Significance
[FeFe]-hydrogenases are H2-forming enzymes with potential in renewable energy applications. Their molecular mechanism of catalysis needs to be understood. A protocol for specific 13CO isotope editing of all carbon monoxide ligands at the six-iron cofactor (H-cluster) was established. Analysis of vibrational modes via quantum chemical calculations implies structural dynamics at the H-cluster in the active-ready state. Site-selective introduction of isotopic reporter groups opens new perspectives to identify intermediates in the catalytic cycle.
Keywords: [FeFe]-hydrogenase, isotope editing, infrared spectroscopy, density functional theory, cofactor dynamics
Abstract
The six-iron cofactor of [FeFe]-hydrogenases (H-cluster) is the most efficient H2-forming catalyst in nature. It comprises a diiron active site with three carbon monoxide (CO) and two cyanide (CN−) ligands in the active oxidized state (Hox) and one additional CO ligand in the inhibited state (Hox-CO). The diatomic ligands are sensitive reporter groups for structural changes of the cofactor. Their vibrational dynamics were monitored by real-time attenuated total reflection Fourier-transform infrared spectroscopy. Combination of 13CO gas exposure, blue or red light irradiation, and controlled hydration of three different [FeFe]-hydrogenase proteins produced 8 Hox and 16 Hox-CO species with all possible isotopic exchange patterns. Extensive density functional theory calculations revealed the vibrational mode couplings of the carbonyl ligands and uniquely assigned each infrared spectrum to a specific labeling pattern. For Hox-CO, agreement between experimental and calculated infrared frequencies improved by up to one order of magnitude for an apical CN− at the distal iron ion of the cofactor as opposed to an apical CO. For Hox, two equally probable isomers with partially rotated ligands were suggested. Interconversion between these structures implies dynamic ligand reorientation at the H-cluster. Our experimental protocol for site-selective 13CO isotope editing combined with computational species assignment opens new perspectives for characterization of functional intermediates in the catalytic cycle.
The reduction of protons to form molecular hydrogen (H2) is catalyzed by [FeFe]-hydrogenases (1, 2). With a turnover rate of up to 10,000 H2 molecules per second in a thermodynamically reversible reaction (3–5), [FeFe]-hydrogenases inspired synthetic hydrogen catalysts (6–8) and renewable fuel technology applications (9, 10). The mechanism of catalysis at the active site cofactor (H-cluster) needs to be elucidated. Further information on functional intermediates is required (11–16) and expected to emerge from spectroscopic studies on H-cluster constructs carrying site-selective isotopic reporter groups (17–20).
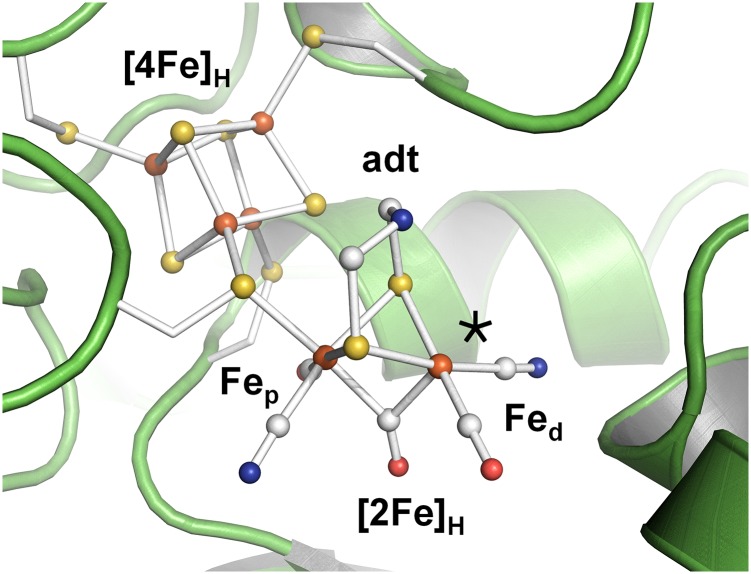
Protein crystallography has identified the H-cluster as a six-iron complex (21–23), in which a canonical cubane cluster ([4Fe4S]H) is linked to a unique diiron moiety ([2Fe]H) (Fig. 1). The two iron ions of [2Fe]H are located in proximal (p) or distal (d) position relative to [4Fe4S]H and carry a bridging amine-dithiolate group [adt; (SCH2)2NH] (19). Both iron ions bind a terminal carbonyl (CO) and a cyanide (CN−) ligand. In crystal structures, the “active-ready,” oxidized state (Hox) of the H-cluster shows a third carbonyl in Fe-Fe bridging position (µCO) and an apical vacancy at Fed (23). On exposure to CO gas, a fourth carbonyl binds at [2Fe]H (24–26) and was modeled in apical position at Fed in Hox-CO (27). Formation of Hox-CO does not affect the formal redox state of the H-cluster, but leads to increased spin delocalization over the diiron site (28). CO binding inhibits H2 turnover and protects the enzyme against O2 and light-induced degradation (24, 29, 30).
Fig. 1.
Crystal structure of [FeFe]-hydrogenase from C. pasteurianum (23). The H-cluster (ball-and-stick) with its cubane ([4Fe4S]H) and diiron subcomplexes ([2Fe]H with an amine-dithiolate = adt bridge) is protein-bound by four cysteine residues. An apical vacant site (*) at Fed was modeled in structures of oxidized enzymes (21–23, 46). The shown CO/CN− ligand orientation herein is annotated standard.
The vibrational modes of the CO and CN− ligands at the diiron site are well accessible by infrared (IR) spectroscopy because they are separated from protein backbone and liquid water bands. Infrared spectroscopy therefore has pioneered elucidation of the molecular structure of the H-cluster and identification of several redox states (24, 31). In particular, the CO stretching frequencies are highly sensitive to structural isomerism, redox transitions, ligand binding, and isotope exchange (11, 12, 15, 18, 24, 31, 32). 13CO editing of the H-cluster has been achieved using 13C-precursors during H-cluster assembly or exposure of [FeFe]-hydrogenases to 13CO gas (18, 24–26, 33). These experiments have yielded either a completely labeled H-cluster, mixtures of labeled species, and mostly the inhibited state. Selective 13CO editing of Hox was hampered by tight binding of exogenous CO, which impaired quantitative regeneration of active enzyme (29, 34). Hox is believed to be the starting state in the H2 conversion cycle of [FeFe]-hydrogenases (1). Selective 13CO editing of Hox thus may provide access to key catalytic H-cluster intermediates (14). Introduction of 13CO groups also facilitates analysis of structure–function relationships using quantum chemical calculations. However, relatively few computational studies to calculate vibrational modes of the diatomic ligands have been carried out (35–39).
We compared three different [FeFe]-hydrogenase proteins, HYDA1, from the green alga Chlamydomonas reinhardtii, and the bacterial enzymes CPI from Clostridium pasteurianum and DDH from Desulfovibrio desulfuricans. HYDA1 represents the “minimal unit” of biological hydrogen turnover as it exclusively binds the H-cluster, whereas CPI and DDH hold accessory iron-sulfur clusters (3, 40). Purified HYDA1 and CPI were reconstituted in vitro with a synthetic diiron site analog to yield the active H-cluster (35, 41, 42), whereas DDH was isolated with a complete cofactor (43). We report the generation of Hox-CO and Hox isotopic species with all possible labeling patterns upon exposure of [FeFe]-hydrogenase protein films to 13CO gas, visible light, and different levels of humidity as monitored by real-time attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR). Density functional theory (DFT) assigned the carbonyl vibrational modes. This approach has established a reaction scheme with 16 options to convert selectively labeled Hox-CO into 8 Hox isotopic species as entry points to the catalytic cycle.
Results
[FeFe]-hydrogenase protein films deposited on an ATR cell were exposed to 12CO, 13CO, or N2 gas with controlled humidity either in darkness or under red or blue light irradiation, exploring the differential wavelength sensitivity of the iron-carbonyl bonds (44). Real-time detection of spectral changes of the stretching vibrations of the diatomic ligands (SI Appendix, Figs. S1–S4) yielded high-quality IR spectra of the thereby derived pure Hox and Hox-CO states (Fig. 2). Frequencies and intensities of IR bands were determined using least-squares fitting. Density functional theory calculations generated geometry-optimized models of the whole H-cluster for Hox-CO and Hox (SI Appendix, Fig. S5). Calculated IR spectra were used for assignment of experimental vibrational bands to individual CO ligands, specific isotopic labeling patterns, and molecular structures.
Fig. 2.
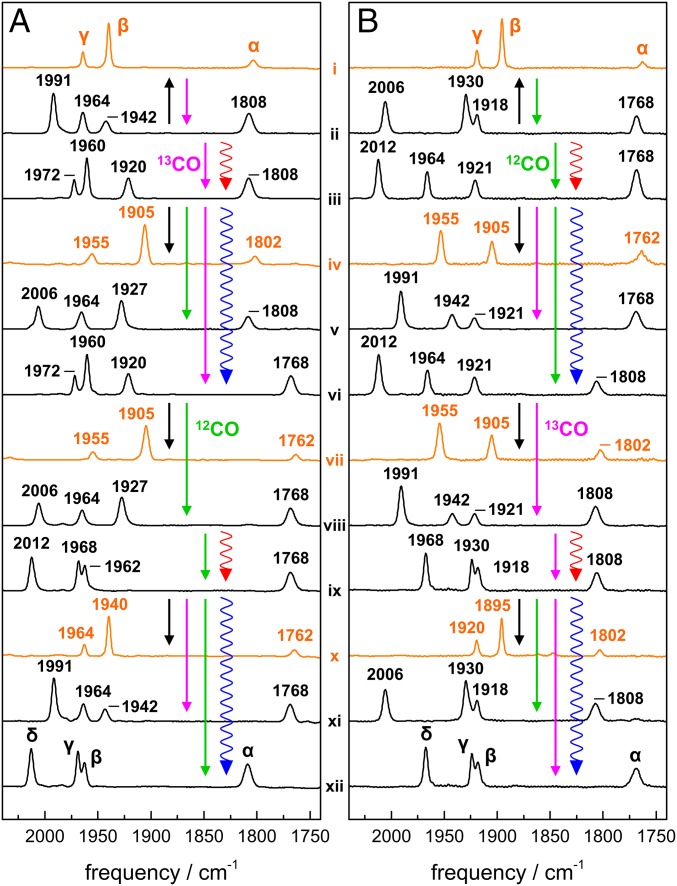
ATR-FTIR spectra of HYDA1 [FeFe]-hydrogenase films. (A) Isotopic species with a p12CO ligand. (B) Isotopic species with a p13CO ligand. IR bands due to stretching vibrations of CO ligands at the H-cluster were normalized to unity area sums. Spectra are attributed to Hox (orange) or Hox-CO (black); CO bands are denoted α, β, γ, and δ. For real-time ATR-FTIR experiments, see SI Appendix, Fig. S3. Straight arrows denote gas exposures (12CO, green; 13CO, magenta; and N2, black), wiggled arrows denote red or blue light irradiation. Numerals i–xii annotate identified spectral species (Table 1).
IR Band Assignment for Unlabeled Hox and Hox-CO.
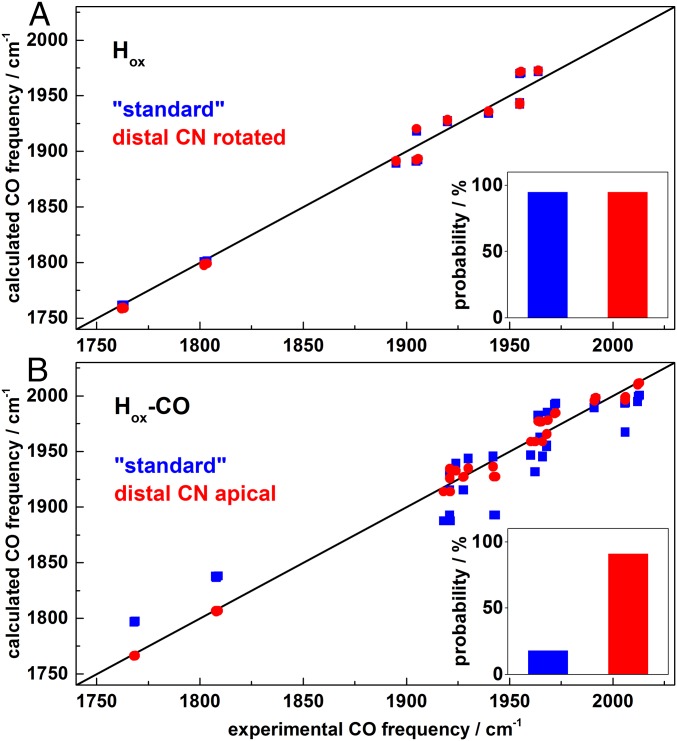
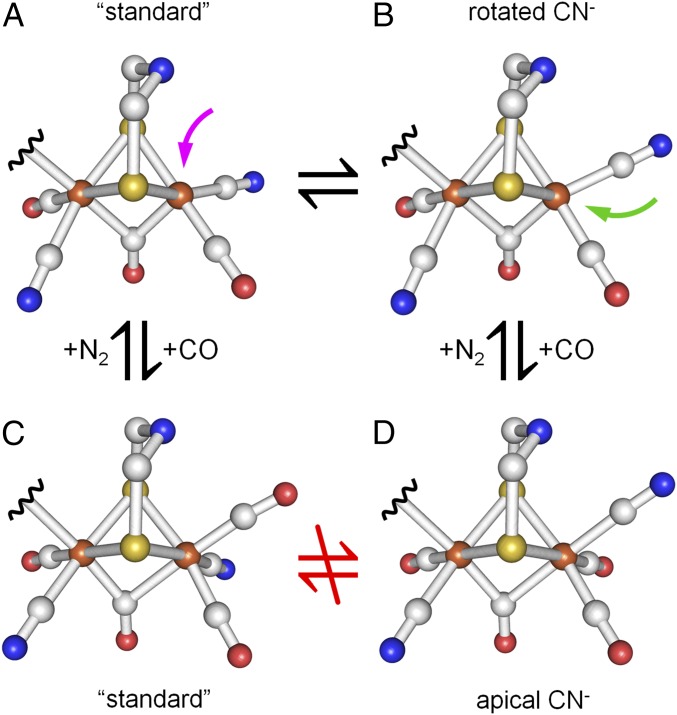
Under an N2 atmosphere HYDA1 showed the typical three CO bands of the Hox state (Fig. 2 A, i). Carbonyl bands shifted by ∼40 cm−1 to lower frequencies due to 13CO isotope editing (see below), whereas the CN− bands shifted less than 1 cm−1 (SI Appendix, Fig. S6) and hence were not decisive for H-cluster species assignment. DFT consistently attributed the CO bands to the largely uncoupled vibrations of the Fe-Fe bridging carbonyl (µCO, band α at 1802 cm−1) and the terminal CO ligands at Fed (dCO, band β at 1940 cm−1) and Fep (pCO, band γ at 1964 cm−1) (SI Appendix, Fig. S9). As a measure for correlation of calculated and experimental CO frequencies, the RMSD (Eq. S1) was calculated (Table 1 and SI Appendix, Table S1 and Fig. S11). A mean RMSD of ∼10 cm−1 was obtained for the four possible Hox rotamers with equatorial CO/CN− ligands at Fep and Fed (Fig. 3A), which indicated good agreement between experimental and calculated CO frequencies. A similar small RMSD was obtained for a Hox rotamer with dCN− rotated toward a more apical position (Fig. 4), whereas a rotated apical dCO was disfavored. In the following, H-cluster rotamer structures are discussed relative to the “standard” model (24, 25, 45) with trans orientation of equatorial CO ligands and apical vacancy at Fed in Hox (Fig. 1).
Table 1.
Assignment of IR spectra to isotopic labeling patterns
| Spectrum | p12CO | RMSD | p13CO | RMSD |
| i | 12 12 12 | 6 (6) | 13 13 13 | 5 (5) |
| iv | 12 12 13 | 12 (12) | 13 13 12 | 12 (10) |
| vii | 12 13 13 | 12 (12) | 13 12 12 | 11 (10) |
| x | 12 13 12 | 6 (6) | 13 12 13 | 6 (6) |
| ii | 12121213 | 11 (31) | 13131312 | 6 (25) |
| iii | 12121313 | 7 (26) | 13131212 | 7 (21) |
| v | 12121312 | 7 (17) | 13131213 | 5 (20) |
| vi | 12131313 | 7 (25) | 13121212 | 8 (20) |
| viii | 12131312 | 7 (17) | 13121213 | 4 (20) |
| ix | 12131212 | 5 (23) | 13121313 | 5 (23) |
| xi | 12131213 | 11 (31) | 13121312 | 6 (25) |
| xii | 12121212 | 5 (24) | 13131313 | 5 (23) |
Experimental IR spectra i–xii are shown in Fig. 2A (p12CO) and Fig. 2B (p13CO). All 12CO/13CO (12/13) labeling patterns are given in the order δ, γ, β, and α (band δ is missing in i, iv, vii, and x). Deviation values (RMSD; Eq. S1) were derived from comparison of experimental and DFT-calculated CO stretch frequencies of rotamers with apical CN− at Fed in Hox-CO and with the distal CN− rotated toward apical position in Hox, compared with standard ligand arrangements (values in parentheses). Correlations of experimental and calculated CO (and CN−) band frequencies and intensities for all 178 studied model structures are shown in SI Appendix, Tables S1–S4 and Figs. S11 and S12. Bold numbers indicate 13CO isotope labeling.
Fig. 3.
Correlation of experimental and calculated CO band frequencies. (A) Hox: standard model (blue) and model with proximal CN− rotated toward apical position (red). (B) Hox-CO: standard model (blue) and model with distal CN− in apical position (red). Diagonals show ideal correlation. Calculated CO frequencies were offset-corrected (31 ± 1 cm−1, Hox; 38 ± 2 cm−1, Hox-CO) for alignment with experimental data (SI Appendix, Tables S1 and S2). (Insets) Approximate rotamer probabilities from IR data analysis (SI Appendix, Table S4).
Fig. 4.
H-cluster rotamer structures of Hox and Hox-CO. A transition from Hox structure (A) to Hox-CO structure (C) is suggested in the standard model where exogenous CO binds at Fed in apical position (magenta arrow). Equilibrium between Hox rotamers A and B facilitates CO binding at Fed in equatorial position (green arrow) and thereby transition to the Hox-CO rotamer with apical CN− at Fed (D). Octahedral coordination of Fed in Hox-CO renders ligand rotation unlikely and prevents a transition between rotamers C and D.
Exchange of N2 by 12CO gas in the headspace above the protein film resulted in the appearance of a forth CO band (δ) at higher IR frequencies due to an additional carbonyl ligand (d2CO) in Hox-CO (Fig. 2 A, xii). We calculated the IR bands of the six possible CO/CN− rotamers. Similar large RMSD values (∼30 cm−1) were observed for the four structures with apical d2CO (SI Appendix, Table S2). An about sixfold improved RMSD (∼5 cm−1) was observed for the Hox-CO structure with apical dCN− and d2CO in the equatorial plane (Fig. 3B). DFT assigned band α to the µCO stretch mode (1808 cm−1) and band β to an anti-symmetric coupled mode with smaller contributions from equatorial d1CO and larger contributions from apical d2CO (1962 cm−1). Band γ was assigned to a coupled mode with similar contributions from the symmetric vibrations of d1CO and d2CO and the antisymmetric stretch mode of pCO (1968 cm−1) and band δ to a coupled symmetric mode with contributions from all four carbonyls (2012 cm−1) in the standard model (SI Appendix, Fig. S9). Except for the energetically separated band α due to the µCO ligand (SI Appendix, Fig. S7), pronounced vibrational coupling of d1CO, d2CO, and pCO precludes a priori assignment of IR bands to specific CO ligands in Hox-CO.
Stepwise 13CO Editing of the H-Cluster.
For HYDA1 protein films, exposure of unlabeled Hox-CO (xii) to 13CO gas caused a >20 cm−1 shift to lower frequencies of bands β and δ, whereas band γ was less affected and α remained unchanged, suggesting a single 13CO ligand at Fed (Fig. 2A, ii). Red light irradiation under 13CO gas resulted in a further >20 cm−1 down-shift of bands β and δ, indicative of a second 13CO ligand at Fed (iii). In the dark, species iii was converted under 12CO gas to a state differing from unlabeled Hox-CO in band β, suggesting a d113CO exchange (v). Blue light irradiation of iii under 13CO caused an exclusive ∼40 cm−1 down-shift of band α, whereas β, γ, and δ remained unchanged. Thus, a state with three shifted CO bands with respect to unlabeled Hox-CO was populated, suggesting two distal 13CO ligands and µ13CO (vi). Exchange to a 12CO atmosphere resulted in a ∼30 cm−1 up-shift of band δ, small shifts to higher frequencies of β and γ, and no change of band α. This pattern agrees with d113CO and μ13CO labeling (viii). Red light irradiation of viii under 12CO yielded a state showing similar β, γ, and δ frequencies as xii, but α remained at its low frequency so that only μ13CO was present (ix). 13CO exposure converted ix to a state reminiscent of spectrum ii, including d213CO and μ13CO labeling (xi). Finally, blue light irradiation of xi under 12CO regained unlabeled Hox-CO (xii). These results suggested that µCO and the distal carbonyls were exchangeable in HYDA1, but not the proximal CO ligand.
At increased humidity of the 13CO aerosol and blue light irradiation, HYDA1 with three 13CO ligands (vi) produced down-shifts of all four CO bands compared with the unlabeled species. This state was assigned to completely 13CO-labeled Hox-CO (33), including the proximal CO ligand (Fig. 2B, xii). Exposure to 12CO caused a ∼40 cm−1 up-shift of δ with only minor changes for γ and β and no difference for α (ii). Further red light irradiation mainly up-shifted band γ by ∼40 cm−1 (iii), which suggested stepwise replacement of the two 13CO ligands at Fed by 12CO in the presence of p13CO. Further 12CO exposure under blue light induced the exchange of µCO as indicated by a ∼40 cm−1 up-shift of band α (vi). Rebinding of 13CO to iii or vi yielded species v or viii, their δ band positions suggesting a single distal 13CO ligand. Red light irradiation under 13CO of viii restored the frequency pattern of xii except for the down-shifted band α (ix). The latter was exchanged only under blue light (xii). 12CO exposure of viii finally regained species ii. Selective 13CO editing of pCO was facilitated only in sufficiently hydrated HYDA1 protein films.
Complementary 13CO editing experiments were performed for CPI and DDH (SI Appendix, Fig. S8). 13CO exchange of the two distal carbonyls was achieved already under red light in these enzymes, possibly related to increased light absorption in the presence of the accessory iron-sulfur clusters, whereas HYDA1 allowed sequential editing with red and blue light. Four of the eight possible Hox-CO isotopic species excluding p13CO were populated in the bacterial enzymes. The CO frequencies, however, were similar in the three enzymes.
Hox-CO Isotopic Species Assignment from DFT.
The IR experiments showed 16 distinct Hox-CO isotopic species with all possible labeling patterns. We calculated IR spectra for 96 Hox-CO models, including 16 possible 13CO-labeling patterns with six CO/CN− rotamers each (SI Appendix, Fig. S12 and Table S2). Similarly large RMSD values (∼30 cm−1) were observed for all isotopic species with an apical dCO, which precluded assignment of the experimental IR spectra for the “standard” Hox-CO geometry. Species with an apical dCN− showed significantly diminished RMSD values for all isotopic labeling patterns. These results facilitated the unambiguous attribution of each IR spectrum to a specific Hox-CO species (Table 1). Both medium and large models showed diminished preference for the dCN− rotamer compared with the small H-cluster model (SI Appendix, Table S2), but still a twofold smaller RMSD was observed for the structure with an apical dCN− ligand. Comprehensive analysis of experimental and calculated IR band frequencies and intensities suggested that Hox-CO structures with proximal CO/CN− inversion were disfavored and further supported an apical dCN− (SI Appendix, Table S4). These results indicated the cyclic isotope editing sequence shown in Fig. 5. The exogenous CO ligand (d2CO) is exchangeable in darkness, red light sensitivity is attributed to the equatorial d1CO, and blue light induces exchange of µCO and pCO, the latter being feasible only in sufficiently hydrated HYDA1 protein films.
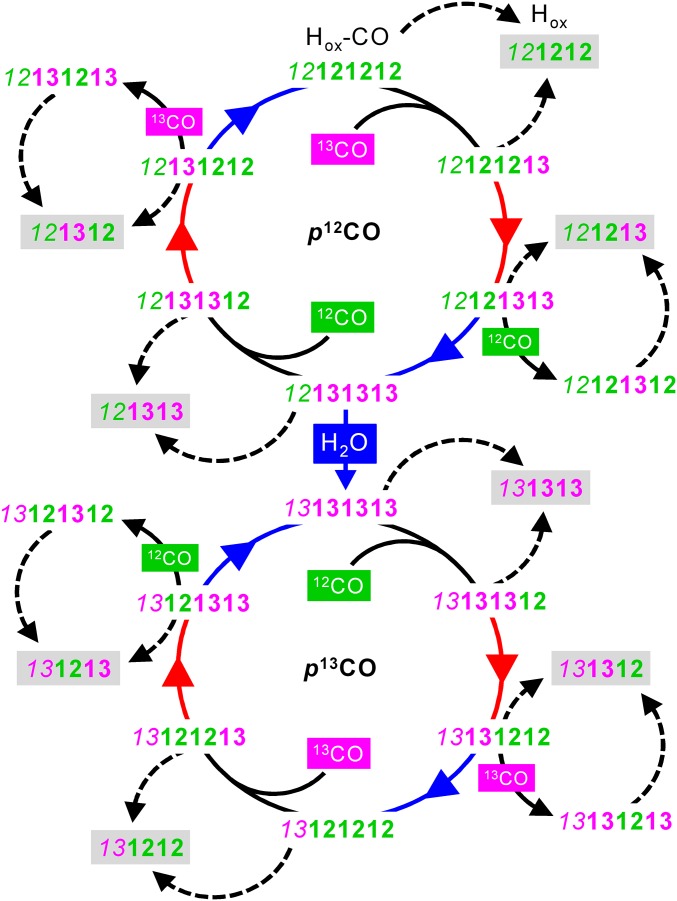
Fig. 5.
Stepwise isotope editing of the H-cluster. Gray shadings highlight eight differently labeled Hox species providing access to the catalytic cycle of hydrogen turnover. Carbonyl ligand patterns are shown in the order p µ d1 d2 (d2 is present only in Hox-CO). Exposure to 13CO (magenta) or 12CO (green) gas is indicated only for the dark steps (solid black arrows) and persisted during the following red or blue light irradiation steps (colored arrows) in the experimental cycle; dashed arrows denote N2 exposure in darkness. The proximal CO ligand is prone to 13CO exchange only in sufficiently hydrated (H2O) protein films.
Site-Selective 13CO Editing and Rotamers of Hox.
Quantitative population of four Hox isotopic species with zero to two 13CO ligands excluding pCO was achieved by N2 gas exposure of HYDA1 protein films at low humidity (Fig. 2A). Hox-CO species xii and ii were converted into unlabeled Hox (i). In comparison with i, Hox-CO species iii and v were converted into a state showing a ∼30 cm−1 down-shift of band β and a smaller shift of γ, implying a single 13CO ligand at Fed (d13CO) (iv). Species vi and viii yielded a Hox state similar to iv, but showing an additional ∼40 cm−1 down-shift of α due μ13CO labeling (vii). Finally, species ix and xi were converted into a state with an exclusive ∼40 cm−1 down-shift of α compared with unlabeled Hox, indicative of μ13CO (x). Starting with completely 13CO-labeled and hydrated HYDA1 in the Hox-CO state, four Hox species with one to three 13CO ligands including pCO were populated by N2 exposure (Fig. 2B). Hox-CO species ii and xii were converted to Hox species i with bands α, γ, and β shifted ∼40 cm−1 to lower frequencies (complete 13CO exchange). Hox-CO species with 13CO at Fep (p13CO) and 12CO at Fed (iv and vii) were converted to Hox species iv and vii showing a γ band intensity (1955 cm−1) exceeding the one of band β (1905 cm−1), which was reversed for Hox species with unlabeled pCO. These are the only Hox isomers with pronounced vibrational coupling of dCO and pCO (SI Appendix, Fig. S9). Hox-CO species ix and xi finally were converted to Hox species x, which resembled species i except for presence of µ12CO.
IR band patterns for the 56 possible Hox structures (7 CO/CN− rotamers with 8 13CO-labeling patterns each) were calculated (SI Appendix, Table S1). Comparison of experimental and calculated CO frequencies revealed by far lowest RMSD values only for isotopic patterns in agreement with the above experimental assignments (Table 1). Analysis of IR band frequencies and intensities of Hox (SI Appendix, Table S1 and Fig. 11) and mutual comparison with the results for Hox-CO (SI Appendix, Table S4) excluded dCO in more apical position. On the other hand, the calculated IR data of a structure with dCN− rotated toward a more apical position were as well in agreement with the experimental data as the standard ligand configuration, for all isotopic species of Hox (Table 1). Both these structures accounted for vibrational coupling of pCO and dCO in the presence of a proximal 13CO (SI Appendix, Fig. S9), which explained the inverted intensity ratio of the β and γ bands in Hox species iv and vii.
Discussion
Our protocol for controlled gas exposure, irradiation, and hydration of [FeFe]-hydrogenase protein films facilitates quantitative population of 8 Hox and 16 Hox-CO species selectively labeled with zero to four 13CO ligands. Fourier-transform IR spectroscopy in ATR configuration facilitates rapid gas exchange for controlled and quantitative state population in [FeFe]-hydrogenase protein films. These experiments have provided an unprecedentedly large IR data set for comparison with quantum chemical calculations. The CO vibrational modes underlying the IR spectra were assigned unambiguously. In Hox, experimentally observed CO stretching frequencies are well separated and differ by at least 24 cm−1 (pCO/dCO). This feature facilitates direct band assignment via 13CO isotope editing. In contrast to Hox, the three terminal carbonyls in Hox-CO show pronounced vibrational coupling that results from changes in ligand geometry and [2Fe]H spin distribution (24–28, 31, 36). Disentangling of spectral shifts as induced by stepwise isotope editing of Hox-CO was achieved via DFT analysis. Our results imply a consistent reaction cycle for isotopic editing of the H-cluster (Fig. 5).
Hox-CO in standard configuration (27, 46) is not in good agreement with the experimental carbonyl vibrations. Models comprising an apical CN− ligand at Fed yielded a vibrationally uncoupled pCO, which is a characteristic feature of the H-cluster (24, 26). Only these models reproduced the altered origin of the pCO vibrational frequency and inverted band intensities for species including d113CO and d213CO. Improved correlation of experimental and calculated IR data for Hox-CO with apical dCN− has been discussed before, but evaluated against insufficiently small experimental IR datasets (39, 47, 48). We prove the effect for 16 Hox-CO species, three phylogenetically distinct [FeFe]-hydrogenases, and varying computational approaches. However, our analysis clearly supports the ligand arrangement at the proximal iron ion in the crystallographic data (21–23).
Available H-cluster structures were modeled with trans equatorial carbonyls and square-pyramidal (Hox) or octahedral geometries (Hox-CO) at the distal iron ion (21–23, 27, 46, 49). At a resolution of ∼1.5 Å or less, however, CO/CN− discrimination remains speculative. These ligands originally were assigned using potential hydrogen bonding of CN− ligands to protein residues (21, 40, 49–51) (SI Appendix, Fig. S10) and before the identity of the adt ligand was unraveled (19). A computational study on the DDH crystal structure preferred the “standard” Hox-CO geometry by ∼6 kJ/mol due to interaction of dCN− with a backbone amine and the conserved Lys237 (39, 48). An interaction between Lys237 and dCN− has also been inferred from EPR but was not supported later (20, 52). Our analysis for all model structures suggests slight distortion of octahedral Fed symmetry in the standard model, whereas for an apical CN− weak H-bonding to the adt nitrogen base occurs (Fig. 4). This geometry was earlier calculated to be stabilized by ∼8 kJ/mol (48). It has been suggested that H2 may form a similar H-bond to adt during the catalytic reaction (37, 49, 51, 53). Substrate (H2) or inhibitor (CO) binding at the active site thus may be governed by intramolecular rather than protein–cofactor interactions. The detailed influence of the protein environment on the fine structure of the H-cluster is difficult to quantify both from experimental and theoretical viewpoints. Our general isotope editing scheme (Fig. 5), however, remains valid irrespective of the precise angular arrangement of the distal ligands.
The Hox standard configuration and a rotamer with more apical CN− and equatorial vacancy at Fed showed similar and superior agreement between experimental and calculated IR data. Accordingly, such structures appear equally probable. Our analysis further favors trans orientation of equatorial carbonyls and a proximal CO/CN− arrangement as in crystallographic assignments (21–23, 49). The HYDA1 and CPI proteins used in this study were activated in vitro with a synthetic diiron site analog (39, 40). We observed no significant differences between our HYDA1 and CPI preparations and the natively maturated DDH so that rotamer formation during in vitro maturation can be excluded (12, 17, 20, 32, 54). The observation that only sufficient hydration of HYDA1 protein films facilitates isotope editing at the proximal iron ion rather indicates that structural flexibility of gas channels (55) is involved in ligand exchange. Under cryogenic conditions (i.e., for diffraction data collection), the standard Hox structure thus dominates. Biologically relevant conditions (i.e., dissolved protein at room temperature as used here) could promote equilibrium between the two ligand geometries at Fed or even dominance of the rotamer with more apical dCN−. Such equilibria exist for diiron compounds in solution (56–58). This view is further reinforced by molecular dynamics simulations on DDH showing that distal ligand rotation is related to motions by up to 2 Å of a nearby phenylalanine side chain (36). Only in the rotated Hox structure, CO can bind in equatorial position at Fed (Fig. 4). Ligand dynamics also impacts on possible motifs of substrate (H2) interactions with the active site.
Hox is the entry point to the hydrogen conversion cycle of [FeFe]-hydrogenases (1). At least two increasingly reduced H-cluster species were derived from Hox; their molecular structures and involvement in catalysis yet remain to be defined (11–15, 36, 59). The fate of the Fe-Fe bridging carbonyl is of particular mechanistic interest. Binding of hydrogen species in apical position at Fed of the H-cluster is believed to be essential for catalysis (1). However, configurations with (semi) bridging or equatorial H-species were considered as well (12, 14, 36) and may result from structural flexibility of the H-cluster (36). Such structural dynamics may facilitate apical or equatorial ligand binding at the distal iron ion and may also be relevant for O2 inactivation of the enzymes via reactive oxygen species formation (24, 29, 30, 60, 61). Our protocol for selective preparation of Hox with eight distinct isotopic labeling patterns introduces spectroscopic probes at individual positions at the cofactor. This approach opens the road for investigations on novel isotopically labeled intermediates in the catalytic cycle to probe structural dynamics during the H2 conversion chemistry of [FeFe]-hydrogenases.
Materials and Methods
HYDA1 Protein Preparation.
[FeFe]-hydrogenase HYDA1 and CPI apo-proteins were overexpressed in Escherichia coli, purified, and quantitatively reconstituted in vitro with a synthetic diiron complex [Fe2(µ-adt)(CO)4(CN)2, adt = (SCH2)2NH] (23, 41). All protein preparation and handling procedures were carried out under strictly anoxic conditions and dim light. DDH was purified from D. desulfuricans with a complete H-cluster (43).
Infrared Spectroscopy.
ATR-FTIR spectroscopy (62) was performed with a Tensor27 spectrometer (Bruker) placed in an anaerobic glovebox and equipped with a mid-IR globar, a liquid-nitrogen–cooled MCT detector, and a silicon prism with two active reflections, which was capped by a sealed PCTFE head-space gas compartment. Infrared spectra were recorded with 1-cm–1 spectral resolution using varying numbers of interferometer scans on thin protein films, corrected for background contributions, and evaluated using a least-squares-fit algorithm. Hydrogenase films were exposed to 13CO, 12CO, or N2 gas by fast exchange of the head-space atmosphere using a multichannel mass flow controller (Sierra Instruments) at room temperature. All gases were sent pro rata through a water-filled wash bottle to create an aerosol that prevents dehydration of protein films. This allowed controlling the water/ protein ratio in the film (hydration) and influenced the velocity of any gas-processing reaction. Humidity refers to the water/gas ratio in the aerosol. A Schott white light source with band pass filters (center wavelengths 640 or 460 nm) was used for irradiation of protein samples. Details on real-time ATR-FTIR experiments and data evaluation are given in SI Appendix, Figs. S1–S4.
Quantum Chemical Calculations.
DFT calculations on H-cluster model structures with 12CO/13CO ligands were carried out using Gaussian09 (63) on the Soroban computer cluster of the Freie Universität Berlin. Starting structures of increasing complexity (SI Appendix, Fig. S5) were constructed using the crystal structure of CO-inhibited CPI [FeFe]-hydrogenase (27) as a template and geometry-optimized using the BP86/TZVP or TPSSh/TZVP functional/basis-set combinations (64–66), and IR spectra were calculated thereafter (67). Details of the computational methods are given in SI Appendix. The calculated structures can be accessed via Dataset S1.
Supplementary Material
Acknowledgments
We thank T. Happe and M. Winkler for generously providing protein samples (HYDA1, CPI) and extensive discussion and J. Fontecilla-Camps for providing a sample of DDH protein. M.S., J.H., and S.T.S. thank the International Max Plank Research School on Multiscale Biosystems and the Focus Area NanoScale (Freie Universität Berlin) for financial support. M.H. gratefully acknowledges funding by Deutsche Forschungsgemeinschaft (DFG) Grant Ha3265/6-1 and Bundesministerium für Bildung und Forschung Grant 05K14KE1. J.D. acknowledges support by the China Scholarship Council and from the DFG, Cluster of Excellence RESOLV, EXC1069. U.-P.A. and F.W. are grateful for financial support by the Fonds of the Chemical Industry (Liebig grant to U.-P.A.) and the DFG (Emmy Noether Grant AP242/2-1 to U.-P.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606178113/-/DCSupplemental.
References
- 1.Lubitz W, Ogata H, Rüdiger O, Reijerse E. Hydrogenases. Chem Rev. 2014;114(8):4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 2.Peters JW, Broderick JB. Emerging paradigms for complex iron-sulfur cofactor assembly and insertion. Annu Rev Biochem. 2012;81:429–450. doi: 10.1146/annurev-biochem-052610-094911. [DOI] [PubMed] [Google Scholar]
- 3.Stripp ST, Happe T. How algae produce hydrogen–news from the photosynthetic hydrogenase. Dalton Trans. 2009;(45):9960–9969. doi: 10.1039/b916246a. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong FA, Fontecilla-Camps JC. Biochemistry. A natural choice for activating hydrogen. Science. 2008;321(5888):498–499. doi: 10.1126/science.1161326. [DOI] [PubMed] [Google Scholar]
- 5.Madden C, et al. Catalytic turnover of [FeFe]-hydrogenase based on single-molecule imaging. J Am Chem Soc. 2012;134(3):1577–1582. doi: 10.1021/ja207461t. [DOI] [PubMed] [Google Scholar]
- 6.Simmons TR, Berggren G, Bacchia M, Fontecave M, Artero V. Mimicking hydrogenases: From biomimetics to artificial enzymes. Coord Chem Rev. 2014;270-271:127–150. [Google Scholar]
- 7.Rauchfuss TB. Chemistry. A promising mimic of hydrogenase activity. Science. 2007;316(5824):553–554. doi: 10.1126/science.1140733. [DOI] [PubMed] [Google Scholar]
- 8.Artero V, et al. From enzyme maturation to synthetic chemistry: The case of hydrogenases. Acc Chem Res. 2015;48(8):2380–2387. doi: 10.1021/acs.accounts.5b00157. [DOI] [PubMed] [Google Scholar]
- 9.Dubini A, Ghirardi ML. Engineering photosynthetic organisms for the production of biohydrogen. Photosynth Res. 2015;123(3):241–253. doi: 10.1007/s11120-014-9991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis NS, Nocera DG. Powering the planet: Chemical challenges in solar energy utilization. Proc Natl Acad Sci USA. 2006;103(43):15729–15735. doi: 10.1073/pnas.0603395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamska-Venkatesh A, et al. New redox states observed in [FeFe] hydrogenases reveal redox coupling within the H-cluster. J Am Chem Soc. 2014;136(32):11339–11346. doi: 10.1021/ja503390c. [DOI] [PubMed] [Google Scholar]
- 12.Adamska A, et al. Identification and characterization of the “super-reduced” state of the H-cluster in [FeFe] hydrogenase: A new building block for the catalytic cycle? Angew Chem Int Ed Engl. 2012;51(46):11458–11462. doi: 10.1002/anie.201204800. [DOI] [PubMed] [Google Scholar]
- 13.Lambertz C, et al. Electronic and molecular structures of the [2Fe] and [4Fe4S] units of the active-site H-cluster in [FeFe]-hydrogenase determined by spin- and site-selective XAE and DFT. Chem Sci (Camb) 2014;5(3):1187–1203. [Google Scholar]
- 14.Chernev P, et al. Hydride binding to the active site of [FeFe]-hydrogenase. Inorg Chem. 2014;53(22):12164–12177. doi: 10.1021/ic502047q. [DOI] [PubMed] [Google Scholar]
- 15.Mulder DW, et al. EPR and FTIR analysis of the mechanism of H2 activation by [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii. J Am Chem Soc. 2013;135(18):6921–6929. doi: 10.1021/ja4000257. [DOI] [PubMed] [Google Scholar]
- 16.De Lacey AL, Fernandez VM, Rousset M, Cammack R. Activation and inactivation of hydrogenase function and the catalytic cycle: Spectroelectrochemical studies. Chem Rev. 2007;107(10):4304–4330. doi: 10.1021/cr0501947. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert-Wilson R, et al. Spectroscopic investigations of [FeFe] hydrogenase maturated with [(57)Fe2(adt)(CN)2(CO)4](2.) J Am Chem Soc. 2015;137(28):8998–9005. doi: 10.1021/jacs.5b03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchenreuther JM, et al. The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science. 2014;343(6169):424–427. doi: 10.1126/science.1246572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silakov A, Wenk B, Reijerse E, Lubitz W. (14)N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: Evidence for a nitrogen in the dithiol bridge. Phys Chem Chem Phys. 2009;11(31):6592–6599. doi: 10.1039/b905841a. [DOI] [PubMed] [Google Scholar]
- 20.Adamska-Venkatesh A, et al. Spectroscopic characterization of the bridging amine in the active site of [FeFe] hydrogenase using isotopologues of the H-cluster. J Am Chem Soc. 2015;137(40):12744–12747. doi: 10.1021/jacs.5b06240. [DOI] [PubMed] [Google Scholar]
- 21.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282(5395):1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 22.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: The structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7(1):13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 23.Esselborn J, et al. A structural view of synthetic cofactor integration into [FeFe]-hydrogenases. Chem Sci (Camb) 2016;7(2):959–968. doi: 10.1039/c5sc03397g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roseboom W, De Lacey AL, Fernandez VM, Hatchikian EC, Albracht SP. The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy. J Biol Inorg Chem. 2006;11(1):102–118. doi: 10.1007/s00775-005-0040-2. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Infrared studies of the CO-inhibited form of the Fe-only hydrogenase from Clostridium pasteurianum I: Examination of its light sensitivity at cryogenic temperatures. Biochemistry. 2002;41(6):2036–2043. doi: 10.1021/bi011510o. [DOI] [PubMed] [Google Scholar]
- 26.De Lacey AL, Stadler C, Cavazza C, Hatchikian EC, Fernandez VM. FTIR characterization of the active site of the Fe-hydrogenase from Desulfovibrio desulfuricans. J Am Chem Soc. 2000;122(45):11232–11233. [Google Scholar]
- 27.Lemon BJ, Peters JW. Binding of exogenously added carbon monoxide at the active site of the iron-only hydrogenase (CpI) from Clostridium pasteurianum. Biochemistry. 1999;38(40):12969–12973. doi: 10.1021/bi9913193. [DOI] [PubMed] [Google Scholar]
- 28.Myers WK, et al. The cyanide ligands of [FeFe] hydrogenase: Pulse EPR studies of (13)C and (15)N-labeled H-cluster. J Am Chem Soc. 2014;136(35):12237–12240. doi: 10.1021/ja507046w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stripp ST, et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc Natl Acad Sci USA. 2009;106(41):17331–17336. doi: 10.1073/pnas.0905343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silakov A, Wenk B, Reijerse E, Albracht SP, Lubitz W. Spin distribution of the H-cluster in the H(ox)-CO state of the [FeFe] hydrogenase from Desulfovibrio desulfuricans: HYSCORE and ENDOR study of (14)N and (13)C nuclear interactions. J Biol Inorg Chem. 2009;14(2):301–313. doi: 10.1007/s00775-008-0449-5. [DOI] [PubMed] [Google Scholar]
- 31.Pierik AJ, Hulstein M, Hagen WR, Albracht SP. A low-spin iron with CN and CO as intrinsic ligands forms the core of the active site in [Fe]-hydrogenases. Eur J Biochem. 1998;258(2):572–578. doi: 10.1046/j.1432-1327.1998.2580572.x. [DOI] [PubMed] [Google Scholar]
- 32.Silakov A, Kamp C, Reijerse E, Happe T, Lubitz W. Spectroelectrochemical characterization of the active site of the [FeFe] hydrogenase HydA1 from Chlamydomonas reinhardtii. Biochemistry. 2009;48(33):7780–7786. doi: 10.1021/bi9009105. [DOI] [PubMed] [Google Scholar]
- 33.Kuchenreuther JM, George SJ, Grady-Smith CS, Cramer SP, Swartz JR. Cell-free H-cluster synthesis and [FeFe] hydrogenase activation: All five CO and CN⁻ ligands derive from tyrosine. PLoS One. 2011;6(5):e20346. doi: 10.1371/journal.pone.0020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldet G, et al. Electrochemical kinetic investigations of the reactions of [FeFe]-hydrogenases with carbon monoxide and oxygen: Comparing the importance of gas tunnels and active-site electronic/redox effects. J Am Chem Soc. 2009;131(41):14979–14989. doi: 10.1021/ja905388j. [DOI] [PubMed] [Google Scholar]
- 35.Siebel JF, et al. Hybrid [FeFe]-hydrogenases with modified active sites show remarkable residual enzymatic activity. Biochemistry. 2015;54(7):1474–1483. doi: 10.1021/bi501391d. [DOI] [PubMed] [Google Scholar]
- 36.Fourmond V, et al. The oxidative inactivation of FeFe hydrogenase reveals the flexibility of the H-cluster. Nat Chem. 2014;6(4):336–342. doi: 10.1038/nchem.1892. [DOI] [PubMed] [Google Scholar]
- 37.Mulder DW, et al. Investigations on the role of proton-coupled electron transfer in hydrogen activation by [FeFe]-hydrogenase. J Am Chem Soc. 2014;136(43):15394–15402. doi: 10.1021/ja508629m. [DOI] [PubMed] [Google Scholar]
- 38.Tye JW, Darensbourg MY, Hall MB. Refining the active site structure of iron-iron hydrogenase using computational infrared spectroscopy. Inorg Chem. 2008;47(7):2380–2388. doi: 10.1021/ic7013732. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, et al. Targeting intermediates of [FeFe]-hydrogenase by CO and CN vibrational signatures. Inorg Chem. 2011;50(9):3888–3900. doi: 10.1021/ic102039z. [DOI] [PubMed] [Google Scholar]
- 40.Winkler M, Esselborn J, Happe T. Molecular basis of [FeFe]-hydrogenase function: An insight into the complex interplay between protein and catalytic cofactor. Biochim Biophys Acta. 2013;1827(8-9):974–985. doi: 10.1016/j.bbabio.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Esselborn J, et al. Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat Chem Biol. 2013;9(10):607–609. doi: 10.1038/nchembio.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berggren G, et al. Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature. 2013;499(7456):66–69. doi: 10.1038/nature12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatchikian EC, Forget N, Fernandez VM, Williams R, Cammack R. Further characterization of the [Fe]-hydrogenase from Desulfovibrio desulfuricans ATCC 7757. Eur J Biochem. 1992;209(1):357–365. doi: 10.1111/j.1432-1033.1992.tb17297.x. [DOI] [PubMed] [Google Scholar]
- 44.Gonzales MA, Mascharak PK. Photoactive metal carbonyl complexes as potential agents for targeted CO delivery. J Inorg Biochem. 2014;133:127–135. doi: 10.1016/j.jinorgbio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 45.van der Spek TM, et al. Similarities in the architecture of the active sites of Ni-hydrogenases and Fe-hydrogenases detected by means of infrared spectroscopy. Eur J Biochem. 1996;237(3):629–634. doi: 10.1111/j.1432-1033.1996.0629p.x. [DOI] [PubMed] [Google Scholar]
- 46.Pandey AS, Harris TV, Giles LJ, Peters JW, Szilagyi RK. Dithiomethylether as a ligand in the hydrogenase h-cluster. J Am Chem Soc. 2008;130(13):4533–4540. doi: 10.1021/ja711187e. [DOI] [PubMed] [Google Scholar]
- 47.Zilberman S, Stiefel EI, Cohen MH, Car R. Resolving the CO/CN ligand arrangement in CO-inactivated [FeFe] hydrogenase by first principles density functional theory calculations. Inorg Chem. 2006;45(15):5715–5717. doi: 10.1021/ic060075p. [DOI] [PubMed] [Google Scholar]
- 48.Greco C, et al. Structural insights into the active-ready form of [FeFe]-hydrogenase and mechanistic details of its inhibition by carbon monoxide. Inorg Chem. 2007;46(18):7256–7258. doi: 10.1021/ic701051h. [DOI] [PubMed] [Google Scholar]
- 49.Nicolet Y, et al. Crystallographic and FTIR spectroscopic evidence of changes in Fe coordination upon reduction of the active site of the Fe-only hydrogenase from Desulfovibrio desulfuricans. J Am Chem Soc. 2001;123(8):1596–1601. doi: 10.1021/ja0020963. [DOI] [PubMed] [Google Scholar]
- 50.Knörzer P, et al. Importance of the protein framework for catalytic activity of [FeFe]-hydrogenases. J Biol Chem. 2012;287(2):1489–1499. doi: 10.1074/jbc.M111.305797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruschi M, et al. Influence of the [2Fe]H subcluster environment on the properties of key intermediates in the catalytic cycle of [FeFe] hydrogenases: Hints for the rational design of synthetic catalysts. Angew Chem Int Ed Engl. 2009;48(19):3503–3506. doi: 10.1002/anie.200900494. [DOI] [PubMed] [Google Scholar]
- 52.Silakov A, Reijerse EJ, Albracht SP, Hatchikian EC, Lubitz W. The electronic structure of the H-cluster in the [FeFe]-hydrogenase from Desulfovibrio desulfuricans: A Q-band 57Fe-ENDOR and HYSCORE study. J Am Chem Soc. 2007;129(37):11447–11458. doi: 10.1021/ja072592s. [DOI] [PubMed] [Google Scholar]
- 53.Rauchfuss TB. Diiron azadithiolates as models for the [FeFe]-hydrogenase active site and paradigm for the role of the second coordination sphere. Acc Chem Res. 2015;48(7):2107–2116. doi: 10.1021/acs.accounts.5b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adamska-Venkatesh A, et al. Artificially maturated [FeFe] hydrogenase from Chlamydomonas reinhardtii: A HYSCORE and ENDOR study of a non-natural H-cluster. Phys Chem Chem Phys. 2015;17(7):5421–5430. doi: 10.1039/c4cp05426a. [DOI] [PubMed] [Google Scholar]
- 55.Cohen J, Kim K, King P, Seibert M, Schulten K. Finding gas diffusion pathways in proteins: Application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects. Structure. 2005;13(9):1321–1329. doi: 10.1016/j.str.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 56.Leidel N, et al. Electronic structure of an [FeFe] hydrogenase model complex in solution revealed by X-ray absorption spectroscopy using narrow-band emission detection. J Am Chem Soc. 2012;134(34):14142–14157. doi: 10.1021/ja304970p. [DOI] [PubMed] [Google Scholar]
- 57.Bethel RD, et al. Regioselectivity in ligand substitution reactions on diiron complexes governed by nucleophilic and electrophilic ligand properties. Inorg Chem. 2015;54(7):3523–3535. doi: 10.1021/acs.inorgchem.5b00072. [DOI] [PubMed] [Google Scholar]
- 58.Barton BE, et al. Isomerization of the hydride complexes [HFe2(SR)2(PR3)(x)(CO)(6-x)]+ (x = 2, 3, 4) relevant to the active site models for the [FeFe]-hydrogenases. Dalton Trans. 2010;39(12):3011–3019. doi: 10.1039/b910147k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hajj V, et al. FeFe hydrogenase reductive inactivation and implication for catalysis. Energy Environ Sci. 2014;7(2):715–719. [Google Scholar]
- 60.Lambertz C, et al. O2 reactions at the six-iron active site (H-cluster) in [FeFe]-hydrogenase. J Biol Chem. 2011;286(47):40614–40623. doi: 10.1074/jbc.M111.283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swanson KD, et al. [FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster. J Am Chem Soc. 2015;137(5):1809–1816. doi: 10.1021/ja510169s. [DOI] [PubMed] [Google Scholar]
- 62.Nyquist RM, Ataka K, Heberle J. The molecular mechanism of membrane proteins probed by evanescent infrared waves. ChemBioChem. 2004;5(4):431–436. doi: 10.1002/cbic.200300687. [DOI] [PubMed] [Google Scholar]
- 63.Frisch MJT, et al. Gaussian 09, Revision D.01. Gaussian; Wallingford, CT: 2009. [Google Scholar]
- 64.Tao J, Perdew JP, Staroverov VN, Scuseria GE. Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys Rev Lett. 2003;91(14):146401. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 65.Schäfer A, Huber C, Ahlrichs R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J Chem Phys. 1994;100(8):5829–5835. [Google Scholar]
- 66.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys. 1988;38(6):3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 67.Ponec R. Structure and bonding in binuclear metal carbonyls. Classical paradigms vs. insights from modern theoretical calculations. Comput Theor Chem. 2015;1053:195–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.