Significance
Spinal muscular atrophy (SMA) is a devastating motor neuron disease, caused by decreased levels of the ubiquitous survival motor neuron (SMN) protein. Despite the well-characterized role of SMN in pre-mRNA splicing, it remains unclear why SMA has a high carrier frequency (∼1:50 Caucasians) and why diminished SMN affects synaptic function. Here, we demonstrate for the first time, to our knowledge, that SMN depletion causes defects in endosomal trafficking that impair synaptic function. Additionally, diminished SMN in human cells reduced endocytosis-dependent viral infection. It is possible that decreased SMN function may increase resistance to infection. Our findings point to endocytic trafficking as a major player in SMA pathogenesis.
Keywords: endocytic trafficking, survival motor neuron, spinal muscular atrophy, C. elegans, infection
Abstract
Spinal muscular atrophy (SMA) is caused by depletion of the ubiquitously expressed survival motor neuron (SMN) protein, with 1 in 40 Caucasians being heterozygous for a disease allele. SMN is critical for the assembly of numerous ribonucleoprotein complexes, yet it is still unclear how reduced SMN levels affect motor neuron function. Here, we examined the impact of SMN depletion in Caenorhabditis elegans and found that decreased function of the SMN ortholog SMN-1 perturbed endocytic pathways at motor neuron synapses and in other tissues. Diminished SMN-1 levels caused defects in C. elegans neuromuscular function, and smn-1 genetic interactions were consistent with an endocytic defect. Changes were observed in synaptic endocytic proteins when SMN-1 levels decreased. At the ultrastructural level, defects were observed in endosomal compartments, including significantly fewer docked synaptic vesicles. Finally, endocytosis-dependent infection by JC polyomavirus (JCPyV) was reduced in human cells with decreased SMN levels. Collectively, these results demonstrate for the first time, to our knowledge, that SMN depletion causes defects in endosomal trafficking that impair synaptic function, even in the absence of motor neuron cell death.
Spinal muscular atrophy (SMA) is one of the most severe neuromuscular diseases of childhood, with an incidence of 1 in 10,000 live births and a high carrier frequency of roughly 1 in 40 Caucasians (1–3). SMA is caused by reduced levels of the ubiquitously expressed survival of motor neuron (SMN) protein and results in degeneration of α-spinal cord motor neurons, muscle weakness, and/or death. Two human genes encode the SMN protein, SMN1 and SMN2. SMA alleles arise at relatively high frequency due to small intrachromosomal de novo rearrangements including the SMN1 locus (4). Patients often carry homozygous SMN1 deletions, although missense and nonsense alleles exist (5). Multiple copies of SMN2 rarely compensate for loss of SMN1 due to a C > T nucleotide change in SMN2 exon 7 that perturbs pre-mRNA splicing and results in a truncated protein of diminished function and stability (SMNΔ7) (5–9).
SMN has numerous roles and interacts with various proteins, yet it remains unclear which interactions are most pertinent to SMA pathogenesis. As a component of the Gemin complex, SMN is required for biogenesis of small nuclear ribonucleoprotein (snRNP) particles critical for pre-mRNA splicing (10–12). Furthermore, SMN is needed for stress granule formation (13, 14), is found in RNP granules moving through neuronal processes, and is part of RNP complexes implicated in synaptic local translation (15–20). Additional roles for SMN, in transcription (21), in the PTEN-mediated protein synthesis pathway (22), in translational control (23), and in cell proliferation/differentiation (24), have been described. Importantly, no consensus has been reached regarding the cellular and molecular pathways whose perturbation results in SMA pathology. Identifying the cellular pathways most sensitive to decreased SMN is essential to understand how SMN depletion causes neuronal dysfunction/death in SMA and to accelerate therapy development.
One of the early events in SMA pathogenesis is the loss of neuromuscular junction (NMJ) function, evidenced by muscle denervation, neurofilament accumulation, and delayed neuromuscular maturation (25–27). In addition, reduced neurotransmitter release and decreased numbers of docked vesicles that precede axonal degeneration and/or motor neuron death have been reported at synapses of severe SMA mouse models (28, 29). Notably, accumulation of synaptic vesicles (SVs) away from release sites was observed in SMA fetal samples (30). The proximate cause of these synaptic changes is unclear. Numerous hypotheses have been proposed, including functional abnormalities in axonal transport and/or calcium channel loss in the nerve terminals (25–30), but none have explained the defects observed in SMA presynaptic regions.
Here, we use a previously established model of SMA in the nematode Caenorhabditis elegans and show, using functional assays, pharmacological challenges, and genetic epistasis, that decreased SMN levels cause endocytic pathway defects. In C. elegans cholinergic motor neurons, decreased SMN levels caused aberrant localization of proteins critical for endocytosis. Further, ultrastructural analysis of endosomal compartments revealed numerous defects when SMN levels were depleted, including loss of synaptic docked vesicles. Endocytic pathway defects were also observed in nonneuronal tissues. Finally, endocytosis-dependent infection by JC polyomavirus (JCPyV) was reduced in human cells with decreased SMN levels. Combined, these results demonstrate for the first time, to our knowledge, that SMN depletion causes widespread defects in endosomal trafficking that impair synaptic function in motor neurons, even in the absence of motor neuron death.
Results
smn-1 Is Required for Neuromuscular Function.
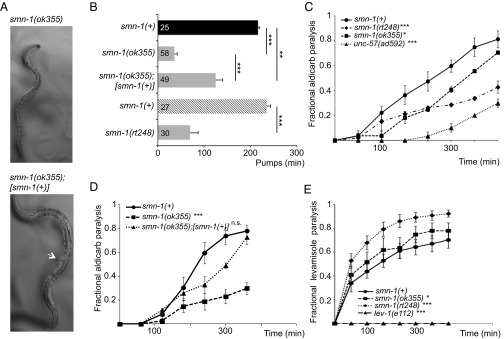
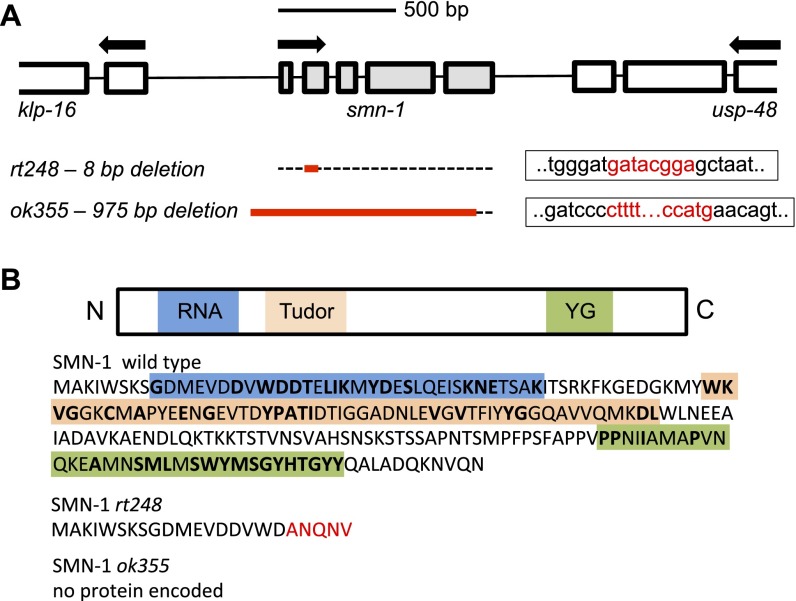
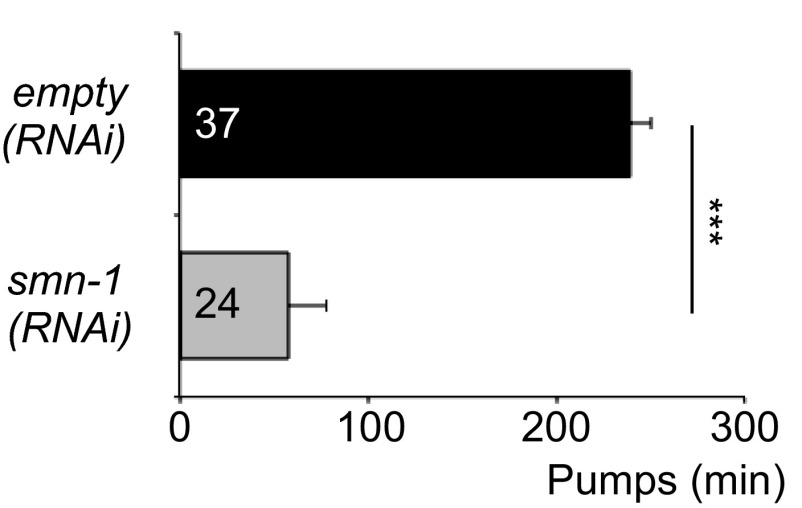
The C. elegans genome encodes a single ortholog of SMN, SMN-1. Animals with a wild-type copy of the endogenous smn-1 gene are referred to herein as smn-1(+) and are used as controls. Diminished smn-1 function causes slow growth and larval lethality and impairs neuromuscular function in pharyngeal pumping during feeding (31, 32) (Fig. 1A). C. elegans feed on microorganisms using a discrete subset of muscles and neurons in the pharynx (33). Animals pump symmetrically and continuously roughly 250 times per minute when food is present. The pumping rates of smn-1 loss-of-function animals [smn-1(ok355)] are significantly reduced (P = 3e-12; Fig. 1B) (31, 32). To confirm that the defects described here are caused by smn-1 loss, we generated a new smn-1 allele, smn-1(rt248), using CRISPR/Cas9-targeted mutagenesis (34, 35). The rt248 allele results in a premature truncation at the 19th amino acid (D19fs) of the SMN-1 protein; disrupts RNA binding, Tudor, and oligomerization domains; and likely eliminates SMN-1 protein function (Fig. S1). The pumping rates of smn-1(rt248) animals were significantly reduced (P = 3e-9; Fig. 1B). This defect was reconfirmed using RNA interference [smn-1(RNAi), P = 2e-09; Fig. S2]. These neuromuscular defects are progressive and not a developmental process; homozygous smn-1(ok355) and smn-1(rt248) larvae initially resemble wild-type animals due to maternal smn-1 loading. Eventually, homozygous smn-1 animals lose maternally loaded smn-1 product, suffer decreased motor neuron function, and usually die as larvae. However, no motor neuron loss is observed (36). The impact of smn-1 loss is recessive; heterozygous animals are overtly normal. Single copy insertion of an smn-1(+) genomic DNA transgene (containing 445 bp of 5′ regulatory sequences, 816 bp of smn-1 genomic DNA, and 558 bp of 3′ noncoding sequences, called rtSi10) fully rescued larval lethality, partially ameliorated pharyngeal pumping defects (P = 0.0004; Fig. 1 A and B), but did not restore fertility of smn-1(ok355) animals. The partial rescue of smn-1(ok355) defects may arise from regulatory sequences required for SMN-1 expression that were not included in the rescue construct. Nevertheless, the results presented here confirm that these defects can be attributed to SMN-1 depletion and that SMN-1 is required for normal neuromuscular function.
Fig. 1.
Decreased SMN function in C. elegans causes defective motor neuron function. (A) Images of age-matched smn-1(ok355) (Top) and smn-1(ok355);[smn-1(+)] (Bottom) animals. Single-copy insertion of an [smn-1(+)] transgene rtSi10 rescued smn-1(ok355) larval lethality and growth but did not restore fertility. Arrow indicates adult vulva, which has not developed in age-matched smn-1(ok355). (B) smn-1(ok355) and smn-1(rt248) animals had reduced pumping rates vs. respective smn-1(+) controls. Studies in B involving smn-1(ok355) were run independently from studies with smn-1(rt248) but combined into one panel for brevity [black column, smn-1(+) control for ok355; cross-hatched, smn-1(+) control for rt248)]. smn-1(ok355) pharyngeal pumping defects (at day 3 posthatching) were ameliorated by introducing an smn-1(+) rescue construct. Mean ± SEM; Mann–Whitney U test, two-tailed: **P < 0.01; ***P < 0.001. (C) smn-1(ok355) and smn-1(rt248) animals were resistant to the acetylcholinesterase inhibitor aldicarb. Time course for paralysis induced by 1 mM aldicarb in smn-1(+), smn-1(ok355), smn-1(rt248), and unc-57(ad592) young L4 hermaphrodites is shown. unc-57 encodes C. elegans endophilin A; unc-57(ad592) causes inappropriate resistance to aldicarb (43). Log-rank test: *P < 0.05; ***P < 0.001. (D) The aldicarb resistance of smn-1(ok355) animals was mostly restored by [smn-1(+)] genomic rescue (rtSi10). Log-rank test: ***P < 0.001; n.s., not significant. (E) smn-1(ok355) and smn-1(rt248) were hypersensitive to levamisole, a nicotinic ACh receptor agonist. smn-1(+), smn-1(ok355), smn-1(rt248), and lev-1(e211) young L4 hermaphrodite paralysis on 0.4 mM levamisole plates is reported. lev-1(e211) animals lack a nicotinic ACh receptor subunit. Error bars indicate ± SEM. Log-rank test: *P < 0.05; ***P < 0.001.
Fig. S1.
Representation of the smn-1 gene and SMN-1 protein domains in the nematode C. elegans. (A) ORF of the smn-1 gene and location of the rt248 and ok355 deletion alleles. Boxes indicate exons, and black lines represent introns. Red letters indicate missing nucleotides. (B) SMN-1 regulatory domains (RNA binding, Tudor, and YG oligomerization) and the putative isoforms encoded by the rt248 and ok355 deletion alleles; rt248 deletion causes a premature stop codon, whereas ok355 deletion does not encode a putative protein. Amino acid similarity between the human and nematode protein sequences is shown in bold. (Scale bar, 500 bp.)
Fig. S2.
C. elegans smn-1(RNAi) knockdown in an RNAi-sensitized background results in decreased pumping rates. Animals sensitive to RNAi in all tissues (KP3948) have decreased pumping rates when fed bacteria expressing double-stranded RNA (dsRNA) against smn-1 [labeled smn-1(RNAi)]. Control is KP3948 animals reared on empty control RNAi [labeled empty(RNAi)]. Error bars indicate ±SEM. Mann–Whitney U test, two-tailed: ***P < 0.001.
smn-1 Regulates Synaptic Transmission.
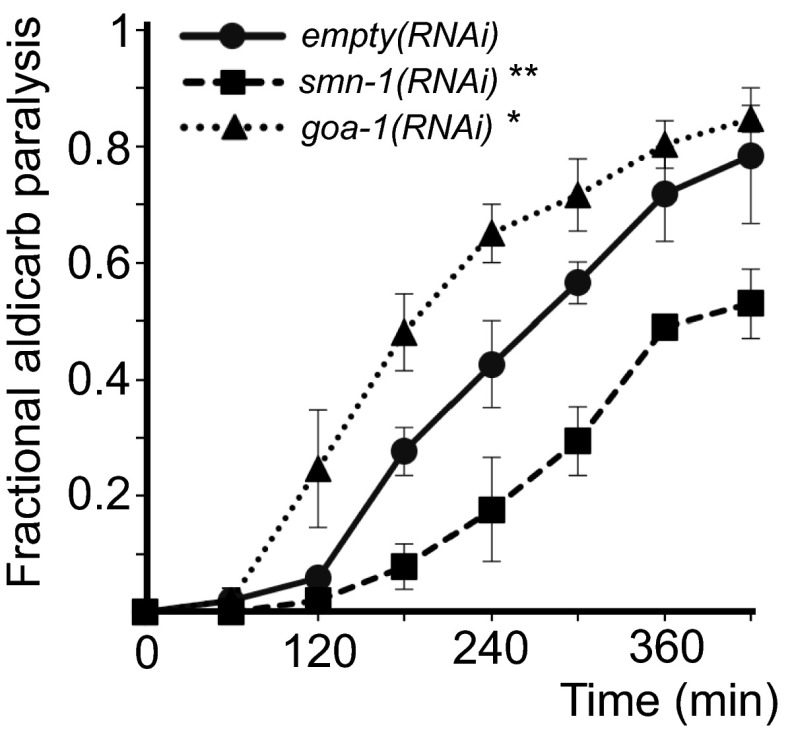
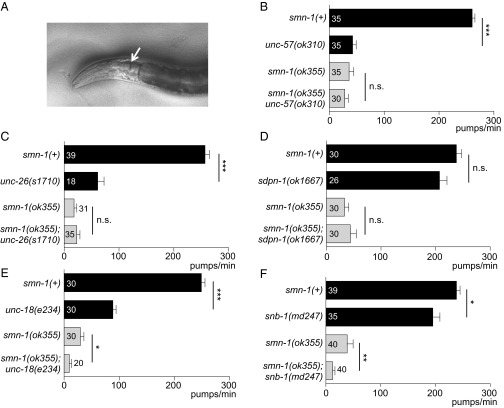
Studies in SMA patients and mice revealed defects in motor neuron synapses when SMN levels are reduced (37). We examined motor neuron function pharmacologically in smn-1(ok355) and smn-1(rt248) animals by assessing sensitivity to aldicarb and levamisole. Aldicarb is an acetylcholinesterase inhibitor that causes muscle hypercontraction and paralysis due to accumulation of acetylcholine (ACh) in the synaptic cleft (38). Decreased ACh release slows the onset of aldicarb-induced paralysis. We found that smn-1(ok355) and smn-1(rt248) animals were resistant to aldicarb compared with animals with normal smn-1 function [P = 0.03 smn-1(ok355) and P = 8e-6 smn-1(rt248) vs. smn-1(+) derived from +/hT2 parents; Fig. 1C]. This increased resistance was due to diminished smn-1, as introducing an smn-1(+) genomic DNA fragment restored aldicarb sensitivity (P = 0.17; Fig. 1D). Resistance to aldicarb was also observed when smn-1 was knocked down by RNAi specifically in cholinergic neurons (P = 0.004; Fig. S3) (39). Therefore, SMN-1 is required for neuromuscular function, likely in cholinergic neurons, which is consistent with a previous study reporting resistance to the acetylcholinesterase inhibitor pyridostigmine bromide (40). smn-1(ok355) and smn-1(rt248) aldicarb resistance could be caused by decreased neurotransmitter release or reduced postsynaptic response to ACh. To discriminate, we assessed smn-1(ok355) and smn-1(rt248) sensitivity to levamisole, an agonist, which induces paralysis by directly activating postsynaptic nicotinic receptors. Levamisole resistance is thought to be purely postsynaptic (41). smn-1(ok355) and smn-1(rt248) were hypersensitive in their response to levamisole [P = 0.02 smn-1(ok355) and P = 5e-4 smn-1(rt248) vs. smn-1(+); Fig. 1E]. According to Miller et al. (42), a normal or hypersensitive response to levamisole suggests that an impaired ACh response at the muscle is not the cause of aldicarb resistance. Together, these data suggest that SMN-1 likely impacts presynaptic function and is required for normal neurotransmitter release.
Fig. S3.
smn-1(RNAi) knockdown in C. elegans cholinergic motor neurons causes resistance to aldicarb. Animals sensitive to RNAi only in cholinergic neurons (XE1581) were resistant to the acetylcholinesterase inhibitor aldicarb when fed bacteria expressing dsRNA against smn-1 [labeled smn-1(RNAi)]. empty(RNAi)-treated XE1581 animals served as controls. Log-rank test: **P < 0.01. Time course for paralysis induced by 1.5 mM aldicarb in smn-1(+), smn-1, and goa-1 RNAi knockdown young adult animals. goa-1 encodes a cholinergic C. elegans GTPase, loss of which causes inappropriate hypersensitivity to aldicarb. goa-1(RNAi) was used here as an internal control (96). Error bars indicate ±SEM. Log-rank test: *P < 0.05.
Genetic Interaction of smn-1 with Endocytic Pathway Genes.
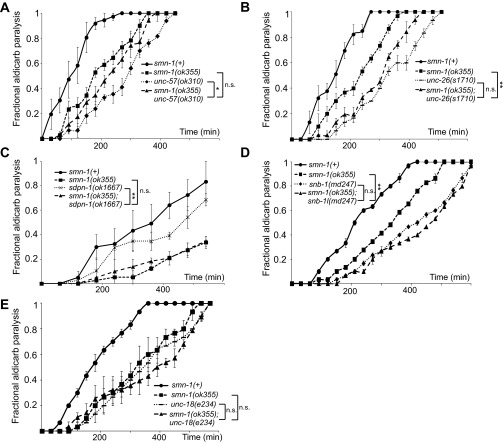
The smn-1(ok355) and smn-1(rt248) synaptic defects could be due to impairments in the SV cycle. Potential functional interactions between smn-1 and genes encoding SV endo- or exocytosis proteins were examined by constructing double mutant strains. For this epistasis analysis, we used two quantitative neuromuscular phenotypes: aldicarb sensitivity and pharyngeal pumping rates. First, double mutant animals were constructed with smn-1(ok355) and complete (null) loss-of-function alleles for unc-57 (endophilin-A) and unc-26 (synaptojanin), which encode proteins in the synaptic clathrin-mediated endocytosis (CME) pathway (43), and sdpn-1 (syndapin), which is required for activity-dependent bulk endocytosis (ADBE) (44). Loss of smn-1 did not exacerbate the aldicarb resistance defects of unc-57(ok310) or unc-26(s1710) compared with each single mutant strain, but rather an intermediate defect was observed (Fig. S4 A and B). The aldicarb response of smn-1(ok355);sdpn-1(ok1667) animals was almost identical to that of smn-1(ok355) animals (P = 0.76; Fig. S4C). When double mutant strains were constructed with complete (null) loss-of-function mutants involved in SV exocytosis, snb-1 (synaptobrevin) or unc-18 (Munc18), the aldicarb response defects were nonadditive [P = 0.55 for snb-1(md247) vs. smn-1(ok355);snb-1(md247) and P = 0.72 for unc-18(e234) vs. smn-1(ok355);unc-18(e234); Fig. S4 D and E]. These results are consistent with smn-1 depletion impairing SV function, but definitive conclusions could not be drawn. Therefore, we turned to pharyngeal pumping rates to assess genetic interactions (Fig. 2A). Loss of C. elegans endophilin, synaptojanin, or syndapin did not exacerbate the decreased pharyngeal pumping rates of smn-1(ok355) [P = 0.34 for smn-1(ok355) unc-57(ok310), P = 0.93 for smn-1(ok355);unc-26(s1710), and P = 0.67 for smn-1(ok355);sdpn-1(ok1667); Fig. 2 B–D]. By contrast, double mutant strains with genes involved in SV exocytosis were additive and had significantly reduced pumping rates compared with each single mutant strain [P = 0.01 for smn-1(ok355);unc-18(e234) and P = 0.008 for smn-1(ok355);snb-1(md247); Fig. 2 E and F]. Overall, these results indicate that SMN-1 depletion impairs synaptic transmission, potentially by decreasing SV recycling.
Fig. S4.
Double mutant analysis using aldicarb resistance is consistent with SV defects. Time course for paralysis induced by 1 mM aldicarb in young L4 hermaphrodites. (A) smn-1(ok355) unc-57(ok310) double mutant animals were resistant to aldicarb and showed an intermediate phenotype in relation to single mutant animals. Log-rank test: P = 0.22 for smn-1(ok355) vs. smn-1(ok355) unc-57(ok310), P = 0.02 for unc-57(ok310) vs. smn-1(ok355) unc-57(ok310). (B) smn-1(ok355);unc-26(s1710) animals also showed an intermediate aldicarb phenotype vs. single mutant animals. Log-rank test: P = 0.004 for smn-1(ok355) vs. smn-1(ok355);unc-26(s1710); P = 0.145 for unc-26(s1710) vs. smn-1(ok355);unc-26(s1710). (C) smn-1(ok355);sdpn-1(ok1667) double mutant animals were as aldicarb-resistant as smn-1(ok355) animals. Log-rank test: P = 0.75 for smn-1(ok355) vs. smn-1(ok355);sdpn-1(ok1667). (D) snb-1(md247) and smn-1(ok355) loss are nonadditive. Log-rank test: P = 0.004 for smn-1(ok355) vs. smn-1(ok355);snb-1(md247); P = 0.55 for snb-1 vs. smn-1(ok355);snb-1(md247). (E) unc-18(e234) and smn-1(ok355) loss are nonadditive. Log-rank test: P = 0.16 for smn-1(ok355) vs. smn-1(ok355);unc-18(e234); P = 0.72 for unc-18(e234) vs. smn-1(ok355);unc-18(e234). Shown is the average fractional paralysis of three independent trials for each time point ± SEM: *P < 0.05; **P < 0.01; n.s., not significant.
Fig. 2.
Double mutant analysis using pharyngeal pumping suggests SV recycling defects. (A) C. elegans neuromuscular function is assessed as pharyngeal grinder (arrow) movement during feeding. (B–D) Complete loss of endophilin-A, synaptojanin, or syndapin (unc-57(ok310), unc-26(s1710), or sdpn-1(ok1667)) did not alter smn-1(ok355) pharyngeal pumping defects. (E and F) Complete loss of Munc-18 or synaptobrevin [unc-18(e234) or snb-1(md247)] exacerbated smn-1(ok355) pharyngeal pumping defects. Additive defects with SV exocytosis loss-of-function mutants suggested that SMN-1 depletion impairs the SV recycling pathway. Total number of animals tested listed for each genotype ± SEM; Mann–Whitney U test, two-tailed: *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
Diminished SMN-1 Alters the Presynaptic Structure of Motor Neurons.
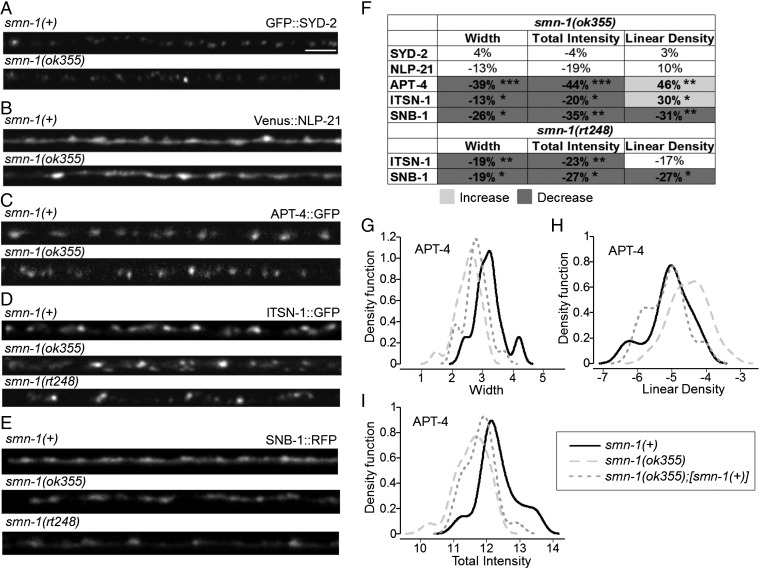
If SV recycling is defective when SMN levels are diminished, then the localization and/or levels of specific presynaptic proteins might change. Synaptic localization was examined in the DA motor neurons of smn-1(ok355) and smn-1(rt248) animals for two fluorescently tagged presynaptic proteins: SNB-1 (synaptobrevin) and ITSN-1 (DAP160/Intersectin) (45, 46). In the dorsal cord, DA motor neurons have no presynaptic inputs and form en passant synapses onto muscle processes (47). This formation results in a punctate pattern of protein localization along the length of the dorsal cord, composed primarily of presynaptic active zones (45). We quantified puncta width, puncta total intensity, and puncta linear density (number per micrometer). In both smn-1(ok355) and smn-1(rt248) animals, puncta of the SV protein SNB-1 were diminished in width, intensity, and density (average puncta width decreased 26% and 19% for ok355 and rt248, respectively, P = 0.02 for ok355 and P = 0.04 for rt248; average total intensity decreased 35% and 27% for ok355 and rt248, respectively, P = 0.005 for ok355 and P = 0.02 for rt248; density decreased roughly 30% for both alleles, P = 0.004 for ok355 and P = 0.01 for rt248) (Fig. 3 E and F). Puncta of the endocytic protein ITSN-1 (DAP160/Intersectin) were also altered in width and intensity in both smn-1(ok355) and smn-1(rt248) animals; linear density changed only in smn-1(ok355) animals (Fig. 3 D and F). ITSN-1 puncta were smaller in size (ITSN-1 average puncta width decreased by 13% and 19% for ok355 and rt248, respectively, P = 0.03 for ok355 and P = 0.008 for rt248; puncta total ITSN-1 intensity was reduced by 20% and 23% for ok355 and rt248, respectively, P = 0.02 for ok355 and P = 0.004 for rt248).
Fig. 3.
Altered localization of presynaptic endocytic proteins in smn-1 loss-of-function animals. (A–E) Representative images of fluorescently tagged, presynaptic proteins expressed in the dorsal nerve cord of cholinergic DA motor neurons of smn-1(+) control, smn-1(ok355), and smn-1(rt248) animals. (Scale bar, 10 μm.) (F) Percent change from control for SYD-2 (α-liprin), NLP-21 (GGARAF neuropeptide family), APT-4 (AP2 α-adaptin), ITSN-1 (DAP160/Intersectin), and SNB-1 (synaptobrevin) in smn-1(ok355) animals is reported for average puncta width, puncta total intensity, and linear density (number per micrometer). ITSN-1 and SNB-1 are reported for smn-1(rt248). Light and dark shading indicate an increase or decrease, respectively, in the smn-1(ok355) and smn-1(rt248) animals compared with smn-1(+) control (see Dataset S1 for extended analysis). Mann–Whitney U test, two-tailed: *P < 0.05; **P < 0.01; ***P < 0.001. (G–I) A single copy of the [smn-1(+)] construct rtSi10 fully rescued APT-4 linear density [control smn-1(+) animals vs. rescued smn-1(ok355);rtSi10[smn-1(+)] P = 0.5; smn-1(ok355) vs. rescued P = 0.002], partially ameliorated puncta width defects [control smn-1(+) vs. rescued P = 0.002; smn-1(ok355) vs. rescued P = 0.03], but may not have improved total intensity [control smn-1(+) vs. rescued P = 5e-04; smn-1(ok355) vs. rescued P = 0.09]. Distributions of APT-4 puncta width, linear density, and puncta total intensity in smn-1(ok355) animals are compared with distributions in control smn-1(+) and rescued animals. Results are presented as kernel density estimates, which convert distribution histograms into smooth, continuous density function curves. The x-axis values were log2-transformed before the calculation of the density function. Linear density (puncta per micrometer) values are less than 1, resulting in negative values after log2 transform (Dataset S1). At least three independent trials were performed (n > 25 animals in total/genotype).
As both smn-1 loss-of-function alleles had the same profile, we also examined three more presynaptic markers in the cholinergic DA motor neurons of smn-1(ok355) animals only: SYD-2 (α-liprin), NLP-21 (GGARAF neuropeptide family), and APT-4 (AP2 α-adaptin). SYD-2 and NLP-21 puncta were unaltered in smn-1(ok355) animals, suggesting that the number of synapses—based on active zones and dense core vesicles (DCVs), respectively—is unchanged when SMN-1 levels drop (Fig. 3 A, B, and F). This is consistent with previous work showing that smn-1(ok355) animals have no overt defects in nervous system morphology and no motor neuron death (31). Puncta of the endocytic APT-4 (AP2 α-adaptin) were altered in intensity and number in smn-1(ok355) animals (Fig. 3 C and F). APT-4 puncta were more numerous along the neuronal processes (APT-4 increased 46%, P = 0.003) but were smaller in size (APT-4: decreased 39%, P = 8e-06). Puncta total intensity was reduced by 44% for APT-4 (P = 3e-05). To further confirm that changes in synaptic protein levels were due to decreased smn-1 function, a wild-type copy of smn-1(+) was introduced to smn-1(ok355) animals expressing APT-4::GFP. Synaptic defects were ameliorated or eliminated (Fig. 3 G–I). Combined, these results suggest that smn-1 loss of function likely causes defects in SV recycling, resulting in aberrant localization of endocytic proteins and fewer functional SVs. To confirm this interpretation, we examined smn-1(ok355) presynaptic specializations at a higher resolution.
Decreased smn-1 Function Causes Defects in Endocytic Compartments.
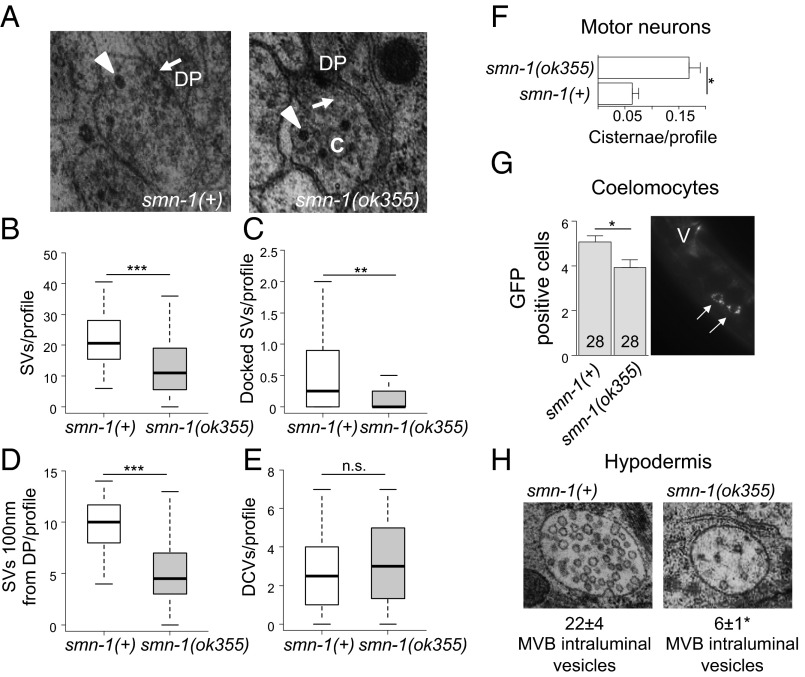
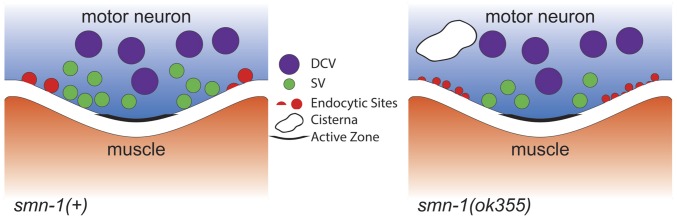
We sectioned and reconstructed part of the ventral nerve cord for both normal smn-1(+) and mutant smn-1(ok355) animals to determine the number and distribution of vesicles in the presynaptic specializations by using transmission electron microscopy (TEM) (Fig. 4A). The total number of SVs per presynaptic region was reduced by 36% in motor neurons of smn-1(ok355) animals vs. smn-1(+) (14 ± 2, n = 45 vs. 22 ± 2, n = 35, respectively, P = 0.0002; Fig. 4B). The average number of docked vesicles at the plasma membrane (P = 0.008) and average number of SVs 100 nm away from the active zone (P = 4e-08) were also decreased in smn-1(ok355) animals vs. smn-1(+) (Fig. 4 C and D). By contrast, the average number of DCVs in each presynaptic profile was not significantly different between smn-1(+) and smn-1(ok355) animals (P = 0.34; Fig. 4E). The changes observed in SVs were consistent with results from the puncta analysis (Fig. 3). TEM studies also revealed an unusual accumulation of cisternae in smn-1(ok355) synapses (Fig. 4F). Cisternae are large and abnormal-sized vesicles that often reflect arrested endocytic vesicle maturation and sorting. Cisternae accumulation was reported in the synapses of C. elegans defective in endocytosis, such as unc-57 and unc-26 animals (43, 48). smn-1(+) animals had 0.06 ± 0.01 cisternae/synaptic specialization, whereas smn-1(ok355) animals exhibited a threefold increase (0.17 ± 0.02 cisternae/profile; P = 0.02). The ultrastructural abnormalities in smn-1(ok355) animals further support a model in which diminished SMN-1 function impairs endosomal trafficking in motor neuron synapses.
Fig. 4.
Endosomal defects are seen in smn-1(ok355) motor neurons and elsewhere. (A) Representative electron micrograph images of smn-1(+) control and smn-1(ok355) NMJs. Arrows point to SV and arrowheads to DCVs. C, cistern; DP, dense projection. (B) SVs per synaptic profile, as defined by the presence of a DP, were reduced in smn-1(ok355) animals. Mann–Whitney U test, two-tailed: ***P < 0.001. (C) Docked SVs in contact with the DP were reduced in smn-1(ok355) animals. Mann–Whitney U test, two-tailed: **P < 0.01. (D) smn-1(ok355) animals had reduced numbers of SVs within the region 100 nm from the synaptic profile/DP. Mann–Whitney U test, two-tailed: ***P < 0.001 (Table S2). (E) The number of DCVs per synaptic profile/DP in smn-1(ok355) animals was not different from control smn-1(+) animals. Mann–Whitney U test, two tailed: n.s., not significant. (F) Cisternae accumulated aberrantly in motor neurons of smn-1(ok355) mutants. Mean ± SEM is shown; Mann–Whitney U test, two-tailed: *P < 0.05. (G) Fluid-phase endocytosis by coelomocytes was decreased in smn-1(ok355) animals, Mann–Whitney U test, two-tailed: *P < 0.05. Image on the Right shows the pair of coelomocyte cells lying just anterior to vulva; arrows indicate GFP accumulation inside the coelomocytes. No GFP accumulation in the body cavity of ok355 animals was observed. (H) Representative electron micrograph images of MVBs in the hypodermis of smn-1(+) control and smn-1(ok355) animals. The MVBs of smn-1(ok355) animals contained fewer intraluminal vesicles compared with smn-1(+) control animals; Mann–Whitney U test, two-tailed: *P < 0.05.
Table S2.
Percentage SVs 100 nm from DP per profile and % docked SVs per profile (related to Fig. 4)
| Strain | Average | SEM |
| % docked SV/profile | ||
| smn-1(+) | 100 | 23.4 |
| smn-1(ok355) | 21.3 | 11.3 |
| % SVs 100 nm from DP/profile | ||
| smn-1(+) | 100 | 5.5 |
| smn-1(ok355) | 42.8 | 5.6 |
Diminished SMN-1 Causes Endosomal Defects in Nonneuronal Tissues.
SV recycling shares common elements with endosomal trafficking in other cells. To determine if smn-1 depletion causes endosomal defects more broadly, other tissues were examined. The C. elegans body cavity contains six coelomocyte cells that rely on fluid-phase endocytosis to clear soluble moieties from the pseudocoelom (49). Animals engineered to secrete GFP from body wall muscles into the body cavity have been used extensively to characterize endocytic activity in coelomocyte cells (50). We compared coelomocyte GFP uptake in smn-1(+) and smn-1(ok355) animals. GFP uptake in smn-1(ok355) animals (P = 0.01; Fig. 4G) is significantly reduced, indicating that diminished SMN-1 causes endocytic pathway defects in these nonneuronal cells. As GFP does not accumulate in the body cavity of smn-1(ok355) animals, the coelomocytes defect is not severe. To confirm that smn-1 loss decreases endocytosis, we used RNAi to knock down SMN-1 specifically in coelomocytes. Loss of SMN-1 resulted in increased GFP intensity in coelomocytes and significant accumulation of GFP in the body cavity, consistent with stalled endocytosis and a cell-autonomous defect. No gross morphology changes or large vacuoles were observed (Figs. S5 and S6).
Fig. S5.
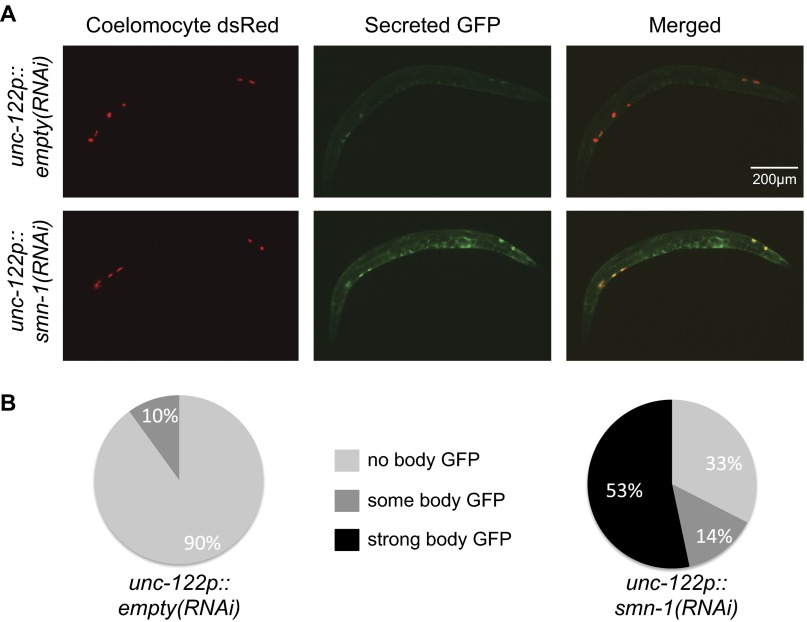
smn-1(RNAi) knockdown causes endocytic defects in coelomocytes. (A) Representative images of animals injected with coelomocyte-specific smn-1 RNAi (unc-122p::smn-1RNAi) or empty control (unc-122p::emptyRNAi). (B) Pie charts illustrate the percentage of animals with indicated level of GFP accumulation in the body cavity. GFP is secreted from muscle cells; coelomocyte-specific knockdown of smn-1 resulted in a significant accumulation of GFP in the body cavity and increased GFP intensity in coelomocytes, consistent with a cell-autonomous endocytic defect. No gross morphology changes and no large vacuoles were observed. Line indicates 200 microns.
Fig. S6.
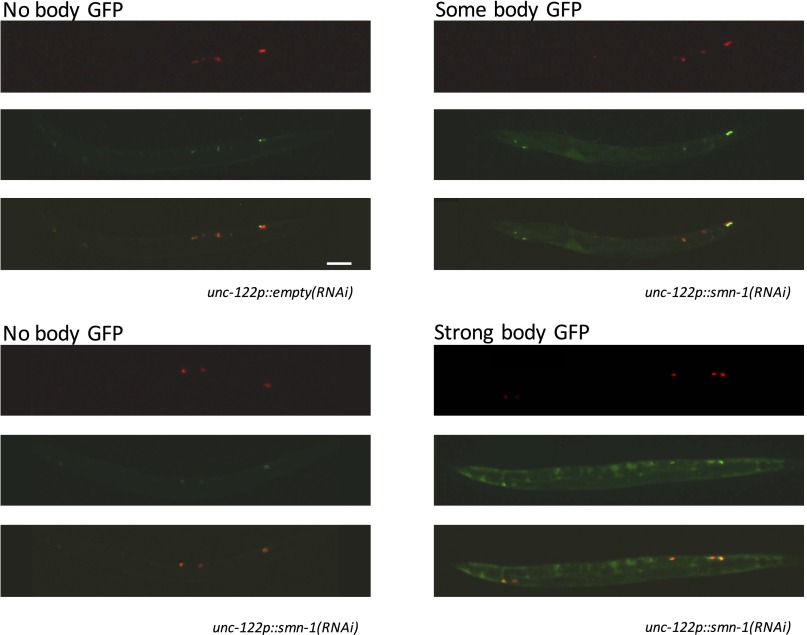
Method for assessing severity of smn-1(RNAi) coelomocyte endocytic defects. In some animals, SMN-1 RNAi knockdown resulted in body cavity accumulation of muscle-secreted GFP; refer to Fig. S5 and Diminished SMN-1 Causes Endosomal Defects in Nonneuronal Tissues for details. Animals expressing coelomocyte-specific smn-1 RNAi, called unc-122p::smn-1(RNAi), are shown here; representative images are for “no body GFP,” “some body GFP,” and “strong body GFP.” Also shown are representative images for animals injected with an empty control (unc-122p::empty RNAi), which display little to no body GFP. Top image, Coelomocyte dsRed; middle image, secreted GFP; bottom image, merged. (Scale bar, 100 μm.)
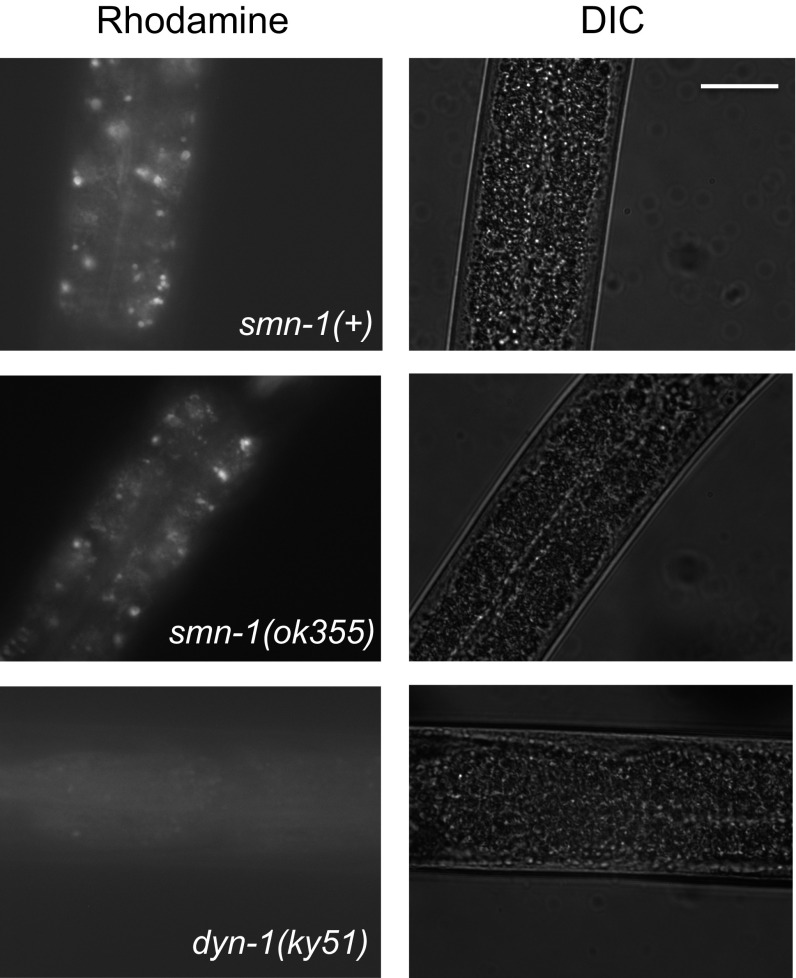
Next, the impact of SMN depletion on late endosomal/lysosomal compartments was examined by assessing multivesicular body (MVB) morphology in TEM sections. MVBs contain internal intraluminal vesicles that can be degraded in the lysosome or transported to the cellular membrane for recycling/secretion. In smn-1(ok355) animals, the average number of MVB intraluminal vesicles per MVB was dramatically decreased compared to smn-1(+) animals (22 ± 4 vs. 6 ± 1, P = 0.002; Fig. 4H), suggesting that diminished SMN-1 impacts either generation or clearance of intraluminal vesicles in this late endosomal compartment. The average diameter of MVBs in smn-1(ok355) animals was unaffected in relation to controls (P = 0.81). Additionally, apical (luminal) endocytosis in the intestine of smn-1(ok355) animals was tested by feeding nematodes rhodamine–dextran. No difference in the endocytosis-dependent accumulation of fluorescence in intestinal cells was observed in smn-1(ok355) vs. smn-1(+) animals (Fig. S7). Overall, these results demonstrate that SMN-1 depletion causes endocytic pathway defects in motor neurons and in other tissues.
Fig. S7.
smn-1(ok355) does not impair uptake of rhodamine dextran via the apical surface of the intestine. Shown are representative images of the intestinal cells of early L4 stage larvae smn-1(ok355) animals fed rhodamine–dextran. No difference was seen in endocytosis-dependent accumulation of rhodamine in intestinal cells. dyn-1(ky51) impairs function of C. elegans dynamin, a GTPase that is critical for endocytosis (97) and serves here as a control. Line indicates 45 microns.
Decreased SMN Impairs Endocytosis-Dependent Viral Infection.
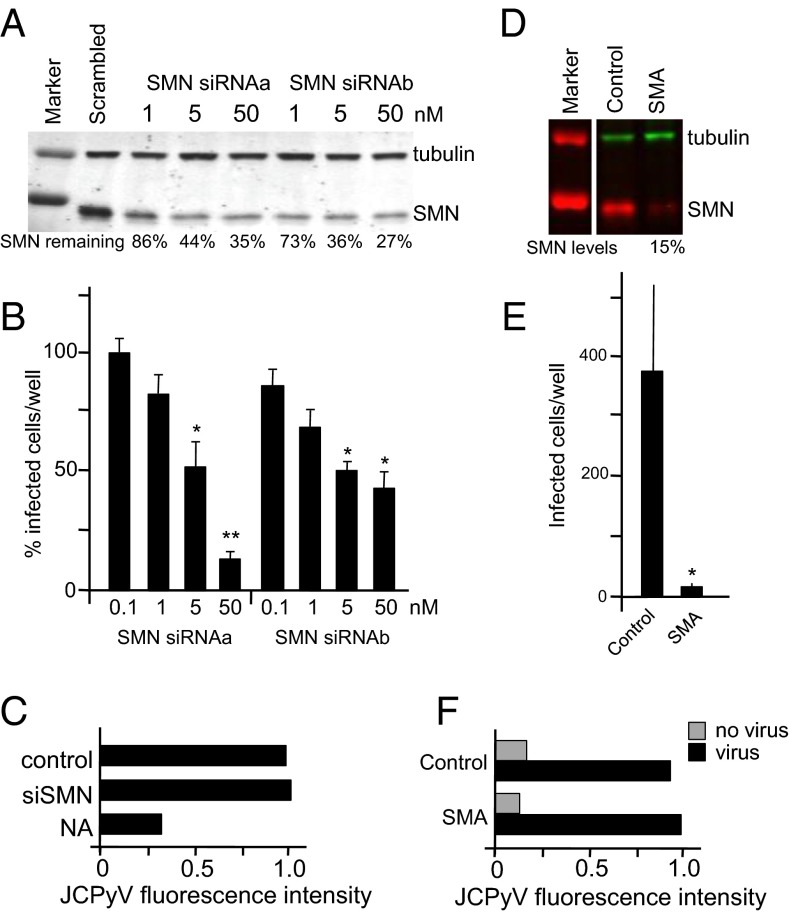
Endocytic trafficking is required for multiple cellular events in both invertebrates and vertebrates, including infection by pathogenic organisms. Many viruses use cellular endocytic pathways to enter cells, and reduction of classical endocytic proteins impairs viral infection in Drosophila (51) and human cells (52–54). To determine whether the defects caused by diminished SMN in C. elegans are conserved in vertebrates, we examined endocytosis-dependent infection of human cells by a virus known to use the endocytic pathway for entry and internalization. JCPyV binds to cell surface receptors and enters cells via CME (52–54). Initially, we tested two different siRNAs and found that both depleted SMN in SVG-A glial cells that support JCPyV infection (Fig. 5A). To determine whether JCPyV infection was reduced by SMN1 knockdown, we compared infection in SMN-depleted cells vs. control scrambled siRNA-treated cells; decreased SMN impaired JCPyV infection (Fig. 5B). To rule out an effect on virus binding before endocytosis, we assessed the binding of fluorescently labeled virus (JCPyV-633) to glial cells treated with SMN-specific or control siRNA by flow cytometry. Binding of JCPyV-633 to glial cells was not affected when SMN levels were decreased by siRNA treatment (Fig. 5C). However, binding was reduced when cells were pretreated with neuraminidase, indicating that JCPyV bound to the appropriate sialic acid receptor (Fig. 5C) (55, 56). To confirm the impact of SMN knockdown on infection, the same experiments were undertaken using commercially available fibroblasts from an unaffected carrier and a type I SMA patient. SMA type I fibroblasts were resistant to infection, whereas virus binding was equivalent to control fibroblasts from a carrier individual with normal SMN levels (Fig. 5 D–F). Collectively, these data suggest that decreased SMN levels result in resistance to JCPyV infection. Overall, SMN depletion may impair infection at the stage of endosomal trafficking and/or at subsequent steps in the infection cycle.
Fig. 5.
Low SMN levels decrease JCPyV infection but not virus binding. (A) SMN siRNA knockdown in SVG-A cells. SVG-A cells reverse-transfected with siRNAs specific for SMN; protein levels were determined by SDS/PAGE and immunoblot analysis using antibodies specific for SMN or tubulin (loading control). Percentage remaining SMN is indicated. (B) JCPyV infection is dependent on SMN. SVG-A cells were reverse-transfected with SMN siRNAs, infected with JCPyV, and quantified based on nuclear VP1 staining. Here, we report the percentage of infected cells, relative to the siRNA scrambled control (n = 3 independent experiments). For each experiment, five fields of view of a 12-well plate were scored for infection, and each was compared with siRNA scrambled control (100%). Error bars indicate SEM; Student’s t test, two-tailed: *P < 0.05; **P < 0.01. (C) JCPyV binding to SVG-A cells is not affected by SMN knockdown. Following siRNA knockdown of SMN, SVG-A cells were either treated or untreated with neuraminidase (NAs). Cells were incubated with JCPyV-633 and analyzed by flow cytometry. Data represent the relative fluorescence intensity of JCPyV-633 binding to cells normalized to control. (D) SMN protein levels in fibroblasts from an individual with SMA and control fibroblasts were determined by SDS/PAGE and immunoblot analysis using antibodies specific for SMN or tubulin (loading control). SMN levels are represented as percentage control SMN. (E) Fibroblasts derived from an SMA patient do not support JCPyV infection. Fibroblast cells were infected with JCPyV and quantified based on nuclear VP1 staining. Bar graphs represent the average number of infected cells per well for three wells and represent data from three independent experiments. Error bars indicate SD; Student’s t test, two-tailed: *P < 0.05. (F) JCPyV binding is not decreased in SMA patient fibroblasts. SMA patient and control fibroblasts were incubated with JCPyV-633 and analyzed by flow cytometry. Relative fluorescence intensity of JCPyV-633 binding to cells normalized to control is indicated.
Discussion
Despite decades of work, we still do not know why decreased SMN levels cause abnormal synaptic organization and defective synaptic neurotransmission in SMA. Here, we report for the first time, to our knowledge, that decreased SMN protein levels impair endocytic pathways, which have not been previously implicated in SMA pathogenesis. The majority of the work presented here is based on a previously defined C. elegans model of SMA in which decreased function of the C. elegans SMN ortholog, SMN-1, results in neuromuscular functional defects, without motor neuron death. We found that the motor neurons of smn-1 loss-of-function animals had reduced numbers of presynaptic docked vesicles, inappropriately high numbers of irregular vesicles called cisternae, aberrant localization of endocytic proteins, and decreased synaptobrevin levels. Additionally, genetic interactions of smn-1 with known components of the SV cycle suggested that endocytosis was impaired. Combined, these results indicate impaired SV recycling and perturbed endocytic pathway function (Fig. 6). We also observed defects in the endosomal compartments of nonneuronal tissues, consistent with the observation that SMA has consequences outside the neuromuscular system. Finally, SMN depletion reduced endocytosis-dependent viral infection in human cells but did not affect virus binding, consistent with results from the C. elegans SMA model. Combined, these results suggest that impaired endocytic trafficking may be a major player in SMA pathology.
Fig. 6.
The motor neurons of smn-1(ok355) animals display endocytic defects. Shown is a schematic summary of puncta and TEM analysis. The number of active zones and DCVs in smn-1(ok355) motor neurons was unchanged, but fewer presynaptic docked vesicles were observed, and more irregular large vesicles, called cisternae, and aberrant localization of both AP2 α-adaptin and Intersectin/DAP160 endocytic proteins were observed in smn-1(ok355) motor neurons.
Depletion of SMN Has Widespread Endocytic Consequences.
Neuromuscular defects are the most obvious hallmarks of SMA, but SMN is ubiquitously expressed and numerous studies have suggested nonneuronal requirements for SMN function in heart (57–59), liver (60), muscle vascular system (61), lung, intestine (62), and pancreatic islets (63). Therefore, it was not surprising that endosomal defects were observed in other C. elegans tissues when SMN levels were compromised. SMN-1 reduction led to impaired endocytic trafficking by coelomocyte cells, which clear the body cavity of small solutes. Also, the number of intraluminal vesicles in hypodermal late endosomes/MVBs was decreased by 72% in smn-1 mutants, suggesting that defects in transmembrane receptor recycling/clearance may occur when SMN levels are diminished. Endosomal pathways play critical roles in protein trafficking and receptor signaling in all tissues. Overall, our results suggest that endosomal defects may contribute to the systemic problems of SMA patients.
SMN-Depleted Cells Are Resistant to Viral Endocytosis-Dependent Infection.
To independently assess the impact of SMN depletion on endocytic pathways, we turned to an established model of endocytosis-dependent viral infection. In Drosophila, heterozygous mutations in genes involved in endocytosis result in resistance to Drosophila C virus infection, consistent with a critical role for endocytosis in infection and pathogenesis (51). Here, we report that SMN depletion results in decreased JCPyV infection both in cells treated with SMN siRNA and in SMA patient-derived cells. There are multiple steps in viral endosomal trafficking and infection that may be impacted by diminished SMN, but viral attachment was not affected. The results presented here are consistent with SMN decrements affecting viral infection that is dependent on endocytosis, but viral infection is complex and further studies will be required to determine how diminished SMN decreases infection. A link between decreased SMN levels and increased resistance to viral endocytosis-dependent infection would be of significant interest and might explain why SMA mutant alleles are common. To our knowledge, a link between SMN levels and infection has not been examined previously. The SMA heterozygosity frequency (1:40–1:60) is similar to the frequency observed for sickle cell anemia and cystic fibrosis alleles, which are known to confer carrier survival benefits in populations at risk (64–66). SMA alleles frequently arise de novo via chromosome rearrangement, and in some cases, the neighboring neuronal apoptosis inhibitory protein (NAIP) gene is deleted (4). Mice lacking the NAIP gene fail to activate a response to Legionella pneumophila infection (67), but results herein suggest a role for SMN should be considered in future studies.
How Could SMN Regulate Endocytic Trafficking?
Two nonexclusive models may explain why diminished SMN causes endocytic pathway defects. In the first model, SMN depletion leads to aberrant trafficking or splicing of mRNAs that encode proteins critical for endocytic function. For example, SMN loss impairs axonal transport of RNP granules containing mRNAs that encode numerous proteins, including annexin 2, annexin 3, and Rab18 (68); their loss may result in endosomal defects. Or more indirectly, loss of SMN may cause missplicing of mRNAs that encode proteins with a role in endocytic trafficking; mRNAs such as annexin 2 are known to be misspliced when SMN levels are depleted (69). In the second model, SMN is part of an RNA/protein complex that promotes endocytic trafficking. SMN directly binds and is transported within axons with the alpha subunit of the coat protein 1 (COPI), a critical player in intracellular vesicular trafficking. COPI can be found on Golgi, ER, and MVB membranes, and COPI loss results in defective endosomal function (70–72). Alternatively, the functional interaction between SMN, Plastin 3, and actin might play a role in the endocytic pathway defects observed when SMN levels are depleted. Plastin 3, an actin-bundling protein, was identified as a gender-protective SMA modifier and was found in a protein complex along with SMN and actin (73). Loss of fimbrin, the yeast PLS3 ortholog, inhibits endocytosis (74), and Plastin 3 interacts with activated Rab5 to facilitate mammalian endocytosis (75). Also, actin filaments can form a critical collar-like structure around the neck of endocytic vesicles as they pull away from the cell membrane (76–78), but it is unclear if SMN or Plastin 3 is associated with these structures. Hence, we consider it possible that SMN and Plastin 3 are essential players of a protein complex required for endocytic trafficking, but additional studies will be needed to determine which of these models best explains the endosomal defects caused by SMN reduction.
Conclusions
The genetic, pharmacological, and functional studies presented here demonstrate that SMN depletion impacts endocytic trafficking. Previous studies connect endocytosis and endosomal trafficking with other neurodegenerative diseases. For example, overexpression of the endosomal protein Rab11 reverses the synaptic transmission and vesicular deficits caused by mutant huntingtin in a Drosophila model of Huntington’s disease (79). Furthermore, CHMP2B mutations cause frontotemporal dementia (FTD) and impair endosome–lysosome fusion (80). Additionally, Farg et al. identified a role in Rab-mediated endosomal trafficking for C9ORF72, a cause of sporadic amyotrophic lateral sclerosis (ALS) (81). Finally, BICD2 mutations are linked to autosomal-dominant SMA (82), and notably loss of the BICD2 Drosophila ortholog impairs CME at presynaptic specializations (83). Identifying the functionally relevant components that connect diminished SMN levels to endocytic pathways could lead to the identification of novel therapeutic interventions for SMA and related neurodegenerative disorders.
Materials and Methods
C. elegans Strains, Constructs, and Transgenes.
Strains listed in Table S1 were maintained at 20 °C under standard conditions (84). For all experiments involving smn-1(ok355) or smn-1(rt248), animals tested were first-generation progeny of parents heterozygous for the hT2 balancer. To maintain a common genetic background, control smn-1(+) animals were similarly derived from +/hT2 parents. Plasmid pHA#582 contains a 1,819-bp fragment corresponding to the smn-1 promoter, coding sequence, and 3′ untranslated region subcloned as an AflII/XhoI product into pCFJ356 (Addgene plasmid 34871) (85). Primers for amplification were as follows: 5-tgatcttaagtctacgagcgacattcatcg and 5-tgatctcgagcagcctctcatcctgattgc. rtSi9[Cb-unc-119(+)]IV and rtSi10[smn-1p::smn-1;Cb-unc-119(+)]IV transgenes were generated by Mos1-mediated single-copy insertion (85, 86). We injected 50 ng/µL of targeting plasmid (pCFJ356 or pHA#582) into EG6703 [unc-119(ed3)III;cxTi10816 IV] animals along with 50 ng/µL pCFJ601 (eft-3p::Mos1 transposase), 10 ng/µL pGH8 (rab-3p::mCherry), 5 ng/µL pCFJ104 (myo-3p::mCherry), and 2.5 ng/µL pCFJ90 (myo-2p::mCherry). Insertion events were identified based on rescue of unc-119 in nonfluorescent animals and confirmed by PCR genotyping, before crossing into smn-1(ok355)/hT2. In Fig. 1 A, B, and D and Fig. 3 G, H, and I, smn-1(ok355);[smn-1(+)] animals carry rtSi10. For these same figures, smn-1(+) and/or smn-1(ok355) animals carry rtSi9, which differs from rtSi10 only in the absence of an smn-1(+) gene copy. The small guide RNA (sgRNA) plasmid targeting the smn-1 gene (pHA#730) for CRISPR/Cas9-mediated genome editing was generated by amplification of PU6::klp-12 (35) and subsequent ligation of the PCR product obtained by the following primers: 5′-AACATCGTCTAAACATTTAGATTTGCAATTCAATTATATAGGGACC-3′ and 5′-TGGGATGATAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3′. The resulting plasmid was injected at 50 ng/μL into wild-type animals following the dpy-10(cn64) coconversion protocol from Arribere et al. (34). After backcross, the resulting smn-1(rt248) allele was balanced over the hT2 chromosomal translocation. smn-1(rt248) creates D19fs and likely eliminates SMN-1 function.
Table S1.
A list of the strains used for this work (related to Materials and Methods)
| Strain | Genotype |
| LM99 | smn-1(ok355)I/hT2(I;III) |
| HA1981 | +/hT2(I;III) |
| DA592 | unc-57(ad592)I |
| CB211 | lev-1(e112)IV |
| HA2251 | smn-1(ok355) unc-57(ok310)I/hT2(I;III) |
| HA2252 | unc-57(ok310)/hT2(I;III) |
| HA1979 | smn-1(ok355)I/hT2(I;III);unc-26(s1710)IV |
| HA1978 | +/hT2(I;III);unc-26(s1710)IV |
| HA2272 | smn-1(ok355)I/hT2(I;III);sdpn-1(ok1667)X |
| HA2271 | +/hT2(I;III);sdpn-1(ok1667)X |
| HA2164 | smn-1(ok355)I/hT2(I;III);unc-18(e234)X |
| HA2165 | +/hT2(I;III);unc-18(e234)X |
| HA2660 | smn-1(ok355)I/hT2(I;III);snb-1(md247)V |
| HA2661 | +/hT2(I;III);snb-1(md247)V |
| HA2131 | smn-1(ok355)I/hT2(I;III);nuIs160[unc-129p::GFP::syd-2]III |
| HA2130 | +/hT2(I;III);nuIs160[unc-129p::GFP::syd-2]III |
| HA2166 | smn-1(ok355)I/hT2(I;III);nuIs183[unc-129p::venus::nlp-21]III |
| HA2167 | +/hT2(I;III);nuIs183[unc-129p::venus::nlp-21]III |
| HA2132 | smn-1(ok355)I/hT2(I;III);nuIs152[unc-129p::GFP::snb-1]II |
| HA2129 | +/hT2(I;III);nuIs152[unc-129p::GFP::snb-1]II |
| HA2491 | +/hT2 (I;III); nuIs214[unc-129p::itsn-1::GFP]III |
| HA2490 | smn-1(ok355)I/hT2(I;III);nuIs214[unc-129p::itsn-1::GFP]III |
| HA2266 | smn-1(ok355)I/hT2(I;III);nuIs184[unc-129p::apt-4::GFP]X |
| HA2265 | +/hT2(I;III);nuIs184[unc-129p::apt-4::GFP]X |
| HA2451 | +/hT2(I;III);rtSi9[Cb-unc-119(+)]IV |
| HA2452 | smn-1(ok355)I/hT2(I;III);rtSi9[Cb-unc-119(+)]IV |
| HA2454 | smn-1(ok355)I/hT2(I;III);rtSi10[smn-1p::smn-1;Cb-unc-119(+)]IV |
| HA2216 | smn-1(ok355)arIs37[myo-3p::ssGFP,dpy-20(+)]I/hT2(I;III) |
| HA2209 | arIs37[myo-3p::ssGFP,dpy-20(+)]I/hT2(I;III) |
| HA2492 | smn-1(ok355)/hT2(I:III);rtSi9[Cb-unc-119(+)]IV;nuIs184[unc-129p::apt-4::GFP]X |
| HA2493 | smn-1(ok355)/hT2(I:III);rtSi10[smn-1p::smn-1;Cb-unc-119(+)]IV;nuIs184[unc-129p::apt-4::GFP]X |
| HA2494 | +/hT2(I;III);rtSi9[Cb-unc-119(+)]IV;nuIs214[unc-129p::itsn-1::GFP]III |
| XE1581 | wpSi10[unc-17p::rde-1::SL2::sid-1+Cb-unc-119(+)]II;eri-1(mg366)IV;rde-1(ne219)V;lin-15B(n744)X |
| KP3948 | eri-1(mg366)IV;lin-15B(n744)X |
| HA2822 | smn-1(rt248)/hT2(I:III);nuIs214[unc-129p::itsn-1::GFP]III |
| HA2824 | smn-1(rt248)/hT2(I:III) |
| HA2823 | smn-1(rt248)/hT2(I:III);nuIs175[myo-2:RFP;unc-129p::RFP::snb-1] |
| HA2826 | smn-1(ok355)I/hT2(I;III);nuIs175[myo-2:RFP;unc-129p::RFP::snb-1] |
| HA2827 | +/hT2(I;III);nuIs175[myo-2:RFP;unc-129p::RFP::snb-1] |
| GS1912 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV |
| HA2748 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV;rtEX845[unc-122p::smn-1RNAi+coel::RFP] |
| HA2749 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV;rtEX846[unc-122p::smn-1RNAi+coel::RFP] |
| HA2750 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV;rtEX847[unc-122p::smn-1RNAi+coel::RFP] |
| HA2751 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV;rtEX848[unc-122p::empty+coel::RFP] |
| HA2752 | arIs37[myo-3p::ssGFP+dpy-20(+)]I;dpy-20(e1282)IV;rtEX849[unc-122p::empty+coel::RFP] |
C. elegans Behavioral Assays.
Pharyngeal pumping assays were performed in the last larval stage as previously described (32). Grinder movement in any axis was scored as a pumping event. Average pumping rates (±SEM) were combined from at least three independent trials (n > 25 animals in total/genotype). Aldicarb and levamisole assays on early L4 stage larval animals were carried out blinded as to genotype in at least three independent trials (n ≥ 30 animals in total/genotype) as described elsewhere (38, 87). Drug-induced paralysis caused by 1 mM aldicarb (Sigma) or 0.4 mM levamisole (Sigma) was scored as inability to move/pump in response to prodding with a metal wire.
C. elegans Light Level Microscopy.
Early L4 stage larval animals were mounted on 2% (vol/vol) agar pads and immobilized using 30 mg/mL 2-3-butanedione monoxime (BDM) (Sigma) in M9 buffer. Images were captured as Z-stacks from the dorsal cord above the posterior gonad reflex (100× objective, Zeiss AxioImager ApoTome and AxioVision software v4.8). At least three independent trials (n > 25 animals in total/genotype) were undertaken. Puncta total intensity, width, and linear density were quantified using the Punctaanalyser program in Matlab (88). Kernel density estimation of the puncta population was determined in R (v3.0.3). Invariant fluorescent illumination was confirmed daily using 0.5 μm fluorescent beads (FluoSpheres, Molecular Probes) (46). Coelomocyte imaging was undertaken blinded as to genotype in three independent trials (n > 25 animals in total/genotype). GFP levels in the six coelomocytes of early L4 stage animals were assessed using a Zeiss V20 stereoscope (50).
C. elegans TEM.
Animals were prepared in parallel for TEM as described (89). Briefly, early L4 nematodes were fixed by a high-pressure freezing apparatus followed by a 2% (vol/vol) osmium in acetone freeze substitution. Ultrathin serial sections (50–60 nm thickness) were collected on Formvar/Pioloform-coated copper slot grids, stained with 4% (vol/vol) uranyl acetate in 70% (vol/vol) methanol, followed by washing and lead citrate incubation. Images were obtained on a Philips CM10 transmission electron microscope using an Olympus Morada camera system driven by the iTEM software (Olympus Soft Imaging Solutions). Image registration and annotation were performed using TrakEM2 (90). We imaged 300 and 500 serial sections for control and smn-1(ok355) animals, respectively. The anterior ventral nerve cord was reconstructed from one animal for each genotype. Synapses were examined from VA and VB cholinergic neurons and the VD ɣ-aminobutyric acid (GABA) neuron (47). Thirty-five control (27 cholinergic and 8 GABAergic) and 45 smn-1(ok355) (25 ACh and 20 GABAergic) neuromuscular synapses were examined. The ratio of GABAergic to cholinergic (ACh) synapses was not significantly different from the control, suggesting that ACh synapses are not preferentially lost (χ2 test, P > 0.05). A synapse was defined as a set of serial sections containing a dense projection. Docked vesicles were defined as those contacting the plasma membrane adjacent to a dense projection. The number of SVs (∼30 nm diameter) and DCVs (∼40 nm diameter) were counted in sections containing a dense projection, and the numbers of each profile were averaged to obtain the final value. The presence of large clear vesicles/cisternae (>40 nm diameter) was analyzed by counting every other serial section within 1 μm to either side of a dense projection. For MVBs, the intraluminal vesicles were counted for >30 cell profiles per genotype.
Cells and Viruses.
Cells were grown at 37 °C in a humidified incubator with 5% (vol/vol) CO2. SVG-A cells are a subclone of the human glial cell line SVG transformed with an origin-defective SV40 mutant and grown in minimum essential medium (MEM) supplemented with 10% (vol/vol) FBS and 1% penicillin–streptomycin (Mediatech, Inc.) (91). Untransformed primary fibroblasts were from a patient with SMA type I (GM09677) and control cells from a disease-free SMA carrier (GM03814) (Coriell Cell Repositories). Fibroblasts were grown in MEM supplemented with 10% (vol/vol) FBS and nonessential amino acids. Generation and propagation of the virus strain Mad-1/SVEΔ were previously described (92).
Transfection.
Cells were reverse-transfected with SMN-specific siRNAs using Lipofectamine RNAiMax (Life Technologies). SMN siRNAs [SMN siRNAa (Hs_SMN1_11), ACGGTTGCATTTACCCAGCTA (cat. no. SI04950932), and SMN siRNAb (Hs-SMN1_12), ATCAGATAACATCAAGCCCAA (cat. no. SI04950939)] from Qiagen were prepared according to the manufacturer’s instructions. Serum-free medium was added to triplicate wells of a 12-well plate, and siRNAs were diluted to a final concentration of 0.1, 1, 5, and 50 nM. Two microliters of RNAiMax Lipofectamine were added, and solutions were mixed and incubated at room temperature (RT) for 20 min. Following incubation, 2 × 105 cells in 1 mL of media containing FBS were added to each well. Cells were incubated at 37 °C for 36 h and then infected or harvested for protein quantitation by immunoblot analysis. Infection data were compared with cells transfected with Allstars siRNA negative control (Qiagen).
Infection.
Patient fibroblasts were plated to 60% (vol/vol) confluency in 12-well plates overnight (O/N). Cells from siRNA transfections were infected at 36 h posttransfection. Media was aspirated and cells were infected with a multiplicity of infection (MOI) of 5 (JCPyV) fluorescent focus units (FFUs) per cell in MEM containing 2% (vol/vol) FBS at 37 °C for 1.5 h. Infected cells were then fed with 2 mL of appropriate media and incubated at 37 °C for 72 h. Cells were washed in 1× PBS, fixed in cold methanol, and incubated at −20 °C. Fixed cells were washed in PBS; permeabilized with 0.5% Triton X-100 (USB Corporation) at RT for 5 min; incubated with PAB597, a hybridoma supernatant that produces a monoclonal antibody against JCPyV VP1 (generously provided by Ed Harlow, Harvard Medical School, Boston) (93), at a 1:10 dilution in PBS at 37 °C for 1 h; washed with PBS; incubated with a goat anti-mouse Alexa Fluor 488-conjugated antibody (1:1,000; 20 µg/mL) (Life Technologies) in PBS at 37 °C for 1 h; and washed again in PBS. Cells were analyzed for VP1 staining in the nucleus under a 20× objective using an Eclipse TE2000-U microscope (Nikon).
Immunoblot.
Cell lysates were prepared by washing cells in 200 µL PBS and scraping and collecting the lysates. Cells were centrifuged at 855 × g for 5 min and pellets were resuspended in 1× RIPA buffer with protease (1:10) and phosphatase (1:100) inhibitors (Sigma) and incubated on ice for 30 min. Cells were pelleted at 18,600 × g, and supernatants were diluted at a 1:1 ratio in SDS loading buffer, boiled at 95 °C for 5 min, and 15 µL was resolved by SDS/PAGE using a 4–15% (vol/vol) Tris·HCl gel (BioRad). Gels were transferred to PVDF membranes (BioRad) using a Transblot system (BioRad) at 10 V for 30 min. Membranes were blocked in 2% (vol/vol) milk in PBS with 0.1% Tween 20% (vol/vol) (PBS-T) O/N, then incubated with a purified mouse anti-SMN primary antibody at 1:5,000 dilution (BD Biosciences; 610647) and an anti-alpha tubulin polyclonal antibody loading control at 1:450 dilution (Abcam; ab4074) at RT for 1 h, washed in PBS-T, and then incubated with a 680 nM goat–anti-mouse secondary antibody at 1:1,000 dilution (Life Technologies) and a 800 nM anti-rabbit secondary antibody at 1:5,000 dilution (LI-COR Biosciences) at RT for 1 h. All antibodies were diluted in 2% (vol/vol) milk. Immunoblots were scanned and analyzed using an Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
Flow Cytometry.
All cells (SVG-A treated with SMN or control siRNAs or fibroblasts) (1 × 106) were washed with PBS, incubated with cell stripper (Cellgro), and removed from plates. Cells were pelleted, washed, and incubated in 100 μL of PBS with 2.5 μg of JCPyV labeled with Alexa Fluor 633 (JCPyV-633) or PBS alone for 2 h on ice. For neuraminidase treatment, cells were first incubated for 45 min at 37 °C in the presence of 5 U/mL type V neuraminidase from Clostridium perfringens (Sigma) or PBS control. Cells were pelleted and washed with PBS twice, resuspended in 1× PBS, and analyzed for virus binding using a BD FACSCalibur equipped with a 633 nm laser line (BD Bioscience). Data were analyzed using FlowJo software (Tree Star, Inc.).
Statistical Analysis.
Two-tailed Mann–Whitney U or log-rank test was used for C. elegans statistical analysis. For JCPyV infection, P values were determined using an unpaired Student’s t test (two-tailed distribution).
SI Text
C. elegans Constructs and Transgenes.
A 797-bp smn-1 genomic fragment was amplified from plasmid pHA#728 and subcloned as an AgeI/NotI product into plasmid coel::RFP (Addgene #8938) to create unc-122p::smn-1(RNAi). Primers for amplification used were as follows: 5′-tgataccggttgatccgctagtgcttgatagtatcc-3′ and 5′-tgatcgcggccgcatggcaaaaatctggtcgaaaagtg-3′. The digested coel::RFP plasmid recircularized after Klenow filling to create the control RNAi plasmid unc-122p::empty (pHA#729). Plasmids were injected into GS1912, separately, at 50 ng/μL alongside coel::RFP also at 50 ng/μL. Table S1 lists the transgenic lines tested (i.e., HA2748, HA2749, HA2750, HA2751, and HA2752).
RNAi Studies.
RNAi studies were performed in an RNAi-enhanced background (KP3948) (94) or cholinergic neuron-specific RNAi-sensitized background (XE1581) (39). Pumping was scored in second-generation animals reared on HT115 bacteria containing control vector L4440, C41G7.1/smn-1(RNAi), or C26C6.2/goa-1(RNAi) (95). The smn-1 RNAi clone contains genomic DNA amplified by primers 5′-GGAGGAATGGAGTGAGAAAACTT-3′ and 5′-GACCAGATTTTTGCCATATTCTG-3′ inserted into L4440 (95). The goa-1 RNAi clone contains genomic DNA amplified by primers 5′-TGATGTACATGACATGCTGTGAA-3′ and 5′-GGAAAAAGTTGCAATTTCAGATG-3′. The clones were verified by sequencing.
Endocytosis Assay.
To test endocytosis by intestinal cell apical membranes, early L4 stage larval smn-1(+), smn-1(ok355), and dyn-1(ky51) animals were placed on nematode growth medium agar plates (+OP50) containing rhodamine–dextran (0.1 mg/mL) (50). Uptake of rhodamine–dextran by feeding was examined in the intestinal cells of early L4 stage F1 progeny. Intestinal imaging was undertaken blinded as to genotype in three independent trials (n > 25 animals in total/genotype).
C. elegans Coelomocyte Imaging.
Accumulation of GFP in the body cavity was examined at 5× magnification using a Zeiss V20 stereoscope. Adult animals were classified as strong–intense GFP, body cavity filled (Fig. S5A, Lower Middle), some–weak GFP, faint in body cavity, or none–no GFP visible in body cavity (Fig. S5A, Top Middle). Accumulation indicates coelomocyte endocytic pathway defects caused by cell-autonomous, cell-specific smn-1(RNAi) (50). The observer was blinded as to RNAi treatment in three independent trials with similar results (n = 40 animals in total/genotype).
Supplementary Material
Acknowledgments
We thank the Kaplan, Jorgensen, and Zhen laboratories for advice on C. elegans fluorescent imaging and quantitation as well as Dr. M. McKeown for helpful discussions. The C. elegans Gene Knockout Consortium (NIH/National Human Genome Research Institute) and the National BioResource Project provided C. elegans strains. Additional strains were provided by the Caenorhabditis elegans consortium (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the SMA Foundation and NIH NINDS Grant NS066888 (to A.C.H.), NIH Grant OD010943 (to D.H.H.), NIH NINDS Grant F31NS089201 (to P.O.), and Institutional Development Award (IDeA) P20GM103423 from the National Institute of General Medical Sciences of the National Institutes of Health (to M.S.M.). Research in the W.J.A. laboratory is funded by Grants P01NS065719 and R01NS043097 (to W.J.A.) and Ruth L. Kirschstein National Research Service Award F32NS064870 (to M.S.M.) from the National Institute of Neurological Disorders and Stroke. Core facilities for the W.J.A. laboratory are supported by Grant P30GM103410 (to W.J.A.) from the National Institute of General Medical Sciences. Core facilities for electron microscopy at the D.H.H. laboratory are supported by NICHD Grant P30 HD71593 for the RFK-IDDRC at Albert Einstein College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600015113/-/DCSupplemental.
References
- 1.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3(2):97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Cusin V, Clermont O, Gérard B, Chantereau D, Elion J. Prevalence of SMN1 deletion and duplication in carrier and normal populations: Implication for genetic counselling. J Med Genet. 2003;40(4):e39. doi: 10.1136/jmg.40.4.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet. 1978;15(6):409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wirth B, et al. De novo rearrangements found in 2% of index patients with spinal muscular atrophy: Mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet. 1997;61(5):1102–1111. doi: 10.1086/301608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 8.Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90(6):1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90(6):1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 12.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95(5):615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 13.Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572(1-3):69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Zou T, et al. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol. 2011;31(4):541–550. doi: 10.1007/s10571-011-9647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akten B, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA. 2011;108(25):10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagliardini S, et al. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum Mol Genet. 2000;9(1):47–56. doi: 10.1093/hmg/9.1.47. [DOI] [PubMed] [Google Scholar]
- 18.Rossoll W, et al. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163(4):801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd AG, et al. SMN, Gemin2 and Gemin3 associate with beta-actin mRNA in the cytoplasm of neuronal cells in vitro. J Mol Biol. 2010;401(5):681–689. doi: 10.1016/j.jmb.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci. 2006;26(33):8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152(1):75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning K, et al. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum Mol Genet. 2010;19(16):3159–3168. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez G, et al. A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet. 2013;22(4):668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 24.Grice SJ, Liu JL. Survival motor neuron protein regulates stem cell division, proliferation, and differentiation in Drosophila. PLoS Genet. 2011;7(4):e1002030. doi: 10.1371/journal.pgen.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kariya S, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(16):2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 27.Lee YI, Mikesh M, Smith I, Rimer M, Thompson W. Muscles in a mouse model of spinal muscular atrophy show profound defects in neuromuscular development even in the absence of failure in neuromuscular transmission or loss of motor neurons. Dev Biol. 2011;356(2):432–444. doi: 10.1016/j.ydbio.2011.05.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong L, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres-Benito L, Neher MF, Cano R, Ruiz R, Tabares L. SMN requirement for synaptic vesicle, active zone and microtubule postnatal organization in motor nerve terminals. PLoS One. 2011;6(10):e26164. doi: 10.1371/journal.pone.0026164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Hernández R, et al. Synaptic defects in type I spinal muscular atrophy in human development. J Pathol. 2013;229(1):49–61. doi: 10.1002/path.4080. [DOI] [PubMed] [Google Scholar]
- 31.Briese M, et al. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18(1):97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimitriadi M, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6(10):e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133(4):897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arribere JA, et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198(3):837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miguel-Aliaga I, et al. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum Mol Genet. 1999;8(12):2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- 37.Monani UR, De Vivo DC. Neurodegeneration in spinal muscular atrophy: From disease phenotype and animal models to therapeutic strategies and beyond. Future Neurol. 2014;9(1):49–65. doi: 10.2217/fnl.13.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahoney TR, Luo S, Nonet ML. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat Protoc. 2006;1(4):1772–1777. doi: 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 39.Firnhaber C, Hammarlund M. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 2013;9(11):e1003921. doi: 10.1371/journal.pgen.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleigh JN, et al. A novel Caenorhabditis elegans allele, smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum Mol Genet. 2011;20(2):245–260. doi: 10.1093/hmg/ddq459. [DOI] [PubMed] [Google Scholar]
- 41.Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95(4):905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller KG, et al. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93(22):12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuske KR, et al. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 2003;40(4):749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 44.Clayton EL, Cousin MA. The molecular physiology of activity-dependent bulk endocytosis of synaptic vesicles. J Neurochem. 2009;111(4):901–914. doi: 10.1111/j.1471-4159.2009.06384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ch’ng Q, Sieburth D, Kaplan JM. Profiling synaptic proteins identifies regulators of insulin secretion and lifespan. PLoS Genet. 2008;4(11):e1000283. doi: 10.1371/journal.pgen.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436(7050):510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 47.White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275(938):327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, et al. The C2H2 zinc-finger protein SYD-9 is a putative posttranscriptional regulator for synaptic transmission. Proc Natl Acad Sci USA. 2006;103(27):10450–10455. doi: 10.1073/pnas.0602073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant B, et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3(6):573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 50.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: Coelomocyte uptake defective mutants. Genetics. 2001;159(1):133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5(1):81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engel S, et al. Role of endosomes in simian virus 40 entry and infection. J Virol. 2011;85(9):4198–4211. doi: 10.1128/JVI.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pho MT, Ashok A, Atwood WJ. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J Virol. 2000;74(5):2288–2292. doi: 10.1128/jvi.74.5.2288-2292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Querbes W, O’Hara BA, Williams G, Atwood WJ. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol. 2006;80(19):9402–9413. doi: 10.1128/JVI.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dugan AS, Gasparovic ML, Atwood WJ. Direct correlation between sialic acid binding and infection of cells by two human polyomaviruses (JC virus and BK virus) J Virol. 2008;82(5):2560–2564. doi: 10.1128/JVI.02123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neu U, et al. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8(4):309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bevan AK, et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet. 2010;19(20):3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heier CR, Satta R, Lutz C, DiDonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19(20):3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shababi M, et al. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19(20):4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- 60.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somers E, Stencel Z, Wishart TM, Gillingwater TH, Parson SH. Density, calibre and ramification of muscle capillaries are altered in a mouse model of severe spinal muscular atrophy. Neuromuscul Disord. 2012;22(5):435–442. doi: 10.1016/j.nmd.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 62.Schreml J, et al. Severe SMA mice show organ impairment that cannot be rescued by therapy with the HDACi JNJ-26481585. Eur J Hum Genet. 2013;21(6):643–652. doi: 10.1038/ejhg.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bowerman M, et al. Defects in pancreatic development and glucose metabolism in SMN-depleted mice independent of canonical spinal muscular atrophy neuromuscular pathology. Hum Mol Genet. 2014;23(13):3432–3444. doi: 10.1093/hmg/ddu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. BMJ. 1954;1(4857):290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allison AC. Malaria in carriers of the sickle-cell trait and in newborn children. Exp Parasitol. 1957;6(4):418–447. doi: 10.1016/0014-4894(57)90032-2. [DOI] [PubMed] [Google Scholar]
- 66.Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ. Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science. 1994;266(5182):107–109. doi: 10.1126/science.7524148. [DOI] [PubMed] [Google Scholar]
- 67.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9(10):1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rage F, et al. Genome-wide identification of mRNAs associated with the protein SMN whose depletion decreases their axonal localization. RNA. 2013;19(12):1755–1766. doi: 10.1261/rna.040204.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133(4):585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peter CJ, et al. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet. 2011;20(9):1701–1711. doi: 10.1093/hmg/ddr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185(2):305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ting CH, et al. The spinal muscular atrophy disease protein SMN is linked to the Golgi network. PLoS One. 2012;7(12):e51826. doi: 10.1371/journal.pone.0051826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oprea GE, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320(5875):524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kübler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12(7):2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagiwara M, et al. Interaction of activated Rab5 with actin-bundling proteins, L- and T-plastin and its relevance to endocytic functions in mammalian cells. Biochem Biophys Res Commun. 2011;407(3):615–619. doi: 10.1016/j.bbrc.2011.03.082. [DOI] [PubMed] [Google Scholar]
- 76.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13(9):1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Collins A, Warrington A, Taylor KA, Svitkina T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 2011;21(14):1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sirotkin V. Cell biology: Actin keeps endocytosis on a short leash. Curr Biol. 2011;21(14):R552–R554. doi: 10.1016/j.cub.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 79.Giorgini F, Steinert JR. Rab11 as a modulator of synaptic transmission. Commun Integr Biol. 2013;6(6):e26807. doi: 10.4161/cib.26807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urwin H, et al. FReJA Consortium Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19(11):2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farg MA, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neveling K, et al. Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am J Hum Genet. 2013;92(6):946–954. doi: 10.1016/j.ajhg.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, et al. Bicaudal-D binds clathrin heavy chain to promote its transport and augments synaptic vesicle recycling. EMBO J. 2010;29(5):992–1006. doi: 10.1038/emboj.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9(2):117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frøkjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40(11):1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato K, et al. Differential requirements for clathrin in receptor-mediated endocytosis and maintenance of synaptic vesicle pools. Proc Natl Acad Sci USA. 2009;106(4):1139–1144. doi: 10.1073/pnas.0809541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim JS, et al. A chemical-genetic strategy reveals distinct temporal requirements for SAD-1 kinase in neuronal polarization and synapse formation. Neural Dev. 2008;3:23. doi: 10.1186/1749-8104-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hall DH, Hartwieg E, Nguyen KC. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- 90.Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7(6):e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Major EO, et al. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA. 1985;82(4):1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vacante DA, Traub R, Major EO. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology. 1989;170(2):353–361. doi: 10.1016/0042-6822(89)90425-x. [DOI] [PubMed] [Google Scholar]
- 93.Atwood WJ, et al. Evaluation of the role of cytokine activation in the multiplication of JC virus (JCV) in human fetal glial cells. J Neurovirol. 1995;1(1):40–49. doi: 10.3109/13550289509111009. [DOI] [PubMed] [Google Scholar]
- 94.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427(6975):645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 95.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 96.van Swinderen B, et al. Goalpha regulates volatile anesthetic action in Caenorhabditis elegans. Genetics. 2001;158(2):643–655. doi: 10.1093/genetics/158.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clark SG, Shurland DL, Meyerowitz EM, Bargmann CI, van der Bliek AM. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc Natl Acad Sci USA. 1997;94(19):10438–10443. doi: 10.1073/pnas.94.19.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.