(13):8556–8564. doi: 10.1093/nar/gku523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jena PV, et al. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J Am Chem Soc. 2009;131(35):12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opresko PL, Sowd G, Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PloS One. 2009;4(3):e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3'-5' DNA helicase. J Biol Chem. 1997;272(49):30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]