Significance
Hydration water around a protein is fundamental to protein’s property and function. How hydration water interacts with protein at their interface has not been fully understood. A universal slaving model that water drives protein fluctuations has been proposed, but direct experimental observation is difficult and challenging. Here, we directly measured hydration water dynamics and protein side-chain relaxations with temperature dependence. With extensive data, we conclude that the surface hydration-shell fluctuations drive protein side-chain motions on the picosecond time scales and thus play a critical role in protein dynamics.
Keywords: hydration shell dynamics, protein side-chain motion, water-driven relaxation, coupled fluctuation, tryptophan scan
Abstract
Protein hydration is essential to its structure, dynamics, and function, but water–protein interactions have not been directly observed in real time at physiological temperature to our awareness. By using a tryptophan scan with femtosecond spectroscopy, we simultaneously measured the hydration water dynamics and protein side-chain motions with temperature dependence. We observed the heterogeneous hydration dynamics around the global protein surface with two types of coupled motions, collective water/side-chain reorientation in a few picoseconds and cooperative water/side-chain restructuring in tens of picoseconds. The ultrafast dynamics in hundreds of femtoseconds is from the outer-layer, bulk-type mobile water molecules in the hydration shell. We also found that the hydration water dynamics are always faster than protein side-chain relaxations but with the same energy barriers, indicating hydration shell fluctuations driving protein side-chain motions on the picosecond time scales and thus elucidating their ultimate relationship.
Water–protein interactions are critical to protein structural stability and flexibility, functional dynamics, and biological activities (1, 2). Various methods such as neutron scattering (3), NMR (4), laser spectroscopy (5, 6), and molecular dynamics (MD) simulations (7) have been used to reveal protein surface hydration and coupled water–protein dynamics on different time and length scales. Hydration water molecules often participate in various protein functions and their motions even directly “control” protein fluctuations (2, 8). Frauenfelder et al. recently proposed a unified model for protein dynamics (8): large-scale protein motions are slaved to the fluctuations of bulk solvent and controlled by solvent viscosity while internal protein motions are slaved to the fluctuations of the hydration shell and controlled by hydration water. However, direct measurements of such coupled fluctuations at physiological temperature are challenging as a result of the ultrafast nature of water motions, and therefore most studies are indirect or at low temperature (3, 4). Here, we used a tryptophan (W) scan to probe global surface hydration (9) and used femtosecond spectroscopy to follow hydration water motions and local side-chain fluctuations in real time. With temperature dependence, we systematically measured their dynamics and thus finally elucidate their ultimate relationship.
Results and Discussion
Tryptophan Scan and Femtosecond Fluorescence Spectroscopy.
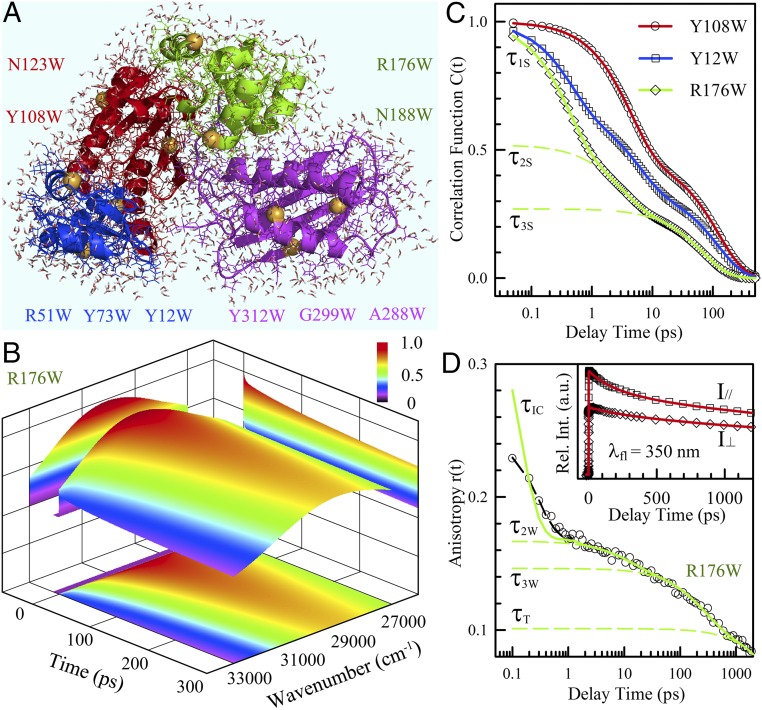
We used DNA polymerase IV (Dpo4) (10), a hyperthermal enzyme without a single tryptophan residue, as a model protein, and designed 10 tryptophan mutants, one at a time, to probe four different domains (Fig. 1A). We performed systematic measurements of tryptophan fluorescence intensity changes with wavelength and time and thereby constructed 3D fluorescence profiles (SI Appendix, SI Note 1). One example of an R176W mutant is shown in Fig. 1B. Based on the methodology we developed to follow the Stokes shifts of tryptophan with time (11, 12), we derived the solvation correlation functions for the 10 mutants. Fig. 1C shows our typical results for three mutants with three exponential decays (τ1S, τ2S, and τ3S, Y12W and R176W) or two exponential decays (τ2S and τ3S, Y108W) from hundreds of femtoseconds to tens of picoseconds. Simultaneously, we measured tryptophan side-chain relaxation through its fluorescence anisotropy dynamics (Fig. 1D, Inset), and the typical result is shown in Fig. 1D for R176W with four exponential decays. The initial component (τIC) is from ultrafast internal conversion of two concurrently excited electronic states (1La and 1Lb) in ∼70 fs (13), and the last decay (τT) represents the protein tumbling motion in ∼20 ns. The two middle decay components (τ2W and τ3W) result from the local wobbling relaxations on picosecond time scales. For nine mutants, we measured protein solvation dynamics and local tryptophan relaxations at seven different temperatures from 1 °C to 60 °C (SI Appendix, Figs. S1–S4).
Fig. 1.
Hydration dynamics and protein side-chain motions detected by tryptophan. (A) A snapshot of MD simulations of solvated Dpo4 (Protein Data Bank ID 2RDI) shown with water molecules within 5 Å to the protein surface. Dpo4 consists of four domains, including thumb (green), palm (red), finger (blue), and little finger (magenta). The yellow spheres indicate the mutation sites replaced by tryptophan. (B) Constructed 3D FRES of tryptophan (R176W) at room temperature. (C) Typical solvation correlation functions of three mutants show the very different dynamics when the probe moves from the buried (Y108W) to exposed (R176W) locations. The symbols are the derived experimental data, and the solid lines are exponential fit. The dashed lines represent three exponential decay components (τ1S, τ2S, and τ3S) for the mutant R176W. (D) The anisotropy decay dynamics show four decay components corresponding to internal conversion (τIC), wobbling (τ2W, τ3W), and tumbling (τT) motions. The symbols are experimental data and the dashed black line is the best fit with the internal conversion model (13), and the solid line is after deconvolution from the instrument response. (Inset) Two parallel and perpendicular fluorescence transients used to calculate the anisotropy decay.
Protein Surface Hydration Dynamics.
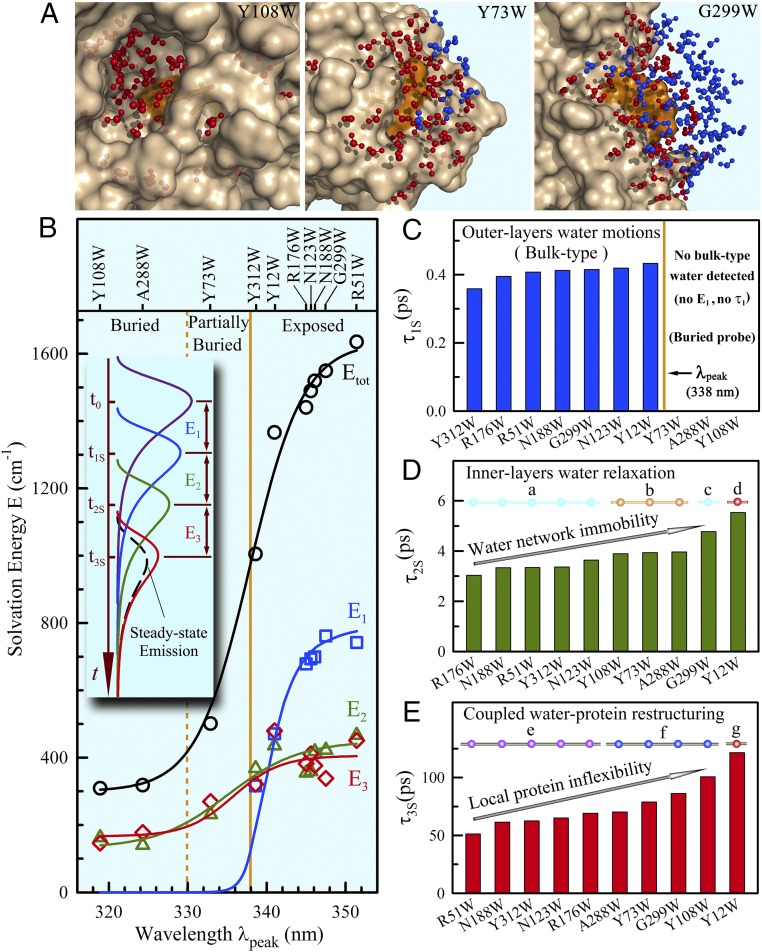
Fig. 2 shows all obtained solvation correlation functions in terms of relaxation energies (Fig. 2B) and times (Fig. 2 C–E) for the 10 mutants at room temperature. Similar to the patterns that observed before in numerous proteins (9, 14–17), the relaxation energies are dominantly from hydration water molecules and the solvation dynamics are highly heterogeneous around the global protein surface (17). In Fig. 2B, the 10 mutants can be divided into two categories, exposed and buried, as usual with the dividing steady-state emission peak (λpeak) of tryptophan at ∼338 nm. The total solvation energy (Etot) shows a sigmoid-type increase along λpeak when the probe moves to the protein surface with more exposure to hydration water molecules. For the exposed probes, we observed the solvation dynamics on three distinct time scales of τ1S in 300–400 fs (Fig. 2C), τ2S in 3–6 ps (Fig. 2D), and τ3S in 50–120 ps (Fig. 2E), corresponding to three relaxation energies of E1, E2, and E3, respectively. For the buried probes, we did not detect first ultrafast solvation dynamics (τ1S) and observed only two solvation dynamics on picosecond scales (τ2S and τ3S).
Fig. 2.
Solvation dynamics of hydration water around the protein surface at 20 °C. (A) Total water molecules within 10 Å to the tryptophan indole ring, separated 5 Å (red) and 5–10 Å (blue) to the protein surface from the buried, to partially buried, and finally to exposed positions from a snapshot of MD simulations. (B) The solvation energy distributions (E1, E2, E3, and the total Etot) of 10 mutants at 20 °C relative to the steady-state emission peaks. The solid lines provide only guidelines for the overall increasing trends. (Inset) Schematic plot displaying the evolution of the emission spectrum by solvation. (C–E) Shown are the solvation relaxation times. The three buried probes did not detect ultrafast component (τ1S and E1). For τ2S and τ3S, the times are strongly correlated with the local chemical identities (a, partial charge; c, dense charge), probe distance (b), topological geometry (d and g), and higher-order structures and tertiary intraprotein interactions (e and f) with the wide heterogeneous dynamics around the protein surface.
Considering the dipole–dipole interactions between tryptophan and neighboring water molecules mainly within ∼10 Å from our MD simulations (SI Appendix, Fig. S5), we found that for the buried probes the 10-Å water molecules are dominantly within 7 Å to the protein surface (Y108W and Y73W in Fig. 2A). Thus, the buried probes dominantly detected one to two inner layers of water molecules in the hydration shell, and the observed two relaxations are mainly from these inner-layer water molecules at the interface. For the exposed probes, besides the inner-layer hydration water response, we also observed ∼30% of 10-Å water molecules beyond 7 Å to the protein surface. Thus, the observed ultrafast bulk-type water relaxation (Fig. 2C) must be from those water molecules in the outer layers of the hydration shell (G299W in Fig. 2A). Consistent with the observations in apomyoglobin (16) and other proteins (18, 19), as well as theoretical work and recent MD simulations (20–24), the water motion in a few picoseconds (Fig. 2D) represents the collective water-network reorientational relaxation at the water–protein interface and significantly slows down, compared with the motions of bulk-type water in the outer layers, as a result of structured water-network collectivity near the protein. The water relaxation in tens of picoseconds results from the cooperative water–protein rearrangements, i.e., the coupled interfacial water–protein restructuring, and directly reflects such cooperativity of coupled water–protein fluctuations (Fig. 2E). Also, similar to our previous results observed in apomyoglobin (16), these water motions are correlated with the local protein’s chemical identities, topological roughness, higher-order structures, and tertiary intraprotein interactions, revealing various water behaviors in the inner hydration layers with wide heterogeneity (Fig. 2D, a–d, and Fig. 2E, e–g). For example, the hydration water near the densely charged protein surface (G299W) relaxes slowly, and the water molecules trapped at the active site and confined in a nanospace (Y12W) show the longest relaxation (Fig. 2 D and E and SI Appendix, Figs. S5 and S6A).
Origin of Coupled Water–Protein Fluctuations.
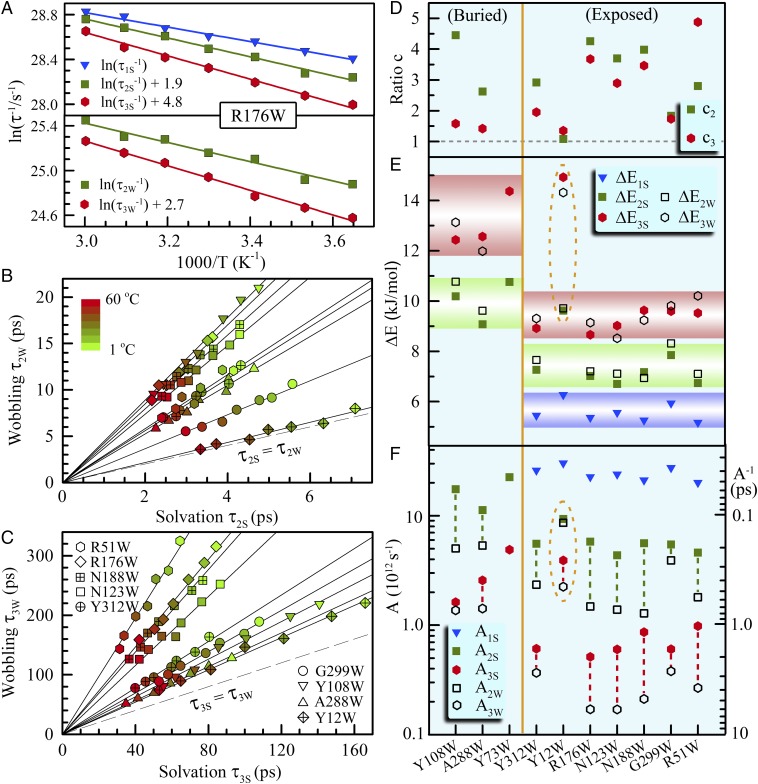
The observed interfacial water motions must interact with the local protein. The observed two relaxations of tryptophan (Fig. 1D) on the picosecond scales should couple with two hydration water motions. Thus, we measured these dynamics with temperature changes from 1 °C to 60 °C for the nine mutants (SI Appendix, Figs. S7–S9), and Fig. 3A shows a typical result for the mutant R176W. We plotted the correlations of τ2W with τ2S in Fig. 3B and of τ3W with τ3S in Fig. 3C. Strikingly, the two sets of data are all linearly correlated through the original point (0,0). Such 18 linear correlations for all of the 9 mutants strongly show that both relaxations of hydration water and tryptophan side-chain are intrinsically connected and must be from one origin. Furthermore, all correlated straight lines lie above the equal line of τiW = τiS (i = 2,3), i.e., all the tryptophan relaxations are always slower than the neighboring hydration water motions (Fig. 3 B and 3C). This observation clearly suggests that such coupled motions are driven by hydration water relaxations, confirming the slaving model of protein side-chain motions controlled by the hydration shell fluctuations (βh-relaxation) (8, 25). Also, the two tryptophan relaxations have very small wobbling semiangles (10°–15°) at room temperature (SI Appendix, Fig. S10), indicating its small-amplitude fluctuations, consistent with the dominant contributions of solvation from hydration water molecules in relaxation energies (Fig. 2B) and times (Fig. 2 C–E).
Fig. 3.
Temperature dependence of hydration water and protein side-chain dynamics. (A) The dynamics of hydration water and tryptophan side chain change with temperature as shown for R176W, following an Arrhenius relation in the experimental temperature range of 1–60 °C (1, 10, 20, 30, 40, 50, and 60 °C). (B and C) Shown are the linear correlations between water solvation and tryptophan relaxation times for the observed two dynamics. The solid lines are linear fits for each mutant. The dashed lines are equal relations between two coupled water/side-chain relaxations. Note that the hydration dynamics are always faster than the tryptophan relaxations. (D) Shown are the ratios (ci; i = 2, 3) of two coupled relaxations, which are also the slopes of the straight lines in B and C. (E and F) Show are the derived activation energies and prefactors by the Arrhenius relation. Hydration water and tryptophan side chain have the same barriers, indicating that the coupled relaxations are from one origin. Note that the exposed probes give the three same barriers.
Fig. 3D shows the ratios of the two relaxation times by defining ci = τiW/τiS, a coefficient that indicates the extent of coupling, i.e., the larger value of ci, the weaker coupling of the hydration water with protein side chain (i.e., tryptophan). All ratios are greater than 1. For the anchored position of R51W, the c3 is the largest, indicating the least coupled water–tryptophan relaxation as a result of the strong cation-π interaction with the neighboring lysine (SI Appendix, Fig. S6B). For the slow water relaxations at the densely charged G299W position and confined Y12W site, the c2 is nearly equal to c3, showing a similar coupling for two types of relaxations. For all other six mutants, the c2 is larger than c3, indicating that at those sites the coupling between hydration water and side chain(s) is weaker in the water–tryptophan reorientational relaxation and stronger in the process of water–tryptophan restructuring.
By using the simple Arrhenius equation of k = Aexp(−ΔE/RT), where k is the rate constant, A is the preexponential factor, and ΔE is the activation energy), we found that all the temperature-dependent relaxations follow a linear relation of ln(k) vs. 1/T as shown for R176W (Fig. 3A), obtaining the prefactors and activation barriers for the three different hydration water motions and two local relaxations of tryptophan. For each linear correlation in Fig. 3 B and C, the hydration water and tryptophan relaxations therefore give similar barriers, only with different prefactors. Fig. 3 E and F show the derived activation barriers and prefactors for all mutants (SI Appendix, Table S1). We obtained a series of new observations. (i) The barriers and prefactors detected by the buried and exposed probes are separately distributed, indicating that the buried probe mainly detected the stronger interfacial water network near the protein with a higher barrier. (ii) The hydration water and tryptophan have similar activation barriers (ΔE2S ∼ ΔE2W and ΔE3S ∼ ΔE3W) within the average SD of 0.4–0.6 kJ/mol and differ only in their prefactors, showing the two relaxations of hydration water and side chain from the same origin. (iii) For the exposed positions, the three barriers of hydration water relaxations are within 5.6 ± 0.3, 7.1 ± 0.4, and 9.2 ± 0.5 kJ/mol for ΔE1S, ΔE2S, and ΔE3S with the corresponding prefactors in time (A−1) of 40 fs, 200 fs, and 1.5 ps, respectively. For the buried positions, the latter two barriers increase to 9.9 ± 0.6 kJ/mol and 13.6 ± 0.6 kJ/mol, also with greater corresponding prefactors. The barrier for the ultrafast motion of the outer-layer water is similar to that of free tryptophan in bulk water (SI Appendix, Fig. S11), indicating a similar relaxation (22, 26). However, the barriers for the inner-layer coupled motions are smaller than those obtained by experiments in several other proteins (27–29). (iv) The prefactors for the hydration water motions are always greater than those of the driven tryptophan relaxations. (v) The water relaxations confined in a nanocavity (SI Appendix, Fig. S5) near the position of Y12W show a special dynamical property of having an ultrafast bulk-type motion but exhibiting two slow behaviors like those probed by a buried tryptophan. These observations clearly showed side-chain relaxations driven by hydration water. With the similar barriers for all the exposed probes in four different domains, the results indicated a significant length scale of collective water network in the hydration shell embracing the protein surface (SI Appendix, Fig. S12). The observed small barriers are only approximately the energy of one hydrogen bond, indicating the nature of collectivity in water/side-chain reorientational relaxation and cooperativity in water/side-chain restructuring.
Conclusion
We have reported our extensive studies on protein hydration dynamics and side-chain motions with the 10 mutants in 4 domains. The surface hydration dynamics are highly heterogeneous over the global protein surface, with two types of relaxations on picosecond time scales. Both hydration water relaxations are coupled with protein side-chain fluctuations with the same energy barriers, elucidating the high cooperativity of such interactions and extended collectivity of the hydration shell. The outer-layer hydration water molecules (beyond 7 Å from the protein surface), although their motions slightly slow down, are mobile and bulk-type. The interfacial hydration water shows significantly slow behaviors by one or two orders of magnitude as a result of the collective nature of coupled water–protein relaxations.
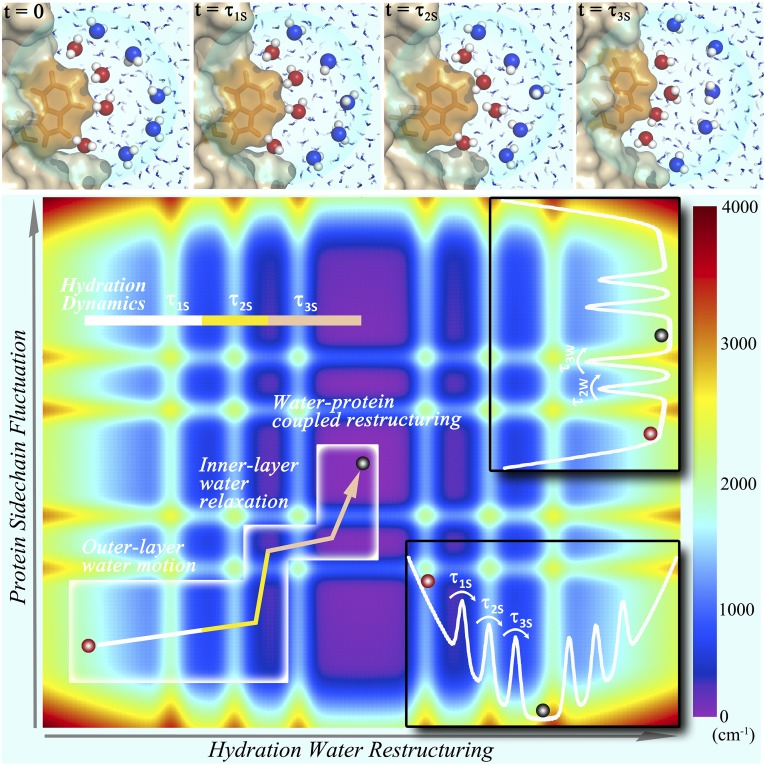
Fig. 4 summarizes our key findings on the coupled dynamics between surface hydration water and protein side chains. Given a local perturbation, the water–protein system in a nonequilibrium configuration relaxes along hydration water and protein coordinates at a potential energy basin with a number of conformational substates (30). The outer-layer water molecules first respond in the femtosecond time regime, passing a small barrier, and then the inner-layer water molecules react on picosecond time scales to drive two coupled relaxations and surmount another two small barriers. Such water-driven relaxations along a small energy funnel can only access a certain limited region in the energy basin (Fig. 4). Hydration layers of water are dynamically ordered to bridge the protein and bulk water and control protein side-chain fluctuations, probably also slaving protein surface and even extending to internal protein motions (8), for a hydrated stable 3D structure with certain dynamic flexibility for function (31–33).
Fig. 4.
Coupled water/side-chain relaxations in a potential energy basin. (Upper) Four panels show the snapshots of MD simulations following several typical inner-layer (red) and outer-layer (blue) water molecules with the relaxation motions corresponding to the observed solvation dynamics. At time τ1S (20 fs), only the outer-layer water molecules locally relax. At time τ2S (130 fs), all water molecules proceed to significant rotational motions. However, all water molecules remain in the local positions, and the protein does not move significantly. At time τ3S (50 ps), all water molecules have made significant rearrangements and also exchanged with bulk water. Meanwhile, the protein surface topology was also altered. Note that the simulated τ1S and τ2S times are significantly shorter than those observed in our experiments. (Lower) Shown are three relaxation processes of hydration water and coupled tryptophan side chain in a potential energy basin with conformational substrates. The arrow in the white box indicates the constrained relaxation pathway with the initial outer-layer ultrafast relaxation (τ1S), which is not coupled to the protein motion, and two water-driven water/side-chain relaxations (τ2S and τ3S), which access only a limited region in the energy basin. (Insets) Two potential energy curves of hydration water and protein side chains used in construction of the contour energy landscape.
Materials and Methods
Protein Preparation.
Sulfolobus solfataricus Dpo4 gene was cloned into the NdeI and XhoI sites of pET22b, and a His6 tag was attached to the C terminal. The mutant plasmids were prepared by site-directed mutagenesis. Each plasmid was individually transformed into Escherichia coli BL21(DE3). All mutants were expressed and purified following procedures reported previously (34). The purified proteins were stored in a storage buffer solution containing 50 mM Tris⋅HCl (pH 7.5), 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, and 50% (vol/vol) glycerol. For fluorescence spectroscopy experiments, the sample was diluted and concentrated again to remove the glycerol. The final concentration of the protein sample was ∼1 mM, and the final buffer condition was 50 mM Tris⋅HCl (pH 7.5), 400 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, and less than 5% (vol/vol) glycerol. l-tryptophan was purchased from Sigma-Aldrich and used as received. The tryptophan was dissolved in the same buffer as the protein, and the final concentration was 3 mM.
Femtosecond Fluorescence Emission Spectroscopy.
All femtosecond-resolved fluorescence measurements were carried out by using the up-conversion method as described before (35). The pump wavelength was centered at 295 nm generated by a series of nonlinear mixing and doubling in 0.2-mm-thick barium borate (BBO) crystals (type I) with a repetition rate of 1 kHz. The pump pulse energy was attenuated to ∼100 nJ before being focused into the motor-controlled rotating sample cell. Use of 295-nm wavelength to excite the sample minimized tyrosine absorption in Dpo4, and the observed fluorescence was essentially from the excited tryptophan emission. The fluorescence emission was collected by a pair of parabolic mirrors and then mixed with a gating pulse (800 nm) in a 0.2-mm-thick BBO crystal through a nonlinear configuration. The up-converted signal ranging from 221 to 253 nm was detected by a photomultiplier coupled with a double-grating monochromator. A single fluorescence transient at the fixed wavelength or a femtosecond-resolved emission spectrum (FRES) at the fixed delay time will be obtained. The instrument response time under the current noncollinear geometry is approximately 400 fs as determined from the water Raman signal around 327 nm. The pump-pulse polarization was set at the “magic angle” (54.7°) with respect to the acceptance axis (vertical) of the up-conversion crystal, and the gating-pulse polarization was set parallel to this axis. For all fluorescence anisotropy measurements, the pump-pulse polarization was rotated parallel or perpendicular to the acceptance axis to obtain the parallel (I//) and perpendicular (I⊥) signals, respectively. The sample cell was mounted in the center of an aluminum block, and its temperature was controlled by a scientific solution bath (VWR). The lifetime of each mutant and tryptophan was determined by time-correlated single-photon counting.
Supplementary Material
Acknowledgments
We thank Prof. James T. Hynes, Dr. Paul Fenimore, and Ms. Jin Yang for helpful discussion and Prof. Zucai Suo for providing the Dpo4 plasmid. This work was supported in part by National Institutes of Health Grants GM095997 and GM118332. The MD simulations were supported in part by an allocation of computing time through the Ohio Supercomputer Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8355.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602916113/-/DCSupplemental.
References
- 1.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108(1):74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 2.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 3.Schirò G, et al. Translational diffusion of hydration water correlates with functional motions in folded and intrinsically disordered proteins. Nat Commun. 2015;6:6490. doi: 10.1038/ncomms7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewandowski JR, Halse ME, Blackledge M, Emsley L. Protein dynamics. Direct observation of hierarchical protein dynamics. Science. 2015;348(6234):578–581. doi: 10.1126/science.aaa6111. [DOI] [PubMed] [Google Scholar]
- 5.Jordanides XJ, Lang MJ, Song X, Fleming GR. Solvation dynamics in proein environments studied by photon echo spectroscopy. J Phys Chem B. 1999;103(37):7995–8005. [Google Scholar]
- 6.Pal SK, Zewail AH. Dynamics of water in biological recognition. Chem Rev. 2004;104(4):2099–2123. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 7.Makarov V, Pettitt BM, Feig M. Solvation and hydration of proteins and nucleic acids: A theoretical view of simulation and experiment. Acc Chem Res. 2002;35(6):376–384. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]
- 8.Frauenfelder H, et al. A unified model of protein dynamics. Proc Natl Acad Sci USA. 2009;106(13):5129–5134. doi: 10.1073/pnas.0900336106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, et al. Mapping hydration dynamics around a protein surface. Proc Natl Acad Sci USA. 2007;104(47):18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W. Damage repair DNA polymerases Y. Curr Opin Struct Biol. 2003;13(1):23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhong D. Hydration dynamics and coupled water-protein fluctuations probed by intrinsic tryptophan. Adv Chem Phys. 2009;143:83–149. [Google Scholar]
- 12.Qin Y, Chang CW, Wang L, Zhong D. Validation of response function construction and probing heterogeneous protein hydration by intrinsic tryptophan. J Phys Chem B. 2012;116(45):13320–13330. doi: 10.1021/jp305118n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Zhang L, Wang L, Zhong D. Femtosecond conical intersection dynamics of tryptophan in proteins and validation of slowdown of hydration layer dynamics. J Am Chem Soc. 2012;134(40):16460–16463. doi: 10.1021/ja305283j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu W, et al. Protein surface hydration mapped by site-specific mutations. Proc Natl Acad Sci USA. 2006;103(38):13979–13984. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen BE, et al. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 2002;296(5573):1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Yang Y, Kao YT, Wang L, Zhong D. Protein hydration dynamics and molecular mechanism of coupled water-protein fluctuations. J Am Chem Soc. 2009;131(30):10677–10691. doi: 10.1021/ja902918p. [DOI] [PubMed] [Google Scholar]
- 17.Zhong D, Pal SK, Zewail AH. Biological water: A critique. Chem Phys Lett. 2011;503(1):1–11. [Google Scholar]
- 18.Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol. 2011;18(2):245–249. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conti Nibali V, Havenith M. New insights into the role of water in biological function: Studying solvated biomolecules using terahertz absorption spectroscopy in conjunction with molecular dynamics simulations. J Am Chem Soc. 2014;136(37):12800–12807. doi: 10.1021/ja504441h. [DOI] [PubMed] [Google Scholar]
- 20.Chandra A, Bagchi B. A molecular theory of collective orientational relaxation in pure and binary dipole liquds. J Chem Phys. 1989;91(3):1829–1842. [Google Scholar]
- 21.Naudi N, Bagchi B. Dielectric relaxation of biological water. J Phys Chem B. 1997;101(50):10954–10961. [Google Scholar]
- 22.Laage D, Hynes JT. A molecular jump mechanism of water reorientation. Science. 2006;311(5762):832–835. doi: 10.1126/science.1122154. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Hassanali AA, Kao YT, Zhong D, Singer SJ. Hydration dynamics and time scales of coupled water-protein fluctuations. J Am Chem Soc. 2007;129(11):3376–3382. doi: 10.1021/ja0685957. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh R, Banerjee S, Hazra M, Roy S, Bagchi B. Sensitivity of polarization fluctuations to the nature of protein-water interactions: Study of biological water in four different protein-water systems. J Chem Phys. 2014;141(22):22D531. doi: 10.1063/1.4902821. [DOI] [PubMed] [Google Scholar]
- 25.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Slaving: Solvent fluctuations dominate protein dynamics and functions. Proc Natl Acad Sci USA. 2002;99(25):16047–16051. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagchi B. Water dynamics in the hydration layer around proteins and micelles. Chem Rev. 2005;105(9):3197–3219. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- 27.Doster W, Settles M. Protein-water displacement distributions. Biochim Biophys Acta. 2005;1749(2):173–186. doi: 10.1016/j.bbapap.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Fenimore PW, Frauenfelder H, McMahon BH, Young RD. Bulk-solvent and hydration-shell fluctuations, similar to alpha- and beta-fluctuations in glasses, control protein motions and functions. Proc Natl Acad Sci USA. 2004;101(40):14408–14413. doi: 10.1073/pnas.0405573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malardier-Jugroot C, Head-Gordon T. Separable cooperative and localized translational motions of water confined by a chemically heterogeneous environment. Phys Chem Chem Phys. 2007;9(16):1962–1971. doi: 10.1039/b616997j. [DOI] [PubMed] [Google Scholar]
- 30.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 31.Zaccai G. How soft is a protein? A protein dynamics force constant measured by neutron scattering. Science. 2000;288(5471):1604–1607. doi: 10.1126/science.288.5471.1604. [DOI] [PubMed] [Google Scholar]
- 32.King JT, Kubarych KJ. Site-specific coupling of hydration water and protein flexibility studied in solution with ultrafast 2D-IR spectroscopy. J Am Chem Soc. 2012;134(45):18705–18712. doi: 10.1021/ja307401r. [DOI] [PubMed] [Google Scholar]
- 33.Henzler-Wildman KA, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450(7171):913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 34.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43(7):2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Kao YT, Qiu W, Wang L, Zhong D. Femtosecond studies of tryptophan fluorescence dynamics in proteins: Local solvation and electronic quenching. J Phys Chem B. 2006;110(37):18097–18103. doi: 10.1021/jp063025e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.